Abstract
The minimal signal required for the cleavage and packaging of replicated concatemeric herpes simplex virus type 1 (HSV-1) DNA corresponds to an approximately 200-bp fragment, Uc-DR1-Ub, spanning the junction of the genomic L and S segments. Uc and Ub occupy positions adjacent to the L and S termini and contain motifs (pac2 and pac1, respectively) that are conserved near the ends of other herpesvirus genomes. We have used homologous Red/ET recombination in Escherichia coli to introduce wild-type and specifically mutated Uc-DR1-Ub fragments into an ectopic site of a cloned HSV-1 genome from which the resident packaging signals had been previously deleted. The resulting constructs were transfected into mammalian cells, and their abilities to replicate and become encapsidated, generate Uc- and Ub-containing terminal fragments, and give rise to progeny virus were assessed. In general, the results obtained agree well with previous observations made using amplicons and confirm roles for the pac2 T element in the initiation of DNA packaging and for the GC-rich motifs flanking the pac1 T element in termination. In contrast to a previous report, the sequence of the DR1 element was also crucial for DNA packaging. Following repair of the resident packaging signals in mammalian cells, recombination occurred at high frequency in progeny virus between the repaired sequences and mutated Uc-DR1-Ub inserts. This restored the ability of mutated Uc-DR1-Ub inserts to generate terminal fragments, although these were frequently larger than expected from simple repair of the original lesion.
Herpesviruses possess linear double-stranded DNA genomes that are circularized early after infection and upon replication generate concatemeric structures. During progeny particle assembly, the cleavage of concatemers at specific sites, corresponding to the genomic termini, is tightly coupled to the insertion of the viral DNA into a preformed structure referred to as the procapsid (reviewed in references 2, 4, and 11). In the case of herpes simplex virus type 1 (HSV-1), a terminally redundant region of the genome, known as the a sequence (Fig. 1a), contains all the cis-acting sequences required for DNA packaging (24, 27). This region, which is 250 to 500 bp in length depending on the virus strain, is present as a single copy at the S terminus and as one or more tandem copies at the L terminus. In addition, one or more copies are present in inverted orientation at the junction between the L and S segments (30, 31).
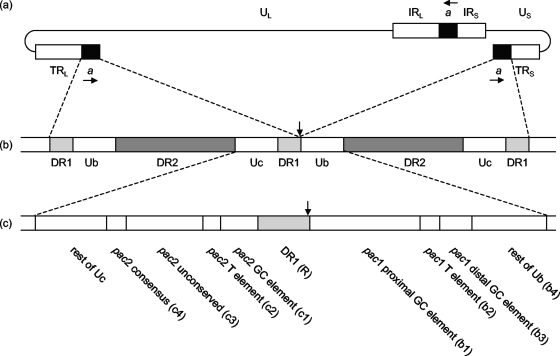
FIG. 1.
Structure of the HSV-1 Uc-DR1-Ub element. (a) Structure of the HSV-1 genome showing the positions and relative orientations (horizontal arrows) of copies of the a sequence. (b) Circularization of linear genomes by direct ligation brings together two copies of the a sequence separated by a single DR1 repeat. The site of ligation, and of cleavage of concatemers, is shown by the vertical arrow. (c) Motifs and regions within the 194-bp Uc-DR1-Ub fragment. To facilitate naming of mutants, component regions of Uc, Ub, and DR1 were also referred to as c1 to c4, b1 to b4, and R, respectively, as indicated in parentheses.
The structure of the HSV-1 a sequence is depicted in Fig. 1b. Each a sequence is flanked by direct repeats (DR1) of 17 to 20 bp, with single copies of DR1 separating tandem a sequences. Genomic termini are generated by a cleavage event toward one end of DR1, and circularization of infecting genomes restores a complete a sequence. The central portion of the a sequence comprises multiple repeats of one or two other short sequences (DR2 and sometimes DR4), while quasi-unique sequences are located between DR1 and either side of the DR2/DR4 repeats. These regions are termed Ub and Uc, and in virion DNA they lie adjacent to the S and L termini, respectively (6, 17, 18).
An approximately 200-bp fragment (Uc-DR1-Ub) spanning the junction between tandem a sequences, such as is generated upon fusion of the genomic ends (Fig. 1b), has been shown to contain all the essential cis-acting sequences necessary for DNA packaging (10, 20). Within the Ub and Uc regions are two domains, pac1 and pac2, respectively, which contain several characteristic sequence motifs that are conserved near the ends of other herpesvirus genomes (3, 8, 15). These motifs, as originally defined by Deiss et al. (8), are illustrated in Fig. 1c. It is now recognized that the major conserved motif within the pac1 region comprises the T-rich element flanked on each side by short G tracts (from the proximal and distal GC-rich regions). In the case of pac2, the T-rich element is most highly conserved with a consensus CGCGGCG motif also frequently being present (32).
Detailed studies, employing primarily HSV-1 and murine cytomegalovirus (MCMV), have highlighted the roles of the major conserved motifs and suggested the following general mechanism by which concatemers are cleaved and packaged (1, 10, 13, 15, 16, 23, 25, 29, 32). Within Uc the most critical sequence is the pac2 T element, which is essential for cleavage to initiate DNA packaging. Cleavage occurs at a fixed distance from the pac2 T element, and the resulting Uc-containing end is inserted into the procapsid. Additional important cis-acting sequences are present further from the cleavage site, possibly including the pac2 consensus motif. Deletion, but not substitution, of the pac2 GC element and unconserved region impaired DNA packaging, suggesting that the relative spacing of the cleavage site, T element, and distal motifs is crucial. Packaging proceeds from pac2 toward the pac1 terminus, and a second cleavage event terminates DNA packaging. This cleavage appears to be directed by, and occurs at a fixed distance from, a single region comprising the pac1 T element and the flanking G tracts. Surprisingly, substitutions within the highly conserved T element are tolerated, but it remains unclear whether this region functions as a spacer element. The UL28 component of the HSV-1 terminase enzyme binds to a specific conformation adopted by the region comprising the T element and G tracts, and this interaction is likely to be crucial for cleavage.
