Abstract
To investigate the response of Streptococcus pneumoniae to three distinct antimicrobial peptides (AMPs), bacitracin, nisin, and LL-37, transcriptome analysis of challenged bacteria was performed. Only a limited number of genes were found to be up- or downregulated in all cases. Several of these common highly induced genes were chosen for further analysis, i.e., SP0385-SP0387 (SP0385-0387 herein), SP0912-0913, SP0785-0787, SP1714-1715, and the blp gene cluster. Deletion of these genes in combination with MIC determinations showed that several putative transporters, i.e., SP0785-0787 and SP0912-0913, were indeed involved in resistance to lincomycin and LL-37 and to bacitracin, nisin, and lincomycin, respectively. Mutation of the blp bacteriocin immunity genes resulted in an increased sensitivity to LL-37. Interestingly, a putative ABC transporter (SP1715) protected against bacitracin and Hoechst 33342 but conferred sensitivity to LL-37. A GntR-like regulator, SP1714, was identified as a negative regulator of itself and two of the putative transporters. In conclusion, we show that resistance to three different AMPs in S. pneumoniae is mediated by several putative ABC transporters, some of which have not been associated with antimicrobial resistance in this organism before. In addition, a GntR-like regulator that regulates two of these transporters was identified. Our findings extend the understanding of defense mechanisms of this important human pathogen against antimicrobial compounds and point toward novel proteins, i.e., putative ABC transporters, which can be used as targets for the development of new antimicrobials.
Increased resistance of bacteria to commonly used antibiotics creates severe problems in treating infectious diseases. The resistance of one of the most important human pathogens, Streptococcus pneumoniae, to commonly used antibiotics has increased significantly in recent decades (15). This bacterium colonizes the nasopharynx and the upper respiratory tract asymptomatically. Nevertheless, under certain circumstances, S. pneumoniae can cause otitis media, meningitis, pneumonia, and sepsis (49). To cause disease, S. pneumoniae has to successfully colonize the mucosal surface of the nasopharynx, followed by dissemination to other parts of the human body. Mucosal surfaces of the human body form the first barrier that protects against pathogens. In this layer, mainly neutrophils and epithelial cells produce antimicrobial peptides (AMPs). Generally, AMPs display a cationic and an amphipathic nature, but they are variable in sequence, secondary structure, size, and mode of action (56). Antimicrobial peptides play an essential role in the host's innate immune response (32).
One human AMP, the 18-kDa human cathelicidin antimicrobial protein hCAP-18 (16), is produced as an inactive preproprotein that consists of a precursor protein, cathelin, and a carboxy-terminal peptide, LL-37 (64). LL-37 is a linear, 37-amino-acid-long cationic peptide with activity against Gram-positive and Gram-negative bacteria (76). It has been shown that the bactericidal action of LL-37 is due to immobilization of the peptide within the membrane lipid bilayer, where, as a consequence, it causes destabilization of the bacterial membrane (53).
In addition to coping with the human immune system, S. pneumoniae has to compete with other bacterial inhabitants, which also produce AMPs as a defense against competitors, to achieve successful colonization of the nasopharynx. AMPs generated by Gram-positive bacteria are named bacteriocins, and one of the best-characterized ones is nisin, produced by Lactococcus lactis and commonly used as a food preservative (59). The antimicrobial activity of nisin is rather broad against Gram-positive bacteria (17, 48). Nisin is able to inhibit peptidoglycan biosynthesis by interaction with lipid II and forms pores in bacterial membranes, which leads to cell death (7, 8, 21). Another attack and defense system used by bacteria is the production of antibiotics such as bacitracin. This toxic compound is a mixture of cyclic polypeptides produced by Bacillus licheniformis (28). Bacitracin is a nonribosomally synthesized antibiotic which, in Gram-positive cocci and bacilli, blocks biosynthesis of the bacterial cell wall by interaction with C55-isoprenyl pyrophosphate (2, 25, 65, 66).
To establish whether S. pneumoniae contains general defense mechanisms against heterologous AMPs, transcriptome analysis of S. pneumoniae D39 was performed upon challenge with three different antimicrobial peptides, i.e., LL-37, nisin, and bacitracin. The transcript levels of genes involved in various processes, such as gene regulation, transport, virulence, fatty acid synthesis, and phosphotransferase systems, had changed significantly. Several highly induced genes were chosen for further analysis. We show, for the first time to our knowledge, that some of these genes, encoding putative ABC transporters, are involved in the defense of S. pneumoniae against multiple antimicrobial compounds, e.g., bacitracin, nisin, LL-37, lincomycin, or Hoechst 33342. Furthermore, we demonstrate that the putative regulatory protein SP1714 is a repressor of its own expression and that of two putative ABC transporter genes, one of which belongs to another operon. In summary, these results give new insight into the transcriptional stress response of S. pneumoniae to structurally different AMPs and enable the identification of common features of the molecular defense mechanisms against various antimicrobial substances in this organism. This will eventually lead to the selection and/or design of more suitable antimicrobial agents and the development of more effective preventive measures.
MATERIALS AND METHODS
Bacteria and growth conditions.
The strains used in this study are listed in Table 1 and were stored in 10% glycerol at −80°C. Streptococcus pneumoniae strains were grown at 37°C in standing Todd-Hewitt (Oxoid) broth supplemented with 0.5% yeast extract (THY) and/or on M17 agar (69) containing 0.25% glucose (GM17) and 3% defibrinated sheep blood (Johnny Rottier, Kloosterzande, The Netherlands). Lactococcus lactis was grown in GM17 without agitation at 30°C. Escherichia coli was grown in TY (tryptone-yeast extract) medium at 37°C with shaking. Where appropriate, media were supplemented with the following antibiotics (final concentrations shown in parentheses): erythromycin and spectinomycin (0.25 μg/ml and 150 μg/ml, respectively, for S. pneumoniae), chloramphenicol (2 μg/ml for S. pneumoniae and 4 μg/ml for L. lactis), tetracycline (2.5 μg/ml for S. pneumoniae), trimethoprim (18 μg/ml for S. pneumoniae), and ampicillin (100 μg/ml for E. coli). Nisin (Sigma) was used for induction of gene expression at a concentration of 5 ng/ml.
TABLE 1.
Strains and plasmids used in this study
| Strain or plasmid | Descriptiona | References or source |
|---|---|---|
| S. pneumoniae strains | ||
| D39 | Serotype 2 strain, cps2 | 1, 39; source was the group of P. W. Hermans |
| D39nisRK | D39 ΔbgaA::nisRK; Tmpr | 29 |
| Δ385-387 | D39 ΔSP0385-0387; Sptr | This work |
| Δ785-787 | D39 ΔSP0785-0787; Eryr | This work |
| Δ912-913 | D39 ΔSP0912-0913; Eryr | This work |
| Δ1714-1715 | D39 ΔSP1714-1715; Eryr | This work |
| Δ1715 | D39 ΔSP1715; Eryr | This work |
| Δblp strain | D39 ΔSPD0473-0476; Eryr | This work |
| OV912 | D39 nisRK/pNZ912; Cmr | This work |
| OV1715 | D39 nisRK/pNZ1715; Cmr | This work |
| CO912 | OV912 Δ912-913 | This work |
| CO1715 | OV1715 Δ1715 | This work |
| CO1716 | OV1715 Δ1714-1715 | This work |
| DM39 | Δ385-387 Δ912-913 | This work |
| DM19 | Δ912-913 Δ1714-1715 | This work |
| PR385 | D39 ΔbgaA::PSP0385-lacZ; Tetr | This work |
| PR785 | D39 ΔbgaA::PSP0785-lacZ: Tetr | This work |
| PR912 | D39 ΔbgaA::PSP0912-lacZ; Tetr | This work |
| PR1714 | D39 ΔbgaA::PSP1714-lacZ; Tetr | This work |
| PR785Δ1714 | PR785/Δ1714-1715 | This work |
| PR912Δ1714 | PR912/Δ1714-1715 | This work |
| PR1714Δ1714 | PR1714/Δ1714-1715 | This work |
| E. coli EC1000 | MC1000 derivative carrying a single copy of the pWV01 repA gene in glgB; Kmr | 40 |
| L. lactis NZ9000 | MG1363 ΔpepN::nisRK | 35 |
| Plasmids | ||
| pPP2 | Promoterless lacZ, for replacement of bgaA (spr0565) with promoter-lacZ fusions, derivative of pPP1; Ampr Tetr | 19 |
| pNZ8048 | Nisin-inducible PnisA; Cmr | 13 |
| pPA1 | pPP2 PSP0385-lacZ | This work |
| pPA2 | pPP2 PSP0785-lacZ | This work |
| pPA3 | pPP2 PSP0912-lacZ | This work |
| pPA4 | pPP2 PSP1714-lacZ | This work |
| pNZ912 | pNZ8048 carrying SP0912-0913 downstream of PnisA | This work |
| pNZ1715 | pNZ8048 carrying SP1715 downstream of PnisA | This work |
Eryr, erythromycin resistance; Cmr, chloramphenicol resistance; Tetr, tetracycline resistance; Sptr, spectinomycin resistance; Tmpr, trimethoprim resistance.
Aliquots of S. pneumoniae D39 cultures were prepared as follows. Overnight cultures of D39 grown in THY were diluted 1:100 in the same medium. Subsequently, the bacteria were grown at 37°C until an optical density at 600 nm (OD600) of ≈0.25 and stored at −80°C in 1-ml aliquots with 10% glycerol (vol/vol).
Antimicrobial agents.
Stock solutions of antimicrobial peptides/agents were stored in aliquots at −20°C. The solutions of bacitracin (Sigma), Hoechst 33342 [2′-(4-ethoxyphenyl)-5-(4-methyl-1-piperazinyl)-2,5′-bi-1H-bezimidazole; Molecular Probes, Inc.], gramicidin (Sigma), lincomycin (Sigma), vancomycin (Sigma), daunomycin (Sigma), and ethidium bromide (Sigma) were prepared in MilliQ water. The stock solution of nisin (Sigma) was prepared in 0.05% acetic acid and that of LL-37 (Innovagen) in 0.01% acetic acid with 0.01% bovine serum albumin (NEB). Dilutions of each antimicrobial compound were always freshly prepared from these stocks.
Strain construction.
Strains, plasmids, and oligonucleotide primers used in this study are listed in Tables 1 and 2. The genome sequence of S. pneumoniae D39 was used to design all primers (39). All the indicated PCR fragments and plasmids were introduced into S. pneumoniae D39 as described previously (29, 58). S. pneumoniae clones were selected on GM17 agar with the appropriate antibiotic(s). L. lactis and E. coli were transformed by electroporation as described before (22). All constructs and deletions were verified by DNA sequencing.
TABLE 2.
