Abstract
Light signals perceived by the phytochromes induce the transition from skotomorphogenic to photomorphogenic development (deetiolation) in dark-germinated seedlings. Evidence that a quadruple mutant (pifq) lacking four phytochrome-interacting bHLH transcription factors (PIF1, 3, 4, and 5) is constitutively photomorphogenic in darkness establishes that these factors sustain the skotomorphogenic state. Moreover, photoactivated phytochromes bind to and induce rapid degradation of the PIFs, indicating that the photoreceptor reverses their constitutive activity upon light exposure, initiating photomorphogenesis. Here, to define the modes of transcriptional regulation and cellular development imposed by the PIFs, we performed expression profile and cytological analyses of pifq mutant and wild-type seedlings. Dark-grown mutant seedlings display cellular development that extensively phenocopies wild-type seedlings grown in light. Similarly, 80% of the gene expression changes elicited by the absence of the PIFs in dark-grown pifq seedlings are normally induced by prolonged light in wild-type seedlings. By comparing rapidly light-responsive genes in wild-type seedlings with those responding in darkness in the pifq mutant, we identified a subset, enriched in transcription factor–encoding genes, that are potential primary targets of PIF transcriptional regulation. Collectively, these data suggest that the transcriptional response elicited by light-induced PIF proteolysis is a major component of the mechanism by which the phytochromes pleiotropically regulate deetiolation and that at least some of the rapidly light-responsive genes may comprise a transcriptional network directly regulated by the PIF proteins.
INTRODUCTION
The role of the phytochromes (phyA through phyE in Arabidopsis thaliana) in controlling seedling deetiolation (the switch from skotomorphogenesis to photomorphogenesis) in response to red and far-red light signals is well established (Rockwell et al., 2006; Schafer and Nagy, 2006). Current evidence regarding the mechanism underlying this switch indicates that photoactivation of the phy molecule results in rapid translocation of the Pfr form from the cytoplasm to the nucleus (Nagatani, 2004) where it interacts, conformer specifically, with a subset of basic helix-loop-helix (bHLH) transcription factors, termed phy-interacting factors (PIFs) (Toledo-Ortiz et al., 2003; Duek and Fankhauser, 2005), initiating the cascade of gene expression changes that direct the overt photomorphogenic response (Quail, 2002; Jiao et al., 2007; Bae and Choi, 2008). The data show further that intranuclear binding of the Pfr form of phyA and/or phyB to several of these proteins, including PIFs 1, 3, 4, and 5, induces rapid (within minutes) phosphorylation and degradation (t1/2, 5 to 30 min) of the transcription factors (Bauer et al., 2004; Al-Sady et al., 2006; Nozue et al., 2007; Shen et al., 2007, 2008; Lorrain et al., 2008), suggesting that these may be primary molecular events in phy signaling. Taken together, the evidence from these and other studies indicates that the PIF proteins accumulate in the cell in young dark-grown seedlings and that photoactivated phy induces their rapid degradation upon initial exposure to light.
Earlier studies aimed at defining the functional role of the PIF proteins in seedling deetiolation and the relevance of phy-induced PIF degradation to this function have provided a complex and sometimes apparently contradictory picture (Monte et al., 2007). However, recently, a certain degree of clarity has emerged. Initially, we reported that an antisense pif3 mutant had a robust etiolated phenotype in the light (Ni et al., 1998). However, this proved to be an artifact due to a mutation in a separate locus, likely the result of insertion of the antisense construct at that site (Monte et al., 2004). Instead, monogenic pif3 mutants, as well as pif1, pif4, and pif5 mutants, all displayed hypersensitive phenotypes (shorter hypocotyls than the wild type) in prolonged light (the opposite to the original antisense line) (Huq and Quail, 2002; Kim et al., 2003; Fujimori et al., 2004; Huq et al., 2004; Monte et al., 2004; Oh et al., 2004; Khanna et al., 2007; Leivar et al., 2008a; Lorrain et al., 2008). These data were broadly interpreted to indicate that the PIFs act negatively in phy signaling (Duek and Fankhauser, 2005; Castillon et al., 2007; Bae and Choi, 2008), although microarray analysis of the pif3 mutant indicated that PIF3 appeared to function positively in the rapid (within 1 h) changes in gene expression induced upon initial exposure of dark-grown seedlings to light (Monte et al., 2004). Subsequently, we showed that the hypersensitive seedling phenotype in prolonged light is largely due to a feedback loop whereby the binding of the PIFs to phyB modulates phyB abundance, and thus global photosensory sensitivity to continuous red light (Rc), as opposed to participating directly in the transduction chain as a signaling intermediate (Khanna et al., 2007; Al-Sady et al., 2008; Leivar et al., 2008a). Therefore, these data called into question the general conclusion from previous experiments that the PIFs act as repressors of phy signaling under these irradiation conditions. Most recently, however, we examined the phenotypes of a series of monogenic, double, triple, and quadruple pif mutants and found a striking constitutively photomorphogenic (cop)-like phenotype in rigorously dark-grown seedlings (Leivar et al., 2008b). This finding provides compelling evidence that the PIFs do indeed function to repress photomorphogenesis in the dark in etiolated seedlings and that the phys act to reverse this phenotypic repression upon light exposure by inducing rapid degradation of the bHLH factors. The data also indicate, however, that there is considerable complexity in the system due to varying degrees of apparent functional redundancy within the PIF family as well as the likely involvement of additional family members. These findings have been confirmed in a recent report (Shin et al., 2009).
During these studies, we discovered another source of potential ambiguity in the interpretation of seedling deetiolation experiments. We found that the practice of irradiating imbibed and stratified pregerminative seed with light, widely used in the Arabidopsis field to stimulate germination, results in formation of the photoactivated Pfr form of the phy molecule in the embryo, as expected. However, this Pfr is then carried over in the seedlings that emerge in subsequent darkness and exerts potentially significant effects on their development (Leivar et al., 2008b). The evidence shows that seedlings produced under these pseudodark conditions can exhibit deetiolation responses during growth in darkness (Leivar et al., 2008b), reminiscent of the well-studied cop mutants (Deng et al., 1991; Jiao et al., 2007), due to the residual Pfr formed in the ungerminated seed. This delayed light effect is most pronounced in the quadruple pif1 pif3 pif4 pif5 (pifq) mutant, where the residual Pfr activity is superimposed on the bona fide partial cop-like phenotype of this mutant caused by genetic removal of these PIFs, referred to above (Leivar et al., 2008b). To circumvent the ambiguity created by the overlap of these two causes of dark deetiolation, it was necessary to develop photobiological procedures, termed true dark conditions, that removed any preformed Pfr from the germinating embryo before any detectable action of the photoreceptor on subsequent development occurred (Leivar et al., 2008b).
Data from multiple genome-wide, microarray-based expression profiling studies indicate that 10 to 20% (>2500) of the genes in the Arabidopsis genome undergo changes in expression during the complete deetiolation process (for review, see Jiao et al., 2007; Quail, 2007). Examination of the effects of mutations in a number of genetically identified signaling components on this global expression pattern revealed varying degrees of qualitative and quantitative perturbation of the profile (for review, see Jiao et al., 2007; Quail, 2007). Of the total light-regulated genes, ∼250 have been identified as early response genes (responding within 1 h of initial exposure of dark-grown seedlings to red light) and are thus candidates for being direct targets of the primary phy signaling pathway (Tepperman et al., 2001, 2004, 2006; Monte et al., 2004). Analysis of phy null mutants showed that phyA and phyB are principally responsible for these expression responses. The data also revealed that transcription factor–encoding genes represent the largest single functional category of these early response genes, suggesting that they may act in cascade fashion to regulate downstream genes in the phy-regulated transcriptional network. A global computational analysis of the early response genes identified a series of sequence motifs potentially involved in the coordinate regulation of these genes, with the G-box motif, CACGTG, being prominently represented (Hudson and Quail, 2003), consistent with the possibility that these are targets of the bHLH transcription factor family (Toledo-Ortiz et al., 2003). Comparison of the photoresponsiveness of the early response genes in a pif3 null mutant with that of the wild type indicated that PIF3 is necessary for the rapid, phy-induced expression of a subset of genes involved in chloroplast development (Monte et al., 2004). On the other hand, comparison of genome-wide gene expression in dark-grown pif1 mutant and wild-type seedlings revealed that PIF1 promotes the expression in darkness of a small subset of genes involved in controlling the chlorophyll biosynthetic pathway (Moon et al., 2008).
In this study, to begin to define the transcriptional network regulated by the PIF protein subfamily in repressing photomorphogenic development in darkness, we examined the genome-wide expression profile of dark-grown pifq mutant seedlings compared with Rc light–grown wild type. During the preparation of this manuscript, another report appeared that also includes a microarray analysis of a pifq mutant (Shin et al., 2009). However, that study was performed using the pseudodark conditions mentioned above (Leivar et al., 2008b), leaving open the possibility that the gene expression patterns interpreted as constitutively induced in that report were instead, to an unknown extent, light-induced responses in the seedlings resulting from the residual Pfr generated in the seed by the white light administered to stimulate germination. In addition, the seedlings were grown on 1% sucrose by Shin et al. (2009). It is well documented that the application of exogenous sucrose significantly perturbs the expression of a spectrum of light-regulated and other genes (Cheng et al., 1992; Dijkwel et al., 1996, 1997; Gibson and Graham, 1999; Graham, 2008), raising the possibility that the expression patterns reported by Shin et al. (2009) deviate from the normal phy-PIF–regulated pattern in young seedlings. We have circumvented these ambiguities here by growing the seedlings in the absence of any exogenous sucrose under the true dark conditions established by Leivar et al. (2008b). In addition, we extended the investigation beyond simply describing a molecular phenotype, to identifying those genes, constitutively altered in the pifq mutant, that also respond rapidly to initial Rc light exposure and are therefore potentially direct targets of these bHLH factors in the primary transcriptional network that initiates and maintains the deetiolation process.
RESULTS
Morphological, Cellular, and Subcellular Phenotypic Responses of Dark- and Light-Grown pifq Mutant Seedlings
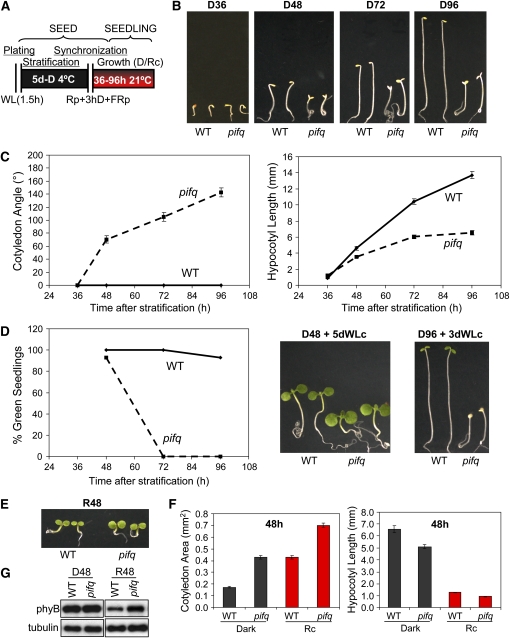
To define the optimal developmental stage to compare the transcriptional profiles of the pifq mutant and wild-type seedlings, we performed a time-course analysis of the visible morphological phenotypes, under true dark conditions, and examined the impact of increasing periods of dark growth on subsequent light responsiveness. Figures 1A to 1C shows that differences in phenotype between the two genotypes in the dark, observable as a cop-like phenotype in pifq, become apparent between 36 and 48 h poststratification and that these differences are amplified with increasing dark growth (up to 4 d here) (see Supplemental Figure 1 online for complete time-course analysis). Our previous microarray studies on phy-regulated gene expression were performed with 4-d-old dark-grown seedlings (Tepperman et al., 2001; Monte et al., 2004; Tepperman et al., 2004, 2006). However, although this was not problematic for wild-type seedlings, we observed that pif1 mutant seedlings grown for increasing periods beyond 2 d in darkness displayed increasing susceptibility to lethal bleaching upon transfer to light, due to accumulation of excess protochlorophyllide (Huq et al., 2004). Subsequently, it became apparent that a similar phenotype occurred to variable extents in other monogenic pif mutants, including pif3, as recently reported (Shin et al., 2009; Stephenson et al., 2009; E. Monte, P. Leivar, and P.H. Quail, unpublished data), raising the question of whether our earlier genome-wide expression analysis of 4-d-old dark-grown pif3 seedlings (Monte et al., 2004) accurately reflected the normal responsiveness of this mutant to phy-mediated light signals or was an indirect effect due to photooxidative damage. To determine whether this problem could be circumvented in the pifq mutant, which contains both pif1 and pif3 mutant loci, we examined the effects of increasing dark-growth periods before exposure to light on the phenotype of these seedlings. As anticipated, pifq seedlings grown for 3 or 4 d in darkness before transfer to light displayed severe bleaching (Figure 1D), an effect that was more dramatic than in any of the pif1 and pif3 single and double mutants (see Supplemental Figure 2 online). By contrast, we found that, as previously observed for the monogenic pif1 (Huq et al., 2004) and pif3 (see Supplemental Figure 2 online) mutants, restricting the dark growth period to only 2 d, precluded bleaching upon subsequent light exposure (Figure 1D; see Supplemental Figure 2 online). Consequently, we have performed all recent phenotypic analyses of this and other pif mutants (Leivar et al., 2008b), as well as the present microarray analysis here, using 2-d-old dark-grown seedlings.
