Abstract
The effects of different structural features on the thermostability of Thermopolyspora flexuosa xylanase XYN10A were investigated. A C-terminal carbohydrate binding module had only a slight effect, whereas a polyhistidine tag increased the thermostability of XYN10A xylanase. In contrast, glycosylation at Asn26, located in an exposed loop, decreased the thermostability of the xylanase. The presence of a substrate increased stability mainly at low pH.
The thermophilic actinomycete Thermopolyspora flexuosa, previously named Nonomuraea flexuosa and before that Actinomadura flexuosa or Microtetraspora flexuosa (15), produces family 11 and family 10 xylanases, which show high thermostability (16, 17, 22). T. flexuosa xylanase XYN10A has a C-terminal family 13 carbohydrate binding module (CBM) (22). Many xylanases have an additional CBM, which can be a cellulose binding domain (CBD) or a xylan binding domain (XBD) (1, 5, 7, 22, 25, 28). XBD typically increases activity against insoluble xylan (1, 5, 24), although some XBDs also bind soluble xylans (21, 25).
We studied the thermostability of T. flexuosa xylanase XYN10A and how CBM and other additional groups affect its thermostability. In addition to confirming the previously described importance of terminal regions, our study identified a loop that is important for the thermostability of T. flexuosa XYN10A. In general, identification of sites important for protein stability is necessary for targeted mutagenesis attempts to increase thermostability.
The T. flexuosa xyn10A gene (GenBank accession no. AJ508953) (22), which encodes the full-length XYN10A xylanase (1-AAST… SYNA-448) containing the catalytic domain and CBM, and a truncated gene, which encodes the catalytic domain only (1-AAST… DALN-301) were expressed in Trichoderma reesei as 3′ fusions to a sequence that encodes the Cel6A CBD (A+B) carrier polypeptide and a Kex2 cleavage site (RDKR) (27). In this article, the catalytic domain and the full-length enzyme are referred to as XYN10A and XYN10A-CBM, respectively. The catalytic domain was also produced in Escherichia coli. For production in E. coli, the sequence encoding the catalytic domain was cloned into a pKKtac vector (33) with and without an additional 3′ sequence encoding a 6×His tag at the protein C terminus (… DALNHHHHHH).
The proteins were purified by hydrophobic interaction chromatography using a Phenyl Sepharose column and by ion-exchange chromatography using a DEAE Sepharose FF column (Amersham Pharmacia Biotech). The 6×His-tagged XYN10A xylanase produced in E. coli was purified by affinity chromatography using Ni-nitrilotriacetic acid (Ni-NTA) agarose beads (Qiagen).
Mass spectrometric (MS) analyses were performed on a high-resolution 4.7-T hybrid quadrupole-Fourier transform ion cyclotron resonance (FT-ICR) instrument (APEX-Qe; Bruker Daltonics), which employs electrospray ionization (ESI) (see supplemental material for details).
Xylanase activity was measured with a 3,5-dinitrosalicylic acid assay by using 1% solubilized birchwood xylan as a substrate (33). The optimum temperature, residual activity, and half-life assays were performed as described earlier (36). SWISS-MODEL (4) was used to automatically model T. flexuosa XYN10A and XYN10A-CBM (PDB codes for the modeling templates are 1v6w and 1e0w, respectively [12, 14]).
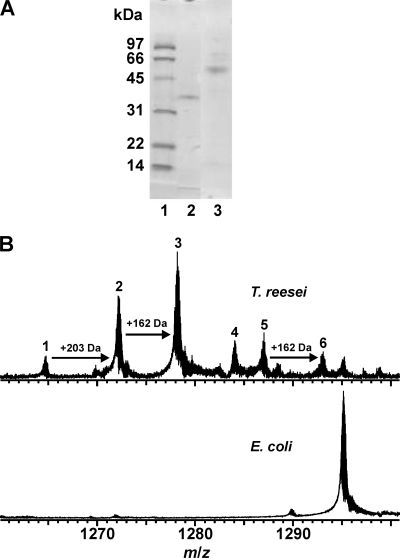
The results of sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) analysis indicated that the masses of XYN10A xylanase and XYN10A-CBM produced in Trichoderma reesei were ∼37 kDa and ∼50 kDa, respectively (Fig. 1A). MS analysis of the 6×His-tagged XYN10A produced in E. coli (SDS-PAGE not shown) indicated the presence of a single protein form (Fig. 1B), with a measured mass of 34,943.25 Da. This is consistent with the theoretical mass of 6×His-tagged XYN10A (34,942.93 Da). In contrast, XYN10A produced in T. reesei was heterogeneously modified, and six protein forms (numbered 1 to 6) were detected (Fig. 1B). The mass of form 1 (34,120.76 Da) is in excellent agreement with the calculated mass of XYN10A (34120.73 Da). The masses of forms 2 and 3, with mass increments of ∼203 and ∼162 Da, respectively, suggested protein glycosylation (+203 Da = GlcNAc; +162 Da = Man). There are two potential sites for N-glycosylation in XYN10A, Asn26 and Asn95. These six protein forms were resolved only by the high-resolution FT-ICR MS technique, not by SDS-PAGE (eluted as a single band [Fig. 1A]).
