Abstract
The replication of positive-strand RNA viruses occurs in cytoplasmic membrane-bound virus replication complexes (VRCs). Depending on the virus, distinct cellular organelles such as the endoplasmic reticulum (ER), chloroplast, mitochondrion, endosome, and peroxisome are recruited for the formation of VRC-associated membranous structures. Previously, the 6,000-molecular-weight protein (6K) of plant potyviruses was shown to be an integral membrane protein that induces the formation of 6K-containing membranous vesicles at endoplasmic reticulum (ER) exit sites for potyvirus genome replication. Here, we present evidence that the 6K-induced vesicles predominantly target chloroplasts, where they amalgamate and induce chloroplast membrane invaginations. The vesicular transport pathway and actomyosin motility system are involved in the trafficking of the 6K vesicles from the ER to chloroplasts. Viral RNA, double-stranded RNA, and viral replicase components are concentrated at the 6K vesicles that associate with chloroplasts in infected cells, suggesting that these chloroplast-bound 6K vesicles are the site for potyvirus replication. Taken together, these results suggest that plant potyviruses sequentially recruit the ER and chloroplasts for their genome replication.
The replication of eukaryotic positive-strand RNA viruses in infected cells is closely associated with unique virus-induced intracellular membranous vesicles (22). These membranous vesicles have been proposed to provide a scaffold for anchoring the virus replication complex (VRC), confine the process of RNA replication to a specific safeguarded cytoplasmic location, and prevent the activation of certain host defense mechanisms that can be triggered by double-stranded RNA (dsRNA) intermediates during virus replication (33, 47). Depending on the type of virus, the virus-induced membranous vesicles are derived from various intracellular organelles in the host. Many plant and animal viruses remodel and utilize the endoplasmic reticulum (ER) in VRCs (1, 6, 17, 33, 34, 36, 38, 39, 46). Other cellular organelles such as endosomes, lysomes, chloroplasts, peroxisomes, and mitochondria have also been suggest to be the replication site for togaviruses, tymoviruses, and tombusviruses, respectively (25, 27, 31). Given that the ER appears to be the site where the host cell translation machinery is hijacked for the biosynthesis of the first set of viral proteins, the subcellular location of virus replication (either in the vicinity of the ER or elsewhere) and the mechanism of transport to locations other than the ER are poorly understood.
Plant potyviruses, accounting for ∼30% of known plant viruses including many agriculturally important viruses, e.g., Turnip mosaic virus (TuMV), Maize dwarf mosaic virus (MDMV), Tobacco etch virus (TEV), and Potato virus Y (PVY), are related to picornaviruses and picorna-like viruses (20, 21, 43). The potyviral genome is a single-stranded positive-sense RNA of about 10 kb in length and encodes at least 11 mature viral proteins (8, 43). Of these 11 proteins, the 6-kDa protein (designated 6K or 6K2) contains a central hydrophobic domain (35). In seminal work, Carrington and colleagues determined that 6K induces the formation of the ER-derived vesicles for TEV replication (35, 38). More recently, viral proteins required for replication and several host factors, namely, eukaryotic initiation factor (isoform) 4E, poly(A)-binding protein, eukaryotic elongation factor 1A, and heat shock cognate 70-3 protein, have been shown to associate with the TuMV 6K-induced vesicles (9, 41), raising the possibility that the potyviral 6K vesicles represent sites of viral genome replication. Furthermore, we have demonstrated that the biogenesis of the potyviral 6K vesicles occurs at COPII-accumulating ER exit sites (ERES) on the ER membrane (45). In this study, we further studied the trafficking of 6K-induced vesicles and found that the 6K-induced mobile vesicles trafficked predominantly from the ER to the periphery of chloroplasts. We show that these 6K vesicles docked on the outer chloroplast envelope and induced chloroplast invaginations. The chloroplast-associated 6K vesicles contained viral replicase components and dsRNA and were concentrated with viral RNA. We provide evidence that the early secretory pathway and actomyosin motility system were required for the trafficking of 6K vesicles from the ER to chloroplasts. These results suggest that plant potyviruses sequentially recruit the ER and chloroplasts for their genome replication.
MATERIALS AND METHODS
Plasmid construction.
Gateway technology (Invitrogen, Burlington, Ontario, Canada) was used to generate all the expression clones in this work. PCR involved in the gateway cloning was carried out with Phusion DNA polymerase (NEB, Pickering, Ontario, Canada). The recombinant TuMV infectious clone tagged with a chimeric gene containing the 6K-coding sequence fused in frame with the green fluorescent protein (GFP)-coding sequence (TuMV::6K-GFP) was described previously (9, 41). The 6K, NIb, and cylindrical inclusion (CI) cistrons of TuMV were amplified from infectious clone TuMV::6K-GFP by PCR. The 6K-NIa cistron of TuMV was amplified from the plasmids containing TuMV 6K-NIa, which had been modified at the junction of 6K2 and the viral genome-linked protein (VPg) and at the active site of NIa-Pro to prevent proteolysis (5, 24). All these fragments were recombined into pDONR221 and then into the binary destination vector pEarleygate101 (12) and pGWB554 (28) to produce plasmids 6K-yellow fluorescent protein (6K-YFP), 6K-NIa-monomeric red fluorescent protein (mRFP), NIb-mRFP, and CI-mRFP, respectively. Plasmids for expressing mouse talin (mTalin)-cyan fluorescent protein (CFP), mTalin-YFP, ERES marker Sar1-CFP, Golgi apparatus marker ERD2-CFP, and untagged Sar1 (H74L) were constructed as described previously (23, 45). Plasmids for expressing YFP-HDEL and CFP-HDEL were ordered from TAIR (http://www.arabidopsis.org).
Agrobacterium-mediated expression.
Four-week-old Nicotiana benthamiana plants were used for Agrobacterium tumefaciens (strain GV3101)-mediated transient expression, essentially as described previously (45). Briefly, binary vectors were transformed into A. tumefaciens GV3101 using electroporation. Agrobacteria were grown overnight in LB medium plus appropriate antibiotics, harvested, and then resuspended in 10 mM MgCl2 containing 100 μM acetosyringone. After incubation for 2 h at room temperature (RT), the culture was diluted to an optical density at 600 nm (OD600) of 0.2 to 0.5 and infiltrated into leaf tissue under gentle pressure using a syringe barrel.
Drug treatments.
Segments of transformed leaves were used for drug treatment. Brefeldin A (BFA) (Sigma-Aldrich, Oakville, Ontario, Canada) was used at a concentration of 10 μg/ml and 50 μg/ml as described previously (10). Latrunculin B (Lat B) (Sigma-Aldrich) was used at a concentration of 25 μM as described previously (45). BFA (50 μg/ml) was infiltrated into an extended leaf of N. benthamiana. After 3 h, the same leaf was agro-infiltrated with TuMV::6K-GFP. The presence of TuMV was monitored 96 h after agro-infiltration by using an enzyme-linked immunosorbent assay (ELISA) kit (Agdia, IN).
Detection of viral RNA and dsRNA.
