Abstract
How the endoplasmic reticulum (ER) and the Golgi apparatus maintain their morphological and functional identity while working in concert to ensure the production of biomolecules necessary for the cell's survival is a fundamental question in plant biology. Here, we isolated and characterized an Arabidopsis thaliana mutant that partially accumulates Golgi membrane markers and a soluble secretory marker in globular structures composed of a mass of convoluted ER tubules that maintain a connection with the bulk ER. We established that the aberrant phenotype was due to a missense recessive mutation in sec24A, one of the three Arabidopsis isoforms encoding the coat protomer complex II (COPII) protein Sec24, and that the mutation affects the distribution of this critical component at ER export sites. By contrast, total loss of sec24A function was lethal, suggesting that Arabidopsis sec24A is an essential gene. These results produce important insights into the functional diversification of plant COPII coat components and the role of these proteins in maintaining the dynamic identity of organelles of the early plant secretory pathway.
INTRODUCTION
The secretory pathway is a complex system of functionally interlinked organelles that communicate with each other to ensure the production of vital cellular constituents, such as lipids, sugars, and proteins, and that enables cells to communicate with the external environment. At the heart of the secretory pathway are the endoplasmic reticulum (ER) and the Golgi apparatus. The plant er and Golgi are arguably the most dynamic of all secretory organelles: ER labeled with green fluorescent protein (GFP) displays remarkable motility (Knebel et al., 1990; Boevink et al., 1998; Brandizzi et al., 2002b; Runions et al., 2006; Tolley et al., 2008; Sparkes et al., 2009). Underneath the plasma membrane, the cortical ER appears as a highly dynamic meshwork of interconnected tubular membranes that are arranged in a polygonal fashion. ER cisternae and tubules also traverse the central vacuole as transvacuolar strands and connect cortical ER at opposite boundaries of the cell. How the plant ER maintains its basic dynamic anatomy is unknown, although actin seems to play a role in the remodeling and movement of the ER and in the general integrity of this organelle (Tamura et al., 2005; Runions et al., 2006). More recently, overexpression of a member of the membrane curvature-inducing reticulon family has been shown to cause constrictions of the ER lumen (Tolley et al., 2008), suggesting that a combination of cytoskeletal elements and noncytoskeletal proteins may be involved in the maintenance of ER structure. In plants, the ER is generally juxtaposed to the Golgi due to the lack of an intermediate compartment (Hawes and Satiat-Jeunemaitre, 2005). The plant Golgi consists of polarized ministacks that are highly motile (moving at rates of up to 4 μm/s) on an actin-myosin system (Boevink et al., 1998; Nebenfuhr et al., 1999). In highly vacuolated cells, Golgi stacks are attached to ER tubules (Sparkes et al., 2009) and to the ER subdomains dedicated to protein export, so-called ER export sites (ERESs) (daSilva et al., 2004; Stefano et al., 2006; Hanton et al., 2009).
The plant ER and Golgi apparatus maintain their distinctive morphology and dynamics even as they entertain a continuous anterograde and retrograde flow of membranes and proteins between each other (Brandizzi et al., 2002b; daSilva et al., 2004). How the two organelles maintain their integrity despite this continuous exchange of molecules is largely unknown. It has been suggested that proteins such as golgins may be involved in the establishment and maintenance of the plant Golgi structure (Latijnhouwers et al., 2005a), but experimental evidence in support of this idea has yet to be produced. Similar to other cellular systems, protein traffic at the plant ER/Golgi interface is regulated by at least two different machineries, called coat protomer complex I (COPI) and COPII (Serafini et al., 1991; Barlowe et al., 1994; Pimpl et al., 2000; Phillipson et al., 2001; daSilva et al., 2004; Stefano et al., 2006). In yeast and mammals, the COPII coat is believed to form transport vesicles on the surface of the ER that shuttle anterograde cargo and trafficking machinery to the Golgi apparatus (Barlowe et al., 1994). The small GTPase Sar1p (for Secretion-Associated, Ras-related protein 1) that regulates COPII coat assembly on the ER is activated by Sec12, an ER membrane-anchored guanine nucleotide exchange factor. Activation of Sar1 is followed by its binding to the ER membrane and recruitment of the COPII coat heteromeric components Sec23/24 and Sec13/31. Each component of the coat has a specific role in protein traffic; for example, Sec23 acts as a Sar1-GAP (GTPase activating protein), and Sec24 is believed to be involved in cargo selection via site-specific recognition of cargo motifs and signals (reviewed in Sato and Nakano, 2007). Protein traffic studies have clearly established that correct assembly of COPII at ERESs plays a critical role in maintaining ER and Golgi membrane integrity. In particular, overexpression of Sec12 or of dominant-negative mutants of Sar1 proteins affects Golgi integrity and results in reabsorption of Golgi membranes into the ER (Andreeva et al., 2000; Takeuchi et al., 2000; daSilva et al., 2004; Hanton et al., 2008).
The Arabidopsis thaliana genome encodes a large number of COPII proteins (Robinson et al., 2007), but the significance of this diversity has yet to be investigated. Functional analyses based on the secretion of bulk flow markers indicated that overexpression of GTP-restricted mutants of two Arabidopsis Sar1 isoforms caused different levels of ER export inhibition in tobacco (Nicotiana tabacum) cells (Hanton et al., 2008). Furthermore, the subcellular distribution of the two Arabidopsis Sar1 proteins was similar, yet nonoverlapping. Importantly, the effect on secretion and the subcellular localization of the two GTPases was influenced by the nature of one amino acid residue at their C terminus (Hanton et al., 2008). These results indicate that functional heterogeneity may exist among plant COPII protein isoforms and that other amino acid changes besides the well-characterized Sar1 dominant-negative mutations can influence the localization and function of the COPII machinery. These findings are intriguing and suggest the exciting possibility that other unidentified COPII mutations may similarly affect ER and Golgi membrane integrity in plants.
To learn more about the mechanisms that regulate the integrity of the early plant secretory pathway, we screened Arabidopsis seedlings from an ethyl methanesulfonate (EMS) population for mutants that showed aberrant distribution of an established Golgi marker, cytosolic tail, and transmembrane domain of a rat sialyl-transferase fused to GFP (ST-GFP; Boevink et al., 1998). This fluorescent marker is normally visible in Golgi stacks; variations in its distribution to distal and proximal locations indicate altered Golgi functional and morphological integrity in relation to membrane retention and/or membrane transport to other secretory organelles (Boulaflous et al., 2008). Here, we report on the identification and characterization of a novel ST-GFP Arabidopsis mutant bearing an amino acid mutation in one of the three Arabidopsis isoforms of the COPII coat protein, Sec24A. This mutation affected the recruitment of Sec24A to ER export sites and was linked to the formation of aberrant tubular clusters of ER and Golgi membranes. Because a sec24A knockout was lethal, the mutant phenotype appears to be linked to partial loss of function of sec24A. These data suggest a model in which COPII coat proteins are important not only for maintaining ER and Golgi membrane integrity in relation to ER protein export in plants but also for helping to maintain the integrity of the ER tubular network.
RESULTS
A Screen of EMS-Mutagenized Arabidopsis Seedlings Expressing a Golgi-GFP Marker Leads to the Identification of a Novel Mutant of Plant Endomembranes
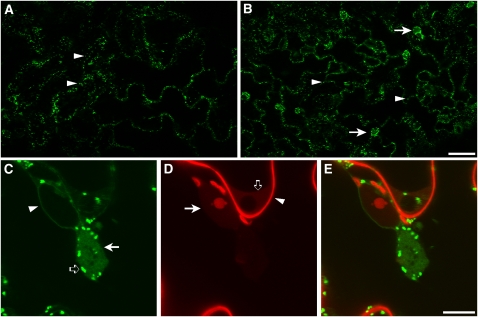
Genetic screens based on the analysis of EMS-treated plants expressing fluorescent protein markers have been successful for the identification of genes regulating the organization and activity of secretory organelles in plant cells (Avila et al., 2003; Tamura et al., 2005; Fuji et al., 2007; Teh and Moore, 2007; Boulaflous et al., 2008). To identify mutants of the plant Golgi apparatus, we analyzed seedlings from an EMS-mutagenized population of Arabidopsis Columbia-0 (Col-0) plants stably expressing the plant Golgi marker ST-GFP (Boevink et al., 1998). This marker is based on the fusion of a transmembrane domain and the cytosolic tail of a rat sialyl-transferase to mGFP5; it is preferentially localized at the Golgi in Nicotiana spp (Boevink et al., 1998; Brandizzi et al., 2002b; Saint-Jore et al., 2002; daSilva et al., 2004; Latijnhouwers et al., 2007) and in Arabidopsis cells (Figure 1A; Peiter et al., 2007). In some Arabidopsis cotyledon epidermal cells, the fusion protein is also visible in the nuclear envelope (see Supplemental Figure 1 online). Low-magnification confocal microscopy analyses of homozygous seedlings of one ST-GFP mutant, hereby named G92, showed that in addition to Golgi stacks, this marker was also localized to large fluorescent globular structures (diameter range 7.5 to 16.5 μm) in most of the cells of the cotyledons (Figure 1B). We did not observe obvious developmental differences between the G92 mutant compared with nonmutagenized ST-GFP and untransformed nonmutagenized wild-type plants (see Supplemental Figure 2 online).
Figure 1.
Identification of a Novel Mutant of the Plant Endomembranes.
(A) and (B) Confocal microscopy images of live cotyledon epidermal cells of Arabidopsis of the control (nonmutagenized ST-GFP; [A]) and of the G92 mutant (B). Arrowheads show the presence of fluorescent spots that correspond to Golgi stacks. Note the presence of additional globular structures in the G92 sample (arrows in [B]) compared with the control (A). Bar = 50 μm.
(C) to (E) High-magnification confocal live-cell images of epidermal cells of the G92 (C) mutant treated with propidium iodide (D), which labels the cell wall ([D], arrowhead) and nucleic acids in the nucleoplasm ([D], arrow) and cytoplasm ([D], open arrow). The cell was imaged with confocal settings for GFP and propidium iodide (see Methods). Note the appearance of the fenestrated G92 globular structures ([C], arrow) that are close to the nucleus and that contain diffused and punctate ST-GFP fluorescence. In (C), an arrowhead and an open arrow point to the nuclear envelope that is also visible in the control (see Supplemental Figure 1 online) and to Golgi stacks, respectively. (E) shows merged images of (C) and (D). Bar = 10 μm.
We generally found one globular structure in the periphery of each cotyledon cell, often juxtaposed to the nucleus in the mid to lower section of the cells (Figures 1C to 1E). In some G92 cotyledon epidermal cells, ST-GFP also labels the nuclear envelope, as in nonmutagenized cotyledons expressing ST-GFP (Figures 1C to 1E). The globular structures were mostly stationary and showed faint, yet fenestrated, GFP fluorescence and enveloped Golgi stacks (Figures 1C to 1E). We also found that Golgi stacks inside the globular structures were not as motile as those distributed at the periphery of the structures and at the cortex of the cell (see Supplemental Movie 1 online).