Oligonucleotide primers used in this study
| Oligonucleotide primer | Nucleotide sequence (5′ to 3′)a | Restriction site |
|---|---|---|
| KN-sp912-913-for-1 | GGAAGCCAGCCACAGGCTGTA | |
| KN-sp912-913-rev-2 | GAGATCTAATCGATGCATGCGTGTCATGAGAATCTCCTTTC | |
| KN-sp912-913-for-3 | AGTTATCGGCATAATCGTTACTTCCTCATCGCCTATGTGCTG | |
| KN-sp912-913-rev-4 | CGTAGATGGTTACCTAAGGGAACC | |
| KN-sp785-787-for-1 | TGACAGGGACTTTGTGAGTGTG | |
| KN-sp785-787-rev-2 | GAGATCTAATCGATGCATGCCCCTCCAGCAAACAATACA | |
| KN-sp785-787-for-3 | AGTTATCGGCATAATCGTCAACAAGATGGACACTCGTCT | |
| KN-sp785-787-rev-4 | GGAAGACTGTTCCATTCCAGAA | |
| KN-sp385-387-for-1 | GTGCCACCATAGCAGATCTACAA | |
| KN-sp385-387-rev-2 | CCTCCTCACTATTTTGATTAGTATGAGAGCAATAATGACATAGGC | |
| KN-sp385-387-for-3 | TGGGAAATATTCATTCTAATTGGCCATTTGGTGGGGCAAGAGGAG | |
| KN-sp385-387-rev-4 | TCACGCTAGAGGTACTTGCTTGC | |
| KN-sp1714-1715-for-1 | TCAGTGCCTCCTGACCGATAATCGGG | |
| KN-sp1714-1715-rev-2 | GAGATCTAATCGATGCATGCTTGGTCTCCTTTCTCTTACCC | |
| KN-sp1714-1715-for-3 | AGTTATCGGCATAATCGTTACTCGGAACCTACTACATCTTGA | |
| KN-sp1714-1715-rev-4 | GTGACAGCTCTAGGTGCAGCT | |
| KN-sp1715-for-1 | CTTGACACAGGACGTTTCTGGGCT | |
| KN-sp1715-rev-2 | GAGATCTAATCGATGCATGCCATTTTCAAATGCTAGTAATGACAT | |
| KN-sp1715-for-3 | AGTTATCGGCATAATCGTTACTCGGAACCTACTACATCTTGA | |
| KN-sp1715-rev-4 | GTGACAGCTCTAGGTGCAGCT | |
| KN-blp-for-1 | CTCATCCAAGATTCCTTGGAGAT | |
| KN-blp-rev-2 | GAGATCTAATCGATGCATGCAGCCACCTCTATTTCAAGCCACC | |
| KN-blp-for-3 | AGTTATCGGCATAATCGTCGAGACAAGTATGGAAAGAG | |
| KN-blp-rev-4 | CAAAGCGTTCTACTGTACCAGACAT | |
| Oversp912-913;fv | CATGCCATGGCACTTTTAGATGTAAAACACG | NcoI |
| Oversp912-913;rev | GCTCTAGAATACCTCGATTTTGAAGTCGAGG | XbaI |
| Oversp1715;fv | CATGCCATGGCATTACTAGCATTTGAAAATG | NcoI |
| Oversp1715;rev | GCTCTAGATGAGTATGTTACATATCTAGG | XbaI |
| Psp385-387-fv | CGGAATTCGTGCCACCATAGCAGATCTACA | EcoRI |
| Psp385-387-rev | GCTCTAGACTCATAGGTTCATCCTCTCCCT | XbaI |
| Psp785-787-fv | CGGAATTCTCCGCTACCTCCACCGATAGCAAT | EcoRI |
| Psp785-787-rev | GCTCTAGACTTCATAATGAAACTCCTTTTC | XbaI |
| Psp912-913-fv | CGGAATTCTGGATGCTGATAACAACTGATAAC | EcoRI |
| Psp912-913-rev | GCTCTAGAGTGTCATGAGAATCTCCTTTCT | XbaI |
| Psp1714-1715-fv | CGGAATTCCTACGAATGGTGTTCCCTTCT | EcoRI |
| Psp1714-1715-rev | GCTCTAGATGTCAAATGTCCAGGACATC | XbaI |
Restriction enzyme sites are underlined.
Construction of deletion strains.
The knockout of the SP0385-SP0387 (SP0385-0387 herein) genes was made with primer pairs KN-sp385-387-for-1/KN-sp385-387-rev-2 and KN-sp385-387-for-3/KN-sp385-387-rev-4 by overlap extension PCR, as described by Song et al. (63), and allelic replacement with a spectinomycin resistance cassette, yielding strain Δ385-387 (63). The deletion strains with an erythromycin resistance cassette and deletion of SP0785-0787 (yielding strain Δ785-787), SP0912-0913 (yielding strain Δ912-913), SP1714-1715 (yielding strain Δ1714-1715; equivalent genes in S. pneumoniae D39 are SPD_1524-1526), SP1715 (yielding strain Δ1715; equivalent genes in S. pneumoniae D39 are SPD_1525-1526), and blp (yielding the Δblp strain; equivalent genes in S. pneumoniae D39 are SPD_0473-0476) were made similarly to the SP0385-0387 mutant, using primer pairs KN-sp785-787-for-1/KN-sp785-787-rev-2 and KN-sp785-787-for-3/KN-sp785-787-rev-4, KN-sp912-913-for-1/KN-sp912-913-rev-2 and KN-sp912-913-for-3/KN-sp912-913-rev-4, KN-sp1714-1715-for-1/KN-sp1714-1715-rev-2 and KN-sp1714-1715-for-3/KN-sp1714-1715-rev-4, KN-sp1715-for-1/KN-sp1715-rev-2 and KN-sp1715-for-3/KN-sp1715-rev-4, and KN-blp-for-1/KN-blp-rev-2 and KN-blp-for-3/KN-blp-rev-4, respectively.
Construction of lacZ fusions.
To construct the chromosomal transcriptional fusions of lacZ to the putative promoters of the presumed operons SP0385-0387, SP0785-0787, SP0912-0913, and SP1714-1715, the putative promoter fragments were amplified from the chromosomal DNA of S. pneumoniae D39 with the primer pairs listed in Table 2. The putative promoter of the SP0385-0387 genes (fragment length, 167 nucleotides [nt]) was amplified with the primer pair Psp385-387-fv/Psp385-387-rev, the putative promoter of the SP0785-0787 genes (fragment length, 311 nt) was amplified with the primer pair Psp785-787-fv/Psp785-787-rev, the putative promoter of the SP0912-0913 genes (fragment length, 585 nt) was amplified with the primer pair Psp912-913-fv/Psp912-913-rev, and the putative promoter of the SP1714-1715 genes (fragment length, 237 nt) was amplified with the primer pair Psp1714-1715-fv/Psp1714-1715-rev. The fragments obtained were cloned into the EcoRI/XbaI sites of pPP2, giving rise to pPA1, pPA2, pPA3, and pPA4. These plasmids were transformed into S. pneumoniae D39 to generate the PR385, PR785, PR912, and PR1714 strains. In addition, the introduction of plasmids PA2, PA3, and PA4 into a Δ1714-1715 mutant resulted in the PR785Δ1714, PR912Δ1714, and PR1714Δ1714 strains, respectively.
Construction of overexpression plasmids.
For overexpression of SP0912-0913 and SP1715 with the nisin-inducible system (13, 35), these genes were amplified with the primer pairs Oversp912-913;fv/Oversp912-913;rev and Oversp1715;fv/Oversp1715;rev, respectively, and were fused to NcoI/XbaI sites in pNZ8048, yielding pNZ912 and pNZ1715. These plasmids were transformed into S. pneumoniae D39, generating the OV912 and OV1715 strains. For the complementation assay, pNZ912 was transformed into the Δ912 strain, yielding strain CO912, and pNZ1715 was transformed into strains Δ1714-1715 and Δ1715, yielding strains CO1716 and CO1715, respectively.
DNA microarrays and transcriptional profiling.
DNA microarrays were produced and analyzed as described before (36, 78).
Experimental design.
One-milliliter aliquots of S. pneumoniae D39 (OD600 of ∼0.25) were used to inoculate 100 ml of THY medium and were grown at 37°C until early logarithmic phase (OD600 of ∼0.25). Subsequently, cultures were split in two and exposed to 0.7 μg/ml bacitracin, 0.1 μg/ml nisin, or 4.5 μg/ml LL-37 (end concentrations) for 15 (early response) and 30 (late response) min. These concentrations of AMPs were chosen based on the results of growth experiments performed with all three AMPs and gave a 10% reduction of the maximal OD compared to that with no AMP. In this manner, the bacteria were stressed with the AMPs but not killed to a great extent, because this would negatively influence the quality of the RNA for the transcriptome experiments. For each AMP, three replicates were performed, and as a control, bacteria without any AMP were used.
RNA isolation, cDNA preparation, and hybridization.
RNA was isolated from 50 ml of three independent cultures for each condition. After centrifugation, pellets were frozen in liquid nitrogen and stored at −80°C. Subsequently, pellets were resuspended in 500 μl of 10 mM Tris-HCl and 1 mM EDTA (pH 8.0), after which 50 μl of 10% sodium dodecyl sulfate, 500 μl of phenol-chloroform-isoamyl alcohol (24:24:1), 500 mg of glass beads (Sigma; 75 to 150 μm), and 175 μl of Macaloid solution (Bentone) were added. RNA was isolated with a High Pure RNA isolation kit (Roche). Subsequently, cDNA was obtained from 15 to 20 μg of total RNA and the Cy3/Cy5-dCTP labeling of cDNA was performed with a CyScribe postlabeling kit (Amersham Biosciences). Hybridization was carried out at 45°C for 16 h in Ambion slide hybridization buffer (Ambion Europe) on superamine glass slides (Array-It; SMMBC). The slides contained replicates of amplicons of 2,087 open reading frames (ORFs) of S. pneumoniae TIGR4 and 184 unique ORFs of S. pneumoniae R6. Amplicon sequences are available on the World Wide Web at http://molgen.biol.rug.nl. Slides were scanned using a GeneTac LSIV confocal laser scanner (Genomics Solutions).
Data analysis.
ArrayPro 4.5 (Media Cybernetics, Inc., Silver Spring, MD) was used to analyze the data. For the processing and normalization of the data, the MicroPrep software was used as described previously (78, 79). Genes with a Bayes P value of <0.0001 and with a differential expression greater than or equal to 1.2 or lower than or equal to 0.8 were considered significantly differentially expressed.
β-Galactosidase assays.