The functional analysis of herpesvirus DNA packaging signals has employed two major approaches. In the first, amplicons (i.e., bacterial plasmids containing a viral DNA replication origin and packaging signal) are transfected into mammalian cells and their ability to be replicated and packaged is assessed following the provision of viral helper functions, either by superinfection with virus particles or by cotransfection of virion DNA (7, 20, 24, 27, 29, 35). The second assay introduces an additional copy of the packaging signal under test at an ectopic site within the viral genome and determines whether it functions as a site for the cleavage of concatemeric DNA and the generation of novel terminal fragments of virion DNA (5, 15, 18, 23, 29, 32). Both these approaches, however, suffer from the disadvantage that recombination occurs between the test packaging signal and the wild-type (wt) signal present either in the helper virus or in its normal location within an ectopic-site recombinant (5, 8, 15, 23, 32). Additionally, concatemers generated following replication of amplicons have a significantly different structure from standard herpesviral genomes in that multiple copies of the packaging signal are present, spaced at regular intervals corresponding to the size of the input plasmid. This raises the possibility that the activity of wt or mutated packaging signals in the amplicon assay may not accurately reflect their behavior in a standard genome.
To avoid these difficulties and allow analysis of mutated packaging signals in the context of the viral genome, we have used a cloned full-length HSV-1 genome, fHSVΔpac, which is complete with the exception that all copies of the a sequence have been deleted (22). This molecule is propagated as a bacterial artificial chromosome (BAC), and specific sequences can be inserted via homologous recombination either in mammalian cells or in the bacterial host. We previously demonstrated that a single copy of the minimal packaging signal Uc-DR1-Ub introduced into the viral thymidine kinase (TK) locus of fHSVΔpac by recombination in mammalian cells was sufficient to allow the products of replication to be packaged in mammalian cells and to allow the generation of viable progeny (28). Here, we describe the introduction of the packaging signal into fHSVΔpac by Red/ET recombination in Escherichia coli (19, 34), allowing previously described (10) and new Uc-DR1-Ub mutants to be screened for their ability to direct encapsidation, generate Uc- and Ub-containing terminal fragments, and give rise to progeny virus.
MATERIALS AND METHODS
Cells and viruses.
Baby hamster kidney 21 clone 13 (BHK) cells were grown in Glasgow minimal essential medium supplemented with 10% newborn calf serum, 10% tryptose phosphate broth, 100 U/ml penicillin, and 100 μg/ml streptomycin (ETC10). After infection or transfection, the cells were maintained in Glasgow minimal essential medium supplemented with 5% newborn calf serum, 100 units of penicillin, and 100 μg streptomycin per ml (EC5). Stocks of virus were prepared and titrated in BHK cells.
Introduction of Uc-DR1-Ub fragments into the full-length HSV-1 genome.
The parental HSV-1 genome used in these studies was the BAC fHSVΔpac (22), which contains a cloned copy of a full-length viral genome from which all the normally resident copies of the a sequence have been removed by means of 1.2-kbp deletions spanning both of the RS-RL junctions (Fig. 2). fHSVΔpac was routinely propagated in the E. coli strain DH10B derivative Genehogs (Invitrogen). The introduction of Uc-DR1-Ub fragments into the TK locus of fHSVΔpac employed in vivo Red/ET recombination using reagents and experimental procedures provided with the counterselection BAC modification kit (Gene Bridges). To facilitate homologous Red/ET recombination, the plasmid pSC101-BAD-gbaA-tet encoding the recombination enzymes was first introduced into Genehogs carrying fHSVΔpac, generating strain fHSVΔpac-Red/ET.
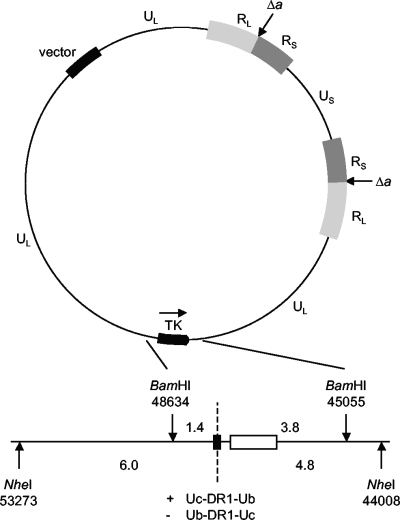
FIG. 2.
Generation of recombinant HSV-1 BACs carrying the Uc-DR1-Ub fragment. The upper part of the figure depicts the BAC fHSVΔpac, which consists of a complete HSV-1 strain 17 genome, from which all copies of the a sequence have been deleted (indicated as Δa), cloned into the vector pBeloBac11 (22). The lower part shows an expansion of the TK locus indicating the positions of flanking BamHI and NheI sites (coordinates taken from reference 14). Uc-DR1-Ub fragments (black box) and the 1.3-kbp rpsL-neo fragment (white box) were inserted separately into the unique SacI and BspEI sites (positions 47358 and 47174, respectively) within a cloned copy of the BamHI P fragment (positions 45055 to 48634). Recombinant BACs were generated by Red/ET recombination following the introduction of the resulting BamHI fragments into bacteria containing fHSVΔpac. The dotted line shows the position at which replicated concatemeric DNA would be cleaved, and the sizes of the predicted terminal fragments are indicated (kbp). The two possible orientations of the Uc-DR1-Ub fragment are designated + and −, as indicated, and allow identification of genomic fragments terminating in Ub or Uc. For example, the 1.4-kbp BamHI fragment from the + orientation and the 3.8-kbp BamHI fragment from the − orientation each represent Uc-containing termini.
The wt Uc-DR1-Ub fragment and the majority of the mutated fragments used in this study were described previously by Hodge and Stow (10). New DR1 substitution and deletion mutants, a new pac1 T-element deletion mutant, and a mutant deleting the “rest of Ub” region were also constructed. The nature and sources of the mutants are outlined in Table 1, and their DNA sequences are shown in Fig. 3. To introduce wt and mutated Uc-DR1-Ub fragments into fHSVΔpac, two fragments were first cloned separately into the TK coding region of plasmid pGX153, which comprises the HSV-1 BamHI P fragment inserted into the corresponding site of the vector pAT153. These fragments were the 1.3-kbp rpsL-neo cassette (Gene Bridges) encoding kanamycin resistance, which was inserted into a unique BspEI site, and the 0.2-kbp Uc-DR1-Ub fragment, which was cloned into a unique SacI site (Fig. 2). The resulting plasmids were cleaved with BamHI, and the viral fragment containing the rpsL-neo and Uc-DR1-Ub inserts was purified. This fragment was electroporated into fHSVΔpac-Red/ET cells, and the induction of the recombination enzymes and selection of kanamycin-resistant cells were performed as described in the Gene Bridges protocol. Subsequent growth at 37°C resulted in the loss of pSC101-BAD-gbaA-tet. The structures of the resulting recombinant BACs were confirmed by restriction enzyme digestion, Southern blotting, and DNA sequencing of the region containing the inserts. It should be noted that the HSV-1 DNA sequences present in fHSVΔpac, pGX153, and the wt Uc-DR1-Ub fragment all originate from HSV-1 strain 17 syn+ (9, 28) and that nucleotide coordinates are taken from the corresponding sequence determined by McGeoch et al. (14).