To establish whether the presence of the aberrant structures was linked to the expression of ST-GFP, we stained cotyledons of G92xLer F2 segregants that did not express ST-GFP with the endomembrane dye DiOC6 (Zheng et al., 2004). Confocal analyses showed the presence of G92 globular structures in DiOC6-treated cells but not in the control (i.e., nonmutagenized ST-GFP; see Supplemental Figure 3A online), thus excluding the possibility that the presence of the globular structures is linked to the overexpression of the ST-GFP transgene.
The G92 Structures Contain Markers Destined for Various Golgi Cisternae
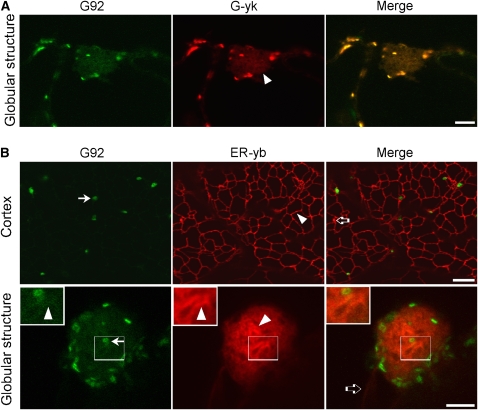
To determine whether the G92 phenotype was linked exclusively to an aberrant distribution of ST-GFP and whether the G92 structures contained proteins destined for cisternae other than the trans-Golgi, where ST-GFP normally resides (Boevink et al., 1998; Latijnhouwers et al., 2005b), we crossed the G92 mutant line with lines stably expressing the cis/medial-Golgi marker, G-yk (Nelson et al., 2007). This marker is based on a yellow fluorescent protein (YFP) fusion of the cytoplasmic tail and transmembrane domain (the first 49 amino acids) of soybean (Glycine max) Man1, soybean α-1,2-mannosidase I (Saint-Jore-Dupas et al., 2006; Nelson et al., 2007). Colocalization analyses were possible because the spectral properties of mGFP5 allow efficient spectral separation from YFP (Brandizzi et al., 2002a). Confocal microscopy analyses of F2 segregants showed that in homozygous G92 plants, the ST-GFP and G-yk signals codistributed at the Golgi and at the G92 globular structures (Figure 2A). Taken together, these data indicate that, compared with a nonmutagenized ST-GFP control, the G92 mutant shows an additional subcellular structure containing multiple distinct Golgi cisternal markers.
Figure 2.
In Addition to Containing the trans-Golgi Marker ST-GFP, the G92 Structures Contain cis- and medial-Golgi Markers and Convoluted ER Tubules.
(A) Confocal live-cell images of a cotyledon epidermal cell of a G92 homozygous F2 plant expressing the cis/medial-Golgi marker G-yk. Note that the G-yk fluorescence in the globular structure (arrowhead) overlaps with that of ST-GFP. Bar = 5 μm.
(B) Cortex panels: Confocal images of the cortex and of a globular structure of a G92 cotyledon cell from a T1 seedling transformed with the ER marker ER-yb. Note the presence of a reticulated ER network of tubules with organization similar to that of a control cell (nonmutagenized ST-GFPx35S:ER-yk; see Supplemental Figure 4 online). Golgi stacks labeled with ST-GFP (G92 panel, arrow) lie on the ER tubules (arrowheads). The open arrow in the merge image shows one of the infrequent punctae that were labeled by ER-yb but not by ST-GFP in the G92 cotyledons. Globular structure panels: The G92 globular structure contains convoluted ER tubules (ER-yb panel, arrowhead) and Golgi (G92 panel, arrow; see also insets and Supplemental Figure 2 online). The ST-GFP fluorescence of the G92 mutant (G92 panel) overlaps with that of the ER marker in the globular structures (arrowheads). Insets show magnified sections of main panels (boxed area × 2). ER tubules extending to the G92 structure and labeled by ER-yb are indicated by an open arrow in (B) in the merged image. Bars in main images = 5 μm.
The G92 Structures Are Clusters of Tubular ER and Golgi Membranes
To further determine the identity of membranes that comprise the G92 structures, we transformed the G92 mutant with the ER marker ER-yb, a soluble marker based on secretory YFP targeted to the ER by virtue of the signal peptide of the Arabidopsis Wall-Associated Kinase 2 and retained in the ER by the tetrapeptide HDEL (Nelson et al., 2007). Analyses of epidermal cells of T1 cotyledons showed that within the globular G92 structures, the ER coalesced into a tangle of tubular membranes (Figure 2B; see Supplemental Movie 2 and Supplemental Figure 4 online). This was markedly different from the network-like appearance of the cortical ER visualized in the same cells (Figure 2B) and in the control (i.e., ST-GFPX35S:ER-yk cotyledons; see Supplemental Figure 4 online). In addition to the ER tubular network of the G92 cotyledons, ER-yb also highlighted occasional punctae of unknown identity that did not colocalize with Golgi stacks (Figure 2B).
In the G92 mutant, faint ST-GFP fluorescence overlapped with that of the ER tubules at the globular structures, resulting in the fenestrated appearance of the G92 structures (Figure 2B, insets).
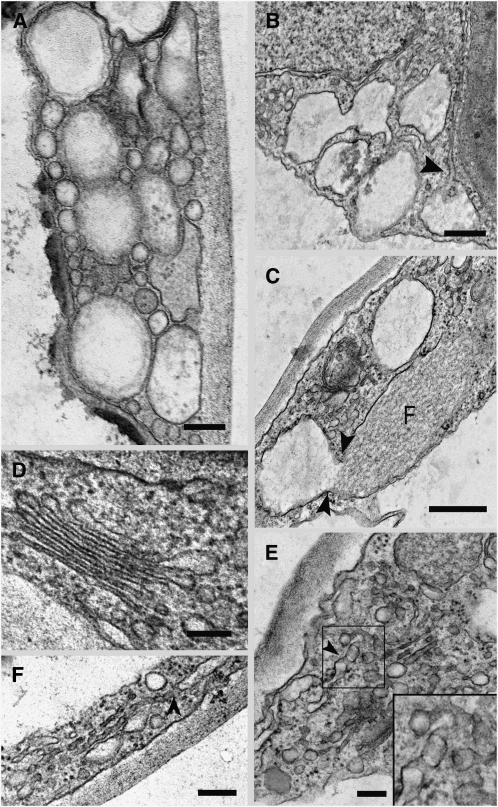
To analyze the globular structures at a submicron resolution, we performed electron microscopy analyses. Epidermal cells of cotyledonal leaves from G92 mutant plants contained characteristic aggregations of dilated vesicles of various sizes of up to 300 nm in diameter (Figure 3A). These appeared to be continuous with the ER (Figure 3B, arrowhead), and this continuity was confirmed by obvious dilation of ER membrane attached to fusiform ER bodies that are known to reside in the lumen of the ER (Figure 3C; Hawes et al., 2001; Matsushima et al., 2003). Compared with Golgi of control plants (Figure 3D) or G92 Golgi distributed outside the globular structures (see Supplemental Figure 5 online), Golgi bodies trapped in the globule structures often showed drastically curved profiles (Figure 3C) or exhibited a pronounced vesicular/tubular profile (Figures 3E and 3F). These Golgi tubules often appeared to be associated with the ER (Figures 3E and 3F, arrowheads).
Figure 3.
Ultrastructure of the G92 Mutant.
Electron microscopy analyses of the G92 cotyledons show alteration of the ER and Golgi in the globular structures.
(A) Aggregation of different sized vesicular clusters in the cytoplasm of a G92 mutant cotyledonal leaf epidermal cell. Bar = 100 nm.
(B) Large vesicular structures in the epidermal cells are connected to the ER (arrowhead). Bar = 200 nm.
(C) Continuity between a fusiform body (F) in the lumen of the ER (Hawes et al., 2001) and a vesicular structure. Bar = 200 nm.
(D) Golgi stack in an epidermal cell of a control cotyledonal leaf from a plant stably transformed with ST-GFP. Bar = 100 nm.
(E) Disrupted Golgi stack. Golgi remnants appear to be vesicular and tubular in structure and can connect with the ER (see arrowhead). Inset shows a detail in the boxed area (magnified × 2) in the main panel. Bar = 100 nm.
(F) Disrupted Golgi stack in a G92 cotyledonal leaf epidermal cell showing apparent continuity with the ER (arrowhead). Bar = 100 nm.
The ER Membranes of the G92 Structures Do Not Form an Isolated Subcellular Compartment
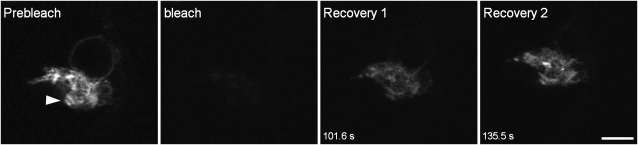
We next sought to further establish whether the membranes of the G92 clusters formed a distinct compartment, separate from the morphologically normal ER. We hypothesized that the G92 membranes would at least be partially continuous with the ER, based on the evidence that Golgi stacks, which are known to be in close association with the ER (Boevink et al., 1998; Sparkes et al., 2009), were able to move to and from the periphery of the clusters (see Supplemental Movie 1 online) and because ER-yb–labeled tubules emanated from the G92 clusters (Figure 2B; see Supplemental Movie 2 online). We therefore performed fluorescence recovery after photobleaching (FRAP) analyses in G92 cotyledonal cells expressing ER-yb to selectively photobleach ER-yb fluorescence (daSilva et al., 2004). FRAP experiments are based on the concept that to maintain steady state fluorescence levels in a cellular compartment, there must be a balance between protein import and export to and from such a compartment (Lippincott-Schwartz et al., 2003). Therefore, we predicted that if the ER tubules of the G92 structures formed isolated clusters, the ER-yb fluorescence within an entire globular structure would not recover after photobleaching. However, as shown in Figure 4, upon bleaching, the fluorescence was recovered in the globular structures. These data confirm that the G92 membranes are continuous with the ER.
Figure 4.
The G92 Globular Structures Do Not Form Isolated Clusters of Membranes.
FRAP analyses of the ER soluble marker, ER-yb, within the entire globular structure (arrowhead) in G92 cells expressing the marker shows recovery of fluorescence upon photobleaching, indicating that the structure is not an isolated mass of membranes and that it is continuous with the ER. Time points of the recovery phase from the bleach event are indicated in the bottom left corner of frames. Bar = 10 μm.