Figure 1.
Time-Course Analysis of pifq Mutant Morphological Phenotypes in Dark and Light.
(A) Schematic representation of the protocol used for seedling growth under true dark (Leivar et al., 2008b) and Rc conditions. Seeds were exposed to 1.5 h of white light (WL) during sterilization and plating and were placed for 5 d at 4°C in darkness (D) (stratification). A poststratification treatment consisting of a 5-min R pulse (Rp) followed by 3 h D incubation and a terminal 5-min far-red (FR) pulse (Rp+3 hD+FRp) was provided before incubation at 21°C in D or Rc (6.7 μmol/m2/s) for the indicated period.
(B) Time-course analysis of the visible morphological phenotypes of pifq mutants grown in darkness for 36 to 96 h (D36-D96). Photos of representative wild-type and pifq mutant seedlings are shown.
(C) Time-course analysis of cotyledon separation (left panel) and hypocotyl length (right panel) phenotypes of wild-type and pifq mutant seedlings grown in darkness.
(D) Time-course analysis of the photobleaching phenotype of wild-type and pifq mutant seedlings grown in darkness for the indicated time before transferring them to continuous WL (WLc) for 3 to 5 d. The percentage of green seedlings was scored from at least 45 seedlings (left panel), and photos of representative wild-type and pifq mutant seedlings are shown (right panel).
(E) Visible morphological phenotypes of pifq mutants grown in Rc for 48 h (R48). Photos of representative wild-type and pifq mutant seedlings are shown.
(F) Quantification of the cotyledon area (left panel) and the hypocotyl length (right panel) of wild-type and pifq mutant seedlings grown in the dark or Rc for 48 h.
(G) Immunoblot analysis of phyB protein levels in wild-type and pifq mutant seedlings grown in the dark or Rc for 48 h. Tubulin was used as loading control. A representative blot is shown. Quantification of replicated experiments is shown in Supplemental Figure 3 online.
Data in (C) and (F) represent the mean and se of at least 20 seedlings.
For comparison, we determined the visible morphological phenotypes of 2-d-old seedlings grown in Rc (Figure 1E; see Supplemental Figure 1 online). The pifq mutant showed a hypersensitive phenotype to Rc relative to the wild type (Figures 1E and 1F; see Supplemental Figure 1 online), consistent with the reported activities of these individual PIFs in Rc (Huq and Quail, 2002; Kim et al., 2003; Huq et al., 2004; Monte et al., 2004; Oh et al., 2004; Khanna et al., 2007). Because PIF3, PIF4, and PIF5 have been shown to downregulate phyB photoreceptor levels under prolonged Rc conditions (Khanna et al., 2007; Al-Sady et al., 2008; Leivar et al., 2008a), we determined the phyB levels in 2-d-old wild-type and pifq seedlings by immunoblots. The results show that pifq mutant seedlings have twofold higher phyB levels than the wild type in Rc but not in darkness (Figure 1G; see Supplemental Figure 3 online). Therefore, the higher levels of phyB could partly explain the hypersensitive phenotype of the pifq mutant to Rc, as has been proposed for pif3, pif4, and pif5 monogenic mutants (Khanna et al., 2007; Al-Sady et al., 2008; Leivar et al., 2008a). By contrast, the repression of photomorphogenesis in the dark by the PIFs must be through a molecular mechanism other than regulating phyB levels.
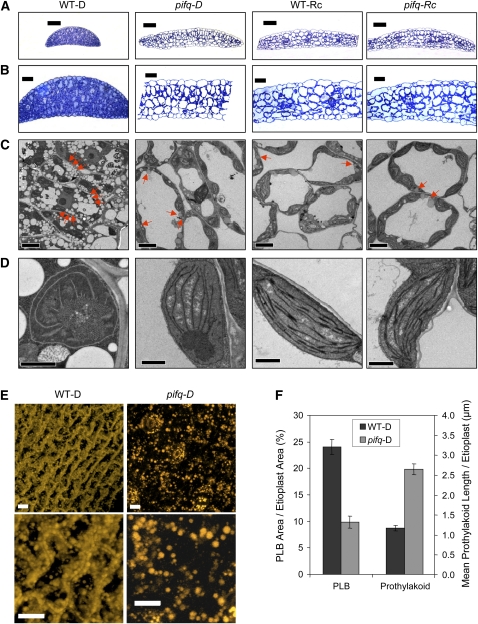
To define the cellular and subcellular phenotypes underlying the visible constitutively deetiolated phenotype of dark-grown pifq mutant seedlings, we performed light and electron microscopy of dark- and Rc-grown seedlings. Cross sections of cotyledons show that the general morphology of dark-grown pifq cotyledons is similar to Rc-grown, wild-type cotyledons and clearly distinct from that of the dark-grown wild type (Figures 2A and 2B). Cell expansion, with increased intercellular air spaces, appears to be the cellular basis for cotyledon enlargement in dark-grown pifq seedlings, as is established for light-induced cotyledon expansion in the wild type (Figures 2A and 2B), and as observed for dark-grown cop1 mutants (Deng et al., 1991).
Figure 2.
Cellular and Subcellular Phenotypes of Dark-Grown pifq Seedlings Phenocopy Rc-Grown Wild Type.
Wild-type and pifq mutant seedlings were grown in D or Rc for 48 h as in Figure 1A.
(A) Toluidine blue staining of semithin (0.5 μm) cross sections of cotyledons. Micrographs were taken by light microscopy. Bars = 50 μm.
(B) Higher-magnification micrographs of samples prepared as in (A). Bars = 20 μm.
(C) Micrographs obtained by transmission electron microscopy of cross sections of cotyledons. High-pressure freezing and microwave processing methods were used for sample preparation, and several micrographs were taken for both methods. Representative images of the cell morphology of dark- and Rc-grown wild-type and pifq seedlings are shown. Red arrows indicate oil bodies. Bars = 5 μm.
(D) Higher-magnification micrographs of samples prepared as in (C). Representative etioplasts (D grown) and chloroplasts (Rc grown) are shown for wild-type and pifq seedlings. Bars = 1 μm.
(E) Nile red staining of wild-type and pifq mutant cotyledons grown in darkness. The cotyledons were stained with Nile Red (Greenspan et al., 1985; Siloto et al., 2006) and examined by confocal microscopy. Maximum intensity projections from 36 slices are shown. The bottom images are higher magnifications of the top ones. Bars = 10 μm.
(F) Quantification of PLB area as a percentage of host etioplast area and mean prothylakoid length per etioplast. Data represent the mean and se of at least 76 etioplasts from several different transmission electron micrographs of wild-type and pifq mutant cotyledons grown in the dark. PLB area and prothylakoid length was measured using Image J.
At the subcellular level, pifq mutants grown in darkness also show features of Rc-grown wild-type seedlings (Figures 2C and 2D). Dark-grown wild-type cotyledonary cells have limited vacuoles and are densely packed with vesicular structures (Figure 2C) that appear to resemble oil bodies (Siloto et al., 2006; Graham, 2008). By contrast, cells in dark-grown pifq seedlings form a large central vacuole, and the abundance and/or density of oil bodies appears to decline, a phenotype similar to the Rc-light grown wild type (Figure 2C). This apparent decrease in density and/or abundance of the oil bodies in the pifq cells compared with the wild type was quite striking when imaged with the lipid stain, Nile Red (Greenspan et al., 1985; Siloto et al., 2006) (Figure 2E). Although some reports indicate that light induces a faster mobilization of fat reserves in the cotyledon cells compared with dark in some plants (Theimer and Rosnitschek, 1978; Davies et al., 1981; Sadeghipour and Bhatla, 2003), we did not attempt to quantify the abundance and density of oil bodies in our samples.
Another feature of cop mutants like cop1 is the partial differentiation of etioplasts into chloroplasts in darkness (Deng et al., 1991), characterized by a reduction or disappearance of the prolamellar body (PLB) and an extension of the prothylakoid membranes. Examination of the ultrastructure of 2-d-old dark-grown wild-type etioplasts showed the typical pattern of large PLB and reduced prothylakoid membranes (Figure 2D), whereas Rc-grown wild-type seedlings displayed a fully developed chloroplast (Figure 2D). The pifq mutant, by contrast, displayed a significant number of etioplasts with reduced or even absent (∼40%) PLBs and increased prothylakoid membranes when grown in darkness (Figure 2D), although we also observed a large number apparently showing a wild-type pattern. Quantification of the apparent differences indicate that pifq mutant etioplasts have more than twofold reduced PLB area relative to the etioplast area compared with the wild type and more than twofold increased prothylakoid length per etioplast (Figure 2F). These results indicate that the pifq mutant already displays a moderate cop-like etioplast phenotype in 2-d-old dark-grown seedlings even in the absence of a noticeable photobleaching phenotype upon transfer to light (Figure 1D). A partial cop-like etioplast phenotype in darkness has also been recently reported in a pif1 pif3 double mutant grown in darkness, although that experiment was done using 4-d-old seedlings grown under pseudodark conditions (Stephenson et al., 2009).
Light-Induced Transcriptome Changes Underlying the Deetiolated State
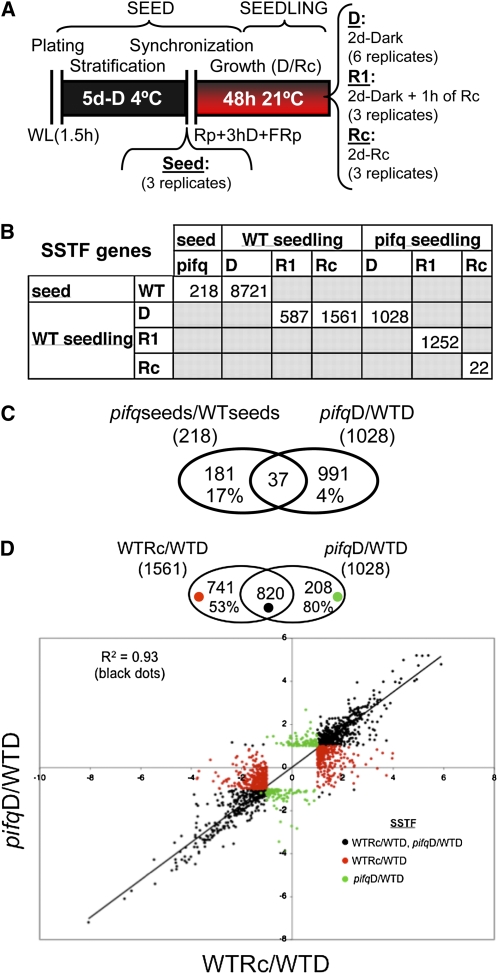
Microarray analysis shows that the expression of 8721 genes changes statistically significantly and by twofold (SSTF; Hu et al., 2009) during 2 d of growth under true dark conditions (Figure 3A; Leivar et al., 2008b) relative to the starting ungerminated seed (wild-type seed versus WT-D, Figure 3B; see Supplemental Data Set 1 online for the gene list). The data also show that growth of wild-type seedlings in Rc light for 2 d postgermination induces SSTF changes in expression of 1561 genes relative to these wild-type seedlings grown in parallel under true dark conditions (WT-Rc versus WT-D, Figure 3B). This observation indicates that ∼7% of the total Arabidopsis genes defined in this way are involved in executing the deetiolation process under these conditions, relatively consistent with earlier studies on older seedlings (Tepperman et al., 2001, 2004; Jiao et al., 2007; Quail, 2007; Hu et al., 2009). By comparison, pifq seedlings grown for 2 d in true dark conditions display SSTF alterations in expression of 1028 genes relative to the dark-grown wild type (WT-D versus pifq-D, Figure 3B). The vast majority of these differences between dark-grown pifq and wild-type seedlings (96%) develop postgerminatively in the seedling, as there are relatively few differences between the pifq and wild-type seed (218 genes, Figure 3B) and only 37 of these overlap with those subsequently different between the two sets of seedlings (Figure 3C; see Supplemental Data Set 2 online).
