Abstract
The hypoxia-inducible transcription factors (HIFs) directly and indirectly mediate cellular adaptation to reduced oxygen tensions. Recent studies have shown that the histone demethylase genes JMJD1A, JMJD2B, and JARID1B are HIF targets, suggesting that HIFs indirectly influence gene expression at the level of histone methylation under hypoxia. In this study, we identify a subset of hypoxia-inducible genes that are dependent on JMJD1A in both renal cell and colon carcinoma cell lines. JMJD1A regulates the expression of adrenomedullin (ADM) and growth and differentiation factor 15 (GDF15) under hypoxia by decreasing promoter histone methylation. In addition, we demonstrate that loss of JMJD1A is sufficient to reduce tumor growth in vivo, demonstrating that histone demethylation plays a significant role in modulating growth within the tumor microenvironment. Thus, hypoxic regulation of JMJD1A acts as a signal amplifier to facilitate hypoxic gene expression, ultimately enhancing tumor growth.
Cellular hypoxia occurs when the demands of growth and metabolism of a tissue surpass the vascular oxygen supply. In response to hypoxia, cells undergo specific alterations in gene expression patterns geared to promote cell survival and maintain homeostasis. This response not only is important in normal development but also is a critical part in the progression of cancers (7). Hypoxia has been implicated in activating the metabolic shift to anaerobic glycolysis, promoting the epithelial-to-mesenchymal transition (EMT), inducing the secretion of proangiogenic factors, and remodeling the extracellular matrix. Although several transcription programs are activated in response to hypoxia, the hypoxia-inducible factors (HIFs) regulate a critical repertoire of genes, making them central regulators of the cellular response to hypoxia (10, 34).
The HIFs are heterodimeric transcription factors consisting of an oxygen-sensitive alpha subunit (HIF-1α, HIF-2α, or HIF-3α) and a constitutively expressed HIF-1β subunit (also known as the arylhydrocarbon nuclear translocator [ARNT]). Under conditions where oxygen concentration is not limiting, HIF-α subunits are hydroxylated by prolyl-hydroxylases, targeting them for ubiquitin-mediated degradation by the von Hippel-Lindau tumor suppressor (VHL) (18, 19). Under hypoxic conditions, HIF-α protein is stabilized, translocates to the nucleus, dimerizes with ARNT, and binds hypoxia-responsive elements (HREs) in the regulatory regions of target genes (51). HIF-1α and HIF-2α will bind the same sequences in cells but do not have completely overlapping abilities to regulate genes (5, 17, 44). Under certain conditions, HIF-3α functions as a dominant negative, antagonizing the activity of HIF-1 and HIF-2 (32).
Several hundred genes are induced in response to hypoxia, and a great deal of research has been focused on identifying direct HIF target genes (34). The massive transcriptional reorganization mediated by hypoxia and HIFs suggests that changes in histone modification would create epigenetic reinforcement of this phenotype (20). HIF-1α function has been shown to influence and be influenced by histone deacetylases (22, 33), yet comparatively little is known regarding HIF-dependent dynamics of histone methylation (8, 21). In a screen for HIF-regulated changes in gene expression, we and others have identified several Jumonji C-domain-containing histone demethylase (JHDM) promoters as direct binding targets of HIF-1α and HIF-2α (3, 43, 53, 55). Histone demethylases constitute a large and diverse family of enzymes, each with a specific ability to influence transcriptional activation or repression that is dependent on the specific histone residue targeted for demethylation (46); however, the specific roles of Jumonjis in modulating transcriptional responses to hypoxia remain unknown.
Using microarray analysis and chromatin immunoprecipitation, we identified HIF targets including the adrenomedullin gene (ADM) and the growth and differentiation factor 15 gene (GDF15) and show that JMJD1A reduces histone H3K9 methylation at these promoters during hypoxia. Furthermore, we demonstrate that loss of JMJD1A is sufficient to reduce tumor growth in vivo, consistent with its role in regulating histone methylation during hypoxia. These studies identify a transcriptional regulatory circuit where induction of JMJD1A by HIF-1α acts as an epigenetic signal amplifier to enhance cellular responses to hypoxia.
MATERIALS AND METHODS
Cell lines and culture conditions.
All cell lines were maintained in Dulbecco modified Eagle medium (DMEM) supplemented with 10% fetal bovine serum (FBS). Small interfering RNA (siRNA) transfections were carried out using Dharmacon Smart Pools with Dharmafect 1 transfection reagent according to manufacturer's protocol (Dharmacon, Laffayette, CO). Short hairpin RNA (shRNA) retroviral constructs against JMJD1A were purchased from Open Biosystems (Huntsville, AL). Retroviral constructs were packaged and introduced into cells as described previously (57). For hypoxia treatments, cells were plated at the desired density 12 h prior to placement in a hypoxia chamber (Invivo2-400; Ruskin Technologies, Leeds, United Kingdom) maintained at 0.5 to 2% oxygen for 0 to 20 h, depending on the experiment. For in vitro growth assays, cells (105) were plated in triplicate in 6-cm plates and counted every 3 days as described previously (52).
ChIP.
Chromatin immunoprecipitation (ChIP) assays were performed as described previously (26) with the following modifications. Hypoxia-treated cells were fixed within the chamber to avoid reoxygenation. For HIF-1α and HIF-2α immunoprecipitations, 120 μg of sonicated chromatin (measured as protein) was incubated with 10 μg of anti-HIF-1α or HIF-2α antibodies, respectively. For histone ChIPs, sonicated chromatin was immunoprecipitated according to guidelines provided by the manufacturer (Abcam). Data were analyzed in a fashion similar to that described by Johnson et al. (21). Relative enrichment was measured by quantitative real-time PCR using a titration of pooled input samples as a standard curve. Histone H3 methylation signals were normalized to input and bulk histone H3 signal, the corresponding IgG/H3 ratio was subtracted, and the fold change in methylation relative to the normoxic control cells was calculated. For calculation of fold difference of methylation (see Fig. 5D), the H3 normalized methylation signal with JMJD1A knockdown was divided by the corresponding signal from the nonsilencing control cells for each time point.
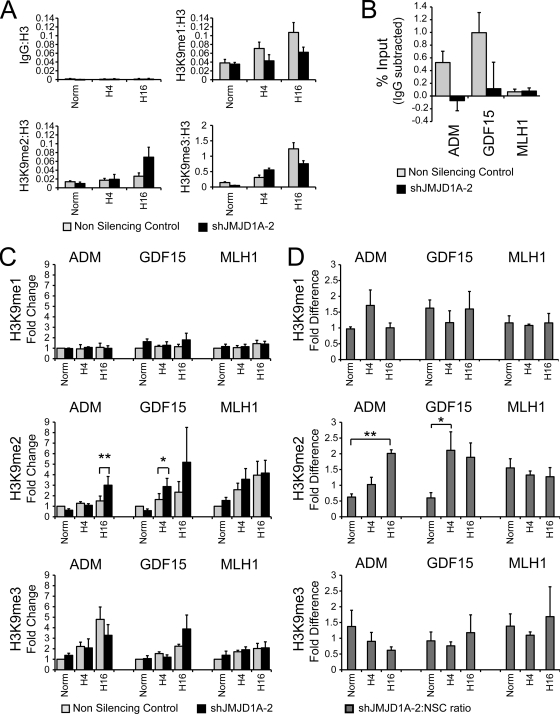
FIG. 5.
