Abstract
Two carbapenem-resistant isolates, one Escherichia coli isolate and one Klebsiella pneumoniae isolate, recovered from an Algerian patient expressed a novel VIM-type metallo-β-lactamase (MBL). The identified blaVIM-19 gene was located on a ca. 160-kb plasmid and located inside a class 1 integron in both isolates. VIM-19 differed from VIM-1 by the Asn215Lys and Ser228Arg substitutions, increasing its hydrolytic activity toward carbapenems. Site-directed mutagenesis experiments showed that both substitutions were necessary for the increased carbapenemase activity of VIM-19. This study indicates that MBLs with enhanced activity toward carbapenems may be obtained as a result of very few amino acid substitutions.
Acquired metallo-β-lactamases (MBLs) are emerging resistance determinants in clinically relevant Gram-negative species (5, 32). These enzymes confer broad-spectrum β-lactam resistance, including resistance to carbapenems (28, 32). In addition, their potential for rapid and wide dissemination make them of great concern (3, 32). Nine types of acquired MBLs have been reported so far, the IMP, VIM, SPM, GIM (25, 32), SIM (14), KHM (29), AIM (D. Yong, T. R. Walsh, J. Bell, B. Ritchie, R. Pratt, and M. A. Toleman, presented at the 46th Interscience Conference on Antimicrobial Agents and Chemotherapy, Washington, DC, 25 to 28 October 2008), NDM (34), and DIM types (L. Poirel, J. M. Rodriguez-Martinez, N. Al Naiemi, Y. Debets-Ossenkopp, and P. Nordmann, presented at the 19th European Congress of Clinical Microbiology and Infectious Diseases, Helsinki, Finland, 16 to 19 May 2009). These MBLs are usually encoded by genes on plasmids and associated with mobile genetic elements, mostly class 1 integrons and ISCR elements (25, 28, 30, 32). They have been identified in Pseudomonas spp. and more rarely in members of the family Enterobacteriaceae and Acinetobacter spp. Those enzymes corresponding to the so-called VIM type have been classified into three clusters according to their amino acid sequences (subgroups VIM-1, VIM-2, and VIM-7) (www.lahey.org/Studies). Those enzymes share similar hydrolytic properties, even if some slight hydrolytic differences have been noticed (6). We report here the identification and characterization of a new VIM variant, belonging to the VIM-1 cluster which exhibits an increased carbapenem-hydrolyzing activity compared with VIM-1.
MATERIALS AND METHODS
Bacterial strains and plasmids.
Identification of Escherichia coli DIH-1 and Klebsiella pneumoniae DIH-2 was performed by using the API 20E system (bioMérieux, Marcy l'Etoile, France). Pseudomonas aeruginosa V4 (4) and Acinetobacter sp. 154 (7) were used as blaVIM-1 and blaVIM-4 positive controls, respectively. E. coli TOP10 was used as the host strain for cloning, and E. coli J53 (resistant to azide) was used as the host for conjugation assays.
Antimicrobial agents and MIC determinations.
The antimicrobial agents and their sources have been described elsewhere (21). Susceptibility testing was performed by disk diffusion assay (Sanofi-Diagnostic Pasteur, Marnes-la-Coquette, France), as previously described. The MICs were determined by Etest (AB Biodisk, Solna, Sweden) on Mueller-Hinton agar plates at 37°C (21). Results of susceptibility testing were recorded according to the CLSI guidelines (2). MBL detection tests were performed using an Etest strip (AB Biodisk).
Cloning experiments, PCR, and DNA sequencing.
Whole-cell DNAs were extracted as previously described (1). PCR screening for MBL-encoding genes blaVIM and blaIMP and for extended-spectrum β-lactamases (ESBLs) encoding genes blaTEM, blaSHV, and blaCTX-M were performed as described previously (26). PCR combinations performed as described previously (26) were performed in order to identify the class 1 integron 5′ and 3′ extremities, including the search for Tn402-related transposon structures described previously (22). In order to express the different blaVIM genes in an identical background, cloning of the blaVIM-1, blaVIM-4, and blaVIM-19 genes was performed in E. coli TOP10 as described previously (23), using the ZeroBlunt TOPO PCR cloning kit (Invitrogen, Cergy-Pontoise, France) followed by selection on plates containing 50 μg/ml of amoxicillin and 30 μg/ml of kanamycin. The PCR amplicon encompassing the entire sequence of the blaVIM genes used for cloning was obtained with primers VIM-CasA (5′-TATGCCGCACCCACCCCTATG-3′) and VIM-CasB (5′-ATGCTACTCGGCGACTGAGC-3′). Those amplicons did not include the original promoter region of the blaVIM genes in order to express those genes under the control of the same promoter provided by plasmid pCR-BluntII-TOPO. The corresponding recombinant strains were used for MIC determinations.
