SUMMARY
Trigger factor (TF) is a molecular chaperone that famously binds to bacterial ribosomes where it contacts emerging nascent chains, but TF is also abundant free in the cytosol where its activity is less well characterized. In vitro studies show that TF promotes protein refolding. We find here that ribosome-free TF stably associates with and rescues from misfolding a large repertoire of full-length proteins. We identify over 170 members of this cytosolic Escherichia coli TF substrate proteome, including ribosomal protein S7. We analyzed the biochemical properties of a TF:S7 complex from Thermotoga maritima and determined its crystal structure. This is the first atomic-level structure of a promiscuous chaperone in complex with a physiological substrate protein. The structure of the complex reveals the molecular basis of substrate recognition by TF, indicates how TF could accelerate protein folding and suggests a role for TF in the biogenesis of protein complexes.
INTRODUCTION
According to Anfinsen's thermodynamic hypothesis, the native structure of a protein achieves the conformation of minimal free energy for the particular polypeptide sequence (Anfinsen, 1973). Thus, many chemically denatured proteins refold spontaneously in the test tube. Protein folding in the cell is often more complicated, however. Whereas newly synthesized proteins emerge from the ribosome vectorially, exposing N-terminal sequences first, the native state may be incompatible with strictly co-translational folding and it may require an intimate co-folding with other components for assembly into multi-component complexes. Folding intermediates can be caught in non-native conformations, and non-native surfaces (charged or hydrophobic) that become exposed to the cytosol are prone to protein aggregation in the crowded cellular environment (Ellis, 2006). Such complications are exacerbated under destabilizing stresses such as thermal shock. The cell employs molecular chaperones to prevent or reverse aggregation or misfolding and to direct inextricably misfolded proteins to degradation (Hartl and Hayer-Hartl, 2002).
The molecular chaperone trigger factor (TF) was first identified as a cytosolic protein in Escherichia coli that stably bound the precursor of outer membrane porin A (proOmpA) (Crooke and Wickner, 1987). It also associates in 1:1 stoichiometry with ribosomes (Lill et al., 1988), where it presumably contacts most nascent chains (Valent et al., 1995). TF protects emergent polypeptides (Hoffmann et al., 2006; Tomic et al., 2006) and is thought to promote their co-translational folding (Stoller et al., 1995; Hesterkamp et al., 1996; Kramer et al., 2004b; Kaiser et al., 2006). Some nascent proteins leave the ribosome sequestered for prolonged periods by TF (Lee and Bernstein, 2002; Kaiser et al., 2006), perhaps to avert aggregation or misfolding (Hesterkamp et al., 1996; Deuerling et al., 1999; Teter et al., 1999; Genevaux et al., 2004), to promote post-translational folding (Lee and Bernstein, 2002; Agashe et al., 2004; Liu et al., 2005) or to facilitate the assembly into complexes (Lee and Bernstein, 2002; Liu et al., 2005).
Cytosolic TF is in 2–3-fold molar excess over ribosomes (Lill et al., 1988). TF in vitro forms stable complexes with chemically denatured full-length proteins and catalyzes refolding (Crooke et al., 1988; Stoller et al., 1995; Scholz et al., 1997; Huang et al., 2000; Kramer et al., 2004b; Liu and Zhou, 2004; Liu et al., 2005; Merz et al., 2006). Despite evident importance of TF chaperone activity, TF is essential only in certain genetic backgrounds such as absence of the Hsp70 chaperone DnaK (Deuerling et al., 1999; Teter et al., 1999; Genevaux et al., 2004; Liu et al., 2005) and at very low temperatures, where cells with reduced TF lose viability at exponential rates (Kandror and Goldberg, 1997). We present a schematic summary of TF activity in Fig. S1.
While preparing TF for structural studies, we discovered that TF interacts with a large repertoire of proteins. We identified 64 cytosolic and 4 presecretory proteins that stably associate with E. coli TF in vivo, including ribosomal protein S7. We found that many TF-associated proteins, including S7, aggregated in a strain lacking the three chaperones TF, DnaK and DnaJ (ΔtigΔdnaKdnaJ) (Genevaux et al., 2004), that these aggregates were efficiently rescued by reintroduction of TF (Deuerling et al., 1999; Deuerling et al., 2003; Genevaux et al., 2004; Merz et al., 2006), and that disaggregation was independent of TF-ribosome association. We also analyzed in vivo and in vitro interactions between TF and S7 from Thermotoga maritima and determined crystal structures of the T. maritima TF:S7 complex and of apo TF. Two native-like S7 molecules are encapsulated in hydrophilic cages formed by two juxtaposed TF molecules. The TF:S7 interface is exceptionally large, highly charged and non-specifically packed. We analyze these characteristics in light of promiscuous activity of TF as a folding chaperone and propose that hydrophilic interactions participate significantly in TF folding activity. We observe masking of exposed interfacial surfaces on S7 by TF, suggesting a role for TF as an assembly factor. Consistent with suggested activities, we find that TF affects ribosome biogenesis under conditions of thermal stress.
RESULTS
Characterization of a Cytosolic TF Substrate Proteome
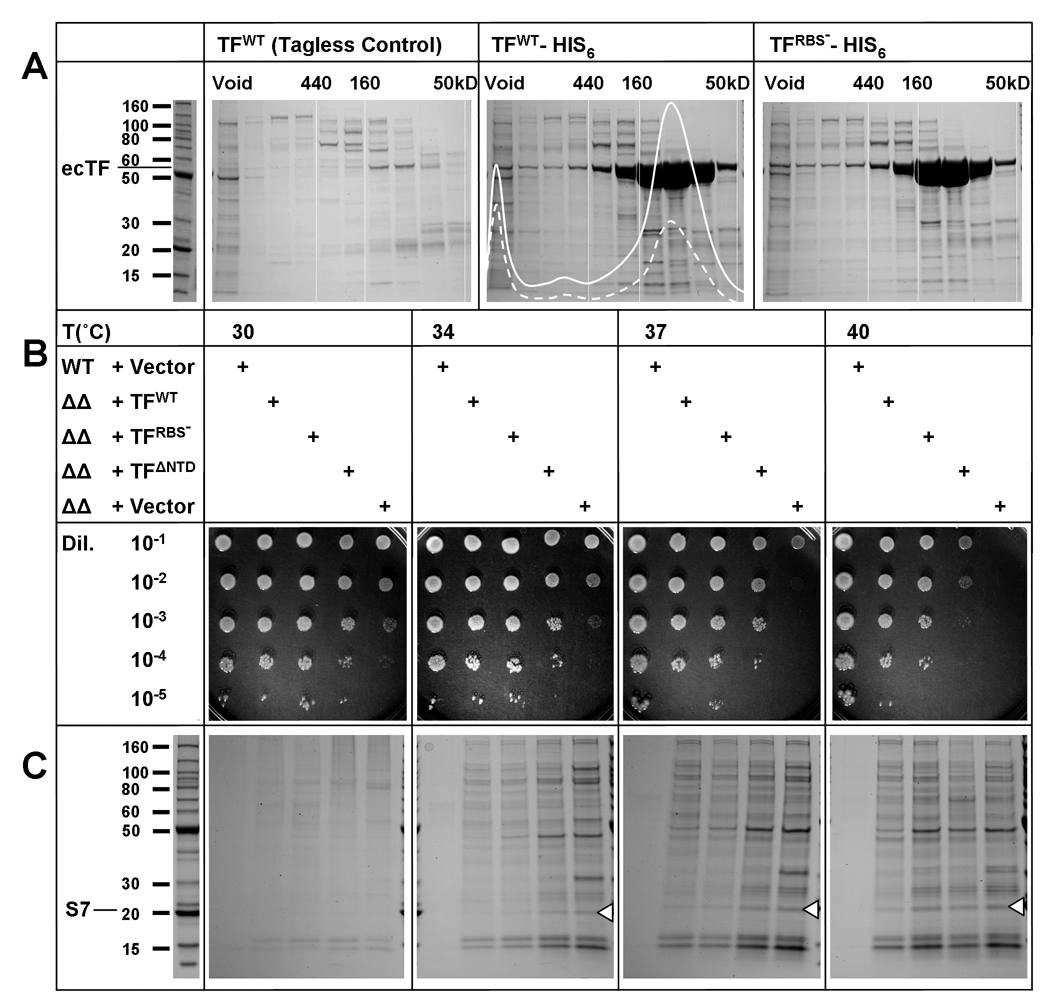
We used a combination of proteomic association and cellular aggregation methods to characterize a TF-substrate proteome. First, we isolated proteins that stably associate with TF in the E. coli cytosol by inducing expression of C-terminally His6-tagged TF for various times, removing membrane components with ultracentrifugation and isolating soluble TF-substrate complexes with metal-affinity chromatography followed by size exclusion chromatography (SEC) (Fig. 1A). Using trypsin-digestion coupled ESI mass spectrometry, we analyzed and compared individual SEC fractions from strains overexpressing His6-tagged and tagless TF to identify bona fide TF-substrate complexes. We found 68 proteins that eluted in SEC fractions containing His6-tagged TF (volumes corresponding to complex sizes between 50 and 200 kDa) but not in corresponding tagless TF controls (Fig. 1A, Table 1, Table S1). Using reverse pull-down assays, we confirmed that interactions persist between E. coli proteins TF and S7 and between TF and proOmpA, two of the identified TF substrates (Fig. S2). Chromosomally TAP-tagged TF also co-purifies with S7 and other proteins identified in our pull-down experiment (Table 1, Table S2), demonstrating that these interactions occur at cellular TF levels (Butland et al., 2005). Ribosomal proteins are abundantly represented among proteins that co-purify with TF. Judging from UV absorbances (Fig. 1A), intact or partially assembled ribosomes are in the SEC void volume, whereas the identified TF-substrate complexes are free of RNA.