JMJD1A binds and demethylates H3K9 on target promoters. HCT116 cells stably expressing shRNA to JMJD1A (shJMJD1A-2) or a nonsilencing control sequence were exposed to 0.5% oxygen for 0 (Norm), 4 (H4), and 16 (16H) hours; fixed in formaldehyde; and interrogated by chromatin immunoprecipitation with antibodies against histone H3K9me1, H3K9me2, and H3K9me3. (A) Representative ChIP of region spanning 250 bp downstream of the ADM transcription start site, demonstrating significant enrichment compared to the IgG control. Data are for a single experiment measured in triplicate and normalized to histone H3 as described in Materials and Methods. (B) Chromatin immunoprecipitated from HCT116 shJMJD1A-2 and nonsilencing control cells (black and gray bars, respectively) with anti-JMJD1A antibody and enrichment on target promoters measured by QPCR. Data represents the average enrichment for three independent experiments after subtraction of IgG signal. (C) Average fold change of H3K9me1, H3K9me2, and H3K9me3 downstream of the ADM, GDF15, and MLH1 transcription start sites. Data represents the averages for four independent ChIP experiments measured in triplicate and normalized to the normoxic nonsilencing control ± standard errors of the means. Statistical significance was calculated using Student's t test: *, P < 0.05; **, P < 0.01. (D). Average fold difference of H3K9 methylation for the experiments of panel C (calculation is described in Materials and Methods). Data represent averages from four independent experiments measured in triplicate ± standard errors of the means. Statistical significance was calculated using Student's t test: *, P < 0.05; **, P < 0.01.
For high-throughput ChIP (ChIP-chip) analysis, immunoprecipitated DNA was amplified as described previously (37) and hybridized to NimbleGen tiled promoter arrays (Roche NimbleGen, Madison, WI) spanning approximately 23,000 promoter regions in the human genome. Promoter summary reports provided by NimbleGen were linked to expression information in Microsoft Access to correlate HIF binding to RCC4 expression (see Table S1 in the supplemental material). Signal Map was used to confirm binding and to extract the coordinates of putative binding peaks for validation experiments. Antibodies used for ChIPs were anti-HIF-1 (BD Biosciences), anti-HIF-2 (Novus), histone H3 (Abcam), histone H3K9me1 (Abcam), histone H3K9me2 (Abcam), histone H3K9me3 (Abcam), and JMJD1A (Abcam). Rabbit IgG (Sigma) was used for a nonspecific IgG control. Primer sequences are available upon request.
Expression microarray analysis.
All microarray sample preparations and hybridizations were carried out with Stanford Human Exonic Evidence Based Oligo (HEEBO) arrays according to protocols available from Pat Brown's lab (http://cmgm.stanford.edu/pbrown/protocols/index.html). HEEBO arrays contain probes for 31,000 unique genes and 8,500 alternate transcripts. Data from scanned microarrays were entered into the Stanford Microarray Database (SMD) for normalization and analysis.
For comparing expression in RCC4 cells to that in RCC4 cells with reconstituted VHL expression (RCC4+VHL cells), amino-allyl-labeled cDNA was reverse transcribed from 25 μg of total RNA and coupled to Cy dyes as described at http://cmgm.stanford.edu/pbrown/protocols/RTaminoAllylCoupling.html. cDNA from RCC4 or RCC4+VHL cells was labeled with Cy5 and hybridized to common reference cDNA from RCC4+VHL cells labeled with Cy3. Labeled cDNA was hybridized and washed as described at http://cmgm.stanford.edu/pbrown/protocols/Direct_Label_Protocol1.html. Expression changes were calculated by averaging data from duplicate RCC4 samples and calculating fold change in expression compared to RCC4+VHL controls.
For profiling the effect of JMJD1A siRNA knockdown in RCC4+VHL cells, RNA was amplified from 1 μg total RNA using the Amino Allyl MessageAmp II aRNA kit (Ambion) and labeled with Cy dyes as described at http://cmgm.stanford.edu/pbrown/protocols/Amplified_Protocol1.html. Samples were from independent triplicate experiments and were labeled with Cy5 and hybridized to common reference cDNA from untreated RCC4+VHL cells. Data were analyzed using multicomponent Significance analysis of microarrays (SAM) (50) to identify genes significantly changed among the siRNA transfections. Genes with a false-discovery rate lower than 0.5% were extracted to calculate fold changes relative to the normoxic siControl samples. Fold change was calculated to identify genes induced greater than 1.5-fold by hypoxia (SiCon-Hypox) and genes where hypoxic expression with siJMJD1A is at least 1.5 times lower than that with the equivalent hypoxic siControl (SiD1A-Hypox). These two groups were imported into GeneVenn (42) to identify regions of overlap (see Table S2 in the supplemental material).
QRT-PCR.
Gene expression was measured using quantitative real-time PCR (QRT-PCR) exactly as described previously (26) using the 18S rRNA or TATA-binding protein (TBP) gene as an internal control. Primer sequences are available upon request.
In vivo experiments.
All animal experiments were performed in accordance with institutional and national guidelines and approved by Stanford University's Administrative Panel on Laboratory Animal Care (APLAC). Male SCID mice, 4 to 6 weeks old, were obtained from Charles River Laboratories. HCT116 cells stably expressing shRNA to JMJD1A or a nonsilencing control sequence were counted and resuspended in sterile phosphate-buffered saline (PBS). Three million cells were injected into the flank of the animal and measured with calipers at regular intervals.
RESULTS
Jumonji domain histone demethylase genes are direct HIF target genes.
To identify direct and functionally relevant HIF target genes involved in chromatin regulation, we performed a directed screen by combining high-throughput chromatin immunoprecipitation (ChIP-chip) with gene expression analysis. The RCC4 clear-cell renal cell carcinoma line was used as a model system because of abundant normoxic expression of HIF-1α and HIF-2α due to genetic inactivation of von Hippel-Lindau (VHL) (35). We probed tiled arrays representing 23,000 human promoter regions for HIF-1α and HIF-2α binding. Using a global average of promoter enrichment (provided as NimbleGen promoter summaries), we biased our search to identify promoter regions with the highest HIF enrichment. Using gene expression profiling, we identified genes induced greater than 1.5-fold in RCC4 cells compared to RCC4 cells with reconstituted VHL expression (RCC4+VHL cells). These genes were then sorted for HIF-1α or HIF-2α binding (Fig. 1A shows a schematic representation; see Table S1 in the supplemental material for complete tabulated results). A common phenomenon with genome-wide ChIP experiments is nonfunctional binding, where a transcription factor is shown to bind but fails to regulate nearby genes (29). Even though our initial filter comparing VHL-repressed expression to HIF binding would be expected to select functional HIF targets, VHL has also been shown to regulate several cellular processes independently of HIF (4, 49). Therefore, we followed up potential HIF targets only if they were also induced by hypoxia in RCC4 cells expressing functional VHL (RCC4+VHL cells).
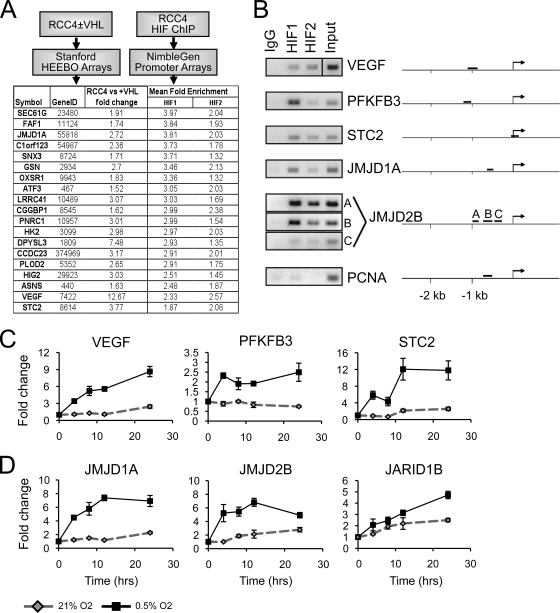
FIG. 1.
Jumonji domain histone demethylases are hypoxia inducible in a HIF-dependent manner. (A) Experimental scheme of the HIF binding to expression screen. RCC4 renal cell carcinoma cells were processed for ChIP-chip as described in Materials and Methods. Promoters corresponding to genes induced greater than 1.5-fold with loss of VHL were sorted by mean HIF binding. An illustrative set of data is shown (the full data set is in Table S1 in the supplemental material). (B) Validation of HIF-1α and HIF-2α binding to selected promoters. VEGF serves as experimental validation control for both HIF-1α and HIF-2α. Multiple amplicons were used to screen promoters containing widely spaced HREs and are denoted A, B, or C as necessary. (C) QRT-PCR analysis or VEGF, PFKFB3, and STC2 expression in RCC4+VHL cells exposed to either 21% or 0.5% O2 for 0, 4, 8, 12, or 24 h. (D) QRT-PCR analysis for JMJD1A, JMJD2B, and JARID1B expression. Data for panel C and D are plotted as fold change normalized to TBP gene expression. Error bars represent standard deviations.