TABLE 1.
BACs used in this study
| BACa | Uc-DR1-Ub fragmentb | Viabilityc | Packagingd | No. of endse |
|---|---|---|---|---|
| P | None (fHSVΔpac) | No | No | 0 |
| O | None (fHSVΔpac + rpsL-neo) | No | No | 0 |
| W (+) | wt Uc-DR1-Ub (pSA1) | Yes | Yes | 2 |
| W (−) | wt Uc-DR1-Ub (pSA1) | Yes | Yes | 2 |
| ΔC (−) | pac2 80-bp deletion (pPH3) | No | No | 0 |
| c4 (+) | pac2 consensus substitution (pPH18) | Yes | Yes | 2 |
| c3 (+) | pac2 unconserved substitution (pPH20) | Yes | Yes | 2 |
| c2 (−) | pac2 T-element substitution (pPH9) | No | No | 0 |
| c1 (−) | pac2 GC-element substitution (pPH16) | Yes | Yes | 2 |
| R (+) | DR1 substitution | No | No | 0 |
| ΔR (+) | DR1 deletion | No | No | 0 |
| ΔB (−) | pac1 69-bp deletion (pPH4) | No | No | 0 |
| b1 (−) | pac1 proximal GC substitution (pPH22) | No | Yes | 1 |
| b2 (−) | pac1 T-element substitution (pPH7) | Yes | Yes | 2 |
| Δb2 (−) | pac1 T-element deletion | No | Yes | 1 |
| b3 (+) | pac1 distal GC substitution (pPH12) | No | Yes | 1 |
| b3 (−) | pac1 distal GC substitution (pPH12) | No | Yes | 1 |
| Δb4 (−) | “Rest of Ub” deletion | Yes | Yes | 2 |
Names assigned to recombinant BACs containing wt and mutated Uc-DR1-Ub fragments. The + and − signs in parentheses indicate the orientations of the Uc-DR1-Ub fragment as designated in Fig. 2. In the text they are retained only where two BACs contain the same fragment in either orientation (i.e., W+, W−, b3+, and b3−).
Where Uc-DR1-Ub fragments have been described previously, the name of the amplicon (10) is indicated in parentheses.
Ability of BAC to give rise to progeny virus when transfected alone.
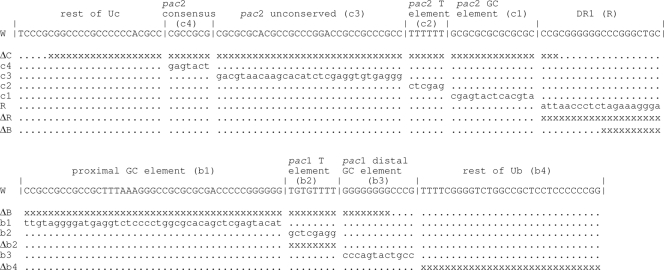
FIG. 3.
Sequences of the HSV-1 Uc-DR1-Ub fragment and of the mutants used in this study. The sequence of the wt fragment (W) is depicted on two lines with the characteristic motifs and regions shown. The site of cleavage of concatemers lies at the right end of the DR1 sequence (6). The substitutions and deletions in the various mutants are shown below the wt sequence. Modified nucleotides are in lowercase, “x” indicates a deleted nucleotide, and “.” indicates an unchanged nucleotide. Only the ΔB mutant is affected at positions within both lines of the wt sequence.
Preparation of BAC DNAs for transfection.
BAC DNAs were prepared from bacteria containing the wt and mutated Uc-DR1-Ub fragments described in Table 1 and also from bacteria harboring the unmodified parental BAC, fHSVΔpac (referred to as BAC P), or a BAC containing the rpsL-neo cassette but no Uc-DR1-Ub fragment (designated BAC O). Four-hundred-milliliter cultures were grown in LB supplemented with 50 μg per ml chloramphenicol, either with or without 15 μg per ml kanamycin, and DNA was purified by alkali lysis followed by cesium chloride-ethidium bromide density gradient centrifugation. DNA concentrations were estimated from absorbance measurements at 260 nm.
Transfection of mammalian cells.
Monolayers of BHK cells in 35-mm petri dishes (2 × 106 cells per plate) were transfected by using the calcium phosphate technique followed by treatment with dimethyl sulfoxide at 4 h as previously described (26). Each monolayer received 0.5 ml of precipitate containing 1 μg BAC DNA and 7 μg calf thymus carrier DNA. In experiments that aimed to repair the deleted a sequences of recombinant BACs, 0.5 μg of BamHI-cleaved plasmid pGX8 was also included in each transfection mix. Plasmid pGX8 contains the 5.9-kbp BamHI K fragment spanning the junction of the L and S segments and entirely covering the 1.2-kbp deletions within fHSVΔpac (Fig. 2), inserted into the BamHI site of pAT153. Virus yields were determined, or plaques were counted, 3 days post-transfection of BAC DNAs either in the presence or in the absence of BamHI K.
DNA isolation and Southern blotting.
Monolayers of BHK cells transfected with recombinant BACs, as described above, were processed 16 h after treatment with dimethyl sulfoxide. The cells from each plate were divided into two equal samples which were used to prepare total cellular and DNase-resistant (encapsidated) DNA as previously described (10). Samples of the DNA corresponding to the yield from 4 × 105 cells were digested with DpnI (which cleaves only unreplicated input BAC molecules) and NheI. The DNA fragments were fractionated by agarose gel electrophoresis and transferred to a Hybond-XL membrane (Amersham). Replicated BAC DNA (DpnI resistant) was detected by hybridization to a 32P-labeled pGX153 probe. NdeI was usually also added to the enzyme digestions to reduce the size of cellular DNA fragments. Since there are no NdeI sites in the BAC DNAs between positions 44008 and 53273 (Fig. 2), the presence of NdeI had no effect on the fragments detected. Phosphorimages of the washed filters were prepared using the Personal Molecular Imager and analyzed with Quantity One software (Bio-Rad).
Analysis of DNAs following virus infection.
Monolayers of BHK cells in 35-mm petri dishes were infected with 3 PFU per cell of virus, and 16 h postinfection total DNA was prepared. Samples were cleaved with BamHI and analyzed by Southern blotting as described above.