Figure 3.
Genetic Removal of PIFs Robustly Phenocopies in Darkness the Sustained Transcriptome Changes Elicited in Wild-Type Seedlings in Rc.
(A) Schematic representation of the protocol for wild-type and pifq seed and seedling growth used for microarray-based transcriptome analysis. Samples were harvested directly at the end of the stratification period (seeds) or after 2 d of growth at 21°C (seedlings) under true dark conditions (D) or in Rc as described in Figure 1A. In addition, 2-d-old true dark–grown seedlings were irradiated with 7.5 μmol/m2/s of R for 1 h (R1). The number of biological replicates for each sample is indicated.
(B) Number of differentially expressed genes that are defined as SSTF different in the pairwise comparisons indicated in the matrix. Expression data and primary analysis for the SSTF genes are reported in Supplemental Data Set 1 online.
(C) Genes that are PIF regulated in both seed and dark-grown seedlings. Venn diagram shows pairwise comparison between SSTF genes differentially expressed in seeds (wild-type seed versus pifq-seed) and seedlings (WT-D versus pifq-D). The percentage of shared genes that are differentially expressed in each set is indicated. The list of shared genes (37) is provided in Supplemental Data Set 2 online.
(D) Comparison of long-term Rc-responsive genes (WT-D versus WT-Rc) and PIF-regulated genes in dark-grown seedlings (WT-D versus pifq-D). Top: Venn diagram compares the SSTF differentially expressed genes in the two genotype growth treatment combinations. The number and percentage of shared genes in the comparison are indicated. Bottom: Scatterplot of log2 fold change values provides a quantitative measure of the correlation in responsiveness for each gene between the two genotype growth treatment combinations. Black dots in the scatterplot represent genes that are shared between the two combinations in the Venn diagram (top), whereas red and green dots represent genes that are specifically present in one of the combinations but not in the other. A trend line and the correlation coefficient for the shared genes (black dots) are indicated.
Of the 1028 SSTF genes differentially expressed in the pifq mutant in the dark, 80% (820 genes) are identical to those altered in expression in the wild type by 2 d in Rc (Figure 3D, top). The strong qualitative and quantitative correlation in the induced and repressed expression responses elicited by these two genotype treatment combinations are shown in the scatterplot in Figure 3D (bottom). This plot reveals that in addition to the high quantitative correlation between the SSTF genes (black symbols; r2 = 0.93), a considerable additional number of the SSTF genes that are induced or repressed by Rc in the wild type also respond to a quantitatively lesser extent, marginally below the twofold cutoff in the pifq mutant in darkness (red symbols). These data indicate that there are extensive similarities in the scope and degree of the gene expression changes induced by genetic removal of the PIF proteins and the normal light-induced changes that establish the deetiolated state in young wild-type seedlings. Of these common 820 SSTF genes, 60% are induced, 38% are repressed (Figure 3D, bottom), and 2% are ambiguous (see below).
In contrast with the large changes in gene expression (1028 genes) observed in the pifq mutant in the dark compared with wild type (WT-D versus pifq-D, Figure 3B), only a small number of genes (22) are differentially expressed between the mutant and the wild type after prolonged Rc irradiation (WT-Rc versus pifq-Rc, Figure 3B). Four out of these 22 genes correspond to PIF1, PIF3, PIF4, and PIF5, whose reduction in expression is due to the insertional mutations in the pifq mutant. Consistent with the described accumulation of the PIF proteins and their established constitutive activity in darkness (Leivar et al., 2008b), the data suggest that whereas these PIFs play a dominant role as transcriptional regulators in the dark, they apparently have a relatively marginal role in this capacity after establishment of the deetiolated state under prolonged Rc irradiation.
Rapidly Light-Responsive PIF-Regulated Genes
The expression changes established in the pifq mutant and the wild type over a 2-d seedling growth period in the dark or Rc, respectively, could be involved primarily in the maintenance of the deetiolated state, or the consequence of it, rather than being causal in the induction of the transition from the etiolated state. Thus, these genes may not all be strong candidates for being direct targets of transcriptional regulation by the PIF proteins under phy control. We therefore also analyzed the rapidly induced expression changes occurring upon initial exposure of 2-d-old true dark–grown seedlings to 1 h of Rc (Figure 3A), a time span over which a rapid and robust decline in PIF levels is known to be induced by the light treatment (Bauer et al., 2004; Monte et al., 2004; Al-Sady et al., 2006; Shen et al., 2007, 2008; Lorrain et al., 2008). This treatment induces a SSTF change in the expression of a total of 587 genes (451 induced and 136 repressed) in wild-type seedlings (WT-D versus WT-R1, Figure 3B). Of these, 291 (50%) sustain a SSTF change in expression after 2 d of Rc (see Supplemental Figure 4A online). This result indicates that half of the early Rc-responsive genes respond transiently to the light signal, whereas the other half display a sustained response to this signal. By comparison, only 196 (33%) of the early response genes exhibit SSTF changes in expression in the 2-d-old dark-grown pifq mutant (see Supplemental Figure 4B online), partly reflecting the lower total number of SSTF genes in the dark-grown pifq mutant than the 2-d-old Rc-light grown wild type (Figures 3B and 3D).
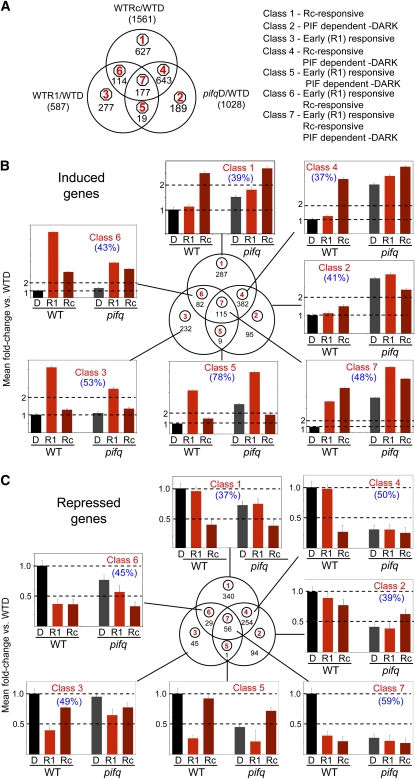
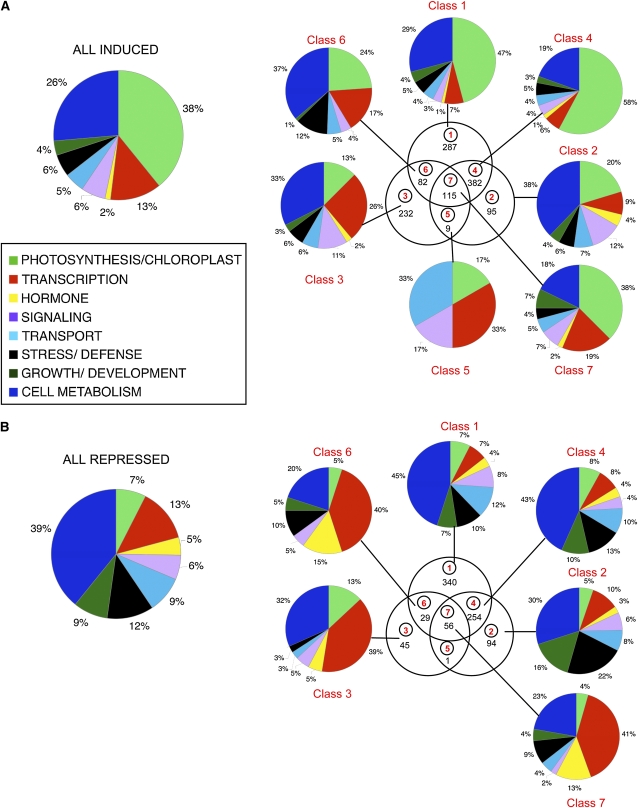
To enhance the probability of identifying genes that are direct targets of phy-induced, PIF-mediated transcriptional regulation, we compared the genes displaying SSTF alterations in expression in all three experimental conditions: the wild type after 2 d Rc (WT-Rc), pifq after 2 d in darkness (pifq-D), and the wild type after 1 h Rc (WT-R1), all relative to WT-D. The three-way Venn diagram of this comparison is shown in Figure 4A. For convenience, the sectors in this diagram are numbered 1 through 7, and the corresponding genes in each sector are designated Class 1 through 7 genes. The patterns of expression regulation for the induced (Figure 4B) and repressed (Figure 4C) genes in each class are shown separately in the corresponding bar graph panels for each sector and in the heat diagrams in Supplemental Figure 5 online. The respective gene lists for each class are presented in the individual work sheets in Supplemental Data Sets 3 and 4 online. The bar graph data in Figure 4 represent the mean expression levels of the genes in each class for each genotype (wild type and pifq) under each of the three experimental growth conditions (D, Rc, and R1). A small number of genes (25) in Classes 4 to 7 that could not be assigned to either the induced or repressed category because they show an inconsistent response to the different treatments (for example, a light-induced gene that is repressed in pifq-D) were excluded from the Venn diagrams and bar graph data in Figures 4B and 4C. A list of these ambiguous genes is shown in Supplemental Data Set 5 online.
Figure 4.
Genome-Wide Patterns of Rapidly Rc Light–Responsive and PIF-Regulated Gene Expression Reveal Pathway Convergence on Potential Target Genes.
Three-way comparison of SSTF genes responding to 1 h Rc (WT-D versus WT-R1), long-term (2d)-Rc (WT-D versus WT-Rc), and to the pifq mutations in darkness (WT-D versus pifq-D). Classification of the genes as induced (B) or repressed (C) is based on the direction of the response, relative to WT-D, elicited by each genotype treatment combination. A small group of 25 genes in Classes 4 to 7 were designated as ambiguous as the direction of the response relative to WT-D differed between WT-R1, WT-Rc, and/or pifqD. These genes are listed in Supplemental Data Set 5 online and were excluded from the analysis shown in (B) and (C). The mean fold change in expression relative to WT-D (set at unity) for all genes in each class is shown in the bar graphs in (B) and (C). Error bars represent the mean standard error for the genes averaged for each genotype treatment combination. Percentage of genes having a G-box in the 3-kb upstream regulatory sequence is indicated in parenthesis.
(A) Venn diagram showing comparison among all genes in the three different sets of SSTF differentially regulated genes (left panel). This comparison between genes responding to WT-R1, WT-Rc, and pifq-D resulted in the definition of seven classes of responsive genes (right panel) corresponding to the sectors of the diagram (circled numbers in red). The number of genes in each sector/class is indicated in black.
(B) Induced genes. The number of induced genes in each class is indicated by the black numbers in each sector of the Venn diagram. The gene lists are in Supplemental Data Set 3 online.
(C) Repressed genes. The number of repressed genes in each class is indicated by the black numbers in each sector of the Venn diagram. The gene lists are in Supplemental Data Set 4 online.