S. pneumoniae strains were incubated at 37°C in THY and grown to early logarithmic phase (OD600 of ∼0.25). Subsequently, D39 derivatives were incubated for 15 (data not shown), 30, and 90 min with or without 0.7 μg/ml bacitracin, 0.1 μg/ml nisin, or 4.5 μg/ml LL-37 (the same end concentrations of these AMPs were used for transcriptome analyses). Next, the pellets were collected and β-galactosidase assays were performed as described previously by Israelsen et al. (26), with the following modifications. Two milliliters of the cell cultures were centrifuged; pellets were resuspended in 250 μl Z buffer (60 mM Na2HPO4·2H2O, 40 mM NaH2PO4·H2O, 10 mM KCl, MgSO4·7H2O) and 15 μl (final concentration, 0.06 mg/ml) cetyltrimethyl ammonium bromide and incubated for 5 min at 30°C. The assay was started by the addition of 50 μl of 4 mg/ml ONPG (O-nitrophenyl β-d-galactopyranoside; Sigma) and stopped by the addition of 250 μl of Na2CO3 (1 M).
Determination of MICs.
Determination of the MICs of the various compounds for S. pneumoniae D39 and the mutants was performed in 96-well microtiter plates. Incubation took place in a microplate reader (GENios; Tecan). Aliquots of strains OV912, OV1715, CO912, CO1715, and CO1716 were made using THY broth with an induction concentration of nisin (5 ng/ml) for the nisin-inducible expression of the genes of interest. For the MIC assays, the aliquots were thawed, spun down, and resuspended in fresh THY broth. The medium of strains OV912, OV1715, CO912, CO1715, and CO1716 was again supplemented with the induction concentration of nisin. Exponentially growing strains (at an OD600 of ∼0.2) were added into the wells of microtiter plates at a total volume of 200 μl/well with increasing concentrations of the antimicrobial substance being tested. The microtiter plates were incubated at 37°C for overnight growth, and the OD600 was measured every 30 min. The MICs were determined when the reference strains (cells without antimicrobial substance) reached half of the maximal optical density. MICs were calculated from the lowest concentration of the antimicrobial substance that was able to inhibit the growth of the tested strain. Strains were grown in the absence of the antibiotics used to select for the genetic modifications of S. pneumoniae to prevent any influence on the MIC. Pneumococcal strains with pNZ8048, a negative control for the overexpression strains, showed no change in susceptibility to the tested drugs (data not shown). To control for a positive influence of the nisin induction on the susceptibility of the strains carrying overexpression vectors, these strains were also examined in the MIC assay without nisin induction, but no change in the susceptibility was observed compared to that with nisin (data not shown). All the susceptibility assays were performed at least in triplicate.
Microarray data accession number.
The DNA microarray data were submitted to the GEO database and are available under accession number GSE16491.
RESULTS
Genome-wide identification of S. pneumoniae genes responding to bacitracin, nisin, or LL-37 challenge.
Nisin, bacitracin, and LL-37 differ in structure and mode of action, but their targets, subunits of the bacterial cell envelope, are thought to be similar. To investigate whether there is a general stress response of S. pneumoniae to different AMPs, transcriptome analyses of strain D39 exposed for 15 and 30 min to sublethal amounts of either bacitracin, nisin, or LL-37 were performed.
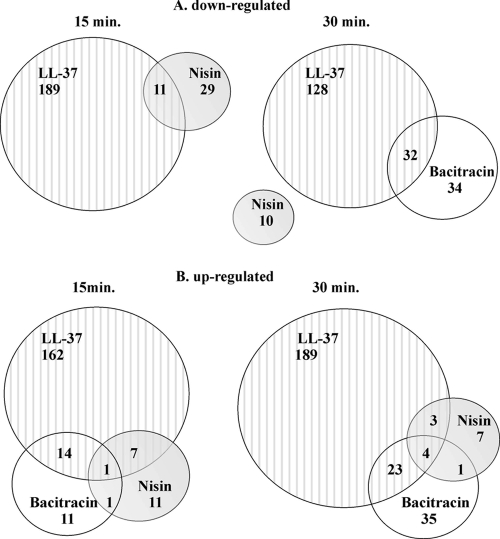
Exposure to all three AMPs resulted in significantly changed (Bayes P value of ≤0.0001 and fold change of ≤0.8 or ≥1.2) transcript levels of genes involved in various processes, such as regulation, transport, fatty acid biosynthesis, virulence, bacteriocin production, metabolic processes, protein fate, and phosphotransferase systems, and many genes encoding hypothetical proteins. LL-37 seemed to have the most profound influence on the transcriptome of S. pneumoniae D39, as the expression of ∼10% of the genome changed upon exposure. A complete overview of significantly up- and downregulated genes are in Table S1 in the supplemental material. The response to each individual AMP had a number of genes in common at both time points (see Table S2, section A, in the supplemental material), and several genes were differentially regulated upon challenge with more than one AMP at both 15 and 30 min (see Table S2, section B). Subsequently, we investigated how many significantly down- and upregulated genes were identified as common in each stress response to bacitracin, nisin, and LL-37 after two time points (Fig. 1; also see Table S3). The data revealed that treatment with nisin and LL-37 for 15 min resulted in only a few (11) downregulated genes in common (Fig. 1; also see Table S1, sections C and E, and Table S3, section A). Prolonging of the time of exposure to these two AMPs to 30 min did not yield any genes in common (Fig. 1A; also see Table S1, sections D and F). Interestingly, there were no downregulated genes identified when strain D39 was exposed for 15 min to bacitracin. After 30 min of treatment with this AMP, 66 genes were downregulated (see Table S1, section B), 32 of which were also downregulated by exposure to LL-37 for 30 min (Fig. 1A; also see Table S1, sections B and F, and Table S3, section A). Treatment with all three AMPs induced the expression of several common genes, the number of which increased with longer exposure (Fig. 1B; also see Table S3).
FIG. 1.
Venn diagrams indicating the numbers of genes downregulated (A) and upregulated (B) in the 15- and 30-min stress response of D39 to bacitracin, nisin, and LL-37. Numbers quantify the genes with significantly altered expression (Bayes P value of ≤0.0001; expression ratio greater than 1.2 or lower than 0.8) that were either shared or exclusive to each D39 response. Lists of genes in common indicated in this figure can be found in Table S3, sections A, B, and C, in the supplemental material.
Although bacitracin, nisin, and LL-37 are distinct antimicrobial compounds, the S. pneumoniae transcriptome response to them revealed certain analogous features. Since we were interested in genes that might be involved in the resistance mechanisms of D39 to two or all three AMPs, which are expected to be upregulated, we focused on the most interesting and prominently induced genes, which are described below.
Genes induced in the response to all three AMPs.
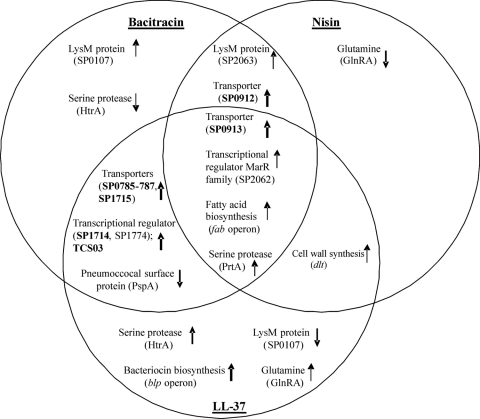
Comparison of the transcriptome profiles of S. pneumoniae D39 in response to bacitracin, nisin, and LL-37 revealed that gene SP0641, encoding the pneumococcal surface serine protease PrtA (6); gene SP2062, a member of the VicRK regulon (50, 52) encoding a putative transcriptional regulator of the MarR (multiple antibiotic resistance regulators) family; and genes SP0419 and SP0422, involved in fatty acid biosynthesis (41), were all moderately (SP0641, 1.3- to ∼3-fold; SP2062, 1.5- to ∼2-fold; SP0419, ∼1.8-fold to 2-fold; and SP0422, 1.4 to ∼2.2-fold) upregulated upon exposure to each AMP at either 15 or 30 min (Fig. 2; also see Table S3 in the supplemental material). Gene SP0913, encoding a permease protein, was induced moderately (2-fold) upon LL-37 treatment and up to 13-fold upon treatment with nisin and bacitracin. SP0912, an ATP-binding protein, was upregulated 9-fold upon nisin and bacitracin exposure, and it probably forms an ABC transporter with SP0913 (3) (Fig. 2 and Table 3). SP0912-0913 share amino acid identity with the known ABC transporters BceAB from Bacillus subtilis and MbrAB from Streptococcus mutans, which are known to be involved in resistance to bacitracin, and the YsaBC transporter from L. lactis that mediates a protective effect against nisin (34, 45, 74). SP0912 shares considerable identity with BceA (52%), MbrA (58%), and YsaC (61%), whereas SP0913 has a moderate identity with BceB (25%), MbrB (30%), and YsaB (31%). Thus, SP0912-0913 were chosen for further study.
FIG. 2.
General comparison of differentially and antagonistically expressed genes/gene products of strain D39 involved in regulation, virulence, and resistance mechanisms upon bacitracin, nisin, and LL-37 stress for 15 and/or 30 min. The direction of the arrow indicates up- or downregulation, and the thickness of the arrow indicates the strength of the differential expression.
TABLE 3.