FIG. 1.
(A) SDS-PAGE of purified XYN10A and XYN10A-CBM produced in Trichoderma reesei. Lane 1, molecular weight markers; lane 2, catalytic domain (XYN10A); lane 3, full-length enzyme (XYN10A-CBM). (B) ESI FT-ICR mass spectra of XYN10A with a 6×His tag produced in E. coli (bottom) and XYN10A produced in T. reesei (top). Only the expanded view at m/z 1260 to 1300, with the signals representing the most abundant protein ion charge state z = 27+, is presented. For the measured and calculated masses of the protein forms identified, see the supplemental material.
In order to locate the glycosylation site or sites, XYN10A proteins produced in E. coli and T. reesei were subjected to on-line pepsin digestion (see supplemental material for details). The sequence coverage for XYN10A xylanase produced in E. coli was 62%. For XYN10A produced in T. reesei, a lower sequence coverage was obtained, but three glycopeptides (residues 20 to 44, 20 to 46, and 20 to 59), carrying one GlcNAc residue, were detected (glycopeptides A to C in Fig. S1B in the supplemental material). A triply charged glycopeptide A was further analyzed by collision-induced dissociation (CID) measurement (see inset in Fig. S1B in the supplemental material). A ladder of b-type fragment ions further identified this peptide and verified Asn26 as the N-glycosylation site in XYN10A, carrying GlcNAc(Man) as a glycan core structure.
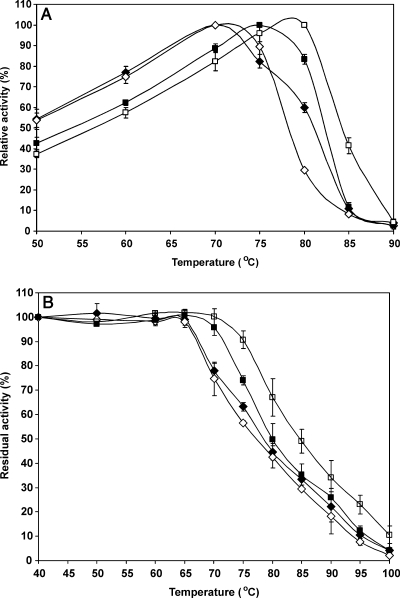
The additional sequences attached to the catalytic domain affected the thermostability of XYN10A xylanase. The deletion of the native C-terminal CBM domain (XYN10A produced in T. reesei) slightly decreased (∼2°C) the apparent temperature optimum in the region of 70 to 75°C (Table 1 and Fig. 2A). However, at 80°C, the deletion of the CBM domain increased the activity (Fig. 2A). Furthermore, the half-life in the presence of the substrate at 80°C was lower when the CBM was present (Table 2).
TABLE 1.
Peaks of the optimum temperatures (30-min assay)a
| Production host | Enzyme | Optimum temp (°C) at: |
||
|---|---|---|---|---|
| pH 5.5 | pH 7 | pH 8.5 | ||
| T. reesei | XYN10A | 70 | 70 | 69 |
| XYN10A-CBM | 70 | 72 | 72 | |
| E. coli | XYN10A | 78 | 75 | 76 |
| XYN10A-6×His | 78 | 78 | 78 | |
One percent solubilized birchwood xylan was used as the substrate in the assay.
FIG. 2.