The screen identified several known HIF target genes, including those for vascular endothelial growth factor (VEGF), 6-phosphofructo-2-kinase/fructose-2,6-biphosphatase 3 (PFKFB3), and stanniocalcin 2 (STC2), providing validity for our ChIP-chip analysis (12, 28, 36). HIF binding at these promoters was confirmed by ChIP-PCR (Fig. 1B). The Jumonji C-terminal-domain-containing histone demethylase (JHDM) gene JMJD1A was also identified as a putative HIF target. JHDMs have received a great deal of attention for their ability to demethylate histones and either activate or repress transcription (46). JMJD1A (also known as JHDM2A and KDM3A) has recently been shown to be a coactivator for nuclear hormone receptors and myogenic factors by demethylating dimethyl lysine 9 on histone H3 (H3K9me2) of target promoters (30, 31, 38, 56).
We then searched through our binding data and identified JMJD2B as another strong target of HIF binding. JMJD2B recognizes tri- and dimethylated lysine 9 (H3K9me3/2) on histone H3, reducing both modifications to the monomethylated state (11, 24, 54). Both HIF-1α and HIF-2α bind near the JMJD1A and JMJD2B promoters (Fig. 1B), confirming recent reports (3, 43, 53, 55).
Specificity of our screen was validated by ChIP-PCR to the promoter of PCNA, a region with no visible HIF enrichment and no hypoxic regulation. We then retrospectively searched our expression data to identify additional Jumonji domain proteins that might be regulated by hypoxia. Expression of the histone H3 lysine 4 (H3K4me3) demethylase JARID1B was increased in RCC4+VHL cells during hypoxia (1.7-fold) (see Table S2 in the supplemental material). Consistent with our finding, recent studies have also identified JARID1B as a hypoxia-inducible gene (16, 55).
Hypoxic transcriptional induction of the JHDMs was confirmed in RCC4+VHL cells. Expression of JMJD1A, JMJD2B, and JARID1B was induced and sustained for at least 24 h upon cellular exposure to hypoxia (Fig. 1D), similar to the case for VEGFA, PFKFB3, and STC2 (Fig. 1C). Our approach combining high-throughput ChIP and gene expression analysis has identified at least three different JHDMs as potential HIF targets, suggesting that HIF plays an important role in regulating widespread epigenetic phenomena through alterations in histone methylation.
Jumonji domain histone demethylases are hypoxia inducible in a HIF-dependent manner.
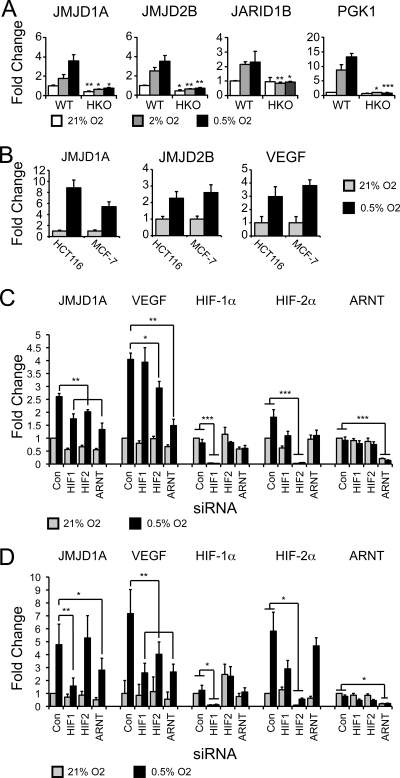
To determine if hypoxic induction of histone demethylases is dependent on HIF-1α, we next utilized wild-type and HIF-1α-deficient mouse embryonic fibroblasts (MEFs). Previous studies have indicated that MEFs have nonfunctional HIF-2α protein (40). Real-time PCR analysis demonstrated that the hypoxic induction of JMJD1A, JMJD2B, and JARID1B was significantly reduced in HIF-1α-deficient MEFs compared to wild-type MEFs, demonstrating dependence on HIF-1α for hypoxic induction (Fig. 2A). JMJD1A expression was also robustly induced in MCF-7 breast cancer and HCT116 colon cancer cells, as was that of JMJD2B and VEGF (Fig. 2B).
FIG. 2.
Jumonji domain histone demethylases are hypoxia inducible in a HIF-dependent manner. (A) QRT-PCR measurement of murine Jmjd1a, Jmjd2b, and Jarid1b in wild-type and HIF-1α knockout MEFs (HKO) exposed to 21%, 2%, and 0.5% O2 for 20 h. Pgk1 serves as a control for a HIF-1α-specific gene. Statistical significance was calculated using Student's t test: *, P < 0.05; **, P < 0.01; ***, P < 0.001. Asterisks refer to comparisons within the respective oxygen concentrations. (B) JMJD1A, JMJD2B, and VEGF expression in HCT116 and MCF-7 cells exposed to 21% or 0.5% oxygen for 20 h. (C) QRT-PCR measurement of hypoxic expression of JMJD1A and VEGF in RCC4+VHL cells after transfection with siRNAs to HIF-1α, HIF-2α, and ARNT. Data represent the averages from six independent experiments. Statistical significance was calculated using Student's t test: *, P < 0.05; **, P < 0.01; ***, P < 0.001. (D) QRT-PCR measurement of hypoxic expression of JMJD1A and VEGF in MCF-7 cells transfected as for panel C. Data represent the averages from three independent experiments, measured in triplicate and normalized to 18S rRNA. Error bars indicate standard errors of the means. Statistical significance was calculated using Student's t test: *, P < 0.05; **, P < 0.01.
Transfection of siRNAs to HIF-1α, HIF-2α, and ARNT reduced the hypoxic induction of JMJD1A in RCC4+VHL cells in a statistically significant manner, indicating that in certain cell types, JMJD1A may be regulated by either HIF-1α or HIF-2α (Fig. 2C). In contrast siRNAs to HIF-1α and ARNT, but not HIF-2α, resulted in a statistically significant reduction of JMJD1A expression under hypoxia in MCF-7 cells. VEGF expression was disrupted by knockdown of HIF-2α and ARNT, confirming functional knockdown of HIF-2 and previous reports that VEGF is primarily a target of HIF-2α in renal cell carcinoma (17, 44). In MCF-7 cells, hypoxic expression of VEGF was reduced in a statistically significant manner by knockdown of HIF-1, HIF-2, and ARNT, demonstrating functional knockdown of HIF-1α and HIF-2α. These experiments demonstrate that the regulation of JMJD1A by HIF-1α is a conserved phenomenon in both mice and humans, while HIF-2α may regulate JMJD1A in certain cellular contexts. Identification of JMJD1A as a HIF-1α target reveals an important mechanistic link between the cellular microenvironment, HIF activation, and epigenetic regulation of biological phenomena.
JMJD1A regulates hypoxic gene expression.
Of the three JHDM genes identified in our experiments, JMJD1A seemed the most likely to have a direct effect on hypoxic gene induction. JMJD1A was one of the strongest HIF-1α peaks identified by the ChIP-chip experiment (Fig. 1A), and its ability to function as a coactivator has been well characterized (56). Since many coactivators identified within specific transcription pathways were later found to enhance HIF-1α activity (6), we hypothesized that JMJD1A might also mediate hypoxic gene expression.