Differential expression of genes selected for further analysis upon S. pneumoniae treatment for different times with bacitracin, nisin, and LL-37a
| TIGR4 locus tag | D39 locus tag | Putative/predicted function | Gene | Fold induction upon exposure for indicated time (min) to: |
|||||
|---|---|---|---|---|---|---|---|---|---|
| Bacitracin |
Nisin |
LL-37 |
|||||||
| 15 | 30 | 15 | 30 | 15 | 30 | ||||
| SP0385 | SPD0350 | Membrane protein | 2.1 | NDE | NDE | NDE | 1.5 | 1.6 | |
| SP0386 | SPD0351 | Sensor histidine kinase | hk03 | 2.4 | NDE | NDE | NDE | 1.5 | 1.7 |
| SP0387 | SPD0352 | DNA-binding response regulator | rr03 | 2.1 | NDE | NDE | NDE | 1.5 | 1.5 |
| SP0525 | SPD0467 | Regulatory protein | blpS | NDE | NDE | NDE | NDE | 1.6 | 1.8 |
| SP0526 | SPD0468 | Response regulator | blpR | NDE | NDE | NDE | NDE | 1.5 | 1.6 |
| SP0527 | SPD0469 | Histidine kinase | blpH | NDE | NDE | NDE | NDE | 1.5 | 1.8 |
| SP0528 | SPD0470 | Peptide pheromone | blpC | NDE | NDE | NDE | NDE | NDE | 1.8 |
| SP0529 | SPD0471 | ABC transporter, permease protein | blpB | NDE | NDE | NDE | NDE | 1.7 | 2.2 |
| SP0530 | SPD0472 | ABC transporter, ATP-binding protein | blpA | NDE | NDE | NDE | NDE | 3.4 | 3.6 |
| SP0533 | SPD0046b | Bacteriocin | blpK | NDE | NDE | NDE | NDE | 1.5 | 1.5 |
| SP0545 | SPD0473 | CAAX protease | blpY | NDE | NDE | NDE | NDE | 5.0 | 5.9 |
| SP0546 | SPD0474 | Immunity protein | blpZ | NDE | NDE | NDE | NDE | 3.5 | 3.8 |
| SP0547 | SPD0475 | CAAX protease | NDE | NDE | NDE | NDE | 3.7 | 4 | |
| SP0785 | SPD0686 | RND efflux-like protein | 1.8 | 2.1 | NDE | NDE | 4.1 | 5.5 | |
| SP0786 | SPD0687 | ABC transporter, ATP-binding protein | 1.9 | 2.3 | NDE | NDE | 5.1 | 5.0 | |
| SP0787 | SPD0688 | ABC transporter, permease protein | 1.9 | 2.4 | NDE | NDE | 4.6 | 6.1 | |
| SP0912 | SPD0804 | ABC transporter, ATP-binding protein | 8.2 | 8.7 | 9.1 | 6 | NDE | NDE | |
| SP0913 | SPD0805 | ABC transporter, permease protein | 12.4 | 9.6 | 13.3 | 11.8 | 1.9 | 1.8 | |
| SP1714 | SPD1524 | GntR transcriptional regulator | 2.9 | NDE | NDE | NDE | 7.2 | 9.1 | |
| SP1715 | SPD1525-1526c | ABC transporter, ATP-binding protein | 2.3 | NDE | NDE | NDE | 11.4 | 13 | |
Genes were selected for further analysis based on a Bayes P value of ≤0.0001 and ≤0.8- and ≥1.2-fold change after 15 min and 30 min with bacitracin, nisin, and LL-37. NDE, not significantly differentially expressed.
blpU encodes a homolog of SP0533. D39 does not posses blpK, but part of the amplicon sequence of SP0533 is identical to that of SPD0046.
The SP1715 amplicon on the array is homologous only to SPD1525; there is no information on the transcript levels of SPD1526.
Genes induced in response to both bacitracin and LL-37.
Bacitracin and LL-37 both induced expression of the SP0385 gene, encoding a putative membrane protein, and the adjacent SP0386-0387 genes, encoding the two-component system 3 (TCS03) (Fig. 2 and Table 3) (38). In addition, several transporters (SP0785-0787 and SP1715) were induced upon exposure to both AMPs, as was a putative transcriptional regulator, SP1714 (Fig. 2 and Table 3; also see Table S1, sections A, B, E, and F, and Table S3 in the supplemental material). TCS03, one of the 13 two-component systems in S. pneumoniae, was upregulated more than 2-fold upon bacitracin treatment and moderately (1.5-fold) upon LL-37 treatment. TCS03 shares amino acid sequence similarity with TCS11 from S. mutans, CesSR from L. lactis, VraRS from Staphylococcus aureus, and LiaRS (YvqEC) from B. subtilis, which have been proposed to be sensors of cell envelope-mediated stresses (27). The transcript level of the adjacent SP0385 gene product changed similarly to that of TCS03. The SP0385 membrane protein, with unknown function, shares 27% sequence identity to LiaF (YvqF), a membrane protein of the liaRS gene cluster. Analysis of the genomic sequence of strain D39 revealed that SP0385 is probably transcribed from the same promoter as the TCS03 genes. To investigate whether the SP0385-0387 genes play a role in S. pneumoniae resistance to AMPs, we chose them for further study.
The expression of the SP0785-0787 genes increased more than 2-fold upon bacitracin stress and more than 4-fold upon LL-37 stress (Fig. 2 and Table 3). Analysis of the D39 genomic sequence indicated that the SP0785-0787 genes might be transcribed from the same promoter, which is in accordance with the transcriptome data. SP0785 encodes a protein annotated in the NCBI database as a membrane fusion protein (MFP) subunit of an efflux transporter. The SP0786-0787 genes are annotated as encoding an ABC transporter, with SP0786 encoding an ATP-binding protein and SP0787 a permease protein with three transmembrane domains. Interestingly, the SP0787 protein showed 34% amino acid sequence identity to BacI, involved in secretion of bacteriocin 21, and 32% amino acid sequence identity to MacB, involved in resistance to macrolides (31, 71). Therefore, we decided to investigate the function of SP0785-0787 further.
The SP1714-1715 genes, presumably in an operon, were upregulated more than 2-fold in response to bacitracin and even 13-fold in response to LL-37 (Table 3) and were chosen for further study. The SP1714 gene encodes a putative transcriptional regulator of, most likely, the GntR (gluconate regulator) family of regulators, while SP1715 encodes a putative ABC transporter.
Genes induced upon challenge with bacitracin and nisin.
Among the genes that were upregulated upon both bacitracin and nisin exposure were the SP0912 gene, described above, and the SP2063 gene (Fig. 2; also see Table S1, sections B and D, and Table S3 in the supplemental material). SP2063, a member of the VicRK regulon (50, 52), was upregulated 7-fold upon bacitracin stress and almost 2-fold upon nisin stress. This gene encodes a protein with a LysM (lysin motif) domain, so it is probably cell wall attached, but otherwise the function is unknown (9) (Fig. 2).
Upregulated genes in common for the nisin and LL-37 response.
Treatment with nisin or LL-37 positively stimulated the expression of several identical genes. Among them was the SP2173 gene, encoding DltD (Fig. 2; also see Table S1, sections C and E, and Table S4, section A, in the supplemental material). Interestingly, all four genes of the dlt operon, dltABCD (SP2173-2176), showed induction upon LL-37 exposure (see Table S1, section E), but only one gene of this operon, dltD, was upregulated upon nisin exposure (Fig. 2; also see Table S1, sections C and E). The dlt operon encodes proteins mediating d-alanylation of the teichoic acids, which improves resistance to neutrophil traps in TIGR4 (80). Furthermore, the dlt operon confers resistance to nisin and gallidermin in strains Rx and D39 and, in S. aureus, to defensins, protegrins, and other cationic AMPs (33, 55). Thus, the upregulation of dlt genes upon LL-37 stress and of dltD upon nisin stress is in accordance with previous data and indicates that this operon also plays a role in the resistance of S. pneumoniae D39 to LL-37.
Differences in the D39 transcriptome response to bacitracin, nisin, and LL-37.
The glnRA (SP0501-0502), htrA (SP2063), SP2240, and blp (SP0525-0529, SP0533, and SP0545-0547) genes had opposite expression levels upon challenge with different AMPs. Surprisingly, the glnRA genes were upregulated upon LL-37 stress, whereas they were downregulated upon challenge with nisin (Fig. 2; also see Table S1, sections C, D, E, and F, in the supplemental material). The glnR gene encodes the repressor of the genes encoding the glutamine synthesis and uptake complex, glnA and glnPQ (30). Although the genes involved in glutamine metabolism are well studied within pathogens (20, 30, 67, 68, 75), it is not clear why glnRA are oppositely expressed upon nisin and LL-37 exposure.
Similarly, the expression of htrA and its adjacent gene SP2240 was antagonistic in the D39 stress response to bacitracin and LL-37 (Fig. 2; also see Table S1, sections B, E, and F, in the supplemental material). These genes were downregulated 2-fold upon bacitracin treatment and upregulated more than 3-fold upon LL-37 exposure. HtrA (high-temperature requirement A), a major virulence factor of S. pneumoniae, is a serine protease that plays a significant role in resistance to high temperatures and oxidative stress and is involved in transformation efficiency (12, 24). One of the pneumococcal TCSs, CiaRH (SP0798-0799), positively controls the expression of htrA and SP2240 (23, 46, 62). Since ciaRH was upregulated upon challenge with LL-37 and not with bacitracin, the induction of htrA and SP2240 expression in response to LL-37 was most likely mediated by CiaRH. The expression of the SP0107 gene, also a member of the VicRK regulon (50, 52), which encodes a protein with a LysM (9) domain and unknown function, increased more than 2-fold upon bacitracin exposure and was reduced approximately 2-fold upon LL-37 exposure (Fig. 2).
One feature completely distinguished the response to LL-37 from that to bacitracin and to nisin; genes of the blp (bacteriocin-like peptide; pnc) locus were induced only upon LL-37 stress (Fig. 2 and Table 3; also see Table S1, sections E and F, in the supplemental material). The blp genes encode proteins for Blp bacteriocin production, regulation, transport, and immunity (11, 12, 14, 42). Since the putative blp immunity genes, SP0545-0547, were strongly induced only upon LL-37 stress (Table 3), we speculated that, in strain D39, they might be involved in a specific resistance mechanism against this AMP, and therefore, blp genes involved in putative bacteriocin production and immunity were selected for further study.
Changes mediated by bacitracin, nisin, and LL-37 on the expression of SP0385-0387, SP0785-0787, SP0912-0913, and SP1714-1715.
In order to confirm the differential patterns of expression upon bacitracin, nisin, and LL-37 challenge, lacZ-promoter fusions of the promoters of the genes selected for further study were made. The same experimental procedure as applied for the transcriptome analysis was used for AMP exposure, with one modification. Exposure of the D39 derivatives to the AMPs for 30 and 90 min resulted in higher β-galactosidase activities than a 15-min exposure. Similar observations were made by R. Bernard et al. for induction of the bceAB promoter with bacitracin (4). The reason for this might be that the bacteria need more than 15 min to fully produce the β-galactosidase enzyme. The expression of one unresponsive promoter under these conditions did not show the same time-dependent increase, indicating that this is not a general effect of the AMPs on the β-galactosidase assay (data not shown). Therefore, we decided to measure the promoter's responses after 30 and 90 min of incubation with each AMP.
The expression of the SP0385-0387 promoter increased upon exposure to all AMPs tested (more than 3-fold upon bacitracin exposure and approximately 2-fold with the other two AMPs), which is in contrast to the transcriptome profiling, where these ORFs were induced only upon bacitracin and LL-37 exposure (Table 4). The activity of the SP0785-0787 promoter increased slightly, approximately 2-fold, upon bacitracin and nisin stimulation, but there was no effect of LL-37 exposure (Table 4), which differs from the results observed in the transcriptome analysis. Induction of PSP0912-0913 activity upon LL-37 stress was not observed, but in response to bacitracin and nisin, its activity was more than 12- and 15-fold higher (Table 4), respectively, which corresponds to the transcriptome data. After 30 min of induction, the expression of SP1714-1715 was enhanced 3-fold upon bacitracin exposure and 6-fold upon LL-37 exposure (Table 4), which is in agreement with the transcriptome data. However, the expression of this promoter also increased approximately 2-fold after 30 min of treatment with nisin, which was not observed for this AMP in the transcriptome analysis. These results demonstrate that the activity of the tested promoters is induced upon exposure to AMPs and corresponds with the transcriptome analysis.