RESULTS
Introduction of a functional DNA packaging signal into fHSVΔpac.
To determine whether wt Uc-DR1-Ub fragments introduced into fHSVΔpac by Red/ET recombination were functional, we first performed a series of transfections with four BAC DNA preparations representing the parent, fHSVΔpac (BAC P), and three recombinants containing the selected rpsL-neo cassette and no Uc-DR1-Ub fragment (BAC O) or the wt fragment in either possible orientation (BACs W− and W+ [Table 1]). When transfected monolayers were incubated for 3 days, plaques were observed on the plates that received W− and W+ but not P or O (data not shown). Over the course of several experiments, W− and W+ yielded similar numbers of plaques corresponding to transfection efficiencies in the range of 50 to 2,000 plaques per μg BAC DNA. To exclude the possibility that BACs P and O might harbor mutations elsewhere in the genome that precluded virus recovery, they were also cotransfected with BamHI-cleaved pGX8 to repair the resident a sequences originally deleted from fHSVΔpac, and in both instances efficient plaque formation resulted.
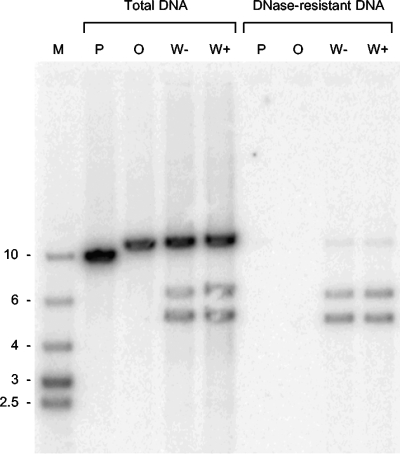
A set of plates transfected for 16 h with BAC P, O, W+, or W− was also examined for the presence of replicated and packaged viral DNA (Fig. 4). Samples of total and DNase-resistant DNA were cleaved with a combination of NheI, NdeI, and DpnI (which cleaves unreplicated input DNA into numerous small fragments) and analyzed by Southern blotting followed by hybridization to labeled pGX153 DNA. This probe would be expected to detect fragments of 10.8, 10.8, 10.6, and 9.3 kbp in concatemeric DNA generated following replication of W+, W−, O, and P, respectively. In addition cleavage at the Uc-DR1-Ub insert of W+ and W− should generate novel fragments of 6.0 and 4.8 kbp (Fig. 2). Examination of the total DNA samples (Fig. 4) indicated that all four BACs replicated to similar levels, but the two smaller fragments resulting from cleavage were observed only with W+ and W−. In contrast, packaged (DNase-resistant) DNA was detected only in cells that received W+ or W−. The predominant species correspond to the expected 6.0- and 4.8-kbp terminal fragments, but a small amount of uncleaved 10.8-kbp fragment was also detected in these samples. This may represent low-level readthrough of the Uc-DR1-Ub signal, perhaps due to segment inversion in concatemers (33) or, as has been previously suggested (21), to the presence of defective genomes or packaged circular molecules.
FIG. 4.
Replication and packaging of BACs containing or lacking the Uc-DR1-Ub fragment. BHK cells were transfected with the indicated BAC, and total and DNase-resistant DNAs were prepared. Samples were cleaved with a combination of NheI, NdeI, and DpnI; fractionated by agarose gel electrophoresis; blotted; and hybridized to labeled pGX153 DNA. Lane M contains molecular size markers of the indicated sizes (kbp).
These data indicate that Red/ET recombination can be successfully employed to introduce a functional ectopic packaging signal into fHSVΔpac, enabling specific cleavage of replicated genomes, DNA packaging, and the recovery of viable virus. Comparison of BACs W+ and W− also indicates that the orientation of the Uc-DR-Ub insert relative to the TK locus has no detectable effect on its function, consistent with the known ability of HSV-1 to package four genomic isomers containing the UL and US regions in either possible orientation.
Ability of mutated Uc-DR1-Ub fragments to support virus replication.
We next used similar approaches to examine the behavior of mutated Uc-DR1-Ub fragments in the context of the fHSVΔpac genome. The mutated fragments and the recombinant BACs are described in Table 1 and Fig. 3 and comprise a series of substitution mutants affecting each of the pac1, pac2, and DR1 motifs at multiple positions plus a selection of deletion mutants.
The BACs were first tested for their ability to generate viable virus when transfected alone or with BamHI-cleaved pGX8. In the absence of pGX8, only five BACs containing mutated Uc-DR1-Ub fragments (c4, c3, c1, b2, and Δb4) gave rise to progeny virus (Table 1), although virus was recovered in every case from the cotransfected cells. The failure of the remaining nine BACs to generate virus is therefore a direct consequence of the mutation introduced into the Uc-DR1-Ub fragment. DNA sequence analysis was performed to confirm that the ability of c4, c3, c1, b2, and Δb4 to give rise to virus was not due to mutation of the test packaging signals. Cells were infected with progeny from the transfected monolayers, and total DNA was prepared. Fragments containing the complete Uc-DR1-Ub fragment were amplified by PCR and sequenced. In each instance the sequence obtained was identical to that of the original transfected BAC, indicating not only that the mutated motifs retain full functionality for DNA cleavage and packaging but also that compensating changes do not occur at the cleavage site.
The behavior of BACs containing substitution mutations indicated that within the Uc region only the sequence of the pac2 T element was critical for virus viability. In contrast, within Ub the pac1 T element, but neither of the flanking GC-rich regions, was tolerant of nucleotide substitutions. The failure of the DR1 substitution mutant (R) to generate virus indicates that DR1 also contains sequences essential for virus growth. Analysis of the deletion mutants identified only one region dispensable for viability, namely, the “rest of Ub” region between the pac1 distal GC element and the DR2 repeat region. Interestingly, deletion of the pac1 T element, in contrast to its substitution, also resulted in a failure to generate virus, suggesting that this motif might serve a spacer function.
Ability of mutated Uc-DR1-Ub fragments to promote DNA packaging.