Enzyme activity and stability profiles. (A) Enzyme activity as a function of temperature. The enzymes were incubated for 30 min at each temperature at pH 7. (B) Enzyme inactivation as a function of temperature. The enzyme samples were incubated without the substrate for 30 min at each temperature (pH 7), and the residual activity was measured at 70°C. Values are means ± standard deviations (error bars) for three experiments. Symbols: ⧫, XYN10A xylanase produced in T. reesei; ⋄, XYN10A-CBM produced in T. reesei; ▪, XYN10A produced in E. coli; □, XYN10A-6×His produced in E. coli.
TABLE 2.
pH-dependent half-life times of a catalytic domain (XYN10A) and a full-length enzyme (XYN10A-CBM) produced in T. reesei
| Enzyme | Half-life (min) of enzyme under various conditions |
|||||||
|---|---|---|---|---|---|---|---|---|
| With substratea |
Without substrate |
|||||||
| pH 4 and 65°C | pH 5.5 and 80°C | pH 7 and 80°C | pH 8.5 and 80°C | pH 4 and 65°C | pH 5.5 and 80°C | pH 7 and 80°C | pH 8.5 and 80°C | |
| XYN10A | 18 | 37 | 37 | 33 | 3.1 | 19 | 23 | 23 |
| XYN10A-CBM | 15 | 17 | 17 | 14 | 1.3 | 33 | 22 | 26 |
One percent solubilized birchwood xylan was used as the substrate in the assay.
Surprisingly, the apparent temperature optimum of XYN10A xylanase produced in E. coli was 4 to 8°C higher than that for XYN10A produced in T. reesei (Fig. 2A and Table 1). In addition, the C-terminal 6×His tag further increased the apparent temperature optimum of XYN10A by ∼3°C at pH 7 and 8.5 (Fig. 2A). The higher stability of XYN10A produced in E. coli was also seen in the residual activity profiles (Fig. 2B). However, the 6×His tag did not elevate the temperature optimum at pH 5.5 (Table 1) and pH 4.0 (not shown).
We also measured the enzyme half-lives with and without substrate (1% solubilized birchwood xylan) at different pH values. Increases of about 5- to 10-fold in the half-lives of both XYN10A xylanase and XYN10A-CBM (produced in T. reesei) were measured at pH 4 in the presence of a substrate (Table 2). The substrate also slightly protected XYN10A in the pH range from pH 5.5 to 8.5. However, no protection by the substrate was detected for XYN10A-CBM at pH 5.5 to 8.5.
By comparing the structures of thermophilic and mesophilic family 10 xylanases, it was suggested that efficient packing of the hydrophobic core, favorable charge interactions with the helix dipole moment, and the presence of prolines at the N termini of alpha-helices are the most probable stabilizing factors (23). Cavity filling and stabilization of loops and N- and C-terminal regions are also important factors (2, 35). By studying chimeric xylanase created by the shuffling of Thermotoga maritima xylanases A and B, it was observed that the N-terminal and C-terminal regions of the xylanase structure formed from the TIM barrel are important for high thermostability (20). Our results also showed that the C-terminal region is important for the thermostability of family 10 xylanases.
An increase in the thermostability of other proteins by a polyhistidine tag has already been demonstrated (8, 9, 10, 19). In T. flexuosa XYN10A xylanase, the 6×His tag had an effect on thermostability only at a neutral or alkaline pH. Since histidine is generally neutral in charge above pH 6.5 (average pKa about 6.5) and positively charged at acidic pH, this suggests that noncharged interactions are critical for the stabilization effect.
The binding of the C-terminal 6×His tag to the surface of XYN10A xylanase probably prevents unfolding from the C terminus. The disulfide bridge between the N and C termini (located close to each other) has previously been demonstrated to increase the melting temperature (Tm) of a family 10 xylanase by 4°C (2, 35). The thermostability increase achieved by the 6×His tag and CBM in T. flexuosa XYN10A was at the same level (in the range of 3°C in the activity assays). Other stabilization mechanisms are also possible, but it seems probable that the role of protein termini is dominant in stabilization by the 6×His tag. The stability of alpha-helices near the C terminus could also be increased by interaction with the 6×His tag (Fig. 3).