The bar graphs and heat diagrams show that the induced Class 1 genes display responsiveness to 2 d of Rc, with little response to 1 h Rc or the absence of the PIFs (Figure 4B; see Supplemental Figure 5 online), indicating that they are late response genes with little indication of being direct targets of the PIFs. Conversely, the induced Class 2 genes display pronounced induction in the absence of the PIF proteins in darkness, with little or no Rc light responsiveness. These genes thus appear to be strongly repressed by the PIF proteins but may not have a direct, major role in the deetiolation process. The induced Class 4 genes show a dual responsiveness to the PIFs and Rc, being strongly derepressed in the dark in the pifq mutant, but exhibiting a delayed response to Rc, as indicated by the absence of a rapid response to R1. This pattern of regulation suggests repression by the PIFs in dark-grown seedlings but that light induction requires additional time-dependent processes beyond the initial period of phy-induced PIF degradation, possibly reflecting indirect PIF regulation of these genes. The induced Class 3 genes exhibit rapid transient Rc light induction only (WT-R1), with no sustained response to 2 d Rc and no response to PIF elimination in the dark-grown pifq seedling. A similar pattern is observed in the induced Class 5 genes (except that relatively weak derepression occurs in the dark-grown pifq mutant) and in the induced Class 6 genes (except that the rapid transient spike in expression is followed by a lower steady state level of expression at 2 d in Rc). In addition, the Class 3 and 6 gene sets display a reduced mean level of transient induction in the pifq mutant in response to 1 h Rc (pifq-R1 compared with WT-R1), indicating a positive function in this rapid phy-induced response, coupled with the apparent absence of a repressive action in dark-grown seedlings. Inspection of the individual gene expression profiles in these classes reveals a subset of genes that exhibit a particularly robust reduction in the level of the transient light induction in the pifq mutant. A similar phenomenon is observed in some ambiguous genes in Class 5 (see Supplemental Data Sets 1 and 5 online), except that relatively weak activity in the dark is detected. These genes include ELIP1 (AT3G022840), ELIP2 (AT4G14690), and SIGE (AT5G24120) previously identified as displaying PIF3-dependent, rapid Rc induction in 4-d-old dark-grown seedlings (Monte et al., 2004). Examples of these individual gene profiles are shown in Supplemental Figure 6 online.
The induced Class 7 genes display a triple response, observable as robust derepression in the dark-grown pifq mutant, rapid Rc-triggered induction, and robust sustained expression after 2 d in Rc (Figure 4B; see Supplemental Figure 5 online). This pattern of regulation is consistent with the possibility of a direct role of the PIFs in repressing expression in the dark, coupled with rapid derepression upon light exposure as a result of phy-induced PIF degradation. These genes are therefore candidates for being direct targets of PIF-regulated deetiolation.
The patterns of regulation of the repressed genes (Figure 4C; see Supplemental Figure 5 online) are generally mirror images of those observed for the induced genes in the corresponding classes described above. This observation implies that the PIFs are capable of acting to coordinately regulate gene expression in a positive and negative manner depending on promoter context. The repressed Class 1, 3, and 6 genes do not display robust changes in expression in the dark-grown pifq mutant compared with the wild type, suggesting the absence of a major role of these genes in promoting skotomorphogenesis. By contrast, the genes in Classes 2, 4, and 7 exhibit pronounced reductions in expression in the dark-grown pifq mutant, suggesting a positive function in maintaining the skotomorphogenic state. However, the Class 2 genes do not respond robustly to Rc light, suggesting the absence of a central role in light-induced deetiolation. The repressed Class 4 genes show a delayed response to Rc light, suggesting either relatively stable transcripts after initial light-induced PIF removal or a possible indirect role for the PIFs in regulating their light-responsive expression.
The repressed Class 7 genes display both rapid and sustained repression in response to Rc in the wild type, as well as in response to the absence of the PIFs in the 2-d-old dark-grown pifq mutant (Figure 4C; see Supplemental Figure 5 online). These genes are therefore candidate direct targets of PIF regulation, whereby the bHLH factors act positively to promote expression in darkness, and this is rapidly and robustly reversed upon light exposure through PIF degradation.
The expression patterns detected by microarray analysis were validated for selected genes by quantitative PCR (qPCR) analysis (see Supplemental Figure 7 online).
Functional Categorization of Gene Sets
The induced and repressed genes in each class are grouped into broad functional categories in Figures 5A and 5B according to their predicted or established molecular or biological function. The data show that, of the induced gene set, photosynthesis/chloroplast (P/C)-related genes are the most abundant in Classes 1 and 4 and the second most abundant in Class 2 (Figure 5A). This molecular phenotype is consistent with the morphological (Figure 1), cellular, and subcellular (Figure 2) manifestations of the establishment of the photomorphogenic state by prolonged Rc light irradiation of the wild-type seedlings, and the constitutively photomorphogenic state established by 2 d of dark growth in the pifq mutant. The dominance of the P/C-related genes is particularly striking in the large, shared Class 4 set, where almost 60% of the annotated genes are induced in both genotypes under these contrasting conditions (WT-Rc and pifq-D). This result indicates that a major component of the light-induced gene network that generates a fully photosynthetically competent plant is strongly phenocopied (although not completely) in the dark by genetic removal of the PIF proteins examined here. Although the P/C-related genes are also prominent in the rapidly Rc light–induced Class 3, 5, and 6 genes, transcription-related genes are overall the most abundant responsive category (Figure 5A), as previously reported (Monte et al., 2004; Tepperman et al., 2006), consistent with a function in transcriptional network regulation. It is notable that a considerable proportion of these genes are only transiently induced by light (Classes 3 and 5), suggesting a possible regulatory function in the initial redirection of transcription during the early steps of deetiolation. The P/C- and transcription-related genes are both abundant in the Class 7 induced set, together dominating this class. These genes are of particular interest because of the potential for the direct involvement of the PIF proteins in both the rapid and sustained response of these genes to Rc.
Figure 5.
Functional Categories of Class 1 to 7 Induced and Repressed Genes.
Induced (A) and repressed (B) genes in each class (1 to 7) were assigned separately to a functional category (color coded). This assignment was based on Gene Ontology annotations for biological and/or molecular function available at TAIR (http://www.Arabidopsis.org). The percentage of the total annotated genes within each class/sector was calculated after excluding the genes with unknown biological or molecular function. The distribution of all the induced (A) and all the repressed (B) genes is also shown for comparison (left).
Of the repressed gene set, genes involved in various aspects of cellular metabolism are prominently represented in Classes 1, 2, and 4 (Figure 5B), consistent with the major change in cellular and subcellular development that underlies the conversion to the fully deetiolated state. By contrast, transcription-related genes dominate the rapidly Rc light–repressed genes in Classes 3 and 6. This result suggests that these genes are likely to function to promote or repress expression of downstream target genes in dark-grown wild-type seedlings and that this function is rapidly abrogated upon exposure to light. Transcription-related genes also dominate the repressed, shared Class 7 set. These genes thus similarly appear to exert their activity in darkness in a light-repressible manner, but do so in a manner that requires the presence of the PIF proteins for expression in the dark-grown seedling. These genes are thus potential candidates for being early components of a phy-modulated, PIF-directed transcriptional network in which they function as transcriptional regulators of downstream targets in the network hierarchy. It is also notable that hormone-related genes are relatively abundant members of the Class 6 and 7 repressed genes compared with the other classes and that auxin-related genes are prominent among this set.
Given that the pifq mutant appears to have reduced abundance and/or altered distribution of oil bodies in darkness compared with the wild type, we searched for altered expression patterns of genes involved in the mobilization of storage lipids in the mutant (Eastmond and Graham, 2001; Graham, 2008). Interestingly, we found five lipase genes (AT1G28570, AT2G42690, AT5G17670, AT1G33811, and AT3G05180) in the induced Class 4 list (i.e., differentially induced both in the wild type in Rc and in the pifq mutant in the dark) that could potentially be involved in the initial breakdown of triacylglycerol to fatty acids (see Supplemental Data Set 3 online). The response profiles for two of these are shown in Supplemental Figure 8A online. Two repressed genes (AT3G48460 and AT5G14180) were also found in the Class 4 list (see Supplemental Data Set 4 online). In addition, we found the MALATE SYNTHASE gene in the repressed Class 4 list (see Supplemental Figure 8B online) and the HYDROXYPYRUVATE REDUCTASE gene in the induced Class 4 list (see Supplemental Figure 8C online), suggesting accelerated glyxosome to peroxisome conversion in both WT-Rc and pifq-D relative to WT-D. These genes are thus candidates for functioning in any light-enhanced storage lipid mobilization that might occur as discussed below (see Discussion).
Promoter Analysis for Potential PIF Protein Target Sites
Given that the PIF proteins have been shown to bind sequence specifically to the core G-box motif, CACGTG (Martinez-Garcia et al., 2000; Huq and Quail, 2002; Shin et al., 2007; de Lucas et al., 2008; Moon et al., 2008; Oh et al., 2009), it might be anticipated that genes that are direct targets of PIF-regulated transcription may harbor this motif in their promoter regions. We therefore examined the genes identified in the above transcriptome analysis for the presence of the G-box core motif within 3 kb upstream of the transcription start site. This analysis is presented in Supplemental Analysis 1 online and associated Figures 4A and 4B and Supplemental Figures 9 and 10 online. Several functional subclasses of genes displayed statistically significant enrichment of G-boxes compared with the average across the ATH1 array (34% of whose promoters contain G-boxes), with the repressed Class 7 transcription factor genes (72% with G-boxes) being particularly notable.
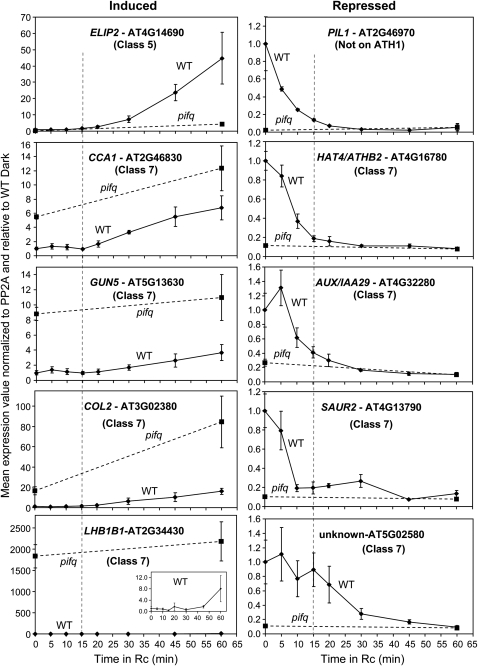
Early Response Kinetics of Rapidly Light-Regulated Genes
To more accurately define the relationship between the rapid decline in PIF protein abundance and the change in expression of early response genes upon initial exposure of dark-grown seedlings to light, we performed detailed, qPCR-based, time-course analysis of the expression of selected (primarily Class 7) induced and repressed genes during the first 1 h of Rc irradiation. For comparison, we also included PIL1 in this analysis, a previously identified, rapidly light-repressed gene (Tepperman et al., 2001; Hwang and Quail, 2008) not present on the ATH1 array. Overall, the repressed genes appear to respond more rapidly to the light signal than the induced genes (Figure 6). Whereas the induced genes display response lag times of 20 to 45 min, four of the five repressed genes have t1/2 times of decline of only 5 to 10 min. These differences in kinetics might be indicative of differences in the directness of the participation of the PIF proteins in each process. The rapidness of the light-triggered decline in expression of the repressed genes correlates strongly with the rate of decline in the abundance of PIF1, PIF3, PIF4, and PIF5 proteins (Bauer et al., 2004; Monte et al., 2004; Al-Sady et al., 2006; Shen et al., 2007, 2008; Lorrain et al., 2008), consistent with possible direct regulation of these genes by the bHLH factors. Conversely, the apparent delay in responsiveness of the induced genes might be consistent with a more indirect involvement of the PIFs in this process.
Figure 6.
Differing Patterns of Rapid Light Responsiveness Suggest Alternate Modes of PIF Regulation of Early Response Genes.