The Globular Structures Are Entwined by Actin Cables, but Their Maintenance Is Not Reliant on the Actin Cytoskeleton
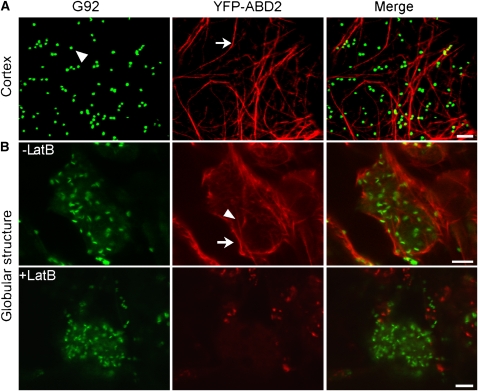
In plant cells, the ER is closely associated with the actin cytoskeleton, and the remodeling and integrity of the ER tubules is reliant on actin (Boevink et al., 1998; Brandizzi et al., 2002b, 2003; Tamura et al., 2005; Runions et al., 2006). To establish whether the presence of G92 structures was due to the absence of actin at the structures, we transformed G92 plants with a YFP fusion to the actin binding domain 2 (ABD2) of Arabidopsis fimbrin, a well-established marker for plant actin (Sheahan et al., 2004). Analyses of T1 G92/35S:YFP-ABD2 cotyledons showed that the actin appeared in clearly defined cable-like bundles at the cell cortex (Figure 5A), as previously reported in Arabidopsis cotyledons (Hardham et al., 2008). Actin cables enveloped the G92 globular structures, and short cables were also present inside the structures (Figure 5B), thus excluding the possibility that the rearrangement of the ER at the G92 structures is linked to a localized absence of the actin cytoskeleton.
Figure 5.
The G92 Globular Structures Are Enwrapped in Actin Cables, and Their Maintenance Is Independent of the Integrity of the Actin Cytoskeleton.
Confocal images of cotyledon epidermal cells of T1 G92 transformants expressing the actin marker YFP-ABD2 (G92/35S:YFP-ABD2). Bars = 5 μm.
(A) Images taken at the cortex of the cell show distended actin cables (arrow) and Golgi bodies labeled with ST-GFP (G92 panel, arrowhead) that appear often in association with actin (see merge panel).
(B) In cross sections of untreated cotyledon cells (-LatB), actin cables appear to form a basket around the G92 structure (arrow) and are also visible inside the structure (arrowhead). The G92 structures withstood treatment with latrunculin B (+LatB) despite the actin cables having lost their organization.
To determine whether the actin cytoskeleton was required for maintenance of the integrity of the G92 structures, we treated cells with latrunculin B, an actin depolymerizing agent (Brandizzi et al., 2002b). We subjected cotyledons to treatment with latrunculin B (25 μM for 1 h) that is known to disrupt the integrity of the actin cytoskeleton without affecting cell viability (Brandizzi et al., 2002b; Stefano et al., 2006). Confocal microscopy analyses showed that the G92 structures largely withstood disruption of the actin cytoskeleton (Figure 5B), indicating that maintenance of the G92 structures is not actin dependent.
G92 Is Allelic to Sec24A, a Component of the COPII Coat
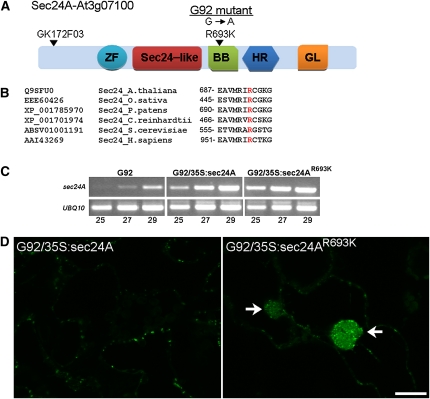
We believe the G92 line represents a novel Arabidopsis mutant with a region of the ER encapsulated in an actin basket and with a unique rearrangement of ER tubular membranes. Also contained within these ER clusters are Golgi markers and enveloped Golgi bodies. To identify the gene affected in this line, we performed map-based cloning with codominant, cleaved amplified polymorphic sequence markers and simple sequence length polymorphism markers (Konieczny and Ausubel, 1993; Bell and Ecker, 1994). We then performed fine mapping using 462 F2 plants that exhibited the G92 phenotype. DNA sequencing showed a single base pair mutation from G to A in one of the three isoforms of the COPII coat component, Sec24-At3g07100, hereby named Sec24A (Figure 6A). The missense mutation causes conversion of an Arg residue 693 to a Lys residue (Sec24AR693K) in a region of the protein conserved among Sec24 proteins from other organisms that is considered to be important for cargo binding (Miller et al., 2003) (Figure 6B).
Figure 6.
The G92 Mutation Affects the Arabidopsis Sec24A Isoform in a Highly Conserved Amino Acid Residue, and It Is Recessive.
(A) Schematic diagram of Arabidopsis Sec24A indicating the insertion of the GABI line GK-172F03. The position of the EMS-induced mutation that results in an amino acid residue change in R693K is also indicated. Protein domains were identified using Conserved Domain Architecture Retrieval Tool: ZF, zinc finger domain Sec23/24; Sec24-like, Sec23/24 α/β trunk domain; BB, Sec23/24 β-barrel domain; HR, Sec23/24 all-helical region; GL, gelsolin-like repeat.
(B) An alignment of the amino acid region containing the Arabidopsis Sec24A R693 residue (highlighted in red) with regions of Sec24 proteins from rice (Oryza sativa, EEE60426), moss (Physcomitrella patens, XP_001785970), algae (Chlamydomonas reinhardtii, XP_001701974), yeast (Saccharomyces cerevisiae_ABSV01001191), and mammals (Homo sapiens, AAI43269) shows that this amino acid residue is highly conserved. (Source: National Center for Biotechnology Information Protein database).
(C) Complementation experiments: A comparison of RT-PCR products (25, 27, and 29 cycles; indicated below the gel) from cotyledons of G92, G92/35S:Sec24A, and G92/35S:Sec24AR693K obtained using primers designed to amplify Sec24A (see Supplemental Table 1 online) shows higher levels of expression of wild-type and mutant Sec24A alleles in transformed lines compared with untransformed G92 seedlings. Amplification was performed on 25 ng of cDNA. Amplification of UBQ10 was used as a control. Additional evidence for plant transformation is provided in Supplemental Figure 6 online.
(D) Confocal images of cross sections of cotyledon epidermal cells of T1 G92/35S:Sec24A and G92/35S:Sec24AR693K plants show presence of the G92 structures in the G92/35S:Sec24AR693K sample (arrows) but not in the G92/35S:Sec24A sample, indicating that the G92 phenotype is complemented by wild-type Sec24A but not Sec24AR693K. Bar = 20 μm.
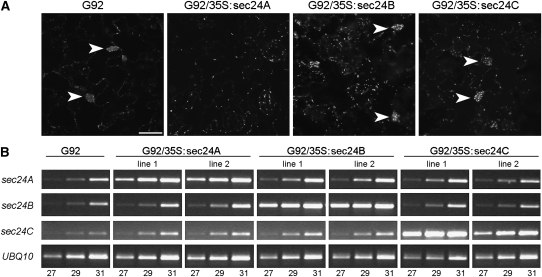
To confirm that the sec24AR693K mutation was indeed responsible for the G92 phenotype, we stably transformed G92 plants with either sec24A or sec24AR693K. Confocal microscopy analyses of the T1 cotyledons showed that the phenotype was complemented by sec24A but not by sec24AR693K (Figures 6C and 6D; see Supplemental Figure 6 online), indicating not only that this mutation behaves in a fully recessive fashion but also that it is responsible for the observed phenotype. To further explore this, we analyzed the subcellular phenotype of G92 plants overexpressing sec24B or sec24C, the two other Arabidopsis sec24 isoforms (Figure 7; see Supplemental Figure 7 online), which are also expressed in cotyledons (see Supplemental Figure 8 online). We established that the G92 phenotype was not complemented by overexpression of either isoform (Figure 7), further supporting the specificity of the mutation in sec24A as a cause for the G92 phenotype.
Figure 7.
sec24B and sec24C Fail to Complement sec24AR693K.
(A) Confocal images of T1 G92 cotyledon epidermal cells alone or transformed with 35S:sec24B or 35S:Sec24C constructs show the presence of globular structures (arrowheads) similar to those in G92 (arrowheads). This is in marked contrast to G92 plants transformed with 35S:Sec24A, which complements the phenotype. Bar = 40 μm.
(B) Expression levels of the sec24 transgenes were analyzed by RT-PCR from two independent T1 G92/35S:Sec24A, Sec24B, and Sec24C lines. Although the transcription levels of sec24B and sec24C in the respective transgenic lines are comparable to sec24A in complemented transgenic plants, sec24B and sec24C failed to complement the sec24AR693K phenotype (A). Amplification was performed on 25 ng of cDNA. Amplification of UBQ10 was used as control. The number of PCR cycles is indicated below the RT-PCR bands. These data are representative of two biological replicates and two technical replicates. Additional evidence for plant transformation is provided in Supplemental Figure 7 online.
Despite the Mutation in the COPII Coat Component Sec24A, the Distribution of Endomembrane Markers Is Compromised at the Aberrant Structures Rather Than at the General ER
Because the G92 mutation was in a component of the COPII coat, we aimed to analyze how the distribution of secretory membrane and soluble cargos destined to non-ER compartments would be affected in the mutant. We first aimed to test the effect of the Sec24AR693K mutation on soluble bulk cargo. To do so, we analyzed the distribution of an ER-targeted red fluorescent protein (secRFP) that is not retained in the ER, which is normally detectable in the apoplast (Samalova et al., 2006). In control plants (i.e., ST-GFP/35S:secRFP; Figure 8A) the marker accumulated in the apoplast and, occasionally, in small punctae, which may correspond to ER bodies where secretory soluble proteins are known to accumulate (Teh and Moore, 2007). In G92 cotyledonal cells, besides the small punctae that resemble those also seen in the control, we did not observe intracellular accumulation of secRFP; we did, however, detect RFP signal in the large globular structures (Figure 8B).
Figure 8.
The Distribution of the Soluble Marker secRFP Is Not Visibly Affected at the Level of the Bulk ER.
(A) Sequential confocal optical sections (8 × 1 μm with 0.0-μm section corresponding to the cell surface [arrow]) of the nonmutagenized T1 ST-GFP line (n.m. ST-GFP; pseudocolored green) expressing the soluble secretory marker secRFP (pseudocolored red) show that the marker reaches the apoplast (open arrowhead in the 4.0 μm panel), and it is not detectable in the cortical ER. secRFP punctae that might correspond to ER bodies are indicated by two arrowheads in the 4.0 and 6.0 μm panels. Bar = 5 μm.
(B) Images of cotyledonal epidermal cells from a G92 T1 plant transformed with secRFP, acquired using the same confocal microscope settings (confocal optical section, laser, photomultipliers, and magnification) as the control (A) show that secRFP is not retained in the cortical ER. secRFP fluorescence is clearly detectable in the apoplast (open arrowhead in the 4.0 μm panel) and at the globular structures (open arrow in the 3.0 μm panel). secRFP was also seen in some cells to accumulate in small globular structures (arrowheads in the 5.0 and 7.0 μm panels) that might either correspond to ER bodies as in the control (A) (Teh and Moore, 2007) or to the punctate structures that are sometimes visible in the ER of the G92 mutant (Figure 2B). An arrow points to the cell surface.