For detection of TuMV RNA using the Pumilio-based system, plasmids CitN-HsPUMHD and HsPUMHD-CitC (where HsPUMHD is the human Pumilio homology domain and CitN and CitC are the N- and C-terminal halves of split mCistrine, respectively) were coexpressed in N. benthamiana epidermal cells infected or not infected with TuMV::6K-GFP, essentially as described previously (42). Briefly, a total of 5 μg of each DNA plasmid of CitN-HsPUMHD and HsPUMHD-CitC was mixed and precipitated onto 1-μm gold particles (Bio-Rad, Mississauga, Ontario, Canada). Fully expanded N. benthamiana leaves on plates (MS medium [Sigma Aldrich] plus 0.8% agar) were bombarded twice using a PDS-1000/He biolistic particle delivery system (Bio-Rad). Bombarded leaves were kept at 25°C with their petioles in water and imaged 24 h postbombardment. For dsRNA detection, N. benthamiana leaves infected with TuMV::6K-GFP 48 h after agro-infiltration were collected, and protoplasts were isolated as described in the following section. Isolated protoplasts were fixed in 4% paraformaldehyde and processed for analysis of immunofluorescence as described previously (9). dsRNA was probed with monoclonal antibody J2 (Scicons, Hungary) and then treated with Alexa Fluor 594-conjugated second antibodies against the mouse IgG (Invitrogen).
Chloroplast preparations and immunoblot analysis.
Intact chloroplasts were isolated from N. benthamiana leaves expressing the YFP-HDEL fusion protein or infected with TuMV::6K-GFP 48 h after agro-infiltration using a chloroplast isolation kit (Sigma-Aldrich) following the supplier's instruction. In brief, after mechanical cell wall breakage and removal of cell debris and unbroken leaf tissues by filtration, total cell chloroplasts were subjected to centrifugation at 5,000 × g for 10 min in a discontinuous Percoll gradient. The intact chloroplasts were collected and resuspended with the incubation buffer containing 330 mM sorbitol, 5 mM MgCl2, and 50 mM HEPES-KOH, pH 7.3. The chloroplast suspension was centrifuged again at 5,000 × g for 3 min and resuspended in the incubation buffer. For salt treatment, the isolated chloroplasts were resuspended in 0.1 M Na2CO3 (pH 10.5) or 1 M NaCl and incubated on ice with occasional mixing for 30 min. The total proteins were extracted from chloroplasts or directly from N. benthamiana infected by TuMV::6K-GFP 48 h after agro-infiltration, essentially as described previously (41). Proteins were separated by sodium dodecyl sulfate (SDS)-polyacrylamide gel electrophoresis (PAGE), transferred onto a polyvinylidene difluoride (PVDF) membrane, and detected by immunoblots using anti-GFP monoclonal antibodies (Sigma-Aldrich). The secondary antibodies were goat anti-mouse-horseradish peroxidase ([HRP] GE Healthcare, Oakville, Ontario, Canada). The proteins recognized by the antibodies were visualized using an ECL development kit (GE Healthcare).
Microscopy.
All imaging analyses of plant tissues using a Leica TCS SP2 inverted confocal microscope with a 63× oil immersion objective were carried out essentially as described previously (45), with the following excitation wavelengths: GFP was excited at 488 nm, and the emitted light was captured at 505 to 525 nm; chlorophyll autofluorescence was emitted at 630 to 680 nm; mCitrine (bimolecular fluorescence complementation [BiFC]) was excited at 514 nm, and the emitted light was captured at 555 to 575 nm; mRFP was excited using 543 nm and captured at 590 to 630 nm. Time-lapse scanning was performed with the Leica TCS SP2 imaging system software. Images were captured digitally and handled using the Leica LCS software. For electron microscopy, the TuMV-infected leaf tissues and healthy leaves were fixed, and transmission electron microscopy was conducted as described previously (31).
RESULTS
Localization of 6K proteins to chloroplasts.
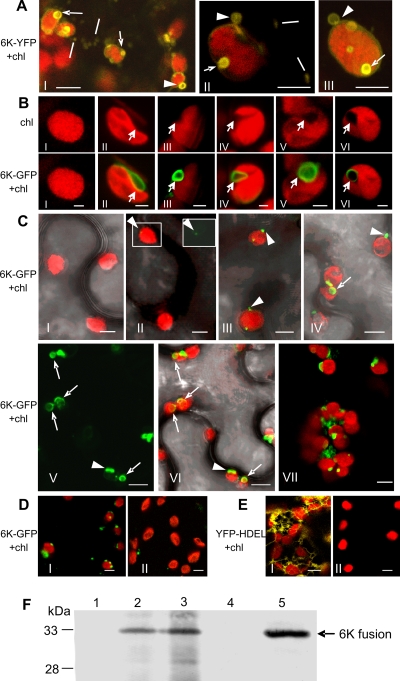
The 6K protein of potyviruses is an integral membrane protein that induces the formation of ER-derived vesicles for virus replication (35, 38). To investigate the subcellular distribution of the 6K-induced vesicles in planta, we transiently expressed the TuMV 6K-YFP in N. benthamiana epidermal cells via agro-infiltration. By 24 h post-agro-infiltration, 6K-YFP induced the formation of heterogeneously sized granular and ring-like vesicles (collectively named 6K bodies) in the cytoplasm, consistent with our recent results using the TEV 6K protein (45). The ring-like vesicles of approximately 1 to 3 μm in diameter were associated with chloroplasts (Fig. 1A). To determine if 6K also targeted chloroplasts during the course of virus infection, TuMV::6K-GFP was agro-infiltrated into N. benthamiana epidermal cells. At 48 h after agro-infiltration, most of 6K-GFP colocalized with chloroplasts, showing discrete patches at the periphery of chloroplasts (Fig. 1B, frames IV, V, and VI). Under the dim background of fluorescence, these patches appeared as ring-like vesicles of approximately 1 to 3 μm in diameter, resembling the appearance and size of vesicles induced by 6K-YFP when expressed alone (Fig. 1A). Furthermore, 6K-GFP frequently induced chloroplast invaginations (Fig. 1B). It appeared that invaginations initiated upon the attachment of 6K to the chloroplast envelope (Fig. 1B, frames II and III), followed by the formation of an elongated arm (Fig. 1B, frame IV) and the uptake and enclosure of the vesicle (Fig. 1B, frames V and VI).
FIG. 1.
The association of 6K proteins with chloroplasts in N. benthamiana leaves. (A) The docking of 6K ring-like vesicles on the chloroplast envelope (arrowheads) or the invaginations of 6K ring-like vesicles into the chloroplast envelope (arrows) when the 6K-YFP was expressed alone 24 h after agro-infiltration. Lines point to mobile granular vesicles in the cytoplasm. (B) Various invaginations in the chloroplast induced by 6K-containing vesicles (arrows). Frame I, chloroplast in healthy cells; frames II and III, attachment of 6K vesicles to the chloroplast envelope; frame IV, 6K vesicle in chloroplast invaginations; frames V and VI, the enclosure of 6K vesicle within chloroplast. (C) The association of 6K vesicles with chloroplasts at different time points after TuMV::6K-GFP infection. Frame I, 0 h after agro-infiltration; frames II and III, 36 h; frames IV, V, and VI, 48 h; frame VII, 60 h. In frame II, the upper-right inset shows the GFP image in the left rectangle. For better clarity, frame V shows only the GFP image, and frame VI overlays frame V with chlorophyll autofluorescence. Arrowheads indicate the adhesions of 6K vesicles to the chloroplast periphery. Arrows indicate the invagination of 6K vesicles into chloroplasts. (D) The effect of salt treatment on the association of 6K vesicles with chloroplasts. Frame I, 6K vesicles associated with chloroplasts isolated from N. benthamiana leaves infected with TuMV::6K-GFP 48 h after agro-infiltration; frame II, upon the treatment of 1 M NaCl, 6K vesicles disassociated with chloroplasts. (E) YFP-labeled ER network was evident in N. benthamiana leaf tissues expressing YFP-HDEL (frame I), and no visible YFP was observed in the chloroplasts purified from N. benthamiana leaves expressing YFP-HDEL (frame II). (F) Immunoblot analysis of GFP or YFP fusion proteins. Lane 1, proteins from chloroplasts isolated from healthy control; lane 2, chloroplasts from leaf tissues infected by TuMV::6K-GFP; lane 3, chloroplasts from leaf tissues expressing 6K-YFP; lane 4, chloroplasts from leaf tissues expressing YFP-HDEL; lane 5, proteins extracted directly from N. benthamiana leaves infected with TuMV::6K-GFP. Anti-GFP monoclonal antibodies were used to detect the 6K-GFP, 6K-YFP, and YFP-HDEL fusion proteins as described previously (9). The molecular masses of 6K-GFP (33 kDa), 6K-YFP (33 kDa), and YFP-HDEL (28 kDa) are indicated. Chl, chlorophyll autofluorescence. Bars, 8 μm (A, C, D, and E) and 2 μm (B).