TABLE 4.
β-Galactosidase activities of the promoters of the SP0385-0387, SP0785-0787, SP0912-0913, and SP1714-1715 genes in the wild-type D39 strain, transcriptionally fused to lacZa
| Strain | Genes regulated by promoter | Activity (Miller units)b of promoter with indicated treatment for indicated time (min) |
|||||||
|---|---|---|---|---|---|---|---|---|---|
| Without AMP |
Bacitracin |
Nisin |
LL-37 |
||||||
| 30 | 90 | 30 | 90 | 30 | 90 | 30 | 90 | ||
| D39 | SP0385-0387 | 32 (6) | 45 (16) | 194 (31) | 259 (46) | 96 (3) | 82 (14) | 99 (36) | 85 (11) |
| SP0785-0787 | 24 (3) | 26 (3) | 45 (6) | 59 (8) | 46 (8) | 38 (1) | 37 (9) | 37 (7) | |
| SP0912-0913 | 4 (1) | 4 (2) | 23 (2) | 52 (12) | 59 (10) | 64 (20) | 4 (0.5) | 3 (0.1) | |
| SP1714-1715 | 25 (4) | 59 (5) | 90 (16) | 114 (24) | 68 (27) | 60 (24) | 146 (40) | 267 (21) | |
| Δ1714-1715 | SP0785-0787 | 126 (26) | ND | 144 (15) | ND | 145 (19) | ND | 180 (15) | ND |
| SP1714-1715 | 539 (137) | ND | 734 (45) | ND | 715 (34) | ND | 656 (8) | ND | |
The activities of the promoters of the SP0785-0787 and SP1714-1715 genes were also studied in a ΔSP1714-1715 strain. In all cases, the bacteria were grown in THY without AMPs or with either 0.7 μg/ml bacitracin, 0.1 μg/ml nisin, or 4.5 μg/ml LL-37.
Values are the averages of the results of five independent experiments, and the standard deviations are indicated in parentheses. ND, not determined.
Determination of MICs for S. pneumoniae mutant derivatives.
In order to determine whether the genes mentioned before play a direct role in the resistance to any of the AMPs used, mutant and/or complementation constructs of these genes were made and the strains obtained were tested for their susceptibility to bacitracin, nisin, and LL-37 (Table 5). To establish whether these genes are also involved in resistance to other antimicrobial agents and could potentially encode multidrug resistance (MDR) transporters, we also exposed the strains to Hoechst 33342, daunomycin, lincomycin, gramicidin, vancomycin, and ethidium bromide (Table 5). None of the mutant strains was more susceptible than the wild type to vancomycin, daunomycin, or ethidium bromide (data not shown). The SP0385-0387 mutant (Δ385-387) was 2-fold more susceptible to bacitracin. The SP0785-0787-deficient strain (Δ785-787) exhibited considerable sensitivity to LL-37 (more than 4-fold) and to lincomycin (∼10-fold). The SP0912-0913 mutant (Δ912-913) showed enhanced sensitivity to nisin, bacitracin, gramicidin, and lincomycin, which could be complemented by expression of the genes in the mutant (CO912). As expected, the blp-deficient strain (Δblp strain) was more sensitive only to LL-37 and not to the other AMPs. Mutation of SP1714-1715 (Δ1714-1715) caused decreased resistance of strain D39 to bacitracin and Hoechst 33342 but, interestingly, increased resistance to LL-37. To exclude a role of the GntR-like regulator (SP1714) in the observed increased susceptibility of the mutant to bacitracin, Hoechst 33342, or LL-37, a mutant of only the putative ABC transporter, SP1715 (Δ1715), was generated. This SP1715 mutant had the same phenotype as the SP1714-1715-deficient strain (Table 5), strongly suggesting that SP1715 encodes a putative ABC transporter that determines resistance to bacitracin and Hoechst 33342 and sensitivity to LL-37. Introduction of either the SP0385-0387 or the SP1714-1715 mutation into the ΔSP0912-0913 background (DM39 and DM19, respectively) did not result in increased sensitivity to bacitracin, nisin, Hoechst 33342, or LL-37 compared to that of the single mutants, indicating that these proteins are functioning in the same pathway. Additionally, overexpression of the SP0912-0913 genes (OV912) increased the resistance of D39 to bacitracin more than 3-fold, whereas it had only a moderate effect on resistance to nisin and gramicidin and no effect on resistance to lincomycin. Overexpression of SP1715 in both mutant and wild-type backgrounds (CO1715, CO1716, and OV1715) increased the sensitivity to LL-37 7-fold compared with that of the wild type, resistance to Hoechst 33342 increased 2-fold, and minor effects were observed for bacitracin. Thus, multiple genes identified in the transcriptome analysis indeed play a role in the resistance of D39 to the tested AMPs. Furthermore, some of these genes also confer resistance to other antimicrobial compounds.
TABLE 5.
MICs for S. pneumoniae D39 and derivatives treated with various antimicrobial substances
| Strain | MIC (μg/ml)a for: |
|||||
|---|---|---|---|---|---|---|
| Bacitracin | Nisin | LL-37 | Hoechst 33342 (μM) | Gramicidin | Lincomycin | |
| D39 | 4 | 0.8 | 14 | 1 | 2.2 | 0.5 |
| Δ385-387 | 1.5 | 0.8 | 14 | 1 | 2.2 | 0.5 |
| Δ785-787 | 4 | 0.8 | 3 | 1 | 1.5 | 0.03 |
| Δ1715 | 1.7 | 0.8 | 30 | 0.5 | 2.2 | 0.5 |
| Δ1714-1715 | 1.7 | 0.8 | 30 | 0.5 | 2.2 | 0.5 |
| CO1715b | 4 | ND | 1 | 1 | ND | ND |
| CO1716c | 5 | ND | 2 | 1 | ND | ND |
| OV1715g | 5 | ND | 2 | 2 | ND | ND |
| Δblp strain | 4 | 0.8 | 3 | 1 | 2.2 | 0.5 |
| Δ912-913 | 0.7 | 0.2 | 14 | 1 | 1 | 0.03 |
| CO912d | 4 | 0.6 | ND | ND | 2 | 4 |
| OV912g | 15 | 1 | ND | ND | 2.5 | 0.5 |
| DM39e | 0.7 | 0.2 | 9 | ND | 1 | 0.5 |
| DM19f | 0.7 | 0.4 | 26 | 0.5 | 2 | ND |
Values are the averages of the results of at least three independent experiments. MICs are given in micrograms per milliliter unless stated otherwise. Bold font indicates a difference of more than approximately 2-fold compared to the MIC for the wild type. ND, not determined.
Strain overexpresses SP1715 in the Δ1715 mutant.
Strain overexpresses SP1715 in the Δ1714-1715 mutant.
Strain overexpresses SP0912-0913 in the Δ912-913 mutant.
Double mutant of Δ385-387 with Δ912-913.
Double mutant of Δ912-913 with Δ1714-1715.
Strain overexpresses SP0912 or SP1715 in wild-type S. pneumoniae D39.
The GntR-like regulator, SP1714, is a repressor of its own expression and that of SP0785-0787.
The SP1714-1715 and SP0785-0787 genes were upregulated upon treatment with bacitracin and LL-37, and mutation of these genes changed the resistance of D39 to these two AMPs. This indicated that the SP1714-1715 and SP0785-0787 genes might belong to the same regulatory pathway. Therefore, we decided to study the influence of the SP1714 regulator on the expression of selected gene promoters (SP0785-0787, SP0912-0913, and SP1714-1715). The activity of PSP1714-1715 in the ΔSP1714-1715 background increased about 6-fold, and this induction was independent of the stress caused by the AMPs (Table 4). Likewise, the activity of PSP0785-0787 in the ΔSP1714-1715 background increased ∼4-fold, which demonstrated that the GntR-like regulator repressed PSP0785-0787 expression, which was again independent of AMP addition (Table 4). Unfortunately, the open reading frame of SP1714 overlaps with that of SP1715, making it difficult to delete only SP1714 without influencing SP1715 expression. Therefore, in order to avoid mutant construction difficulties and to exclude the possibility that SP1715 played a part in the observed regulatory effects, we examined the expression from these promoters in a ΔSP1715 mutant. As expected, there was no effect of SP1715 deletion on the expression of the promoters of SP0785-0787 and SP1714-1715. Likewise, there was no effect of either SP1714-1715 or SP1715 deletion on the expression of the SP0912-0913 promoter (data not shown). These data suggest that SP1714, encoding a GntR-like regulator, is a repressor of its own expression, as well as that of SP1715 and SP0785-0787.
DISCUSSION
The objective of this study was to investigate whether the stress response of S. pneumoniae D39 to bacitracin, nisin, and LL-37 would reveal common features. A second objective was to determine whether any genes identified have a direct role in conferring resistance to these and various other antimicrobial compounds. Bacitracin, nisin, and LL-37 differ in structure and mode of action, but their targets, subunits of the bacterial cell envelope, are similar. Comparison of the transcriptome response to each compound revealed that they had a low number of significantly differentially expressed genes in common (Fig. 1 and 2). The response of strain D39 to LL-37 was rather broad compared to that to bacitracin or nisin. This extensive reaction to LL-37 suggests a more general response of D39 to human peptides than to bacterial compounds, i.e., bacitracin and nisin (Fig. 1). Analysis of the differentially expressed genes after either 15 min or 30 min of exposure to the tested AMPs showed little overlap in downregulated genes in comparison to that in induced genes (Fig. 1). Comparison of the early response (15 min) to the late one (30 min) for each AMP showed that there was little overlap of commonly up- or downregulated genes (see Fig. S1 and Table S2, section A, in the supplemental material). However, among these commonly induced genes, we identified several, SP0912-0913, SP0785-0787, and SP1714-1715, that were involved in the resistance of D39 to the AMPs tested, as shown by susceptibility assays. Thus, the transcriptome response of D39 to the AMPs changes with time but genes determining resistance are induced in both the early (15 min) and the late (30 min) responses. Interestingly, the reaction of D39 to LL-37 and bacitracin had more genes in common than the reaction to LL-37 and nisin or to bacitracin and nisin (Fig. 1), which might suggest a more similar general stress response to bacitracin and LL-37.
The genes SP0385-0387, SP0912-0913, SP0785-0787, SP1714-1715, and blp had large changes in expression upon challenge with one or more AMPs; therefore, they were characterized in more detail since they could be alternative candidates for resistance inhibition by specific drugs. Notably, transcription of homologues of some of the genes identified in this study, e.g., SP0386-0387, SP0912-0913, SP0785-0787, and SP1714-1715, has also been found to be affected in response to various antimicrobial compounds, including bacitracin, nisin, or LL-37, in several other Gram-positive bacteria, i.e., L. lactis, B. subtilis, and B. licheniformis (34, 45, 57).