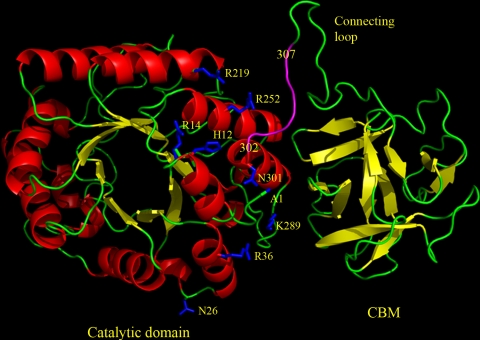
FIG. 3.
Modeled structure of full-length XYN10A xylanase. The model was created by SWISS-MODEL using 1v6w as a template, and the figure was made using PyMOL (11). The residue Asn301 is the C terminus of the expressed catalytic core. The residue Ala1 (A1) shows the position of the N terminus. The glycosylation site Asn26 and the positively charged residues (His12, Arg14, Arg36, Arg219, Arg252, and Lys289) in the range of the 6×His tag are shown as one-letter codes. The sequence positions corresponding to the 6×His tag (positions 302 to 307 in full-length XYN10A) are shown in magenta, although the conformation of the 6×His tag is not known. The active site is located on the other side of the barrel.
Structural modeling was used to examine the regions potentially binding the 6×His tag. In the crystal and nuclear magnetic resonance (NMR) structures 1ddf, 1jt3, and 1zu2, the length of the 6×His tag varies between 12 and 20 Å, since the conformation of the freely protruding 6×His tag may vary significantly. Thus, the 6×His tag forms a rather large binding surface with much variation in the conformation. Since the stabilizing effect of the 6×His tag is pH dependent, it could be that the nearby arginines, having positive charges, have a role in breaking the interactions of the polyhistidine when it becomes positively charged at low pH (Fig. 3). Three nearby arginines (Arg14, Arg219, and Arg252) and a histidine (His12) in the 12-Å distance range from the first histidine in the 6×His tag might cause charge repulsion, and Arg36 and Lys289 at a distance of 17 to 20 Å in the opposite direction might also cause similar repulsion (Fig. 3).
The glycosylation site (Asn26) is located in a well-exposed loop (amino acids 21 to 28) between a beta-strand (amino acids 15 to 20) and alpha-helix (amino acids 29 to 37). Glycosylation can increase the thermostability (6, 18, 29). It can also destabilize, and, according to molecular dynamics simulations, increased mobility correlates with the destabilization caused by glycosylation (31). Glycosylation in a well-exposed loop in XYN10A xylanase could increase local mobility or destabilize the enzyme by affecting the local conformation.
The presence of a substrate increased the stability of both the core and full-length XYN10A xylanase under stronger acidic conditions of pH 4 (Table 2). At pH 5.5 to 8.5, the relative effect was smaller for the XYN10A core and missing in XYN10A-CBM. Protection by a substrate, especially at acidic pH, was observed by Xiong et al. (36) for a family 11 xylanase produced by Thermomyces lanuginosus. A possible explanation for this is that the substrate changes the structure of the enzyme or is involved in hydrogen bonding in the active site in a pH-dependent manner. At pH 4, in which the carboxylic acids start to become on average protonated and the ion pair networks are therefore disturbed, the thermostability of the enzyme is lower than at higher pH. Thus, the substrate could partially neutralize the lower thermostability at low pH by providing new stabilizing interactions. These results suggest that the active site canyon is also important for the stability of xylanases.
The effect of the CBM on the thermostability of XYN10A xylanase was twofold; under some conditions, it increased the thermostability, and under other conditions, it decreased the thermostability. Thus, there is no strong thermostabilizing effect by the CBM on T. flexuosa XYN10A. It was observed earlier that the additional domains may function as thermostabilizing domains, because their deletion often decreased the stability of xylanases (3, 30, 32). However, an increase in thermostability has also been observed when a CBM has been deleted (3, 22, 23a, 26). Thus, the effect of a CBM on thermostability varies, and the reason could be that the primary function of a CBM is to bind polysaccharide fibers and not thermostabilization. In general, the high thermostability of xylanases is not dependent on CBMs, and in fact, they might have diverse effects. The same holds true for protein glycosylations.