Both strands of the cloned DNA inserts of recombinant plasmids were sequenced by using an Applied Biosystems sequencer (ABI 377). The nucleotide and deduced protein sequences were analyzed with software available over the Internet from the National Center for Biotechnology Information website (http://www.ncbi.nlm.nih.gov/BLAST/).
Site-directed mutagenesis.
The VIM-19 β-lactamase identified contained a single substitution (Asn215Lys) compared to the sequence of VIM-4 and two substitutions (Asn215Lys and Ser228Arg) compared to the sequence of VIM-1. Therefore, we aimed to evaluate the role of the single Asn215Lys substitution in increased carbapenemase activity. For this purpose, a site-directed mutagenesis protocol was used as described by the manufacturer (QuikChange site-directed mutagenesis kit; Stratagene). Recombinant plasmid pVIM-1 was used as the template in PCR amplification with primers VIM-1-N215K-F (5′-GTCCCGTCAGCGAAAGTGCTATACGG-3′) and VIM-1-N215K-R (5′-CCGTATAGCACTTTCGCTGACGGGAC-3′). It gave rise to recombinant plasmid pVIM-1-N215K, which was subsequently transformed into E. coli TOP10. Sequence analysis of the inserts confirmed the presence of the expected mutation, which led to the N215K replacement in the mature β-lactamase VIM-1-N215K.
IEF analysis.
The β-lactamase extract from a culture of E. coli TOP10 harboring recombinant plasmid pVIM-19 was subjected to analytical isoelectric focusing (IEF) analysis as described previously (15). The focused β-lactamases were detected by overlaying the gel with 1 mM nitrocefin (Calbiochem, La Jolla, CA).
β-Lactamase purification.
Cultures of E. coli TOP10 harboring recombinant plasmid pVIM-19 were grown overnight at 37°C in 4 liters of Trypticase soy broth containing amoxicillin (100 μg/ml) and kanamycin (30 μg/ml). β-Lactamase VIM-19 was purified by ion-exchange chromatography. Briefly, the bacterial suspension was pelleted, resuspended in 50 ml of 100 mM sodium phosphate buffer (pH 7.0) plus 50 μM ZnCl2 and 1 mM MgCl2, sonicated, cleared by ultracentrifugation, and treated with DNase. The extract was then dialyzed against 50 mM Bis-Tris buffer (pH 6.5) plus 50 μM ZnCl2 and 1 mM MgCl2 and loaded onto a preequilibrated Q-Sepharose column. The β-lactamase-containing fractions were eluted with a linear NaCl gradient (0 to 1 M). The same procedure was repeated using a 30 mM cacodylate buffer (pH 6.5) plus 50 μM ZnCl2 and 1 mM MgCl2. Finally, fractions containing the highest β-lactamase activities were pooled and subsequently dialyzed overnight against 50 mM HEPES buffer (pH 7.5) including 50 μM ZnCl2 and 1 mM MgCl2. The β-lactamase activity was determined qualitatively using nitrocefin hydrolysis (Oxoid, Dardilly, France). The protein content was measured using the Bio-Rad DC protein assay. The purification factor was measured by comparing the activities of the VIM-19 crude extract and purified enzyme using 100 μM imipenem as the substrate.
Kinetic studies.
Kinetic measurements (kcat and Km) of purified β-lactamase VIM-19 were performed spectrophotometrically as described previously (24).
Plasmid content and conjugation assays.
Plasmid DNAs of E. coli DIH-1 and K. pneumoniae DIH-2 were extracted by using the Kieser method (16). E. coli NCTC50192 harboring four plasmids of 154, 66, 48, and 7 kb was used as the size marker for plasmids. Plasmid DNAs were analyzed by agarose gel electrophoresis as described previously (16). Direct transfer of the β-lactam resistance markers into E. coli J53 was attempted by liquid mating-out assays at 37°C. Selection was performed on agar plates supplemented with amoxicillin (50 μg/ml) and azide (100 μg/ml).
Nucleotide sequence accession number.
The nucleotide sequence reported in this work has been deposited in the GenBank nucleotide database under accession no. FJ822963.
RESULTS
Characteristics of E. coli and K. pneumoniae isolates.