Figure 1. In Vivo TF Function.
(A) Proteomic analysis of TF interactions in the E. coli cytosol. SDS PAGE analyses are shown for fractions of TF complexes first purified by metal-affinity chromatography from cytosolic lysates from TF-expressing cells and then separated by size exclusion chromatography. Separations from His6-tagged wild-type ecTF (center) are compared with those from a tagless control (left) and from a ribosome-binding deficient (RBS−) mutant (right), His6-tagged TF(FRK44-46AAA). Samples were purified with identical protocols. UV-absorbances at 280nm (straight line) and at 254nm (dashed line) are overlaid on the SEC profile of His6-tagged TF, and gel lanes are from corresponding fractions. Elution volumes of markers are shown (Void, 440kD, 160kD and 50kD). (B) Cell viability dependence on TF. Overnight cultures of wild-type MC4100 and the derivative ΔtigΔdnaKdnaJ mutant cell line transformed with vectors harboring TF variants or the empty control were serially diluted and spotted on LB agar plates at indicated temperatures. (C) TF protection against cellular protein aggregation. The indicated strains of variously transformed cells were grown for approximately 4 h at specified temperatures. Aggregates were isolated as described (Tomoyasu et al., 2001) and visualized on 4–20% SDS–PAGE gels stained with Coomassie blue. White triangles mark the position of S7 as verified by mass spec sequencing of five S7 peptides from an excised gel band.
Table 1.
E. coli TF interacting proteins.
| TF-sensitive Aggregation | |||
|---|---|---|---|
| TF Co-purification | |||
| 30S Subunit | S11, S10, S14, S19,S21 | S21, S3, S51, S71, S9, S11 | S4, S6, S12 |
| 50S Subunit | L10, L16, L17, L22, L28 | L1, L2, L31, L41, L5, L6, L12, L13, L14, L15 | L11, L18, L231, L27 |
| Protein-nucleic acid | MutM, TopA, TrmA | Crp, CysS, RlmN | |
| Homomultimer | AsnB, ArgD, AvtA, CbpA, FtnA, GatD, GldA, Glk, HisB, IspD, LacA, LuxS, Maa, NanK, NudC, PdxH, Rnc, Tdk, ThrA | FabA, FruR, NusD | AsnS, AccC1, AdhE, AldA, DrpA, EnvM/FabI, FabZ, FbaA, GapA, GlmS, GlnA, GlpK*, GltA, GuaA, Icd, KatE, KatG, Kbl, MaeB, MalT, Mdh, MetG, Ndk, PflB, PpsA, PrsA, PssA, Pta, PyrG, RecA, RihA, RmlB, Rph, SeqA, SpeD, SthA, TalB, ThrS, TktA, TnaA |
| Heteromultimer | CarA, CarB, GatA, HybO, NuoE | DadA, GatZ, MreB, SdhA, UvrA, SucB | AceE, AceF, AspA1, AtpA, AtaD, BipA, CysJ, CysI, CysK, DeaD, DnaB, FadB, FfH, GatY, GlyQ, GyrA, GyrB, InfB, InfC, LepA, Lon, LpdA, Mfd, NrdA, NuoC, NuoG, Pnp, PyrB, RpoB, RpoC, RpoD, SucA, SucC, SucD |
| Monomer | AcnB, AlaS, GlcB, TrxA | ||
| No assembly information | YiiD, YfiF | AceB, BglA, FadE, IntF, MurC, TreC, Ugd, YbeD, YbeZ, YeiQ, YheS, YjhC, YjjK | |
| Inner Membrane | HemY, HflB, HflX, TreB | ||
| Outer Membrane | AfaC | OmpA | OmpC, OmpF, OmpT, OmpX |
| Secreted | OpgG, PstS | Lpp, SpeA | |
Detected with chromosomally tap-tagged TF with > 80% confidence score (Butland et al., 2005)
TF chaperone action is thought to require association with the ribosome at the polypeptide exit tunnel and is attributed to TF interactions with emergent nascent polypeptides (Kramer et al., 2002; Ferbitz et al., 2004; Hoffmann et al., 2006; Kaiser et al., 2006; Raine et al., 2006; Merz et al., 2008; Rutkowska et al., 2008). We therefore contemplated the possibility that some or all of our identified TF substrates might have persisted from nascent chain binding followed by release of the TF-substrate complex from the ribosome. Thus, we tested cells expressing His6-tagged TF FRK44-46AAA, a ribosome-binding-site variant (RBS−) that shows negligible ribosome binding (Kramer et al., 2002; Maier et al., 2003; Kaiser et al., 2006). To our surprise, we found that the interaction profiles of His6-tagged RBS−-TF are indistinguishable from those of His6-tagged WT-TF (Fig. 1A); thus, the TF-substrate complexes that we observe arise independently from TF association with ribosomes. As was first observed with proOmpA (Crooke and Wickner, 1987), TF forms stable post-translational associations in the cytosol and it does so with a large repertoire beyond proOmpA.
It is well established that the aggregation and growth phenotype of the ΔtigΔdnaKdnaJ mutant E. coli cell line can be rescued by expression of either DnaK and DnaJ or TF alone (Genevaux et al., 2004; Kramer et al., 2004b; Merz et al., 2006). Thus, we systematically tested the effects of TF expression in ΔtigΔdnaKdnaJ cells at various temperatures. Since we found in our proteomic studies that ribosome association is not needed for TF-substrate association, we also tested two ribosome-binding deficient mutants (FRK44-46AAA and ΔNTD). Moreover, Genevaux et al. (2004) had noted that RBS− mutants exert only mild effects on TF function in the cell. TF constructs were expressed at approximately wild-type levels from a low-copy plasmid harboring the wild-type TF promoter (Fig. S3) as overexpression of TF is deleterious.
We find that ΔtigΔdnaKdnaJ cells (ΔΔ + Vector) are viable at 30°C (although less so than wild-type cells), but viability decreases progressively as temperature increases (Fig. 1B). Protein aggregation in ΔtigΔdnaKdnaJ follows in parallel (Fig. 1C), but with greater overall sensitivity than for cell viability. Expression of either WT-TF or RBS−-TF in ΔtigΔdnaKdnaJ restores cell viability and alleviates aggregation equivalently at 34° and 37°C. Even TF-ΔNTD proved quite effective for cell viability. At 40°C, RBS−-TF becomes less effective than WT-TF suggesting a role for ribosome-associated activity. Strikingly, TF has much greater effect on cell viability than on aggregation, especially at 37°C, which suggests that TF may also be resolving toxic solubly malfolded proteins, perhaps by directing them to degradation. Taken together, these data demonstrate that TF can rescue the ΔtigΔdnaKdnaJ phenotype and that these in vivo chaperone functions of TF are independent of ribosomal association.
The physiological function of TF is intimately associated with its ability to resolve protein aggregates (Fig. 1B,C). We therefore undertook to identify putative TF substrates from these protein aggregates, which we isolated using a well established protocol (Tomoyasu et al., 2001) and analyzed by trypsin-digestion coupled ESI mass spectrometry. We found 136 proteins that aggregated in the ΔtigΔdnaKdnaJ cell at non-permissive temperatures (Table 1) including many that also co-purify with TF, notably ribosomal protein S7 (Fig. 1C). These results are consistent with but go beyond other reports (Deuerling et al., 2003; Maisonneuve et al., 2008).
We identified a total of 178 putative cytosolic TF substrates by TF co-purification or by TF-resolvable aggregation; 42 proteins exclusively co-purify with TF, 110 proteins exclusively aggregate in the ΔtigΔdnaKdnaJ mutant cell line, and 26 proteins are in both datasets (Table 1). Proteins that co-purify with TF range in size from 8.4 kDa (S21) to 117.8 kDa (CarB) with a mean of 36.5 kDa, similar to the average for all E. coli proteins. We find that a large number of TF substrates form oligomers or macromolecular assemblages where they must engage other components. Ribosomal proteins found associated with TF, for example, typically contain long N- or C-terminal tails or extended internal loops that interact extensively with RNA inside the ribosome, whereas these elements are often disordered in crystal structures of the isolated proteins. Non-globularity of ribosomal proteins in situ is correlated significantly with TF association (Fig. S4). Folding to the ‘native’ state for such proteins requires association with their partners; and, by Anfinsen’s hypothesis, only together do these achieve the minimal Gibbs free energy as determined by the sum of all of interatomic interactions (Anfinsen, 1973). Thus, the free energy minimum of a ribosome-bound protein is lower than that of its isolated form, however native-like that conformation may be. Compensation for this free energy difference could provide a thermodynamic basis for the extended association between TF and some cytosolic proteins.
