Abstract
The hop plant, Humulus lupulus L., has an exceptionally high content of secondary metabolites, the hop α-acids, which possess a range of beneficial properties, including antiseptic action. Studies performed on the mode of action of hop iso-α-acids have hitherto been restricted to lactic acid bacteria. The present study investigated molecular mechanisms of hop iso-α-acid resistance in the model eukaryote Saccharomyces cerevisiae. Growth inhibition occurred at concentrations of hop iso-α-acids that were an order of magnitude higher than those found with hop-tolerant prokaryotes. Chemostat-based transcriptome analysis and phenotype screening of the S. cerevisiae haploid gene deletion collection were used as complementary methods to screen for genes involved in hop iso-α-acid detoxification and tolerance. This screening and further analysis of deletion mutants confirmed that yeast tolerance to hop iso-α-acids involves three major processes, active proton pumping into the vacuole by the vacuolar-type ATPase to enable vacuolar sequestration of iso-α-acids and alteration of cell wall structure and, to a lesser extent, active export of iso-α-acids across the plasma membrane. Furthermore, iso-α-acids were shown to affect cellular metal homeostasis by acting as strong zinc and iron chelators.
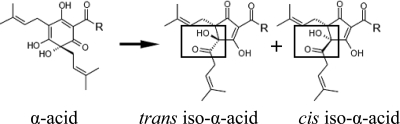
The hop plant, Humulus lupulus L., belongs to the Cannabaceae family (36). Hop cones have an exceptionally high content of secondary metabolites, the hop acids, which account for up to 25% of their dry weight (13). The hop acids consist of two related groups of compounds, the α-acids and the β-acids. Three major types of α-acids (humulone, cohumulone, and adhumulone) are water extracted from the resin and isomerized to six different iso-α-acid isomers (cis,trans-humulone, cis,trans-cohumulone, and cis,trans-adhumulone, Fig. 1). These iso-α-acids possess antiseptic properties and impart the typical bitter flavor of beer (28). Their average concentration is 20 to 30 mg liter−1 in lager beer (28). In addition to the α-acids, hop-derived polyphenols, flavonoids, and prenylated flavonoids are present in trace amounts in beer (13).
FIG. 1.
Isomerization of α-acids to cis- and trans-iso-α-acids during wort boiling. Humulone [R = -(CH)2CH(CH3)2] is converted to cis- and trans-isohumulone, cohumulone [R = -CH(CH3)2] is converted to cis- and trans-isocohumulone, and adhumulone [R = -CH(CH3)CH2CH3] is converted to cis- and trans-isoadhumulone.
The antiseptic properties of hop extracts have been widely studied because of their relevance to beer production. In 1888, Hayduck showed that the antiseptic properties of the hop plant were due to the soft resins (24). Later studies identified humulone as a hop antiseptic agent. Gram-positive, but not Gram-negative, bacteria are sensitive to α- and iso-α-acids. This sensitivity increases with decreasing pH (52, 53), thus resembling classic weak organic acid stress, where uncoupling of the plasma membrane pH gradient causes an increased ATP requirement for proton export (48). Indeed, potentiometric studies support the notion that undissociated trans-isohumulone acts as a protonophore that dissipates the transmembrane pH gradient (1). Lactic acid bacteria that occur as common beer spoilage organisms, such as Lactobacillus brevis, have developed resistance to hop iso-α-acids. The mechanisms responsible include expulsion of iso-α-acids by the cytoplasmic membrane ABC transporter HorA (4, 5, 48). Increased activity of H+-ATPase has also been implicated in tolerance, via the generation of a proton motive force that can be used to energize export of Mn-chelated hop iso-α-acids from the cell (55, 56).
Beneficial health effects have been attributed to hop compounds since medieval times (65), and recent experimental research has indicated anticancer potential for selected hop-derived compounds (reviewed in references 18 and 57). However, the molecular mechanisms that underlie these effects and possible mechanisms of hop acid tolerance in eukaryotic cells remain largely unknown. The present study aimed to investigate mechanisms of hop iso-α-acid toxicity and tolerance in a eukaryotic organism. The availability of a well-annotated genome sequence (19), its excellent genetic accessibility, and the importance of Saccharomyces yeasts in beer fermentation make Saccharomyces cerevisiae an ideal model organism for such a study. Two complementary genome-wide approaches were employed to investigate cellular responses of S. cerevisiae to hop extracts enriched in iso-α-acids. Microarray-based transcriptome analysis was performed on chemostat cultures of an S. cerevisiae reference strain grown in the presence or absence of iso-α-acids. In addition, screening of the nearly complete set of yeast open reading frame haploid knockouts generated by the Saccharomyces Genome Deletion Project (Open Biosystems) (67) identified specific mutants with increased hop sensitivity. Subsequently, the involvement of selected genes and cellular processes in hop acid sensitivity and tolerance was analyzed by construction and detailed analysis of selected mutant strains.
MATERIALS AND METHODS
Strains.
The S. cerevisiae strains used in this study are listed in Table 1. Deletion mutants were constructed by using standard yeast medium and genetic techniques and were routinely grown at 30°C on complex (YPD) or defined medium (11). Gene deletions were introduced into strain CEN.PK113-5D (provided by P. Kötter, Frankfurt, Germany), which carries an auxotrophy for uracil. The Geneticin (G418) resistance cassette was amplified by using the pUG6 vector as the template (22), while the genes for hygromycin B (hphNT1) and nourseothricin (natNT2) resistance were amplified from pAG32 and pAG25, respectively (20). Dominant selection markers were removed by the Cre-loxP recombination system as previously described (22). For chemostat cultivation, strains IMK133 (MATa MAL2-8c SUC2 ura3 pdr3::loxP pdr1::loxP kanMX loxP) and IMK246 (MATa MAL2-8c SUC2 ura3 pdr3::loxP pdr1::loxP kanMX loxP yrr1::hphNT1 yrm1::natNT2) were transformed with plasmid p426-GPD (40) carrying URA3, resulting in strains IMK159 and IMZ110, respectively (Table 1). Strains expressing C-terminal green fluorescent protein (GFP) fusions were selected on hygromycin-containing plates. The hygromycin resistance cassette containing the GFP tag was amplified from the pYM25 vector as the template (30). For the oligonucleotide primer sequences used in this study, see Table S1 in the supplemental material.
TABLE 1.
Strains used in this study
| Strain | Genotypea | Source or reference |
|---|---|---|
| Open Biosystems | MATahis3Δ1leu2Δ0met15Δ0ura3Δ0gene::loxP-kanMX-loxP | Open Biosystems |
| Knockout collection | ||
| CEN.PK113-7D | MATaMAL2-8cSUC2 | P. Kötterb |
| CEN.PK113-5D | MATaMAL2-8cSUC2ura3 | P. Kötter |
| IMK123 | MATaMAL2-8cSUC2ura3pdr1::loxP-kanMX-loxP | This study |
| IMK124 | MATaMAL2-8cSUC2ura3pdr3::loxP-kanMX-loxP | This study |
| IMK130 | MATaMAL2-8cSUC2ura3snq2::loxP-kanMX-loxP | This study |
| IMK131 | MATaMAL2-8cSUC2ura3pdr5::loxP-kanMX-loxP | This study |
| IMK132 | MATaMAL2-8cSUC2ura3pdr15::loxP-kanMX-loxP | This study |
| IMK133 | MATaMAL2-8cSUC2ura3pdr3::loxPpdr1::loxP-kanMX-loxP | This study |
| IMK150 | MATaMAL2-8cSUC2ura3pdr5::loxPpdr15::loxPsnq2::loxP-kanMX-loxP | This study |
| IMK241 | MATaMAL2-8cSUC2ura3pdr3::loxPpdr1::loxP-kanMX-loxPyrm1::hphNT1 | This study |
| IMK246 | MATaMAL2-8cSUC2ura3pdr3::loxPpdr1::loxP-kanMX-loxPyrm1::hphNT1yrr1::natNT2 | This study |
| IMK159 | MATaMAL2-8cSUC2ura3pdr3::loxPpdr1::loxP-kanMX-loxP p426GPD(URA3) | This study |
| IMZ110 | MATaMAL2-8cSUC2ura3pdr3::loxPpdr1::loxP-kanMX-loxPyrm1::hphNT1yrr1::natNT2 p426GPD (URA3) | This study |
| IMN008 | MATaMAL2-8cSUC2ura3pdr5::yEGFP-hph | This study |
| IMN009 | MATaMAL2-8cSUC2ura3pdr15::yEGFP-hph | This study |
| IMN010 | MATaMAL2-8cSUC2ura3snq2::yEGFP-hph | This study |
| IMN011 | MATaMAL2-8cSUC2ura3tpo1::yEGFP-hph | This study |
Knocked-out genes are in boldface. yEGFP, yeast enhanced GFP; gene, any of the 4,786 open reading frames that comprise the MATa yeast knockout collection (Open Biosystems, Huntsville, AL).
