Abstract
Despite increasing applications of mass spectrometry (MS) to characterize post-translational modifications (PTMs) on histone proteins, most existing protocols are not properly suited to robustly measure them in a high-throughput quantitative manner. In this work, we expand on current protocols and describe improved methods for quantitative Bottom Up characterization of histones and their PTMs with comparable sensitivity, but much higher throughput than standard MS approaches. This is accomplished by first bypassing off-line fractionation of histone proteins and working directly with total histones from a typical nuclei acid extraction. Next, using a chemical derivatization procedure that is combined with stable-isotope labeling in a two-step process, we can quantitatively compare samples using nanoLC-MS/MS. We show that our method can successfully detect 17 combined H2A/H2B variants and over 25 combined histone H3 and H4 PTMs in a single MS experiment. We test our method by quantifying differentially expressed histone PTMs from wild-type yeast and a methyltransferase knockout strain. This improved methodology establishes that time and sample consuming off-line HPLC or SDS-PAGE purification of individual histone variants prior to MS interrogation as commonly performed is not strictly required. Our protocol significantly streamlines the analysis of histone PTMs and will allow for studies of differentially expressed PTMs between multiple samples during biologically relevant processes in a rapid and quantitative fashion.
Keywords: Histone, post-translational modification, electron transfer dissociation, mass spectrometry, quantitative, stable isotope labeling, proteomics
Introduction
Histone proteins, particularly their N-terminal tails, are decorated with a myriad of post-translational modifications (PTMs) including phosphorylation, methylation and acetylation.1 These PTMs occur in multiple but specific amino acid residues,2 and have been linked to several important cellular events or disease.1,3–5 The biological diversity and specificity associated with histone modification patterns has led to the ‘Histone Code’ hypothesis,3 which proposes that multiple co-existing histone PTMs form “codes” that function to dynamically regulate gene expression.4 Mass spectrometry (MS) has emerged as a powerful method, complimentary to antibody approaches to characterize histone PTMs.2 Top 5, 6 and Middle Down7, 8 MS methods analyze the concurrent modifications of intact proteins or large histone polypeptides respectively. In contrast, the Bottom Up approach enzymatically digests histones into short peptides prior to MS analysis.2 Several Bottom Up methods allowing for both the characterization and quantification of histone modified forms have been developed (for a detailed review of these methods see Trelle and Jensen).9
Nevertheless, the high abundance of Arg and Lys residues on histones is problematic for most Bottom Up analyses, as digestion with standard proteases such as trypsin yields small, irreproducible peptides that are often difficult to analyze by MS.10 Although it is possible to quantify histone modified forms through tryptic Bottom Up MS in combination with the use of stable isotope labeling of amino acids in cell culture (SILAC),17 not all histone samples are easily amenable to such labeling as those from tissues and fluids. Label free methods have been also used for histone quantification studies. However, these methods are still somewhat problematic in the case of the highly modified histone H3 and H4, again due to the Lys and Arg residues, and typically only endogenously fully modified “blocked” peptides are normally observed.11–13 To circumvent these issues, the use of proteases that cleave after only one of the two basic amino acids have been employed; for example, Arg-C has been used to cleave only after Arg residues in histone H3.14 However, Arg-C appears to be a much less efficient and specific than trypsin.2 Alternatively, several methods capable of generating uniform tryptic-like peptides first through chemical modification of lysine residues before trypsin digestion have been developed.10, 15 One such method involves the use of a propionylation reagent, and this reaction has been widely adapted by several research groups.10, 16–21 Propionylation of histones converts the free amino group in the N-terminus and endogenously unmodified or monomethylated internal lysines to propionyl amides causing a mass shift of +56 Da and protecting these residues from tryptic digestion. For quantitative comparison of two histone samples, propionic anhydride derivatization followed by trypsin digestion was subsequently combined with an esterification reaction introducing a stable isotope label to modify carboxylic acid groups.22–25 Although this protocol has greatly facilitated histone PTM analysis, the secondary esterification reaction it involves has considerable drawbacks including its particular sensitivity to moisture and the subsequent sample losses incurred in eliminating water from the reaction mixture.10
In this study, we expand on the propionylation procedures and present an improved cost-effective quantitative method for Bottom Up for characterization of histone modifications using a robust, straight-forward stable isotope derivatization procedure. In our experiments, unfractionated total acid extracted histones are first propionylated through reaction with d0-propionic anhydride26 and are subsequently digested using trypsin. Then, a second derivatization with either d0- or d10-propionic anhydride can be used to incorporate a stable isotope label on the newly formed free N-terminal amino groups. Resulting peptides from two samples are mixed and analyzed through nanoLC-MS/MS experiments. Relative variations on histone PTM levels between samples can be directly detected as histone peptides from the d0-propionyl and d5-propionyl labeled samples will appear as peak doublets separated by a +5 Da mass shift. Our protocol is successful in analyzing bulk histones from unfractionated acid extracts, as many histone H2B and H2A variants and histone H3 and H4 PTMs can be detected in a single experiment. These results demonstrate that we can bypass the need for time and sample consuming off-line HPLC or SDS-PAGE purification of histones as is typically performed. Our platform considerably streamlines quantitative analysis of histone modified forms, and we anticipate that these methods will facilitate future studies on differentially expressed histone PTMs and their role throughout physiologically relevant processes.
Experimental Methods
Mammalian and Yeast Cell Culture, Nuclei Isolation and Histone Extraction
HeLa S3 cells were grown and harvested as previously described by Thomas et al.27 Nuclei were isolated and histone proteins were extracted as described by Garcia et al.7 Briefly, histones were acid extracted from nuclei with 0.4 N H2SO4 and precipitated with trichloroacetic acid (TCA), followed by washes with acetone containing 0.1% HCl and then pure acetone. The resulting pellets were redissolved in deionized water prior to further processing. Total protein concentrations of each acid extract were determined using the Bradford assay. Yeast strains were grown in 1 L of YPD media (1% yeast extract, 2% peptone, 2% glucose) to OD600 = 1.5–1.8. Cells were then harvested and nuclei were prepared as previously described.28 Histones were isolated from yeast nuclei essentially as described before,29 with the following changes. The prepared nuclei were extracted with high salt wash buffer (40 mM HEPES-KOH, pH = 7.5, 350 mM NaCl, 0.1% Tween 20, 10% glycerol, 1 µg/ml leupeptin, aprotinin, and pepstatin A, 1 mM PMSF) followed by centrifugation at 4,000 g. Histones were extracted using 0.4 N H2SO4 from nuclei pellets as described by von Holt.30 Histones were quantified by measuring OD218.