In the course of virus infection, chloroplasts initially showed peripheral punctate GFP signals 36 h after agro-infiltration (Fig. 1C, frames II and III; see also Movie S1 in the supplemental material), indicative of the adhesions of 6K vesicles to chloroplasts. Of approximately 500 chloroplasts observed under the confocal microscope, all of them were found to associate with 6K-GFP vesicles that docked on the chloroplast envelope membrane. At 48 h after agro-infiltration, 6K vesicles expanded, apparently through the homotypic fusion, and induced chloroplast invaginations (Fig. 1C, frames IV, V, and VI). Approximately 40% of chloroplasts displayed different levels of invaginations. At a relatively later stage of virus infection, i.e., 60 h after agro-infiltration, chloroplasts gradually aggregated to form clumps (Fig. 1C, frame VIII). In this infection period, in addition to the formation of large chloroplast-associated vesicles, 6K vesicles aggregated at the junctions between adjacent chloroplasts (Fig. 1C, frame VIII; see also Movie S2 in the supplemental material).
To confirm the association of 6K vesicles with chloroplasts, chloroplasts were purified from N. benthamiana leaves infected with TuMV::6K-GFP or expressing YFP-HDEL (HDEL is an ER retention signal) 48 h after agro-infiltration. Confocal microscopy imaging showed that almost all chloroplasts from TuMV::6K-GFP-infected tissues contained GFP signals (Fig. 1D, frame I), whereas no visible YFP fluorescence was found in chloroplasts purified from YFP-HDEL-expressing leaves (Fig. 1E, frame II). Western blotting was conducted to detect GFP or YFP fusion proteins in these purified chloroplasts (Fig. 1F). No proteins were detected in chloroplasts purified from healthy control (Fig. 1F, lane 1) or chloroplasts from YFP-HDEL-expressing cells (Fig. 1F, lane 4). In contrast, a protein corresponding to the predicted size of 6K-GFP or 6K-YFP was detected in chloroplasts purified from leaves infected by TuMV::6K-GFP (Fig. 1F, lane 2) or expressing 6K-YFP (Fig. 1F, lane 3) or in proteins extracted directly from leaves infected by TuMV::6K-GFP (Fig. 1F, lane 5). The association of 6K vesicles with chloroplasts was further subjected to salt treatments. Incubation of purified chloroplasts with either 1 M NaCl (Fig. 1D, frame II) or 0.1 M Na2CO3 (pH 10.5) (data not shown) efficiently disassociated 6K vesicles from chloroplasts. Taken together, these data suggest that 6K vesicles, either expressed alone or in TuMV-infected leaves, associate peripherally with chloroplasts.
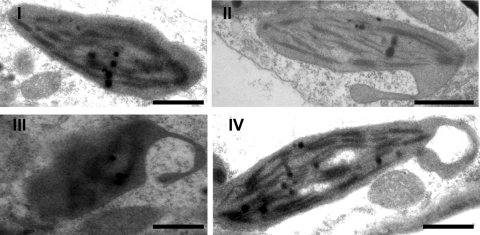
Electron microscopy was conducted to further investigate the association of chloroplasts with potyvirus infection. Electron microscopy of thin sections of TuMV-infected leaves showed that chloroplasts were abnormally distorted and frequently formed pseudopodium-like extrusions (Fig. 2, panel II). These “pseudopodia” of chloroplasts appeared to engulf the cytoplasm, leading to the formation of membrane-bound vesicles within the chloroplast (Fig. 2, panels III and IV). The size of these double-membrane-bound vesicles was approximately 1 to 2 μm in diameter (Fig. 1D, panels III and IV). This result suggests that these membrane-bound vesicles form through invaginations of the chloroplast envelope. The apparently similar process of chloroplast invaginations and the similar sizes of the 6K vesicles observed with electron and confocal microscopy support the notion that 6K bodies target the periphery of the chloroplast where they induce the formation of chloroplast extrusions and, ultimately, are engulfed by chloroplasts.
FIG. 2.
Transmission electron micrographs of abnormally distorted chloroplasts containing membrane-bound vesicles in TuMV-infected leaves. (I) Chloroplast in healthy plant. (II) Chloroplast with membrane-bound extrusion. (III and IV) Amoeboid shaping of chloroplasts, showing the extrusions of chloroplast encircling a large vesicle. Bars, 1 μm.
Colocalization of virus replication complexes to 6K vesicles in association with chloroplasts.
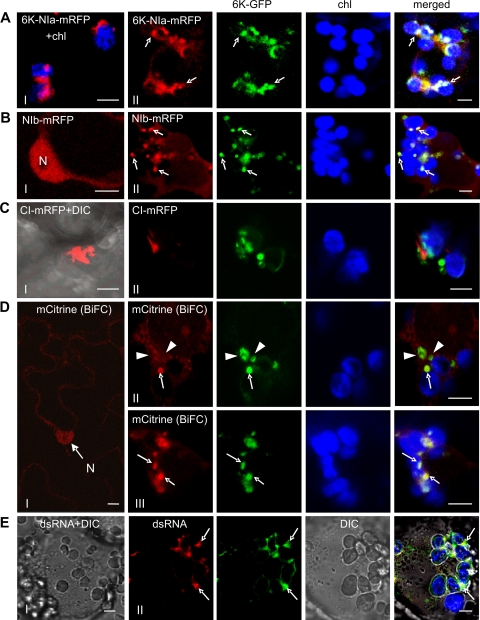
NIa, 6K-NIa, Nib, and CI have been considered essential for viral genome replication (43) and have been found to associate with 6K-containing vesicles in virus-infected plant tissues (9). To determine if the chloroplast-bound 6K vesicles recruited these replicase components during virus infection, plant expression vectors containing chimeric genes encoding each of these viral proteins tagged with mRFP were coinfiltrated into N. benthamiana with TuMV::6K-GFP. When expressed alone (48 h after agro-infiltration), 6K-NIa-mRFP formed chloroplast-bound structures (Fig. 3A, frame I), NIb-mRFP accumulated in the nucleus (Fig. 3B, frame I), and CI-mRFP formed aggregates in the cytoplasm (Fig. 3C, frame I). During virus infection, at the same time point, the discrete patches of chloroplast-bound 6K-GFP colocalized with the 6K-NIa-mRFP (Fig. 3A, frame II) and NIb-mRFP (Fig. 3B, frame II). In addition to the formation of structures penetrating the cell wall (data not shown), CI-mRFP accumulated as spike-like structures in proximity to the 6K-GFP vesicles in close association with chloroplast-bound 6K vesicles (Fig. 3C, frame II).