This study was initiated by the isolation of carbapenem-resistant E. coli and K. pneumoniae isolates in January 2008 in our hospital. They had been recovered from rectal swabs obtained at the hospital entrance (systematic screening for multidrug-resistant bacteria) from a 30-year-old patient transferred from Algiers, Algeria, where he had been hospitalized 10 days after injuries caused by a terrorist attack in December 2007.
Isolates E. coli DIH-1 and K. pneumoniae DIH-2 were resistant to most β-lactams, including imipenem and ertapenem (Table 1). These isolates were also resistant to all aminoglycosides and to chloramphenicol, tetracycline, trimethoprim, and sulfonamides; they were susceptible only to fluoroquinolones and colistin (data not shown). MBL detection tests were positive for both isolates, and PCR screening for MBL-encoding genes identified a novel blaVIM type in both isolates that was defined as blaVIM-19 (http://www.lahey.org/Studies/). Synergy tests performed with clavulanic acid and cefepime showed the production of an ESBL only in K. pneumoniae DIH-2. PCR for ESBL genes followed by sequencing identified the blaCTX-M-3 gene in K. pneumoniae DIH-2.
TABLE 1.
MICs of β-lactams for E. coli and K. pneumoniae isolates and strainsa
| β-Lactam(s)b | MIC (μg/ml) of β-lactam for: |
||||||||
|---|---|---|---|---|---|---|---|---|---|
| E. coli DIH-1 (VIM-19) | K. pneumoniae DIH-2 (VIM-19) | E. coli TOP10 (pVIM-19) | E. coli TOP10 (pVIM-4) | E. coli TOP10 (pVIM-1) | E. coli TOP10 (pVIM1-N215K) | E. coli J53(pR3) (VIM-19) | E. coli TOP10 | E. coli J53 | |
| Amoxicillin | >512 | >512 | >512 | >512 | >512 | >512 | >512 | 4 | 4 |
| Amoxicillin + CLA | >512 | >512 | >512 | >512 | >512 | >512 | >512 | 4 | 4 |
| Ticarcillin | >512 | >512 | >512 | >512 | >512 | >512 | >512 | 4 | 4 |
| Ticarcillin + CLA | >512 | >512 | >512 | >512 | >512 | >512 | >512 | 4 | 4 |
| Piperacillin | >512 | >512 | 256 | 256 | 256 | 256 | 256 | 1 | 1 |
| Piperacillin + TZB | >512 | >512 | 256 | 256 | 256 | 256 | 256 | 1 | 1 |
| Cefuroxime | >512 | >512 | >512 | >512 | >512 | >512 | >512 | 2 | 4 |
| Ceftazidime | >512 | 128 | 32 | 32 | 512 | 512 | 32 | 0.06 | 0.06 |
| Cefotaxime | >512 | >512 | 64 | 64 | 32 | 64 | 64 | 0.12 | 0.12 |
| Cefepime | 16 | 16 | 2 | 1 | 16 | 16 | 2 | 0.06 | 0.06 |
| Cefpirome | 32 | 64 | 2 | 2 | 32 | 32 | 2 | 0.06 | 0.12 |
| Cefoxitin | >512 | >512 | 256 | 256 | 256 | 256 | 256 | 4 | 4 |
| Aztreonam | 4 | 2 | 0.12 | 0.12 | 0.12 | 0.12 | 0.12 | 0.12 | 0.12 |
| Imipenem | 8 | 8 | 8 | 4 | 1 | 1 | 4 | 0.06 | 0.06 |
| Meropenem | 4 | 4 | 1 | 0.5 | 0.25 | 0.25 | 0.5 | 0.016 | 0.016 |
| Ertapenem | 16 | 16 | 2 | 0.75 | 0.125 | 0.125 | 1.5 | 0.006 | 0.006 |
The E. coli and K. pneumoniae isolates and strains follow: E. coli DIH-1 and K. pneumoniae DIH-2 clinical isolates; E. coli TOP10 strains harboring recombinant plasmid pVIM-19, pVIM-4, pVIM-1-N215K, or pVIM-1 expressing β-lactamase VIM-19, VIM-4, VIM-1-N215K, or VIM-1, respectively; E. coli J53 transconjugant containing the natural plasmid expressing VIM-19 from E. coli DIH-1; and E. coli TOP10 and E. coli J53 reference strains.
CLA, clavulanic acid at a fixed concentration of 4 μg/ml; TZB, tazobactam at a fixed concentration of 4 μg/ml.