Wild-type (solid curve) and pifq mutant (dashed curve) seedlings were grown for 2 d in the dark as indicated in Figure 3A and then exposed to Rc (7.5 μmol/m2/s) for increasing periods from 0 (dark control) to 60 min. Expression of the indicated genes was determined by qPCR, and PP2A was used as a normalization control as described (Shin et al., 2007). Data are presented relative to the mean of WT-D set at unity and represent the mean and se of three independent biological replicates. Vertical dashed line marks the 15-min Rc time point.
The magnitude of the effect of the genetically imposed absence of the PIF proteins on the expression of the light-repressed genes in 2-d-old dark-grown seedlings is shown for the pifq mutant at time zero (unirradiated samples) in Figure 6. In addition, the data show that the light-triggered decline in expression levels rapidly reaches the basal level detected in this quadruple null mutant, suggesting that any residual steady state levels of the PIF proteins in the light are ineffective in supporting detectable expression of these genes.
The reciprocal effect (compared with the repressed genes) of the genetically imposed absence of the PIFs on the expression of the light-induced Class 7 genes (CCA1, GUN5, COL2, and LHB1B1) in 2-d-old dark-grown seedlings is shown by the time zero data points for these genes in Figure 6. These data suggest that the PIFs act to repress these genes in darkness and that the light-triggered degradation of these proteins, directly or indirectly, results in derepression of expression. By contrast, the expression of the single Class 5 induced gene in this set, ELIP2, is only relatively weakly affected in 2-d-old dark-grown seedlings, but the magnitude of the light-induced increase in expression is severely reduced by the absence of the PIFs. This result is consistent with a positive role of these bHLH proteins, directly or indirectly, in light-dependent steps in the induction process, as previously concluded (Monte et al., 2004), a role not achievable by simple genetic removal of a repressive function.
Comparative Expression Analysis
To provide a broader framework for discussion of the roles of the PIF proteins in regulating seedling gene expression, we have provided a comparative analysis of our present data on the pifq mutant with those of other genome-wide studies on the pif3 (Monte et al., 2004), pif1 (Moon et al., 2008), and pifq (Shin et al., 2009) mutants and a more limited analysis of photosynthetic genes in the pif1, pif3, and pif1 pif3 mutants (Stephenson et al., 2009) (see Supplemental Analysis 2 and associated data in Supplemental Figures 11 to 16 and Supplemental Data Sets 6 to 9 online). After completion of this manuscript, a microarray analysis was published of a pif4 pif5 double mutant showing differential expression under pseudodark conditions of a small subset of genes compared with the wild type (Lorrain et al., 2009).
DISCUSSION
Previously, we provided genetic evidence that the PIF1, 3, 4, and 5 phy-interacting bHLH transcription factors act to repress premature photomorphogenic seedling development in postgerminative darkness and that light reverses this repression via photoactivation of phy photoreceptor family members that then bind to and induce degradation of the PIF proteins (Leivar et al., 2008b). Here, we have identified several of the major parameters of cellular architecture and subcellular differentiation that underlie the PIF-regulated aspects of this global process and have defined the transcriptional network that drives these morphogenic responses. In addition, we have shown that the differential patterns of transcriptional regulation elicited by the different combinations of light treatment and PIF genotype investigated here provide insight into the molecular mode of participation of the PIF proteins in the transcriptional regulation of the different categories of target genes.
The cytological evidence presented here shows that the cellular and subcellular reorganization induced in the dark by the genetically imposed absence of the PIFs largely phenocopies the process normally induced by light in wild-type seedlings. The cotyledonary cells of the pifq mutant display the cell expansion responsible for cotyledon expansion, the increased vacuolization associated with this expansion, a reduced abundance and/or altered distribution of storage oil bodies, and induction of partial conversion of the etioplasts to chloroplasts, characteristic of the morphological transition of wild-type cells during normal deetiolation (Figure 2). These changes are consistent with the well-known fundamental qualitative switch in cellular function from heterotrophic to photoautotrophic growth displayed by these cells in response to initial exposure of wild-type seedlings to light. The data indicate, therefore, that these four PIF transcription factors pleiotropically control the circuitry that regulates a major fraction of the cellular processes involved in deetiolation.
The transcriptome analysis strongly supports this conclusion. The data show that 80% of the genes that are misregulated in pifq mutant seedlings grown in the dark are normally regulated by red light in the wild type (Figure 3D). This finding, together with the robust quantitative correlation in expression of the overlapping genes between these two sets of seedlings (Figure 3D), indicates that it is a central function of these PIFs to control the expression of phy-regulated genes during normal seedling development. The function of the remaining 20% that are PIF regulated but not apparently robustly light regulated (Figure 3D) remains to be determined. Conversely, because only 53% of the long-term Rc light–regulated genes are also misregulated in the pifq mutant in the dark, the light regulation of the remaining 47% would appear to involve other components. This result is consistent with the observation that the extent of constitutive deetiolation reached in the dark-grown mutant is less than that of the wild type in the light (Figure 1F; see Supplemental Figure 1 online). Other candidate regulatory components potentially responsible for the residual light responsiveness include other bHLH PIFs not yet investigated in this respect, such as PIF6/PIL2 and PIF7 (Khanna et al., 2004; Leivar et al., 2008a). An intriguing additional observation is that only 22 genes are differentially regulated between the wild type and pifq mutants grown in Rc for 2 d (Figure 3B), despite the hypersensitive phenotype of the mutant under this light treatment (Figure 1F; see Supplemental Figure 1 online). Although PIF-induced overexpression of phyB (Figure 1G; see Supplemental Figure 3 online) can account for this response (Khanna et al., 2007; Al-Sady et al., 2008; Leivar et al., 2008a), the apparent absence of major gene expression differences requires further exploration.
Our data more than double the number of genes reported recently to be regulated in common by Rc light in the wild type and by the absence of these PIFs in a dark-grown pifq mutant (Shin et al., 2009) (see Supplemental Analysis 2, Supplemental Figure 16, and Supplemental Data Set 9 online). Moreover, only 22% of the genes we identified overlap with those reported by these authors. The reasons for these differences have not been determined. One possibility is that, whereas we used Affymetrix ATH1 microarrays, Shin et al. used an Agilent platform. Although the obvious difference of nonoverlapping gene coverage on the two microarrays was eliminated by removing those genes from this analysis (see Supplemental Figure 16 and Supplemental Data Set 9 online), differences in detection sensitivity on the two platforms are possible. However, two other critical differences in experimental design between the two studies may have contributed. First, Shin et al. provided high levels of exogenous sucrose to their seedlings, whereas we do not. Second, these authors grew their seedlings under pseudodark conditions, whereas we grew ours under true dark conditions. Both of these parameters are likely to substantially modify the gene expression patterns (Cheng et al., 1992; Dijkwel et al., 1996, 1997; Gibson and Graham, 1999; Graham, 2008; Leivar et al., 2008b). We demonstrated previously (Leivar et al., 2008b) that the residual Pfr in dark-grown seedlings under pseudodark conditions induces partial deetiolation not observed under true dark conditions. Therefore, it is possible that unanticipated gene expression changes in the pseudodark-grown wild-type controls (used as the baseline for calculating the fold changes used to identify differentially regulated genes in the pifq mutant) distort the data such that expression changes in an extensive number of genes are masked. Similarly, because sucrose is known to function as a signaling molecule that can alter light-induced gene expression (Cheng et al., 1992; Dijkwel et al., 1996, 1997; Gibson and Graham, 1999; Graham, 2008), it is possible that the growth of seedlings on high levels of exogenous sucrose results in significant changes in gene expression that alter the apparent PIF- and phy-regulated genome-wide pattern (see below).
Not unexpectedly, given that development of the capacity to photosynthesize is well known to be a central parameter of the deetiolation process, genes associated with chloroplast function are observed to dominate the induced gene set regulated in common by light and the PIFs (Figure 5A). This dominance is particularly striking in the large shared Class 4 gene set where close to 60% are in this functional category and is consistent with the observed incipient chloroplast development in the dark-grown pifq mutant (Figure 2D). A similar pattern was reported by Shin et al. (2009). However, by comparison, the additional 636 shared light- and PIF-regulated genes identified here results in an ∼3.5-fold increase in the total number of chloroplast-associated genes detected as being regulated by the phy-PIF signaling pathway (see Supplemental Analysis 2, Supplemental Figure 16, and Supplemental Data Set 9 online).
The apparent reduced abundance of oil bodies in the dark-grown pifq mutant compared with the wild type (Figure 2) suggests an enhanced mobilization of storage lipids in this mutant similar to light-grown wild-type seedlings. Light-enhanced mobilization of storage lipids during postgerminative growth appears to be a species-specific response, since it has been observed in several plant species such as rapeseed (Brassica napus), cucumber (Cucumis sativus), and sunflower (Helianthus annuus; Theimer and Rosnitschek, 1978; Davies et al., 1981; Sadeghipour and Bhatla, 2003) but not in others like mustard (Sinapis alba; Bajracharya and Schopfer, 1979). The regulatory mechanisms for such enhanced oil body mobilization remain unknown. Some reports indicate light regulation of the components involved in the hydrolysis and metabolism of lipids, such as light induction of lipase enzymatic activity (Smolenska and Lewak, 1974; Davies et al., 1981). Similarly, light-induced conversion of glyoxysomes to peroxisomes through repression of glyoxysomal-specific enzyme activities (MALATE SYNTHASE and ISOCITRATE LYASE) and activation of peroxisomal-specific enzyme activities (GLYCOLATE OXIDASE and HYDROXYPYRUVATE REDUCTASE) are well established (Theimer and Rosnitschek, 1978; Bajracharya and Schopfer, 1979; Davies et al., 1981; Nishimura et al., 1996; Graham, 2008). The light-induced conversion of glyoxysomes to peroxisomes is considered to be involved in the differential metabolic use of storage lipids during the transition from heterotrophic growth in darkness to autotrophic growth in light (Eastmond et al., 2000; Eastmond and Graham, 2001; Graham, 2008). We have identified several genes potentially involved in different aspects of the mobilization of oil bodies whose expression is both light responsive and misregulated in the pifq mutant in the dark (see Results and Supplemental Figure 8 online), all of them being Class 4. None of these genes were detected as PIF regulated in the recent report by Shin et al. (2009), possibly because the exogenous source of carbon that these authors used might have altered the metabolic use of storage lipids in the dark-grown seedlings, as has been demonstrated (Eastmond et al., 2000; Eastmond and Graham, 2001; Graham, 2008). Collectively, therefore, our data indicate that the transcriptional changes regulated by the PIF proteins in response to light signals through the phy system are substantially broader, both quantitatively and qualitatively as regards identification of additional responsive pathways, than previously reported.
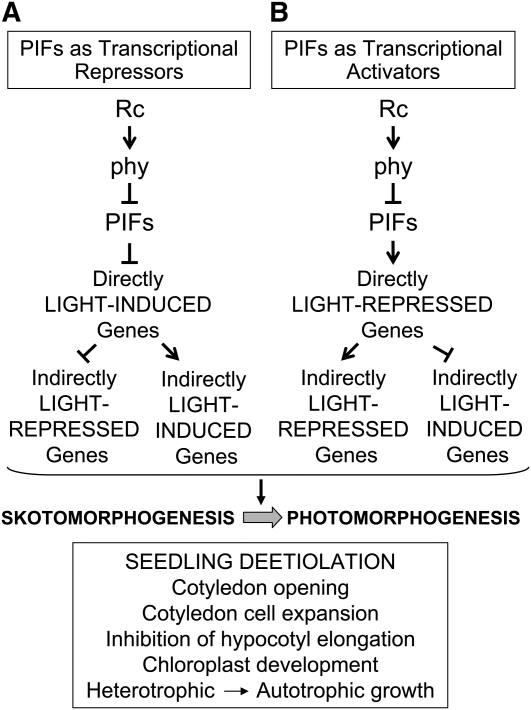
Our study extends beyond only describing the molecular phenotype of the pifq mutant established after 2 d of growth in the dark by using a strategy aimed at distinguishing genes that are potentially directly regulated by the PIF quartet from those more likely to be indirectly regulated in response to the light signal. Figure 7 provides a simplified, formal schematic summary of the alternate direct and indirect control modes by which responsive genes might be transcriptionally activated or repressed by the PIF proteins, in regulating seedling deetiolation in response to light activation of the phy pathway. The scheme illustrates that the PIFs could act directly to constitutively activate or repress target genes in the dark and that this action is reversed by photoactivated phy in response to light. These directly PIF-regulated genes could then potentially act secondarily to either activate or repress downstream targets in the transcriptional cascade. By including identification of genes that respond rapidly (within 1 h) to Rc upon initial irradiation of dark-grown seedlings, we have been able to define seven classes of genes displaying different patterns of regulation in response to the different light treatment genotype combinations (Figure 4). These different regulatory patterns suggest that the PIFs exert mechanistically different modes of transcriptional control in regulating these different classes of target genes.