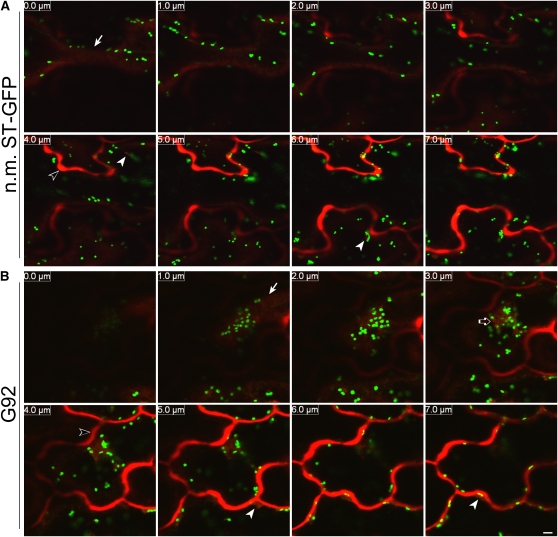
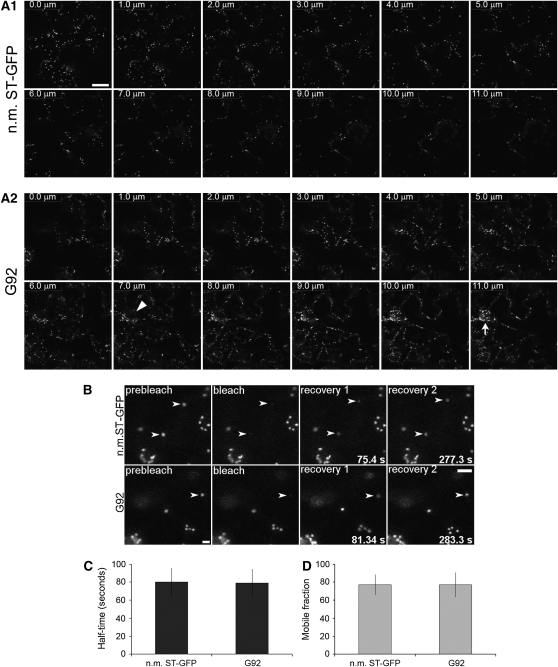
We next sought to establish whether line G92 showed defects in membrane cargo distribution as well as membrane protein traffic at the ER/Golgi interface. ST-GFP appeared to be a good marker for this, as it is localized preferentially in the Golgi. Furthermore, the absence of known ER export motifs that may be selectively recognized by Sec24 (Wendeler et al., 2007) in the nine–amino acid (MIHTNLKKK) cytosolic tail of ST-GFP suggested that the marker would be appropriate for tracking bulk membrane cargo. In our microscopy observations of G92 cotyledon cells presented above, we did not detect the ST-GFP signal in the ER, except for labeling of the globular structures and of the nuclear envelope (Figures 1, 2, 5, and 6). To confirm the accuracy of this observation, we performed optical slicing and three-dimensional reconstructions of sequential optical sections (>12 sections × 1-μm thickness) through G92 cotyledon epidermal cells and compared them with optical sections through the corresponding cells of nonmutagenized ST-GFP cotyledons. These analyses showed a lack of obvious redistribution of ST-GFP to the ER outside the G92 structures (Figure 9A; see Supplemental Movie 3 online).
Figure 9.
Membrane Cargo Distribution and Protein Dynamics in the G92 Mutant.
(A) Sequential confocal optical sections (12 × 1 μm thickness) of cotyledon epidermal cells of nonmutagenized ST-GFP (n.m. ST-GFP; control; panel A1) and G92 seedlings (panel A2). Section 1 corresponds to the top section at the cell's cortex, and section 12 represents the innermost section. Consecutive slices are indicated by numbers at the top left corner. Note that there is no obvious redistribution of ST-GFP fluorescence outside the Golgi stacks (punctae) in the control. In the G92 sample, ST-GFP fluorescence is visible at the G92 structure (arrow); otherwise, it is contained within the Golgi stacks. A nuclear envelope proximal to the G92 structure is indicated by an arrowhead. See Supplemental Movie 3 online for a three-dimensional reconstruction of the optical slices. Bar = 20 μm.
(B) to (D) FRAP analyses on Golgi stacks at the cortex of cotyledon epidermal cells of nonmutagenized ST-GFP (n.m. ST-GFP) and G92 seedlings. Time of acquisition of individual frames after bleaching is indicated at the bottom right corner of frames (B). Bar = 5 μm. There was no significant difference (P < 0.05) in fluorescence recovery half times (C) and mobile fraction (%) between samples (D). Sample size (i.e., number of bleached Golgi stacks) = 13 for each sample. Error bars represent sd of the mean.
To test whether, in the G92 mutant, protein exchange between the ER and the Golgi, where COPII operates, occurs in a similar manner as in nonmutagenized ST-GFP plants, we performed FRAP analyses of Golgi stacks in G92 plants using Golgi of nonmutagenized ST-GFP cotyledons as controls. If the ER/Golgi interface was functionally affected by the Sec24A mutation, we would expect significant differences in half-time of fluorescence recovery and mobile fraction (i.e., the fraction of fluorescent molecules that move between bleached and unbleached regions) upon photobleaching of G92 Golgi stacks in comparison to nonmutagenized ST-GFP Golgi (Brandizzi et al., 2002b). As shown in Figure 9B, photobleaching of the fluorescence in Golgi stacks at the cell cortex was followed by recovery in both G92 and nonmutagenized ST-GFP cotyledon cells without significant differences in the fluorescence recovery half-times (Figure 9C; P < 0.05) and in the mobile fraction in bleached G92 and control Golgi (Figure 9D; P < 0.05). Taken together, these data indicate that the G92 allele does not significantly affect protein exchange between the ER and Golgi, at least not at the cell cortex. A direct comparison of the recovery rates between the Golgi at the cell cortex and Golgi within the globular structures was not feasible because of the low signal-to-noise fluorescence level in the Golgi in the clusters in comparison to Golgi at the cell cortex. However, qualitative photobleaching experiments of the Golgi stacks within G92 structures showed that the fluorescence of the Golgi membranes inside the structures recovered upon bleaching (see Supplemental Movie 4 online); thus, the Golgi bodies inside the G92 structures could also exchange membrane cargo with the ER.
These data indicate that, except for a partial retention in the globular structures of markers that are normally destined to proximal or distal locations, the Sec24AR693K mutation does not result in detectable differences in the distribution of these markers compared with a control or in a significant disruption of the ER/Golgi interface activities outside the globular structures.
Arabidopsis Sec24A Is Important for Viability
We next wanted to establish whether the G92 phenotype was linked to a total or a partial loss of function of Sec24A in COPII-mediated protein transport. Because wild-type sec24A did not complement a yeast sec24 temperature-sensitive mutant (see Supplemental Figure 9 online), we tested for linkage to the G92 phenotype using segregation analysis. We obtained GK-172F03, an allele that contains a T-DNA insertion located in the second exon of sec24A and is most likely null. Genomic DNA analysis of 72 GK-172F03 seedlings did not yield any homozygous progeny. Based on these data, we concluded that sec24A is likely an essential gene. To further test this possibility, we propagated three heterozygous plants and genotyped 48 10-d-old seedlings from three individuals using PCR (see Supplemental Figure 10 online). The total of 144 (i.e., 3 × 48) included 56 wild-type and 88 heterozygous seedlings. None of the progeny were homozygous for this sec24A allele in all 144 seedlings analyzed.
These data indicate that sec24A is an essential gene and that the other Sec24 isoforms cannot compensate for the loss of sec24A, at least not at the early stages of plant growth and development.
The R693K Mutation Affects the Distribution of Sec24A
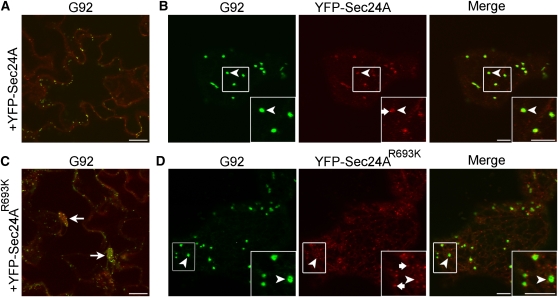
The R693K mutation of Arabidopsis Sec24A occurs at a position corresponding to R561 in yeast Sec24 (Figure 6), which has been implicated in cargo binding (Miller et al., 2003). Therefore, we hypothesized that the R693K mutation could affect the behavior of Sec24A at ERESs where Sec24 would bind cargo. To test this, we compared the fluorescence distribution of YFP-Sec24 and YFP-Sec24AR693K in cotyledons of stably transformed G92 plants. We reasoned that YFP-Sec24A and YFP-Sec24AR693K would compete for ERES binding with the endogenous Sec24AR693K protein and, therefore, that differences in the localization would be specific for each Sec24A protein. Confocal analyses showed that, similar to the situation for untagged Sec24A proteins (Figure 6), expression of wild-type YFP-Sec24A, but not YFP-Sec24AR693K, complemented the mutant G92 cluster phenotype (Figures 10A and 10B). As shown in Figure 10B, YFP-Sec24A was localized at Golgi-associated ERESs (inset, arrowhead), in addition to small punctae (inset, arrow) and the cytosol, consistent with previous observations (Hanton et al., 2009). This distribution was in clear contrast with that of YFP-Sec24AR693K (Figure 10D). Indeed, we could not verify the presence of well-defined Golgi-associated ERESs similar to those labeled by YFP-Sec24A when using YFP-Sec24AR693K (inset, arrowhead); rather, the fluorescence of this marker appeared to be predominantly distributed in the cytosol and in dispersed bright punctae (Figure 10D, inset arrows) in contrast with the distribution of YFP-Sec24A fluorescence, suggesting that the distribution of YFP-Sec24A was affected by the mutation.
Figure 10.
Localization of YFP-Sec24A Is Affected by the R693K Mutation.
Confocal images of epidermal cells of cotyledons of T1 G92 plants expressing either 35S:YFP-Sec24 ([A] and [B]) or 35S:YFP-Sec24AR693K ([C] and [D]).
(A) The YFP-Sec24A construct complements the Sec24AR693K phenotype.
(B) At a higher magnification, YFP-Sec24 fluorescence is clearly visible at ERESs (main panels and insets, arrowheads) that are associated with the Golgi (main panels and insets, arrowheads). YFP-Sec24 fluorescence is also visible in the cytosol and at some small punctae (inset, arrow), consistent with previous reports (Stefano et al., 2006; Hanton et al., 2007, 2009).
(C) In contrast with YFP-Sec24A, YFP-Sec24AR693K did not complement the G92 mutant phenotype.