Institut für Mikrobiologie der J. W. Goethe Universität.
Media.
The synthetic medium (59, 64) for shake flask and chemostat cultivation contained 5 g liter−1 (NH4)2SO4, 3 g liter−1 KH2PO4, 0.5 g liter−1 MgSO4, and trace elements. The pH of the medium was adjusted to 4.0, 5.0, or 6.0, and it was sterilized by autoclaving at 120°C. Glucose (20 g liter−1 for shake flask cultivation and 25 g liter−1 for chemostat cultivation), vitamins and, for chemostat cultivation, the anaerobic growth factors Tween 80 and ergosterol were added as supplements after sterilization (64). The glucose was autoclaved separately at 110°C, and the vitamins were filter sterilized. When necessary, uracil was added to solid medium to a final concentration of 0.02 g liter−1. Phenotype screening and plate assays were performed with synthetic agar medium containing all of the auxotrophic requirements (yeast nitrogen base at 6.7 g liter−1 and yeast dropout supplement at 1.92 g liter−1 [11]) with an initial pH of 6.0. For anaerobic growth, 420 μg/ml Tween 80 and 10 μg/ml ergosterol were added to the medium. The assays at alkaline pH were performed with synthetic agar medium containing 100 mM Trizma buffer, and the pH was adjusted to 7.5 or 8.0.
Preisomerized hop extract.
Hop extract enriched in iso-α-acids was obtained from Joh. Barth & Sohn (Nuremberg, Germany). The extract consisted of a mixture of six different iso-α-acids. Purification of iso-α-acids from hop extracts was performed according to the method of G. Aerts and J. De Clippeleer (personal communication). Three purified fractions were obtained: a total iso-α-acid purified fraction, a cis-enriched fraction, and a trans-enriched fraction. The four different hop solutions were analyzed by high-performance liquid chromatography (HPLC) with a Symmetry C18 column (Waters Chromatography, Milford, MA) using 48.6% (vol/vol) acetonitrile as the mobile phase. The compositions of the hop extracts used in this study are indicated in Table 2. All hop fractions were stored at 4°C, protected from light, and filter sterilized before use.
TABLE 2.
Compositions of the four hop solutions used in this study and modifications introduced into the medium to normalize the hop iso-α-acid stress level
| Solution | Total iso-α-acids (g liter−1) | Individual iso-α-acid concn (g liter−1)a |
% (concn [g liter−1]) |
Amt (μl) for 100 ml medium 0.2 g liter−1 total iso-α-acidsb | ||||||
|---|---|---|---|---|---|---|---|---|---|---|
| TIC | CIC | TIH | CIH | TIA | CIA | cis-iso-α-acids | trans-iso-α-acids | |||
| Iso-α-acid-enriched hop extract | 160 | 14.7 | 38.2 | 24.3 | 62.4 | 5.6 | 14.6 | 64 (102.4) | 28 (44.8) | 135 |
| Purified hop extract | 175.9 | 14.7 | 39.0 | 26.9 | 73.3 | 5.9 | 16.1 | 72 (126.7) | 27 (47.5) | 120 |
| cis-enriched fraction | 75.6 | 3.3 | 15.7 | 5.1 | 39.1 | 1.1 | 8.3 | 86 (62.4) | 13 (9.4) | 220 |
| trans-enriched fraction | 18.5 | 44.9 | 2.8 | 10.8 | 22.1 | 5.9 | 1.2 | 14 (2.6) | 86 (15.9) | 370 |
Abbreviations: TIC, trans-isocohumulone; CIC, cis-isocohumulone; TIH, trans-isohumulone; CIH, cis-isohumulone; TIA, trans-isoadhumulone; CIA, cis-isoadhumulone.
cis, 0.14 g liter−1; trans, 0.06 g liter−1.
Shake flask cultivation.
Growth experiments were performed with 500-ml flasks containing 100 ml of mineral medium supplemented with glucose, vitamins, and iso-α-acid-enriched hop extract at an initial pH of 4.0, 5.0, or 6.0. The flasks were incubated at 30°C on an orbital shaker set at 200 rpm. The growth of different strains was monitored by measurement of optical density at 660 nm.
Phenotype screening and plate assays.
The Open Biosystems haploid deletion collection (BY4741) (http://www.openbiosystems.com/GeneExpression/Yeast/YKO/) (67) was screened for hop acid sensitivity. Liquid cultures from 96-well plates were used as the inoculum for growth in plate assays on glucose synthetic medium supplemented with 0, 0.2, or 1 g liter−1 of iso-α-acid-enriched hop extract. Agar plates were incubated at 30°C, and growth was scored after a maximum of 48 h. Hop extract susceptibility of yeast mutants was confirmed by spotting 105 cells ml−1 (and three serial 10-fold dilutions) of exponentially growing cultures onto synthetic medium plates supplemented with 0.2 or 1 g liter−1 hop extract (iso-α-acid-enriched preparation and purified fraction). The mixture consisted of approximately 70% cis- and 30% trans-iso-α-acids (Table 2), meaning that 0.2 g liter−1 total iso-α-acids was equivalent to 0.14 g liter−1 cis- and 0.06 g liter−1 trans-iso-α-acids. The supplementation of the medium with cis- and trans-iso-α-acid-enriched solutions was normalized so as to obtain concentrations equivalent to those of the initial fractions (Table 2). Anaerobic cultures were incubated in a Bactron Anaerobic Environmental Chamber (Sheldon Manufacturing, Inc., Cornelius, OR).
Chemostat cultivation.
Anaerobic glucose-limited steady-state chemostat cultures at a dilution rate of 0.10 h−1 of S. cerevisiae strains CEN.PK113-7D, IMK159 (pdr3::loxP pdr1::loxP kanMX loxP), and IMZ110 (pdr3::loxP pdr1::loxP kanMX loxP yrr1::hphNT1 yrm1::natNT2) were prepared as previously described (64). Iso-α-acid-enriched hop extracts were filter sterilized and added at a concentration of 0.2 g liter−1. Medium reservoirs containing iso-α-acid-enriched hop extracts were stored at 4°C and protected from light.
Analytical methods.
Culture supernatants were obtained after centrifugation of samples from the chemostats. For glucose determination and carbon recovery, culture supernatants and media were analyzed by HPLC on an AMINEX HPX-87H ion-exchange column using 5 mM H2SO4 as the mobile phase. Culture dry weights were determined via filtration as described by Postma et al. (45).
Microarray analysis.
Sampling of cells from chemostats, total RNA extraction, cDNA synthesis, and hybridization on the arrays were performed as previously described (14).
Transcriptomic data acquisition and statistical analysis.
Acquisition and quantification of array images and data filtering were performed using Affymetrix GeneChip Operating Software version 1.2. To eliminate insignificant variations, genes with expression values below 12 were set to 12 as previously described (9). The Significance Analysis of Microarrays (version 1.12) (61) add-in to Microsoft Excel was used for comparison of replicate array experiments using a fold change threshold of two and an expected false-discovery rate of 0.125%. Transcript data can be downloaded from the Genome Expression Omnibus database (3) (http://www.ncbi.nlm.nih.gov/geo/) under series accession number GSE15094.
Fisher's exact test.
The genes differentially expressed were examined for enrichment in functional annotation in the MIPS (39) and GO (17) databases as previously described (33).
Motif discovery.
Promoter analysis was performed using the web-based software Regulatory Sequence Analysis tools (62, 63). The promoters (from −800 to −1) of each set of coregulated genes were analyzed for overrepresented hexanucleotides.
Fluorescence microscopy.
Strains containing GFP-fused ABC transporters were grown in shake flasks containing mineral medium supplemented with glucose, vitamins, and uracil in the presence or absence of 0.2 g liter−1 iso-α-acids. The cells were washed once and observed under a microscope. Phase-contrast and fluorescence microscopy was performed with a Zeiss Imager.D1 microscope equipped with a 100× Plan Neofluor oil immersion lens. Zeiss filter set FS10 (excitation bandpass, 450 to 490 nm; emission bandpass, 515 to 565 nm) was used for GFP studies. Images were taken with a Zeiss Axiocam MRc (exposure time, 547 ms) and analyzed using the Axiovision 4.5 software.
Differential vacuolar and cytosolic pool fractionation.