RP-HPLC Fractionation of Bulk Histones
Acid extracted bulk histones were separated as described by Garcia et al.25 Briefly, acid extracts from nuclei were fractionated on a C18 column (4.6 mm i.d. × 250 mm, Vydac, Hesperia, CA) using an Beckman Coulter System Gold HPLC (Fullerton, CA) with a gradient of 30–60% B in 100 minutes, followed by 60–100%B in 20 minutes (A = 5% MeCN in 0.2% TFA, B = 90% acetonitrile in 0.188% TFA). Fractions were collected in 1 minute time intervals, pooled and dried to completion in a SpeedVac. An aliquot of the protein fractions were checked for quality using 15% SDS-PAGE.
Histone Sample Preparation for Mass Spectrometry
Bulk acid extracted histones from HeLa or yeast cells (~50 µg) or HPLC purified histone variants from HeLa cells (<5 µg) were derivatized by treatment with propionyl anhydride reagent essentially as described before.10 Briefly, this reagent was created using 75 µL of MeOH and 25 µL of propionic anhydride (Sigma Aldrich, St. Louis, MO). Equal volumes of reagent and histone protein were mixed and allowed to react at 37 °C for 15 minutes and reduced to near dryness using a SpeedVac concentrator for removal of reaction remnants. Propionylated histones were then digested with trypsin (Promega, Madison, WI) at a substrate/enzyme ratio of 20:1 for 6 hours at 37°C after dilution of the sample with 100 mM ammonium bicarbonate buffer solution (pH = 8.0). The reaction was quenched by the addition of concentrated acetic acid and freezing (−80°C). A second round of propionylation was then performed to propionylate the newly created peptide N-termini. For quantification studies, samples were stable isotope labeled using d10-propionic anhydride (Cambridge Isotope Laboratories, Inc., Andover, MA).26 For example, one sample was derivatized using d0-propionic anhydride both before and after trypsin digestion, while a second sample was derivatized using d0-propionic anhydride before trypsin digestion and derivatized with d10 reagent after trypsin digestion (introducing a +5 Da mass shift). For comparative MS analysis, protein concentrations of each sample were determined using Bradford assays and then samples were accordingly mixed for equal protein quantity.
NanoLC-MS/MS
A small aliquot of the histone digests were desalted using in-house made C18 STAGE Tips prepared as previously described, 31 and loaded by an Eksigent AS-2 autosampler (Eksigent Technologies Inc., Dublin, CA) onto a fused silica microcapillary (75 µm) column constructed with an ESI tip and packed in-house with 5um C18 YMC ODS-A resin. Peptides were HPLC separated with an Agilent 1200 series binary pump with an in-line flow splitter across a 110 minute linear gradient ranging from 2% to 35% buffer B (Buffer A = 0.1 M acetic acid, Buffer B = 70% acetonitrile in 0.1 M acetic acid) with a constant flow of approximately 100–200 nL/min. The HPLC system was coupled to an LTQ-Orbitrap mass spectrometer (ThermoFisher Scientific, San Jose, CA) taking a full scan MS spectrum (m/z 290–1650) in the Orbitrap with a resolution of 30,000 after accumulation of approximately 500,000 ions followed by collisionally activated dissociation (CAD) of the seven most intense ions in the LTQ after accumulation of approximately 10,000 ions. All data was collected in centroid mode. Maximum filling time was 500 ms for the full scans. The decision-tree algorithm was used to perform concurrent CAD and electron transfer dissociation (ETD) fragmentation in the same experiment, deciding in real time which fragmentation method to employ based on the charge state and m/z of the precursor as previously described.32 For ETD, an automatic gain control value of 3E6 for the reagent anion and a reaction time of 80 ms were used. Precursor ion charge state screening was enabled and all unassigned charge states as well as singly charged species were rejected. The dynamic exclusion list was restricted to a maximum of 500 entries with a maximum retention period of 120 seconds and a relative mass window of <1 Da.
Data Analysis
CAD and ETD mass spectra were searched using the SEQUEST algorithm within the Bioworks Browser (Version 3.3.1 SP1, Thermo Fisher Scientific Inc., San Jose, CA) against both human or yeast protein databases and human and yeast histone protein database derived from sequences obtained from the National Center for Biotechnology Information (NCBI) database (August 2008). Trypsin protein sequence was also included in the databases. Enzyme specificity was set to trypsin, fully enzymatic, allowing for up to 3 missed cleavage sites (since endogenous and chemical modification of lysine residues hinders enzymatic digestion). Propionylation (+56.026 Da) on the N-terminus of the peptides was set as a fixed modification, while oxidation of methionine (+15.995 Da) was set as a variable modification for all searches. For histone PTM searches, propionylation (+56.026 Da), acetylation (+42.010 Da), mono- (+70.042 Da), di- (+28.031 Da) and trimethylation (+42.046 Da) of lysine residues were selected as variable modifications. Histone monomethylation was searched as the sum of the masses for propionylation (+56.026 Da) and methylation (+14.016 Da) because monomethylated residues can still be propionylated. Parent mass tolerance was set to 0.1 Da and fragment ion tolerance was set to 0.5 Da. Resulting peptide lists were filtered using standard criteria as previously used.33 Additionally, we also used a peptide probability cutoff of 1×10−3 as calculated by the Bioworks program. The false discovery rate was estimated to be 1% for peptide IDs after searching reverse databases. All MS/MS spectra from modified peptides were also manually inspected for accurate mass and correct fragment assignment. Relative quantification of histone modifications was determined by measuring the area under the extracted ion chromatogram peak corresponding to a specific modified peptide normalized to the sum of the peak areas corresponding to all observed modified forms of such peptide.