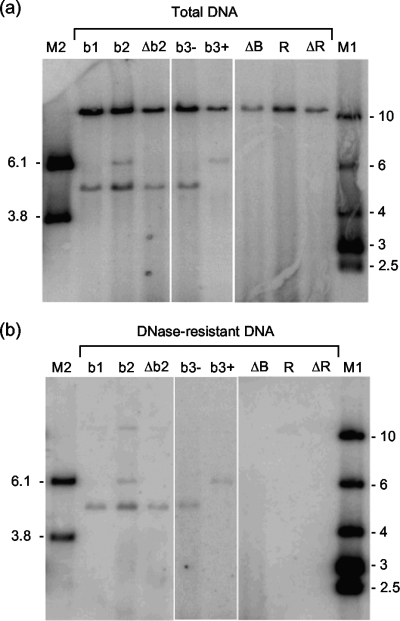
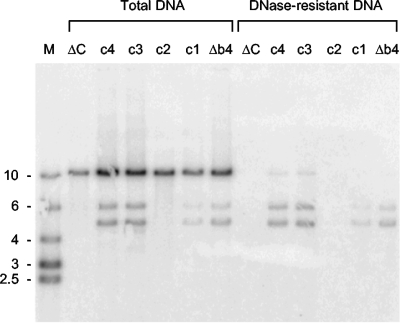
The BACs containing mutated Uc-DR1-Ub fragments were then tested for their ability to support DNA packaging. The results are presented in Fig. 5 and 6 and summarized in Table 1. As expected, all the BACs were replicated following transfection, but differences were observed in the generation of cleavage products. Five BACs, b2 (Fig. 5) and Δb4, c1, c3, and c4 (Fig. 6), yielded two terminal fragments as previously observed for the wt Uc-DR1-Ub fragment (Fig. 4), and there was a complete correlation with their ability to generate infectious progeny when transfected alone. As expected, packaged DNA was also detected with this group of BACs. The sizes of both the Ub- and Uc-containing terminal fragments of the viruses derived from these five BACs (and also the BACs containing the wt Uc-DR1-Ub fragment) remained stable on passage. This contrasts with wt HSV-1, in which amplification of the a sequence at the Uc terminus results in fragment size heterogeneity (30, 31), and indicates that such modification of the Uc terminus is not essential for viability.
FIG. 5.
Replication and packaging of BACs containing mutated Uc-DR1-Ub fragments. BHK cells were transfected with the indicated BACs, and samples of total (a) and DNase-resistant (b) DNA were analyzed as described for Fig. 4. Lanes M1 and M2 contain molecular size markers whose sizes (kbp) are indicated.
FIG. 6.
Replication and packaging of BACs containing mutated Uc-DR1-Ub fragments. BHK cells were transfected with the indicated BAC, and samples of total and DNase-resistant DNA were analyzed as described for Fig. 4. Lane M contains molecular size markers of the indicated sizes (kbp).
The remaining BACs fell into two classes, with either one or no terminal fragment being detectable in the total DNA samples. No fragment was observed with BACs ΔB, R, and ΔR (Fig. 5) and ΔC and c2 (Fig. 6), and in each of these instances packaged DNA was also undetectable.
The remaining four BACs, b1, Δb2, b3−, and b3+ (Fig. 5), yielded a single cleaved product. In the case of b1, Δb2, and b3− this was the smaller 4.8-kbp NheI terminal fragment, whereas b3+ yielded only the larger 6.0-kbp terminal fragment. BACs b3− and b3+ differ only in having the mutated fragment inserted in opposite orientations, while b1 and Δb2 have the fragment inserted in the same orientation as that in b3−. From Fig. 2 it is apparent that in all four cases the detected fragment corresponds to that containing Uc at its terminus. These four fragments, but not the corresponding Ub-containing termini, were also detected in the DNase-resistant DNA sample, suggesting that following initial cleavage of the Uc-DR1-Ub fragment, the Uc end is inserted into the capsid and packaging is initiated, but there is a failure to cleave the concatemer at the next Ub sequence to terminate packaging. The absence of the Ub-containing fragment from total DNA suggests that, following initiation of packaging, the free Ub end is degraded.
In the absence of a functional termination signal, it might have been expected that packaging of b1, Δb2, b3−, and b3+ DNA would progress beyond the next Uc-DR1-Ub fragment in the concatemer. However, the absence of the 10.8-kbp junction fragment from DNase-resistant DNA indicates that this is not the case. Samples of DNase-resistant DNA were therefore cleaved with BamHI plus DpnI and hybridized to a probe representing the whole HSV-1 genome. The results indicated that in each case fragments representing approximately 50% of the genome were present (data not shown). It seems probable that at some stage a full-length genome may have been inserted into the capsid but, in the absence of successful events to complete the packaging process, the high internal pressure resulted in the ejection of a significant proportion of the DNA. Whether this occurred in vivo (for example due to the terminase ceasing to hydrolyze ATP or falling off the DNA) or during cell lysis and the preparation of DNase-resistant DNA is not clear. Although the failure to package a full-length genome resembles the phenotype observed with a null mutant containing a lesion in the UL25 DNA packaging protein (25), further work is required to determine whether UL25 might play a role in the pac1-mediated cleavage event.
Analysis of viruses generated following repair of a sequence deletions.
The above experiments identified five mutated Uc-DR1-Ub fragments that were incapable of being cleaved or supporting DNA packaging (ΔC, c2, R, ΔR, and ΔB). Because these fragments did not allow packaging to be initiated, it was not possible to determine whether the mutations might also affect the cleavage event that terminates DNA encapsidation. A possible way of addressing this question was to analyze whether the same Uc-DR1-Ub could be cleaved following initiation of DNA packaging elsewhere in the genome. This was done by using progeny derived following the repair of the resident a sequences in the BACs that failed to generate virus when transfected alone.
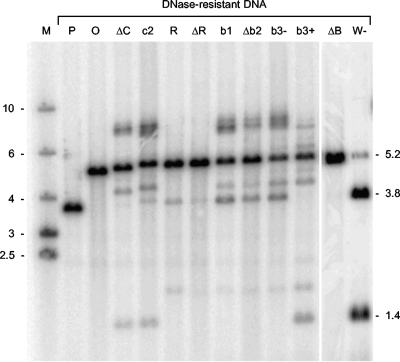
BHK cell monolayers were infected with virus obtained following the rescue of each of the 11 nonviable BACs (Table 1) or with a control virus derived from cells transfected with a BAC (W−) containing the wt Uc-DR-Ub fragment. DNase-resistant DNA was prepared, and the BamHI digestion products were analyzed by hybridization to a probe covering the BamHI P fragment. The results are shown in Fig. 7. If cleavage does not occur within the Uc-DR1-Ub insert, a fragment of 5.2 kbp is expected, whereas cleavage generates possible terminal fragments of 1.4 and 3.8 kbp (Fig. 2). BAC W− generated predominantly the two terminal fragments, but as observed previously (Fig. 4), a small amount of the uncleaved fragment was also detected. As expected, no cleavage within the BamHI P locus was observed with progeny derived from BACs O and P, which lack any Uc-DR1-Ub insert and generate only the uncleaved fragments of 4.9 and 3.6 kbp, respectively. Taking into account the orientations of the inserts in BACs ΔC, c2, R, ΔR, and ΔB, if cleavage to terminate packaging can occur within the Uc-DR1-Ub fragment of the rescued progeny, it would generate Ub-containing termini of 1.4, 1.4, 3.8, 3.8, and 1.4 kbp, respectively. These fragments were observed in all cases except ΔB, where no terminal fragment was detected. It therefore seems probable that the mutations within Uc and DR1 that prevent initiation of DNA packaging do not affect the termination process, and only ΔB appears to be deficient in both cleavage events.