Figure 7.
Simplified Schematic Model Depicting the Formal Alternate Direct and Indirect Control Modes Potentially Exercised by the PIF Proteins to Transcriptionally Activate or Repress Genes Regulating Seedling Deetiolation in Response to phy-Mediated Light Signals.
The PIFs are shown as acting constitutively in darkness as either transcriptional repressors (A) or activators (B) of direct target genes. Light-activated phy molecules trigger reversal of each potential activity through induced proteolytic degradation of the PIF factors, thereby derepressing light-induced direct target genes (A) and inactivating light-repressed direct target genes (B). Regulatory genes (such as transcription factor genes) within this direct target category then act either positively or negatively on downstream genes in the transcriptional network, thereby indirectly inducing or repressing their expression in response to the light signal.
The rapid responsiveness of the Class 3, 5, 6, and 7 genes to light (Figure 4) renders them potential direct targets of phy signaling. However, 67% of these (Classes 3 and 6) display no dependence at 2 d on the PIF quartet for this responsiveness in darkness (Figure 4A). Although this absence of evidence is not necessarily evidence of the absence of PIF involvement in the light-induced response, because that involvement could be transient, direct tests of this possibility are currently lacking. Certainly the apparent requirement for PIF activity for the maximum transient light induction or repression at 1 h displayed by the Class 3 and 6 genes (Figure 4) is consistent with this possibility. This is especially so in the case of the robustly dependent examples of ELIP1, SIGE, and the Class 5 gene ELIP2 (Figure 6; see Supplemental Figure 6 online). In addition, these Classes are enriched in genes encoding transcription-related factors, especially the repressed subset (Figure 5), consistent with a regulatory role in the transcriptional cascade. Nevertheless, the question of whether these genes are likely direct targets of PIF activity remains unclear from these data. By contrast, the delayed light responsiveness of the Class 1 and 4 genes (Figure 4) indicates that they are less likely to be direct PIF targets.
The remaining core Class 7 genes currently appear to represent the most likely direct targets of PIF transcriptional regulatory activity, as these respond both rapidly to the light signal and to the genetically generated absence of the PIF quartet (Figures 4 and 6). Consistent with this possibility, this class is enriched in genes that contain G-box motifs in their promoter regions (see Supplemental Figure 9 online), thus providing potential binding sites for these bHLH proteins. In addition, the Class 7 genes, and especially the repressed subset, are also strongly enriched in transcription factor genes (Figure 5), suggesting potential roles in the downstream transcriptional network (Figure 7). Interestingly, important components of the circadian clock and/or shade avoidance response, such as CCA1, PIL1, PIL2/PIF6, and ATHB2 (Franklin, 2008; Harmer, 2009), are included in these Class 7 transcription factor genes and are thus potential direct PIF targets.
The rapid responsiveness of the Class 7 genes to the light signal (Figure 6) is strongly correlated with the rapid light-induced degradation of the PIF proteins (Bauer et al., 2004; Al-Sady et al., 2006; Nozue et al., 2007; Shen et al., 2007, 2008; Lorrain et al., 2008) consistent with a close causal connection. If both the induced and repressed genes are directly regulated by the PIF proteins, this regulation would by definition require contrasting mechanisms of transcriptional control in the two cases. For the induced genes, the enhanced expression induced in the dark in the pifq mutant (Figure 6) implies that the PIFs act in darkness as transcriptional repressors of these genes and that light induction results from degradation-triggered derepression (Figure 7A). By contrast, for the repressed genes, the loss of expression in the dark in the pifq mutant (Figure 6) implies that the PIFs act as transcriptional activators of these genes and that light-imposed repression results from degradation-triggered loss of these activators (Figure 7B). If, on the other hand, there is only a single mechanism of PIF regulation of transcription during seedling deetiolation, and only one of these two gene subsets is directly regulated, the light-repressed genes may be the more likely of the two to be direct targets, for two reasons. First, where examined, robust light repression of expression is observed within 15 min of initiation of the light signal for these genes, whereas the induced genes appear to display a lag in responsiveness of 15 min or more (Figure 6). Second, 72% of the repressed transcription factor genes in Class 7 have G-box motifs in their promoters, a highly significant overrepresentation, whereas the comparable induced subset do not (see Supplemental Figure 10 online). Further analysis will be necessary to resolve this question.
Although some recent studies aimed at identifying PIF target genes by chromatin immunoprecipitation (ChIP) have been reported, the majority have been of limited scope, using PCR analysis of preselected genes, have been performed on materials other than etiolated seedlings, and/or have been performed under pseudodark conditions, making direct comparison with our present data difficult (Shin et al., 2007; de Lucas et al., 2008; Feng et al., 2008; Moon et al., 2008; Oh et al., 2009). Nevertheless, some of the findings support the proposition that some of our Class 7 genes may be direct PIF targets. de Lucas et al. (2008) identified the G-box–containing, Class 7 repressed genes PIL1 (AT2G46970) and XTR7 (AT4G14130) as being targets of PIF4 in dark-adapted green seedlings harvested at the end of the night (Figure 6; see Supplemental Data Set 4 online). Similarly, one out of the six PIF3 target genes reported by Shin et al. (2007) in seedlings grown under continuous far-red light conditions, CHI (AT3G55120), is a G-box–containing Class 7 induced gene (see Supplemental Data Set 3 online). In a ChIP-chip analysis of PIF1 targets in ungerminated seed, Oh et al. (2009) identified one G-box–containing Class 7 induced gene (HYDROLASE, AT4G37470; see Supplemental Data Set 3 online) and 10 G-box–containing Class 7 repressed genes (CKX5, AT1G75450; FHL, AT5G02200; PIL2/PIF6, AT3G62090; UNKNOWN, AT5G02580; ATGH9C3, AT4G11050; ARF18, AT3G61830; PATHOGENESIS-RELATED, AT4G36010; SULPHOTRANSFERASE, AT5G07010; CYSTEINE PROTEASE INHIBITOR-RELATED, AT2G31980; and DRM1, AT1G28330; see Supplemental Data Set 4 online). Future genome-wide chromatin immunoprecipitation assays for targets of the individual PIF factors in etiolated seedlings, coupled with functional analyses of putative target promoters, will be required to definitively map the phy-PIF–regulated transcriptional network.
METHODS
Seedling and Plant Growth and Measurements
Wild-type and pifq mutant seeds (Leivar et al., 2008b) were plated on GM medium without sucrose at room temperature as described (Monte et al., 2003). During this procedure, the seeds were routinely exposed to white light for a total of 1.5 h after imbibition. Seeds were then stratified for 5 d at 4°C in darkness, induced to germinate with a 5-min Rp (46 μmol/m2/s) and then incubated in the dark for 3 h at 21°C before exposure to a terminal 5-min FRp (58 μmol/m2/s) to suppress pseudodark effects as reported (Leivar et al., 2008b). With few exceptions that are indicated, seeds were then placed in either dark or Rc (6.7 μmol/ m2/s) at 21°C for 45 h (2-d-old seedlings). Where indicated, 2-d-old dark-grown seedlings were treated with 1 h of R (7.5 μmol/m2/s).
Measurements of light, hypocotyl length, cotyledon area, and cotyledon angle were as described (Monte et al., 2003; Leivar et al., 2008b). Measurements were typically done with at least 25 to 30 seedlings. Cotyledon area was measured for 35 to 40 cotyledons.
For analysis of the photobleaching phenotype, seedlings were grown for 2 to 4 d in the dark as described above and then transferred to white light for 3 to 5 d as described (Huq et al., 2004). The number of seedlings with green cotyledons was scored. Bleached seedlings were considered those that didn't turn green at all or that showed intermediate phenotypes (pale green or green with bleached patches).
Protein Extraction, Immunoblots, and Quantification
Protein extracts of Arabidopsis thaliana seedlings were prepared as described (Leivar et al., 2008a). Total protein was quantified using a Protein DC kit (Bio-Rad), and β-mercaptoethanol was added just before loading. Aliquots from each sample containing equal amounts of protein were then subjected to polyacrylamide gel electrophoresis. Mouse monoclonal anti-phyB (B1 and B7) antibodies were used to immunodetect phyB. A mouse monoclonal antibody against α-tubulin (Sigma-Aldrich) was used as a control for loading. Anti-mouse-horseradish peroxidase was used as secondary antibody (Promega), and an ECL plus chemiluminescence kit (Amersham) was used for detection. phyB signal normalized to tubulin from three biological replicates was quantified from the blots using Image J software. The linearity of the signal was assessed running a parallel protein extract dilution curve as described (Leivar et al., 2008a).
Light and Transmission Electron Microscopy
Details on the standard procedure followed for light and electron microscopy of cotyledons from 2-d-old dark- and Rc-grown seedlings (see above) as well as the quantification of prolamellar body and mean prothylakoid length per etioplast can be found in Supplemental Method 1 online.
Visualization of Oil Bodies Using Nile Red and Confocal Microscopy
Seedlings were grown in the dark for 2 d as indicated above and stained with Nile red (Invitrogen) to visualize neutral lipids (Greenspan et al., 1985). The protocol for Nile red staining of Arabidopsis seedlings was as described (Siloto et al., 2006). Images were acquired using a confocal laser scanning Zeiss LSM510 Meta microscope using a ×40 PlanNeofluor 1.3 oil objective (excitation = 488 nm; emission = 560 to 615 band-pass). Maximum intensity projections were obtained with Imaris software (Bitplane).
Gene Expression Analysis by qPCR
Two-day-old dark-grown seedlings were grown as indicated above and then treated with red light (7.5 μmol/m2/s) for up to 1 h. qPCR analysis was performed as described (Khanna et al., 2007) with minor variations. Briefly, 1 μg of total RNA extracted using the RNeasy Plus plant mini kit (Qiagen) was treated with DNase I (Invitrogen) to further eliminate genomic DNA according to the manufacturer's instructions. First-strand cDNA synthesis was done using the Super-Script First-Strand cDNA synthesis kit for RT-PCR (Invitrogen) and oligo (dT20) as a primer. cDNA was treated with RNase H before 1:30 dilution with water, and 10 μL were used for real-time PCR (MyIQ single-color real-time PCR detection system; Bio-Rad). Eva-green (Biotium) was used for detection, and 0.1% Tween 20, 0.1 mg/mL BSA, and 5% DMSO were added to the PCR mix as described (Khanna et al., 2007). Each PCR was repeated at least two times, and the mean expression values from these technical replicates were used for further calculations. Gene expression was measured from three biological replicates, and PP2A (AT1G13320) was used as a normalization control as described (Shin et al., 2007). Normalized gene expression was represented relative to the dark-grown wild-type set as a unity. Primer sequences for qPCR can be found in Supplemental Table 1 online.