TF Interactions with Ribosomal Protein S7
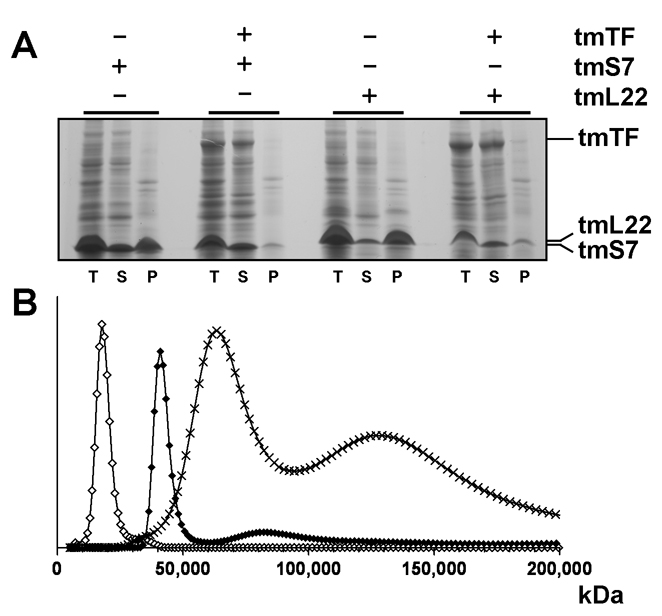
E. coli TF (ecTF) associates with S7 and other ribosomal proteins in the cell (Fig. 1A, Table 1). S7 occurs primarily in ribosome fractions and is very sparse alone in soluble form (Fredrick et al., 2000); thus, TF:S7 co-purification is not simply effected by high abundance. Moreover, tagless TF co-purifies with His6-tagged S7 (Fig. S2), and TF suppresses S7 aggregation in ΔtigΔdnaKdnaJ cells (Fig. 1C, Table 1). When T. maritima S7 (tmS7) is overexpressed in E. coli, it predominantly accumulates as insoluble aggregates, but the precipitated fraction is mostly resolved when tmTF is co-expressed with tmS7; as is also true for another putative ecTF substrate, ribosomal protein L22 (Fig. 2A). These results are consistent with tmTF acting as a molecular chaperone to prevent tmS7 and tmL22 aggregation in vivo and confirm our observations of E. coli S7 aggregation in a chaperone deficient genetic background. Soluble components were not increased, however; perhaps TF helps direct aggregation-prone proteins to degradation (S7 is an SOS-enhanced substrate for the ClpXP protease system (Neher et al., 2006)).
Figure 2. Characteristics of TF:Substrate Interactions in Vivo and in Vitro.
(A) Expression of tmS7 and tmL22 in E. coli with and without tmTF. Gel lanes show proteins in total (T), supernatant (S), and pellet (P) fractions. Lanes correspond to: 1–3 (tmS7), 4–6 (tmS7 + tmTF), 7–9 (tmL22), 10–12 (tmL22 + tmTF). (B) Size-distribution analysis of the T. maritima TF:S7 complex by sedimentation velocity ultracentrifugation and Lamm equation modelling. Open diamonds correspond to tmS7 (Mw 17kD), full diamonds to tmTF (Mw 48kD) and crosses to the TF:S7 complex (Mw 65kD and 130kD).
To characterize the T. maritima TF:S7 interaction in solution, we performed velocity and equilibrium sedimentation analyses. Our data revealed that both tmTF and tmS7 when alone migrate with sedimentation velocities correspondent to the monomeric species, 48 kDa and 17 kDa, respectively, but that mixtures of tmTF with tmS7 show associations between the two proteins in vitro. Oligomeric species migrate with apparent molecular masses of 65 kDa and 130 kDa, corresponding to tmTF:tmS7 complexes with stoichiometries of 1:1 and 2:2, respectively (Fig. 2B). Sedimentation equilibrium experiments suggest that TF:S7 exists in monomer-dimer-tetramer equilibrium with an overall dissociation constant of K (AABB) of 4.7 × 10−10 M3 (Fig. S5).
Structure Determinations
To analyze the structure of T. maritima TF, we produced and purified the full-length protein (tmTF425), a slightly truncated variant (tmTF404) and also nine other domain constructs. We solved high-resolution crystal structures from domains encompassing residues 1–116 (tmTFN) and residues 243–404 (tmTFC) (Martinez-Hackert and Hendrickson, 2007). Large single crystals of intact TF grew readily, but typically diffracted to 8Å spacings at best; however, one orthorhombic crystal (C2221) of tmTF404 diffracted to ~3.5Å spacings after dehydration. Full-length tmTF425 complexed with ribosomal protein tmS7 reproducibly formed tetragonal crystals (P43212) that diffracted anisotropically to 3.5/4.0Å spacings (Table S3).
We solved the TF:S7 crystal structure using selenomethionyl (SeMet) substituted protein and MAD phasing. The experimental electron-density map is clear and continuous for both TF and S7, secondary structure was readily interpretable and domains were instantly recognizable; however, as a result of limited resolution, correct side chain rotamers were in some instances difficult to ascertain (Fig. S6). We placed high-resolution crystallographic structures of T. maritima TF domains and Bacillus stearothermophilus S7 into appropriate experimental electron densities to assist in model building (Fig. S7), manually modeled the intermediate PPIase domain, and thoroughly rebuilt and refined the full atomic model (R = 25.7% and Rfree = 32.8%, Table S4). We solved the crystal structure of apo TF by molecular replacement from the refined TF complex structure (R = 24.9% and Rfree = 35.8%, Table S4).
Structures within the TF:S7 Complex
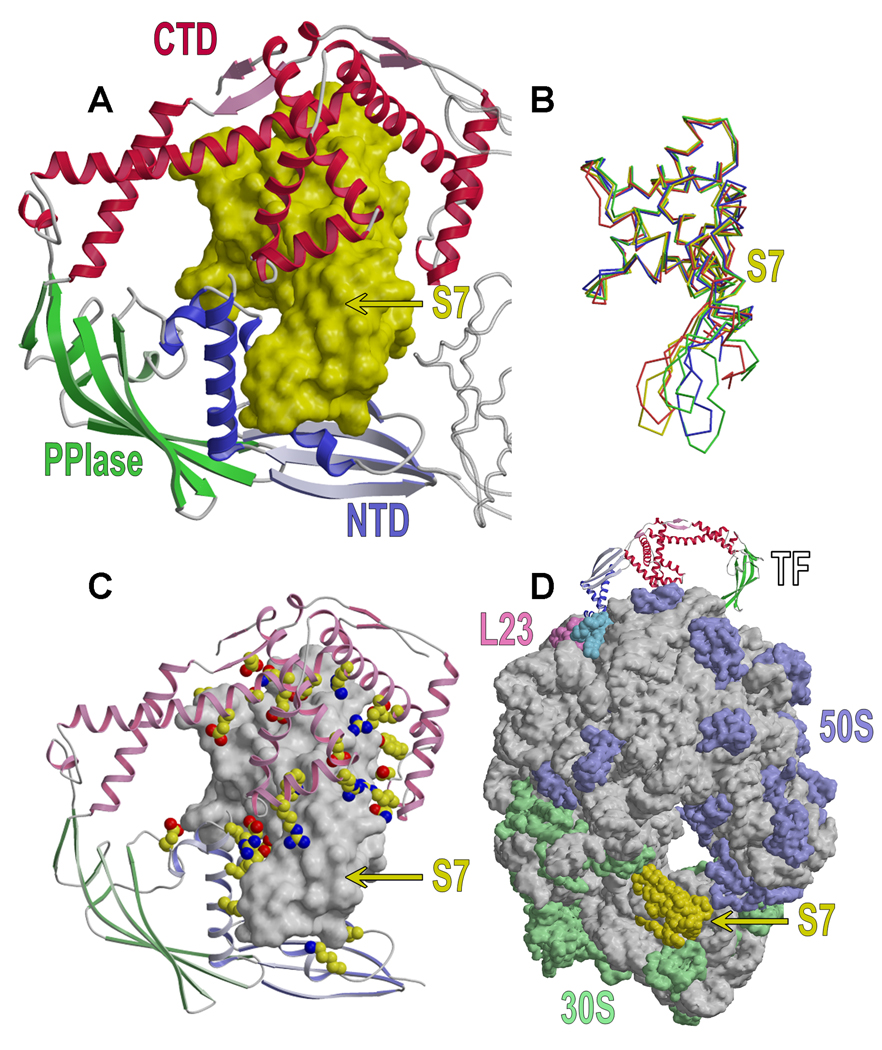
TF within the complex comprises three domains (Fig. 3A–C,Fig. S6–S8), each homologous to a known structure: the N-terminal domain (NTD), residues 1–110, which harbors the ribosome binding loop, resembles a motif ensconced within the redox-regulated chaperone Hsp33; the middle domain, residues 147–229, is an atypical member of the peptidyl-prolyl cis/trans isomerase (PPIase) FK506-binding-protein family; the C-terminal domain (CTD), residues 111–146 and 230–425, is structurally similar to a domain of the chaperone SurA and to all of Mpn555, a protein of unknown function (Figs. S9,S10). The NTD and PPIase domains are connected via an extended linker (residues 111–146). This segment is an integral element of the CTD and is presumably required for the correct folding and activity of the TF CTD structure (Merz et al., 2006; Martinez-Hackert and Hendrickson, 2007). Thereby, in three dimensions, CTD is in the middle of the TF structure with NTD and PPIase domains at opposite ends.
Figure 3. Structure of TF in Complex with Ribosomal Protein S7.