Genetic support of blaVIM-19.
Analysis of the genetic environment of blaVIM-19 in isolates DIH-1 and DIH-2 showed that it was part of a gene cassette located at the first position of a class 1 integron. However, PCR combinations failed to identify the 3′ extremity (either 3′ CS or Tn402-like backbone) of the class 1 integron and consequently possible additional gene cassettes. Conjugation experiments produced E. coli transconjugants exhibiting an MBL phenotype using both E. coli DIH-1 and K. pneumoniae DIH-2 as donors, containing a single 160-kb plasmid (named pR3) harboring the blaVIM-19 gene. Both transconjugants additionally expressed resistance to kanamycin, tobramycin, gentamicin, chloramphenicol, trimethoprim, tetracycline, and sulfonamides. This result suggested strongly that the blaVIM-19-positive plasmids identified in E. coli DIH-1 and K. pneumoniae DIH-2 were identical. In addition, an E. coli transconjugant expressing VIM-19 in addition to an ESBL (evidenced through a slight synergy between clavulanate and aztreonam) was obtained only with K. pneumoniae DIH-2 as the donor strain, in which a 70-kb plasmid was identified that harbored the blaCTX-M-3 gene. No transconjugant expressing the ESBL CTX-M-3 only was obtained, suggesting that the plasmid harboring blaCTX-M-3 might be mobilizable by that carrying the blaVIM-19 gene.
Characterization of the VIM-19 MBL.
β-Lactamase VIM-19 differed from VIM-4 by a single amino acid substitution (Asn215Lys) and from VIM-1 by two substitutions (Asn215Lys and Ser228Arg) (Fig. 1). As expected, expression of the blaVIM-1, blaVIM-4, and blaVIM-19 genes in E. coli TOP10 conferred resistance or reduced susceptibility to all β-lactams except to aztreonam (Table 1). However, the MICs of imipenem, meropenem, and ertapenem were higher for VIM-19 than those for VIM-1 and VIM-4 once cloned in E. coli, suggesting the involvement of residues Lys215 and Arg228 in higher carbapenemase activity. In contrast, the MICs of ceftazidime and cefepime for E. coli carrying pVIM-19 [E. coli(pVIM-4)] were lower than those obtained for E. coli(pVIM-1), but similar to those for E. coli(pVIM-4) (Table 1). Those results strengthened the role of the Ser228 residue in higher hydrolysis of expanded-spectrum cephalosporins. The resistance pattern of the E. coli recombinant strain expressing the Asn215Lys-mutated VIM-1 was similar to the pattern of a strain expressing VIM-1, thus ruling out the possibility that Lys215 alone could play a role in the higher carbapenemase activity of VIM-19 (Table 1).
FIG. 1.
Comparison of the amino acid sequences of VIM-19, VIM-1, VIM-4, VIM-1 (N215K), and VIM-2. Dashes indicate conserved residues. The differences found between VIM-1 and VIM-19 are boxed. Residues in boldface type belong to the His and Cys active sites of MBLs. The numbering is according to the updated BBL scheme (10). The vertical arrow indicates the signal peptide cleavage site.
IEF analysis identified a pI of 5.2 for β-lactamase VIM-19 that was purified to near homogeneity (>95%) as determined by sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) analysis (data not shown), and the purification factor was estimated to be 40-fold. β-Lactamase VIM-19 hydrolyzed all tested β-lactams except aztreonam. Kinetic data showed that VIM-19 hydrolyzed imipenem and meropenem at higher levels than VIM-1 did (6- and 7-fold, respectively), although cefepime was less hydrolyzed (Table 2). VIM-19 hydrolyzed imipenem more efficiently than it hydrolyzed meropenem, as observed for VIM-1 (Table 2). Those data are in accordance with the resistance phenotype observed for the E. coli recombinant strains. Higher catalytic efficiencies were also observed for VIM-19 compared to VIM-1 with benzylpenicillin, piperacillin, and cefotaxime (100-, 8-, and 30-fold, respectively).
TABLE 2.