For the determination of the subcellular localization of hop iso-α-acids in yeast cells, shake flask cultures were grown on glucose synthetic medium with 0.2 g liter−1 hop extract. The equivalent of 3 × 108 cells were harvested by centrifugation (4,500 rpm, 5 min) and washed thrice with water. Differential fractionation of vacuolar and cytosolic pools from total cells was performed as previously described (32, 43). Total cell extract and vacuolar and cytosolic pools were analyzed for iso-α-acid content by HPLC as described above. Arginine was measured by HPLC using a Biochrom20 amino acid analyzer from Biochrom Ltd. (Cambridge, England) with Li-citrate buffer as the mobile phase. Detection was performed after derivatization with ninhydrin at 570 nm according to the procedure developed by Ansynth Service B.V. (Roosendaal, The Netherlands).
RESULTS
Effects of hop iso-α-acids on specific growth rates and biomass yields on glucose.
To assess the impact of hop iso-α-acids on specific growth rates, S. cerevisiae CEN.PK113-7D was grown in shake flask cultures on glucose synthetic medium (pH 6.0) in the presence of different concentrations of iso-α-acid-enriched hop extract. At 0.2 g liter−1 of iso-α-acids, S. cerevisiae CEN.PK113-7D exhibited a specific growth rate close to that of reference cultures grown in the absence of hop acids (μmax = 0.38 h−1, Table 3). A further increase in the iso-α-acid concentration to 0.5 g liter−1 led to a decrease in the growth rate to 0.32 h−1. Growth inhibition was more pronounced at a low initial culture pH (pH 5.0 and pH 4.0, Table 3), suggesting that the undissociated forms of the iso-α-acids are primarily responsible for growth inhibition.
TABLE 3.
Maximum specific growth rate of CEN.PK113-7D grown in shake flask cultures in the presence of increasing concentrations of iso-α-acid-enriched hop extract as a function of pH
| Growth condition | μmax (h−1)a at pH of: |
||
|---|---|---|---|
| 6.0 | 5.0 | 4.0 | |
| Reference (no hops) | 0.39 ± 0.00 | 0.36 ± 0.01 | 0.33 ± 0.01 |
| 0.2 g liter−1 iso-α-acid-enriched hop extract | 0.39 ± 0.01 (1.01)b | 0.34 ± 0.01 (0.95) | 0.28 ± 0.01 (0.87) |
| 0.5 g liter−1 iso-α-acid-enriched hop extract | 0.32 ± 0.02 (0.81) | 0.28 ± 0.01 (0.77) | 0.20 ± 0.01 (0.62) |
Values represent the mean ± standard deviation of data from triplicate independent experiments.
In parentheses is the decay ratio versus the reference, which was set to 1.
Effects of hop iso-α-acids on biomass yield were analyzed in anaerobic glucose-limited chemostat cultures (dilution rate, 0.10 h−1) of S. cerevisiae CEN.PK113-7D grown in the presence or absence of 0.2 g liter−1 iso-α-acid-enriched hop extract. Under these conditions, the biomass yield (Yx/s) in the presence of hop acids was slightly but significantly (Student's t test, P value = 0.015 < 0.05) lower than in the reference cultures (Table 4). An uncoupling effect, possibly at the plasma membrane, was further supported by slight increases in the specific rates of ethanol and carbon dioxide production in the cultures supplemented with hop extracts (Table 4).
TABLE 4.
Physiology of CEN.PK113-7D in glucose-limited, anaerobic chemostat cultures at a dilution rate of 0.10 h−1 in the presence or absence of 0.2 g liter−1 iso-α- acid-enriched hop extracta
| Growth condition | YX/S (g g−1) | Production rate (mmol g−1 h−1) |
Carbon recovery (%) | ||
|---|---|---|---|---|---|
| Glucose | Ethanol | CO2 | |||
| Reference (no hops) | 0.10 ± 0.0 | 5.6 ± 0.3 | 8.8 ± 0.1 | 10.1 ± 0.3 | 102 ± 7 |
| 0.2 g liter−1 iso-α-acid-enriched hop extract | 0.08 ± 0.0 | 6.3 ± 0.1 | 10.3 ± 0.4 | 11.2 ± 0.7 | 99 ± 1 |
Values represent the mean ± standard deviation of data from triplicate independent steady-state chemostat cultivations.
Genome-wide analysis of hop iso-α-acid tolerance mechanisms.
To identify molecular mechanisms involved in hop iso-α-acid resistance in S. cerevisiae, a haploid deletion collection in the BY4741 strain background was screened for resistance to iso-α-acid-enriched hop extracts. This screen revealed 20 hop extract-sensitive mutants (Table 5), of which 14 were inhibited by 0.2 g liter−1 of hop iso-α-acids while 6 only showed reduced growth in the presence of 1 g liter−1 hop iso-α-acids (see Fig. S1 in the supplemental material). To verify that the observed phenotype was specific to iso-α-acids (and not by contaminants that might be present in the hop extracts), three additional hop fractions were tested: a purified iso-α-acid extract and two preparations that were enriched with the cis or trans isomers of iso-α-acids. The sensitivity of these 20 mutants was unaltered under both aerobic and anaerobic conditions, thus confirming that they were indeed sensitive to cis and trans isomers of iso-α-acids. When the 20 deletion mutations were introduced into the CEN.PK113-5D genetic background, the resulting mutants all exhibited the same phenotype as the BY4741-derived mutants (results not shown). The main cellular functions affected in the 20 sensitive mutants (Table 5) were vacuolar acidification (10 genes), vacuolar protein sorting (5 genes), and homeostasis of iron and zinc (2 genes). Three additional mutations were identified as causing hop acid sensitivity: rnr4Δ (involved in DNA repair), fun32Δ (functions in signal transduction for shmoo formation in response to ceramide), and och1Δ (α-1,6-mannosyltransferase).
TABLE 5.
Hop-sensitive mutants identified by screening the Open Biosystems deletion collection
| Cellular function and gene(s) | Description |
|---|---|
| Vacuolar acidification | |
| VMA6, VMA9 | V0 domain subunits |
| VMA1, VMA5, VMA8, VMA10, VMA13 | V1 domain subunits |
| VPH2, VMA21, VMA22 | V-ATPase assembly factors |
| Protein trafficking | |
| VPS16 | Vacuolar protein sorting |
| CHC1a | Clathrin coat required for internalization step of endocytosis |
| VPS52a, VPS53a, VPS54a | Vacuolar protein sorting |
| Metal homeostasis | |
| AFT1 | Iron-responsive transcription factor |
| ZAP1a | Zinc-activated protein |
| Other | |
| OCH1 | α-1,6-Mannosyltransferase, initiation of mannose outer chain elongation |
| FUN32 | Regulatory A subunit of protein serine-threonine phosphatase 2A, activated by ceramide |
| RNR4a | Ribonucleotide reductase 4, required for normal levels of translesion synthesis DNA repair |
The corresponding mutant was only sensitive to 1 g liter−1 hop iso-α-acids. All other mutants were sensitive to 0.2 g liter−1 hop iso-α-acids.
Genome-wide transcriptional responses to hop acids were studied by microarray analysis of anaerobic, glucose-limited chemostat cultures of S. cerevisiae CEN.PK113-7D grown in the presence or absence of 0.2 g liter−1 iso-α-acid-enriched hop extract. Based on a fold change threshold of two and a false-discovery rate of 0.125%, 120 genes were transcriptionally upregulated and 198 genes were downregulated in the presence of hop iso-α-acids (see Table S2 in the supplemental material). Fisher's exact test was applied to the sets of up- and downregulated genes to assess enrichment for functional categories from two publicly available ontology databases: MIPS (39) and GO (2). No enrichment of functional categories was found among the downregulated genes. However, analysis of the 120 upregulated genes revealed overrepresentation of the MIPS category “cell rescue, defense, and virulence” and, more specifically, the subcategories “stress response” (17 genes), “osmotic and salt stress responses” (7 genes), and “detoxification” (9 genes) (Table 6). Most of the upregulated genes belonging to the category “stress response” encoded cell wall proteins (PIR1, PIR3, TIR3, SED1, YGP1, PAU1, PAU2, and PAU7). Furthermore, six multidrug resistance (MDR) transporters were significantly upregulated. Of these, the largest change (12-fold) was observed for PDR5, while transcript levels of SNQ2, TPO1, and PDR15 increased by 2- to 4-fold. The transcript levels of two additional genes encoding MDR transporters, PDR16 and AZR1, also increased significantly, but their expression levels remained low (see Table S2 in the supplemental material).
TABLE 6.