FIG. 7.
Analysis of DNase-resistant DNA from cells infected with rescued progeny. BHK cell monolayers were infected with virus stocks derived from cells cotransfected with the indicated BAC and pGX8 DNA or, in the case of W−, with BAC DNA alone. DNA-resistant DNA was isolated and, following cleavage with BamHI, was analyzed as described for Fig. 4. Lane M contains molecular size markers of the indicated sizes (kbp), and the positions of the 3.8- and 1.4-kbp terminal fragments and 5.2-kbp uncleaved fragment are indicated on the right side.
This conclusion must, however, be interpreted with caution since the experiment provides clear evidence that other recombinational events are occurring. In contrast to the situation when ΔC, c2, R, and ΔR were transfected alone, it was apparent that Uc-containing fragments were also generated. However, these were predominantly a few hundred base pairs larger than the expected sizes of 3.8, 3.8, 1.4, and 1.4 kbp, respectively. The structure of these termini has not been analyzed in detail, but it is possible that a full-length a sequence (approximately 400 bp) has been added at the terminus which provides a functional Uc region to initiate cleavage. Smiley et al. (23) described similar observations for HSV-1 recombinants into which mutated a sequences had been introduced at an ectopic site and proposed a repair process involving recombination with copies of the a sequence at the L-S junction.
Figure 7 also shows that the BACs with inserts that impair termination and produce only one terminal fragment when transfected alone (b1, Δb2, b3−, and b3+) similarly yielded two terminal fragments from the BamHI locus following rescue. The predicted Ub-containing termini of b1, Δb2, b3−, and b3+ are 1.4, 1.4, 1.4, and 3.8 kbp, respectively, and in each case it is apparent that these fragments were replaced by fragments a few hundred base pairs larger, consistent with the addition of an intact a sequence to the terminus providing a functional pac1 region. Following the introduction of a full-length a sequence into the rescued viruses, it is probable that the mechanisms responsible for a sequence amplification in wt HSV-1 can operate, and this likely explains the faint ladder of larger bands apparent at the Uc termini of these recombinants. Prominent fragments larger than 6 kb are also generated in most cases. The origin of these fragments was not investigated, but they may result from the additional segment inversion events that can occur when copies of the a sequence are present both in their normal locations and at an ectopic site (23).
DISCUSSION
This paper describes an improved approach for analyzing the cis-acting signals that participate in the cleavage and packaging of herpesvirus DNA. By introducing single copies of test packaging signals into an HSV-1 genome lacking the normal resident copies, we have overcome one of the major drawbacks of the previously used amplicon and ectopic-site approaches, namely, the high-frequency recombination between the fragment under test and wt copies of the a sequence present either in the helper virus or elsewhere in the genome of the ectopic-site recombinant. The severity of this problem is clearly illustrated by the high frequency with which novel terminal fragments of slightly increased size were generated from the region containing the Uc-DR1-Ub insert following repair of the resident a sequences (Fig. 7). Furthermore, in contrast to the amplicon assay but like replicated HSV-1 DNA, pairs of similarly orientated cleavage sites defining the two ends of a packaged molecule should mainly occur at genome-length intervals in concatemers. However, due to the occurrence of segment inversion in concatemeric HSV-1 DNA, the relative orientation of the inserted packaging signals can become changed, resulting in a proportion of the DNA becoming incapable of generating full-length encapsidated genomes (12, 28, 33).
Analysis of a series of mutated Uc-DR1-Ub fragments (Fig. 5 and 6; Table 1) confirmed and extended our previous observations made using the amplicon assay (10). Of the mutants that had been previously tested, ΔC (containing a large deletion within Uc extending into DR1), c2 (pac2 T-element substitution), and ΔB (containing a large deletion within Ub extending into DR1), which were all highly impaired in the amplicon assay, were completely defective in DNA cleavage and packaging. The four mutated fragments that supported generation of virus progeny, c1, c3, and c4 (substitutions of the pac2 GC element, unconserved region, and consensus sequence, respectively) and b2 (pac1 T-element substitution), showed no significant differences in DNA packaging, the ability to be propagated, or the generation of terminal fragments compared to wt Uc-DR1-Ub in the amplicon assay. Mutants b1 and b3 (pac1 proximal and distal GC region substitutions, respectively), which supported DNA packaging but not progeny production, exhibited normal DNA packaging but a reduced ability to be serially propagated in the amplicon assay. Moreover, b1 (but not b3) was previously shown to be defective in the generation of an Ub-containing terminus in cleaved amplicon DNA. Thus, for the nine mutated fragments that have been tested in both assays there is very good correspondence between the results obtained. The approach described in this paper does, however, appear to be more discriminating in certain respects: for example, no cleavage or packaging is detected with the most impaired mutants, whereas low levels are apparent in the amplicon assay. Similarly, the remaining mutants fall clearly into either the viable or the nonviable class when assayed in the context of a viral genome but differ only in their relative efficiency to be serially propagated in the amplicon assay. It is likely that the occurrence of recombinational repair in the amplicon assay accounts in part for these differences and also for our previous detection of a Ub-containing terminus with b3 (10). The tandem repeat structure of replicated amplicon DNA may also permit packaging to be terminated by an “illegitimate” non-sequence-specific mechanism, resulting in the generation of particles containing genomes that can reinitiate infection following circularization by homologous recombination. If such an illegitimate mechanism were to operate in the context of the viral genome, it is probable that the resulting packaged molecules would either lack essential genes or be incapable of circularization to initiate infection.
Data were also presented for four new Uc-DR1-Ub mutants, allowing the development of a more complete picture of the roles of the various motifs within this region. The five mutants affecting the pac2 region had all been examined previously (10). The results reported here confirm that the pac2 region is essential for the initiation of the cleavage-packaging process and that while the sequence of the T element is critical, extensive nucleotide substitutions within the consensus, unconserved, and GC elements are tolerated. It has recently been reported that in MCMV a second essential packaging element, which is GC rich, is present within pac2 located further from the cleavage site than the T element (32). It remains possible that a corresponding signal is present in HSV-1 within the region designated “rest of Uc” and is supported by our previous observation that in the amplicon assay, deletion but not substitution of the intervening unconserved region caused a significant impairment in DNA packaging (10).
