Abstract
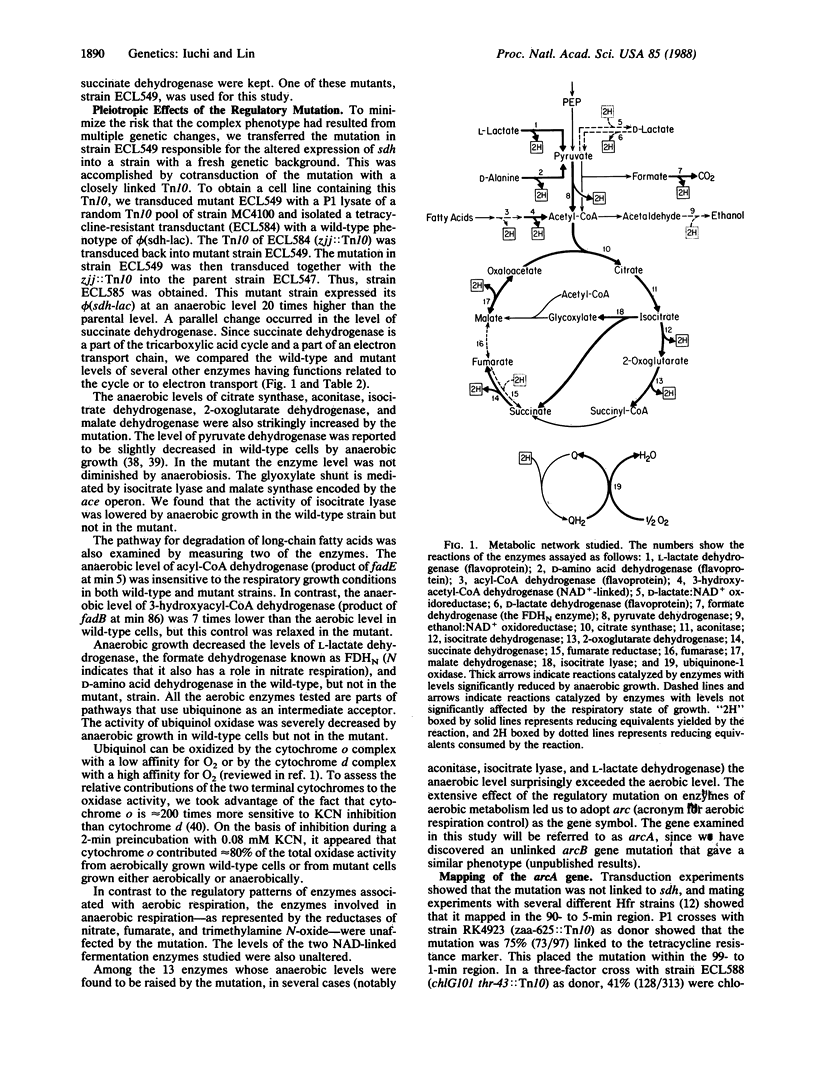
In Escherichia coli the levels of numerous enzymes associated with aerobic metabolism are decreased during anaerobic growth. In an arcA mutant the anaerobic levels of these enzymes are increased. The enzymes, which are encoded by different regulons, include members that belong to the tricarboxylic acid cycle, the glyoxylate shunt, the pathway for fatty acid degradation, several dehydrogenases of the flavoprotein class, and the cytochrome o oxidase complex. Transductional crosses placed the arcA gene near min O on the chromosomal map. Complementation tests showed that the arcA gene corresponded to the dye gene, which is also known as fexA, msp, seg, or sfrA because of various phenotypic properties [Bachmann, B. (1983) Microbiol. Rev. 47, 180-230]. A dye-deletion mutant was derepressed in the aerobic enzyme system. The term modulon is proposed to describe a set of regulons that are subject to a common transcriptional control.