(A) Ribbon diagram of tmTF colored by domains. The N-terminal domain (NTD) is colored blue; the PPIase domain green; the C-terminal domain (CTD) red. NTD and PPIase are connected via a linker colored yellow here and subsequently colored red as part of CTD. Disordered TF residues shown as a blue dotted line correspond to the ribosome binding loop. (B) Ribbon diagram of TF colored by domains as in A except for linker, now red. The yellow ribbon corresponds to tmS7 as it is bound inside the CTD cleft. (C) TF:S7 complex. Two TF molecules (ribbons colored by domains as in b) encapsulate two S7 molecules (yellow molecular surfaces). (D) Surface representation of the TF:S7 complex, TF is colored red and blue, S7 is colored yellow. (E) Ribbon diagram of substrate free tmTF colored by domains as in B. The yellow ribbon represents a symmetry related tmTF with its NTD (solid) bound inside the CTD cleft. (F) Ribbon diagram of substrate free ecTF colored by domains as in B. The yellow ribbon represents a symmetry related ecTF with its NTD (solid) bound inside the CTD cleft.
The NTD and CTD domains contain helical segments that resemble protruding limbs. These helical protrusions and the PPIase domain point in the same direction, thereby creating a hollow, sickle-like structure. The outer surface of this structure is smoothly convex; the concave, highly irregular inner surface forms an extended cleft. In this, the TF structure vaguely resembles a miniature form of prefoldin, a chaperone that harbors a voluminous cleft bordered by six helical coils (Siegert et al., 2000).
The TF:S7 complex is organized as a symmetric heterotetramer, consistent with ultracentrifugation results, and it is centered on a two-fold axis in the crystal. Two TF molecules join to encapsulate two S7 molecules within their apposed extended clefts (Fig. 3C,D,Fig. S6,S8). Most of the contacts presented by TF to S7 involve the limb-like helical protrusions (Fig. 3B–D, Fig. 4A,Fig. S8). NTD contacts S7 primarily through its third α-helix, around which the flexible β-hairpin structure of S7 appears to adapt, and CTD effectively envelops one hemisphere of S7 with its three helical protrusions. This is consistent with an observed importance of CTD in TF chaperone activity (Genevaux et al., 2004; Kramer et al., 2004b; Merz et al., 2006). Interactions between the PPIase domain and S7 are negligible, which agrees with experiments showing that the PPIase domain of TF is not required for its chaperone activity (Li et al., 2001; Genevaux et al., 2004; Kramer et al., 2004a). The PPIase domain is intimately involved in forming the TF:S7 tetrameric complex, however, making contacts through conserved residues to NTD′ from the apposed TF (Fig. 3C,Fig. S9C).
Figure 4. S7 Interactions with TF.
(A) S7 (yellow molecular surface) is encapsulated by NTD and CTD of apposed TFs. TF (colored by domains as in 3B) is rotated by approximately 180° along a vertical axis relative to 3B. (B) Superposition of S7 structures oriented as in A. tmTF bound tmS7 is colored red, ribosome bound Thermus thermophilus S7 (pdbid: 1FJG, chain G) green, isolated T. thermophilus S7 (pdbid: 1RSS) yellow and isolated B. stearothermophilus S7 (pdbid: 1HUS) blue. (C) Ribbon diagram of TF oriented as in A with the molecular surface of S7 colored grey. Included are hydrophilic and polar TF residues that contact S7 with hydrogen bonds or salt bridges. Carbon atoms are colored yellow, nitrogen atoms blue and oxygen atoms red. (D) TF in the ribosome-bound state. Ribosomal RNA is shown as a grey surface, 50S proteins are colored blue, 30S proteins are colored green. TF docks on the 50S subunit by binding to proteins L23 (pink) and L29 (cyan). TF is shown as a ribbon diagram with S7 as a yellow surface. S7 as bound to 30S is also shown as yellow surface.
Ribosomal protein S7 assumes a native-like conformation in the complex with TF. This consists of six α-helices and a highly twisted β-hairpin extended from between helices 3 and 4. Helices 1–5 cluster to form the core of the S7 structure and C-terminal helix 6 extends alongside the β-hairpin. The helical core is practically invariant among several known S7 structures (Fig. 4B,Fig. S7B); the average RMSD between isolated, TF-bound and ribosome-bound S7 core is 0.65Å. However, S7 exhibits significant plasticity outside of the helical core. The most prominent differences, up to 12Å among the diverse S7 structures, are seen in the β-hairpin (Fig. 4B,Fig. S7B). This appears to reflect intrinsic flexibility since the isolated and ribosome-bound S7 structures from the same organism differ substantially. Ten N- and eight C-terminal residues are even more flexible and are disordered in the isolated and TF-bound structures. The β-hairpin and the termini are clearly structured in the ribosome complex, where the N-terminus and β-hairpin contact 16S RNA extensively and the C-terminus interacts with the ribosomal protein S11 and 16S RNA.
Structure of Substrate-free TF
ApoTF (substrate-free) is structurally similar to TF as complexed with S7 (Fig. 3B,E, Fig. S11), but with substantial interdomain flexion (Table S5, Fig. S11, Supplemental Text). Our structures of tmTF are also similar overall to the substrate-free structure of E. coli TF (ecTF) (Ferbitz et al., 2004)(Fig. 3B,E,F,Fig. S10–S12), but differences in interdomain dispositions are greater than those observed between the two states of tmTF (Fig. S11E, Table S5). Clearly, intrinsic segmental flexibility accounts for much of the differences between TF structures.
Both apoTF structures are, in a sense, not strictly substrate-free. It turns out that in each of apo tmTF and apo ecTF the crystal lattice has the NTD′ domain of a symmetry-related TF molecule bound within the S7 binding cleft of the CTD (Figs. 3B,E,F). Relative NTD′ orientations are different, but NTD′-CTD contacts are substantial for both (total buried areas of 1280Å2 for tmTF and 2800Å2 for ecTF). As for S7, these NTDs retain their native conformations. These associations are consistent with the CTD cavity having a role in non-specific binding as found in our substrate proteome for E. coli TF, and they probably also relate to TF ‘dimerization’ (Patzelt et al., 2002; Kaiser et al., 2006).
Properties of the TF:S7 Interface
The highly convoluted TF:S7 interface is extensive; a total of 4,520Å2 of solvent-accessible surface area is buried into the interface formed with each S7, including 795Å2 from NTD, 1,415Å2 from CTD, a bit from the PPIase, and 2,260Å2 from the substrate protein S7 (25% of its surface). The substantial involvement of the NTD and CTD domains in the interaction with the substrate protein is consistent with the observed contribution of each domain to TF chaperone activity in vivo and in vitro (Genevaux et al., 2004; Kramer et al., 2004b; Merz et al., 2006) (Figs. 5A). Additional TF:TF interactions, dominated by contacts from juxtaposed NTD and PPIase domains, contribute 1,095Å2 of buried surface area per TF protomer. In sum, approximately 11,230Å2 of solvent-accessible surface area is buried from the components upon formation of the heterotetrameric TF:S7 complex.
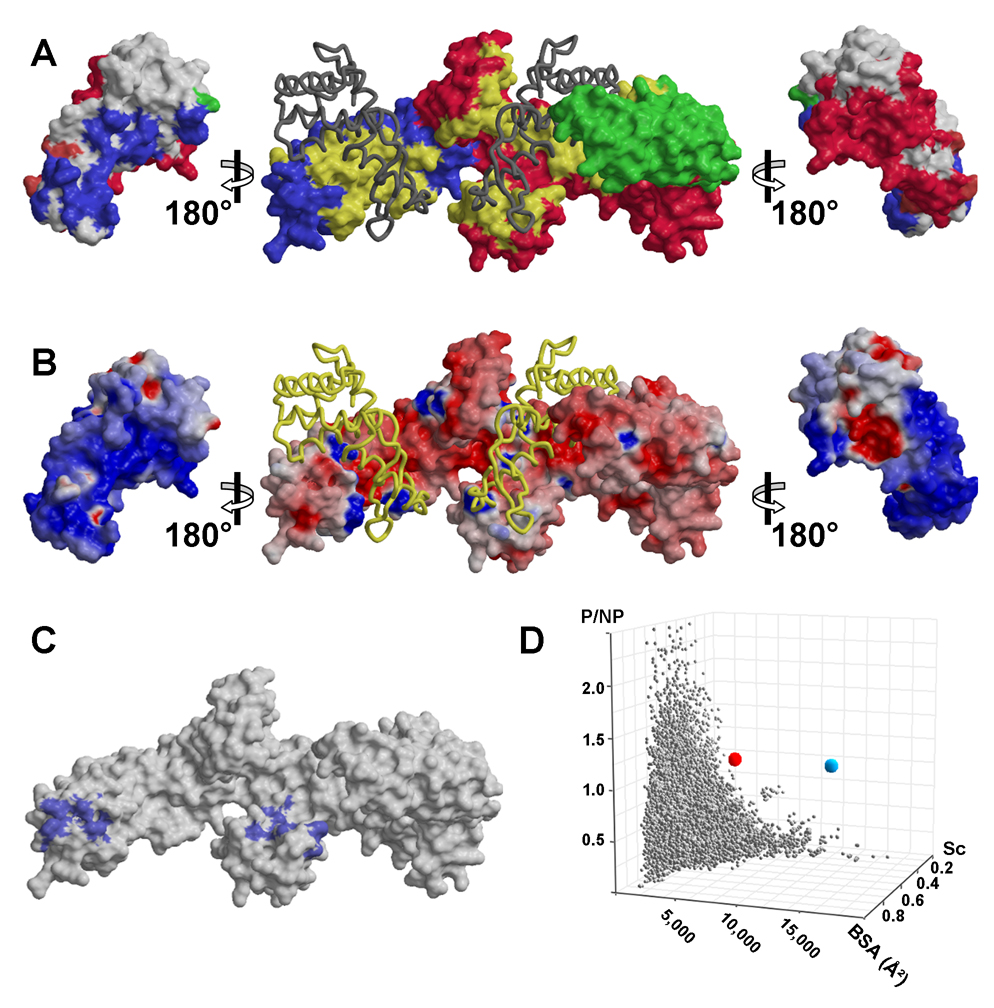
Figure 5. Surface Representations of T. maritima and E. coli TF.