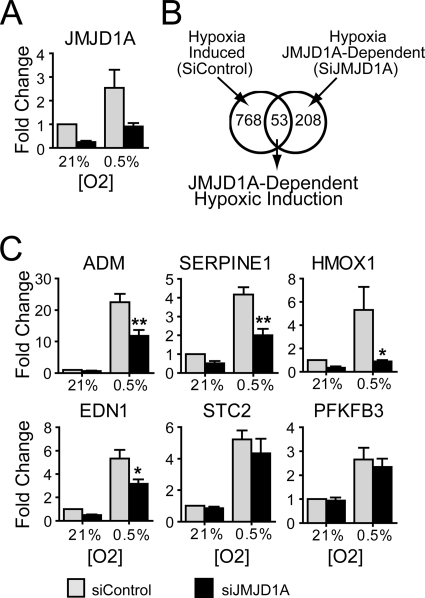
In order to identify potential JMJD1A targets during hypoxia, RCC4+VHL cells were transfected with siRNA to JMJD1A or control siRNA and exposed to hypoxia (0.5% O2) or normoxia (21% O2), and gene expression was analyzed using Stanford HEEBO microarrays (see Materials and Methods). Knockdown of JMJD1A mRNA was approximately 3-fold under normoxia and hypoxia (Fig. 3A). We identified 821 genes induced >1.5-fold by hypoxia in cells transfected with control siRNA (Fig. 3B, left circle), a result similar to those in earlier reports (34). Of these genes, 53 were dependent on JMJD1A for full hypoxic induction (region of overlap in Fig. 3B). Included in this data set were a number of known HIF targets such as ADM, EDN1, SERPINE1, PLAUR, and HMOX1 (complete lists are in Table S2 in the supplemental material). We also identified 208 genes with decreased hypoxic expression with loss of JMJD1A but were not induced in the control cell line, implying that hypoxic induction of JMJD1A plays a broader role in maintaining gene expression during hypoxia (Fig. 3B, right circle; see Table S2 in the supplemental material).
FIG. 3.
JMJD1A regulates hypoxic induction of a subset of hypoxia-regulated genes in RCC4+VHL cells. (A) Quantitative RT-PCR of JMJD1A expression in RCC4+VHL cells transfected with siRNA to JMJD1A (siD1A) and exposed to 21% or 0.5% oxygen for 16 h. (B) Venn diagram graphically showing the intersection between genes induced greater than 1.5-fold by hypoxia in the siControl-transfected cells (left circle) and genes reduced at least 1.5-fold under hypoxia with siJMJD1A compared to the hypoxic siControl (right circle). Genes dependent on JMJD1A for hypoxic induction are depicted in the overlap. (C) RCC4+VHL cells were transfected as for panel A, and the expression of genes identified in panel B (ADM, SERPINE1, EDN1, and HMOX) was measured by QRT-PCR. STC2 and PFKFB3 serve as examples of two JMJD1A-independent hypoxia inducible genes. Data are from six experiments measured in triplicate (± standard errors of the means) and normalized to the normoxic control siRNA (SiCon). Statistical significance was calculated using Student's t test: *, P < 0.05; **, P < 0.01.
Validation of JMJD1A-dependent expression was confirmed using QRT-PCR (Fig. 3C). ADM, SERPINE1 (PAI1), EDN1, and HMOX1 had statistically significant dependence on JMJD1A for hypoxic induction. These genes were chosen for further analysis because they were known HIF targets and had known functions, making them more fruitful targets for later functional analysis. Though not statistically significant, JMJD1A increased hypoxic expression of SERPINB8, EDN2, and GDF15 (data not shown), confirming a trend of regulation by JMJD1A. The majority of hypoxia-induced genes, including STC2 and PFKFB3, showed little or no dependence on JMJD1A (Fig. 3C; see Table S2 in the supplemental material). The specific regulation of some but not all hypoxia-induced genes by JMJD1A suggests that multiple mechanisms regulate hypoxic gene expression, highlighting the importance of promoter-specific recruitment of transcription regulators.
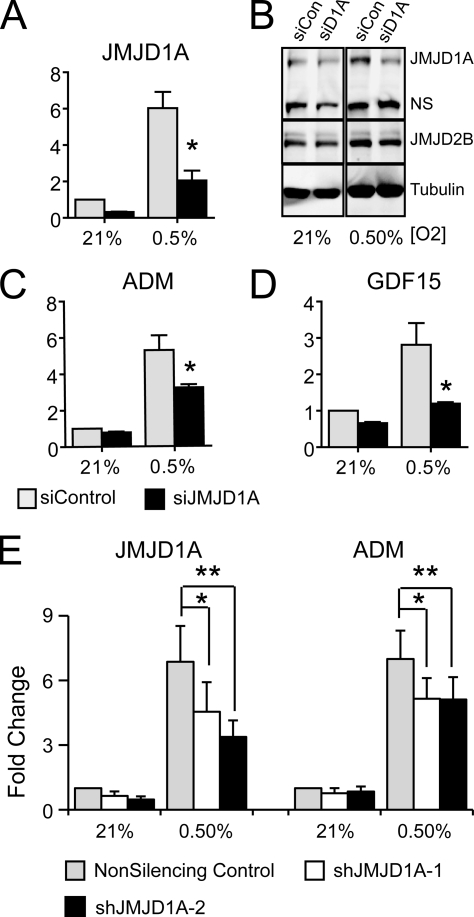
Because of the clear link between VHL loss, HIF activation, and progression of renal cell carcinoma, cell lines like RCC4 are ideal systems for the study of HIF activity and hypoxic transcription phenomena. A significant disadvantage of using RCC-derived cells is the generally poor rate of tumor initiation in xenograft experiments, making it difficult to evaluate the functional aspects of gene expression. We found that JMJD1A is also robustly induced in HCT116 colon carcinoma cells (Fig. 2B), which grow well in tumor xenograft experiments, making them a better system for studying in vivo function of JMJD1A. In HCT116 cells transfected with siRNA to JMJD1A, there was a robust knockdown of JMJD1A RNA and protein (Fig. 4A and B). Accordingly, the hypoxic induction of ADM and GDF15 was robustly and significantly dependent on hypoxic JMJD1A expression (Fig. 4C and D). For use in longer-term studies, we also established two HCT116-derived cell lines expressing shRNA to JMJD1A. Compared to a line expressing a nonsilencing control construct, JMJD1A mRNA was reduced approximately 2- to 3-fold in both cell lines in normoxia (Fig. 4E). While both shJMJD1A-1 and shJMJD1A-2 significantly suppressed hypoxic induction of ADM by approximately 25 to 30% compared to the nonsilencing control (Fig. 4E), cells expressing shJMJD1A-2 maintained effective knockdown more consistently over time, making it a more useful line for further experiments (data not shown). These results demonstrate that hypoxic induction of JMJD1A is crucial for optimal hypoxic expression of targets and not solely a result of reduced expression under normoxia.
FIG. 4.
JMJD1A regulates ADM and GDF15 in HCT116 cells. (A) Quantitative RT-PCR of JMJD1A expression in HCT116 cells transfected with siRNA to JMJD1A (siD1A) and exposed to 21% or 0.5% oxygen for 16 h. Data are the averages from three experiments measured in triplicate (± standard errors of the means), normalized to the normoxic siControl-transfected cells. Statistical significance was calculated using Student's t test: *, P < 0.05. (B) Western blot analysis of whole-cell lysates harvested from HCT116 colon carcinoma cells transfected with siRNA to JMJD1A and exposed to normoxia (21% O2) and hypoxia (0.5% O2). Both a nonspecific band (NS) and tubulin serve as loading controls. (C and D) QRT-PCR of ADM (C) and GDF15 (D) expression in HCT116 cells transfected as for panel A. Data are the averages from three experiments measured in triplicate (± standard errors of the means), normalized to the normoxic siControl-transfected cells. Statistical significance was calculated using Student's t test: *, P < 0.05. (E) HCT116 cells stably expressing two different shRNA constructs and a nonsilencing control were exposed to 0.5% or 21% oxygen for 16 h, and expression of JMJD1A and ADM was analyzed by QRT-PCR. Fold change was calculated relative to normoxic nonsilencing control cells. Data represent average fold changes for five independent replicates measured in triplicate ± standard errors of the means. Expression normalized to statistical significance was calculated using Student's t test: *, P < 0.05; **, P < 0.01.
Transcriptional regulation by JMJD1A during hypoxia is due to changes in histone methylation of target promoters.