Enrichment analysis for functional categoriesa among the 120 transcriptionally upregulated genes in response to 0.2 g liter−1 iso-α-acid-enriched hop extract in anaerobic carbon-limited chemostat culture
| Database and functional category | No. of genes in: |
P value | Genes | |
|---|---|---|---|---|
| Module (total n = 120) | Genome (total n = 6,384) | |||
| GO, iron ion transport | 6 | 32 | 2.1 × 10−5 | AFT1, SIT1, FIT1, FIT2, FIT3, FET3 |
| MIPS | ||||
| 32. Cell rescue, defense, and virulence | 3 | 559 | 5.2 × 10−8 | RTA1, PDR16, FET3 |
| 32.01. Stress response | 17 | 454 | 1.3 × 10−5 | AHP1, PIR1, PIR3, HMS2, PAU7, CLN3, TIR3, SOD2, PAU1, WWM1, PAU2, SED1, SNQ2, RAD59, YBR016W, PIN4, YGP1 |
| 32.01.03. Osmotic and salt stress responses | 7 | 59 | 1.8 × 10−4 | VRP1, RRD1, HSP12, ISC1, GPD1, LAS17, GRE2 |
| 32.07. Detoxification | 9 | 119 | 7.0 × 10−4 | AHP1, TPO1, SOD2, AZR1, SIT1, SNQ2, SRL1, PDR5, GRE2 |
The PDRE regulatory network participates in hop iso-α-acid tolerance.
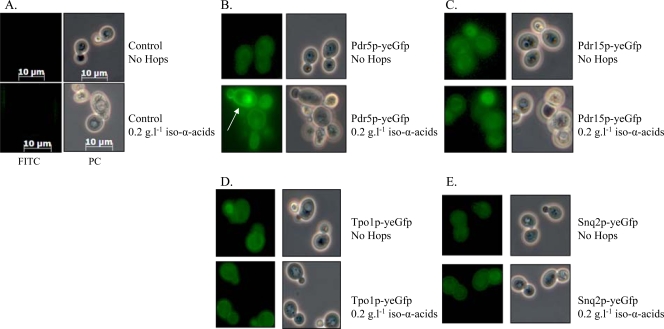
Chemostat-based transcriptome analysis revealed a significant upregulation of several MDR transporters in the presence of iso-α-acids. To verify that this response was not limited to transcription, C-terminal GFP fusions of PDR5, PDR15, TPO1, and SNQ2 were constructed. Yeast strains expressing these gene fusions were grown in shake flasks on glucose synthetic medium with and without 0.2 g liter−1 iso-α-acid-enriched hop extract. Subsequently, the expression and cellular localization of the fusion proteins were examined by fluorescence microscopy. The fusion proteins Pdr5p, Pdr15p, Tpo1p, and Snq2p were expressed in the absence, as well as in the presence, of hop extract (Fig. 2). Pdr5p, which was transcribed at the highest levels, was clearly localized to the plasma membrane and strongly induced in response to hop iso-α-acids (Fig. 2). The profile of the Tpo1p fusion was the same, albeit less pronounced. Due to low expression levels, no clear indication could be retrieved from the Pdr15p and Snq2 localization assays.
FIG. 2.
Fluorescence microscopy of cells containing GFP-fused ABC transporters grown in the absence or presence of 0.2 g liter−1 iso-α-acid-enriched hop extract. Cells were grown in shake flasks containing mineral medium supplemented with glucose, vitamins, and uracil in the presence or absence of iso-α-acid-enriched hop extract. Zeiss filter set FS10 (excitation bandpass, 450 to 490 nm; emission bandpass, 515 to 565 nm) was used for GFP studies, and images were taken with a Zeiss Axiocam MRc (exposure time, 547 ms). The arrow indicates the localization of Pdr5p in the plasma membrane of yeast cells grown in the presence of 200 ppm hops. Panels: A, isogenic reference CEN.PK113-5D; B, IMN008 (Pdr5::Gfp); C, IMN009 (Pdr15::Gfp); D, IMN011 (Tpo1::Gfp); E, IMN010 (Snq2::Gfp). FITC, fluorescein isothiocyanate; PC, phase-contrast.
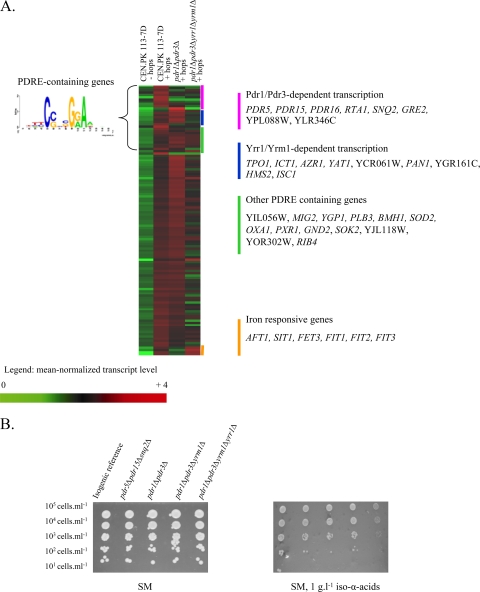
Although transcriptome analysis suggested involvement of MDR transporters in iso-α-acid resistance, the corresponding genes were not identified in the screening of the yeast deletion collection. Analysis of upstream cis-regulatory sequences revealed that 30 hop-upregulated genes (25%) contained a PDRE (pleiotropic drug resistance element) (23), CSNBGRA (Fig. 3A), that can be recognized by five functionally redundant transcription factors (Pdr1p, Pdr3p, Pdr8p, Yrr1p, and Yrm1p). All of the MDR transporter genes discussed in the preceding section were previously shown to be part of this regulon. To study the possibility that the absence of a hop acid sensitivity phenotype in strains carrying individual deletions in transporter genes was due to functional redundancy of MDR transporters, a pdr5 pdr15 snq2 triple-deletion mutant was constructed and all of the strains were tested under aerobic and anaerobic conditions. Although expression of these three genes was clearly induced by hop iso-α-acids and fusion proteins of Pdr5 were located at the plasma membrane, the triple-deletion strain was not more sensitive to hop extracts than the corresponding reference strain (Fig. 3B). In addition, despite the clear induction of the PDRE regulon by hop extracts, single (pdr1Δ or pdr3Δ)- and double (pdr1Δ pdr3Δ)-deletion mutants did not exhibit an increased sensitivity to hop extracts. However, when the structural genes for all four transcriptional activators (PDR1, PDR3, YRR1, and YRM1) were deleted, increased sensitivity was observed in medium containing 1 g liter−1 hop iso-α-acids (Fig. 3B). This result confirmed both the involvement and functional redundancy of the PDRE regulon in hop iso-α-acid resistance.
FIG. 3.
Involvement of the PDRE regulatory network in hop iso-α-acid tolerance. (A) Heat map and identities of genes with a hop iso-α-acid-responsive transcription profile. Transcript levels were determined with Affymetrix Genechips YG-98 and are represented as the average of two or three independent experiments. Shown are the mean normalized transcript levels of 120 genes found to be upregulated when comparing the presence of 0.2 or 1 g liter−1 hop iso-α-acids to the reference condition in CEN.PK113-7D. The transcript levels of these genes in the pdr1Δ pdr3Δ and pdr1Δ pdr3Δ yrr1Δ yrm1Δ mutant strains grown in the presence of hop iso-α-acids are also indicated. (B) Phenotypic analysis of PDR-deficient mutants: the isogenic reference strain and the pdr5Δ pdr15Δ snq2Δ, pdr1Δ pdr3Δ, pdr1Δ pdr3Δ yrr1Δ, and pdr1Δ pdr3Δ yrr1Δ yrm1Δ mutant strains were spotted at 105 cells ml−1 (and three 10-fold serial dilutions) on synthetic agar medium (SM) in the presence or absence of 1 g liter−1 hop iso-α-acids under aerobic conditions (similar results were obtained under anaerobic conditions [data not shown]).
To investigate the transcriptional changes caused by deletion of PDR1 and PDR3, and subsequently YRR1 and YRM1, the pdr1Δ pdr3Δ and pdr1Δ pdr3Δ yrr1Δ yrm1Δ mutant strains were grown in chemostat cultures (experiments were performed in duplicate) in the presence of 0.2 g liter−1 iso-α-acid-enriched hop extract. Under these conditions, the physiological data for both strains were similar to those of the isogenic reference strain (result not shown). Comparison of transcriptome data with those of hop extract-supplemented cultures of reference strain CEN.PK113-7D (see Table S3 in the supplemental material) showed that only one-third of the PDRE-containing genes were solely dependent on Pdr1 and Pdr3 (among which are PDR5, PDR15, PDR16, RTA1, and SNQ2) (Fig. 3). In addition, we found that two MDR transporter-encoding genes (TPO1 and AZR1) were only downregulated upon the deletion of all four transcriptional activators. Hence, the sensitivity of the pdr1Δ pdr3Δ yrr1Δ yrm1Δ quadruple mutant to hop extract in comparison to that of the pdr1Δ pdr3Δ double mutant can be attributed to the transcriptional activation of fewer plasma membrane MDR transporters, thus resulting in less-efficient removal of iso-α-acids from the cells.
Acidification of intracellular compartments: a key mechanism.