FIG. 3.
Colocalization of virus replication complexes with 6K bodies associated with chloroplasts during TuMV::6K-GFP infection 48 h (all panels except D, frame I) or 60 h (D, frame III) after agro-infiltration. (A) When expressed alone, 6K-NIa-mRFP formed chloroplast-bound structures (frame I). In virus-infected cells, 6K-NIa-mRFP colocalized with 6K-GFP vesicles (frame II, arrows). (B) When expressed alone, NIb-mRFP accumulated in the nucleus (N) (frame I). NIb-mRFP was recruited to 6K-GFP vesicles (frame II, arrows). (C) When expressed alone, CI-mRFP formed aggregates in the cytoplasm (frame I). The spike-like structures of CI-mRFP were localized in proximity to the 6K-GFP vesicles (frame II). (D) The HsPUMHD-based BiFC background in the nucleus (N) and cytoplasm in the uninfected tissue (frame I). The HsPUMHD-based BiFC labeling of virus replication complexes associated with chloroplast-bound 6K-GFP vesicles (arrows) (frames II and III). Arrowheads correspond to the 6K vesicles that were poorly labeled with HsPUMHD-based BiFC signals. (E) The association of dsRNA with chloroplast-bound 6K vesicles. Detection of dsRNA in protoplasts isolated from TuMV::6K-GFP-infected N. benthamiana leaves. Arrows indicate the colocalization of dsRNA-containing foci and 6K-containing vesicles. DIC, differential interference contrast; Chl, chlorophyll autofluorescence (in blue). Bar, 8 μm.
Recently, a novel technology for localizing plant viral RNAs and VRCs in planta using human Pumilio, an RNA-binding protein, coupled to bimolecular fluorescence complementation (BiFC) has been developed (42). The system consists of two human Pumilio homology domains (HsPUMHD) fused to the N- or C-terminal halves of split mCistrine (CitN-HsPUMHD and HsPUMHD-CitC). Both domains bind to the canonical UGUA(C/U)AUA sequence (7). This technology successfully localized VRCs of Potato virus X (PVX) and Tobacco mosaic virus (TMV), with each viral genome containing a single binding site (42). Sequence analysis of the TuMV genome revealed the presence of a binding site (nucleotides [nt]nt 2187 to 2194; UGUACAUA). When the two domains were coexpressed in cells preinfected with TuMV::6K-GFP, the BiFC fluorescent signals were detectable 48 h after agro-infiltration, highlighting the chloroplast-associated 6K-GFP vesicles (Fig. 3D, frame II). However, the HsPUMHD-based BiFC signal only poorly labeled the 6K vesicles that did not associate with chloroplasts (Fig. 3D, frame II), indicating that there is a scarcity of the viral RNA. At 60 h after agro-infiltration, the chloroplast-bound 6K vesicles were enriched with very strong BiFC signals (Fig. 3D, frame III), suggesting the presence of the bulk of the viral RNA in these 6K vesicles. Notably, in comparison with coexpression in the uninfected tissue (Fig. 3D, frame I), the fluorescent signal was not obvious in the nucleus, suggesting a redistribution of HsPUMHD (Fig. 3D, frames I, II, and III). These results indicate that viral RNA mainly localized to the 6K vesicles.
The use of dsRNA-specific antibodies in immunofluorescence microscopy has provided information about the putative replication site for several positive-strand RNA viruses (9, 11, 19, 27, 40). To determine whether the chloroplast-bound 6K vesicles contained dsRNA, protoplasts were isolated from N. benthmiana leaves infected by TuMV::6K-GFP and probed with the monoclonal antibody J2 that recognizes RNA duplexes larger than 40 bp. At 48 h after agro-infiltration, dsRNA-containing discrete foci colocalized with distinct patches of 6K-GFP in close association with chloroplasts (Fig. 3E, frame II). No immunofluorescence was visible in uninfected cells processed and imaged under the same condition (Fig. 3E, frame I). All the above data support the idea that the chloroplast-bound 6K vesicles are the site for TuMV replication.
The involvement of the vesicular transport pathway in the formation and translocation of 6K vesicles.
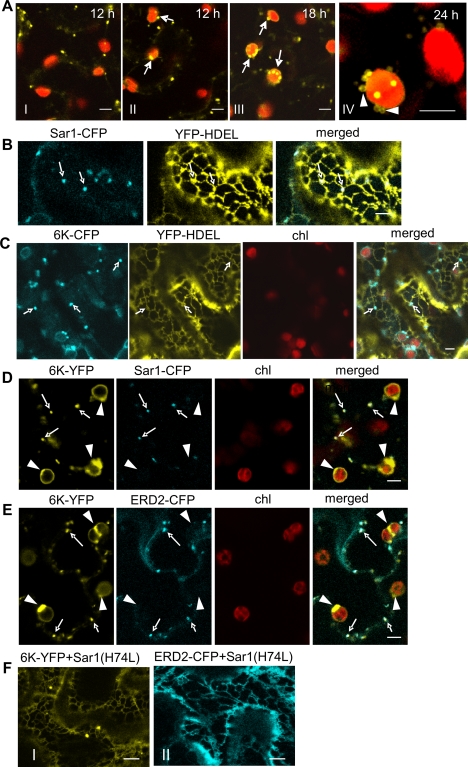
The localization of 6K-YFP was extensively examined in planta. 6K-YFP was first detectable 12 h after agro-infiltration (Fig. 4A, frames I and II). It formed mobile granular vesicles in the cytoplasm (Fig. 4A, frames I and II; see also Movie S3 in the supplemental material). Some 6K-YFP granular vesicles trafficked around the periphery of the chloroplasts and appeared to land on and exit from the chloroplast surface (see Movie S4). At 18 h after agro-infiltration, more 6K vesicles bound to the chloroplasts (Fig. 4A, frame III; see also Movie S5). By 18 and 24 h after agro-infiltration, chloroplast-bound 6K proteins became large ring-like vesicles (Fig. 4A, frames III and IV; see also Movie S6 in the supplemental material).
FIG. 4.
The ER-to-Golgi transport of the 6K protein is COPII-dependent. (A) The localization of mobile granular vesicles induced by 6K-YFP to chloroplasts in leaf epidermal cells. Arrows (frames II and III) point to the association of granular vesicles with the chloroplast. Arrowheads (frame IV) correspond to the ring-like vesicles. The times of acquisition of frames are indicated in the upper corner of each panel. (B and C) Confocal images of cells expressing YFP-HDEL (an ER maker) and Sar1-CFP, an ERES marker (B) or 6K-CFP (C) in N. benthamiana leaves 24 h after agro-infiltration. Arrows indicated the localization of ERES (B) or granular vesicles of 6K-CFP(C) at the cortical ER membrane. (D) Confocal images of cells expressing Sar1-CFP and 6K-YFP in leaf tissues 14 h after agro-infiltration. Arrows indicate the colocalization of granular vesicles of 6K-YFP with the ERES marker Sar1-CFP. Arrowheads correspond to the chloroplast-associated ring-like structures of 6K-YFP that did not colocalize with Sar1-CFP. (E) Confocal images of cells expressing Golgi marker ERD2-CFP and 6K-YFP 14 h after agro-infiltration. Arrows indicate the colocalization of granular vesicles of 6K-YFP with Golgi marker ERD2-CFP. Arrowheads point to the chloroplast-associated 6K-YFP ring-like structures that did not colocalize with ERD2-CFP. (F) Upon coexpression of ERD2-CFP or 6K-YFP with an Sar1 mutant [Sar1(H74L)], a pronounced ER staining was induced 14 h after agro-infiltration. Bars, 8 μm.