Since JMJD1A induces gene expression by specifically reducing the methylation of histone H3K9me2 and H3K9me1 to the unmethylated state in vitro and in vivo (56), reduction of JMJD1A expression would be expected to increase H3K9 dimethylation on target promoters such as ADM and GDF15. HCT116 cells expressing shJMJD1A-2 shRNA or a nonsilencing control construct were exposed to hypoxia for 4 or 16 h and fixed with formaldehyde for chromatin immunoprecipitation. Cells grown in normoxia were fixed at time zero as a control for hypoxic changes. Using chromatin immunoprecipitation followed by QPCR analysis, we measured the ratio of H3K9me1, H3K9me2, and H3K9me3 to bulk histone H3 on regions approximately 200 bp downstream of the transcription start sites of ADM and GDF15. A representative ChIP on the ADM promoter is depicted in Fig. 5A, demonstrating robust enrichment over the IgG control for all histone antibodies tested. After 16 h of hypoxia, H3K9me2 was approximately two times higher in the shJMJD1A-2 cells than in the nonsilencing control, proportional to the reduction of JMJD1A (Fig. 4C). This result was confirmed by calculating the fold change in histone methylation compared to the normoxic, nonsilencing control cells for four independent experiments and averaging the data at each time point (Fig. 5C). While there was no statistically significant difference in HeK9me1 or H3K9me3 for either ADM or GDF15, H3K9me2 was increased in a statistically significant manner after 4 h and 16 h for GDF15 (P < 0.05) and ADM (P < 0.01), respectively (Fig. 5C). On the MLH1 promoter, a gene previously shown to be repressed by hypoxic activation of G9a and regulated independently of JMJD1A (reference 8 and data not shown), H3K9me2 increased with hypoxia in both the nonsilencing control and shJMJD1A-2 cells, but there was no significant difference between the two cell lines.
Because individual variation in cell density, hypoxic treatments, and ChIP procedure created some variability in the fold changes measured (see H3K9me2 for GDF15 at 16 h), we also calculated the individual ratio of histone methylation between the two cell lines at each time point and averaged the data for the same four independent replicates (Fig. 5D). This analysis highlights the overall trends in JMJD1A-dependent methylation during hypoxia, confirming the results obtained by comparing fold change. With reduced JMJD1A expression, there is a clear relative increase in H3K9me2 in hypoxia for ADM and GDF15, but not for MLH1, while there are no statistically significant differences for H3K9me1. For H3K9me3, there appears to be a relative decrease in H3K9me3 for ADM with loss of JMJD1A, but it was not statistically significant because of high variability in the ratios at time zero.
H3K9 trimethylation showed little dependence on JMJD1A expression, but did show some differences between the different promoters. In the nonsilencing control cells, H3K9me3 was induced higher on ADM than on GDF15 or MLH1, suggesting that H3K9 methylation on ADM is regulated by a methylase with terminal trimethylation activity such as ESET, while GDF15 may be regulated by a methyltransferase that terminates with H3K9me2 such as G9a (Fig. 5B). MLH1 has previously been shown to be regulated by G9a under hypoxia (8). Reduction of JMJD1A in the knockdown line also reduced JMJD1A binding on the ADM and GDF15 promoters, but not MLH1 (Fig. 5B), confirming that ADM and GDF15 are direct targets of demethylation by JMJD1A.
JMJD1A enhances tumor growth in vivo.
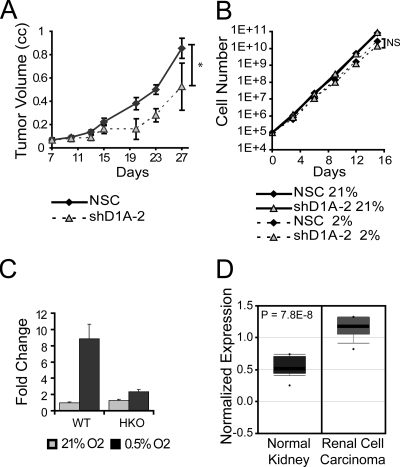
We have demonstrated in vitro that in both clear-cell renal cell carcinoma and colon carcinoma cell lines, JMJD1A regulates genes implicated in cancer cell growth, invasion, and survival (Fig. 3 and 4; see Table S2 in the supplemental material). Subcutaneous injection of nonsilencing control and shJMJD1A-2 HCT116 cells into the flanks of SCID mice resulted in a significantly reduced rate of tumor growth for the shJMJD1A line (Fig. 6A). Tumor xenografts are hypoxic (1), and the reduction in tumor growth resulting from reduced JMJD1A expression is consistent with the reduced hypoxic expression of protumorigenic factors such as ADM and GDF15 in vitro. In contrast, culturing HCT116 cells in monolayers in 2% oxygen reduced the growth rate compared to normoxia (21% oxygen), but loss of JMJD1A expression had little to no effect (Fig. 6B), suggesting that JMJD1A influences tumor growth within the context of the tumor microenvironment. In HCT116 cells, JMJD1A is robustly induced during hypoxia in a HIF-1α-dependent manner (Fig. 6C), consistent with a report that colorectal carcinomas have a hypoxic signature (13). JMJD1A expression is also significantly higher in renal cell carcinoma (14, 45), a cancer type associated with loss of the VHL tumor suppressor and constitutive activation of HIF and its transcriptional targets (Fig. 6D). JMJD1A therefore plays an important part in mediating the protumorigenic effects of the hypoxic microenvironment and HIF activation by demethylating the chromatin near key target promoters (Fig. 7).
FIG. 6.
JMJD1A promotes tumor growth. (A) Loss of JMJD1A reduces tumor growth rate. A total of 3 × 106 HCT116 cells expressing shRNA to JMJD1A (shD1A-2; n = 3) or a nonsilencing control (NSC; n = 5) were injected into the flanks of SCID mice and the volume measured at the indicated intervals. Data are the averages of tumor volumes ± standard errors of the means. *, P < 0.05 as determined by nonlinear regression. (B) Loss of JMJD1A has no effect on in vitro growth rate. A total of 105 HCT116 cells expressing shRNA to JMJD1A or a nonsilencing control were grown as described in Materials and Methods in 21% oxygen or 0.5% oxygen. NS indicate a nonsignificant difference in growth rate as measured by nonlinear regression. (C) Hypoxic JMJD1A expression is abolished in HIF-1α knockout (HKO) HCT116 cells exposed to 21% and 0.5% O2 for 16 h. Expression is normalized to that in normoxic wild-type cells (WT). (D) Box-and-whisker plot of JMJD1A expression in renal cell carcinoma. The plot is a scalable vector graphic derived from a microarray analysis by Gumz et al. (14) downloaded from Oncomine (http://www.oncomine.org/) with text labels modified for clarity.
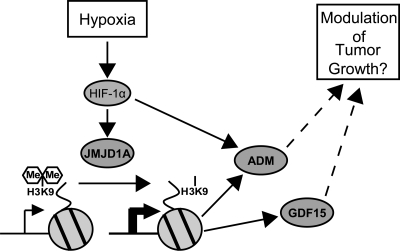
FIG. 7.
Model for JMJD1A action during hypoxia. Cellular hypoxia stabilizes HIF-1α, which induces several genes, including JMJD1A. Increased JMJD1A expression decreases H3K9 methylation on the promoters of target genes such as ADM and GDF15, enhancing hypoxic expression and possibly modulating tumor growth.
DISCUSSION
When cells are exposed to hypoxic conditions, they undergo a fundamental shift in gene expression patterns that permits adaptation and survival (27). The hypoxia-inducible factors (HIFs) are key regulators of the hypoxic response, regulating transcription of genes involved in metabolism, angiogenesis, differentiation, invasion, and metastasis. In this report, we identify HIF-1α-dependent expression of three known JHDM genes: JMJD1A, JMJD2B, and JARID1B. Recent studies have also shown that JMJD1A, JMJD2B, and JARID1B are direct HIF targets; however, the roles of JHDMs in the cellular response to hypoxia have not been identified (3, 43, 53, 55). In this report, we demonstrate that JMJD1A regulates a subset of hypoxia-induced genes, including ADM, EDN1, HMOX1, and GDF15, and that JMJD1A is important for tumor growth in the hypoxic microenvironment of tumor xenografts.