(D) Rather than being at Golgi-associated ERESs (main panels and insets, arrowheads), the fluorescence of YFP-Sec24AR693K appears to be distributed mostly at numerous small disperse punctae (inset, arrows). Insets show magnified sections of boxed regions in main images (2×). Bars in panels and insets = 5 μm.
DISCUSSION
Identification of a Mutation of the ER Export Machinery That Induces a Novel Phenotype of the Early Secretory Membranes
Here, we show a novel mutation in the COPII subunit Sec24A that partially affects the morphology of the ER tubular network as well as the Golgi membrane and soluble secretory protein distribution. In contrast with previously reported mutations of the G-protein Sar1 that induce extensive reabsorbance of Golgi membranes into the ER (Takeuchi et al., 2000; daSilva et al., 2004), the main effect of the Sec24A allele described in our work is a partial deformation of the ER network to create a localized membranous cluster containing convoluted ER tubules and proteins destined to other secretory compartments. Therefore, our results add to the repertoire of functions attributable to COPII by indicating that, in addition to protein export from the ER, this machinery is also required for maintenance of the ER tubular network.
The G92 Phenotype May Be Linked to Impaired Export of Isoform-Specific Cargo Necessary for the Maintenance of ER and Golgi Membrane Integrity
The basic machinery required for budding and cargo sorting during ER protein export is the COPII coat, composed of the small GTPase Sar1, the Sec23/24 complex, and the Sec13/31 complex (Barlowe et al., 1994). Over the past few years, several lines of evidence have implicated the Sec24 subunit in the process of cargo recruitment based on the recognition of cytosolic export signals on membrane-anchored proteins (Aridor et al., 1998; Miller et al., 2002; Mancias and Goldberg, 2008). Analyses of crystal structures of the human and yeast Sec24 proteins have established the structural determinants for discrimination among these transport signals (Mancias and Goldberg, 2008). The R693K residue in Arabidopsis Sec24A corresponds to the R561 position in budding yeast Sec24, which is part of the B-site pocket that binds several cargos, including the vesicle-SNARE Bet1 (Miller et al., 2003). Our data show that general protein traffic is not visibly perturbed in the G92 mutant. Furthermore, visualization of the G92 structures in plants that do not express ST-GFP exclude the possibility that the defects in endomembrane organization are linked to overexpression of the transgene. Instead, it is possible that the Sec24A mutant is perturbed in its ability to export an isoform-specific cargo that functions in ER/Golgi organization. This hypothesis is supported by the evidence that, contrarily to sec24A, sec24B and sec24C do not complement the G92 phenotype. SNAREs or other structurally important proteins, including those that recruit cytoskeletal elements, may be the Sec24A-specific cargo. For example, it has been shown that absence of the mouse p31, the mammalian SNARE ortholog of yeast Use1p/Slt1p, causes extensive ER vesiculation and subsequent fusion of the ER structures; the disorganization of the ER in p31 null cells also retarded general ER-to-Golgi transport and caused accumulation of proteins in the ER (Uemura et al., 2009). These findings suggest that it is possible that the G92 mutant lacks (1) efficient export of SNARE that is a Sec24A-specific cargo with similar function to p31 in the regulation of ER tubules and, consequently, (2) the ability to prevent random fusion of ER structures. If constitutive traffic is disrupted, inappropriate fusion of vesicles between the ER and the Golgi may occur, creating an aberrant compartment. This could also explain the redistribution of Golgi markers and secRFP in the ER of the G92 clusters.
Our data do not exclude the possibility that links between the ER and Golgi membranes and the cytoskeleton may not be maintained correctly in the G92 mutant. It has been proposed that the action of the cytoskeleton and associated proteins may be the basis of mechanisms maintaining the tubular structure of the ER in animal cells (Feiguin et al., 1994; Klopfenstein et al., 1998; Vedrenne et al., 2005; Vedrenne and Hauri, 2006). In plant cells, functionally similar links appear to exist: in Arabidopsis, the Golgi protein KATAMARI1/MURUS3 has been shown to serve as an important component for the Golgi-mediated organization of the actin cytoskeleton (Tamura et al., 2005). Although in contrast with G92 the macro phenotype of katamari1 showed obvious growth and developmental defects, subcellular analyses of katamari1 showed alteration of the endomembrane organization of Arabidopsis cotyledons with the appearance of structures similar to the G92 clusters (Tamura et al., 2005). Although the ER of the G92 structures is enwrapped in an actin cage, it is possible that the links between the ER and Golgi membranes are compromised. Based on this hypothesis, the Sec24AR693K mutation may selectively lead to impaired traffic of proteins with similar functions to KATAMARI1 at the ER/Golgi-cytoskeletal interface, with consequent formation of the aberrant G92 membrane clusters.
The localized appearance of the G92 clusters could be the manifestation of a stress response of the entire ER to the effect of the Sec24A mutation, although it is also possible that the Sec24A mutation affects mostly a subdomain of the ER. Since accumulation of soluble and membrane markers in the ER can alter the morphology of this organelle (Crofts et al., 1999; Lee et al., 2002), the Sec24A mutation may affect protein export in a region of the ER and induce formation of the globular structure. In support of this hypothesis, live cell images have shown that Sec24A interacts with ER export motifs exposed on cytosolic tails of membrane proteins in the perinuclear area in broad bean (Vicia faba) guard cells (Sieben et al., 2008).
Simultaneous Partial Redundancy and Specificity of Sec24/Cargo Interaction May Explain Tolerance of Partial Loss of Function of Sec24A
The Arabidopsis genome contains several COPII genes: two Sec12, five Sar1, seven Sec23, three Sec24, two Sec13, and two Sec31 (Robinson et al., 2007). The reason for this variety is yet to be determined. With the exception of a role for the plant Sec24 isoform CEF (Clone Eighty Four) in increasing tolerance to hydroperoxides when overexpressed in yeast (Belles-Boix et al., 2000), functional studies for the other plant Sec24 proteins have yet to be proposed, especially in relation to an endogenous system. Our data support the hypothesis that the G92 phenotype is linked to a partial loss of function of sec24A, which may affect Sec24A distribution, and we have evidence that absence of Sec24A is lethal, at least in early stages of plant growth. Sec24A partial loss of function may be tolerated by plants on the basis of partial functional redundancy among the Arabidopsis Sec24 isoforms. Partial Sec24 redundancy occurs also in nonplant species. Studies in yeast have shown that a knockout of the essential SEC24 gene can be partially rescued by overexpression of the nonessential Sec24 homolog ISS1 (Kurihara et al., 2000). On the other hand, Lst1p, another yeast Sec24 homolog, is specifically required for efficient traffic of Gas1p, a GPI-anchored protein, and Pma1p, a plasma membrane proton ATPase (Peng et al., 2000; Shimoni et al., 2000). Similarly, studies using single knockdowns showed a high degree of redundancy for all mammalian Sec24 isoforms in recognizing cargo proteins and a selective requirement for Sec24A in the traffic of transmembrane cargo with a dileucine signal in the cytosolic tail (Wendeler et al., 2007). It has been suggested that mammalian Sec24 isoforms have evolved by gene duplication events that led to redundancy in cargo binding; selective evolution would then have enhanced the cargo preference of different Sec24 isoforms, without loss of the binding sites for other cargo (Wendeler et al., 2007). This scenario would ensure ER export independently of the availability of a specific isoform (Wendeler et al., 2007). Consistent with this hypothesis, partial loss of function of Sec24A may be compensated for by the other Sec24 isoforms to favor general ER protein export in the G92 mutant; a reduced export of specific Sec24A cargo due to the R693K mutation would induce the formation of the membrane clusters. This is further supported by the evidence that overexpression of either sec24B or sec24C does not complement the sec24AR693K mutation. These data present functional insights in the plant Sec24 family of proteins, and our lab is currently exploring further the degree of functional overlap among Arabidopsis Sec24 isoforms in a homozygous sec24A knockout genetic background.
Our analysis of a sec24A knockout mutant suggests that Sec24A is required in the early stages of plant growth, although we do not exclude the possibility that Sec24A is also required for pollen viability or functionality. Lethality of a sec24A knockout may be linked to two scenarios that are not mutually exclusive: there could be a total loss of export of vital factors from the ER due to (1) Sec24A cargo specificity and/or (2) translationally and/or posttranslationally regulated activity of the Sec24 isoforms (i.e., Sec24A may be the only Sec24 isoform active in the early stages of plant growth or during pollen development and growth).
Concluding Remarks
Compared with yeast and animals, much less is known about mechanisms for ER and Golgi integrity in plants. Yeast conditional mutants and cell-free analyses have led to the identification of numerous genes involved in membrane traffic at the Golgi (Tschopp et al., 1984; Baker and Schekman, 1989). A small interfering RNA genome-wide screen in Drosophila melanogaster has led to the identification of mutants of Golgi architecture (fragmented or swollen Golgi) and mutants with partial redistribution of Golgi proteins into the ER (Bard et al., 2006). Our work shows that a forward genetics approach based on a fluorescent fusion Golgi reporter is an excellent strategy for the identification of mutants of the plant Golgi in relation to other secretory organelles. Through this study, we have also provided functional insights into the Sec24 family of plant proteins and ascribed a novel role to COPII in the regulation of membrane morphology and maintenance. Our data provide the foundation for functional analyses of the other members of the plant Sec24-family, and because the R693K mutation occurs in a conserved residue, translation of our findings to other systems will allow testing of the degree of conservation of fundamental mechanisms for endomembrane integrity.
METHODS
Fluorescent Proteins and Molecular Cloning
The fluorescent proteins used in this study were based on fusions with mGFP5 (Haseloff et al., 1997) and EYFP (Clontech). secRFP was generated by fusion of a sporamin signal sequence to monomeric RFP (Campbell et al., 2002). YFP-ABD2 was generated by splicing the ABD2 coding sequence (Sheahan et al., 2004) downstream of YFP. YFP-ABD2 was subcloned in the pEARLY-Gate 104 binary vector and expressed under the control of the cauliflower mosaic virus 35S promoter. Untagged sec24A, sec24B, sec24C, and sec24AR693K were generated by PCR of cDNA sequences followed by subcloning in the binary vector pEARLY-Gate 100 for 35S-driven expression. YFP-tagged Sec24A and YFP-tagged Sec24AR693K used for Arabidopsis thaliana transformation were generated by subcloning the wild-type Sec24A and Sec24AR693K cDNA into pEARLY-Gate 104 and pENTR3C binary vectors, respectively, for 35S-driven expression. ER-yb and ER-yk refer to the same marker subcloned in two vectors bearing different antibiotic resistance (Nelson et al., 2007). Primer sequences used in this work are listed in Supplemental Table 1 online.
RNA Extraction and PCR Analysis
RNA extraction was performed using the RNeasy plant mini kit (Qiagen). Reverse transcription experiments were performed using the Superscript III first-strand synthesis kit (Invitrogen). PCR experiments were performed in standard conditions and were performed using 0.2 mM deoxynucleotide triphosphate, 0.2 μM primers, and 1 unit of Taq polymerase (Promega).