Previously, the mobile 6K granular vesicles accumulated at ERES on the ER membrane (45). Indeed, the mobile 6K granular vesicles localized at the cortical ER membrane (Fig. 4C), where ERES marker Sar1-CFP associated (Fig. 4B). Sar1, also known as the small GTPase Sar1, is one of three essential cytosolic components of the COPII complex which accumulate at ERES on the ER membrane (15). To determine if the chloroplast-bound 6K vesicles also associated with ERES, Sar1-CFP was coexpressed with 6K-YFP. The mobile 6K granular vesicles colocalized with Sar1-CFP (Fig. 4D). However, the chloroplast-bound 6K bodies did not colocalize with the ERES (Fig. 4D). In N. benthamiana leaf epidermal cells, COPII-accumulating ERES and Golgi bodies are in close vicinity (10, 45). Our results confirmed this and showed that mobile 6K granular vesicles also associated with the Golgi marker ERD2-CFP (Fig. 4E). ERD2 is a well-described secretory marker whose translocation to the Golgi apparatus depends on the functional COPII secretory pathway (37). Export of membrane proteins from the ER occurs via COPII vesicles for the delivery of their cargo to the Golgi apparatus (10). Thus, the trafficking of 6K granular vesicles to the Golgi apparatus is likely dependent on COPII vesicles. To test this, 6K was coexpressed with an untagged GTP-restricted mutant of Sar1 [Sar1(H74L)], which blocks the COPII-dependent ER-to-Golgi apparatus transport (10). Coexpression of this Sar1 mutant redistributed the 6K localization and led to the accumulation of 6K in the ER (Fig. 4F, frame I), similar to the effect shown on ERD2-CFP (Fig. 4F, frame II).
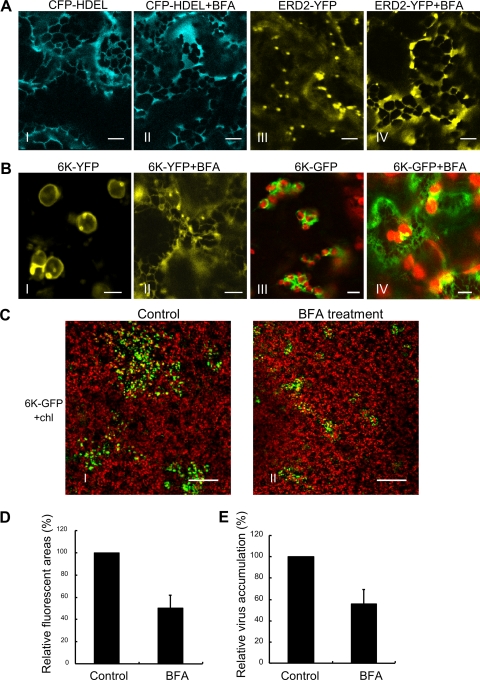
To investigate whether 6K was transported to chloroplasts through the ER-Golgi system, we analyzed the effect of BFA on the distribution of 6K-YFP 24 h after agro-infiltration. BFA is known to perturb the ER-Golgi vesicular transport pathway (10). As expected, BFA did not affect the distribution of the ER-labeling CFP-HDEL but did disrupt the transport of ERD2-YFP (Fig. 5A). The drug treatment redistributed a remarkable portion of 6K-YFP into the ER network (Fig. 5B, frames I and II). A similar redistribution was also observed in TuMV::6K-GFP-infected cells treated with BFA, resulting in the visualization of the weak ER network (Fig. 5B, frames III and IV). In BFA-treated leaves, TuMV accumulation decreased by approximately 50% in comparison with that in the control leaves (Fig. 5C, D, and E), suggesting that the ER-Golgi secretory pathway is required for the formation and translocation of 6K vesicles and for TuMV multiplication.
FIG. 5.
Targeting of 6K-YFP to chloroplasts requires the vesicular transport pathway in N. benthamiana. (A) A strong ER labeling is observed after the treatment of leaf tissues expressing CFP-HDEL (frames I and II) or ERD2-YFP (frames III and IV) with BFA (50 μg/ml) for 30 min. (B) The treatment of BFA (50 μg/ml) redistributes 6K-YFP into the ER network (frames I and II). The effects of BFA treatment on the distribution of 6K-GFP vesicles during virus infection 72 h after agro-infiltration are shown (frames III and IV). A weak ER staining was induced. (C to E) BFA treatment reduces TuMV infection. (C) Confocal micrographs of N. benthamiana leaves that were preinfiltrated with BFA (50 μg/ml) for 3 h and then agro-infiltrated with TuMV::6K-GFP. Photographs show the typical TuMV::6K-GFP infection represented by the sizes of GFP foci 4 days postinfection in BFA-treated or control leaves. GFP foci are shown in green, and chloroplast autofluorescence is shown in red. (D and E) Quantification of the effect of BFA treatment on TuMV::6K-GFP infection using the relative average areas of GFP foci and the relative average values of ELISA, respectively. The mean values with standard error (SE) are given as a percentage relative to the control. Bars, 8 μm (A to C) and 300 μm (D).
The requirement of the actomyosin motility system for the trafficking of 6K vesicles to chloroplasts.
The actomyosin motility system, empowered by myosin motors, has been implicated in transport of subcellular organelles and viral proteins (2, 3, 18, 32). At 20 h after agro-infiltration, we observed that the granular vesicles of 6K-YFP coaligned with and trafficked along actin filaments labeled by mTalin-CFP, targeting the periphery of chloroplast with a mean velocity of 0.5 μm/s (Fig. 6A and B; see also Movie S7 in the supplemental material). This value is similar to the velocity for the motility of Golgi stacks on dynamic actin cytoskeleton (Fig. 6C) (30). The intracellular mobility of 6K granular vesicles was arrested after treatment for 2 h with Lat B, an inhibitor of actin polymerization (Movie S8), consistent with the recent report (9).
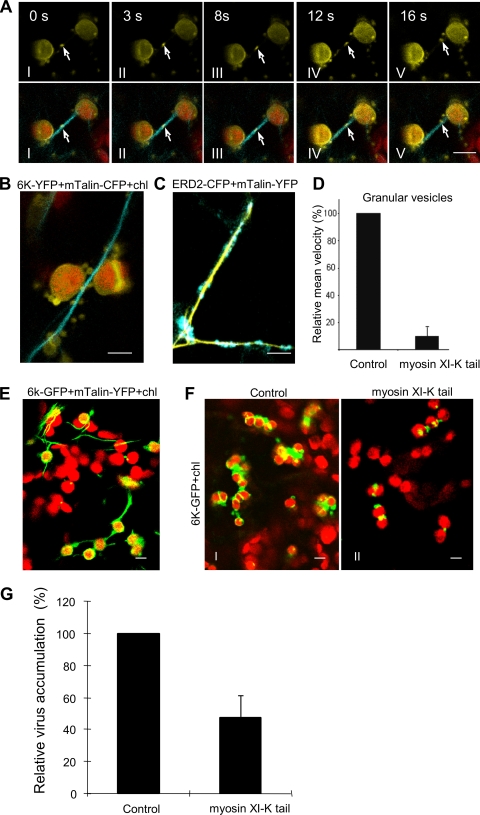
FIG. 6.