Results and Discussion
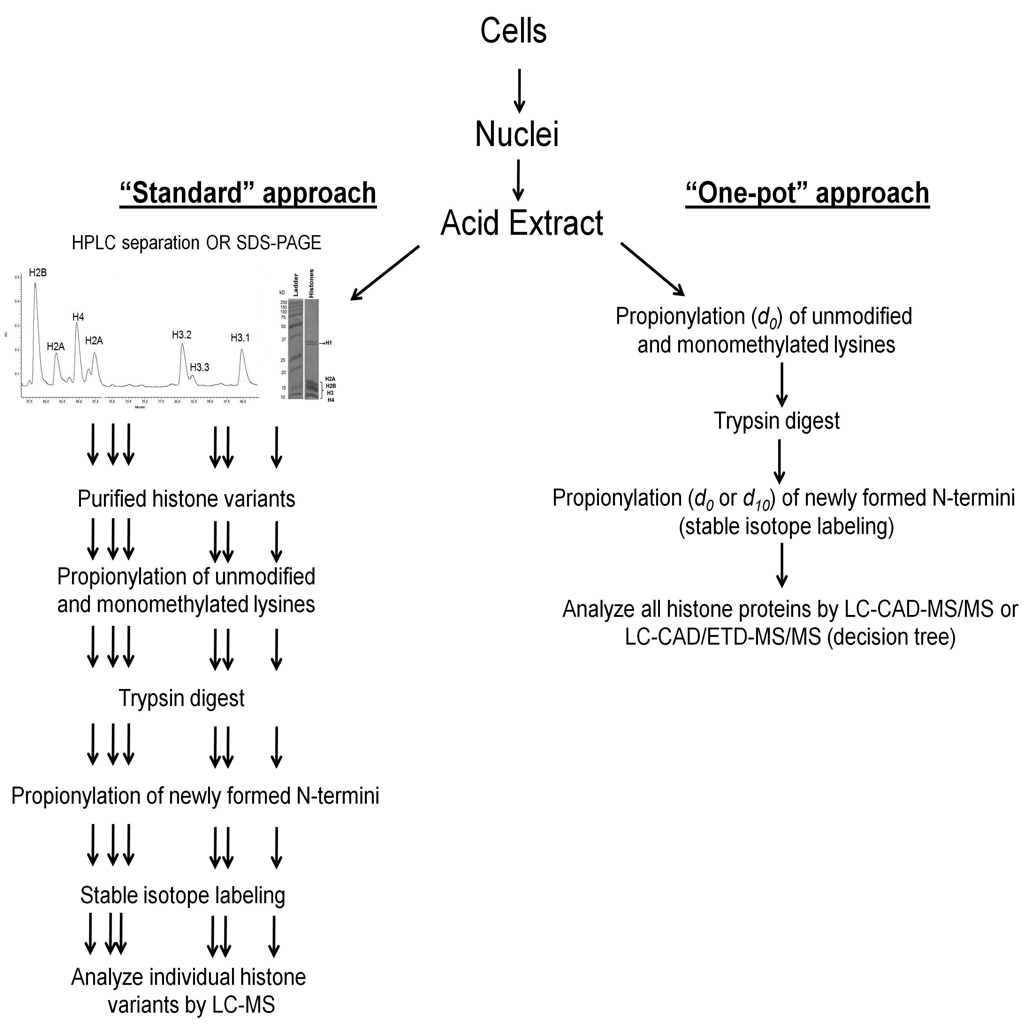
In this report, we detail a one-pot method to quantitatively analyze the majority of histone proteins and PTMs in a single MS experiment (Figure 1). Traditional methods involve extensive purification through HPLC or SDS-PAGE prior to MS analysis of each individual histone variant (Figure 1, left panel). While very effective, these methods are lengthy and prone to several drawbacks such as inherent sample loss in HPLC methods, while SDS-PAGE suffers from being more laborious work for extracting peptides as well as potentially inducing exogenous chemical modification artifacts on proteins that may be mistaken for endogenous modification.34 These off-line fractionation methods prior to MS analysis also vastly reduce the high-throughput abilities of the entire platform and increase the starting material amount needed, especially for HPLC purification. Our streamlined protocol entails direct derivatization and stable isotope labeling of bulk histones in unfractionated total acid extracts by two rounds of propionylation using either d0-or d10-propionyl anhydride followed by comparative analysis through LC-MS/MS using CAD alone or together with ETD fragmentation (“decision tree” driven fragmentation used to choose in real time whether to perform either ETD or CAD based on precursor m/z and charge state) 32 to select the optimum fragmentation method for all peptides (Figure 1, right panel). Through our method, we are able to reduce sample loss, preparation time and MS acquisition time.
Figure 1.
Flowchart showing the characterization of histone PTMs through chemical derivatization and stable isotope peptide labeling. Histones are acid extracted from nuclei and can be processed through two different methods. One of these methods (standard approach) involves the purification of histone variants through reverse-phase HPLC or SDS-PAGE, and then each individual variant is chemically derivatized (i.e. propionylated), enzymatically digested, and either chemically modified again and isotopically labeled before peptides are finally analyzed by LC-MS/MS. A newer alternative approach involves the propionylation of unfractionated bulk histones, followed by tryptic digestion and a second propionylation step. An isotope label for relative quantification can be included in the second propionylation step by the use of d10-propionic anhydride. Aliquots of each peptide solution (labeled and unlabeled) are mixed equally for comparative analysis by MS.
To test this one-pot approach, we compared MS experiments from HPLC purified individual histones against unfractionated bulk histones isolated from total acid extracts. Identical acid extracts derived from the same number of HeLa cells were subjected to either HPLC purification or directly processed by our one-pot propionylation method. For one-pot propionylation, ~5% of the total amount of acid extract (~ 5 µg) was used to obtain sufficient material for MS analysis. In the traditional approach, 100% of the acid extract (~ 100 µg) was used for analytical scale HPLC purification and approximately 1 µg of individual histone was used for further analysis. Our initial objective was to determine if we could detect the same post-translational modifications on histones H3 and H4, as well as a similar number of histone H2A and H2B variants using both approaches. We chose these goals, as the vast majority of histone PTMs are higher in number and abundance on histones H3 and H4, while the complexity on histones H2A and H2B is derived from the multiple variant family members with modest to low level PTMs. Overall, our one-pot approach on total acid extracts is able to detect a similar number of histone H2A and H2B variants identified through analysis of HPLC purified histones (Supplemental Table 1). Our results show that our method can identify many histone H2A/H2B variants in a single two hour run demonstrating that the one-pot approach does not significantly qualitatively suffer from a predicted decrease in dynamic range or sensitivity. Additionally, and of equal importance, we demonstrate that HPLC or SDS-PAGE purification of individual histone family members prior to Bottom Up MS analysis is no longer absolutely required for histone analysis. With regards to histone H3 PTMs, our new protocol is able to detect virtually all of the methylation and acetylation sites that are detected on individual histone H3.2 purified through the traditional HPLC-purification method (Supplemental Table 2). We are also able to detect all acetylation sites on histone H4, plus all methylation states on K20 (data not shown). In agreement with prior reports,35 we find ETD MS/MS are more effective than CAD for characterizing longer or higher charged peptides (data not shown).