Two new mutants replacing or deleting the DR1 region (R and ΔR, respectively) revealed that the sequence of this region is, like the pac2 T element, crucial for the initiation of cleavage and packaging. This result initially appears to conflict with previous observations from Smiley's group (23, 29), who demonstrated that the sites of the cleavage events defining either end of the viral genome were not dependent upon the presence of a complete DR1 region but rather occurred at fixed distances from the pac1 and pac2 T elements. However, it is noteworthy that their construct retained 13 bp of DR1 at its Uc end (although not the actual site of cleavage), which may be required to initiate the cleavage and packaging process. This interpretation is consistent with our previous observation that the pac2 GC element appears to play an important spacer role in DNA packaging (10) and points to sequences within DR1 being a third component of the signal required for the initiation of DNA packaging.
The construct analyzed by Varmuza and Smiley (29) lacked most of the proximal GC element and all of DR1 from its Ub terminus but was still cleaved at the standard distance from the pac1 T element, suggesting that DR1 does not play a role in termination of packaging. Since initiation of DNA packaging did not occur with mutants R and ΔR, we could not directly assess whether DR1 plays a role in the terminating cleavage event. The analysis of the progeny resulting from repair of the resident a sequences in BACs containing the R or ΔR fragment (Fig. 7) nevertheless lends some support to the hypothesis that DR1 may contain sequences important for initiation of cleavage and packaging but be dispensable for termination. In both instances a normally sized Uc terminus (the 1.4-kbp BamHI fragment) was replaced by a fragment approximately 400 bp larger, possibly reflecting the addition of a full-length a sequence. In contrast, a corresponding change in the size of the 3.8-kbp Ub-containing terminal fragment was not observed, suggesting that similar modification of this end of the inserted Uc-DR1-Ub fragment may not be required for the terminating cleavage event.
Turning to the Ub region, a new mutant, Δb4, revealed that no essential sequences are present within the “rest of Ub” region and indicated that the key pac1 elements must be confined to the remaining 64 bp of Ub. This corresponds very closely with the minimal essential region for pac1 function identified in MCMV (32). The results obtained with the pac1 substitution mutants are in good agreement with previous studies using HSV-1 amplicons and ectopic-site MCMV recombinants (10, 15, 32). These confirm that important sequences are present within the proximal and distal GC-rich regions but that the sequence of the T element is not crucial. Although not addressed in the current study, McVoy and colleagues have identified the conserved homopolymer tracts immediately flanking the T element as crucial sequences within the GC-rich regions (15, 32). Interestingly, a newly constructed pac1 deletion mutant, Δb2, exhibited behavior similar to that of the substitution mutants affecting the GC-rich regions (b1 and b3), indicating that the T element may function as a spacer to maintain the relative positioning of the flanking homopolymer tracts.
We previously noted that while deletions individually affecting the three pac1 regions did not prevent initiation of packaging, the larger deletion in ΔB did (10). It now appears likely that this is because the deletion not only affects the pac1 motifs but extends into the DR1 region. Irrespective of this, ΔB appears to be the only one of our mutants deficient in both cleavage events. Interestingly, this is also the only mutated fragment that does not acquire the ability to be cleaved when the resident a sequences are repaired (Fig. 7). This observation provides support for the hypothesis of Smiley et al. (23) that recombinational repair in viruses containing ectopic packaging signals is stimulated by DNA breaks induced by the cleavage-packaging system.
As described above, to determine whether mutations that abolished the ability to initiate packaging might also prevent termination, we analyzed progeny obtained following the repair of the a sequence deletions in recombinant BACs. Although tentative conclusions could be drawn, the data confirmed that recombination is a major complicating factor in such an approach (Fig. 7). Future experiments are therefore aimed at spatially separating the initiation and termination cleavage events by introducing two separate fragments into fHSVΔpac, which should allow the sequence requirements for each process to be independently assessed.
Acknowledgments
We thank Cornel Fraefel (University of Zurich) for provision of fHSVΔpac and are grateful to Duncan McGeoch and Andrew Davison for helpful comments on the manuscript.
Footnotes
Published ahead of print on 28 October 2009.
REFERENCES
- 1.Adelman, K., B. Salmon, and J. D. Baines. 2001. Herpes simplex virus DNA packaging sequences adopt novel structures that are specifically recognized by a component of the cleavage and packaging machinery. Proc. Natl. Acad. Sci. USA 98:3086-3091. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Baines, J. D., and S. K. Weller. 2005. Cleavage and packaging of herpes simplex virus 1 DNA, p. 135-150. In C. E. Catalano (ed.), Viral genome packaging machines: genetics, structure and mechanism. Kluwer Academic/Plenum Publishers, New York, NY.
- 3.Broll, H., H.-J. Buhk, W. Zimmermann, and M. Goltz. 1999. Structure and function of the prDNA and genomic termini of the γ2-herpesvirus bovine herpesvirus type 4. J. Gen. Virol. 80:979-986. [DOI] [PubMed] [Google Scholar]
- 4.Brown, J. C., M. A. McVoy, and F. L. Homa. 2002. Packaging DNA into herpesvirus capsids, p. 111-153. In A. Holzenburg and E. Bogner (ed.), Structure-function relationships of human pathogenic viruses. Kluwer Academic/Plenum Publishers, New York, NY.