No fewer than 10 genes encoding subunits of the vacuolar-type ATPase (V-ATPase), VMA1, VMA5, VMA6, VMA8, VMA9, VMA10, VMA13, VMA21, VMA22, and VPH2, were essential for survival in the presence of hop iso-α-acids (Table 5). They encode subunits of both the V0 (VMA6 and VMA9) and V1 domains (VMA1, VMA5, VMA8, VMA10, and VMA13), along with three assembly factors (VPH2, VMA21, and VMA22) involved in the early-stage assembly of the complex in the endoplasmic reticulum (21). Microarray analysis showed that the transcript levels of these genes were not affected by hop extract addition.
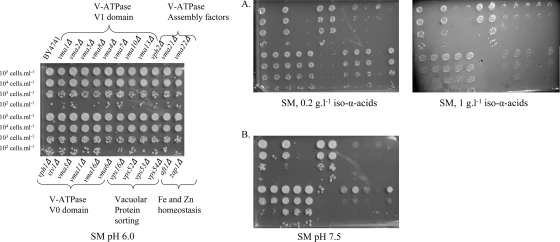
The identification of only 10 of the 17 vma mutants in this screen suggested some specificity for the subunits of the V-ATPase required for hop resistance. To corroborate this, all 17 vma mutants were retrieved from the collection and submitted to a phenotypic analysis at different concentrations of hop iso-α-acids (Fig. 4). The sensitivity of the vma mutants to hop iso-α-acids was 40-fold higher; vma mutants were already sensitive to 0.005 g/liter iso-α-acids. In vivo V-ATPase activity was assessed by growing the vma mutants under alkaline conditions (a well-known phenotype associated with the loss of V-ATPase activity [31]), using synthetic medium agar plates buffered to pH 7.5 with Trizma buffer (Fig. 4). While confirming the results of the initial screening, this further phenotype analysis also showed that the seven hop extract-insensitive vma mutants (the vma2Δ, vma4Δ, vma7Δ, vph1Δ, stv1Δ, vma3Δ, vma11Δ, and vma16Δ mutants) were also insensitive to medium buffered at pH 7.5 (Fig. 4). This result indicates a different contribution of individual subunits to the total in vivo V-ATPase activity which perfectly correlates with their involvement in hop acid tolerance.
FIG. 4.
Phenotypic analysis of mutants of the V-ATPases, vacuolar protein sorting, and ion homeostasis. The strains (BY4741 genetic background) were spotted at 105 cells ml−1 (and three 10-fold serial dilutions) on synthetic agar medium (SM). Panel A, supplemented with 0.2 or 1 g liter−1 hop iso-α-acids; panel B, buffered to pH 7.5 under aerobic conditions (similar results were obtained under anaerobic conditions [data not shown]).
At a hop iso-α-acid concentration of 1 g liter−1, mutants affected in vacuolar protein sorting (the vps16Δ, vps52Δ, vps53Δ, and vps54Δ mutants) and endocytosis (the chc1Δ mutant) showed clear sensitivity (Table 5). vps deletion strains are also sensitive to alkaline conditions (12) (Fig. 4), suggesting an involvement of vacuolar protein sorting in the maintenance of acidic conditions in intracellular compartments. Together, these data indicate that acidification of the vacuole, the Golgi apparatus, and/or the endosomal system is a key mechanism for yeast resistance to hop iso-α-acids.
Hop iso-α-acids are localized in the yeast vacuole.
To investigate whether hop iso-α-acids are targeted to the vacuole, S. cerevisiae CEN.PK113-7D was grown in shake flasks on glucose synthetic medium with 0.2 g liter−1 hop iso-α-acids. Analysis of culture medium and supernatant samples showed that 50% of the iso-α-acids supplied in the medium were taken up by the yeast cells. When analyzing the biomass samples, 70% of the iso-α-acids supplied in the medium were recovered in both biomass and supernatant, indicating possible chemical or biochemical degradation or conversion of iso-α-acids during the incubation period.
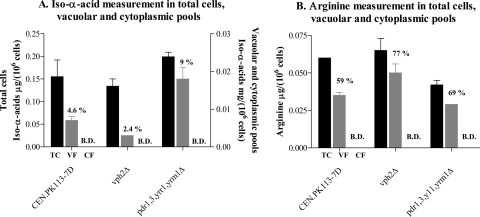
After fractionation, the total biomass and cytoplasmic and vacuolar fractions were analyzed for iso-α-acid content. Distribution of arginine, which is known to be stored in the vacuole (32, 51), was measured as a reference. While the concentration of iso-α-acids and arginine in cytosolic fractions was below the detection limit, a substantial 5% of the total cellular iso-α-acids colocalized with arginine in the vacuolar fraction (Fig. 5A and B). Ninety-five percent of the cellular iso-α-acids could not be retraced in either the cytoplasmic or the vacuolar fraction, suggesting a high localization of iso-α-acids in insoluble parts such as the cell wall and/or cellular membranes. To assess the role of vacuolar acidification and the impact of a higher accumulation of iso-α-acids in the cell, vph2Δ (encoding the assembly factor of the V-ATPase complex) and pdr1Δ pdr3Δ yrr1Δ yrm1Δ mutant strains were included in the analysis. Quantification of iso-α-acid accumulation in the PDR-defective (pdr1Δ pdr3Δ yrr1Δ yrm1Δ) mutant showed a twofold increase in vacuolar iso-α-acid content. In contrast, a mutant defective in the assembly of the V-ATPase complex, the vph2Δ mutant, showed a twofold decrease in the vacuolar content of iso-α-acids relative to its isogenic reference (Fig. 5A). These data indicate that the extent of vacuolar accumulation of hop iso-α-acids depends on influx of extracellular hop acids into the cells, as well as on the generation of a proton motive force by the V-ATPase.
FIG. 5.
Differential fractionation of vacuolar and cytoplasmic pools for iso-α-acid (A) and arginine (B) determination. Values are given in μg/106 cells and are the average of two independent experiments. The percentages are relative to the total biomass determinations. B.D., below detection. Total cells (TC) are represented by black bars, vacuolar fractions (VF) by gray bars, and cytosolic fractions (CF) by white bars.
Hop extracts affect iron and zinc status.
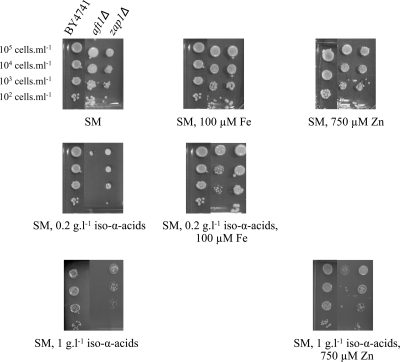
Both the screening of the yeast deletion collection (Table 5) and microarray studies (Table 6) suggested that hop iso-α-acids affected metal ion homeostasis. Among the genes that were transcriptionally upregulated by iso-α-acid-enriched hop extracts, the functional category “iron ion transport” was significantly overrepresented (Table 6). Not only AFT1, encoding the main transcriptional activator of genes involved in iron homeostasis and transport, but also five of its target genes (SIT1, FIT1, FIT2, FIT3, and FET3) were upregulated up to 20-fold in cultures supplemented with hop extract (Table 6). Expression of these iron-related genes was downregulated in a pdr1Δ pdr3Δ mutant strain (Fig. 3), indicating indirect activation via the PDR network. Consistent with the transcriptome analysis, an aft1Δ mutant was sensitive to 0.2 g liter−1 iso-α-acid-enriched hop extract (Table 5). At a hop extract concentration of 1 g liter−1, deletion of ZAP1, which encodes a transcriptional activator of genes involved in zinc homeostasis and transport (68), also resulted in hop iso-α-acid sensitivity. To test whether the observed hop sensitivities of the aft1Δ and zap1Δ mutants were indeed caused by metal ion deficiency, iron and zinc supplementation studies were performed with both mutants. In the presence of 0.2 g liter−1 α-acid-enriched iso-α-acids, addition of 100 μM FeSO4 (10-fold higher than the standard concentration) restored the hop tolerance of an aft1Δ mutant (Fig. 6). The same result was obtained with the purified hop fraction, indicating that the effect was specific to the iso-α-acids. Similarly, hop extract tolerance of a zap1Δ mutant strain at an iso-α-acid concentration of 1 g liter−1 was restored by the addition of 750 μM ZnSO4 (50-fold higher than the reference concentration; Fig. 6). These results are compatible with the chelation of iron and zinc by hop acids, thereby limiting the bioavailability of these essential elements.
FIG. 6.
Phenotypic analysis of aft1Δ and zap1Δ mutants. The strains (BY4741 and the aft1Δ and zap1Δ mutants) were spotted at 105 cells ml−1 (and three 10-fold serial dilutions) on synthetic agar medium (SM; containing 10 μM Fe and 15 μM Zn) supplemented with either 100 μM iron or 750 μM zinc in the presence of 0.2 or 1 g liter−1 hop iso-α-acids.