(A) Contact surfaces. The molecular surface of TF (middle) is colored by domains as in 3B but with the imprint of bound S7 colored in yellow. The dark ribbon shows S7. Molecular surfaces of S7 (outside) are rotated to display the imprint of bound TF. Blue residues contact NTD, green contact PPIase, red contact CTD and magenta contact both NTD and CTD. (B) Electrostatic potential of TF and S7. Molecular surfaces are oriented as in A and S7 is drawn as a yellow ribbon. Surfaces are colored in degrees of positive (blue) and negative (red) potential. (C) Hydrophobic patches on the tmTF surface. Vicinal apolar atoms that form continuous hydrophobic surfaces are colored blue. (D) Comparison of the TF:S7 interface with approximately 44,000 structurally defined interfaces between pairs of protein domains catalogued in PYBASE (Davis and Sali, 2005). Properties of the TF:S7 interface are represented by the red and blue spheres (dimer and tetramer, respectively). PYBASE interaction sets are represented by grey spheres. Sc corresponds to Shape Complementarity Value, BSA corresponds to Buried Surface Area and P/NP corresponds to the ratio of polar versus non-polar interfacial residues.
The electrostatic surface potentials of the chaperone (both substrate-bound and substrate-free) and substrate reveal a predominately hydrophilic TF:S7 interface (Fig. 5B). Fifteen negatively and seven positively charged TF residues and five negatively and fourteen positively charged S7 residues contribute salt bridges and hydrogen bonds to the interface (Fig. 4C). Additional hydrogen bonds are provided by multiple polar residues. Hydrophobic contacts in the TF:S7 interface occur primarily between the aliphatic components of the interacting arginines, lysines and glutamates; but some non-polar residues, including leucines, isoleucines and valines, also contribute. Our resolution is too limited to define the role of water molecules in the TF:S7 interaction.
We analyzed the tmTF and ecTF structures to identify explicit hydrophobic surface patches consisting of vicinal apolar atoms (Fig. 5C). There are two conserved hydrophobic patches on the TF molecules, each substantially larger for ecTF than tmTF: 990Å2 and 750Å2 versus 475Å2 and 460Å2, respectively. The most conspicuous hydrophobic patch, located at the tip of NTD near the ribosome-binding loop, may be in position to contact hydrophobic residues of emerging nascent proteins. The second major patch covers the tip of one helical protrusion from CTD. Neither patch features prominently in the TF:S7 interaction, contributing only a fraction of each area to the complex interface.
TF as a Ribosome Assembly Factor
S7 is an integral component of the 30S ribosome subunit, where it interacts extensively with 16S RNA and proteins S9 and S11 in situ (Fig. 6A, Table S6) (Brodersen et al., 2002). The surface area buried upon incorporation of S7 into the 30S subunit is 4,580Å2, similar to the area buried between S7 and TF (Table S6). A fraction of the S7 surface is contacted by both 16S RNA and TF (Fig. 6B). This overlapping surface contributes approximately 1,800Å2 of buried surface area to each complex. As a result, TF can mask a significant portion of the S7:30S contact surface (~40%).
Figure 6. TF and Ribosome Biogenesis.
(A) Surfaces of T. thermophilus 30S (tt30S) protein components excluding S7 are shown in red, 16S RNA in blue; S7 is colored grey. (B) Contact imprints on the surface of T. thermophilus S7 (ttS7) oriented as in 5a. Surfaces colored blue uniquely contact tt30S, tmS7 homologs of surfaces colored yellow uniquely contact tmTF, and surfaces colored magenta contact both tt30S and tmTF. (C) Polysome profiles of wild-type cells grown at 30°C and wild-type and Δtig cells grown at 44°C. (D) Average relative peak-height ratios of 70S/30S (light-gray), 70S/50S (dark-blue) and 50S/30S (light-blue) from the indicated strains including TF overexpression (OX) at 44°C are shown. (E) Model of cytosolic TF function. TF could sequester nascent proteins or bind fully synthesized but unstable subunits, like S7, and remain stably associated with these subunits until productive folding or assembly occurs.
The contacting surfaces between S7 and 30S are well matched with a shape complementarity (Sc) index (Lawrence and Colman, 1993) of 0.71 as contrasted with the poorly packed interface of the TF:S7 complex (Sc=0.46). Contact specificity at the S7-TF interface is much lower than at the S7-30S interface (Table S6). Therefore, while TF may effectively mask interacting surfaces on S7, TF may not effectively compete with S7:30S complex formation.
As many ribosomal proteins co-purify with TF (Table 1) and TF masks ribosome interacting sites on S7 (Fig. 6A,B), we hypothesized that TF could be involved in ribosome biogenesis. We examined whether deletion of TF resulted in polysome defects as assayed by sucrose density centrifugation. Deletion of TF causes an evident increase in 50S particles at elevated temperatures (Figs. 6C,D); although the relative ratio of 70S to 30S particles was little affected, the ratio of 70S to 50S subunits decreased from 7.2 at 30°C to 4.4 at 44°C and futher to 2.0 in Δtig cells growing at 44°C. TF overexpression at 44°C restored this ratio to the 30°C WT level (6.5). The 30S:50S ratio shows a similar pattern (Fig. 6D). Neither TF deletion nor overexpression affected the polysome profile at 37°C (data not shown), whereas S7 overexpression is harmful (Fredrick et al., 2000). We conclude that deletion of TF, at least under conditions of stress, such as elevated temperature, results in a distinct ribosome assembly defect.
Attributes of Promiscuity in TF Binding
Trigger factor is a promiscuous chaperone and S7 is representative of its cytoplasmic substrates. Accordingly, the TF:S7 interface reflects a non-specific interaction and its properties are extraordinary when compared to specific complexes, as for S7 in the ribosome. Specific interfaces, exemplified by antibody-antigen and enzyme-inhibitor complexes, are generally characterized by substantial interfacial areas, high shape complementarity and predominance of non-polar contacts (Lo Conte et al., 1999). An average, ‘standard-size’ interface buries ~1,500Å2 of surface area; its Sc index is ~0.7 where 1.0 represents a perfect match; and non-polar character is characterized by a paucity of charged residues, reflected in buried-charged densities of 0.4 and 0.6 charges/nm2 for enzyme-inhibitor and antibody-antigen complexes, respectively (Q.R.F. and W.A.H., in preparation).
The TF:S7 interface presents a combination of properties entirely different from those of specific interfaces (Fig. 5D). First, this interface is very large. Its buried surface area (4,520Å2) is approximately three times that of a ‘standard-size’ specific interface. Second, the TF:S7 interface is poorly packed. Its Sc value of 0.46 is among the lowest in the published literature. Finally, the interface between TF and S7 is dominantly polar. It has a buried charge density far greater than other known protein-protein and protein-DNA complexes (1.46 charges/nm2) (Q.R.F. and W.A.H., in preparation).
Inter-domain flexibility (Fig. S11) likely facilitates TF adaptation to diverse substrates. Flexibility in the long-side-chained charged residues that predominate at TF binding surfaces (Figs. 4C) could add to plasticity for promiscuous binding. We envision a complex equilibrium of states in TF-substrate complexes (Fig. 6E), including variations in stoichiometry (Fig. 2).
To test the unusual binding interfaces of the TF:S7 crystal structure, we designed a set of mutations aiming to disrupt by charge repulsion and bulk; we made multiple arginine replacements at various TF contacts with the electropositive S7 (Fig. 5B), largely replacing TF carboxylates (Table S7, Fig. S13A). One of three interfacial Arg-cluster mutations is highly defective in S7 interaction and one of two double-cluster mutations (+9 charge change) is completely devoid S7 binding (Fig. S13B). These results are consistent with a flexibly promiscuous TF binding surface that prefers basic substrates. We also used cross-linking experiments to examine the roles of different TF domains. Whereas WT-TF shows bands consistent with 1:1, 1:2 and 2:2 TF:S7 cross-linked products, TF-ΔNTD, TF-ΔPPIase and TF-CTD (ΔNΔP) each showed only 1:1 cross-linked products (Fig. S13C). These results are consistent (a) with WT 2:2 complexes forming in solution, as also seen by ultracentrifugation (Fig. 2B & Fig. S5), (b) with S7 being needed for the TF:TF interaction, (c) with sufficiency of CTD alone for S7 binding, and (d) with essentiality of NTD-PPIase contacts for TF:TF complexation. Residues at the NTD-PPIase contacts are poorly conserved, however; and, since TF:TF contacts are substrate-mediated and substrates vary, it is likely that many TF-substrate complexes will have 1:1 stoichiometry even though T. maritima TF can bind S7 in a 2:2 manner.