We showed that the SP0912-0913 genes, encoding a putative ABC transporter, were induced upon exposure to all three AMPs tested (Fig. 2 and Table 3) and that the mutant was more sensitive to bacitracin and nisin and, additionally, to lincomycin and gramicidin (Table 5). The finding that SP0912-0913 is involved in resistance to lincomycin, nisin, and bacitracin is in accordance with previous data for the SP0912-0913 homologue from B. subtilis, BceAB (formerly YtsCD), which was induced upon bacitracin and LL-37 challenge and which conferred resistance to bacitracin in this bacterium (5, 57). The other homologues of SP0912-0913, MbrAB from S. mutans and YsaBC from L. lactis, modulated bacitracin and nisin resistance, respectively (34, 74). Although it was shown that SP0912-0913 genes were induced in S. pneumoniae TIGR4 and Tupelo strains upon vancomycin challenge (18), we have not seen increased sensitivity of the SP0912-0913 mutant to this antibiotic (data not shown). Thus, the SP0912-0913 transporter does not appear to be directly involved in resistance to vancomycin. The finding that SP0912-0913 is involved in resistance to antimicrobial compounds acting on the cell envelope, i.e., nisin and bacitracin, and antimicrobial compounds involved in protein synthesis inhibition, i.e., lincomycin (10), strongly suggests that the ABC transporter might be of the MDR type. Recently, Becker et al. showed that this ABC transporter is indeed involved in resistance of S. pneumoniae R6 to bacitracin (3).
Both the TCS03 gene and the upstream gene SP0385, which probably forms an operon with TCS03, were induced upon bacitracin and LL-37 challenge (Fig. 2 and Table 3). The exact function of TCS03 in S. pneumoniae is not yet known, but it has been shown that the expression of the SP0385-0387 genes was positively affected upon vancomycin stress but repressed during invasive disease in cerebrospinal fluid (CSF) (18, 54). TCS03 shares significant amino acid sequence similarity to TCS11 from S. mutans, to CesSR from L. lactis, to VraRS from S. aureus, and to LiaRS (YvqEC) from B. subtilis. It has been shown that these homologous TCSs are induced upon challenge with various AMPs, although they did not confer significant resistance to the antimicrobial agents tested (44, 45, 57, 77). This study also showed that SP0385-0387 did not confer significant resistance to the compounds tested, except for bacitracin (Table 5), which corresponds to the phenotype of TCS03 homologues in L. lactis CesSR, S. aureus VraRS, and B. subtilis LiaRS (37, 44, 45, 47). Therefore, it has been proposed that these TCSs are the sensors of cell envelope-mediated stresses, but their exact role in the response to AMPs remains unclear (27). Interestingly, three genes that belong to the VicRK regulon (SP0107, SP2062, and SP0203) were induced by AMP in our study. The VicRK TCS and its homologues in other Gram-positive bacteria regulate, among others, genes involved in murein biosynthesis and are essential (81); in S. pneumoniae this is due to its regulation of PcsB (51). In S. mutans, it was shown that the VicRK homologue is under the positive control of the LiaRS system (72). Thus, it might well be that to withstand exposure to AMPs and the subsequent stress on the cell wall, the VicRK regulon is also necessary.
The SP0785-0787 genes, encoding a putative ABC transporter, were induced in response to both bacitracin and LL-37 (Fig. 2 and Table 3), and the SP0785-0787-deficient strain was significantly more sensitive to LL-37 and lincomycin and moderately sensitive to gramicidin (Table 4). The SP0785-0787 genes were upregulated upon vancomycin stress (18), but the susceptibility assay did not show increased sensitivity of the SP0785-0787 mutant to this antibiotic. Interestingly, Marrer et al. (43) demonstrated that the SP0785-0787 genes were induced upon bacitracin, chloramphenicol, and fusidic acid exposure but repressed by actinomycin and ciprofloxacin challenges (43). These data indicate that SP0785-0787 might be involved in S. pneumoniae resistance to even more antimicrobial compounds than were tested here, which could imply that the SP0785-0787 proteins display some characteristics of MDR and are of direct importance for the global defense mechanism against antimicrobial compounds in S. pneumoniae.
The SP1714 and SP1715 genes, encoding a GntR-like regulator and a putative ABC transporter, respectively, were considerably upregulated upon challenge with LL-37 and bacitracin (Fig. 2 and Table 3). Strains deficient in SP1715 and both SP1714 and SP1715 were more sensitive to Hoechst 33342 and bacitracin. Surprisingly, the SP1715 and SP1714-1715 mutants were more resistant to LL-37 than the wild type, whereas complementation and overexpression of SP1715 increased the sensitivity of strain D39 to LL-37. These data indicate that, on the one hand, SP1715 is involved in D39's sensitivity to LL-37 and, on the other, in D39's resistance to bacitracin and Hoechst 33342. Furthermore, we show that SP1714 is a negative regulator of its own gene and, most likely, also of SP1715 that determines sensitivity to LL-37 and seems to be in the same operon and of SP0785-0787, which protect against LL-37. Since SP1714 was upregulated upon challenge with LL-37 and bacitracin, we speculate that the stress caused by these antimicrobial compounds induces an unknown factor which subsequently interacts with SP1714. This interaction might cause release of SP1714 from a dedicated promoter site and, consequently, derepression of genes regulated negatively by SP1714, i.e., SP1714-1715 and SP0785-0787.
Most of the described GntR-like regulators are repressors of various bacterial metabolic pathways, such as gluconate, histidine, and arabinose biosynthesis (61). Recently, Truong-Bolduc and Hooper identified a new GntR-like regulator, NorG, that regulates the expression of quinolone and β-lactam multidrug efflux pumps (73). In previous studies, the expression profile of SP1714-1715 increased after induction with vancomycin (18), but treatment with penicillin had an opposite effect (60). In addition, these genes were downregulated in the CSF fraction during a transcriptome study of S. pneumoniae during invasive disease (54). These data could imply that the expression of SP1714-1715 depends on external stimuli and that the GntR-like protein, SP1714, might regulate the response to a wide variety of toxic components, most likely via an additional regulatory mechanism. The exact function of the GntR-like regulator, SP1714, remains to be determined and is the subject of ongoing studies.
Interestingly, the blp genes were induced only upon stimulation with LL-37 (Fig. 2 and Table 3). Notably, from the eight TCS mutants tested for growth efficiency in a respiratory tract infection (RTI) model, only a BlpR mutant was attenuated, indicating that it is an essential TCS under these conditions (70). The reason why BlpRH was essential for pneumococcal survival within the RTI remained unclear. Our transcriptome data showed that the presence of LL-37 induced the entire blp locus, especially the putative blp immunity genes. Previously, it has been demonstrated that the chemically synthesized peptide pheromone BlpC first induces the two-component system, BlpRH, which subsequently leads to upregulation of the complete blp gene cluster (14). Since LL-37 and BlpC are short linear cationic peptides, we hypothesize that like BlpC, LL-37 could interact with BlpH and, consequently, through BlpR, activate the entire blp locus. We also speculate that the blp immunity proteins could confer D39 resistance to LL-37, which is strongly supported by the finding that the blp-deficient strain was sensitive to LL-37. This would explain why BlpRH is essential in the RTI, where many AMPs, such as LL-37, are present. In order to confirm our hypothesis, we will continue to evaluate whether LL-37 can induce the expression of BlpRH and if the expression of the entire blp locus will be enhanced in consequence. In addition, we will examine whether the blp mutant is more sensitive to other human AMPs.
To conclude, the transcriptional response of S. pneumoniae D39 to three distinct AMPs, bacitracin, nisin, and LL-37, was diverse and complex and revealed that only a few genes were differentially expressed in response to all three. Most importantly, mutants of some of these genes in D39, i.e., SP0912-0913, SP0785-0787, and SP1714-1715, exhibited cross-sensitivity/resistance to several antimicrobial substances, including some that were not used in the initial challenge experiments, which, to our knowledge, has not been shown before. Additionally, we showed that the blp locus is involved in determining the resistance of D39 to a human AMP, LL-37. Therefore, some of these genes might be interesting candidates for inhibition by specific blocking reagents, which would result in novel medicines for the prevention and treatment of pneumococcal diseases.
Supplementary Material
Acknowledgments
We thank Rachel Hamer for her technical help in conducting some of the experiments presented in this study. We thank Rutger Brouwer and Anne de Jong for their help with the submission of the array data to the GEO database.
Footnotes
Published ahead of print on 16 November 2009.
Supplemental material for this article may be found at http://aac.asm.org/.