DISCUSSION
In beer-spoiling lactic acid bacteria such as L. brevis, subculturing in hop acid-containing medium is required to develop resistance to the concentrations of hop acids that occur in beer fermentation (48). In the present study, we investigated the tolerance mechanisms of laboratory strains of S. cerevisiae that had not been previously adapted to hop acids. The lowest concentration of hop iso-α-acids used in the present study is ca. 10-fold higher than the MIC reported for hop-resistant L. brevis (ranging from 0.03 to 0.05 g liter−1) (50) and than in regular lager fermentations (28), and even at these concentrations, the effects of hop iso-α-acids on specific growth rate and biomass yield were moderate. The molecular mechanisms underlying this innate tolerance were investigated by two approaches, which suggested that S. cerevisiae uses complementary cellular mechanisms to defend itself against hop iso-α-acids. All of the mechanisms identified in this study suggest that depletion of hop iso-α-acids from the cytosol is essential in hop iso-α-acid resistance. First, the cell wall composition is modified in response to hop acids, which is likely to contribute to their binding to cell wall constituents and/or to function as a “molecular sieve” of the cell wall that reduces the access of hop iso-α-acids. Membrane transporters belonging to the PDRE regulon act as a second layer of defense and export hop acids from the cytosol to the external medium and, possibly, into the vacuole. Vacuolar accumulation of hop acids, which requires energizing of the vacuolar membrane by the constitutively expressed V-ATPase, is a third key process in protecting cells from the negative effects of hop acids. Finally, upregulation of Aft1p- and Zap1p-regulated genes involved in the homeostasis of cellular Fe and Zn levels appears to counteract the chelation of metal ions by hop iso-α-acids (8).
The two genome-wide screening approaches were found to be highly complementary. Of the three processes that were investigated in detail, the role of the vacuole was only found in the screening of the deletion library, while the PDRE regulon was only implicated in hop acid tolerance through transcriptome analysis. Metal homeostasis and cell wall modification were identified by both approaches, although in the latter case, only a single gene was found in the screening of the mutant library. Many genes involved in the early steps of N-glycosylation of cell wall proteins are essential for yeast growth, thus precluding phenotypic analysis in haploid deletion strains (25). Of the nonessential mannosyltransferases that function in later steps of N-glycosylation, Och1, which was found to be involved in hop resistance, initiates the addition of mannose sugars to the core N-linked oligosaccharides in the cis-Golgi apparatus (41). The Mnn1-10 proteins function later in the glycosylation process, specifically, in side chain branches of linear mannose chains (after Och1-mediated elongation of the core oligosaccharide) (42). The fact that only the och1Δ mutant was sensitive to hop acids indicates that linear mannosylation of glycoproteins is necessary but sufficient to ensure yeast hop resistance.
In contrast to the situation in lactic acid bacteria, where a single ABC transporter (HorA) confers hop resistance (49), multiple transporters belonging to the PDR regulon contributed to hop acid tolerance in S. cerevisiae. The observed functional redundancy of this network and of its transcriptional regulation, which contributes to yeast tolerance to a wide range of organic compounds, confirms and extends the results from previous studies (10, 15, 16, 26, 34, 37, 44, 60). An important difference between prokaryotic and yeast iso-α-acid resistance relates to intracellular compartmentalization and, especially, the role of the vacuole. V-ATPases acidify intracellular compartments such as the Golgi apparatus, endosomes of the secretory pathway, and the vacuole (21, 31). Organelle acidification has been implicated in protein sorting in the biosynthetic and endocytic pathways, proteolytic activation of zymogen precursors, and transmembrane transport of viral contents and toxins (47, 50, 54, 58). Our results presented here indicate that vacuolar acidification by V-ATPases provides the driving force for iso-α-acid accumulation. If analogous to hop acid export in prokaryotes (55, 56), hop acids might be transported into the yeast vacuole as metal anion chelates, which would contribute to the iron and zinc homeostasis responses observed in the presence of hop acids.
In cellular fractionation studies, a large part of the cell-associated iso-α-acids could not be retraced to either cytoplasmic or vacuolar fractions, suggesting their possible localization in the insoluble fractions of the cell, most probably in the cell wall. Cellular retention of hop acids in the cell wall and in the vacuoles is relevant for the brewing process, as it decreases the concentration of these compounds in the final product. Furthermore, brewing yeast strains are commonly reused 4 to 10 times for inoculation in succeeding brews (38). Serial repitching results in a loss of vitality and viability (38), which, based on our results, may at least partly be due to hop iso-α-acid stress in the vacuole. In addition, colocalization of hop iso-α-acids and zinc in the vacuole (6) may decrease zinc bioavailability, another common problem related to beer fermentation (29).
Involvement of the vacuole and intracellular trafficking pathways has been recognized in mechanisms of eukaryote resistance to many toxic compounds (7, 27, 66). These results are consistent with recent findings in mammalian cells that drug resistance is correlated with changes in intracellular trafficking (35, 46), although the exact contribution of these pathways is not known. Identification of the key mechanisms of yeast hop iso-α-acid tolerance will aid the analysis of similar systems in human cells, which is highly relevant for the therapeutic application of hop acids (18, 57). Furthermore, based on the results of this study, it may be possible to engineer hop acid-hypersensitive yeast strains. Such strains would provide a very interesting experimental platform to study molecular targets for hop acid cytotoxicity in yeast and, via heterologous expression studies, in human cells.
Supplementary Material
Acknowledgments
We thank Erwin Suir, Yigen Huang, and Ruben Esse for their technical assistance. We thank G. Aerts (Katholieke Hogeschool St-Lieven, Ghent, Belgium) for kindly supplying the purified and cis- and trans-iso-α-acid-enriched solutions. We also thank Heineken Supply Chain (Zoeterwoude, the Netherlands) for performing HPLC measurement of iso-α-acids. Finally, we thank C. De Virgilio (University of Fribourg, Fribourg, Switzerland) for his invaluable advice on the differential fractionation of cytoplasmic and vacuolar pools.
The research group of J.T.P. is part of the Kluyver Centre for Genomics of Industrial Fermentation, which is supported by the Netherlands Genomics Initiative.
Footnotes
Published ahead of print on 13 November 2009.
Supplemental material for this article may be found at http://aem.asm.org/.
The authors have paid a fee to allow immediate free access to this article.
REFERENCES
- 1.Archibald, F. S., and M. N. Duong. 1984. Manganese acquisition by Lactobacillus plantarum. J. Bacteriol. 158:1-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Ashburner, M., C. A. Ball, J. A. Blake, D. Botstein, H. Butler, J. M. Cherry, A. P. Davis, K. Dolinski, S. S. Dwight, J. T. Eppig, M. A. Harris, D. P. Hill, L. Issel-Tarver, A. Kasarskis, S. Lewis, J. C. Matese, J. E. Richardson, M. Ringwald, G. M. Rubin, and G. Sherlock. 2000. Gene ontology: tool for the unification of biology. Nat. Genet. 25:25-29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Barrett, T., D. B. Troup, S. E. Wilhite, P. Ledoux, D. Rudnev, C. Evangelista, I. F. Kim, A. Soboleva, M. Tomashevsky, K. A. Marshall, K. H. Phillippy, P. M. Sherman, R. N. Muertter, and R. Edgar. 2009. NCBI GEO: archive for high-throughput functional genomic data. Nucleic Acids Res. 37:D885-D890. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Behr, J., M. G. Ganzle, and R. F. Vogel. 2006. Characterization of a highly hop-resistant Lactobacillus brevis strain lacking hop transport. Appl. Environ. Microbiol. 72:6483-6492. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Behr, J., L. Israel, M. G. Ganzle, and R. F. Vogel. 2007. Proteomic approach for characterization of hop-inducible proteins in Lactobacillus brevis. Appl. Environ. Microbiol. 73:3300-3306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Bilinski, C. A., and J. J. Miller. 1983. Translocation of zinc from vacuole to nucleus during yeast meiosis. Can. J. Genet. Cytol. 25:415-419. [DOI] [PubMed] [Google Scholar]
- 7.Blackburn, A. S., and S. V. Avery. 2003. Genome-wide screening of Saccharomyces cerevisiae to identify genes required for antibiotic insusceptibility of eukaryotes. Antimicrob. Agents Chemother. 47:676-681. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Blanco, C. A., I. Caballero, A. Rojas, M. Gomez, and J. Alvarez. 2003. Chelation of aqueous iron(III) by 2-acetyl-1,3-cyclohexanedione and beer ageing. Food Chem. 81:561-568. [Google Scholar]
- 9.Boer, V. M., J. H. de Winde, J. T. Pronk, and M. D. Piper. 2003. The genome-wide transcriptional responses of Saccharomyces cerevisiae grown on glucose in aerobic chemostat cultures limited for carbon, nitrogen, phosphorus, or sulfur. J. Biol. Chem. 278:3265-3274. [DOI] [PubMed] [Google Scholar]
- 10.Brôco, N., S. Tenreiro, C. A. Viegas, and I. Sa-Correia. 1999. FLR1 gene (ORF YBR008c) is required for benomyl and methotrexate resistance in Saccharomyces cerevisiae and its benomyl-induced expression is dependent on pdr3 transcriptional regulator. Yeast 15:1595-1608. [DOI] [PubMed] [Google Scholar]
- 11.Burke, D., D. Dawson, and T. Stearns. 2000. Methods in yeast genetics: edition 2000. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY.