Full text
PDF



Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Amarasingham C. R., Davis B. D. Regulation of alpha-ketoglutarate dehydrogenase formation in Escherichia coli. J Biol Chem. 1965 Sep;240(9):3664–3668. [PubMed] [Google Scholar]
- Bachmann B. J. Linkage map of Escherichia coli K-12, edition 7. Microbiol Rev. 1983 Jun;47(2):180–230. doi: 10.1128/mr.47.2.180-230.1983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beutin L., Achtman M. Two Escherichia coli chromosomal cistrons, sfrA and sfrB, which are needed for expression of F factor tra functions. J Bacteriol. 1979 Sep;139(3):730–737. doi: 10.1128/jb.139.3.730-737.1979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beutin L., Manning P. A., Achtman M., Willetts N. sfrA and sfrB products of Escherichia coli K-12 are transcriptional control factors. J Bacteriol. 1981 Feb;145(2):840–844. doi: 10.1128/jb.145.2.840-844.1981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bremer E., Silhavy T. J., Weinstock G. M. Transposable lambda placMu bacteriophages for creating lacZ operon fusions and kanamycin resistance insertions in Escherichia coli. J Bacteriol. 1985 Jun;162(3):1092–1099. doi: 10.1128/jb.162.3.1092-1099.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Buxton R. S., Drury L. S., Curtis C. A. Dye sensitivity correlated with envelope protein changes in dye (sfrA) mutants of Escherichia coli K12 defective in the expression of the sex factor F. J Gen Microbiol. 1983 Nov;129(11):3363–3370. doi: 10.1099/00221287-129-11-3363. [DOI] [PubMed] [Google Scholar]
- Buxton R. S., Drury L. S. Identification of the dye gene product, mutational loss of which alters envelope protein composition and also affects sex factor F expression in Escherichia coli K-12. Mol Gen Genet. 1984;194(1-2):241–247. doi: 10.1007/BF00383523. [DOI] [PubMed] [Google Scholar]
- Buxton R. S., Hammer-Jespersen K., Hansen T. D. Insertion of bacteriophage lambda into the deo operon of Escherichia coli K-12 and isolation of plaque-forming lambdadeo+ transducing bacteriophages. J Bacteriol. 1978 Nov;136(2):668–681. doi: 10.1128/jb.136.2.668-681.1978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clark D. Regulation of fatty acid degradation in Escherichia coli: analysis by operon fusion. J Bacteriol. 1981 Nov;148(2):521–526. doi: 10.1128/jb.148.2.521-526.1981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cocks G. T., Aguilar T., Lin E. C. Evolution of L-1, 2-propanediol catabolism in Escherichia coli by recruitment of enzymes for L-fucose and L-lactate metabolism. J Bacteriol. 1974 Apr;118(1):83–88. doi: 10.1128/jb.118.1.83-88.1974. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Comeau D. E., Ikenaka K., Tsung K. L., Inouye M. Primary characterization of the protein products of the Escherichia coli ompB locus: structure and regulation of synthesis of the OmpR and EnvZ proteins. J Bacteriol. 1985 Nov;164(2):578–584. doi: 10.1128/jb.164.2.578-584.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Drury L. S., Buxton R. S. DNA sequence analysis of the dye gene of Escherichia coli reveals amino acid homology between the dye and OmpR proteins. J Biol Chem. 1985 Apr 10;260(7):4236–4242. [PubMed] [Google Scholar]
- Gaffney D., Skurray R., Willetts N. Regulation of the F conjugation genes studied by hybridization and tra-lacZ fusion. J Mol Biol. 1983 Jul 25;168(1):103–122. doi: 10.1016/s0022-2836(83)80325-8. [DOI] [PubMed] [Google Scholar]
- HIRSCH C. A., RASMINSKY M., DAVIS B. D., LIN E. C. A FUMARATE REDUCTASE IN ESCHERICHIA COLI DISTINCT FROM SUCCINATE DEHYDROGENASE. J Biol Chem. 1963 Nov;238:3770–3774. [PubMed] [Google Scholar]
- Hall M. N., Silhavy T. J. Genetic analysis of the ompB locus in Escherichia coli K-12. J Mol Biol. 1981 Sep 5;151(1):1–15. doi: 10.1016/0022-2836(81)90218-7. [DOI] [PubMed] [Google Scholar]