Kinetic parameters of VIM-19 and VIM-1 enzymes
| β-Lactam | Kinetic parameter of enzyme |
kcat/Km (μM−1s−1) ratio for VIM-19/VIM-1 | |||||
|---|---|---|---|---|---|---|---|
| VIM-19a |
VIM-1b |
||||||
| Km (μM) | kcat (s−1) | kcat/Km (μM−1s−1) | Km (μM) | kcat (s−1) | kcat/Km (μM−1s−1) | ||
| Benzylpenicillin | 300 | 1,300 | 5 | 850 | 30 | 0.05 | 100 |
| Piperacillin | 250 | 860 | 4 | 3,500 | 1,860 | 0.5 | 8 |
| Cefoxitin | 60 | 30 | 0.5 | 130 | 25 | 0.2 | 2.5 |
| Cefotaxime | 30 | 900 | 30 | 250 | 170 | 0.7 | 30 |
| Ceftazidime | >1,000 | >20 | 0.02 | 800 | 60 | 0.1 | |
| Cefepime | 350 | 100 | 0.5 | 145 | 550 | 4 | 0.12 |
| Aztreonam | NDc | ND | ND | >1,000 | <0.01 | <0.0001 | |
| Imipenem | 40 | 250 | 6 | 1.5 | 2 | 1 | 6 |
| Meropenem | 15 | 25 | 2 | 50 | 15 | 0.3 | 7 |
| Ertapenem | 200 | 15 | 0.1 | ||||
Data are the means of three independent experiments. Standard deviations were within 15% of the means.
VIM-1 values were reported by Franceschini et al. (8).
ND, no detectable hydrolysis (<0.01 s−1) for a maximum amount of 5 μg of purified enzyme and up to 200 nmol of substrate.
DISCUSSION
We identified here a novel MBL, VIM-19, that had a higher ability to hydrolyze carbapenems associated with a lower ability to hydrolyze several expanded-spectrum cephalosporins. Differences in the hydrolysis parameters of β-lactams had been already noticed for some distantly related VIM enzymes, especially between VIM-1 and VIM-2, but interestingly here the peculiar properties of VIM-19 are based on only two substitutions.
The Lys215 residue in VIM-19 is unique compared to all VIM-type amino acid sequences and is located outside the active site. We showed here that this specific substitution alone was not responsible for the higher carbapenemase activity observed with VIM-19, even if a slight increased carbapenemase activity was noticed. However, the association of amino acids Lys215 together with Arg228 significantly increased the activity of VIM-19 against carbapenems compared to VIM-1 and VIM-4. That observation might be paralleled with that made for VIM-11 by Marchiaro et al. (18), indicating that amino acid substitutions located outside the active site may modulate the hydrolytic properties of the enzyme.
The Arg228 residue is the second residue leading to a higher carbapenemase activity of VIM-19. It has been shown that this same residue, which is present in the VIM-2 sequence, defined a precise positively charged space for substrate binding by forming with its side chain a kind of “wall” that encloses one side of the active site (11). The presence at position 228 of a Ser residue instead of an Arg residue may eliminate this physical restraint, consequently opening that area (11). This might explain the lower affinity for imipenem (higher Km values) of VIM-19 and VIM-2 compared to VIM-1, but the paradoxical better affinity of VIM-19 for meropenem compared to VIM-1 remains unexplained (Table 2 and Fig. 1). Note that our results also showed that the Arg228 residue in VIM-19 is responsible for a lower hydrolytic activity toward ceftazidime and cefepime.
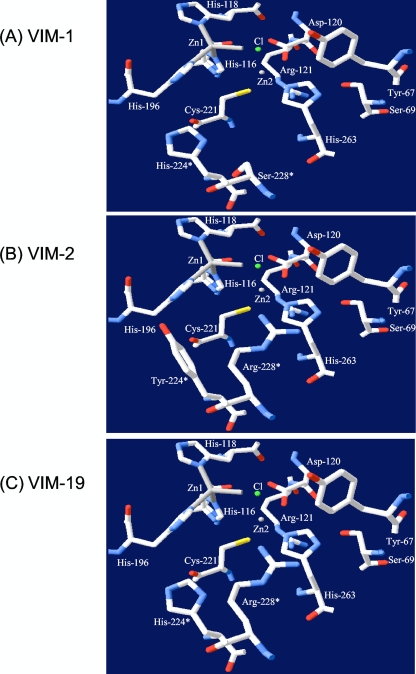
The VIM-1 and VIM-2 enzymes have been shown to possess two distinct three-dimensional (3D) structures, and their primary structures diverge significantly with 25 different amino acids (11). Those two enzymes exhibit differences in their respective His and Cys active sites (His224 and Ser228 for VIM-1 and Tyr224 and Arg228 for VIM-2). The corresponding residues in VIM-19 consist of His224 and Arg228, therefore being an hybrid active site between VIM-1 and VIM-2, which is also shared with VIM-4 and VIM-7 (27, 31). Considering previous kinetic studies comparing VIM-2 and VIM-1 (6, 8, 11, 24), it has been shown that the affinities of both enzymes for penicillins are different, but also that their affinities for carbapenems may differ, with VIM-1 hydrolyzing imipenem, cefepime, and cefpirome better than VIM-2, and conversely VIM-2 hydrolyzing meropenem more efficiently than VIM-1 (32). The data obtained for VIM-19 compared to VIM-1 are in accordance with those of previous observations, considering that the active site of VIM-19 resembles the active site of VIM-2 more than that of VIM-1 (Fig. 2).