All of the Jumonji domain histone demethylases (JHDMs) currently described utilize 2-oxoglutarate and molecular oxygen to hydroxylate and remove the methyl groups from lysine residues of modified histones (25, 39, 46). Consistent with the observed mechanism of JHDMs, hypoxia has been reported to increase bulk histone methylation, presumably through inhibition of JHDMs or increased methyltransferase activity (8, 21). However, in these same studies, reduced methylation is maintained on the promoters of hypoxia-inducible genes, implying that something is actively demethylating H3 at those loci or is blocking the activity of histone methyltransferases. In this report, we demonstrate that JMJD1A is important for increasing or maintaining the expression of specific genes under hypoxia by maintaining a lower level of H3K9 dimethylation. Reduced JMJD1A expression resulted in increased H3K9me2 directly downstream of the ADM and GDF15 promoters during hypoxia, demonstrating that JMJD1A is enzymatically active in cells even under restrictive oxygen concentrations. Hypoxic induction of JMJD1A may increase the concentration of histone demethylase activity to promoters to compensate for reduced oxygen during hypoxia. shRNA-mediated knockdown of JARID1B resulted in global increase in H3K4me3 in HepG2 cells treated with hypoxia, providing another demonstration that JHDMs can function despite reduced oxygen concentrations (55).
Our results are consistent with the observation that exogenous overexpression of JMJD1A in hypoxia does not affect global H3K9 methylation (3). While the transcriptional mechanisms inhibited by H3K9 dimethylation during hypoxia remain to be elucidated, JMJD1A-dependent demethylation on specific promoters is very consistent with the more selective regulation by JMJD1A observed in other cell systems (31, 38, 48, 56). Our observation that hypoxic regulation of JMJD1A has specific effects on histone methylation and gene expression provides a direct and specific mechanism for maintaining a derepressed pattern of histone methylation on target promoters during hypoxia. More comprehensive analysis of histone methylation phenomena during hypoxia will be important for determining the extent of hypoxic JMJD1A localization and activity. Additionally, identifying the functional overlap and interactions between the other hypoxia-regulated JHDMs will be crucial for understanding how global regulation of histone demethylation mediates hypoxic gene expression.
Many of the genes that we identified as JMJD1A targets were factors involved in vascular dilation (ADM) (23), iron metabolism (GDF15) (47), and angiogenesis (ADM, EDN1, SERPINE1, and HMOX1) (27). This is consistent with the finding that loss of JMJD1A had an effect on tumor xenograft growth but not in vitro cell growth. Regulation of JMJD1A by HIF-1α may represent a feed-forward mechanism for minimizing the energy required to reactivate or maintain expression transcriptional targets after exposure to hypoxia. Additionally, hypoxia and JMJD1A have separately been shown to maintain stem cell populations (9, 31). While the effects of JMJD1A had relatively modest effects on tumor growth, hypoxic regulation of JMJD1A may help maintain the undifferentiated phenotype characteristic of cancer cells.
A key goal for future studies will be to understand the mechanism that targets JMJD1A to specific hypoxia-inducible targets and not others. JMJD1A has recently been shown to regulate expression of adipogenic genes by facilitating peroxisome proliferator-activated receptor gamma (PPARγ) binding and transcriptional activity on the Ucp1 promoter (48). JMJD1A also acts as a coactivator for the androgen receptors (56), presumably through an LXXLL motif common to many coactivators for nuclear hormone receptors (15). While the estrogen-related receptors (ERRs) have been shown to cooperate with HIF (2), ERR-α was shown to bind the promoters of glycolytic genes, which are not JMJD1A targets in our analysis (see Table S2 in the supplemental material). It remains to be determined what proportion of hypoxic expression depends on combinatorial action of nuclear hormone receptors and HIFs. Although the PAS domain of ARNT has recently been shown to interact with coactivators containing LXXLL motifs (41), the selective regulation of hypoxic targets by JMJD1A suggests that it is not generally recruited through HIFs or ARNT. A recent study has also demonstrated recruitment of JMJD1A to myocardin factors (30), suggesting that multiple regulatory pathways can recruit JMJD1A to target promoters. The specific recruitment of JMJD1A to target promoters during hypoxia therefore remains an open question and one of active study.
In conclusion, we have dissected a pathway linking oxygen sensing to induction of JHDM expression and alterations in chromatin modification during hypoxia (modeled in Fig. 7). In hypoxia, stabilized HIF-1α induces expression of JMJD1A, reducing H3K9 dimethylation on specific hypoxia-responsive promoters, including ADM, EDN1, HMOX1, and GDF15, resulting in increased expression and subsequent adaptive responses favoring tumor cell growth. Since JMJD1A also regulates normoxic expression of target genes, hypoxic induction may feed forward to maintain expression of key hypoxia-regulated genes after an initial exposure to reduced oxygen. Hypoxic regulation of JMJD1A likely serves as a “signal amplifier” to enhance and prolong the protumorigenic phenotype of cells exposed to a hypoxic microenvironment.
Supplementary Material
Acknowledgments
We thank members of the Giaccia lab for critical reading and discussion.
This work was supported by an NIH grant (CA 88480) awarded to A.J.G., by an NIH-NRSA award (CA124082) to A.J.K., by an NIH Training Grant (CA09151-32) to E.B.R., and by an Amgen Foundation Summer Research Award to S.F.
Footnotes
Published ahead of print on 26 October 2009.
Supplemental material for this article may be found at http://mcb.asm.org/.
REFERENCES
- 1.Adam, M. F., M. J. Dorie, and J. M. Brown. 1999. Oxygen tension measurements of tumors growing in mice. Int. J. Radiat. Oncol. Biol. Phys. 45:171-180. [DOI] [PubMed] [Google Scholar]
- 2.Ao, A., H. Wang, S. Kamarajugadda, and J. Lu. 2008. Involvement of estrogen-related receptors in transcriptional response to hypoxia and growth of solid tumors. Proc. Natl. Acad. Sci. U.S.A. 105:7821-7826. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Beyer, S., M. M. Kristensen, K. S. Jensen, J. V. Johansen, and P. Staller. 2008. The histone demethylases JMJD1A and JMJD2B are transcriptional targets of hypoxia-inducible factor HIF. J. Biol. Chem. 283:36542-36552. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bommi-Reddy, A., I. Almeciga, J. Sawyer, C. Geisen, W. Li, E. Harlow, W. G. Kaelin, Jr., and D. A. Grueneberg. 2008. Kinase requirements in human cells. III. Altered kinase requirements in VHL−/− cancer cells detected in a pilot synthetic lethal screen. Proc. Natl. Acad. Sci. U.S.A. 105:16484-16489. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Bracken, C. P., A. O. Fedele, S. Linke, W. Balrak, K. Lisy, M. L. Whitelaw, and D. J. Peet. 2006. Cell-specific regulation of hypoxia-inducible factor (HIF)-1alpha and HIF-2alpha stabilization and transactivation in a graded oxygen environment. J. Biol. Chem. 281:22575-22585. [DOI] [PubMed] [Google Scholar]