The requirement of the actomyosin motility system for the trafficking of 6K vesicles to chloroplasts and the formation of chloroplast clumps. (A) The trafficking of granular vesicles of 6K-YFP (arrows) along actin filaments labeled by mTalin-CFP to the periphery of chloroplasts (red fluorescence) 24 h after agro-infiltration. The times of acquisition are indicated in the upper corners. (B and E) 6K-associated chloroplasts were closely aligned with the actin filaments when 6K was expressed alone 24 h after agro-infiltration (B) or during TuMV::6K-GFP infection 48 h after agro-infiltration (E). (C) The trafficking of the Golgi apparatus of ERD2-CFP along actin filaments labeled by mTalin-YFP 24 h after agro-infiltration. (D) Mean velocities of the punctate inclusions in cells overexpressing the myosin XI-K tail to the control cells. Values representing means with SE are given as a percentage relative to the control. (F) The typical images of 6K-associated chloroplasts in the absence (control) or in the presence of myosin XI-K tail during TuMV::6K-GFP infection (6K-GFP) 72 h after agro-infiltration. (G) N. benthamiana leaves were preinfiltrated with Agrobacterium strains carrying plasmids encoding myosin XI-K tail for 24 h, agro-infiltrated with TuMV::6K-GFP, and assayed for accumulation of TuMV by ELISA. The values representing means with SE are given as a percentage relative to the control. Bar, 8 μm.
As myosin XI-K is essential for the movement of subcellular organelles (3, 32), we examined whether it was also required for the trafficking of 6K vesicles. Overexpression of the myosin XI-K tail, a dominant negative mutant of myosin XI-K (3, 32), nearly halted the trafficking of 6K granular vesicles, resulting in an approximately 15-fold reduction in the mean velocity (Fig. 6D; see Movie S9 in the supplemental material). In cells infected by TuMV::6K-GFP, many 6K-bound chloroplasts either aligned along or closely associated with the actin microfilaments (Fig. 6E), followed by the formation of chloroplast aggregates (Fig. 6F, frame I). Overexpression of the myosin XI-K tail dramatically inhibited the association of 6K-GFP with chloroplasts and the aggregation of chloroplasts in cells infected by TuMV::6K-GFP (Fig. 6F, frame II). To test if myosin XI-K was required in virus infection, the myosin XI-K tail was expressed in the epidermal cells of N. benthamiana, followed by infection with TuMV::6K-GFP. TuMV infection was significantly reduced in the leaves expressing the myosin XI-K tail (Fig. 6G). Taken together, these data suggest that the actomyosin motility system is required for the trafficking of 6K vesicles to chloroplasts and may play an important role in potyvirus infection.
DISCUSSION
The ER has long been recognized as the organelle that the potyviral 6K protein associates with for viral genome replication (9, 35, 38, 45). In the present study, we show that during TuMV infection, the 6K-GFP-induced vesicles trafficked from the ER to the outer chloroplast envelope and induced chloroplast membrane invaginations (Fig. 1 and 2). We demonstrate that the chloroplast-associated 6K vesicles colocalized with viral replicase components (Fig. 3A, B, and C) and dsRNA (Fig. 3E) and were enriched with the bulk of viral RNA (Fig. 3D). Thus, we suggest these chloroplast-associated 6K vesicles are the site for potyvirus replication. This suggestion is supported by several previous observations on potyviruses including TuMV, MDMV, TEV, and PVY. First, 6K-GFP-induced vesicles have been shown to contain viral and cellular proteins required for TuMV replication (9). Second, viral RNA-replicative intermediates are present in chloroplasts of MDMV-infected maize leaf tissues (26). Third, the full-length negative-strand viral RNA was shown to be associated with the chloroplast-enriched fraction from TEV-infected tobacco (13). Finally, the viral genomic RNA was found in chloroplasts of tobacco plants infected with PVY (14). Collectively, these findings indicate that plant potyviruses initiate viral genome translation on the ER and form the 6K vesicles at ERES that traffic to chloroplasts for virus replication (Fig. 7).
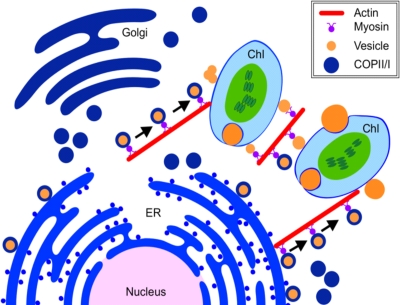
FIG. 7.
A model of the intracellular activities of the potyviral 6K vesicles in plant cells. The potyviral 6K proteins accumulate at the ERES on the ER membrane and transport to the Golgi apparatus via the COPII-mediated early secretory pathway. The Golgi-associated 6K proteins traffic along the actin filaments and target the chloroplast. After the disassociation from the Golgi apparatus, the 6K bodies reside on the chloroplast envelope for virus replication.
In addition to the ER and chloroplasts, single-stranded positive-sense RNA viruses also employ other organelles as replication sites. For instance, the replication of Tomato bushy stunt virus (TBSV), a tombusvirus, takes place on the surface of peroxisomal membranes (27). The TBSV p33 protein is responsible for targeting and anchoring VRCs to mitochondria and peroxisomes. In host cells where peroxisome biogenesis was defective, TBSV targeted the ER for replication (16). Thus, it is possible that this virus (and other positive-sense RNA viruses) also initiates viral genome translation on the ER, induces the formation of viral membrane protein-associated vesicles therein, and further targets their preferential organelles for robust virus replication.
To investigate how potyviral 6K vesicles arrived at chloroplasts, we examined whether the 6K membrane protein contains a plastid-targeting signal. As the 6K protein is predicted to be only an ER-associated membrane protein by iPSORT (4), we looked into the role of the ER-Golgi vesicular transport pathway in the transport process. We found that the 6K-induced vesicles formed at the ERES on the ER membrane, trafficked to the Golgi apparatus via COPII vesicles, and subsequently targeted the periphery of chloroplasts through the actomyosin motility system (Fig. 4, 5 and 6; see also Movie S3 in the supplemental material). Indeed, disruption of the ER-Golgi secretory trafficking by BFA confined 6K within the endomembrane system and obstructed the targeting of 6K to chloroplasts (Fig. 5). This is in agreement with previous findings that two chloroplast precursors, though containing an ER-targeting signal, translocate into chloroplasts through the BFA-sensitive ER-Golgi secretory pathway in plants (29, 44). In this study, we show that Lat B, a drug inhibiting actin polymerization, arrested the intracellular mobility of 6K granular vesicles (see Movie S8) and that overexpression of the myosin XI-K tail, a dominant negative mutant of myosin XI-K, nearly halted the trafficking of 6K granular vesicles (Fig. 6D; see Movie S9 in the supplemental material). Together with recent evidence showing that the ER is tightly associated with the actin network in plant cells (48), these results support the notion that the trafficking of the 6K vesicles from the ER to chloroplasts is modulated by the actin/ER network.
Based on the above discussion, we propose a model for the intracellular accumulation and trafficking of 6K vesicles (Fig. 7). The potyviral 6K proteins accumulate at the ERES on the ER membrane and then migrate to the Golgi apparatus in a COPII-dependent manner. The Golgi-associated 6K vesicles move along the actin filaments and target the chloroplast. After disassociation with the Golgi apparatus, the 6K vesicles on the chloroplast envelope fuse with each other into larger vesicles, and some of them induce chloroplast invaginations. Further molecular identification and characterization of the components of the chloroplast-bound 6K vesicles will certainly shed light on the replication mechanism of potyviruses and related viruses.