In conclusion, we identified several regions in T. flexuosa XYN10A xylanase that affect the protein's thermostability. The effects of the additional groups were either stabilizing or destabilizing. This information can be used in the design of stabilizing mutations. Our study also showed that the production system can considerably affect the properties of the enzymes produced, e.g., due to glycosylation, and that when adding purification tags in recombinant proteins, their potential effects should be considered.
Supplementary Material
Acknowledgments
We thank Johanna Aura for technical assistance, Antti Nyyssölä for assistance in protein purification, and Jarno Kallio, Sanna Hiljanen-Berg, and Sirpa Okko for T. reesei strain fermentation.
Financial support from the Research Foundation of the Helsinki University of Technology and the Academy of Finland is gratefully acknowledged.
Footnotes
Published ahead of print on 23 October 2009.
Supplemental material for this article may be found at http://aem.asm.org/.
REFERENCES
- 1.Ali, M. K., H. Hayashi, S. Karita, M. Goto, T. Kimura, K. Sakka, and K. Ohmiya. 2001. Importance of the carbohydrate-binding module of Clostridium stercorarium Xyn10B to xylan hydrolysis. Biosci. Biotechnol. Biochem. 65:41-47. [DOI] [PubMed] [Google Scholar]
- 2.Andrews, S. R., E. J. Taylor, G. Pell, F. Vincent, V. M.-A. Ducros, G. J. Davies, J. H. Lakey, and H. J. Gilbert. 2004. The use of forced protein evolution to investigate and improve stability of family 10 xylanases. J. Biol. Chem. 279:54369-54379. [DOI] [PubMed] [Google Scholar]
- 3.Araki, R., S. Karita, A. Tanaka, T. Kimura, and K. Sakka. 2006. Effect of family 22 carbohydrate-binding module on the thermostability of Xyn10B catalytic module from Clostridium stercorarium. Biosci. Biotechnol. Biochem. 70:3039-3041. [DOI] [PubMed] [Google Scholar]
- 4.Arnold, K., L. Bordoli, J. Kopp, and T. Schwede. 2006. The SWISS-MODEL workspace: a web-based environment for protein structure homology modelling. Bioinformatics 22:195-201. [DOI] [PubMed] [Google Scholar]
- 5.Black, G. W., G. P. Hazlewood, S. J. Millward-Sadler, J. I. Laurie, and H. J. Gilbert. 1995. A modular xylanase containing a novel non-catalytic xylan-specific binding domain. Biochem. J. 307:191-195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Broersen, K., A. G. Voragen, R. J. Hamer, and H. H. De Jongh. 2004. Glycoforms of beta-lactoglobulin with improved thermostability and preserved structural packing. Biotechnol. Bioeng. 86:78-87. [DOI] [PubMed] [Google Scholar]
- 7.Charnock, S. J., D. N. Bolam, J. P. Turkenburg, H. J. Gilbert, L. M. Ferreira, G. J. Davies, and C. M. Fontes. 2000. The X6 “thermostabilizing” domains of xylanases are carbohydrate-binding modules: structure and biochemistry of the Clostridium thermocellum X6b domain. Biochemistry 39:5013-5021. [DOI] [PubMed] [Google Scholar]
- 8.Chen, B., R. Bautista, K. Yu, G. A. Zapata, A. G. Mulkerrin, and S. M. Chamow. 2003. Influence of histidine on the stability and physical properties of a fully human antibody in aqueous and solid forms. Pharm. Res. 20:1952-1960. [DOI] [PubMed] [Google Scholar]
- 9.Dabrowski, S., and J. Kur. 1998. Recombinant His-tagged DNA polymerase I. Cloning, purification and partial characterization of Thermus thermophilus recombinant DNA polymerase. Acta Biochim. Pol. 45:653-660. [PubMed] [Google Scholar]
- 10.Dabrowski, S., and B. K. Ahring. 2003. Cloning, expression and purification of the His6-tagged hyper-thermostable dUTPase from Pyrococcus woesei in Escherichia coli: application in PCR. Protein Expr. Purif. 31:72-78. [DOI] [PubMed] [Google Scholar]
- 11.DeLano, W. L. 2002. The PyMOL molecular graphics system. Delano Scientific, San Carlos, CA.