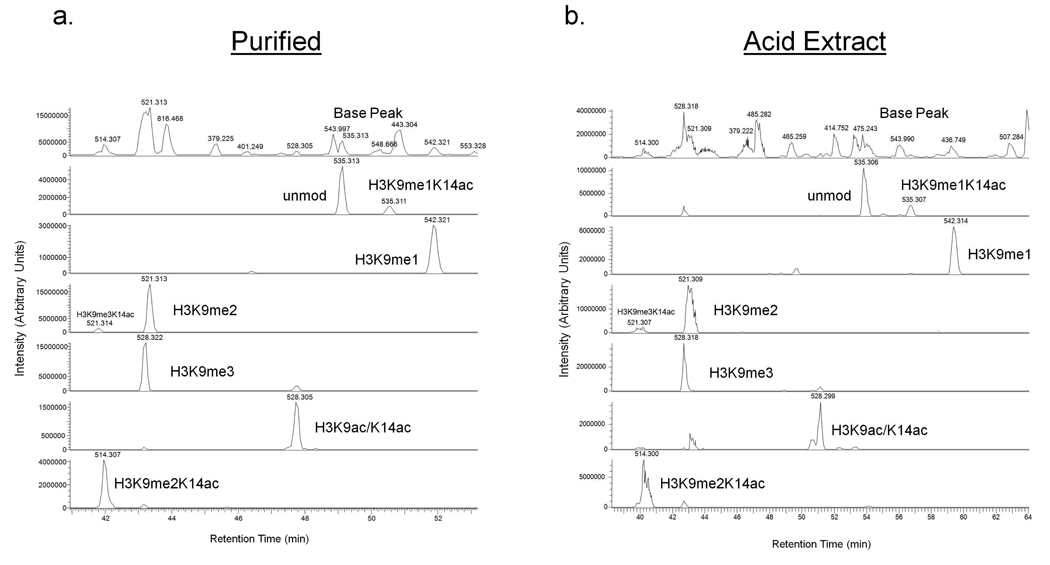
Another more analytical objective of our experiments was to make sure that we could obtain the same quantitative content from the one-pot approach as could be obtained from analysis of purified histones. This was an initial concern for us as in the one-pot approach we have a more complex mixture of peptides generated from many histone proteins potentially resulting in ion suppression effects, especially for less abundant modified peptides. To examine this possibility, we decided to quantitate the various modified forms of a histone H3 peptide. Figure 2 shows data from the 9–17 residue fragment (KSTGGKAPR) from histone H3 obtained through both the one-pot and standard MS approaches. This particular peptide is somewhat challenging because it spans two modification sites (K9 and K14) that can be modified with all possible degrees of methylation on K9 and acetylation on K9 or K14 in all several combinations. Furthermore, these modified peptides are usually in lower abundance (~5X) than most other H3 peptides due to signal dilution across the many modified forms. Shown in Figure 2 are the base peak and extracted ion chromatograms (XICs) for several modified forms of the 9–17 peptide obtained through the standard (HPLC purified H3.2 variant, Figure 2a) and one-pot MS analyses (Acid Extract, Figure 2b). The H3.2 variant was chosen, as it is arguably the most abundant H3 variant in human cells. As can be seen, the base peak chromatogram of purified histone H3.2 compared to a raw acid extract is markedly different, and as expected the acid extract chromatogram has many abundant peaks resulting from other non-histone H3 peptides (Figure 2a–b, first panel). Nevertheless, the peaks for the particular histone modified forms of the H3 9–17 peptides have similar retention time patterns (Figure 2, lower panels). As mentioned before, all modified forms of this peptide observed in the purified sample are also detected in the total acid extracted sample. Despite some variation in the elution times, the overall retention time pattern for the various modified forms persists between samples.
Figure 2.
Total ion chromatogram of the base peak and extracted ion chromatograms for various modified peptides ([M+2H]2+ ions) spanning the 9–17 residues KSTGGKAPR for (a) HPLC purified histone H3.2 and (b) whole acid total histone extracts after chemical derivatization by propionylation. Labels indicate the particular modified form eluting in that peak as determined after inspection of the MS/MS spectra (Supplemental Data). As can be seen, very similar relative retention time patterns and peaks can be detected in both sample sets.
Relative quantification of the histone peptides can be accomplished by measuring the area under the XIC peak corresponding to a specific peptide and expressing that as a fraction of the total sum of the peak areas corresponding to all observed modified forms. The relative abundance values for K9 and K14 modifications are shown in Table 1. Abundance values for all the modified forms of the 9–17 peptide of histone H3 overlap well between samples within a standard deviation. The average standard deviation of peptide abundances across all modified 9–17 peptides is ±1.76 for the HPLC purified H3 sample, and ± 1.87 for the total acid extracted one-pot sample. Similar results were obtained for many other histone H3 peptides (data not shown). Therefore, we feel that we can obtain similar quantitative information for histone peptides from acid extracted total histones analyzed by our one-pot shotgun approach as one would through the analysis of purified histones. However, it is important to note that our method is unable to link PTMs to specific variants for some histones, including some H2A and all H3 members. Consequently, our one-pot protocol is not appropriate for applications in which this information is sought, and thus the traditional method must be used to separate out specific H2A, and H3 variants followed by MS interrogation to gather this PTM information. Additionally, if endogenous histone propionylation36 is the main research emphasis, then a different approach should be employed.
Table 1.