Plant Materials and Growth Conditions
For Arabidopsis work, we used wild-type plants of Arabidopsis (ecotypes Col and Landsberg erecta) and a transgenic Arabidopsis line (ecotype Col) expressing ST-GFP. We also used the Arabidopsis mutant sec24-1 (GK-172F03) from the GABI-KAT consortium (Max Planck Institut, Cologne, Germany). Surface-sterilized seeds were sown onto 0.8% agar in Murashige and Skoog (MS) medium supplemented with Gamborg's B5 vitamins and 1% (w/v) sucrose and were grown at 21°C under 16-h-light/8-h-dark conditions. Cotyledons used in this work were harvested from 10- to 14-d-old seedlings. For antibiotic selection of primary transformants, surface-sterilized seeds were stratified on MS medium (see above) for 7 d with appropriate selection. Selected seedlings were then transferred to MS medium (see above) without antibiotics for at least 4 d prior to analyses.
Isolation of the G92 Mutant and Genetic Analyses
M1 ST-GFP seeds were soaked for 16 h in 0.25% (v/v) methanesulfonic acid ethyl ester (Sigma-Aldrich) and then washed for 11 h in running water. The M1 seeds were grown after self-fertilization, and the M2 seeds were collected from individual M1 plants to generate M2 lines (800 independent lines). Thirty seeds from each M2 line were grown for 7 to 10 d and analyzed for displacement of the ST-GFP marker. The G92 homozygous mutant was crossed with Landsberg erecta to generate a mapping population. The polymorphism between the two ecotypes was analyzed using a combination of cleaved amplified polymorphic sequence markers and simple sequence length polymorphism markers (Konieczny and Ausubel, 1993; Bell and Ecker, 1994). F2 plants exhibiting the G92 phenotype were selected and the DNA analyzed. For rough mapping, 20 F2 plants were isolated. Fine mapping was performed upon DNA isolated from 462 F2 plants. Nucleotide sequences were determined from both strands using the ABI Prism Big Dye Terminator Cycle Sequence Reaction Kit (Applied Biosystems) and a DNA sequencer (Prism 3100; Applied Biosystems).
Arabidopsis Stable Transformation and Complementation
Arabidopsis plants were transformed by the floral dip method (Clough and Bent, 1998), and transformants were obtained on MS medium supplemented with glufosinate ammonium salt (BASTA; final concentration 20 μg/mL) and 0.8% (w/v) agar. For the G92 complementation, the cDNA sequences of untagged sec24 isoforms and sec24R693K were subcloned under the control of a cauliflower mosaic virus 35S in the binary vector pEARLY-Gate100.
Confocal Laser Scanning Microscopy
An inverted laser scanning confocal microscope (LSM510 META; Carl Zeiss) was used for confocal analyses. For GFP5 and YFP imaging, settings were as described by Brandizzi et al. (2002a) and Hanton et al. (2007), with excitation of argon laser lines at 458 and 514 nm and a 1-μm pinhole. Imaging of G92 cells labeled with propidium iodide (PI) was performed using 488-nm excitation of an argon laser line for GFP5 and 543-nm He/Ne for PI; a 488/543 beam splitter and BP505-530 and LP 560 emission filters were used for acquisition of the GFP and PI signal, respectively. DiOC6-stained cells were imaged with excitation of the argon laser line at 488 nm and a 505- to 575-nm emission filter. FRAP analyses were conducted as described earlier (Brandizzi et al., 2002b). Quantitative photobleaching was performed in cells treated with latrunculin to disrupt actin (Brandizzi et al., 2002b). Statistical analyses included the Student's two-tailed t test, assuming equal variance, and data with P < 0.05 were considered significant. Qualitative spot bleaching was performed in untreated cells. Postacquisition analyses were performed with the Zeiss AIM software. PaintShop Pro and Photoshop Imaging Suite were used for further image handling. Images reported in microscopy figures are representative of at least five independent experiments.
Fluorescent Dyes and Drug Treatments
Cellular nucleic acids were stained by immersing cotyledons in a solution with PI (Invitrogen; working solution: 1 μg/mL) in water for 15 min. Actin depolymerization was induced with latrunculin B (Sigma-Aldrich), at a final concentration of 25 μM (Calbiochem; stock solution, 10 mM in DMSO), for 1 h, as previously described (Brandizzi et al., 2002b). Endomembranes were stained with DiOC6 (Molecular Probes; working solution: 1.8 μM) in water for 30 min, as described earlier (Zheng et al., 2004). All stock solutions were kept at −20°C, and working solutions were prepared fresh just before use. For analysis and observation at the microscope, samples were mounted on a slide with the solution in which they were last treated.
Electron Microscopy
Cotyledons were fixed in 1% glutaraldehyde and 1% paraformaldehyde in 0.1 M sodium cacodylate buffer, pH 6.9, washed three times in buffer and postfixed in 2% aqueous osmium tetroxide for 90 min (Satiat-Jeunemaitre et al., 1996; Langhans et al., 2007). Samples were washed four times in water and subsequently block stained overnight in 1% aqueous uranylacetate (Hayat, 1975). Samples were dehydrated in acetone and embedded in Spurr resin (Spurr, 1969) and sectioned with a RMC PowerTome XL. Poststaining was performed in 1 to 3% uranyl acetate in methanol followed by treatment with lead citrate for 5 to 10 min (Robinson et al., 1985). Sections were observed with a JEOL 1200EX or Hitachi H-7650 transmission electron microscope.
Yeast Complementation
To create LMY864, the sec24Δ strain used in yeast complementation studies strain YPH499 (MATa, ade2-101, his3Δ200, leu2Δ1, lys2-801, trp1Δ63, ura3-52) (Johnston and Davis, 1984) was first transformed with pLM22 (CEN SEC24-URA3; Miller et al., 2003) through standard lithium acetate yeast transformation methods (Kasier et al., 1994). The endogenous SEC24 locus was then disrupted by integrating a PCR product containing the NatMX cassette amplified from pAG25 with flanking ends homologous to the 5′ and 3′ regions of the endogenous SEC24 locus. Genomic DNA was prepared from colonies resistant on rich media (YPD: 1% yeast extract, 2% peptone, and 2% glucose) containing 100 μg/mL nourseothricin (clonNAT; Werner BioAgents) and confirmed for deletion of the SEC24 locus by PCR. LMY864 was transformed with the following plasmids expressing various forms of Atsec24A or yeast SEC24 from single-copy (CEN) or multicopy (2 μ) vectors: p425GPD-AtSec24A (2 μ GPDpr-AtSEC24A-LEU2), p425GPD-Sec24AR693K (2 μ GPDpr-AtSEC24-G92-LEU2), p425GPD, pLM23 (CEN SEC24-HIS3; Miller et al., 2003), and pLM128 (CEN sec24-R561M-HIS3; Miller et al., 2003). Transformants were plated on synthetic complete media (SC: 0.67% yeast nitrogen base and 2% glucose, supplemented with amino acids according to the specific auxotrophies required for plasmid selection and grown for 3 d at 30°C. Transformants were tested for complementation of sec24Δ by streaking onto SC plates containing 5-fluororotic acid (0.1% final concentration) to select against the URA3 bearing plasmid containing the wild-type yeast SEC24 gene. Plates were scanned after growth at 30°C for 3 d.
Real-Time Quantitative PCR
Real-time quantitative PCR was performed on the ABI 7500 Fast real-time PCR system using SYBR green assays. Primers were designed for the amplification of sec24A, sec24B, sec24C, ubiquitin10, actin2, and β-6-tubulin by real-time PCR analysis using Primer Express software version 3 (Applied Biosystems) (see Supplemental Table 1 online). Total RNA was prepared from cotyledons and true leaves harvested from 19-d-old ST-GFP and G92 seedlings using TRIzol reagent (Invitrogen) and quantified by UV spectroscopy. Total RNA (5 μg) was treated with Turbo DNA-free DNase (Ambion) to remove traces of genomic DNA contamination. First-strand cDNA was prepared from DNA-free RNA (600 ng) with random hexamer primers using the Superscript III kit (Invitrogen) in a reaction volume of 20 μL. The resulting cDNA was diluted with nuclease-free water to a final concentration of 2.5 ng μL−1. All PCR reactions (10 μL) were assembled in triplicate and contained 5 μL of 2× Fast-SYBR Green Master Mix (Applied Biosystems), 2.5 pmol of forward and reverse primer, and 2 μL of template. “No RT” and “no template” control reactions were also included for each target gene. PCR reactions were cycled 10 min at 95°C followed by 40 cycles of 95°C for 15 s and then 60°C for 1min. A dissociation curve was added to the end of each assay to ensure that single products were amplified. Baseline and cycle thresholds were automatically assigned using Applied Biosystems 7500 Fast System software, and each amplification plot and dissociation curve was visually inspected for integrity. Cts were exported into Microsoft Excel, and averages of triplicate reactions were determined. Relative expression was determined using the ΔΔCt method corrected for primer efficiencies (Pfaffl, 2001). Amplification efficiencies were determined for each primer pair, and only pairs having efficiencies >90% were used for quantitation. Amplification of the ubiquitin10, actin2, and β-6-tubulin housekeeping genes was assayed, and it was determined that actin2 expression was the most consistent across all cDNA templates. Data were expressed relative to the expression of sec24A in ST-GFP cotyledons (calibrator) and normalized to the expression of actin2.
Accession Numbers
Sequence data from this article can be found in the Arabidopsis Genome Initiative or GenBank/EMBL databases under the following accession numbers: sec24A (At3g07100), sec24B (At3g44340), sec24C (At4g32640), ubiquitin10 (At4g05320), actin2 (At3g18780), and β-6-tubulin (At5g12250).
Supplemental Data
The following materials are available in the online version of this article.
Supplemental Figure 1. ST-GFP Is Localized in the Nuclear Envelope of Cotyledonal Epidermal Leaves.
Supplemental Figure 2. G92 Plants Do Not Show Obvious Developmental or Growth Phenotypes.
Supplemental Figure 3. The Globular Structures Are Present Even in the Absence of ST-GFP Expression.
Supplemental Figure 4. Cortical ER Network of Control Cells.
Supplemental Figure 5. Electron Micrographs of G92 Golgi at the Cell Cortex.
Supplemental Figure 6. The G92/35S:Sec24 Transformants Bear the Glufosinate Resistance Gene Encoded in the Transforming Binary Vector pEARLYGate 100.
Supplemental Figure 7. The G92/35S:Sec24 Isoform Transformants Bear the Glufosinate Resistance Gene Encoded in the Transforming Binary Vector pEARLYGate 100.
Supplemental Figure 8. The Sec24 Isoforms Are Expressed in Cotyledons and Rosette Leaves of G92 and Control Plants.
Supplemental Figure 9. Arabidopsis Sec24A Does Not Complement Yeast Sec24.