- 12.Ducros, V., S. J. Charnock, U. Derewenda, Z. S. Derewenda, Z. Dauter, C. Dupont, F. Shareck, R. Morosoli, D. Kluepfel, and G. J. Davies. 2000. Substrate specificity in glycoside hydrolase family 10. Structural and kinetic analysis of the Streptomyces lividans xylanase 10A. J. Biol. Chem. 275:23020-23026. [DOI] [PubMed] [Google Scholar]
- 13.Reference deleted.
- 14.Fujimoto, Z., S. Kaneko, A. Kuno, H. Kobayashi, I. Kusakabe, and H. Mizuno. 2004. Crystal structures of decorated xylooligosaccharides bound to a family 10 xylanase from Streptomyces olivaceoviridis E-86. J. Biol. Chem. 279:9606-9614. [DOI] [PubMed] [Google Scholar]
- 15.Goodfellow, M., L. A. Maldonado, and E. T. Quintana. 2005. Reclassification of Nonomuraea flexuosa (Meyer 1989) Zhang et al. 1998 as Thermopolyspora flexuosa gen. nov., comb. nov., nom. rev. Int. J. Syst. Evol. Microbiol. 55:1979-1983. [DOI] [PubMed] [Google Scholar]
- 16.Hakulinen, N., O. Turunen, J. Jänis, M. Leisola, and J. Rouvinen. 2003. Three-dimensional structures of thermophilic β-1,4-xylanases from Chaetomium thermophilum and Nonomuraea flexuosa: comparison of twelve xylanases in relation to their thermal stability. Eur. J. Biochem. 270:1399-1412. [DOI] [PubMed] [Google Scholar]
- 17.Holtz, C., H. Kaspari, and J. H. Klemme. 1991. Production and properties of xylanases from thermophilic actinomycetes. Antonie Van Leeuwenhoek 59:1-7. [DOI] [PubMed] [Google Scholar]
- 18.Jafari-Aghdam, J., K. Khajeh, B. Ranjbar, and M. Nemat-Gorgani. 2005. Deglycosylation of glucoamylase from Aspergillus niger: effects on structure, activity and stability. Biochim. Biophys. Acta 1750:61-68. [DOI] [PubMed] [Google Scholar]
- 19.Job, V., G. Molla, M. S. Pilone, and L. Pollegioni. 2002. Overexpression of a recombinant wild-type and His-tagged Bacillus subtilis glycine oxidase in Escherichia coli. Eur. J. Biochem. 269:1456-1463. [DOI] [PubMed] [Google Scholar]
- 20.Kamondi, S., A. Szilágyi, L. Barna, and P. Závodszky. 2008. Engineering the thermostability of a TIM-barrel enzyme by rational family shuffling. Biochem. Biophys. Res. Commun. 374:725-730. [DOI] [PubMed] [Google Scholar]
- 21.Kittur, F. S., S. L. Mangala, A. A. Rusd, M. Kitaoka, H. Tsujibo, and K. Hayashi. 2003. Fusion of family 2b carbohydrate-binding module increases the catalytic activity of a xylanase from Thermotoga maritima to soluble xylan. FEBS Lett. 549:147-151. [DOI] [PubMed] [Google Scholar]
- 22.Leskinen, S., A. Mäntylä, R. Fagerstörm, J. Vehmaanperä, R. Lantto, M. Paloheimo, and P. Suominen. 2005. Thermostable xylanases, Xyn10A and Xyn11A, from the actinomycete Nonomuraea flexuosa: isolation of the genes and characterization of recombinant Xyn11A polypeptides produced in Trichoderma reesei. Appl. Microbiol. Biotechnol. 67:495-505. [DOI] [PubMed] [Google Scholar]
- 23.Lo Leggio, L., S. Kalogiannis, M. K. Bhat, and R. W. Pickersgill. 1999. High resolution structure and sequence of T. aurantiacus xylanase I: implications for the evolution of thermostability in family 10 xylanases and enzymes with βα-barrel architecture. Proteins Struct. Funct. Genet. 36:295-306. [PubMed] [Google Scholar]
- 23a.Mamo, G., R. Hatti-Kaul, and B. Mattiasson. 2007. Fusion of carbohydrate binding modules from Thermotoga neopolitana with a family 10 xylanase from Bacillus halodurans S7. Extremophiles 11:169-177. [DOI] [PubMed] [Google Scholar]