FIG. 2.
Comparative lateral view of the active site (His and Cys sites) of VIM-1 (A), VIM-2 (B) (Protein Data Bank entry 1ko3) (11), and VIM-19 (C). The asterisks indicate the positions of the two residues which differ between VIM-1 (His224 and Ser228), VIM-2 (Tyr224 and Arg228), and VIM-19 (His224 and Arg228). The representation has been determined using the software Swiss-Pdb Viewer available at www.expasy.org/spdbv/ (12, 19, 20). The colors of the atoms are standard for the program and are as follows: white, carbon; red, oxygen; blue, nitrogen; yellow, sulfur; gray, zinc ion; green sphere, chloride ion. Residues are numbered according to the BBL standard numbering scheme (9).
This study identified MBL producers for the first time in Algeria, thus indicating a likely dissemination in North Africa, whereas VIM-2 and VIM-4 have been reported in Tunisia (13, 17). Its plasmid and integron location may be the source of its spread as exemplified by its identification in two different enterobacterial species. The identification of VIM-19 constitutes the first example of an evolution of a VIM-type enzyme toward increased hydrolytic activity, which results from just a single amino acid substitution. A similar observation had been made among the IMP-type MBL family, with the IMP-6 variant differing from IMP-1 by a Ser196Gly substitution (thus another location inside the protein structure) and consequently possessing the ability to hydrolyze meropenem more efficiently (33).
Acknowledgments
This work was funded mostly by grants from the European Community (LSHM-CT-2005-018705 and TROCAR, HEALTH-F3-2008-223031) and INSERM, but it was also funded by a grant from the Ministère de l'Education Nationale et de la Recherche (UPRES-EA3539), Université Paris XI, Paris, France. J.-M.R.-M. was funded by a postdoctoral grant from the Ministerio de Educación y Ciencia from Spain (2007/0292).
We thank S. Bernabeu for excellent technical assistance.
Footnotes
Published ahead of print on 16 November 2009.
REFERENCES
- 1.Bellais, S., D. Aubert, T. Naas, and P. Nordmann. 2000. Molecular and biochemical heterogeneity of class B carbapenem-hydrolyzing β-lactamases in Chryseobacterium meningosepticum. Antimicrob. Agents Chemother. 44:1878-1886. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Clinical and Laboratory Standards Institute. 2008. Performance standards for antimicrobial susceptibility testing. 18th informational supplement. Clinical and Laboratory Standards Institute, Wayne, Pa.
- 3.Cornaglia, G., M. Akova, G. Amicosante, R. Canton, R. Cauda, J.-D. Docquier, M. Edelstein, J.-M. Frère, M. Fuzi, M. Galleni, H. Giamarellou, M. Gniadkowski, R. Koncan, B. Libisch, F. Luzzaro, V. Miriagou, F. Navarro, P. Nordmann, L. Pagani, L. Peixe, L. Poirel, M. Souli, E. Tacconelli, A. Vatopoulos, and G. M. Rossolini. 2007. Metallo-β-lactamases as emerging resistance determinants in Gram-negative pathogens: open issues. Int. J. Antimicrob. Agents 29:380-388. [DOI] [PubMed] [Google Scholar]
- 4.Corvec, S., L. Poirel, J.-W. Decousser, P.-Y. Allouch, H. Drugeon, and P. Nordmann. 2006. Emergence of carbapenem-hydrolysing metallo-β-lactamase VIM-1 in Pseudomonas aeruginosa isolates in France. Clin. Microbiol. Infect. 12:941-942. [DOI] [PubMed] [Google Scholar]
- 5.Crowder, M. W., J. Spencer, and A. J. Vila. 2006. Metallo-β-lactamases: novel weaponry for antibiotic resistance in bacteria. Acc. Chem. Res. 39:721-728. [DOI] [PubMed] [Google Scholar]
- 6.Docquier, J.-D., J. Lamotte-Brasseur, M. Galleni, G. Amicosante, J.-M. Frère, and G. M. Rossolini. 2003. On functional and structural heterogeneity of VIM-type metallo-β-lactamases. J. Antimicrob. Chemother. 51:257-266. [DOI] [PubMed] [Google Scholar]