REFERENCES
- 1.Avery, O. T., C. M. MacLeod, and M. McCarty. 1944. Studies of the chemical nature of the substance inducing transformation of pneumococcal types. Induction of transformation by a desoxyribonucleic acid fraction isolated from Pneumococcus type III. Mol. Med. 1:344-365. [PMC free article] [PubMed] [Google Scholar]
- 2.Azevedo, E. C., E. M. Rios, K. Fukushima, and G. M. Campos-Takaki. 1993. Bacitracin production by a new strain of Bacillus subtilis. Extraction, purification, and characterization. Appl. Biochem. Biotechnol. 42:1-7. [DOI] [PubMed] [Google Scholar]
- 3.Becker, P., R. Hakenbeck, and B. Henrich. 2009. An ABC transporter of Streptococcus pneumoniae involved in susceptibility to vancoresmycin and bacitracin. Antimicrob. Agents Chemother. 53:2034-2041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bernard, R., A. Guiseppi, M. Chippaux, M. Foglino, and F. Denizot. 2007. Resistance to bacitracin in Bacillus subtilis: unexpected requirement of the BceAB ABC transporter in the control of expression of its own structural genes. J. Bacteriol. 189:8636-8642. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Bernard, R., P. Joseph, A. Guiseppi, M. Chippaux, and F. Denizot. 2003. YtsCD and YwoA, two independent systems that confer bacitracin resistance to Bacillus subtilis. FEMS Microbiol. Lett. 228:93-97. [DOI] [PubMed] [Google Scholar]
- 6.Bethe, G., R. Nau, A. Wellmer, R. Hakenbeck, R. R. Reinert, H. P. Heinz, and G. Zysk. 2001. The cell wall-associated serine protease PrtA: a highly conserved virulence factor of Streptococcus pneumoniae. FEMS Microbiol. Lett. 205:99-104. [DOI] [PubMed] [Google Scholar]
- 7.Breukink, E., and B. de Kruijff. 1999. The lantibiotic nisin, a special case or not? Biochim. Biophys. Acta 1462:223-234. [DOI] [PubMed] [Google Scholar]
- 8.Breukink, E., I. Wiedemann, C. van Kraaij, O. P. Kuipers, H. Sahl, and B. de Kruijff. 1999. Use of the cell wall precursor lipid II by a pore-forming peptide antibiotic. Science 286:2361-2364. [DOI] [PubMed] [Google Scholar]
- 9.Buist, G., A. Steen, J. Kok, and O. P. Kuipers. 2008. LysM, a widely distributed protein motif for binding to (peptido)glycans. Mol. Microbiol. 68:838-847. [DOI] [PubMed] [Google Scholar]
- 10.Chang, F. N., C. J. Sih, and B. Weisblum. 1966. Lincomycin, an inhibitor of aminoacyl sRNA binding to ribosomes. Proc. Natl. Acad. Sci. U. S. A. 55:431-438. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Dawid, S., A. M. Roche, and J. N. Weiser. 2007. The blp bacteriocins of Streptococcus pneumoniae mediate intraspecies competition both in vitro and in vivo. Infect. Immun. 75:443-451. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Dawid, S., M. E. Sebert, and J. N. Weiser. 2009. Bacteriocin activity of Streptococcus pneumoniae is controlled by the serine protease HtrA via posttranscriptional regulation. J. Bacteriol. 191:1509-1518. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.de Ruyter, P. G., O. P. Kuipers, and W. M. de Vos. 1996. Controlled gene expression systems for Lactococcus lactis with the food-grade inducer nisin. Appl. Environ. Microbiol. 62:3662-3667. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.de Saizieu, A., C. Gardes, N. Flint, C. Wagner, M. Kamber, T. J. Mitchell, W. Keck, K. E. Amrein, and R. Lange. 2000. Microarray-based identification of a novel Streptococcus pneumoniae regulon controlled by an autoinduced peptide. J. Bacteriol. 182:4696-4703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Doern, G. V., S. S. Richter, A. Miller, N. Miller, C. Rice, K. Heilmann, and S. Beekmann. 2005. Antimicrobial resistance among Streptococcus pneumoniae in the United States: have we begun to turn the corner on resistance to certain antimicrobial classes? Clin. Infect. Dis. 41:139-148. [DOI] [PubMed] [Google Scholar]
- 16.Durr, U. H., U. S. Sudheendra, and A. Ramamoorthy. 2006. LL-37, the only human member of the cathelicidin family of antimicrobial peptides. Biochim. Biophys. Acta 1758:1408-1425. [DOI] [PubMed] [Google Scholar]
- 17.Gross, E., and J. L. Morell. 1971. The structure of nisin. J. Am. Chem. Soc. 93:4634-4635. [DOI] [PubMed] [Google Scholar]
- 18.Haas, W., D. Kaushal, J. Sublett, C. Obert, and E. I. Tuomanen. 2005. Vancomycin stress response in a sensitive and a tolerant strain of Streptococcus pneumoniae. J. Bacteriol. 187:8205-8210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Halfmann, A., R. Hakenbeck, and R. Bruckner. 2007. A new integrative reporter plasmid for Streptococcus pneumoniae. FEMS Microbiol. Lett. 268:217-224. [DOI] [PubMed] [Google Scholar]
- 20.Harth, G., S. Maslesa-Galic, M. V. Tullius, and M. A. Horwitz. 2005. All four Mycobacterium tuberculosis glnA genes encode glutamine synthetase activities but only GlnA1 is abundantly expressed and essential for bacterial homeostasis. Mol. Microbiol. 58:1157-1172. [DOI] [PubMed] [Google Scholar]
- 21.Hasper, H. E., N. E. Kramer, J. L. Smith, J. D. Hillman, C. Zachariah, O. P. Kuipers, B. de Kruijff, and E. Breukink. 2006. An alternative bactericidal mechanism of action for lantibiotic peptides that target lipid II. Science 313:1636-1637. [DOI] [PubMed] [Google Scholar]
- 22.Holo, H., and I. F. Nes. 1995. Transformation of Lactococcus by electroporation. Methods Mol. Biol. 47:195-199. [DOI] [PubMed] [Google Scholar]
- 23.Ibrahim, Y. M., A. R. Kerr, J. McCluskey, and T. J. Mitchell. 2004. Control of virulence by the two-component system CiaR/H is mediated via HtrA, a major virulence factor of Streptococcus pneumoniae. J. Bacteriol. 186:5258-5266. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Ibrahim, Y. M., A. R. Kerr, J. McCluskey, and T. J. Mitchell. 2004. Role of HtrA in the virulence and competence of Streptococcus pneumoniae. Infect. Immun. 72:3584-3591. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Ishihara, H., M. Takoh, R. Nishibayashi, and A. Sato. 2002. Distribution and variation of bacitracin synthetase gene sequences in laboratory stock strains of Bacillus licheniformis. Curr. Microbiol. 45:18-23. [DOI] [PubMed] [Google Scholar]
- 26.Israelsen, H., S. M. Madsen, A. Vrang, E. B. Hansen, and E. Johansen. 1995. Cloning and partial characterization of regulated promoters from Lactococcus lactis Tn917-lacZ integrants with the new promoter probe vector, pAK80. Appl. Environ. Microbiol. 61:2540-2547. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Jordan, S., M. I. Hutchings, and T. Mascher. 2008. Cell envelope stress response in Gram-positive bacteria. FEMS Microbiol. Rev. 32:107-146. [DOI] [PubMed] [Google Scholar]
- 28.Kang, J. W., G. de Reymaeker, A. van Schepdael, E. Roets, and J. Hoogmartens. 2001. Analysis of bacitracin by micellar electrokinetic capillary chromatography with mixed micelle in acidic solution. Electrophoresis 22:1356-1362. [DOI] [PubMed] [Google Scholar]
- 29.Kloosterman, T. G., J. J. Bijlsma, J. Kok, and O. P. Kuipers. 2006. To have neighbour's fare: extending the molecular toolbox for Streptococcus pneumoniae. Microbiology 152:351-359. [DOI] [PubMed] [Google Scholar]
- 30.Kloosterman, T. G., W. T. Hendriksen, J. J. Bijlsma, H. J. Bootsma, S. A. van Hijum, J. Kok, P. W. Hermans, and O. P. Kuipers. 2006. Regulation of glutamine and glutamate metabolism by GlnR and GlnA in Streptococcus pneumoniae. J. Biol. Chem. 281:25097-25109. [DOI] [PubMed] [Google Scholar]
- 31.Kobayashi, N., K. Nishino, and A. Yamaguchi. 2001. Novel macrolide-specific ABC-type efflux transporter in Escherichia coli. J. Bacteriol. 183:5639-5644. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Koczulla, A. R., and R. Bals. 2003. Antimicrobial peptides: current status and therapeutic potential. Drugs 63:389-406. [DOI] [PubMed] [Google Scholar]
- 33.Kovacs, M., A. Halfmann, I. Fedtke, M. Heintz, A. Peschel, W. Vollmer, R. Hakenbeck, and R. Bruckner. 2006. A functional dlt operon, encoding proteins required for incorporation of d-alanine in teichoic acids in gram-positive bacteria, confers resistance to cationic antimicrobial peptides in Streptococcus pneumoniae. J. Bacteriol. 188:5797-5805. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Kramer, N. E., S. A. van Hijum, J. Knol, J. Kok, and O. P. Kuipers. 2006. Transcriptome analysis reveals mechanisms by which Lactococcus lactis acquires nisin resistance. Antimicrob. Agents Chemother. 50:1753-1761. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Kuipers, O., P. de Ruyter, M. Kleerebezem, and W. de Vos. 1998. Quorum sensing-controlled gene expression in lactic acid bacteria. J. Biotechnol. 64:15-21. [Google Scholar]
- 36.Kuipers, O. P., A. de Jong, R. J. Baerends, S. A. van Hijum, A. L. Zomer, H. A. Karsens, C. D. den Hengst, N. E. Kramer, G. Buist, and J. Kok. 2002. Transcriptome analysis and related databases of Lactococcus lactis. Antonie Van Leeuwenhoek 82:113-122. [PubMed] [Google Scholar]
- 37.Kuroda, M., H. Kuroda, T. Oshima, F. Takeuchi, H. Mori, and K. Hiramatsu. 2003. Two-component system VraSR positively modulates the regulation of cell-wall biosynthesis pathway in Staphylococcus aureus. Mol. Microbiol. 49:807-821. [DOI] [PubMed] [Google Scholar]
- 38.Lange, R., C. Wagner, A. de Saizieu, N. Flint, J. Molnos, M. Stieger, P. Caspers, M. Kamber, W. Keck, and K. E. Amrein. 1999. Domain organization and molecular characterization of 13 two-component systems identified by genome sequencing of Streptococcus pneumoniae. Gene 237:223-234. [DOI] [PubMed] [Google Scholar]
- 39.Lanie, J. A., W. L. Ng, K. M. Kazmierczak, T. M. Andrzejewski, T. M. Davidsen, K. J. Wayne, H. Tettelin, J. I. Glass, and M. E. Winkler. 2007. Genome sequence of Avery's virulent serotype 2 strain D39 of Streptococcus pneumoniae and comparison with that of unencapsulated laboratory strain R6. J. Bacteriol. 189:38-51. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Leenhouts, K., G. Buist, A. Bolhuis, A. ten Berge, J. Kiel, I. Mierau, M. Dabrowska, G. Venema, and J. Kok. 1996. A general system for generating unlabelled gene replacements in bacterial chromosomes. Mol. Gen. Genet. 253:217-224. [DOI] [PubMed] [Google Scholar]
- 41.Lu, Y. J., and C. O. Rock. 2006. Transcriptional regulation of fatty acid biosynthesis in Streptococcus pneumoniae. Mol. Microbiol. 59:551-566. [DOI] [PubMed] [Google Scholar]