- 5.Chou, J., and B. Roizman. 1985. Isomerization of herpes simplex virus 1 genome: identification of the cis-acting and recombination sites within the domain of the a sequence. Cell 41:803-811. [DOI] [PubMed] [Google Scholar]
- 6.Davison, A. J., and N. M. Wilkie. 1981. Nucleotide sequences of the joint between the L and S segments of herpes simplex virus types 1 and 2. J. Gen. Virol. 55:315-331. [DOI] [PubMed] [Google Scholar]
- 7.Deiss, L. P., and N. Frenkel. 1986. Herpes simplex virus amplicon: cleavage of concatemeric DNA is linked to packaging and involves amplification of the terminally reiterated a sequence. J. Virol. 57:933-941. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Deiss, L. P., J. Chou, and N. Frenkel. 1986. Functional domains within the a sequence involved in the cleavage-packaging of herpes simplex virus DNA. J. Virol. 59:605-618. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Fraefel, C., S. Song, F. Lim, P. Lang, L. Yu, Y. Wang, P. Wild, and A. I. Geller. 1996. Helper virus-free transfer of herpes simplex virus type 1 plasmid vectors into neural cells. J. Virol. 70:7190-7197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Hodge, P. D., and N. D. Stow. 2001. Effects of mutations within the herpes simplex virus type 1 DNA encapsidation signal on packaging efficiency. J. Virol. 75:8977-8986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Homa, F. L., and J. C. Brown. 1997. Capsid assembly and DNA packaging in herpes simplex virus. Rev. Med. Virol. 7:107-122. [DOI] [PubMed] [Google Scholar]
- 12.Martin, D. W., and P. C. Weber. 1996. The a sequence is dispensable for isomerization of the herpes simplex virus type 1 genome. J. Virol. 70:8801-8812. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Martinez, R., R. T. Sarisky, P. C. Weber, and S. K. Weller. 1996. Herpes simplex virus type 1 alkaline nuclease is required for efficient processing of viral DNA replication intermediates. J. Virol. 70:2075-2085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.McGeoch, D. J., M. A. Dalrymple, A. J. Davison, A. Dolan, M. C. Frame, D. McNab, L. J. Perry, J. E. Scott, and P. Taylor. 1988. The complete DNA sequence of the long unique region in the genome of herpes simplex virus type 1. J. Gen. Virol. 69:1531-1574. [DOI] [PubMed] [Google Scholar]
- 15.McVoy, M. A., D. E. Nixon, S. P. Adler, and E. S. Mocarski. 1998. Sequences within the herpes-conserved pac1 and pac2 motifs are required for cleavage and packaging of the murine cytomegalovirus genome. J. Virol. 72:48-56. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.McVoy, M. A., D. E. Nixon, J. K. Hur, and S. P. Adler. 2000. The ends of herpesvirus DNA replicative concatemers contain pac2 cis cleavage/packaging elements and their formation is controlled by terminal cis sequences. J. Virol. 74:1587-1592. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Mocarski, E. S., and B. Roizman. 1981. Site-specific inversion sequence of the herpes simplex virus genome: domain and structural features. Proc. Natl. Acad. Sci. U. S. A. 78:7047-7051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Mocarski, E. S., and B. Roizman. 1982. Structure and role of the herpes simplex virus DNA termini in inversion, circularization and generation of virion DNA. Cell 31:89-97. [DOI] [PubMed] [Google Scholar]
- 19.Muyrers, J. P. P., Y. Zhang, G. Testa, and A. F. Stewart. 1999. Rapid modification of bacterial artificial chromosomes by ET-recombination. Nucleic Acids Res. 27:1555-1557. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Nasseri, M., and E. S. Mocarski. 1988. The cleavage recognition signal is contained within sequences surrounding an a-a junction in herpes simplex virus DNA. Virology 167:25-30. [DOI] [PubMed] [Google Scholar]
- 21.Poffenberger, K. L., and B. Roizman. 1985. A noninverting genome of a viable herpes simplex virus 1: presence of head-to-tail linkages in packaged genomes and requirements for circularization after infection. J. Virol. 53:587-595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Saeki, Y., T. Ichikawa, A. Saeki, E. A. Chiocca, K. Tobler, M. Ackermann, X. O. Breakfield, and C. Fraefel. 1998. Herpes simplex virus type 1 DNA amplified as bacterial artificial chromosome in Escherichia coli: rescue of replication-competent virus progeny and packaging of amplicon vectors. Hum. Gene Ther. 9:2787-2794. [DOI] [PubMed] [Google Scholar]
- 23.Smiley, J. R., J. Duncan, and M. Howes. 1990. Sequence requirements for DNA rearrangements induced by the terminal repeat of herpes simplex virus type 1 KOS DNA. J. Virol. 64:5036-5050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Spaete, R. R., and N. Frenkel. 1982. The herpes simplex virus amplicon: a new eukaryotic defective-virus cloning-amplifying vector. Cell 30:295-304. [DOI] [PubMed] [Google Scholar]
- 25.Stow, N. D. 2001. Packaging of genomic and amplicon DNA by the herpes simplex virus type 1 UL25-null mutant KUL25NS. J. Virol. 75:10755-10765. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Stow, N. D., and N. M. Wilkie. 1976. An improved technique for obtaining enhanced infectivity with herpes simplex virus type 1 DNA. J. Gen. Virol. 33:447-458. [DOI] [PubMed] [Google Scholar]
- 27.Stow, N. D., E. C. McMonagle, and A. J. Davison. 1983. Fragments from both termini of the herpes simplex virus type 1 genome contain signals required for the encapsidation of viral DNA. Nucleic Acids Res. 11:8205-8220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Strang, B. L., and N. D. Stow. 2005. Circularization of the herpes simplex virus type 1 genome upon lytic infection. J. Virol. 79:12487-12494. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Varmuza, S. L., and J. R. Smiley. 1985. Signals for site-specific cleavage of HSV-1 DNA: maturation involves two separate cleavage events at sites distal to the recognition sequences. Cell 41:793-802. [DOI] [PubMed] [Google Scholar]
- 30.Wadsworth, S., G. S. Hayward, and B. Roizman. 1976. Anatomy of herpes simplex virus DNA. V. Terminally repetitive sequences. J. Virol. 17:503-512. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Wagner, M. J., and W. C. Summers. 1978. Structure of the joint region and the termini of the DNA of herpes simplex virus type 1. J. Virol. 27:374-387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Wang, B. D., D. E. Nixon, and M. A. McVoy. 2008. Definition of the minimal cis-acting sequences necessary for genome maturation of the herpesvirus murine cytomegalovirus. J. Virol. 82:2394-2404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Zhang, X., S. Efstathiou, and A. Simmons. 1994. Identification of novel herpes simplex virus replicative intermediates by field inversion gel electrophoresis: implications for viral DNA amplification strategies. Virology 202:530-539. [DOI] [PubMed] [Google Scholar]
- 34.Zhang, Y., J. P. P. Muyrers, G. Testa, and A. F. Stewart. 2000. DNA cloning by homologous recombination in Escherichia coli. Nat. Biotechnol. 18:1314-1317. [DOI] [PubMed] [Google Scholar]
- 35.Zimmermann, J., and W. Hammerschmidt. 1995. Structure and role of the terminal repeats of Epstein-Barr virus in processing and packaging of virion DNA. J. Virol. 69:3147-3155. [DOI] [PMC free article] [PubMed] [Google Scholar]