Supplemental Figure 10. No Homozygous Plants Were Identified in a Screen of a sec24A Insertion Mutant.
Supplemental Table 1. List of Primers Used in This Work.
Supplemental Movie 1. Golgi Stacks within the G92 Structures Are Relatively Immobile in Comparison to Those Outside the Structures and at the Cortex of the Cell.
Supplemental Movie 2. Three-Dimensional Analyses of the G92 Globular Structures.
Supplemental Movie 3. ST-GFP Fluorescence Is Found in the Golgi Stacks and in the G92 Clusters, but Not in the Cortical ER.
Supplemental Movie 4. The Golgi Stacks within the G92 Structures Can Exchange Membrane Cargo with Neighboring Membranes.
Supplemental Movie Legends.
Supplementary Material
Acknowledgments
We acknowledge support by the Chemical Sciences, Geosciences, and Biosciences Division, Office of Basic Energy Sciences, Office of Science, U.S. Department of Energy (Award DE-FG02-91ER20021) and National Science Foundation MCB 0841594 (F.B.) and the Biotechnology and Biological Science Research Council (C.H.). We also acknowledge support by the Japan Society for the Promotion of Science Postdoctoral Fellowships for Research Abroad (K.T.) for work in C.H.'s laboratory. We acknowledge the kind gift of the G-yk and ER-yk plants and of the ER-yb DNA construct from Andreas Nebenführ (University of Tennessee) and of the ABD2 coding sequence from Chris Staiger (Purdue University). We thank Linda Danhof for technical help and Karen Bird for editing the manuscript.
The author responsible for distribution of materials integral to the findings presented in this article in accordance with the policy described in the Instructions for Authors (www.plantcell.org) is: Federica Brandizzi (brandizz@msu.edu).
Online version contains Web-only data.
References
- Andreeva, A.V., Zheng, H., Saint-Jore, C.M., Kutuzov, M.A., Evans, D.E., and Hawes, C.R. (2000). Organization of transport from endoplasmic reticulum to Golgi in higher plants. Biochem. Soc. Trans. 28 505–512. [PubMed] [Google Scholar]
- Aridor, M., Weissman, J., Bannykh, S., Nuoffer, C., and Balch, W.E. (1998). Cargo selection by the COPII budding machinery during export from the ER. J. Cell Biol. 141 61–70. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Avila, E.L., Zouhar, J., Agee, A.E., Carter, D.G., Chary, S.N., and Raikhel, N.V. (2003). Tools to study plant organelle biogenesis. Point mutation lines with disrupted vacuoles and high-speed confocal screening of green fluorescent protein-tagged organelles. Plant Physiol. 133 1673–1676. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baker, D., and Schekman, R. (1989). Reconstitution of protein transport using broken yeast spheroplasts. Methods Cell Biol. 31 127–141. [DOI] [PubMed] [Google Scholar]
- Bard, F., Casano, L., Mallabiabarrena, A., Wallace, E., Saito, K., Kitayama, H., Guizzunti, G., Hu, Y., Wendler, F., Dasgupta, R., Perrimon, N., and Malhotra, V. (2006). Functional genomics reveals genes involved in protein secretion and Golgi organization. Nature 439 604–607. [DOI] [PubMed] [Google Scholar]
- Barlowe, C., Orci, L., Yeung, T., Hosobuchi, M., Hamamoto, S., Salama, N., Rexach, M.F., Ravazzola, M., Amherdt, M., and Schekman, R. (1994). COPII: A membrane coat formed by Sec proteins that drive vesicle budding from the endoplasmic reticulum. Cell 77 895–907. [DOI] [PubMed] [Google Scholar]
- Bell, C.J., and Ecker, J.R. (1994). Assignment of 30 microsatellite loci to the linkage map of Arabidopsis. Genomics 19 137–144. [DOI] [PubMed] [Google Scholar]
- Belles-Boix, E., Babiychuk, E., Montagu, M.V., Inze, D., and Kushnir, S. (2000). CEF, a sec24 homologue of Arabidopsis thaliana, enhances the survival of yeast under oxidative stress conditions. J. Exp. Bot. 51 1761–1762. [DOI] [PubMed] [Google Scholar]
- Boevink, P., Oparka, K., Santa Cruz, S., Martin, B., Betteridge, A., and Hawes, C. (1998). Stacks on tracks: The plant Golgi apparatus traffics on an actin/ER network. Plant J. 15 441–447. [DOI] [PubMed] [Google Scholar]
- Boulaflous, A., Faso, C., and Brandizzi, F. (2008). Deciphering the Golgi apparatus: From imaging to genes. Traffic 9 1613–1617. [DOI] [PubMed] [Google Scholar]
- Brandizzi, F., Fricker, M., and Hawes, C. (2002. a). A greener world: The revolution in plant bioimaging. Nat. Rev. Mol. Cell Biol. 3 520–530. [DOI] [PubMed] [Google Scholar]
- Brandizzi, F., Saint-Jore, C., Moore, I., and Hawes, C. (2003). The relationship between endomembranes and the plant cytoskeleton. Cell Biol. Int. 27 177–179. [DOI] [PubMed] [Google Scholar]
- Brandizzi, F., Snapp, E.L., Roberts, A.G., Lippincott-Schwartz, J., and Hawes, C. (2002. b). Membrane protein transport between the endoplasmic reticulum and the Golgi in tobacco leaves is energy dependent but cytoskeleton independent: Evidence from selective photobleaching. Plant Cell 14 1293–1309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Campbell, R.E., Tour, O., Palmer, A.E., Steinbach, P.A., Baird, G.S., Zacharias, D.A., and Tsien, R.Y. (2002). A monomeric red fluorescent protein. Proc. Natl. Acad. Sci. USA 99 7877–7882. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clough, S.J., and Bent, A.F. (1998). Floral dip: A simplified method for Agrobacterium-mediated transformation of Arabidopsis thaliana. Plant J. 16 735–743. [DOI] [PubMed] [Google Scholar]
- Crofts, A.J., Leborgne-Castel, N., Hillmer, S., Robinson, D.G., Phillipson, B., Carlsson, L.E., Ashford, D.A., and Denecke, J. (1999). Saturation of the endoplasmic reticulum retention machinery reveals anterograde bulk flow. Plant Cell 11 2233–2248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- daSilva, L.L., Snapp, E.L., Denecke, J., Lippincott-Schwartz, J., Hawes, C., and Brandizzi, F. (2004). Endoplasmic reticulum export sites and Golgi bodies behave as single mobile secretory units in plant cells. Plant Cell 16 1753–1771. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feiguin, F., Ferreira, A., Kosik, K.S., and Caceres, A. (1994). Kinesin-mediated organelle translocation revealed by specific cellular manipulations. J. Cell Biol. 127 1021–1039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fuji, K., Shimada, T., Takahashi, H., Tamura, K., Koumoto, Y., Utsumi, S., Nishizawa, K., Maruyama, N., and Hara-Nishimura, I. (2007). Arabidopsis vacuolar sorting mutants (green fluorescent seed) can be identified efficiently by secretion of vacuole-targeted green fluorescent protein in their seeds. Plant Cell 19 597–609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hanton, S.L., Chatre, L., Matheson, L.A., Rossi, M., Held, M.A., and Brandizzi, F. (2008). Plant Sar1 isoforms with near-identical protein sequences exhibit different localisations and effects on secretion. Plant Mol. Biol. 67 283–294. [DOI] [PubMed] [Google Scholar]
- Hanton, S.L., Chatre, L., Renna, L., Matheson, L.A., and Brandizzi, F. (2007). De novo formation of plant endoplasmic reticulum export sites is membrane cargo induced and signal mediated. Plant Physiol. 143 1640–1650. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hanton, S.L., Matheson, L.A., Chatre, L., and Brandizzi, F. (2009). Dynamic organization of COPII coat proteins at endoplasmic reticulum export sites in plant cells. Plant J. 57 963–974. [DOI] [PubMed] [Google Scholar]
- Hardham, A.R., Takemoto, D., and White, R.G. (2008). Rapid and dynamic subcellular reorganization following mechanical stimulation of Arabidopsis epidermal cells mimics responses to fungal and oomycete attack. BMC Plant Biol. 8 63. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haseloff, J., Siemering, K.R., Prasher, D.C., and Hodge, S. (1997). Removal of a cryptic intron and subcellular localization of green fluorescent protein are required to mark transgenic Arabidopsis plants brightly. Proc. Natl. Acad. Sci. USA 94 2122–2127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hawes, C., Saint-Jore, C., Martin, B., and Zheng, H.Q. (2001). ER confirmed as the location of mystery organelles in Arabidopsis plants expressing GFP! Trends Plant Sci. 6 245–246. [DOI] [PubMed] [Google Scholar]
- Hawes, C., and Satiat-Jeunemaitre, B. (2005). The plant Golgi apparatus – Going with the flow. Biochim. Biophys. Acta 1744 93–107. [DOI] [PubMed] [Google Scholar]
- Hayat, M.A. (1975). Positive Staining for Electron Microscopy. (New York: Van Nostrand Rheinold).
- Johnston, M., and Davis, R.W. (1984). Sequences that regulate the divergent GAL1-GAL10 promoter in Saccharomyces cerevisiae. Mol. Cell. Biol. 4 1440–1448. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kasier, C.A., Michaelis, S., and Mitchell, A. (1994). Methods in Yeast Genetics. (Cold Spring Harbor, NY: Cold Spring Harbor Laboratory Press).