DISCUSSION
Implications for Folding
Many studies show that TF assists in the folding of nascent and full-length proteins in vivo and in vitro, but the molecular basis for its chaperone activity has been unclear. This work reveals atomic details of an interaction between TF and a full-length substrate. To understand how TF promotes protein folding, we compare our observations on TF with the GroEL/GroES chaperonin system. The mechanism of GroEL/GroES activity involves encapsulation of a single substrate protein in an isolated chamber, also termed the ‘Anfinsen cage’ (Ellis, 1994; Fenton and Horwich, 2003; Tang et al., 2006). The confined Anfinsen cage environment disfavors the extended conformations of unfolded proteins and stabilizes the compact structures of native-like proteins (Fenton and Horwich, 2003; Tang et al., 2006), as seen for a GroEL/GroES encapsulated substrate in a recent cryoEM structure (Clare et al., 2009). Moreover, the hydrophilic environment lining the GroEL/GroES Anfinsen cage could contribute to chaperonin activity by favoring the burial of hydrophobic and exposure of hydrophilic residues (Fenton and Horwich, 2003) and catalyzing the structural annealing of some unfolded proteins (Tang et al., 2006).
Apposed TF molecules of the TF:S7 complex encapsulate the substrate S7 in a compact, native-like conformation within an ‘Anfinsen-cage’-like chamber (Fig. 4A). Importantly, as in the GroEL/GroES chamber, the physicochemical environment of the TF Anfinsen cage could enable TF to facilitate folding. We envision that the hydrophilic lining of the TF Anfinsen cage could attract the hydrophilic groups of non-native proteins, as it does with folded S7, thus provoking their externalization and concomitant burial of hydrophobic amino acids in the confined folding substrate. We suggest that such encapsulation reduces the conformational entropy of the folding protein by stabilizing compact, constructive folding intermediates to catalyze the folding process (Takagi et al., 2003). A single TF molecule presents an open cavity, but we know from TF:S7 centrifugation results (Fig. 2B) and from self-associated TF in crystal lattices (Figs. 3E,F) that 1:1 complexes also form and can do so with portions of multi-domain proteins. The analogy of TF to GroEL/GroES is certainly inexact, however, as the mechanisms for substrate capture and release differ greatly and are poorly understood for TF.
TF as bound to the ribosome appears poised to have its substrate cleft oriented toward the ribosome surface (Merz et al., 2008); thus, the ribosome itself should help to complete a hydrophilic Anfinsen cage. We have modeled such a complex (Fig. 5D, S14). At the ribosome, TF interacts physically with nascent polypeptides as they emerge from the polypeptide exit tunnel where, it is suggested, hydrophobic segments preferentially contact TF to facilitate folding (Kaiser et al., 2006; Raine et al., 2006; Lakshmipathy et al., 2007; Rutkowska et al., 2008). Nonetheless, many features of TF action on the ribosome are consistent with a hydrophilic Anfinsen-cage mechanism: TF binds full-length folding proteins more tightly (Scholz et al., 1997; Maier et al., 2001) than short peptides (Patzelt et al., 2001), TF binding to translating ribosomes increases with nascent chain length (Kaiser et al., 2006; Raine et al., 2006; Rutkowska et al., 2008), association rates are quite similar for TF binding to ribosome-nascent-chain complexes carrying matched folded or unfolded nascent chains (Rutkowska et al., 2008), TF protects nascent proteins of up to 280 residues against proteolytic degradation (Hoffmann et al., 2006; Tomic et al., 2006), TF accommodates and protects small folded domains within its internal substrate-binding cavity (Merz et al., 2008) and, importantly, Raine et al. (2006) demonstrate that TF affinity for ribosome-nascent chain complexes of matched chain lengths increases with the fraction of hydrophilic residues in the nascent chain (Fig S15, Table S8).
Implications for Holding
Besides catalyzing protein folding, TF stably binds a number of proteins. This was originally observed with proOmpA (Crooke et al., 1988) and is evident from our E. coli TF substrate proteome. A clue to a role for TF holding activity is that most identified cytosolic TF substrates are subunits of multimeric complexes, including many ribosomal proteins. This leads us to think that TF could take part in regulating ribosome biogenesis as nucleoplasmin does in nucleosome assembly. Indeed, Laskey et al. (Laskey et al., 1978) coined the term ‘molecular chaperone’ to describe this role for nucleoplasmin binding to histones H2A and H2B. Similarly, TF may be a chaperone for proper ribosome biogenesis as it binds ribosomal proteins such as S7.
This idea of TF as an assembly chaperone has support from several observations. Ribosomal proteins in our TF proteome correlate with non-globularity of these proteins in situ in the ribosome, consistent with a TF role in protecting thermodynamically non-native, unassembled states (Fig. S4); TF-associated ribosomal proteins correlate with the 30S assembly map, excluding those that incorporate most rapidly into 30S particles (Talkington et al., 2005) (Fig. S4); TF in our TF:S7 complex masks 16S RNA binding sites on S7 (Fig. 6A,B); and TF deletion results in defective ribosome assembly under thermal stress (Fig. 6C,D). Ribosome assembly in eukaryotic cells is coordinated by some 170 trans-acting accessory proteins, including transport factors and assembly chaperones, and bacterial ribosome assembly factors have also been identified, including DnaK (El Hage and Alix, 2004) and S7 itself, a repressor of its own translation (Dean et al., 1981). We submit that TF may shelter the cell from aberrant complex assembly and untimely exposure of interfacial surfaces by holding assembly intermediates, like isolated S7, until productive folded integration can occur. Here, release is “triggered” not by ATP but by the free energy landscape of the substrate itself: S7 reaches its state of minimal Gibbs free energy only after it assembles into the 30S subunit; in turn, the TF:S7 complex is more stable than isolated S7 free in solution (Fig. 6E).
EXPERIMENTAL PROCEDURES
Bacterial Strains and Plasmids
Genetic analyses were carried out in E. coli MC4100 ΔtigΔdnaKdnaJ:Kanr:Cmr (Genevaux et al., 2004). For complementation, the wild-type tig gene and derivatives including the native tig promoter were cloned into the low-copy pACYC177 vector using the HindIII and XhoI restriction sites found within the Kanr cassette.
Bacterial Viability Assays
Bacterial cultures for electro-competent cells were grown from single colonies at 20°C in LB. Competent cells were electroporated with the appropriate plasmid constructs and plated at 20°C for 2 days on LB agar containing Ampicillin (100 µg/ml). Fresh colonies were grown at 20°C to an OD of approximately 1.5, serially diluted and spotted on LB agar plates at indicated temperatures.
Detection of Aggregated Proteins
Fresh colonies were grown at 20°C in LB ampicillin, diluted 1:50 in the same medium and incubated for approximately 4 hours at different temperatures. Aggregated proteins were isolated as described (Tomoyasu et al., 2001) and analyzed by SDS–PAGE.
Protein Expression and Purification
T. maritima TF and S7 were cloned into pet24d (Novagen) and produced in E. coli BL21-Codon Plus RIL. TF was purified using metal-affinity, anion-exchange after release of contaminating substrates in 8M urea at 65°C and size exclusion chromatography. Insoluble S7 was resuspended in 8 M urea and purified using cation-exchange and size exclusion chromatography.
The two-promoter pRSFDuet-1 expression vector (Novagen) was used to co-produce T. maritima TF and S7 in E. coli Rosetta cells (Novagen). Bacteria were grown in auto-induction media to an OD of 10. Cells were sonicated in 20 mM Tris–HCl (pH 8.0) and 200 mM NaCl. Lysates were centrifuged and analyzed by SDS-PAGE.
Proteome-wide Analysis of E. coli TF Complexes
Tagged and untagged ecTF were cloned into pet24d+, induced with IPTG for 10, 30 and 60 minutes and harvested by sonication and centrifugation. Cleared cell lysates were loaded onto a HiTrap Chelating column, eluted with a linear EDTA gradient. TF-containing peak fractions were further purified with Superdex 200 16/60 gel filtration. Resulting fractions were individually analyzed with LC-MS-MS. Peptides were identified with Mascot (www.matrixscience.com) and data were analyzed with ProteoIQ (BioInquire).
Analytical Ultracentrifugation
TF, S7 and the TF:S7 complex were analyzed with sedimentation velocity at 42K rpm and with sedimentation equilibrium at 10K, 12.5K, and 15K rpm and six different concentrations using a Beckman XLA analytical ultracentrifuge. Data were processed using the programs SEDFIT and SEDPHAT.
Analysis of Cell Extracts
Ribosome subunits were separated by layering 1000 µg RNA on 12 ml 5–45% sucrose gradients. Gradients were centrifuged at 41,000 rpm for 2.15 h.
Structural Analysis
The crystal lattice of tmTF404 is in space group C2221 with a = 95.39Å, b = 114.49Å, c = 94.56Å and that of tmTF:tmS7 is in space group P43212 with a = b = 94.16Å, c = 193.07Å. X-ray data were collected at 100 K at NSLS beam line X4A (Table S3). Structures of the TF:S7 complex and of apo TF were solved, respectively, by MAD phasing from the selenomethionyl proteins and by molecular replacement. Statistics for the refined structures are in Table S4.
Asphericity Calculations
Molecular surfaces and volumes of 30S proteins from recently deposited E. coli ribosome coordinates (1VS5) were calculated with PROGEOM. Asphericity was defined as the inverse of sphericity (ψ) (Wadell, 1935):
where Vp is the molecular volume and Ap the molecular surface of a protein.