- 12.Conboy, M. J., and M. S. Cyert. 2000. Luv1p/Rki1p/Tcs3p/Vps54p, a yeast protein that localizes to the late Golgi and early endosome, is required for normal vacuolar morphology. Mol. Biol. Cell 11:2429-2443. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.De Keukeleire, D., L. De Cooman, H. Rong, A. Heyerick, J. Kalita, and S. R. Milligan. 1999. Functional properties of hop polyphenols, p. 739-760. In G. G. Gross, R. W. Hemingway, and T. Yoshida (ed.), Plant polyphenols 2: chemistry, biology, pharmacology, ecology. Kluwer Academics/Plenum Publishers, New York, NY. [DOI] [PubMed]
- 14.De Nicola, R., L. A. Hazelwood, E. A. De Hulster, M. C. Walsh, T. A. Knijnenburg, M. J. Reinders, G. M. Walker, J. T. Pronk, J. M. Daran, and P. Daran-Lapujade. 2007. Physiological and transcriptional responses of Saccharomyces cerevisiae to zinc limitation in chemostat cultures. Appl. Environ. Microbiol. 73:7680-7692. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.DeRisi, J., B. van den Hazel, P. Marc, E. Balzi, P. Brown, C. Jacq, and A. Goffeau. 2000. Genome microarray analysis of transcriptional activation in multidrug resistance yeast mutants. FEBS Lett. 470:156-160. [DOI] [PubMed] [Google Scholar]
- 16.Devaux, F., E. Carvajal, S. Moye-Rowley, and C. Jacq. 2002. Genome-wide studies on the nuclear PDR3-controlled response to mitochondrial dysfunction in yeast. FEBS Lett. 515:25-28. [DOI] [PubMed] [Google Scholar]
- 17.Eilbeck, K., S. E. Lewis, C. J. Mungall, M. Yandell, L. Stein, R. Durbin, and M. Ashburner. 2005. The Sequence Ontology: a tool for the unification of genome annotations. Genome Biol. 6:R44. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Gerhäuser, C. 2005. Beer constituents as potential cancer chemopreventive agents. Eur. J. Cancer 41:1941-1954. [DOI] [PubMed] [Google Scholar]
- 19.Goffeau, A., B. G. Barrell, H. Bussey, R. W. Davis, B. Dujon, H. Feldmann, F. Galibert, J. D. Hoheisel, C. Jacq, M. Johnston, E. J. Louis, H. W. Mewes, Y. Murakami, P. Philippsen, H. Tettelin, and S. G. Oliver. 1996. Life with 6000 genes. Science 274:546, 563-567. [DOI] [PubMed] [Google Scholar]
- 20.Goldstein, A. L., and J. H. McCusker. 1999. Three new dominant drug resistance cassettes for gene disruption in Saccharomyces cerevisiae. Yeast 15:1541-1553. [DOI] [PubMed] [Google Scholar]
- 21.Graham, L. A., A. R. Flannery, and T. H. Stevens. 2003. Structure and assembly of the yeast V-ATPase. J. Bioenerg. Biomembr. 35:301-312. [DOI] [PubMed] [Google Scholar]
- 22.Güldener, U., S. Heck, T. Fielder, J. Beinhauer, and J. H. Hegemann. 1996. A new efficient gene disruption cassette for repeated use in budding yeast. Nucleic Acids Res. 24:2519-2524. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Harbison, C. T., D. B. Gordon, T. I. Lee, N. J. Rinaldi, K. D. Macisaac, T. W. Danford, N. M. Hannett, J. B. Tagne, D. B. Reynolds, J. Yoo, E. G. Jennings, J. Zeitlinger, D. K. Pokholok, M. Kellis, P. A. Rolfe, K. T. Takusagawa, E. S. Lander, D. K. Gifford, E. Fraenkel, and R. A. Young. 2004. Transcriptional regulatory code of a eukaryotic genome. Nature 431:99-104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Hayduck, F. 1888. Making tetrahydroisoalpha acids and hexahydroisoalpha acids by catalytic reduction and isomerization. Wochenschr. Brau. 5:937. [Google Scholar]
- 25.Herscovics, A., and P. Orlean. 1993. Glycoprotein biosynthesis in yeast. FASEB J. 7:540-550. [DOI] [PubMed] [Google Scholar]
- 26.Hikkel, I., A. Lucau-Danila, T. Delaveau, P. Marc, F. Devaux, and C. Jacq. 2003. A general strategy to uncover transcription factor properties identifies a new regulator of drug resistance in yeast. J. Biol. Chem. 278:11427-11432. [DOI] [PubMed] [Google Scholar]
- 27.Hillenmeyer, M. E., E. Fung, J. Wildenhain, S. E. Pierce, S. Hoon, W. Lee, M. Proctor, R. P. St. Onge, M. Tyers, D. Koller, R. B. Altman, R. W. Davis, C. Nislow, and G. Giaever. 2008. The chemical genomic portrait of yeast: uncovering a phenotype for all genes. Science 320:362-365. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Hough, J. S., D. E. Briggs, R. Stevens, and T. M. Young. 1982. Malting and brewing science, 2nd ed. Chapman & Hall, New York, NY.
- 29.Jacobsen, T., R. Volden, S. Engan, and O. Aubert. 1979. A chemometric study of some beer flavour components. J. Inst. Brew. 89:265-270. [Google Scholar]
- 30.Janke, C., M. M. Magiera, N. Rathfelder, C. Taxis, S. Reber, H. Maekawa, A. Moreno-Borchart, G. Doenges, E. Schwob, E. Schiebel, and M. Knop. 2004. A versatile toolbox for PCR-based tagging of yeast genes: new fluorescent proteins, more markers and promoter substitution cassettes. Yeast 21:947-962. [DOI] [PubMed] [Google Scholar]
- 31.Kane, P. M. 2006. The where, when, and how of organelle acidification by the yeast vacuolar H+-ATPase. Microbiol. Mol. Biol. Rev. 70:177-191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Kitamoto, K., K. Yoshizawa, Y. Ohsumi, and Y. Anraku. 1988. Dynamic aspects of vacuolar and cytosolic amino acid pools of Saccharomyces cerevisiae. J. Bacteriol. 170:2683-2686. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Kresnowati, M. T., W. A. van Winden, M. J. Almering, A. ten Pierick, C. Ras, T. A. Knijnenburg, P. Daran-Lapujade, J. T. Pronk, J. J. Heijnen, and J. M. Daran. 2006. When transcriptome meets metabolome: fast cellular responses of yeast to sudden relief of glucose limitation. Mol. Syst. Biol. 2:49. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Le Crom, S., F. Devaux, P. Marc, X. Zhang, W. S. Moye-Rowley, and C. Jacq. 2002. New insights into the pleiotropic drug resistance network from genome-wide characterization of the YRR1 transcription factor regulation system. Mol. Cell. Biol. 22:2642-2649. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Liang, X. J., S. Mukherjee, D. W. Shen, F. R. Maxfield, and M. M. Gottesman. 2006. Endocytic recycling compartments altered in cisplatin-resistant cancer cells. Cancer Res. 66:2346-2353. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Lowson, J. 1962. Textbook of botany. University Tutorial Press, London, United Kingdom.
- 37.Lucau-Danila, A., T. Delaveau, G. Lelandais, F. Devaux, and C. Jacq. 2003. Competitive promoter occupancy by two yeast paralogous transcription factors controlling the multidrug resistance phenomenon. J. Biol. Chem. 278:52641-52650. [DOI] [PubMed] [Google Scholar]
- 38.Martin, V., D. E. Quain, and K. A. Smart. 2003. Brewing yeast oxidative stress responses: impact of brewery handling, p. 61-74. In K. A. Smart (ed.), Brewing yeast fermentation performance. Blackwell Science, Oxford, United Kingdom.