Microarray-Based Expression Profiling: RNA Isolation, cRNA Synthesis, and Hybridizations
Wild-type and pifq mutant seedlings were grown in the dark or in Rc (6.7 μmol/m2/s) for 2 d before harvesting as indicated above. In addition, 2-d-old dark-grown seedlings were irradiated with red light (7.5 μmol/m2/s) for 1 h (R1). Seed samples were harvested after stratification. Three different biological replicates of each treatment were grown separately and extracted, processed, and analyzed independently, except for the dark-treated samples (D), where six biological replicates were analyzed. Total RNA isolation, cRNA synthesis, and microarray hybridizations were performed mainly as described (Monte et al., 2004). Briefly, total RNA was prepared using the method of Chang et al. (1993) followed by further purification using RNeasy columns (Qiagen). Probe synthesis was performed as described in the GeneChip Expression Analysis Technical Manual (http://www.affymetrix.com; Affymetrix), following the One-Cycle Target Labeling protocol. ATH1 microarrays (Affymetrix) were used for gene expression detection. Hybridization and washes were performed as described by Affymetrix in the Functional Genomics Laboratory facility at UC Berkeley (http://microarrays.berkeley.edu/affy_services.php).
Microarray Data Analysis
Microarray data analysis was performed as described (Monte et al., 2004) with minor modifications. Log scale gene expression values were calculated using a robust multiarray analysis. Fold change values between various genotypes and treatments were calculated using the mean expression value of the replicate samples. Statistically significant differential expression was determined using the PLM and LIMMA packages (http://www.bioconductor.org). SSTF genes were defined as those that differ by ≥2-fold with P values (adjusted for false discovery rate) ≤ 0.05 as described (Hu et al., 2009).
To simplify the functional classification analysis, a single functional category was assigned to each locus as indicated. Functional designations for each locus were determined using a recent annotation of the Arabidopsis genome (TAIR8) as well as Gene Ontology information. Any gene product targeted to the chloroplast was assigned to the Photosynthesis/Chloroplast category. Gene products with predicted or established transcription or DNA binding activity were assigned to the Transcription category.
For the G-box enrichment analysis, the total number of genes containing at least one G-box within 3 kb upstream of the translation initiation site was determined using the Patmatch tool found at the TAIR website (http://www.Arabidopsis.org/cgi-bin/patmatch/nph-patmatch.pl). The number of gene promoters with G-boxes on the Arabidopsis ATH1 array was then derived from this list. Statistically significant (P ≤ 0.05) enrichment of G-boxes in the various gene subsets compared with genes represented in the ATH1array was then calculated from the hypergeometric distribution.
Accession Numbers
The microarray data reported in this publication have been deposited in the National Center for Biotechnology Information Gene Expression Omnibus (GEO; http://www.ncbi.nlm.nih.gov/geo/) under the GEO Series accession number GSE17159. Sequence data can be found in the Arabidopsis Genome Initiative database under accession numbers AT2G20180 (PIF1/PIL5), AT1G09530 (PIF3), AT2G43010 (PIF4), and AT3G59060 (PIF5/PIL6).
Supplemental Data
The following materials are available in the online version of this article.
Supplemental Figure 1. Complete Time-Course Analysis of Morphological Phenotypes of pifq Mutant Seedlings Grown in the Dark and in Rc.
Supplemental Figure 2. Photobleaching Phenotype of pif1, pif3, pif1 pif3, and pifq Mutants Grown under True Dark Conditions before Transfer to Light.
Supplemental Figure 3. Quantification of phyB Levels in pifq Mutant Seedlings Grown in the Dark or in Rc for 2 d.
Supplemental Figure 4. Comparison of Early R-Responsive Genes with Long-Term R-Responsive Genes and PIF-Regulated Genes in the Dark.
Supplemental Figure 5. Hierarchical Cluster Analysis of Class 1 to 7 SSTF Genes Responding to WT-R1, WT-Rc, and/or pifq-D Reported in Figure 4A.
Supplemental Figure 6. Expression Profiles of Examples of Early-R1 Induced Genes That Are Dependent on the PIFs for This R Light Induction.
Supplemental Figure 7. qPCR Validation of the Expression of Selected Early R1-Responsive Genes That Are PIF Dependant in the Dark.
Supplemental Figure 8. Expression Profiles of phy-PIF–Regulated Genes Potentially Involved in Enhanced Mobilization of Oil Bodies.
Supplemental Figure 9. Statistical analysis for G-box enrichment in the 3Kbp-upstream putative promoter regions of SSTF genes by Class.
Supplemental Figure 10. Statistical Analysis for G-Box Enrichment in the 3-kb Upstream Putative Promoter Regions of SSTF Genes by Functional Category.
Supplemental Figure 11. Comparison of 2-d-Old and 4-d-Old Early R1-Responsive Genes in Wild-Type Seedlings.
Supplemental Figure 12. Comparative Analysis of the Expression Profiles in the pifq (Supplemental Data Set 1) and pif3 (Monte et al., 2004) Mutants of Genes Defined as Early R1 Induced That Are PIF3 Dependent for This R Light Induction.
Supplemental Figure 13. Comparative Analysis of the Expression Profiles in pifq (Supplemental Data Set 1) and pif3 (Monte et al., 2004) Mutants of Genes Described as Early R1 Repressed That Are PIF3 Dependent for This R Light Repression.
Supplemental Figure 14. Comparative Analysis of the Expression Profiles in pifq (Supplemental Data Set 1) and pif3 (Monte et al., 2004) Mutants of Genes That Are PIF3 Regulated in the Dark (WT-D versus pif3-D).
Supplemental Figure 15. Comparative Analysis of the Expression Profiles in the pifq (Supplemental Data Set 1) and pif3 (Monte et al., 2004) Mutants of PIF3-Regulated Tetrapyrrole Biosynthesis Genes Reported by Stephenson et al. (2009).
Supplemental Figure 16. Comparison of SSTF Genes Described as Responsive to Both WT-Rc and pifq-D Compared with WT-D in This Study (Figure 3D; Supplemental Data Set 1) and by Shin et al. (2009).
Supplemental Table 1. Primer Sequences Used for qPCR.
Supplemental Analysis 1. Promoter Analysis for Potential PIF Protein Target Sites.
Supplemental Analysis 2. Comparative Expression Analysis.
Supplemental Method 1. Light and Transmission Electron Microscopy.
Supplemental Data Set 1. Expression Data and Primary Analysis for the SSTF Genes Reported in Figure 3B.
Supplemental Data Set 2. PIF-Regulated Genes in Both Seed and Dark-Grown Seedlings.
Supplemental Data Set 3. SSTF Class 1 to 7 Genes That Are Induced in WT-R1, WT-Rc, and/or pifq-D Relative to WT-D.
Supplemental Data Set 4. SSTF Class 1 to 7 Genes That Are Repressed in WT-R1, WT-Rc, and/or pifq-D Relative to WT-D.
Supplemental Data Set 5. List of SSTF Genes Defined as Ambiguous since They Show Converse Responses (Induction or Repression) in WT-R1, WT-Rc, and/or pifq-D Relative to WT-D.
Supplemental Data Set 6. Comparative Analysis of SSTF Genes Responding to R1 (WTR1 versus WTD) in 2- and 4-d-Old Wild-Type Seedlings.
Supplemental Data set 7. Comparative Analysis between pifq and pif3 (Monte et al., 2004) Mutants of the Expression Profiles of Genes Described as Early R1 Responsive That Are PIF3 Dependent for This R Light Response (Monte et al., 2004).
Supplemental Data Set 8. Comparative Analysis between pifq and pif3 (Monte et al., 2004) Mutants of the Expression Profiles of Genes Defined as PIF3-Regulated Genes in the Dark (WT-D versus pif3-D).
Supplemental Data Set 9. Comparison of SSTF Genes Described as Responsive to Both WT-Rc and pifq-D Compared with WT-D in This study (Figure 3D; Supplemental Data Set 1) and by Shin et al. (2009).
Supplementary Material
Acknowledgments
We thank the Functional Genomics Laboratory at UC Berkeley for hybridizations of the microarrays, K. McDonald and R. Zalpuri (Electron Microscopy Lab, UC Berkeley) for electron microscopy analysis, S. Ruzin (Biological Imaging Facility, UC Berkeley) for confocal microscopy analysis, M. Casey (Plant Gene Expression Center, UC Berkeley) for technical support, A. Smith (Plant Gene Expression Center, UC Berkeley) for making media and solutions, and M. Hudson (University of Illinois at Urbana-Champaign) for advice on statistics. This work was supported by a postdoctoral fellowship from the Spanish Ministry of Education and Science to P.L. and by National Institutes of Health Grant GM-47475, Department of Energy Grant DEFG03-87ER13742, and USDA Agricultural Research Service Current Research Information System Grant 5335-21000-027-00D to P.H.Q.
The author responsible for distribution of materials integral to the findings presented in this article in accordance with the policy described in the Instructions for Authors (www.plantcell.org) is: Peter H. Quail (quail@nature.berkeley.edu).
Online version contains Web-only data.