Relative quantification of individual post-translational modifications on the histone H3 peptide (K9STGGK14APR) from HPLC purified and one-pot methods shown in Figure 2. Relative quantification of histone modifications was achieved by measuring the area under the chromatogram peak corresponding to a specific modified peptide normalized to the sum of the peak areas corresponding to all observed modified forms of such peptide. Standard errors were obtained from the standard deviation from duplicate experiments for each sample.
| Histone H3 Peptide | HPLC Purified | One-pot Shotgun |
|---|---|---|
| KSTGGKAPR | ||
| Unmodified | 12.51 ± 3.41 | 11.11 ± 1.06 |
| K9me1 | 10.90 ± 1.64 | 9.27 ± 1.67 |
| K9me2 | 38.66 ± 2.07 | 42.45 ± 4.54 |
| K9me3 | 24.05 ± 3.20 | 25.64 ± 2.20 |
| K9ac | 0.20 ± 0.05 | 0.17 ± 0.02 |
| K14ac | 13.68 ± 0.16 | 11.36 ± 1.71 |
We then desired to improve the relative quantification of histone peptides through Bottom Up analysis across multiple samples by the integration of a stable isotope labeling step into the second propionylation derivatization. Normally, a second round of propionylation derivatization using d0-propionic anhydride is performed after trypsin digestion to cap the newly generated N-termini of the peptides with a propionyl amide bond that improves retention on C18 columns and helps limit the charge on the peptides (a very good procedure for producing mostly 2+ peptides for CAD fragmentation). For a comparative one-pot shotgun approach, we use d10-propionic anhydride on one of the samples in the second round of derivatization to incorporate a stable isotope d5-propionyl amide label on the newly formed free peptide N-termini. The resulting peptides from two samples (d0-and d5-propionyl amides) can be mixed and analyzed together through LC-MS/MS. Histone PTM levels between samples can be directly compared as peptides from the d0-propionyl and d5-propionyl will appear as peak doublet pairs separated by a +5 Da mass difference. For doubly and triply charged peptides, this mass difference translates into a 2.5 or 1.67 m/z shifts, respectively. This labeling has been previously used to measure phosphorylation stoichiometry through propionylation of the N-termini of all peptides in a mixture.26 Our labeling improves on traditional methods by combining the second propionylation step with the isotopic-labeling step, as previous methods involve a second d0-propionylation step followed then by an isotopic labeling esterification step of carboxylic acid groups.10
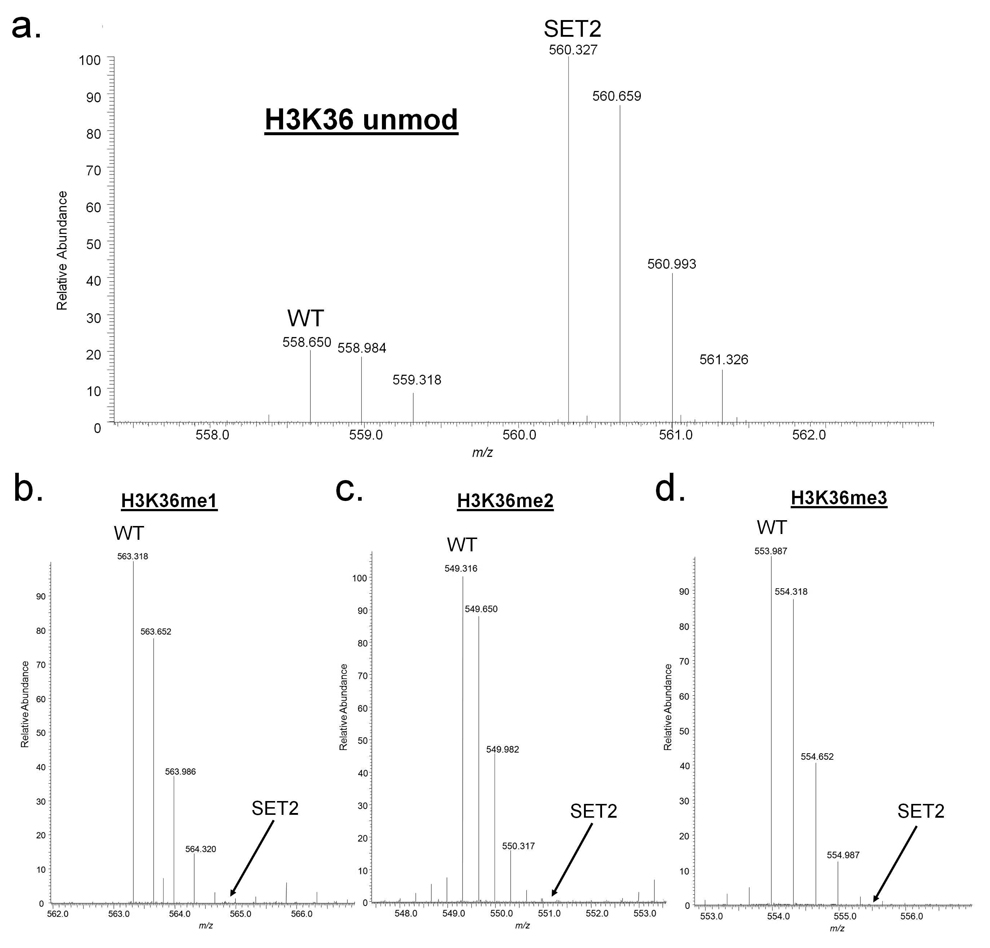
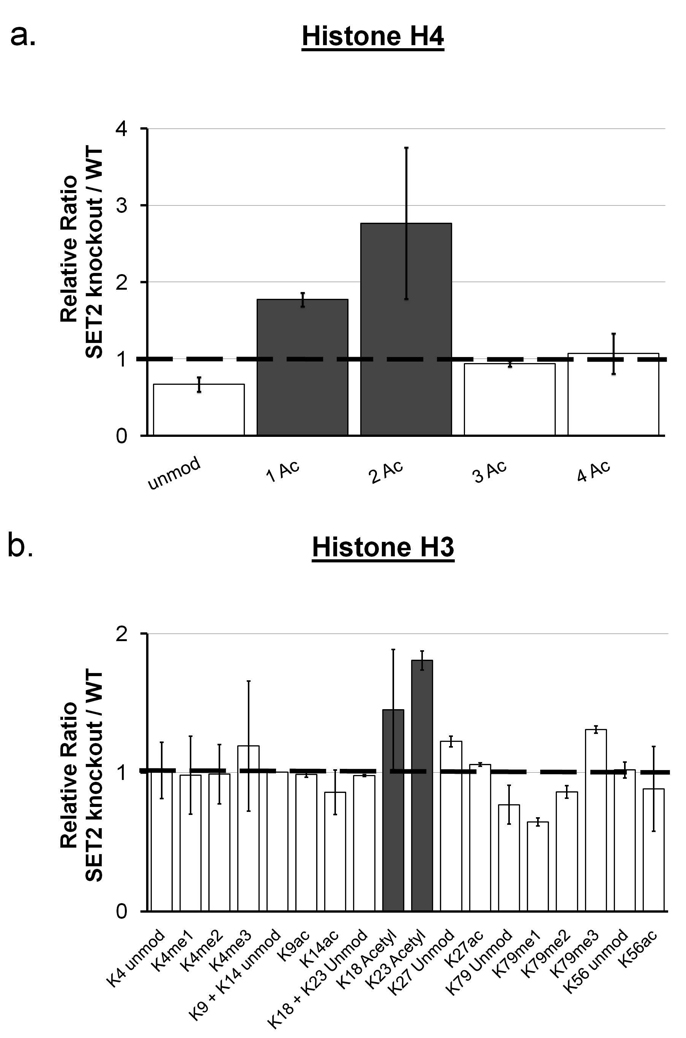
We demonstrate the utility of this labeling to investigate the differences in histone PTMs profiles from wild-type and set2 deletion Saccharomyces cerevisiae strains. Set2 is the only methyltransferase responsible for the mono-, di- and trimethylation of histone H3 K36 in yeast.37, 38 We used our one-pot approach to explore the effects of set2 deletion on K36 methylation levels for wild type (d0-labeled) and mutant (-set2, d5-labeled) yeast strains. Unmodified K36 levels are found to be higher in the Set2 knockout strain (Figure 3a). Unsurprisingly, we observe that all degrees of methylation on K36 are abolished in the set2 deletion strain compared to the wild type yeast sample (Figure 3b–d). Also as expected, we did not detect any other significant changes in the modification level of other well known methylation sites on yeast histone H3 at either K4 or K79 (Figure 4b). In yeast, these sites are methylated by methyltransferases Set1 and Dot1 respectively, and thus deletion of Set2 does not affect modification of these sites.39 In contrast, we observe changes in the extent of acetylation on particular sites on both histone H3 and H4. Through our quantitative procedures, we determined that Set2 knockout also results in a 2-fold and 3-fold increase for the mono-and diacetylated forms of the histone H4 4–17 peptide respectively (Figure 4a). A smaller but