- 6.Carrero, P., K. Okamoto, P. Coumailleau, S. O'Brien, H. Tanaka, and L. Poellinger. 2000. Redox-regulated recruitment of the transcriptional coactivators CREB-binding protein and SRC-1 to hypoxia-inducible factor 1alpha. Mol. Cell. Biol. 20:402-415. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Chan, D. A., and A. J. Giaccia. 2007. Hypoxia, gene expression, and metastasis. Cancer Metastasis Rev. 26:333-339. [DOI] [PubMed] [Google Scholar]
- 8.Chen, H., Y. Yan, T. L. Davidson, Y. Shinkai, and M. Costa. 2006. Hypoxic stress induces dimethylated histone H3 lysine 9 through histone methyltransferase G9a in mammalian cells. Cancer Res. 66:9009-9016. [DOI] [PubMed] [Google Scholar]
- 9.Covello, K. L., J. Kehler, H. Yu, J. D. Gordan, A. M. Arsham, C. J. Hu, P. A. Labosky, M. C. Simon, and B. Keith. 2006. HIF-2alpha regulates Oct-4: effects of hypoxia on stem cell function, embryonic development, and tumor growth. Genes Dev. 20:557-570. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Elvidge, G. P., L. Glenny, R. J. Appelhoff, P. J. Ratcliffe, J. Ragoussis, and J. M. Gleadle. 2006. Concordant regulation of gene expression by hypoxia and 2-oxoglutarate-dependent dioxygenase inhibition: the role of HIF-1alpha, HIF-2alpha, and other pathways. J. Biol. Chem. 281:15215-15226. [DOI] [PubMed] [Google Scholar]
- 11.Fodor, B. D., S. Kubicek, M. Yonezawa, R. J. O'Sullivan, R. Sengupta, L. Perez-Burgos, S. Opravil, K. Mechtler, G. Schotta, and T. Jenuwein. 2006. Jmjd2b antagonizes H3K9 trimethylation at pericentric heterochromatin in mammalian cells. Genes Dev. 20:1557-1562. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Forsythe, J. A., B. H. Jiang, N. V. Iyer, F. Agani, S. W. Leung, R. D. Koos, and G. L. Semenza. 1996. Activation of vascular endothelial growth factor gene transcription by hypoxia-inducible factor 1. Mol. Cell. Biol. 16:4604-4613. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Furlan, D., N. Sahnane, I. Carnevali, R. Cerutti, F. Bertoni, I. Kwee, S. Uccella, V. Bertolini, A. M. Chiaravalli, and C. Capella. 2008. Up-regulation of the hypoxia-inducible factor-1 transcriptional pathway in colorectal carcinomas. Hum. Pathol. 39:1483-1494. [DOI] [PubMed] [Google Scholar]
- 14.Gumz, M. L., H. Zou, P. A. Kreinest, A. C. Childs, L. S. Belmonte, S. N. LeGrand, K. J. Wu, B. A. Luxon, M. Sinha, A. S. Parker, L. Z. Sun, D. A. Ahlquist, C. G. Wood, and J. A. Copland. 2007. Secreted frizzled-related protein 1 loss contributes to tumor phenotype of clear cell renal cell carcinoma. Clin. Cancer Res. 13:4740-4749. [DOI] [PubMed] [Google Scholar]
- 15.Heery, D. M., E. Kalkhoven, S. Hoare, and M. G. Parker. 1997. A signature motif in transcriptional co-activators mediates binding to nuclear receptors. Nature 387:733-736. [DOI] [PubMed] [Google Scholar]
- 16.Holmquist-Mengelbier, L., E. Fredlund, T. Lofstedt, R. Noguera, S. Navarro, H. Nilsson, A. Pietras, J. Vallon-Christersson, A. Borg, K. Gradin, L. Poellinger, and S. Pahlman. 2006. Recruitment of HIF-1alpha and HIF-2alpha to common target genes is differentially regulated in neuroblastoma: HIF-2alpha promotes an aggressive phenotype. Cancer Cell 10:413-423. [DOI] [PubMed] [Google Scholar]
- 17.Hu, C. J., S. Iyer, A. Sataur, K. L. Covello, L. A. Chodosh, and M. C. Simon. 2006. Differential regulation of the transcriptional activities of hypoxia-inducible factor 1 alpha (HIF-1alpha) and HIF-2alpha in stem cells. Mol. Cell. Biol. 26:3514-3526. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Ivan, M., K. Kondo, H. Yang, W. Kim, J. Valiando, M. Ohh, A. Salic, J. M. Asara, W. S. Lane, and W. G. Kaelin, Jr. 2001. HIFalpha targeted for VHL-mediated destruction by proline hydroxylation: implications for O2 sensing. Science 292:464-468. [DOI] [PubMed] [Google Scholar]
- 19.Jaakkola, P., D. R. Mole, Y. M. Tian, M. I. Wilson, J. Gielbert, S. J. Gaskell, A. Kriegsheim, H. F. Hebestreit, M. Mukherji, C. J. Schofield, P. H. Maxwell, C. W. Pugh, and P. J. Ratcliffe. 2001. Targeting of HIF-alpha to the von Hippel-Lindau ubiquitylation complex by O2-regulated prolyl hydroxylation. Science 292:468-472. [DOI] [PubMed] [Google Scholar]
- 20.Johnson, A. B., and M. C. Barton. 2007. Hypoxia-induced and stress-specific changes in chromatin structure and function. Mutat. Res. 618:149-162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Johnson, A. B., N. Denko, and M. C. Barton. 2008. Hypoxia induces a novel signature of chromatin modifications and global repression of transcription. Mutat. Res. 640:174-179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Kato, H., S. Tamamizu-Kato, and F. Shibasaki. 2004. Histone deacetylase 7 associates with hypoxia-inducible factor 1alpha and increases transcriptional activity. J. Biol. Chem. 279:41966-41974. [DOI] [PubMed] [Google Scholar]
- 23.Kitamura, K., K. Kangawa, M. Kawamoto, Y. Ichiki, S. Nakamura, H. Matsuo, and T. Eto. 1993. Adrenomedullin: a novel hypotensive peptide isolated from human pheochromocytoma. Biochem. Biophys. Res. Commun. 192:553-560. [DOI] [PubMed] [Google Scholar]
- 24.Klose, R. J., K. Yamane, Y. Bae, D. Zhang, H. Erdjument-Bromage, P. Tempst, J. Wong, and Y. Zhang. 2006. The transcriptional repressor JHDM3A demethylates trimethyl histone H3 lysine 9 and lysine 36. Nature 442:312-316. [DOI] [PubMed] [Google Scholar]
- 25.Klose, R. J., and Y. Zhang. 2007. Regulation of histone methylation by demethylimination and demethylation. Nat. Rev. Mol. Cell Biol. 8:307-318. [DOI] [PubMed] [Google Scholar]
- 26.Krieg, A. J., E. M. Hammond, and A. J. Giaccia. 2006. Functional analysis of p53 binding under differential stresses. Mol. Cell. Biol. 26:7030-7045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Le, Q. T., N. C. Denko, and A. J. Giaccia. 2004. Hypoxic gene expression and metastasis. Cancer Metastasis Rev. 23:293-310. [DOI] [PubMed] [Google Scholar]
- 28.Leonard, M. O., D. C. Cottell, C. Godson, H. R. Brady, and C. T. Taylor. 2003. The role of HIF-1 alpha in transcriptional regulation of the proximal tubular epithelial cell response to hypoxia. J. Biol. Chem. 278:40296-40304. [DOI] [PubMed] [Google Scholar]
- 29.Li, X. Y., S. MacArthur, R. Bourgon, D. Nix, D. A. Pollard, V. N. Iyer, A. Hechmer, L. Simirenko, M. Stapleton, C. L. Luengo Hendriks, H. C. Chu, N. Ogawa, W. Inwood, V. Sementchenko, A. Beaton, R. Weiszmann, S. E. Celniker, D. W. Knowles, T. Gingeras, T. P. Speed, M. B. Eisen, and M. D. Biggin. 2008. Transcription factors bind thousands of active and inactive regions in the Drosophila blastoderm. PLoS Biol. 6:e27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Lockman, K., J. M. Taylor, and C. P. Mack. 2007. The histone demethylase, Jmjd1a, interacts with the myocardin factors to regulate SMC differentiation marker gene expression. Circ. Res. 101:e115-e123. [DOI] [PubMed] [Google Scholar]