- Hall M. N., Silhavy T. J. The ompB locus and the regulation of the major outer membrane porin proteins of Escherichia coli K12. J Mol Biol. 1981 Feb 15;146(1):23–43. doi: 10.1016/0022-2836(81)90364-8. [DOI] [PubMed] [Google Scholar]
- Hathaway B. G., Bergquist P. L. Temperature-sensitive mutations affecting the replication of F-prime factors in Escherichia coli K 12. Mol Gen Genet. 1973 Dec 31;127(4):297–306. doi: 10.1007/BF00267100. [DOI] [PubMed] [Google Scholar]
- Herbert A. A., Guest J. R. Two mutations affecting utilization of C4-dicarboxylic acids by Escherichia coli. J Gen Microbiol. 1970 Oct;63(2):151–162. doi: 10.1099/00221287-63-2-151. [DOI] [PubMed] [Google Scholar]
- Iuchi S., Kuritzkes D. R., Lin E. C. Escherichia coli mutant with altered respiratory control of the frd operon. J Bacteriol. 1985 Mar;161(3):1023–1028. doi: 10.1128/jb.161.3.1023-1028.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Iuchi S., Kuritzkes D. R., Lin E. C. Three classes of Escherichia coli mutants selected for aerobic expression of fumarate reductase. J Bacteriol. 1986 Dec;168(3):1415–1421. doi: 10.1128/jb.168.3.1415-1421.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Iuchi S., Lin E. C. Molybdenum effector of fumarate reductase repression and nitrate reductase induction in Escherichia coli. J Bacteriol. 1987 Aug;169(8):3720–3725. doi: 10.1128/jb.169.8.3720-3725.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Iuchi S., Lin E. C. The narL gene product activates the nitrate reductase operon and represses the fumarate reductase and trimethylamine N-oxide reductase operons in Escherichia coli. Proc Natl Acad Sci U S A. 1987 Jun;84(11):3901–3905. doi: 10.1073/pnas.84.11.3901. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jamieson A. F., Bergquist P. L. Genetic mapping of chromosomal mutations affecting the replication of the F-factor of Escherichia coli. Mol Gen Genet. 1976 Oct 18;148(2):221–223. doi: 10.1007/BF00268388. [DOI] [PubMed] [Google Scholar]
- Kita K., Konishi K., Anraku Y. Terminal oxidases of Escherichia coli aerobic respiratory chain. II. Purification and properties of cytochrome b558-d complex from cells grown with limited oxygen and evidence of branched electron-carrying systems. J Biol Chem. 1984 Mar 10;259(5):3375–3381. [PubMed] [Google Scholar]
- Kranz R. G., Gennis R. B. Immunological characterization of the cytochrome o terminal oxidase from Escherichia coli. J Biol Chem. 1983 Sep 10;258(17):10614–10621. [PubMed] [Google Scholar]
- LIN E. C., KOCH J. P., CHUSED T. M., JORGENSEN S. E. Utilization of L-alpha-glycerophosphate by Escherichia coli without hydrolysis. Proc Natl Acad Sci U S A. 1962 Dec 15;48:2145–2150. doi: 10.1073/pnas.48.12.2145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- LIN E. C., LERNER S. A., JORGENSEN S. E. A method for isolating constitutive mutants for carbohydrate-catabolizing enzymes. Biochim Biophys Acta. 1962 Jul 2;60:422–424. doi: 10.1016/0006-3002(62)90423-7. [DOI] [PubMed] [Google Scholar]
- Langley D., Guest J. R. Biochemical genetics of the alpha-keto acid dehydrogenase complexes of Escherichia coli K12: genetic characterization and regulatory properties of deletion mutants. J Gen Microbiol. 1978 May;106(1):103–117. doi: 10.1099/00221287-106-1-103. [DOI] [PubMed] [Google Scholar]
- Lerner T. J., Zinder N. D. Another gene affecting sexual expression of Escherichia coli. J Bacteriol. 1982 Apr;150(1):156–160. doi: 10.1128/jb.150.1.156-160.1982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lester R. L., DeMoss J. A. Effects of molybdate and selenite on formate and nitrate metabolism in Escherichia coli. J Bacteriol. 1971 Mar;105(3):1006–1014. doi: 10.1128/jb.105.3.1006-1014.1971. [DOI] [PMC free article] [PubMed] [Google Scholar]
- MAAS W. K. STUDIES ON THE MECHANISM OF REPRESSION OF ARGININE BIOSYNTHESIS IN ESCHERICHIA COLI. II. DOMINANCE OF REPRESSIBILITY IN DIPLOIDS. J Mol Biol. 1964 Mar;8:365–370. doi: 10.1016/s0022-2836(64)80200-x. [DOI] [PubMed] [Google Scholar]