reproducible effect occurs on histone H3 K23 and K18 acetylation, which increase in the Set2 deletion strain as well (Figure 4b). Set2 has been recently implicated in the regulation of histone deacetylation, as the HDAC Rpd3S recognizes the Set2 methylated histones and deacetylates histones within transcribed sequences.38, 40 Rpd3S is one of two forms of Rpd3, and S. cerevisiae Rpd3 is involved in global, untargeted histone deacetylation.38, 41, 42 Accordingly, set2 deletion strains deficient in K36 methylation have higher histone acetylation amounts resulting from the lack of recruitment of Rpd3 to nucleosomes.43 The increased histone H4 and H3 acetylation levels we observe in set2 deletion strains are consistent with this previous observation. Through this example, we show that our streamlined method is capable of detecting direct and even minor secondary histone PTM changes in biologically complex samples.
Figure 3.
Side-by-side comparison of K36 methylation levels for wild type (d0-labeled) and Set2 knockout (d5-labeled) yeast strains as determined through a one-pot shotgun approach. Full Mass spectrum for the [M+3H]3+ peptide ions (27–40 residues, KSAPSTGGVKKPHR) corresponding to intrinsically unmodified K36 is shown in (a). Mass spectra for individual methylation degrees as labeled are shown in panels (b–d). Labels specify the particular modified form which is present or missing (all methyl states absent in Set2 knockout).
Figure 4.
Relative abundance of post-translational modified histone peptides for Set2 knockout and wild type yeast strains on histone (a) H3 and (b) H4. As before, quantification of histone modifications was achieved by measuring the area under the chromatogram peak corresponding to a specific modified peptide normalized to the sum of the peak areas corresponding to all observed modified forms of such peptide. The relative abundance ratio was calculated as the ratio of such quantities for the Set2 knockout and wild type strains. For histone H3, K36 was not included as a possible modification because it is completely absent in the Set2 knockout strain (thus a ratio could not be calculated). Error bars of two standard deviations are shown.
Conclusions
In this paper, we describe a one-pot shotgun method for Bottom Up characterization of all histones and their modifications with increased throughput, and reduced analysis time and sample requirements by using a straight-forward chemical derivatization/stable-isotope labeling of the acid extracted total histones followed by LC-MS/MS analysis. We demonstrate that our method can successfully identify and quantify several histone H2A/H2B variants and H3/H4 PTMs in a single experiment and bypasses the need for the customary laborious and problematic off-line HPLC or SDS-PAGE purification of histones prior to MS analysis. Furthermore, resulting peptides from two samples can be quantitatively analyzed allowing for direct detection of differentially expressed histone PTMs between different cellular states. Our protocol greatly simplifies the analysis of histone PTMs and we hope that this methodology will permit the study of differentially expressed histone marks and their role throughout physiologically relevant epigenetic processes in expedited fashion.
Supplementary Material
Acknowledgments
This work was supported by Princeton University and an American Society for Mass Spectrometry Research award to B.A.G and a grant from the National Institutes of Health to S.D.B. (GM74183). We thank Adam Henry and Christina Velasquez for technical help.
Footnotes
Supporting Information Available. Tables listing histone H2A and H2B variants and histone H3 (H4) modifications detected through purified histone and our one-pot MS analyses are presented. Representative tandem mass spectra for all histone H3 and H4 PTMs detected in mammalian and yeast histones are presented as Supplementary Data. This material is available free of charge via the Internet at http://pubs.acs.org.