- 42.Lux, T., M. Nuhn, R. Hakenbeck, and P. Reichmann. 2007. Diversity of bacteriocins and activity spectrum in Streptococcus pneumoniae. J. Bacteriol. 189:7741-7751. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Marrer, E., K. Schad, A. T. Satoh, M. G. Page, M. M. Johnson, and L. J. Piddock. 2006. Involvement of the putative ATP-dependent efflux proteins PatA and PatB in fluoroquinolone resistance of a multidrug-resistant mutant of Streptococcus pneumoniae. Antimicrob. Agents Chemother. 50:685-693. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Martinez, B., A. L. Zomer, A. Rodriguez, J. Kok, and O. P. Kuipers. 2007. Cell envelope stress induced by the bacteriocin Lcn972 is sensed by the lactococcal two-component system CesSR. Mol. Microbiol. 64:473-486. [DOI] [PubMed] [Google Scholar]
- 45.Mascher, T., N. G. Margulis, T. Wang, R. W. Ye, and J. D. Helmann. 2003. Cell wall stress responses in Bacillus subtilis: the regulatory network of the bacitracin stimulon. Mol. Microbiol. 50:1591-1604. [DOI] [PubMed] [Google Scholar]
- 46.Mascher, T., D. Zahner, M. Merai, N. Balmelle, A. B. de Saizieu, and R. Hakenbeck. 2003. The Streptococcus pneumoniae cia regulon: CiaR target sites and transcription profile analysis. J. Bacteriol. 185:60-70. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Mascher, T., S. L. Zimmer, T. A. Smith, and J. D. Helmann. 2004. Antibiotic-inducible promoter regulated by the cell envelope stress-sensing two-component system LiaRS of Bacillus subtilis. Antimicrob. Agents Chemother. 48:2888-2896. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Mattick, T. R., and A. Hirsch. 1944. A powerful inhibitory substance produced by group N streptococci. Nature 154:551. [Google Scholar]
- 49.Mitchell, T. J. 2003. The pathogenesis of streptococcal infections: from tooth decay to meningitis. Nat. Rev. Microbiol. 1:219-230. [DOI] [PubMed] [Google Scholar]
- 50.Mohedano, M. L., K. Overweg, A. de la Fuente, M. Reuter, S. Altabe, F. Mulholland, D. de Mendoza, P. Lopez, and J. M. Wells. 2005. Evidence that the essential response regulator YycF in Streptococcus pneumoniae modulates expression of fatty acid biosynthesis genes and alters membrane composition. J. Bacteriol. 187:2357-2367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Ng, W. L., K. M. Kazmierczak, and M. E. Winkler. 2004. Defective cell wall synthesis in Streptococcus pneumoniae R6 depleted for the essential PcsB putative murein hydrolase or the VicR (YycF) response regulator. Mol. Microbiol. 53:1161-1175. [DOI] [PubMed] [Google Scholar]
- 52.Ng, W. L., H. C. Tsui, and M. E. Winkler. 2005. Regulation of the pspA virulence factor and essential pcsB murein biosynthetic genes by the phosphorylated VicR (YycF) response regulator in Streptococcus pneumoniae. J. Bacteriol. 187:7444-7459. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Oren, Z., J. C. Lerman, G. H. Gudmundsson, B. Agerberth, and Y. Shai. 1999. Structure and organization of the human antimicrobial peptide LL-37 in phospholipid membranes: relevance to the molecular basis for its non-cell-selective activity. Biochem. J. 341:501-513. [PMC free article] [PubMed] [Google Scholar]
- 54.Orihuela, C. J., J. N. Radin, J. E. Sublett, G. Gao, D. Kaushal, and E. I. Tuomanen. 2004. Microarray analysis of pneumococcal gene expression during invasive disease. Infect. Immun. 72:5582-5596. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Peschel, A., M. Otto, R. W. Jack, H. Kalbacher, G. Jung, and F. Götz. 1999. Inactivation of the dlt operon in Staphylococcus aureus confers sensitivity to defensins, protegrins, and other antimicrobial peptides. J. Biol. Chem. 274:8405-8410. [DOI] [PubMed] [Google Scholar]
- 56.Peschel, A., and H. G. Sahl. 2006. The co-evolution of host cationic antimicrobial peptides and microbial resistance. Nat. Rev. Microbiol. 4:529-536. [DOI] [PubMed] [Google Scholar]
- 57.Pietiainen, M., M. Gardemeister, M. Mecklin, S. Leskela, M. Sarvas, and V. P. Kontinen. 2005. Cationic antimicrobial peptides elicit a complex stress response in Bacillus subtilis that involves ECF-type sigma factors and two-component signal transduction systems. Microbiology 151:1577-1592. [DOI] [PubMed] [Google Scholar]
- 58.Pozzi, G., L. Masala, F. Iannelli, R. Manganelli, L. S. Havarstein, L. Piccoli, D. Simon, and D. A. Morrison. 1996. Competence for genetic transformation in encapsulated strains of Streptococcus pneumoniae: two allelic variants of the peptide pheromone. J. Bacteriol. 178:6087-6090. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Rayman, M. K., B. Aris, and A. Hurst. 1981. Nisin: a possible alternative or adjunct to nitrite in the preservation of meats. Appl. Environ. Microbiol. 41:375-380. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Rogers, P. D., T. T. Liu, K. S. Barker, G. M. Hilliard, B. K. English, J. Thornton, E. Swiatlo, and L. S. McDaniel. 2007. Gene expression profiling of the response of Streptococcus pneumoniae to penicillin. J. Antimicrob. Chemother. 59:616-626. [DOI] [PubMed] [Google Scholar]
- 61.Sa-Nogueira, I., and L. J. Mota. 1997. Negative regulation of l-arabinose metabolism in Bacillus subtilis: characterization of the araR (araC) gene. J. Bacteriol. 179:1598-1608. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Sebert, M. E., L. M. Palmer, M. Rosenberg, and J. N. Weiser. 2002. Microarray-based identification of htrA, a Streptococcus pneumoniae gene that is regulated by the CiaRH two-component system and contributes to nasopharyngeal colonization. Infect. Immun. 70:4059-4067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Song, J. H., K. S. Ko, J. Y. Lee, J. Y. Baek, W. S. Oh, H. S. Yoon, J. Y. Jeong, and J. Chun. 2005. Identification of essential genes in Streptococcus pneumoniae by allelic replacement mutagenesis. Mol. Cells 19:365-374. [PubMed] [Google Scholar]
- 64.Sorensen, O. E., P. Follin, A. H. Johnsen, J. Calafat, G. S. Tjabringa, P. S. Hiemstra, and N. Borregaard. 2001. Human cathelicidin, hCAP-18, is processed to the antimicrobial peptide LL-37 by extracellular cleavage with proteinase 3. Blood 97:3951-3959. [DOI] [PubMed] [Google Scholar]
- 65.Stone, K. J., and J. L. Strominger. 1971. Mechanism of action of bacitracin: complexation with metal ion and C(55)-isoprenyl pyrophosphate. Proc. Natl. Acad. Sci. U. S. A. 68:3223-3227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Storm, D. R., and J. L. Strominger. 1973. Complex formation between bacitracin peptides and isoprenyl pyrophosphates. The specificity of lipid-peptide interactions. J. Biol. Chem. 248:3940-3945. [PubMed] [Google Scholar]
- 67.Tamura, G. S., D. S. Bratt, H. H. Yim, and A. Nittayajarn. 2005. Use of glnQ as a counterselectable marker for creation of allelic exchange mutations in group B streptococci. Appl. Environ. Microbiol. 71:587-590. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Tamura, G. S., A. Nittayajarn, and D. L. Schoentag. 2002. A glutamine transport gene, glnQ, is required for fibronectin adherence and virulence of group B streptococci. Infect. Immun. 70:2877-2885. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Terzaghi, B. E., and W. E. Sandine. 1975. Improved medium for lactic streptococci and their bacteriophages. Appl. Microbiol. 29:807-813. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Throup, J. P., K. K. Koretke, A. P. Bryant, K. A. Ingraham, A. F. Chalker, Y. Ge, A. Marra, N. G. Wallis, J. R. Brown, D. J. Holmes, M. Rosenberg, and M. K. Burnham. 2000. A genomic analysis of two-component signal transduction in Streptococcus pneumoniae. Mol. Microbiol. 35:566-576. [DOI] [PubMed] [Google Scholar]
- 71.Tomita, H., S. Fujimoto, K. Tanimoto, and Y. Ike. 1997. Cloning and genetic and sequence analyses of the bacteriocin 21 determinant encoded on the Enterococcus faecalis pheromone-responsive conjugative plasmid pPD1. J. Bacteriol. 179:7843-7855. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Tremblay, Y. D., H. Lo, Y. H. Li, S. A. Halperin, and S. F. Lee. 2009. Expression of the Streptococcus mutans essential two-component regulatory system VicRK is pH and growth-phase dependent and controlled by the LiaFSR three-component regulatory system. Microbiology 155:2856-2865. [DOI] [PubMed] [Google Scholar]
- 73.Truong-Bolduc, Q. C., and D. C. Hooper. 2007. The transcriptional regulators NorG and MgrA modulate resistance to both quinolones and beta-lactams in Staphylococcus aureus. J. Bacteriol. 189:2996-3005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Tsuda, H., Y. Yamashita, Y. Shibata, Y. Nakano, and T. Koga. 2002. Genes involved in bacitracin resistance in Streptococcus mutans. Antimicrob. Agents Chemother. 46:3756-3764. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Tullius, M. V., G. Harth, and M. A. Horwitz. 2003. Glutamine synthetase GlnA1 is essential for growth of Mycobacterium tuberculosis in human THP-1 macrophages and guinea pigs. Infect. Immun. 71:3927-3936. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Turner, J., Y. Cho, N. N. Dinh, A. J. Waring, and R. I. Lehrer. 1998. Activities of LL-37, a cathelin-associated antimicrobial peptide of human neutrophils. Antimicrob. Agents Chemother. 42:2206-2214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Utaida, S., P. M. Dunman, D. Macapagal, E. Murphy, S. J. Projan, V. K. Singh, R. K. Jayaswal, and B. J. Wilkinson. 2003. Genome-wide transcriptional profiling of the response of Staphylococcus aureus to cell-wall-active antibiotics reveals a cell-wall-stress stimulon. Microbiology 149:2719-2732. [DOI] [PubMed] [Google Scholar]
- 78.van Hijum, S. A., A. de Jong, R. J. Baerends, H. A. Karsens, N. E. Kramer, R. Larsen, C. D. den Hengst, C. J. Albers, J. Kok, and O. P. Kuipers. 2005. A generally applicable validation scheme for the assessment of factors involved in reproducibility and quality of DNA-microarray data. BMC Genomics 6:77. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.van Hijum, S. A., J. Garcia de la Nava, O. Trelles, J. Kok, and O. P. Kuipers. 2003. MicroPreP: a cDNA microarray data pre-processing framework. Appl. Bioinformatics 2:241-244. [PubMed] [Google Scholar]
- 80.Wartha, F., K. Beiter, B. Albiger, J. Fernebro, A. Zychlinsky, S. Normark, and B. Henriques-Normark. 2007. Capsule and d-alanylated lipoteichoic acids protect Streptococcus pneumoniae against neutrophil extracellular traps. Cell. Microbiol. 9:1162-1171. [DOI] [PubMed] [Google Scholar]
- 81.Winkler, M. E., and J. A. Hoch. 2008. Essentiality, bypass, and targeting of the YycFG (VicRK) two-component regulatory system in gram-positive bacteria. J. Bacteriol. 190:2645-2648. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.