References
- Al-Sady, B., Kikis, E.A., Monte, E., and Quail, P.H. (2008). Mechanistic duality of transcription factor function in phytochrome signaling. Proc. Natl. Acad. Sci. USA 105 2232–2237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Al-Sady, B., Ni, W., Kircher, S., Schafer, E., and Quail, P.H. (2006). Photoactivated phytochrome induces rapid PIF3 phosphorylation prior to proteasome-mediated degradation. Mol. Cell 23 439–446. [DOI] [PubMed] [Google Scholar]
- Bae, G., and Choi, G. (2008). Decoding of light signals by plant phytochromes and their interacting proteins. Annu. Rev. Plant Biol. 59 281–311. [DOI] [PubMed] [Google Scholar]
- Bajracharya, D., and Schopfer, P. (1979). Effect of light on the development of glyoxysomal functions in the cotyledons of mustard (Sinapis alba L.) seedlings. Planta 145 181–186. [DOI] [PubMed] [Google Scholar]
- Bauer, D., Viczian, A., Kircher, S., Nobis, T., Nitschke, R., Kunkel, T., Panigrahi, K.C., Adam, E., Fejes, E., Schafer, E., and Nagy, F. (2004). Constitutive photomorphogenesis 1 and multiple photoreceptors control degradation of phytochrome interacting factor 3, a transcription factor required for light signaling in Arabidopsis. Plant Cell 16 1433–1445. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Castillon, A., Shen, H., and Huq, E. (2007). Phytochrome interacting factors: Central players in phytochrome-mediated light signaling networks. Trends Plant Sci. 12 514–521. [DOI] [PubMed] [Google Scholar]
- Chang, S., Puryear, J., and Cairney, J. (1993). A simple and efficient method for isolating RNA from pine trees. Plant Mol. Biol. Rep. 11 113–116. [Google Scholar]
- Cheng, C.L., Acedo, G.N., Cristinsin, M., and Conkling, M.A. (1992). Sucrose mimics the light induction of Arabidopsis nitrate reductase gene transcription. Proc. Natl. Acad. Sci. USA 89 1861–1864. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davies, H.V., Gaba, V., Black, M., and Chapman, J.M. (1981). The control of food mobilisation in seeds of Cucumis sativus L. Planta 152 70–73. [DOI] [PubMed] [Google Scholar]
- de Lucas, M., Daviere, J.M., Rodriguez-Falcon, M., Pontin, M., Iglesias-Pedraz, J.M., Lorrain, S., Fankhauser, C., Blazquez, M.A., Titarenko, E., and Prat, S. (2008). A molecular framework for light and gibberellin control of cell elongation. Nature 451 480–484. [DOI] [PubMed] [Google Scholar]
- Deng, X.W., Caspar, T., and Quail, P.H. (1991). cop1: A regulatory locus involved in light-controlled development and gene expression in Arabidopsis. Genes Dev. 5 1172–1182. [DOI] [PubMed] [Google Scholar]
- Dijkwel, P.P., Huijser, C., Weisbeek, P.J., Chua, N.H., and Smeekens, S.C. (1997). Sucrose control of phytochrome A signaling in Arabidopsis. Plant Cell 9 583–595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dijkwel, P.P., Kock, P., Bezemer, R., Weisbeek, P.J., and Smeekens, S. (1996). Sucrose represses the developmentally controlled transient activation of the plastocyanin gene in Arabidopsis thaliana seedlings. Plant Physiol. 110 455–463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Duek, P.D., and Fankhauser, C. (2005). bHLH class transcription factors take centre stage in phytochrome signalling. Trends Plant Sci. 10 51–54. [DOI] [PubMed] [Google Scholar]
- Eastmond, P.J., and Graham, I.A. (2001). Re-examining the role of the glyoxylate cycle in oilseeds. Trends Plant Sci. 6 72–78. [DOI] [PubMed] [Google Scholar]
- Eastmond, P.J., Germain, V., Lange, P.R., Bryce, J.H., Smith, S.M., and Graham, I.A. (2000). Postgerminative growth and lipid catabolism in oilseeds lacking the glyoxylate cycle. Proc. Natl. Acad. Sci. USA 97 5669–5674. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feng, S., et al. (2008). Coordinated regulation of Arabidopsis thaliana development by light and gibberellins. Nature 451 475–479. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Franklin, K.A. (2008). Shade avoidance. New Phytol. 179 930–944. [DOI] [PubMed] [Google Scholar]
- Fujimori, T., Yamashino, T., Kato, T., and Mizuno, T. (2004). Circadian-controlled basic/helix-loop-helix factor, PIL6, implicated in light-signal transduction in Arabidopsis thaliana. Plant Cell Physiol. 45 1078–1086. [DOI] [PubMed] [Google Scholar]
- Gibson, S.I., and Graham, I.A. (1999). Another player joins the complex field of sugar-regulated gene expression in plants. Proc. Natl. Acad. Sci. USA 96 4746–4748. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Graham, I.A. (2008). Seed storage oil mobilization. Annu. Rev. Plant Biol. 59 115–142. [DOI] [PubMed] [Google Scholar]
- Greenspan, P., Mayer, E.P., and Fowler, S.D. (1985). Nile red: A selective fluorescent stain for intracellular lipid droplets. J. Cell Biol. 100 965–973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harmer, S.L. (2009). The circadian system in higher plants. Annu. Rev. Plant Biol. 60 357–377. [DOI] [PubMed] [Google Scholar]
- Hu, W., Su, Y.S., and Lagarias, J.C. (2009). A light-independent allele of phytochrome B faithfully recapitulates photomorphogenic transcriptional networks. Mol. Plant 2 166–182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hudson, M.E., and Quail, P.H. (2003). Identification of promoter motifs involved in the network of phytochrome A-regulated gene expression by combined analysis of genomic sequence and microarray data. Plant Physiol. 133 1605–1616. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huq, E., Al-Sady, B., Hudson, M., Kim, C., Apel, K., and Quail, P.H. (2004). Phytochrome-interacting factor 1 is a critical bHLH regulator of chlorophyll biosynthesis. Science 305 1937–1941. [DOI] [PubMed] [Google Scholar]
- Huq, E., and Quail, P.H. (2002). PIF4, a phytochrome-interacting bHLH factor, functions as a negative regulator of phytochrome B signaling in Arabidopsis. EMBO J. 21 2441–2450. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hwang, Y.S., and Quail, P.H. (2008). Phytochrome-regulated PIL1 derepression is developmentally modulated. Plant Cell Physiol. 49 501–511. [DOI] [PubMed] [Google Scholar]
- Jiao, Y., Lau, O.S., and Deng, X.W. (2007). Light-regulated transcriptional networks in higher plants. Nat. Rev. Genet. 8 217–230. [DOI] [PubMed] [Google Scholar]
- Khanna, R., Huq, E., Kikis, E.A., Al-Sady, B., Lanzatella, C., and Quail, P.H. (2004). A novel molecular recognition motif necessary for targeting photoactivated phytochrome signaling to specific basic helix-loop-helix transcription factors. Plant Cell 16 3033–3044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Khanna, R., Shen, Y., Marion, C.M., Tsuchisaka, A., Theologis, A., Schafer, E., and Quail, P.H. (2007). The basic helix-loop-helix transcription factor PIF5 acts on ethylene biosynthesis and phytochrome signaling by distinct mechanisms. Plant Cell 19 3915–3929. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim, J., Yi, H., Choi, G., Shin, B., and Song, P.S. (2003). Functional characterization of phytochrome interacting factor 3 in phytochrome-mediated light signal transduction. Plant Cell 15 2399–2407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leivar, P., Monte, E., Al-Sady, B., Carle, C., Storer, A., Alonso, J.M., Ecker, J.R., and Quail, P.H. (2008. a). The Arabidopsis phytochrome-interacting factor PIF7, together with PIF3 and PIF4, regulates responses to prolonged red light by modulating phyB levels. Plant Cell 20 337–352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leivar, P., Monte, E., Oka, Y., Liu, T., Carle, C., Castillon, A., Huq, E., and Quail, P.H. (2008. b). Multiple phytochrome-interacting bHLH transcription factors repress premature seedling photomorphogenesis in darkness. Curr. Biol. 18 1815–1823. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lorrain, S., Allen, T., Duek, P.D., Whitelam, G.C., and Fankhauser, C. (2008). Phytochrome-mediated inhibition of shade avoidance involves degradation of growth-promoting bHLH transcription factors. Plant J. 53 312–323. [DOI] [PubMed] [Google Scholar]
- Lorrain, S., Trevisan, M., Pradervand, S., and Fankhauser, C. (2009). Phytochrome interacting factors 4 and 5 redundantly limit seedling de-etiolation in continuous far-red light. Plant J., in press. [DOI] [PubMed]
- Martinez-Garcia, J.F., Huq, E., and Quail, P.H. (2000). Direct targeting of light signals to a promoter element-bound transcription factor. Science 288 859–863. [DOI] [PubMed] [Google Scholar]
- Monte, E., Alonso, J.M., Ecker, J.R., Zhang, Y., Li, X., Young, J., Austin-Phillips, S., and Quail, P.H. (2003). Isolation and characterization of phyC mutants in Arabidopsis reveals complex crosstalk between phytochrome signaling pathways. Plant Cell 15 1962–1980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Monte, E., Al-Sady, B., Leivar, P., and Quail, P.H. (2007). Out of the dark: How the PIFs are unmasking a dual temporal mechanism of phytochrome signalling. J. Exp. Bot. 58 3125–3133. [DOI] [PubMed] [Google Scholar]
- Monte, E., Tepperman, J.M., Al-Sady, B., Kaczorowski, K.A., Alonso, J.M., Ecker, J.R., Li, X., Zhang, Y., and Quail, P.H. (2004). The phytochrome-interacting transcription factor, PIF3, acts early, selectively, and positively in light-induced chloroplast development. Proc. Natl. Acad. Sci. USA 101 16091–16098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moon, J., Zhu, L., Shen, H., and Huq, E. (2008). PIF1 directly and indirectly regulates chlorophyll biosynthesis to optimize the greening process in Arabidopsis. Proc. Natl. Acad. Sci. USA 105 9433–9438. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nagatani, A. (2004). Light-regulated nuclear localization of phytochromes. Curr. Opin. Plant Biol. 7 708–711. [DOI] [PubMed] [Google Scholar]
- Ni, M., Tepperman, J.M., and Quail, P.H. (1998). PIF3, a phytochrome-interacting factor necessary for normal photoinduced signal transduction, is a novel basic helix-loop-helix protein. Cell 95 657–667. [DOI] [PubMed] [Google Scholar]
- Nishimura, M., Hayashi, M., Kato, A., Yamaguchi, K., and Mano, S. (1996). Functional transformation of microbodies in higher plant cells. Cell Struct. Funct. 21 387–393. [DOI] [PubMed] [Google Scholar]
- Nozue, K., Covington, M.F., Duek, P.D., Lorrain, S., Fankhauser, C., Harmer, S.L., and Maloof, J.N. (2007). Rhythmic growth explained by coincidence between internal and external cues. Nature 448 358–361. [DOI] [PubMed] [Google Scholar]
- Oh, E., Kang, H., Yamaguchi, S., Park, J., Lee, D., Kamiya, Y., and Choi, G. (2009). Genome-wide analysis of genes targeted by PHYTOCHROME INTERACTING FACTOR 3-LIKE5 during seed germination in Arabidopsis. Plant Cell 21 403–419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oh, E., Kim, J., Park, E., Kim, J.I., Kang, C., and Choi, G. (2004). PIL5, a phytochrome-interacting basic helix-loop-helix protein, is a key negative regulator of seed germination in Arabidopsis thaliana. Plant Cell 16 3045–3058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Quail, P.H. (2002). Phytochrome photosensory signalling networks. Nat. Rev. Mol. Cell Biol. 3 85–93. [DOI] [PubMed] [Google Scholar]
- Quail, P.H. (2007). Phytochrome-regulated gene expression. J. Integr. Plant Biol. 49 11–20. [Google Scholar]
- Rockwell, N.C., Su, Y.S., and Lagarias, J.C. (2006). Phytochrome structure and signaling mechanisms. Annu. Rev. Plant Biol. 57 837–858. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sadeghipour, H.R., and Bhatla, S.C. (2003). Light-enhanced oil body mobilization in sunflower seedlings accompanies faster protease action on oleosins. Plant Physiol. Biochem. 41 309–316. [Google Scholar]
- Schafer, E., and Nagy, F. (2006). Photomorphogenesis in Plants and Bacteria. (Dordrecht, The Netherlands: Springer).
- Shen, Y., Khanna, R., Carle, C.M., and Quail, P.H. (2007). Phytochrome induces rapid PIF5 phosphorylation and degradation in response to red-light activation. Plant Physiol. 145 1043–1051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shen, H., Zhu, L., Castillon, A., Majee, M., Downie, B., and Huq, E. (2008). Light-induced phosphorylation and degradation of the negative regulator PHYTOCHROME-INTERACTING FACTOR1 from Arabidopsis depend upon its direct physical interactions with photoactivated phytochromes. Plant Cell 20 1586–1602. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shin, J., Kim, K., Kang, H., Zulfugarov, I.S., Bae, G., Lee, C.H., Lee, D., and Choi, G. (2009). Phytochromes promote seedling light responses by inhibiting four negatively-acting phytochrome-interacting factors. Proc. Natl. Acad. Sci. USA 106 7660–7665. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shin, J., Park, E., and Choi, G. (2007). PIF3 regulates anthocyanin biosynthesis in an HY5-dependent manner with both factors directly binding anthocyanin biosynthetic gene promoters in Arabidopsis. Plant J. 49 981–994. [DOI] [PubMed] [Google Scholar]
- Siloto, R.M., Findlay, K., Lopez-Villalobos, A., Yeung, E.C., Nykiforuk, C.L., and Moloney, M.M. (2006). The accumulation of oleosins determines the size of seed oil bodies in Arabidopsis. Plant Cell 18 1961–1974. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smolenska, G., and Lewak, S. (1974). The role of lipases in the germination of dormant apple embryos. Planta 116 361–370. [DOI] [PubMed] [Google Scholar]
- Stephenson, P.G., Fankhauser, C., and Terry, M.J. (2009). PIF3 is a repressor of chloroplast development. Proc. Natl. Acad. Sci. USA 106 7654–7659. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tepperman, J.M., Hudson, M.E., Khanna, R., Zhu, T., Chang, S.H., Wang, X., and Quail, P.H. (2004). Expression profiling of phyB mutant demonstrates substantial contribution of other phytochromes to red-light-regulated gene expression during seedling de-etiolation. Plant J. 38 725–739. [DOI] [PubMed] [Google Scholar]
- Tepperman, J.M., Hwang, Y.S., and Quail, P.H. (2006). phyA dominates in transduction of red-light signals to rapidly responding genes at the initiation of Arabidopsis seedling de-etiolation. Plant J. 48 728–742. [DOI] [PubMed] [Google Scholar]
- Tepperman, J.M., Zhu, T., Chang, H.S., Wang, X., and Quail, P.H. (2001). Multiple transcription-factor genes are early targets of phytochrome A signaling. Proc. Natl. Acad. Sci. USA 98 9437–9442. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Theimer, R.R., and Rosnitschek, I. (1978). Development and intracellular localization of lipase activity in rapeseed (Brassica napus L.) cotyledons. Planta 139 249–256. [DOI] [PubMed] [Google Scholar]
- Toledo-Ortiz, G., Huq, E., and Quail, P.H. (2003). The Arabidopsis basic/helix-loop-helix transcription factor family. Plant Cell 15 1749–1770. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.