References
- 1.Cheung P, Allis CD, Sassone-Corsi P. Signaling to chromatin through histone modifications. Cell. 2000;103(2):263–271. doi: 10.1016/s0092-8674(00)00118-5. [DOI] [PubMed] [Google Scholar]
- 2.Garcia BA, Shabanowitz J, Hunt DF. Characterization of histones and their post-translational modifications by mass spectrometry. Curr Opin Chem Biol. 2007;11(1):66–73. doi: 10.1016/j.cbpa.2006.11.022. [DOI] [PubMed] [Google Scholar]
- 3.Strahl BD, Allis CD. The language of covalent histone modifications. Nature. 2000;403(6765):41–45. doi: 10.1038/47412. [DOI] [PubMed] [Google Scholar]
- 4.Cosgrove MS. Histone proteomics and the epigenetic regulation of nucleosome mobility. Expert Rev Proteomics. 2007;4(4):465–478. doi: 10.1586/14789450.4.4.465. [DOI] [PubMed] [Google Scholar]
- 5.Pesavento JJ, Mizzen CA, Kelleher NL. Quantitative analysis of modified proteins and their positional isomers by tandem mass spectrometry: human histone H4. Anal Chem. 2006;78(13):4271–4280. doi: 10.1021/ac0600050. [DOI] [PubMed] [Google Scholar]
- 6.Boyne MT, 2nd, Pesavento JJ, Mizzen CA, Kelleher NL. Precise characterization of human histones in the H2A gene family by top down mass spectrometry. J Proteome Res. 2006;5(2):248–253. doi: 10.1021/pr050269n. [DOI] [PubMed] [Google Scholar]
- 7.Garcia BA, Pesavento JJ, Mizzen CA, Kelleher NL. Pervasive combinatorial modification of histone H3 in human cells. Nat Methods. 2007;4(6):487–489. doi: 10.1038/nmeth1052. [DOI] [PubMed] [Google Scholar]
- 8.Phanstiel D, Brumbaugh J, Berggren WT, Conard K, Feng X, Levenstein ME, McAlister GC, Thomson JA, Coon JJ. Mass spectrometry identifies and quantifies 74 unique histone H4 isoforms in differentiating human embryonic stem cells. Proc Natl Acad Sci U S A. 2008;105(11):4093–4098. doi: 10.1073/pnas.0710515105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Trelle MB, Jensen ON. Functional proteomics in histone research and epigenetics. Expert Rev Proteomics. 2007;4(4):491–503. doi: 10.1586/14789450.4.4.491. [DOI] [PubMed] [Google Scholar]
- 10.Garcia BA, Mollah S, Ueberheide BM, Busby SA, Muratore TL, Shabanowitz J, Hunt DF. Chemical derivatization of histones for facilitated analysis by mass spectrometry. Nat Protoc. 2007;2(4):933–938. doi: 10.1038/nprot.2007.106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Lee AY, Paweletz CP, Pollock RM, Settlage RE, Cruz JC, Secrist JP, Miller TA, Stanton MG, Kral AM, Ozerova ND, Meng F, Yates NA, Richon V, Hendrickson RC. Quantitative analysis of histone deacetylase-1 selective histone modifications by differential mass spectrometry. J Proteome Res. 2008;7(12):5177–5186. doi: 10.1021/pr800510p. [DOI] [PubMed] [Google Scholar]
- 12.Beck HC, Nielsen EC, Matthiesen R, Jensen LH, Sehested M, Finn P, Grauslund M, Hansen AM, Jensen ON. Quantitative proteomic analysis of post-translational modifications of human histones. Mol Cell Proteomics. 2006;5(7):1314–1325. doi: 10.1074/mcp.M600007-MCP200. [DOI] [PubMed] [Google Scholar]
- 13.Miao J, Fan Q, Cui L, Li J. The malaria parasite Plasmodium falciparum histones: organization, expression, and acetylation. Gene. 2006;369:53–65. doi: 10.1016/j.gene.2005.10.022. [DOI] [PubMed] [Google Scholar]
- 14.McKittrick E, Gafken PR, Ahmad K, Henikoff S. Histone H3.3 is enriched in covalent modifications associated with active chromatin. Proc Natl Acad Sci U S A. 2004;101(6):1525–1530. doi: 10.1073/pnas.0308092100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Smith CM, Gafken PR, Zhang Z, Gottschling DE, Smith JB, Smith DL. Mass spectrometric quantification of acetylation at specific lysines within the amino-terminal tail of histone H4. Anal Biochem. 2003;316(1):23–33. doi: 10.1016/s0003-2697(03)00032-0. [DOI] [PubMed] [Google Scholar]
- 16.Robin P, Fritsch L, Philipot O, Svinarchuk F, Ait-Si-Ali S. Post-translational modifications of histones H3 and H4 associated with the histone methyltransferases Suv39h1 and G9a. Genome Biol. 2007;8(12):R270. doi: 10.1186/gb-2007-8-12-r270. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Drogaris P, Wurtele H, Masumoto H, Verreault A, Thibault P. Comprehensive profiling of histone modifications using a label-free approach and its applications in determining structure-function relationships. Anal Chem. 2008;80(17):6698–6707. doi: 10.1021/ac800739d. [DOI] [PubMed] [Google Scholar]
- 18.Peters AH, Kubicek S, Mechtler K, O'Sullivan RJ, Derijck AA, Perez-Burgos L, Kohlmaier A, Opravil S, Tachibana M, Shinkai Y, Martens JH, Jenuwein T. Partitioning and plasticity of repressive histone methylation states in mammalian chromatin. Mol Cell. 2003;12(6):1577–1589. doi: 10.1016/s1097-2765(03)00477-5. [DOI] [PubMed] [Google Scholar]
- 19.Ouvry-Patat SA, Schey KL. Characterization of antimicrobial histone sequences and posttranslational modifications by mass spectrometry. J Mass Spectrom. 2007;42(5):664–674. doi: 10.1002/jms.1200. [DOI] [PubMed] [Google Scholar]
- 20.Bonaldi T, Imhof A, Regula JT. A combination of different mass spectroscopic techniques for the analysis of dynamic changes of histone modifications. Proteomics. 2004;4(5):1382–1396. doi: 10.1002/pmic.200300743. [DOI] [PubMed] [Google Scholar]
- 21.Mandava V, Fernandez JP, Deng H, Janzen CJ, Hake SB, Cross GA. Histone modifications in Trypanosoma brucei. Mol Biochem Parasitol. 2007;156(1):41–50. doi: 10.1016/j.molbiopara.2007.07.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Garcia BA, Joshi S, Thomas CE, Chitta RK, Diaz RL, Busby SA, Andrews PC, Ogorzalek Loo RR, Shabanowitz J, Kelleher NL, Mizzen CA, Allis CD, Hunt DF. Comprehensive phosphoprotein analysis of linker histone H1 from Tetrahymena thermophila. Mol Cell Proteomics. 2006;5(9):1593–1609. doi: 10.1074/mcp.M600086-MCP200. [DOI] [PubMed] [Google Scholar]