- 31.Loh, Y. H., W. Zhang, X. Chen, J. George, and H. H. Ng. 2007. Jmjd1a and Jmjd2c histone H3 Lys 9 demethylases regulate self-renewal in embryonic stem cells. Genes Dev. 21:2545-2557. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Makino, Y., R. Cao, K. Svensson, G. Bertilsson, M. Asman, H. Tanaka, Y. Cao, A. Berkenstam, and L. Poellinger. 2001. Inhibitory PAS domain protein is a negative regulator of hypoxia-inducible gene expression. Nature 414:550-554. [DOI] [PubMed] [Google Scholar]
- 33.Maltepe, E., G. W. Krampitz, K. M. Okazaki, K. Red-Horse, W. Mak, M. C. Simon, and S. J. Fisher. 2005. Hypoxia-inducible factor-dependent histone deacetylase activity determines stem cell fate in the placenta. Development 132:3393-3403. [DOI] [PubMed] [Google Scholar]
- 34.Manalo, D. J., A. Rowan, T. Lavoie, L. Natarajan, B. D. Kelly, S. Q. Ye, J. G. Garcia, and G. L. Semenza. 2005. Transcriptional regulation of vascular endothelial cell responses to hypoxia by HIF-1. Blood 105:659-669. [DOI] [PubMed] [Google Scholar]
- 35.Maxwell, P. H., M. S. Wiesener, G. W. Chang, S. C. Clifford, E. C. Vaux, M. E. Cockman, C. C. Wykoff, C. W. Pugh, E. R. Maher, and P. J. Ratcliffe. 1999. The tumour suppressor protein VHL targets hypoxia-inducible factors for oxygen-dependent proteolysis. Nature 399:271-275. [DOI] [PubMed] [Google Scholar]
- 36.Obach, M., A. Navarro-Sabate, J. Caro, X. Kong, J. Duran, M. Gomez, J. C. Perales, F. Ventura, J. L. Rosa, and R. Bartrons. 2004. 6-Phosphofructo-2-kinase (pfkfb3) gene promoter contains hypoxia-inducible factor-1 binding sites necessary for transactivation in response to hypoxia. J. Biol. Chem. 279:53562-53570. [DOI] [PubMed] [Google Scholar]
- 37.Oberley, M. J., J. Tsao, P. Yau, and P. J. Farnham. 2004. High-throughput screening of chromatin immunoprecipitates using CpG-island microarrays. Methods Enzymol. 376:315-334. [DOI] [PubMed] [Google Scholar]
- 38.Okada, Y., G. Scott, M. K. Ray, Y. Mishina, and Y. Zhang. 2007. Histone demethylase JHDM2A is critical for Tnp1 and Prm1 transcription and spermatogenesis. Nature 450:119-123. [DOI] [PubMed] [Google Scholar]
- 39.Ozer, A., and R. K. Bruick. 2007. Non-heme dioxygenases: cellular sensors and regulators jelly rolled into one? Nat. Chem. Biol. 3:144-153. [DOI] [PubMed] [Google Scholar]
- 40.Park, S. K., A. M. Dadak, V. H. Haase, L. Fontana, A. J. Giaccia, and R. S. Johnson. 2003. Hypoxia-induced gene expression occurs solely through the action of hypoxia-inducible factor 1alpha (HIF-1alpha): role of cytoplasmic trapping of HIF-2alpha. Mol. Cell. Biol. 23:4959-4971. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Partch, C. L., P. B. Card, C. A. Amezcua, and K. H. Gardner. 2009. Molecular basis of coiled coil coactivator recruitment by the aryl hydrocarbon receptor nuclear translocator (ARNT). J. Biol. Chem. 284:15184-15192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Pirooznia, M., V. Nagarajan, and Y. Deng. 2007. GeneVenn—a web application for comparing gene lists using Venn diagrams. Bioinformation 1:420-422. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Pollard, P. J., C. Loenarz, D. Mole, M. A. McDonough, J. M. Gleadle, C. Schofield, and P. J. Ratcliffe. 2008. Regulation of Jumonji-domain containing histone demethylases by hypoxia inducible factor (HIF) 1-alpha. Biochem. J. 416:387-394. [DOI] [PubMed] [Google Scholar]
- 44.Raval, R. R., K. W. Lau, M. G. Tran, H. M. Sowter, S. J. Mandriota, J. L. Li, C. W. Pugh, P. H. Maxwell, A. L. Harris, and P. J. Ratcliffe. 2005. Contrasting properties of hypoxia-inducible factor 1 (HIF-1) and HIF-2 in von Hippel-Lindau-associated renal cell carcinoma. Mol. Cell. Biol. 25:5675-5686. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Rhodes, D. R., J. Yu, K. Shanker, N. Deshpande, R. Varambally, D. Ghosh, T. Barrette, A. Pandey, and A. M. Chinnaiyan. 2004. ONCOMINE: a cancer microarray database and integrated data-mining platform. Neoplasia 6:1-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Shi, Y., and J. R. Whetstine. 2007. Dynamic regulation of histone lysine methylation by demethylases. Mol. Cell 25:1-14. [DOI] [PubMed] [Google Scholar]
- 47.Tanno, T., N. V. Bhanu, P. A. Oneal, S. H. Goh, P. Staker, Y. T. Lee, J. W. Moroney, C. H. Reed, N. L. Luban, R. H. Wang, T. E. Eling, R. Childs, T. Ganz, S. F. Leitman, S. Fucharoen, and J. L. Miller. 2007. High levels of GDF15 in thalassemia suppress expression of the iron regulatory protein hepcidin. Nat. Med. 13:1096-1101. [DOI] [PubMed] [Google Scholar]
- 48.Tateishi, K., Y. Okada, E. M. Kallin, and Y. Zhang. 2009. Role of Jhdm2a in regulating metabolic gene expression and obesity resistance. Nature 458:757-761. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Turcotte, S., D. A. Chan, P. D. Sutphin, M. P. Hay, W. A. Denny, and A. J. Giaccia. 2008. A molecule targeting VHL-deficient renal cell carcinoma that induces autophagy. Cancer Cell 14:90-102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Tusher, V. G., R. Tibshirani, and G. Chu. 2001. Significance analysis of microarrays applied to the ionizing radiation response. Proc. Natl. Acad. Sci. U.S.A. 98:5116-5121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Wang, G. L., B. H. Jiang, E. A. Rue, and G. L. Semenza. 1995. Hypoxia-inducible factor 1 is a basic-helix-loop-helix-PAS heterodimer regulated by cellular O2 tension. Proc. Natl. Acad. Sci. U.S.A. 92:5510-5514. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Welford, S. M., B. Bedogni, K. Gradin, L. Poellinger, M. Broome Powell, and A. J. Giaccia. 2006. HIF1alpha delays premature senescence through the activation of MIF. Genes Dev. 20:3366-3371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Wellmann, S., M. Bettkober, A. Zelmer, K. Seeger, M. Faigle, H. K. Eltzschig, and C. Buhrer. 2008. Hypoxia upregulates the histone demethylase JMJD1A via HIF-1. Biochem. Biophys. Res. Commun. 372:892-897. [DOI] [PubMed] [Google Scholar]
- 54.Whetstine, J. R., A. Nottke, F. Lan, M. Huarte, S. Smolikov, Z. Chen, E. Spooner, E. Li, G. Zhang, M. Colaiacovo, and Y. Shi. 2006. Reversal of histone lysine trimethylation by the JMJD2 family of histone demethylases. Cell 125:467-481. [DOI] [PubMed] [Google Scholar]
- 55.Xia, X., M. E. Lemieux, W. Li, J. S. Carroll, M. Brown, X. S. Liu, and A. L. Kung. 2009. Integrative analysis of HIF binding and transactivation reveals its role in maintaining histone methylation homeostasis. Proc. Natl. Acad. Sci. U.S.A. 106:4260-4265. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Yamane, K., C. Toumazou, Y. Tsukada, H. Erdjument-Bromage, P. Tempst, J. Wong, and Y. Zhang. 2006. JHDM2A, a JmjC-containing H3K9 demethylase, facilitates transcription activation by androgen receptor. Cell 125:483-495. [DOI] [PubMed] [Google Scholar]
- 57.Yun, Z., H. L. Maecker, R. S. Johnson, and A. J. Giaccia. 2002. Inhibition of PPAR gamma 2 gene expression by the HIF-1-regulated gene DEC1/Stra13: a mechanism for regulation of adipogenesis by hypoxia. Dev. Cell 2:331-341. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.