- 24.Mangala, S. L., F. S. Kittur, M. Nishimoto, K. Sakka, K. Ohmiya, M. Kitaoka, and K. Hayashi. 2003. Fusion of family VI cellulose-binding domains to Bacillus halodurans xylanase increases its catalytic activity and substrate binding capacity to insoluble xylan. J. Mol. Catal. B Enzymol. 21:221-230. [Google Scholar]
- 25.Meissner, K., D. Wassenberg, and W. Liebl. 2000. The thermostabilizing domain of the modular xylanase XynA of Thermotoga maritima represents a novel type of binding domain with affinity for soluble xylan and mixed-linkage β-1,3/β-1,4-glucan. Mol. Microbiol. 36:898-912. [DOI] [PubMed] [Google Scholar]
- 26.Morris, D. D., M. D. Gibbs, C. W. J. Chin, M.-H. Koh, K. K. Y. Wong, R. W. Allison, P. J. Nelson, and P. L. Bergquist. 1998. Cloning of the xynB gene from Dictyoglomus thermophilum Rt46B.1 and action of the gene product on kraft pulp. Appl. Environ. Microbiol. 64:1759-1765. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Paloheimo, M., A. Mäntylä, J. Kallio, and P. Suominen. 2003. High-yield production of a bacterial xylanase in the filamentous fungus Trichoderma reesei requires a carrier polypeptide with an intact domain structure. Appl. Environ. Microbiol. 69:7073-7082. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Rixon, J. E., J. H. Clarke, G. P. Hazelwood, R. W. Hoyland, A. J. McCarthy, and H. J. Gilbert. 1996. Do the non-catalytic polysaccharide-binding domains and linker regions enhance the biobleaching properties of modular xylanases? Appl. Microbiol. Biotechnol. 46:514-520. [DOI] [PubMed] [Google Scholar]
- 29.Sevo, M., G. Degrassi, N. Skoko, V. Venturi, and G. Ljubijankić. 2002. Production of glycosylated thermostable Providencia rettgeri penicillin G amidase in Pichia pastoris. FEMS Yeast Res. 1:271-277. [DOI] [PubMed] [Google Scholar]
- 30.Shin, E. S., M. J. Yang, K. Hwa Jung, E. J. Kwon, J. S. Jung, S. K. Park, J. Kim, H. D. Yun, and H. Kim. 2002. Influence of the transposition of the thermostabilizing domain of Clostridium thermocellum xylanase (XynX) on xylan binding and thermostabilization. Appl. Environ. Microbiol. 68:3496-3501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Spiriti, J., F. Bogani, A. van der Vaart, and G. Ghirlanda. 2008. Modulation of protein stability by O-glycosylation in a designed Gc-MAF analog. Biophys. Chem. 134:157-167. [DOI] [PubMed] [Google Scholar]
- 32.Sunna, A., M. D. Gibbs, and P. L. Bergquist. 2000. The thermostabilizing domain, XynA, of Caldibacillus cellulovorans xylanase is a xylan binding domain. Biochem. J. 346:583-586. [PMC free article] [PubMed] [Google Scholar]
- 33.Turunen, O., K. Etuaho, F. Fenel, J. Vehmaanperä, X. Wu, J. Rouvinen, and M. Leisola. 2001. Combination of weakly stabilizing mutations with a disulfide-bridge in the α-helix region of Trichoderma reesei endo-1,4-β-xylanase II increases the thermal stability through synergism. J. Biotechnol. 88:37-46. [DOI] [PubMed] [Google Scholar]
- 34.Reference deleted.
- 35.Xie, H., J. Flint, M. Vardakou, J. H. Lakey, R. J. Lewis, H. J. Gilbert, and C. Dumon. 2006. Probing the structural basis for the difference in thermostability displayed by family 10 xylanases. J. Mol. Biol. 360:157-167. [DOI] [PubMed] [Google Scholar]
- 36.Xiong, H., A. Nyyssölä, J. Jänis, H. Santa, O. Pastinen, N. von Weymarn, M. Leisola, and O. Turunen. 2004. Characterization of the xylanase produced by submerged cultivation of Thermomyces lanuginosus DSM 10635. Enzyme Microb. Technol. 35:93-99. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.