- 7.Figueiredo, S., L. Poirel, A. Papa, V. Koulourida, and P. Nordmann. 2008. First identification of VIM-4 metallo-beta-lactamase in Acinetobacter spp. Clin. Microbiol. Infect. 14:289-290. [DOI] [PubMed] [Google Scholar]
- 8.Franceschini, N., B. Caravelli, J.-D. Docquier, M. Galleni, J.-M. Frère, G. Amicosante, and G. M. Rossolini. 2000. Purification and biochemical characterization of the VIM-1 metallo-β-lactamase. Antimicrob. Agents Chemother. 44:3003-3007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Galleni, M., J. Lamotte-Brasseur, G. M. Rossolini, J. Spencer, O. Dideberg, and J.-M. Frère. 2001. Standard numbering scheme for class B β-lactamases. Antimicrob. Agents Chemother. 45:660-663. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Garau, G., I. García-Sáez, C. Bebrone, C. Anne, P. Mercuri, M. Galleni, J.-M. Frère, and O. Dideberg. 2004. Update of the standard numbering scheme for class B β-lactamases. Antimicrob. Agents Chemother. 48:2347-2349. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Garcia-Saez, I., J.-D. Docquier, G. M. Rossolini, and O. Dideberg. 2008. The three-dimensional structure of VIM-2, a Zn-β-lactamase from Pseudomonas aeruginosa in its reduced and oxidised form. J. Mol. Biol. 375:604-611. [DOI] [PubMed] [Google Scholar]
- 12.Guex, N., and M. C. Peitsch. 1997. SWISS-MODEL and the Swiss-PdbViewer: an environment for comparative protein modeling. Electrophoresis 18:2714-2723. [DOI] [PubMed] [Google Scholar]
- 13.Ktari, S., G. Arlet, B. Mnif, V. Gautier, F. Mahjoubi, J. M. Ben, M. Bouaziz, and A. Hammami. 2006. Emergence of multidrug-resistant Klebsiella pneumoniae isolates producing VIM-4 metallo-β-lactamase, CTX-M-15 extended-spectrum β-lactamase, and CMY-4 AmpC β-lactamase in a Tunisian university hospital. Antimicrob. Agents Chemother. 50:4198-4201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Lee, K., J. H. Yum, D. Yong, H. M. Lee, H. D. Kim, J.-D. Docquier, G. M. Rossolini, and Y. Chong. 2005. Novel acquired metallo-β-lactamase gene, blaSIM-1, in a class 1 integron from Acinetobacter baumannii clinical isolates from Korea. Antimicrob. Agents Chemother. 49:4485-4491. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Mammeri, H., L. Poirel, and P. Nordmann. 2003. In vivo selection of a chromosomally encoded β-lactamase variant conferring ceftazidime resistance in Klebsiella oxytoca. Antimicrob. Agents Chemother. 47:3739-3742. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Mammeri, H., M. Van de Loo, L. Poirel, L. Martinez-Martinez, and P. Nordmann. 2005. Emergence of plasmid-mediated quinolone resistance in Escherichia coli in Europe. Antimicrob. Agents Chemother. 49:71-76. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Mansour, W., L. Poirel, D. Bettaieb, O. Bouallegue, N. Boujaafar, and P. Nordmann. 2009. Metallo-β-lactamase-producing Pseudomonas aeruginosa isolates in Tunisia. Diagn. Microbiol. Infect. Dis. 64:458-461. [DOI] [PubMed] [Google Scholar]
- 18.Marchiaro, P., P. E. Tomatis, M. A. Mussi, F. Pasteran, A. M. Viale, A. S. Limansky, and A. J. Vila. 2008. Biochemical characterization of metallo-β-lactamase VIM-11 from a Pseudomonas aeruginosa clinical strain. Antimicrob. Agents Chemother. 52:2250-2252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Peitsch, M. C. 1996. ProMod and Swiss-Model: Internet-based tools for automated comparative protein modelling. Biochem. Soc. Trans. 24:274-279. [DOI] [PubMed] [Google Scholar]
- 20.Peitsch, M. C., T. N. Wells, D. R. Stampf, and J. L. Sussman. 1995. The Swiss-3DImage collection and PDB-Browser on the World-Wide Web. Trends Biochem. Sci. 20:82-84. [DOI] [PubMed] [Google Scholar]