Supplementary Material
Supplemental data include fifteen figures, eight tables and supplemental experimental procedures, and can be found with this article online at xxx.
ACKNOWLEDGMENTS
We are grateful to M. Floer and C.J. Lusty for comments. We thank M. A. Gawinowicz for mass spectrometry; T. Mareeva and Y. Sykulev for Biacore measurements; H. Bernstein, P. Genevaux, C. Georgoplous and F.U. Hartl for strains; P. Genevaux for anti-TF; G. Boel, R. Gonzales, J. Hunt and A. Tzagoloff for help with polysome fractionation; M. Babu, A. Emili and J. Greenblatt for TAP-tag TF data; N. Belgado, J. Cheung, M. Collins, A. Duran, Q. Fan, G. Gregorio, J. Lidestri, Q. Liu, A. Korkut, A. Marina, C. Min, J. Moore, H. Xie, R. Xu and Z. Zhang for assistance. This work was supported in part by fellowships from the Leukemia and Lymphoma Society of America (E.M.-H.) and the Jane Elissa/Charlotte Meyers Endowment Fund (E.M.-H.), and by NIH grant GM34102 (W.A.H.). Beamline X4A at the National Synchrotron Light Source (NSLS) is supported by the New York Structural Biology Center.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Accession Numbers
Atomic coordinates and diffraction data are deposited into the Protein Data Bank with accession codes 3GTY (TF:S7) and 3GU0 (apoTF).
REFERENCES
- Agashe VR, Guha S, Chang HC, Genevaux P, Hayer-Hartl M, Stemp M, Georgopoulos C, Hartl FU, Barral JM. Function of trigger factor and DnaK in multidomain protein folding: increase in yield at the expense of folding speed. Cell. 2004;117:199–209. doi: 10.1016/s0092-8674(04)00299-5. [DOI] [PubMed] [Google Scholar]
- Anfinsen CB. Principles that govern the folding of protein chains. Science. 1973;181:223–230. doi: 10.1126/science.181.4096.223. [DOI] [PubMed] [Google Scholar]
- Brodersen DE, Clemons WM, Jr, Carter AP, Wimberly BT, Ramakrishnan V. Crystal structure of the 30 S ribosomal subunit from Thermus thermophilus: structure of the proteins and their interactions with 16 S RNA. J. Mol. Biol. 2002;316:725–768. doi: 10.1006/jmbi.2001.5359. [DOI] [PubMed] [Google Scholar]
- Butland G, Peregrin-Alvarez JM, Li J, Yang W, Yang X, Canadien V, Starostine A, Richards D, Beattie B, Krogan N, et al. Interaction network containing conserved and essential protein complexes in Escherichia coli. Nature. 2005;433:531–537. doi: 10.1038/nature03239. [DOI] [PubMed] [Google Scholar]
- Clare DK, Bakkes PJ, van Heerikhuizen H, van der Vies SM, Saibil HR. Chaperonin complex with a newly folded protein encapsulated in the folding chamber. Nature. 2009;457:107–110. doi: 10.1038/nature07479. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Crooke E, Guthrie B, Lecker S, Lill R, Wickner W. ProOmpA is stabilized for membrane translocation by either purified E. coli trigger factor or canine signal recognition particle. Cell. 1988;54:1003–1011. doi: 10.1016/0092-8674(88)90115-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Crooke E, Wickner W. Trigger factor: a soluble protein that folds pro-OmpA into a membrane-assembly-competent form. Proc. Natl. Acad. Sci. U. S. A. 1987;84:5216–5220. doi: 10.1073/pnas.84.15.5216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davis FP, Sali A. PIBASE: a comprehensive database of structurally defined protein interfaces. Bioinformatics. 2005;21:1901–1907. doi: 10.1093/bioinformatics/bti277. [DOI] [PubMed] [Google Scholar]
- Dean D, Yates JL, Nomura M. Identification of ribosomal protein S7 as a repressor of translation within the str operon of E. coli. Cell. 1981;24:413–419. doi: 10.1016/0092-8674(81)90331-7. [DOI] [PubMed] [Google Scholar]
- Deuerling E, Patzelt H, Vorderwulbecke S, Rauch T, Kramer G, Schaffitzel E, Mogk A, Schulze-Specking A, Langen H, Bukau B. Trigger Factor and DnaK possess overlapping substrate pools and binding specificities. Mol. Microbiol. 2003;47:1317–1328. doi: 10.1046/j.1365-2958.2003.03370.x. [DOI] [PubMed] [Google Scholar]
- Deuerling E, Schulze-Specking A, Tomoyasu T, Mogk A, Bukau B. Trigger factor and DnaK cooperate in folding of newly synthesized proteins. Nature. 1999;400:693–696. doi: 10.1038/23301. [DOI] [PubMed] [Google Scholar]
- El Hage A, Alix JH. Authentic precursors to ribosomal subunits accumulate in Escherichia coli in the absence of functional DnaK chaperone. Mol. Microbiol. 2004;51:189–201. doi: 10.1046/j.1365-2958.2003.03813.x. [DOI] [PubMed] [Google Scholar]
- Ellis RJ. Molecular chaperones. Opening and closing the Anfinsen cage. Curr. Biol. 1994;4:633–635. doi: 10.1016/s0960-9822(00)00140-8. [DOI] [PubMed] [Google Scholar]
- Ellis RJ. Molecular chaperones: assisting assembly in addition to folding. Trends Biochem. Sci. 2006;31:395–401. doi: 10.1016/j.tibs.2006.05.001. [DOI] [PubMed] [Google Scholar]
- Fenton WA, Horwich AL. Chaperonin-mediated protein folding: fate of substrate polypeptide. Q. Rev. Biophys. 2003;36:229–256. doi: 10.1017/s0033583503003883. [DOI] [PubMed] [Google Scholar]
- Ferbitz L, Maier T, Patzelt H, Bukau B, Deuerling E, Ban N. Trigger factor in complex with the ribosome forms a molecular cradle for nascent proteins. Nature. 2004;431:590–596. doi: 10.1038/nature02899. [DOI] [PubMed] [Google Scholar]
- Fredrick K, Dunny GM, Noller HF. Tagging ribosomal protein S7 allows rapid identification of mutants defective in assembly and function of 30 S subunits. J. Mol. Biol. 2000;298:379–394. doi: 10.1006/jmbi.2000.3563. [DOI] [PubMed] [Google Scholar]
- Genevaux P, Keppel F, Schwager F, Langendijk-Genevaux PS, Hartl FU, Georgopoulos C. In vivo analysis of the overlapping functions of DnaK and trigger factor. EMBO Rep. 2004;5:195–200. doi: 10.1038/sj.embor.7400067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hartl FU, Hayer-Hartl M. Molecular chaperones in the cytosol: from nascent chain to folded protein. Science. 2002;295:1852–1858. doi: 10.1126/science.1068408. [DOI] [PubMed] [Google Scholar]
- Hesterkamp T, Hauser S, Lutcke H, Bukau B. Escherichia coli trigger factor is a prolyl isomerase that associates with nascent polypeptide chains. Proc. Natl. Acad. Sci. U. S. A. 1996;93:4437–4441. doi: 10.1073/pnas.93.9.4437. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoffmann A, Merz F, Rutkowska A, Zachmann-Brand B, Deuerling E, Bukau B. Trigger factor forms a protective shield for nascent polypeptides at the ribosome. J. Biol. Chem. 2006;281:6539–6545. doi: 10.1074/jbc.M512345200. [DOI] [PubMed] [Google Scholar]
- Huang GC, Li ZY, Zhou JM, Fischer G. Assisted folding of D-glyceraldehyde-3-phosphate dehydrogenase by trigger factor. Protein Sci. 2000;9:1254–1261. doi: 10.1110/ps.9.6.1254. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaiser CM, Chang HC, Agashe VR, Lakshmipathy SK, Etchells SA, Hayer-Hartl M, Hartl FU, Barral JM. Real-time observation of trigger factor function on translating ribosomes. Nature. 2006;444:455–460. doi: 10.1038/nature05225. [DOI] [PubMed] [Google Scholar]
- Kandror O, Goldberg AL. Trigger factor is induced upon cold shock and enhances viability of Escherichia coli at low temperatures. Proc. Natl. Acad. Sci. U. S. A. 1997;94:4978–4981. doi: 10.1073/pnas.94.10.4978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kramer G, Patzelt H, Rauch T, Kurz TA, Vorderwulbecke S, Bukau B, Deuerling E. Trigger factor peptidyl-prolyl cis/trans isomerase activity is not essential for the folding of cytosolic proteins in Escherichia coli. J. Biol. Chem. 2004a;279:14165–14170. doi: 10.1074/jbc.M313635200. [DOI] [PubMed] [Google Scholar]
- Kramer G, Rauch T, Rist W, Vorderwulbecke S, Patzelt H, Schulze-Specking A, Ban N, Deuerling E, Bukau B. L23 protein functions as a chaperone docking site on the ribosome. Nature. 2002;419:171–174. doi: 10.1038/nature01047. [DOI] [PubMed] [Google Scholar]