- 23.Johnson L, Mollah S, Garcia BA, Muratore TL, Shabanowitz J, Hunt DF, Jacobsen SE. Mass spectrometry analysis of Arabidopsis histone H3 reveals distinct combinations of post-translational modifications. Nucleic Acids Res. 2004;32(22):6511–6518. doi: 10.1093/nar/gkh992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Hake SB, Garcia BA, Duncan EM, Kauer M, Dellaire G, Shabanowitz J, Bazett-Jones DP, Allis CD, Hunt DF. Expression patterns and post-translational modifications associated with mammalian histone H3 variants. J Biol Chem. 2006;281(1):559–568. doi: 10.1074/jbc.M509266200. [DOI] [PubMed] [Google Scholar]
- 25.Garcia BA, Busby SA, Shabanowitz J, Hunt DF, Mishra N. Resetting the epigenetic histone code in the MRL-lpr/lpr mouse model of lupus by histone deacetylase inhibition. J Proteome Res. 2005;4(6):2032–2042. doi: 10.1021/pr050188r. [DOI] [PubMed] [Google Scholar]
- 26.Zhang X, Jin QK, Carr SA, Annan RS. N-Terminal peptide labeling strategy for incorporation of isotopic tags: a method for the determination of site-specific absolute phosphorylation stoichiometry. Rapid Commun Mass Spectrom. 2002;16(24):2325–2332. doi: 10.1002/rcm.864. [DOI] [PubMed] [Google Scholar]
- 27.Thomas CE, Kelleher NL, Mizzen CA. Mass spectrometric characterization of human histone H3: a bird's eye view. J Proteome Res. 2006;5(2):240–247. doi: 10.1021/pr050266a. [DOI] [PubMed] [Google Scholar]
- 28.Edmondson DG, Smith MM, Roth SY. Repression domain of the yeast global repressor Tup1 interacts directly with histones H3 and H4. Genes Dev. 1996;10(10):1247–1259. doi: 10.1101/gad.10.10.1247. [DOI] [PubMed] [Google Scholar]
- 29.Fukuma M, Hiraoka Y, Sakurai H, Fukasawa T. Purification of yeast histones competent for nucleosome assembly in vitro. Yeast. 1994;10(3):319–331. doi: 10.1002/yea.320100305. [DOI] [PubMed] [Google Scholar]
- 30.von Holt C, Brandt WF, Greyling HJ, Lindsey GG, Retief JD, Rodrigues JD, Schwager S, Sewell BT. Isolation and characterization of histones. Methods Enzymol. 1989;170:431–523. doi: 10.1016/0076-6879(89)70061-6. [DOI] [PubMed] [Google Scholar]
- 31.Rappsilber J, Ishihama Y, Mann M. Stop and go extraction tips for matrix-assisted laser desorption/ionization, nanoelectrospray, and LC/MS sample pretreatment in proteomics. Anal Chem. 2003;75(3):663–670. doi: 10.1021/ac026117i. [DOI] [PubMed] [Google Scholar]
- 32.Swaney DL, McAlister GC, Coon JJ. Decision tree-driven tandem mass spectrometry for shotgun proteomics. Nat Methods. 2008;5(11):959–964. doi: 10.1038/nmeth.1260. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Washburn MP, Wolters D, Yates JR., 3rd Large-scale analysis of the yeast proteome by multidimensional protein identification technology. Nat Biotechnol. 2001;19(3):242–247. doi: 10.1038/85686. [DOI] [PubMed] [Google Scholar]
- 34.Jung SY, Li Y, Wang Y, Chen Y, Zhao Y, Qin J. Complications in the assignment of 14 and 28 Da mass shift detected by mass spectrometry as in vivo methylation from endogenous proteins. Anal Chem. 2008;80(5):1721–1729. doi: 10.1021/ac7021025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Mikesh LM, Ueberheide B, Chi A, Coon JJ, Syka JE, Shabanowitz J, Hunt DF. The utility of ETD mass spectrometry in proteomic analysis. Biochim Biophys Acta. 2006;1764(12):1811–1822. doi: 10.1016/j.bbapap.2006.10.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Chen Y, Sprung R, Tang Y, Ball H, Sangras B, Kim SC, Falck JR, Peng J, Gu W, Zhao Y. Lysine propionylation and butyrylation are novel post-translational modifications in histones. Mol Cell Proteomics. 2007;6(5):812–819. doi: 10.1074/mcp.M700021-MCP200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Fingerman IM, Du HN, Briggs SD. Controlling histone methylation via trans-histone pathways. Epigenetics. 2008;3(5):237–242. doi: 10.4161/epi.3.5.6869. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Lee JS, Shilatifard A. A site to remember: H3K36 methylation a mark for histone deacetylation. Mutat Res. 2007;618(1–2):130–134. doi: 10.1016/j.mrfmmm.2006.08.014. [DOI] [PubMed] [Google Scholar]
- 39.Bannister AJ, Kouzarides T. Reversing histone methylation. Nature. 2005;436(7054):1103–1106. doi: 10.1038/nature04048. [DOI] [PubMed] [Google Scholar]
- 40.Carrozza MJ, Li B, Florens L, Suganuma T, Swanson SK, Lee KK, Shia WJ, Anderson S, Yates J, Washburn MP, Workman JL. Histone H3 methylation by Set2 directs deacetylation of coding regions by Rpd3S to suppress spurious intragenic transcription. Cell. 2005;123(4):581–592. doi: 10.1016/j.cell.2005.10.023. [DOI] [PubMed] [Google Scholar]
- 41.Kurdistani SK, Grunstein M. Histone acetylation and deacetylation in yeast. Nat Rev Mol Cell Biol. 2003;4(4):276–284. doi: 10.1038/nrm1075. [DOI] [PubMed] [Google Scholar]
- 42.Kurdistani SK, Robyr D, Tavazoie S, Grunstein M. Genome-wide binding map of the histone deacetylase Rpd3 in yeast. Nat Genet. 2002;31(3):248–254. doi: 10.1038/ng907. [DOI] [PubMed] [Google Scholar]
- 43.Du HN, Fingerman IM, Briggs SD. Histone H3 K36 methylation is mediated by a trans-histone methylation pathway involving an interaction between Set2 and histone H4. Genes Dev. 2008;22(20):2786–2798. doi: 10.1101/gad.1700008. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.