- 21.Philippon, L. N., T. Naas, A. T. Bouthors, V. Barakett, and P. Nordmann. 1997. OXA-18, a class D clavulanic acid-inhibited extended-spectrum β-lactamase from Pseudomonas aeruginosa. Antimicrob. Agents Chemother. 41:2188-2195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Pitout, J. D., G. Revathi, B. L. Chow, B. Kabera, S. Kariuki, P. Nordmann, and L. Poirel. 2008. Metallo-β-lactamase-producing Pseudomonas aeruginosa isolated from a large tertiary centre in Kenya. Clin. Microbiol. Infect. 14:755-759. [DOI] [PubMed] [Google Scholar]
- 23.Poirel, L., A. Carrër, J. D. Pitout, and P. Nordmann. 2009. Integron mobilization unit as a source of mobility of antibiotic resistance genes. Antimicrob. Agents Chemother. 53:2492-2498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Poirel, L., T. Naas, D. Nicolas, L. Collet, S. Bellais, J.-D. Cavallo, and P. Nordmann. 2000. Characterization of VIM-2, a carbapenem-hydrolyzing metallo-β-lactamase and its plasmid- and integron-borne gene from a Pseudomonas aeruginosa clinical isolate in France. Antimicrob. Agents Chemother. 44:891-897. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Poirel, L., J. D. Pitout, and P. Nordmann. 2007. Carbapenemases: molecular diversity and clinical consequences. Future Microbiol. 2:501-512. [DOI] [PubMed] [Google Scholar]
- 26.Poirel, L., G. F. Weldhagen, T. Naas, C. De Champs, M. G. Dove, and P. Nordmann. 2001. GES-2, a class A β-lactamase from Pseudomonas aeruginosa with increased hydrolysis of imipenem. Antimicrob. Agents Chemother. 45:2598-2603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Pournaras, S., A. Tsakris, M. Maniati, L. S. Tzouvelekis, and A. N. Maniatis. 2002. Novel variant (blaVIM-4) of the metallo-β-lactamase gene blaVIM-1 in a clinical strain of Pseudomonas aeruginosa. Antimicrob. Agents Chemother. 46:4026-4028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Rossolini, G. M., E. Mantengoli, J. D. Docquier, R. A. Musmanno, and G. Coratza. 2007. Epidemiology of infections caused by multiresistant gram-negatives: ESBLs, MBLs, panresistant strains. New Microbiol. 30:332-339. [PubMed] [Google Scholar]
- 29.Sekiguchi, J., K. Morita, T. Kitao, N. Watanabe, M. Okazaki, T. Miyoshi-Akiyama, M. Kanamori, and T. Kirikae. 2008. KHM-1, a novel plasmid-mediated metallo-β-lactamase from a Citrobacter freundii clinical isolate. Antimicrob. Agents Chemother. 52:4194-4197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Toleman, M. A., P. M. Bennett, and T. R. Walsh. 2006. ISCR elements: novel gene-capturing systems of the 21st century? Microbiol. Mol. Biol. Rev. 70:296-316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Toleman, M. A., K. Rolston, R. N. Jones, and T. R. Walsh. 2004. blaVIM-7, an evolutionarily distinct metallo-β-lactamase gene in a Pseudomonas aeruginosa isolate from the United States. Antimicrob. Agents Chemother. 48:329-332. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Walsh, T. R., M. A. Toleman, L. Poirel, and P. Nordmann. 2005. Metallo-β-lactamases: the quiet before the storm? Clin. Microbiol. Rev. 18:306-325. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Yano, H., A. Kuga, R. Okamoto, H. Kitasato, T. Kobayashi, and M. Inoue. 2001. Plasmid-encoded metallo-β-lactamase (IMP-6) conferring resistance to carbapenems, especially meropenem. Antimicrob. Agents Chemother. 45:1343-1348. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Yong, D., M. A. Toleman, C. G. Giske, H. S. Cho, K. Sundman, K. Lee, and T. R. Walsh. 21 September 2009. Characterization of a new metallo-β-lactamase gene, blaNDM-1, and a novel erythromycin esterase gene carried on a unique genetic structure in Klebsiella pneumoniae sequence type 14 from India. Antimicrob. Agents Chemother. doi: 10.1128/AAC.00774-09. [DOI] [PMC free article] [PubMed]