Supplementary Material
Acknowledgments
We are grateful to K. Andrew White and Hélène Sanfaçon for critical reading of the manuscript, to Karl J. Oparka and Jens Tilsner for plasmids and advice in the HsPUMHD BiFC assay, to Valerian V. Dolja for a plasmid containing the myosin XI-K tail, to Nam-Hai Chua for plasmid pYSC14, to Tsuyoshi Nakagawa for vector pGWB554, and to Alex Molnar and Changwei Zhang for their assistance.
This work was supported by Agriculture and Agri-Food Canada and by the Natural Sciences and Engineering Research Council of Canada.
Footnotes
Published ahead of print on 11 November 2009.
Supplemental material for this article may be found at http://jvi.asm.org/.
REFERENCES
- 1.Ahlquist, P. 2006. Parallels among positive-strand RNA viruses, reverse-transcribing viruses and double-stranded RNA viruses. Nat. Rev. Microbiol. 4:37-382. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Avisar, D., A. I. Prokhnevsky, and V. V. Dolja. 2008. Class VIII myosins are required for plasmodesmatal localization of a closterovirus Hsp70 homolog. J. Vriol. 82:2836-2843. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Avisar, D., A. I. Prokhnevsky, K. S. Makarova, E. V. Koonin, and V. V. Dolja. 2008. Myosin XI-K is required for rapid trafficking of Golgi stacks, peroxisomes, and mitochondria in leaf cells of Nicotiana benthamiana. Plant Physiol. 146:1098-1108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bannai, H., Y. Tamada, O. Maruyama, K. Nakai, and S. Miyano. 2002. Extensive feature detection of N-terminal protein sorting signals. Bioinformatics 18:298-305. [DOI] [PubMed] [Google Scholar]
- 5.Beauchemin, C., and J. F. Laliberte. 2007. The poly(A) binding protein is internalized in virus-induced vesicles or redistributed to the nucleolus during Turnip mosaic virus infection. J. Virol. 81:10905-10913. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Carette, J. E., J. van Lent, S. A. MacFarlane, J. Wellink, and A. van Kammen. 2002. Cowpea mosaic virus 32- and 60-kilodalton replication proteins target and change the morphology of endoplasmic reticulum membranes. J. Virol. 76:6293-6301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Cheong, C. G., and T. M. Tanaka Hall. 2006. Engineering RNA sequence specificity of Pumilio repeats. Proc. Natl. Acad. Sci. U. S. A. 103:13635-13639. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Chung, B. Y., W. A. Miller, J. F. Atkins, and A. E. Firth. 2008. An overlapping essential gene in the Potyviridae. Proc. Natl. Acad. Sci. U. S. A. 105:5897-5902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Cotton, S., R. Grangeon, K. Thivierge, I. Mathieu, C. Ide, T. Wei, A. Wang, and J. F. Laliberté. 2009. Turnip mosaic virus RNA replication complex vesicles are mobile, align with microfilaments and are each derived from a single viral genome. J. Virol. 83:10460-10471. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.daSilva, L. L., E. L. Snapp, J. Denecke, J. Lippincott-Schwartz, C. Hawes, and F. Brandizzi. 2004. Endoplasmic reticulum export sites and Golgi bodies behave as single mobile secretory units in plant cells. Plant Cell 16:1753-1771. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Dunoyer, P., C. Ritzenthaler, O. Hemmer, P. Michler, and C. Fritsch. 2002. Intracellular localization of the Peanut clump virus replication complex in tobacco BY-2 protoplasts containing green fluorescent protein-labeled endoplasmic reticulum or Golgi apparatus. J. Virol. 76:865-874. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Earley, K. W., J. R. Haag, O. Pontes, K. Opper, T. Juehne, K. Song, and C. S. Pikaard. 2006. Gateway-compatible vectors for plant functional genomics and proteomics. Plant J. 45:616-629. [DOI] [PubMed] [Google Scholar]
- 13.Gadh, I. P. S., and V. Hari. 1986. Association of tobacco etch virus related RNA with chloroplasts in extracts of infected plants. Virology 150:304-307. [DOI] [PubMed] [Google Scholar]
- 14.Gunasinghe, U. B., and P. H. Berger. 1991. Association of potato virus Y gene products with chloroplasts in tobacco. Mol. Plant Microbe Interact. 4:452-457. [Google Scholar]
- 15.Hanton, S. L., L. Chatre, L. A. Matheson, M. Rossi, M. A. Held, and F. Brandizzi. 2008. Plant Sar1 isoforms with near-identical protein sequences exhibit different localisations and effects on secretion. Plant Mol. Biol. 67:283-294. [DOI] [PubMed] [Google Scholar]
- 16.Jonczyk, M., K. B. Pathak, M. Sharma, and P. D. Nagy. 2007. Exploiting alternative subcellular location for replication: tombusvirus replication switches to the endoplasmic reticulum in the absence of peroxisomes. Virology 362:320-330. [DOI] [PubMed] [Google Scholar]
- 17.Ju, H. J., T. D. Samuls, Y. S. Wang, E. Blancaflor, M. Payton, R. Mitra, K. Krishnamurthy, R. S. Nelson, and J. Verchot-Lubicz. 2005. The Potato virus X TGBp2 movement protein associates with endoplasmic reticulum-derived vesicles during virus infection. Plant Physiol. 138:1877-1895. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Kawakami, S., Y. Watanabe, and R. N. Beachy. 2004. Tobacco mosaic virus infection spreads cell to cell as intact replication complexes. Proc. Natl. Acad. Sci. U. S. A. 101:6291-6296. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Knoops, K., M. Kikkert, S. H. E. Van Den Worm, J. C. Zevenhoven-Dobbe, Y. Van Der Meer, A. J. Koster, A. M. Mommaas, and R. J. Snijder. 2008. SARS-coronavirus replication is supported by a reticulovesicular network of modified endoplasmic reticulum. PLoS. Biol. 6:e226. doi: 10.1371/journal.pbio.0060226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Koonin, E. V., and V. V. Dolja. 1993. Evolution and taxonomy of positive-strand RNA viruses: implications of comparative analysis of amino acid sequences. Crit. Rev. Biochem. Mol. Biol. 28:375-430. [DOI] [PubMed] [Google Scholar]
- 21.Koonin, E. V., Y. I. Wolf, K. Nagasaki, and V. V. Dolja. 2008. The Big Bang of picorna-like virus evolution antedates the radiation of eukaryotic supergroups. Nat. Rev. Microbiol. 6:925-939. [DOI] [PubMed] [Google Scholar]
- 22.Kopek, B. G., G. Perkins, D. J. Miller, M. H. Ellisman, and P. Ahlquist. 2007. Three-dimensional analysis of a viral RNA replication complex reveals a virus-induced mini-organelle. PLoS. Biol. 5:e220. doi: 10.1371/journal.pbio.0050220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Kost, B., P. Spielhofer, and N. H. Chua. 1998. A GFP-mouse talin fusion protein labels plant actin filaments in vivo and visualizes the actin cytoskeleton in growing pollen tubes. Plant J. 16:393-401. [DOI] [PubMed] [Google Scholar]