- Maloy S. R., Bohlander M., Nunn W. D. Elevated levels of glyoxylate shunt enzymes in Escherichia coli strains constitutive for fatty acid degradation. J Bacteriol. 1980 Aug;143(2):720–725. doi: 10.1128/jb.143.2.720-725.1980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matsushita K., Patel L., Gennis R. B., Kaback H. R. Reconstitution of active transport in proteoliposomes containing cytochrome o oxidase and lac carrier protein purified from Escherichia coli. Proc Natl Acad Sci U S A. 1983 Aug;80(16):4889–4893. doi: 10.1073/pnas.80.16.4889. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McEwen J., Silverman P. Chromosomal mutations of Escherichia coli that alter expression of conjugative plasmid functions. Proc Natl Acad Sci U S A. 1980 Jan;77(1):513–517. doi: 10.1073/pnas.77.1.513. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McPhedran P., Sommer B., Lin E. C. CONTROL OF ETHANOL DEHYDROGENASE LEVELS IN AEROBACTER AEROGENES. J Bacteriol. 1961 Jun;81(6):852–857. doi: 10.1128/jb.81.6.852-857.1961. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Olsiewski P. J., Kaczorowski G. J., Walsh C. Purification and properties of D-amino acid dehydrogenase, an inducible membrane-bound iron-sulfur flavoenzyme from Escherichia coli B. J Biol Chem. 1980 May 25;255(10):4487–4494. [PubMed] [Google Scholar]
- Overath P., Raufuss E. M. The induction of the enzymes of fatty acid degradation in Escherichia coli. Biochem Biophys Res Commun. 1967 Oct 11;29(1):28–33. doi: 10.1016/0006-291x(67)90535-9. [DOI] [PubMed] [Google Scholar]
- Raunio R. P., Jenkins W. T. D-alanine oxidase form Escherichia coli: localization and induction by L-alanine. J Bacteriol. 1973 Aug;115(2):560–566. doi: 10.1128/jb.115.2.560-566.1973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roeder W., Somerville R. L. Cloning the trpR gene. Mol Gen Genet. 1979 Nov;176(3):361–368. doi: 10.1007/BF00333098. [DOI] [PubMed] [Google Scholar]
- Smith M. W., Neidhardt F. C. 2-Oxoacid dehydrogenase complexes of Escherichia coli: cellular amounts and patterns of synthesis. J Bacteriol. 1983 Oct;156(1):81–88. doi: 10.1128/jb.156.1.81-88.1983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spencer M. E., Guest J. R. Isolation and properties of fumarate reductase mutants of Escherichia coli. J Bacteriol. 1973 May;114(2):563–570. doi: 10.1128/jb.114.2.563-570.1973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stewart V., MacGregor C. H. Nitrate reductase in Escherichia coli K-12: involvement of chlC, chlE, and chlG loci. J Bacteriol. 1982 Aug;151(2):788–799. doi: 10.1128/jb.151.2.788-799.1982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stewart V. Requirement of Fnr and NarL functions for nitrate reductase expression in Escherichia coli K-12. J Bacteriol. 1982 Sep;151(3):1320–1325. doi: 10.1128/jb.151.3.1320-1325.1982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tarmy E. M., Kaplan N. O. Chemical characterization of D-lactate dehydrogenase from Escherichia coli B. J Biol Chem. 1968 May 25;243(10):2579–2586. [PubMed] [Google Scholar]
- Wanner B. L. Novel regulatory mutants of the phosphate regulon in Escherichia coli K-12. J Mol Biol. 1986 Sep 5;191(1):39–58. doi: 10.1016/0022-2836(86)90421-3. [DOI] [PubMed] [Google Scholar]
- Wood J. M. Membrane association of proline dehydrogenase in Escherichia coli is redox dependent. Proc Natl Acad Sci U S A. 1987 Jan;84(2):373–377. doi: 10.1073/pnas.84.2.373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu T. T. A model for three-point analysis of random general transduction. Genetics. 1966 Aug;54(2):405–410. doi: 10.1093/genetics/54.2.405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yanisch-Perron C., Vieira J., Messing J. Improved M13 phage cloning vectors and host strains: nucleotide sequences of the M13mp18 and pUC19 vectors. Gene. 1985;33(1):103–119. doi: 10.1016/0378-1119(85)90120-9. [DOI] [PubMed] [Google Scholar]