- Kramer G, Rutkowska A, Wegrzyn RD, Patzelt H, Kurz TA, Merz F, Rauch T, Vorderwulbecke S, Deuerling E, Bukau B. Functional dissection of Escherichia coli trigger factor: unraveling the function of individual domains. J. Bacteriol. 2004b;186:3777–3784. doi: 10.1128/JB.186.12.3777-3784.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lakshmipathy SK, Tomic S, Kaiser CM, Chang HC, Genevaux P, Georgopoulos C, Barral JM, Johnson AE, Hartl FU, Etchells SA. Identification of nascent chain interaction sites on trigger factor. J. Biol. Chem. 2007;282:12186–12193. doi: 10.1074/jbc.M609871200. [DOI] [PubMed] [Google Scholar]
- Laskey RA, Honda BM, Mills AD, Finch JT. Nucleosomes are assembled by an acidic protein which binds histones and transfers them to DNA. Nature. 1978;275:416–420. doi: 10.1038/275416a0. [DOI] [PubMed] [Google Scholar]
- Lawrence MC, Colman PM. Shape complementarity at protein/protein interfaces. J. Mol. Biol. 1993;234:946–950. doi: 10.1006/jmbi.1993.1648. [DOI] [PubMed] [Google Scholar]
- Lee HC, Bernstein HD. Trigger factor retards protein export in Escherichia coli. J. Biol. Chem. 2002;277:43527–43535. doi: 10.1074/jbc.M205950200. [DOI] [PubMed] [Google Scholar]
- Li ZY, Liu CP, Zhu LQ, Jing GZ, Zhou JM. The chaperone activity of trigger factor is distinct from its isomerase activity during co-expression with adenylate kinase in Escherichia coli. FEBS Lett. 2001;506:108–112. doi: 10.1016/s0014-5793(01)02896-4. [DOI] [PubMed] [Google Scholar]
- Lill R, Crooke E, Guthrie B, Wickner W. The "trigger factor cycle" includes ribosomes, presecretory proteins, and the plasma membrane. Cell. 1988;54:1013–1018. doi: 10.1016/0092-8674(88)90116-x. [DOI] [PubMed] [Google Scholar]
- Liu CP, Perrett S, Zhou JM. Dimeric trigger factor stably binds folding-competent intermediates and cooperates with the DnaK-DnaJ-GrpE chaperone system to allow refolding. J. Biol. Chem. 2005;280:13315–13320. doi: 10.1074/jbc.M414151200. [DOI] [PubMed] [Google Scholar]
- Liu CP, Zhou JM. Trigger factor-assisted folding of bovine carbonic anhydrase II. Biochem. Biophys. Res. Commun. 2004;313:509–515. doi: 10.1016/j.bbrc.2003.11.142. [DOI] [PubMed] [Google Scholar]
- Lo Conte L, Chothia C, Janin J. The atomic structure of protein-protein recognition sites. J. Mol. Biol. 1999;285:2177–2198. doi: 10.1006/jmbi.1998.2439. [DOI] [PubMed] [Google Scholar]
- Maier R, Scholz C, Schmid FX. Dynamic association of trigger factor with protein substrates. J. Mol. Biol. 2001;314:1181–1190. doi: 10.1006/jmbi.2000.5192. [DOI] [PubMed] [Google Scholar]
- Maier R, Eckert B, Scholz C, Lilie H, Schmid FX. Interaction of trigger factor with the ribosome. J. Mol. Biol. 2003;326:585–592. doi: 10.1016/s0022-2836(02)01427-4. [DOI] [PubMed] [Google Scholar]
- Maisonneuve E, Fraysse L, Moinier D, Dukan S. Existence of abnormal protein aggregates in healthy Escherichia coli cells. J. Bacteriol. 2008;190:887–893. doi: 10.1128/JB.01603-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martinez-Hackert E, Hendrickson WA. Structures of and interactions between domains of trigger factor from Thermotoga maritima. Acta Crystallogr. D. 2007;63:536–547. doi: 10.1107/S090744490700964X. [DOI] [PubMed] [Google Scholar]
- Merz F, Boehringer D, Schaffitzel C, Preissler S, Hoffmann A, Maier T, Rutkowska A, Lozza J, Ban N, Bukau B, Deuerling E. Molecular mechanism and structure of Trigger Factor bound to the translating ribosome. EMBO J. 2008;27:1622–1632. doi: 10.1038/emboj.2008.89. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Merz F, Hoffmann A, Rutkowska A, Zachmann-Brand B, Bukau B, Deuerling E. The C-terminal domain of Escherichia coli trigger factor represents the central module of its chaperone activity. J. Biol. Chem. 2006;281:31963–31971. doi: 10.1074/jbc.M605164200. [DOI] [PubMed] [Google Scholar]
- Neher SB, Villen J, Oakes EC, Bakalarski CE, Sauer RT, Gygi SP, Baker TA. Proteomic profiling of ClpXP substrates after DNA damage reveals extensive instability within SOS regulon. Mol. Cell. 2006;22:193–204. doi: 10.1016/j.molcel.2006.03.007. [DOI] [PubMed] [Google Scholar]
- Patzelt H, Kramer G, Rauch T, Schonfeld HJ, Bukau B, Deuerling E. Three-state equilibrium of Escherichia coli trigger factor. Biol. Chem. 2002;383:1611–1619. doi: 10.1515/BC.2002.182. [DOI] [PubMed] [Google Scholar]
- Patzelt H, Rudiger S, Brehmer D, Kramer G, Vorderwulbecke S, Schaffitzel E, Waitz A, Hesterkamp T, Dong L, Schneider-Mergener J, et al. Binding specificity of Escherichia coli trigger factor. Proc. Natl. Acad. Sci. U. S. A. 2001;98:14244–14249. doi: 10.1073/pnas.261432298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Raine A, Lovmar M, Wikberg J, Ehrenberg M. Trigger factor binding to ribosomes with nascent peptide chains of varying lengths and sequences. J. Biol. Chem. 2006;281:28033–28038. doi: 10.1074/jbc.M605753200. [DOI] [PubMed] [Google Scholar]
- Rutkowska A, Mayer MP, Hoffmann A, Merz F, Zachmann-Brand B, Schaffitzel C, Ban N, Deuerling E, Bukau B. Dynamics of trigger factor interaction with translating ribosomes. J. Biol. Chem. 2008;283:4124–4132. doi: 10.1074/jbc.M708294200. [DOI] [PubMed] [Google Scholar]
- Scholz C, Stoller G, Zarnt T, Fischer G, Schmid FX. Cooperation of enzymatic and chaperone functions of trigger factor in the catalysis of protein folding. EMBO J. 1997;16:54–58. doi: 10.1093/emboj/16.1.54. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Siegert R, Leroux MR, Scheufler C, Hartl FU, Moarefi I. Structure of the molecular chaperone prefoldin: unique interaction of multiple coiled coil tentacles with unfolded proteins. Cell. 2000;103:621–632. doi: 10.1016/s0092-8674(00)00165-3. [DOI] [PubMed] [Google Scholar]
- Stoller G, Rucknagel KP, Nierhaus KH, Schmid FX, Fischer G, Rahfeld JU. A ribosome-associated peptidyl-prolyl cis/trans isomerase identified as the trigger factor. EMBO J. 1995;14:4939–4948. doi: 10.1002/j.1460-2075.1995.tb00177.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takagi F, Koga N, Takada S. How protein thermodynamics and folding mechanisms are altered by the chaperonin cage: molecular simulations. Proc. Natl. Acad. Sci. U. S. A. 2003;100:11367–11372. doi: 10.1073/pnas.1831920100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Talkington MW, Siuzdak G, Williamson JR. An assembly landscape for the 30S ribosomal subunit. Nature. 2005;438:628–632. doi: 10.1038/nature04261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tang YC, Chang HC, Roeben A, Wischnewski D, Wischnewski N, Kerner MJ, Hartl FU, Hayer-Hartl M. Structural features of the GroEL-GroES nano-cage required for rapid folding of encapsulated protein. Cell. 2006;125:903–914. doi: 10.1016/j.cell.2006.04.027. [DOI] [PubMed] [Google Scholar]
- Teter SA, Houry WA, Ang D, Tradler T, Rockabrand D, Fischer G, Blum P, Georgopoulos C, Hartl FU. Polypeptide flux through bacterial Hsp70: DnaK cooperates with trigger factor in chaperoning nascent chains. Cell. 1999;97:755–765. doi: 10.1016/s0092-8674(00)80787-4. [DOI] [PubMed] [Google Scholar]
- Tomic S, Johnson AE, Hartl FU, Etchells SA. Exploring the capacity of trigger factor to function as a shield for ribosome bound polypeptide chains. FEBS Lett. 2006;580:72–76. doi: 10.1016/j.febslet.2005.11.050. [DOI] [PubMed] [Google Scholar]
- Tomoyasu T, Mogk A, Langen H, Goloubinoff P, Bukau B. Genetic dissection of the roles of chaperones and proteases in protein folding and degradation in the Escherichia coli cytosol. Mol. Microbiol. 2001;40:397–413. doi: 10.1046/j.1365-2958.2001.02383.x. [DOI] [PubMed] [Google Scholar]
- Valent QA, Kendall DA, High S, Kusters R, Oudega B, Luirink J. Early events in preprotein recognition in E. coli: interaction of SRP and trigger factor with nascent polypeptides. EMBO J. 1995;14:5494–5505. doi: 10.1002/j.1460-2075.1995.tb00236.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wadell H. Volume, Shape and Roundness of Quartz Particles. J. Geol. 1935;43:250–280. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplemental data include fifteen figures, eight tables and supplemental experimental procedures, and can be found with this article online at xxx.