- 24.Laliberté, J. F., O. Nicolas, C. Chatel, C. Lazure, and R. Morosoli. 1992. Release of a 22-kDa protein derived from the amino-terminal domain of the 49-kDa NIa of turnip mosaic potyvirus in Escherichia coli. Virology 190:510-514. [DOI] [PubMed] [Google Scholar]
- 25.Magliano, D., J. A. Marshall, D. S. Bowden, N. Vardaxis, J. Meanger, and J. Y. Lee. 1998. Rubella virus replication complexes are virus-modified lysosomes. Virology 240:57-63. [DOI] [PubMed] [Google Scholar]
- 26.Mayhew, D. E., and R. E. Ford. 1974. Detection of ribonuclease-resistant RNA in chloroplasts of corn leaf tissue infected with maize dwarf mosaic virus. Virology 57:503-509. [DOI] [PubMed] [Google Scholar]
- 27.McCartney, A. W., J. S. Greenwood, M. R. Fabian, K. A. White, and R. T. Mullen. 2005. Localization of the tomato bushy stunt virus replication protein p33 reveals a peroxisome-to-endoplasmic reticulum sorting pathway. Plant Cell 17:3513-3531. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Nakagawa, T., T. Suzuki, S. Murata, S. Nakamura, T. Hino, K. Maeo, R. Tabata, T. Kawai, K. Tanaka, Y. Niwa, Y. Watanabe, K. Nakamura, T. Kimura, and S. Ishiguro. 2007. Improved gateway binary vectors: high-performance vectors for creation of fusion constructs in transgenic analysis of plants. Biosci. Biotechnol. Biochem. 71:2095-2100. [DOI] [PubMed] [Google Scholar]
- 29.Nanjo, Y., H. Oka, N. Ikarashi, K. Kaneko, A. Kitajima, T. Mitsui, F. J. Muñoz, M. Rodríguez-López, E. Baroja-Fernández, and J. Pozueta-Romero. 2006. Rice plastidial N-glycosylated nucleotide pyrophosphatase/phosphodiesterase is transported from the ER-Golgi to the chloroplast through the secretory pathway. Plant Cell 18:2582-2592. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Nebenführ, A., L. Gallagher, T. Dunahay, J. Frohlick, A. Mazurkiewicz, J. Meehl, and L. Staehelin. 1999. Stop-and-go movements of plant Golgi stacks are mediated by the acto-myosin system. Plant Physiol. 121:1127-1142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Prod'homme, D., S. Le Panse, G. Drugeon, and I. Jupin. 2001. Detection and subcellular localization of the Turnip yellow mosaic virus 66K replication protein in infected cells. Virology 281:88-101. [DOI] [PubMed] [Google Scholar]
- 32.Prokhnevsky, A. I., V. V. Peremyslov, and V. V. Dolja. 2008. Overlapping functions of the four class XI myosins in Arabidopsis growth, root hair elongation and organelle motility. Proc. Natl. Acad. Sci. U. S. A. 105:19744-19749. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Reichel, C., and R. N. Beachy. 1998. Tobacco mosaic virus infection induces severe morphological changes of the endoplasmic reticulum. Proc. Natl. Acad. Sci. U. S. A. 95:11169-11174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Restrepo-Hartwig, M., and P. Ahlquist. 1996. Brome mosaic virus helicase- and polymerase-like proteins colocalize on the endoplasmic reticulum at sites of viral RNA synthesis. J. Virol. 70:8908-8916. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Restrepo-Hartwig, M. A., and J. C. Carrington. 1994. The tobacco etch potyvirus 6-kilodalton protein is membrane associated and involved in viral replication. J. Virol. 68:2388-2397. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Ritzenthaler, C., C. Laporte, F. Gaire, P. Dunoyer, C. Schmitt, S. Duval, A. Piéquet, A. M. Loudes, O. Rohfritsch, C. Stussi-Garaud, and P. Pfeiffer. 2002. Grapevine fanleaf virus replication occurs on endoplasmic reticulum-derived membranes. J. Virol. 76:8808-8819. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Saint-Jore, C. M., J. Evins, H. Batoko, F. Brandizzi, I. Moore, and C. Hawes. 2002. Redistribution of membrane proteins between the Golgi apparatus and endoplasmic reticulum in plants is reversible and not dependent on cytoskeletal networks. Plant J. 29:661-678. [DOI] [PubMed] [Google Scholar]
- 38.Schaad, M. C., P. E. Jensen, and J. C. Carrington. 1997. Formation of plant RNA virus replication complexes on membranes: role of an endoplasmic reticulum-targeted viral protein. EMBO. J. 16:4049-4059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Suhy, D. A., T. H. Giddings, Jr., and K. Kirkegaard. 2000. Remodeling the endoplasmic reticulum by poliovirus infection and by individual viral proteins: an autophagy-like origin for virus-induced vesicles. J. Virol. 74:8953-8965. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Targett-Adams, P., S. Boulant, and J. McLauchlan. 2008. Visualization of double-stranded RNA in cells supporting hepatitis C virus RNA replication. J. Virol. 82:2182-2195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Thivierge, K., S. Cotton, P. J. Dufresne, I. Mathieu, C. Beauchemin, C. Ide, M. G. Fortin, and J. F. Laliberté. 2008. Eukaryotic elongation factor 1A interacts with Turnip mosaic virus RNA-dependent RNA polymerase and VPg-Pro in virus-induced vesicles. Virology 377:216-225. [DOI] [PubMed] [Google Scholar]
- 42.Tilsner, J., O. Linnik, N. M. Christensen, K. Bell, I. M. Roberts, C. Lacomme, and K. J. Oparka. 2009. Live-cell imaging of viral RNA genomes using a Pumilio-based reporter. Plant J. 57:758-770. [DOI] [PubMed] [Google Scholar]
- 43.Urcuqui-Inchima, S., A. L. Haenni, and F. Bernardi. 2001. Potyvirus proteins: a wealth of functions. Virus Res. 74:157-175. [DOI] [PubMed] [Google Scholar]
- 44.Villarejo, A., S. Burén, S. Larsson, A. Déjardin, M. Monné, C. Rudhe, J. Karlsson, S. Jansson, P. Lerouge, N. Rolland, G. von Heijne, M. Grebe, L. Bako, and G. Samuelsson. 2005. Evidence for a protein transported through the secretory pathway en route to the higher plant chloroplast. Nat. Cell Biol. 7:1224-1231. [DOI] [PubMed] [Google Scholar]
- 45.Wei, T., and A. Wang. 2008. Biogenesis of cytoplasmic membranous vesicles for plant potyvirus replication occurs at the endoplasmic reticulum exit sites in a COPI- and COPII-dependent manner. J. Virol. 82:12252-12264. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Welsch, S., S. Miller, I. Romero-Brey, A. Merz, C. K. Bleck, P. Walther, S. D. Fuller, C. Antony, J. Krijnse-Locker, and R. Bartenschlager. 2009. Composition and three-dimensional architecture of the dengue virus replication and assembly sites. Cell Host Microbe 5:365-375. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Wileman, T. 2006. Aggresomes and autophagy generate sites for virus replication. Science 312:875-878. [DOI] [PubMed] [Google Scholar]
- 48.Yokota, E., S. Ueda, K. Tamura, H. Orii, S. Uchi, S. Sonobe, I. Hara-Nishimura, and T. Shimmen. 2009. An isoform of myosin XI is responsible for the translocation of endoplasmic reticulum in tobacco cultured BY-2 cells. J. Exp. Bot. 60:197-212. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.