- 39.Mewes, H. W., K. Albermann, K. Heumann, S. Liebl, and F. Pfeiffer. 1997. MIPS: a database for protein sequences, homology data and yeast genome information. Nucleic Acids Res. 25:28-30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Mumberg, D., R. Muller, and M. Funk. 1995. Yeast vectors for the controlled expression of heterologous proteins in different genetic backgrounds. Gene 156:119-122. [DOI] [PubMed] [Google Scholar]
- 41.Nakanishi-Shindo, Y., K. Nakayama, A. Tanaka, Y. Toda, and Y. Jigami. 1993. Structure of the N-linked oligosaccharides that show the complete loss of alpha-1,6-polymannose outer chain from och1, och1 mnn1, and och1 mnn1 alg3 mutants of Saccharomyces cerevisiae. J. Biol. Chem. 268:26338-26345. [PubMed] [Google Scholar]
- 42.Nakayama, K., Y. Nakanishi-Shindo, A. Tanaka, Y. Haga-Toda, and Y. Jigami. 1997. Substrate specificity of alpha-1,6-mannosyltransferase that initiates N-linked mannose outer chain elongation in Saccharomyces cerevisiae. FEBS Lett. 412:547-550. [DOI] [PubMed] [Google Scholar]
- 43.Ohsumi, Y., K. Kitamoto, and Y. Anraku. 1988. Changes induced in the permeability barrier of the yeast plasma membrane by cupric ion. J. Bacteriol. 170:2676-2682. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Onda, M., K. Ota, T. Chiba, Y. Sakaki, and T. Ito. 2004. Analysis of gene network regulating yeast multidrug resistance by artificial activation of transcription factors: involvement of Pdr3 in salt tolerance. Gene 332:51-59. [DOI] [PubMed] [Google Scholar]
- 45.Postma, E., A. Kuiper, W. F. Tomasouw, W. A. Scheffers, and J. P. van Dijken. 1989. Competition for glucose between the yeasts Saccharomyces cerevisiae and Candida utilis. Appl. Environ. Microbiol. 55:3214-3220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Rajagopal, A., and S. M. Simon. 2003. Subcellular localization and activity of multidrug resistance proteins. Mol. Biol. Cell 14:3389-3399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Ramsay, L. M., and G. M. Gadd. 1997. Mutants of Saccharomyces cerevisiae defective in vacuolar function confirm a role for the vacuole in toxic metal ion detoxification. FEMS Microbiol. Lett. 152:293-298. [DOI] [PubMed] [Google Scholar]
- 48.Sakamoto, K., and W. N. Konings. 2003. Beer spoilage bacteria and hop resistance. Int. J. Food Microbiol. 89:105-124. [DOI] [PubMed] [Google Scholar]
- 49.Sakamoto, K., A. Margolles, H. W. van Veen, and W. N. Konings. 2001. Hop resistance in the beer spoilage bacterium Lactobacillus brevis is mediated by the ATP-binding cassette multidrug transporter HorA. J. Bacteriol. 183:5371-5375. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Sami, M., K. Suzuki, K. Sakamoto, H. Kadokura, K. Kitamoto, and K. Yoda. 1998. A plasmid pRH45 of Lactobacillus brevis confers hop resistance. J. Gen. Appl. Microbiol. 44:361-363. [DOI] [PubMed] [Google Scholar]
- 51.Shimazu, M., T. Sekito, K. Akiyama, Y. Ohsumi, and Y. Kakinuma. 2005. A family of basic amino acid transporters of the vacuolar membrane from Saccharomyces cerevisiae. J. Biol. Chem. 280:4851-4857. [DOI] [PubMed] [Google Scholar]
- 52.Shimwell, J. L. 1937. On the relation between the staining properties of bacteria and their reaction towards hop antiseptic. Part I, II. J. Inst. Brew. 43:111-118. [Google Scholar]
- 53.Shimwell, J. L. 1937. On the relation between the staining properties of bacteria and their reaction towards hop antiseptic. Part III. J. Inst. Brew. 43:191-195. [Google Scholar]
- 54.Sidhu, G. S., A. K. Singh, R. N. Sundarrajan, S. V. Sundar, and R. K. Maheshwari. 1999. Role of vacuolar H(+)-ATPase in interferon-induced inhibition of viral glycoprotein transport. J. Interferon Cytokine Res. 19:1297-1303. [DOI] [PubMed] [Google Scholar]
- 55.Simpson, W. J. 1993. Ionophoric action of trans-isohumulone on Lactobacillus brevis. J. Gen. Microbiol. 139:1041-1045. [Google Scholar]
- 56.Simpson, W. J., and J. L. Fernandez. 1994. Mechanism of resistance of lactic acid bacteria to trans-isohumulone. J. Am. Soc. Brew. Chem. 52:9-11. [Google Scholar]
- 57.Stevens, J. F., C. L. Mirand, D. R. Buhler, and M. L. Deinzer. 1998. Chemistry and biology of hop flavonoids. J. Am. Soc. Brew. Chem. 56:136-145. [Google Scholar]
- 58.Szczypka, M. S., Z. Zhu, P. Silar, and D. J. Thiele. 1997. Saccharomyces cerevisiae mutants altered in vacuole function are defective in copper detoxification and iron-responsive gene transcription. Yeast 13:1423-1435. [DOI] [PubMed] [Google Scholar]
- 59.Tai, S. L., V. M. Boer, P. Daran-Lapujade, M. C. Walsh, J. H. de Winde, J. M. Daran, and J. T. Pronk. 2005. Two-dimensional transcriptome analysis in chemostat cultures. Combinatorial effects of oxygen availability and macronutrient limitation in Saccharomyces cerevisiae. J. Biol. Chem. 280:437-447. [DOI] [PubMed] [Google Scholar]
- 60.Teixeira, M. C., P. J. Dias, T. Simoes, and I. Sa-Correia. 2008. Yeast adaptation to mancozeb involves the up-regulation of FLR1 under the coordinate control of Yap1, Rpn4, Pdr3, and Yrr1. Biochem. Biophys. Res. Commun. 367:249-255. [DOI] [PubMed] [Google Scholar]
- 61.Tusher, V. G., R. Tibshirani, and G. Chu. 2001. Significance analysis of microarrays applied to the ionizing radiation response. Proc. Natl. Acad. Sci. U. S. A. 98:5116-5121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.van Helden, J., B. Andre, and J. Collado-Vides. 1998. Extracting regulatory sites from the upstream region of yeast genes by computational analysis of oligonucleotide frequencies. J. Mol. Biol. 281:827-842. [DOI] [PubMed] [Google Scholar]
- 63.van Helden, J., A. F. Rios, and J. Collado-Vides. 2000. Discovering regulatory elements in non-coding sequences by analysis of spaced dyads. Nucleic Acids Res. 28:1808-1818. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Verduyn, C., E. Postma, W. A. Scheffers, and J. P. van Dijken. 1990. Energetics of Saccharomyces cerevisiae in anaerobic glucose-limited chemostat cultures. J. Gen. Microbiol. 136:405-412. [DOI] [PubMed] [Google Scholar]
- 65.Verzele, M. 1991. Chemistry and analysis of hop and beer bitter acids, p. 1-16. Elsevier, Amsterdam, the Netherlands.
- 66.Wagner, M. C., E. E. Molnar, B. A. Molitoris, and M. G. Goebl. 2006. Loss of the homotypic fusion and vacuole protein sorting or Golgi-associated retrograde protein vesicle tethering complexes results in gentamicin sensitivity in the yeast Saccharomyces cerevisiae. Antimicrob. Agents Chemother. 50:587-595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Winzeler, E. A., D. D. Shoemaker, A. Astromoff, H. Liang, K. Anderson, B. Andre, R. Bangham, R. Benito, J. D. Boeke, H. Bussey, A. M. Chu, C. Connelly, K. Davis, F. Dietrich, S. W. Dow, M. El Bakkoury, F. Foury, S. H. Friend, E. Gentalen, G. Giaever, J. H. Hegemann, T. Jones, M. Laub, H. Liao, N. Liebundguth, D. J. Lockhart, A. Lucau-Danila, M. Lussier, N. M'Rabet, P. Menard, M. Mittmann, C. Pai, C. Rebischung, J. L. Revuelta, L. Riles, C. J. Roberts, P. Ross-Macdonald, B. Scherens, M. Snyder, S. Sookhai-Mahadeo, R. K. Storms, S. Veronneau, M. Voet, G. Volckaert, T. R. Ward, R. Wysocki, G. S. Yen, K. Yu, K. Zimmermann, P. Philippsen, M. Johnston, and R. W. Davis. 1999. Functional characterization of the S. cerevisiae genome by gene deletion and parallel analysis. Science 285:901-906. [DOI] [PubMed] [Google Scholar]
- 68.Zhao, H., and D. J. Eide. 1997. Zap1p, a metalloregulatory protein involved in zinc-responsive transcriptional regulation in Saccharomyces cerevisiae. Mol. Cell. Biol. 17:5044-5052. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.