- Klopfenstein, D.R., Kappeler, F., and Hauri, H.P. (1998). A novel direct interaction of endoplasmic reticulum with microtubules. EMBO J. 17 6168–6177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Knebel, W., Quader, H., and Schnepf, E. (1990). Mobile and immobile endoplasmic reticulum in onion bulb epidermis cells: short- and long-term observations with a confocal laser scanning microscope. Eur J Cell Biol. 52 328–340. [PubMed] [Google Scholar]
- Konieczny, A., and Ausubel, F.M. (1993). A procedure for mapping Arabidopsis mutations using co-dominant ecotype-specific PCR-based markers. Plant J. 4 403–410. [DOI] [PubMed] [Google Scholar]
- Kurihara, T., Hamamoto, S., Gimeno, R.E., Kaiser, C.A., Schekman, R., and Yoshihisa, T. (2000). Sec24p and Iss1p function interchangeably in transport vesicle formation from the endoplasmic reticulum in Saccharomyces cerevisiae. Mol. Biol. Cell 11 983–998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Langhans, M., Hawes, C., Hillmer, S., Hummel, E., and Robinson, D.G. (2007). Golgi regeneration after brefeldin A treatment in BY-2 cells entails stack enlargement and cisternal growth followed by division. Plant Physiol. 145 527–538. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Latijnhouwers, M., Gillespie, T., Boevink, P., Kriechbaumer, V., Hawes, C., and Carvalho, C.M. (2007). Localization and domain characterization of Arabidopsis golgin candidates. J. Exp. Bot. 58 4373–4386. [DOI] [PubMed] [Google Scholar]
- Latijnhouwers, M., Hawes, C., and Carvalho, C. (2005. a). Holding it all together? Candidate proteins for the plant Golgi matrix. Curr. Opin. Plant Biol. 8 632–639. [DOI] [PubMed] [Google Scholar]
- Latijnhouwers, M., Hawes, C., Carvalho, C., Oparka, K., Gillingham, A.K., and Boevink, P. (2005. b). An Arabidopsis GRIP domain protein locates to the trans-Golgi and binds the small GTPase ARL1. Plant J. 44 459–470. [DOI] [PubMed] [Google Scholar]
- Lee, M.H., Min, M.K., Lee, Y.J., Jin, J.B., Shin, D.H., Kim, D.H., Lee, K.H., and Hwang, I. (2002). ADP-ribosylation factor 1 of Arabidopsis plays a critical role in intracellular trafficking and maintenance of endoplasmic reticulum morphology in Arabidopsis. Plant Physiol. 129 1507–1520. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lippincott-Schwartz, J., Altan-Bonnet, N., and Patterson, G.H. (2003). Photobleaching and photoactivation: Following protein dynamics in living cells. Nat. Cell Biol. (Suppl): S7–14. [PubMed]
- Mancias, J.D., and Goldberg, J. (2008). Structural basis of cargo membrane protein discrimination by the human COPII coat machinery. EMBO J. 27 2918–2928. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matsushima, R., Hayashi, Y., Yamada, K., Shimada, T., Nishimura, M., and Hara-Nishimura, I. (2003). The ER body, a novel endoplasmic reticulum-derived structure in Arabidopsis. Plant Cell Physiol. 44 661–666. [DOI] [PubMed] [Google Scholar]
- Miller, E., Antonny, B., Hamamoto, S., and Schekman, R. (2002). Cargo selection into COPII vesicles is driven by the Sec24p subunit. EMBO J. 21 6105–6113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miller, E.A., Beilharz, T.H., Malkus, P.N., Lee, M.C., Hamamoto, S., Orci, L., and Schekman, R. (2003). Multiple cargo binding sites on the COPII subunit Sec24p ensure capture of diverse membrane proteins into transport vesicles. Cell 114 497–509. [DOI] [PubMed] [Google Scholar]
- Nebenfuhr, A., Gallagher, L.A., Dunahay, T.G., Frohlick, J.A., Mazurkiewicz, A.M., Meehl, J.B., and Staehelin, L.A. (1999). Stop-and-go movements of plant Golgi stacks are mediated by the acto-myosin system. Plant Physiol. 121 1127–1142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nelson, B.K., Cai, X., and Nebenfuhr, A. (2007). A multicolored set of in vivo organelle markers for co-localization studies in Arabidopsis and other plants. Plant J. 51 1126–1136. [DOI] [PubMed] [Google Scholar]
- Peiter, E., Montanini, B., Gobert, A., Pedas, P., Husted, S., Maathuis, F.J., Blaudez, D., Chalot, M., and Sanders, D. (2007). A secretory pathway-localized cation diffusion facilitator confers plant manganese tolerance. Proc. Natl. Acad. Sci. USA 104 8532–8537. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peng, R., De Antoni, A., and Gallwitz, D. (2000). Evidence for overlapping and distinct functions in protein transport of coat protein Sec24p family members. J. Biol. Chem. 275 11521–11528. [DOI] [PubMed] [Google Scholar]
- Pfaffl, M.W. (2001). A new mathematical model for relative quantification in real-time RT-PCR. Nucleic Acids Res. 29 e45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Phillipson, B.A., Pimpl, P., daSilva, L.L., Crofts, A.J., Taylor, J.P., Movafeghi, A., Robinson, D.G., and Denecke, J. (2001). Secretory bulk flow of soluble proteins is efficient and COPII dependent. Plant Cell 13 2005–2020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pimpl, P., Movafeghi, A., Coughlan, S., Denecke, J., Hillmer, S., and Robinson, D.G. (2000). In situ localization and in vitro induction of plant COPI-coated vesicles. Plant Cell 12 2219–2236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robinson, D.G., Ehlers, U., Herken, R., Hermann, B., Mayer, F., and Schuermann, F.W. (1985). Praeparationsmethodik in der Elektronenmikroskopie. (Berlin, Heidelberg: Springer).
- Robinson, D.G., Herranz, M.C., Bubeck, J., Pepperkok, R., and Ritzenthaler, C. (2007). Membrane dynamics in the early secretory pathway. Crit. Rev. Plant Sci. 26 199–225. [Google Scholar]
- Runions, J., Brach, T., Kuhner, S., and Hawes, C. (2006). Photoactivation of GFP reveals protein dynamics within the endoplasmic reticulum membrane. J. Exp. Bot. 57 43–50. [DOI] [PubMed] [Google Scholar]
- Saint-Jore, C.M., Evins, J., Batoko, H., Brandizzi, F., Moore, I., and Hawes, C. (2002). Redistribution of membrane proteins between the Golgi apparatus and endoplasmic reticulum in plants is reversible and not dependent on cytoskeletal networks. Plant J. 29 661–678. [DOI] [PubMed] [Google Scholar]
- Saint-Jore-Dupas, C., Nebenfuhr, A., Boulaflous, A., Follet-Gueye, M.L., Plasson, C., Hawes, C., Driouich, A., Faye, L., and Gomord, V. (2006). Plant N-glycan processing enzymes employ different targeting mechanisms for their spatial arrangement along the secretory pathway. Plant Cell 18 3182–3200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Samalova, M., Fricker, M., and Moore, I. (2006). Ratiometric fluorescence-imaging assays of plant membrane traffic using polyproteins. Traffic 7 1701–1723. [DOI] [PubMed] [Google Scholar]
- Satiat-Jeunemaitre, B., Cole, L., Bourett, T., Howard, R., and Hawes, C. (1996). Brefeldin A effects in plant and fungal cells: something new about vesicle trafficking? J. Microsc. 181 162–177. [DOI] [PubMed] [Google Scholar]
- Sato, K., and Nakano, A. (2007). Mechanisms of COPII vesicle formation and protein sorting. FEBS Lett. 581 2076–2082. [DOI] [PubMed] [Google Scholar]
- Serafini, T., Stenbeck, G., Brecht, A., Lottspeich, F., Orci, L., Rothman, J.E., and Wieland, F.T. (1991). A coat subunit of Golgi-derived non-clathrin-coated vesicles with homology to the clathrin-coated vesicle coat protein beta-adaptin. Nature 349 215–220. [DOI] [PubMed] [Google Scholar]
- Sheahan, M.B., Staiger, C.J., Rose, R.J., and McCurdy, D.W. (2004). A green fluorescent protein fusion to actin-binding domain 2 of Arabidopsis fimbrin highlights new features of a dynamic actin cytoskeleton in live plant cells. Plant Physiol. 136 3968–3978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shimoni, Y., Kurihara, T., Ravazzola, M., Amherdt, M., Orci, L., and Schekman, R. (2000). Lst1p and Sec24p cooperate in sorting of the plasma membrane ATPase into COPII vesicles in Saccharomyces cerevisiae. J. Cell Biol. 151 973–984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sieben, C., Mikosch, M., Brandizzi, F., and Homann, U. (2008). Interaction of the K(+)-channel KAT1 with the coat protein complex II coat component Sec24 depends on a di-acidic endoplasmic reticulum export motif. Plant J. 56 997–1006. [DOI] [PubMed] [Google Scholar]
- Sparkes, I.A., Ketelaar, T., Ruijter, N.C., and Hawes, C. (2009). Grab a Golgi: Laser trapping of Golgi bodies reveals in vivo interactions with the endoplasmic reticulum. Traffic, in press. [DOI] [PubMed]
- Spurr, A.R. (1969). A low-viscosity epoxy resin embedding medium for electron microscopy. J. Ultrastruct. Res. 26 31–43. [DOI] [PubMed] [Google Scholar]
- Stefano, G., Renna, L., Chatre, L., Hanton, S.L., Moreau, P., Hawes, C., and Brandizzi, F. (2006). In tobacco leaf epidermal cells, the integrity of protein export from the endoplasmic reticulum and of ER export sites depends on active COPI machinery. Plant J. 46 95–110. [DOI] [PubMed] [Google Scholar]
- Takeuchi, M., Ueda, T., Sato, K., Abe, H., Nagata, T., and Nakano, A. (2000). A dominant negative mutant of sar1 GTPase inhibits protein transport from the endoplasmic reticulum to the Golgi apparatus in tobacco and Arabidopsis cultured cells. Plant J. 23 517–525. [DOI] [PubMed] [Google Scholar]
- Tamura, K., Shimada, T., Kondo, M., Nishimura, M., and Hara-Nishimura, I. (2005). KATAMARI1/MURUS3 is a novel Golgi membrane protein that is required for endomembrane organization in Arabidopsis. Plant Cell 17 1764–1776. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Teh, O.K., and Moore, I. (2007). An ARF-GEF acting at the Golgi and in selective endocytosis in polarized plant cells. Nature 448 493–496. [DOI] [PubMed] [Google Scholar]
- Tolley, N., Sparkes, I.A., Hunter, P.R., Craddock, C.P., Nuttall, J., Roberts, L.M., Hawes, C., Pedrazzini, E., and Frigerio, L. (2008). Overexpression of a plant reticulon remodels the lumen of the cortical endoplasmic reticulum but does not perturb protein transport. Traffic 9 94–102. [DOI] [PubMed] [Google Scholar]
- Tschopp, J., Esmon, P.C., and Schekman, R. (1984). Defective plasma membrane assembly in yeast secretory mutants. J. Bacteriol. 160 966–970. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Uemura, T., Sato, T., Aoki, T., Yamamoto, A., Okada, T., Hirai, R., Harada, R., Mori, K., Tagaya, M., and Harada, A. (2009). p31 deficiency influences endoplasmic reticulum tubular morphology and cell survival. Mol. Cell Biol. 29 1869–1881. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vedrenne, C., and Hauri, H.P. (2006). Morphogenesis of the endoplasmic reticulum: Beyond active membrane expansion. Traffic 7 639–646. [DOI] [PubMed] [Google Scholar]
- Vedrenne, C., Klopfenstein, D.R., and Hauri, H.P. (2005). Phosphorylation controls CLIMP-63-mediated anchoring of the endoplasmic reticulum to microtubules. Mol. Biol. Cell 16 1928–1937. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wendeler, M.W., Paccaud, J.P., and Hauri, H.P. (2007). Role of Sec24 isoforms in selective export of membrane proteins from the endoplasmic reticulum. EMBO Rep. 8 258–264. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zheng, H., Kunst, L., Hawes, C., and Moore, I. (2004). A GFP-based assay reveals a role for RHD3 in transport between the endoplasmic reticulum and Golgi apparatus. Plant J. 37 398–414. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.