Abstract
We identified an Arabidopsis (Arabidopsis thaliana) ethyl methanesulfonate mutant, modified vacuole phenotype1-1 (mvp1-1), in a fluorescent confocal microscopy screen for plants with mislocalization of a green fluorescent protein-δ tonoplast intrinsic protein fusion. The mvp1-1 mutant displayed static perinuclear aggregates of the reporter protein. mvp1 mutants also exhibited a number of vacuole-related phenotypes, as demonstrated by defects in growth, utilization of stored carbon, gravitropic response, salt sensitivity, and specific susceptibility to the fungal necrotroph Alternaria brassicicola. Similarly, crosses with other endomembrane marker fusions identified mislocalization to aggregate structures, indicating a general defect in protein trafficking. Map-based cloning showed that the mvp1-1 mutation altered a gene encoding a putative myrosinase-associated protein, and glutathione S-transferase pull-down assays demonstrated that MVP1 interacted specifically with the Arabidopsis myrosinase protein, THIOGLUCOSIDE GLUCOHYDROLASE2 (TGG2), but not TGG1. Moreover, the mvp1-1 mutant showed increased nitrile production during glucosinolate hydrolysis, suggesting that MVP1 may play a role in modulation of myrosinase activity. We propose that MVP1 is a myrosinase-associated protein that functions, in part, to correctly localize the myrosinase TGG2 and prevent inappropriate glucosinolate hydrolysis that could generate cytotoxic molecules.
The plant endomembrane system is a complex network of subcellular compartments that includes the endoplasmic reticulum (ER), Golgi apparatus, vacuole, plasma membrane, secretory vesicles, and numerous intermediary compartments. Protein trafficking through the endomembrane system requires specific cargo recognition and delivery mechanisms that are mediated by a series of highly specific targeting signals (Surpin and Raikhel, 2004), whose proper recognition is critical for the function of numerous downstream processes, such as floral development (Sohn et al., 2007), gravitropism (Kato et al., 2002; Surpin et al., 2003; Yano et al., 2003), abiotic stress tolerance (Zhu et al., 2002), autophagy (Surpin et al., 2003; Bassham., 2007), pathogen defense (Robatzek, 2007), and turgor pressure and growth (De, 2000).
The importance of protein trafficking for plant survival was demonstrated by the identification of the essential Arabidopsis (Arabidopsis thaliana) gene VACUOLELESS1 (VCL1; Rojo et al., 2001). VCL1 was identified as a homolog of Saccharomyces cerevisiae VPS16, which is critical for yeast vacuole biogenesis. Knockouts of yeast VPS16 lack discernible vacuoles but survive despite their severe phenotype. The absence of vacuoles in Arabidopsis vcl1-1 mutants results in embryo lethality (Rojo et al., 2001). The essential nature of trafficking in plants was also demonstrated by insertional mutagenesis of syntaxin genes, where lethality was observed after disruption of single genes in families with highly homologous members (Lukowitz et al., 1996; Sanderfoot et al., 2001). Thus, despite large families of endomembrane components with many homologous genes, many are not redundant in Arabidopsis.
Although embryo-lethal mutations provide critical data, it is difficult to obtain additional information. Less severe mutations have proven successful for functional genetics studies of endomembrane trafficking proteins. For example, point mutations in the KATAMARI1/MURUS3 (KAM1/MUR3; Tamura et al., 2005) and KATAMARI2/GRAVITROPISM DEFECTIVE2 (KAM2/GRV2; Tamura et al., 2007; Silady et al., 2008) genes lead to disruption of endomembranes, resulting in the formation of perinuclear aggregates containing organelles. Nonlethal trafficking disruptions have also been generated using chemical genomics, where small molecules were used to perturb trafficking of a soluble cargo protein (Zouhar et al., 2004) and localization of endomembrane markers (Surpin et al., 2005; Robert et al., 2008). Such studies have provided valuable clues about these essential cellular processes.
In order to obtain less severe, viable mutants with defects in endomembrane protein trafficking, we previously identified point mutants with defects in localization of a tonoplast reporter protein, GFP:δ-TIP (Avila et al., 2003). Two hundred one putative mutants were grouped into four categories based on the nature of their defects. One unique mutant, cell shape phenotype1, was recently characterized as a trehalose-6-phosphate synthase with roles in regulation of plant architecture, epidermal pavement cell shape, and trichome branching (Chary et al., 2008).
Here, we describe an endomembrane trafficking mutant categorized by perinuclear aggregates of GFP:δ-TIP fluorescence (Avila et al., 2003). We refer to this mutant as modified vacuole phenotype1-1 (mvp1-1). At least five endomembrane fusion proteins are partially relocalized to these structures. Positional cloning identified MVP1 as a myrosinase-associated protein (MyAP) localized previously to the tonoplast by proteomics (Carter et al., 2004). mvp1-1 mutants showed reduced endomembrane system functionality, as demonstrated by defects in growth, utilization of stored carbon, gravitropic responsiveness, salt sensitivity, and increased susceptibility to a fungal necrotroph. MVP1 interacted specifically with THIOGLUCOSIDE GLUCOHYDROLASE2 (TGG2), a known myrosinase protein in Arabidopsis, and the mvp1-1 mutation had a significant effect on nitrile production during glucosinolate hydrolysis, suggesting a role in myrosinase function. Furthermore, MVP1 may function in quality control of glucosinolate hydrolysis by contributing to the proper tonoplast localization of TGG2.
RESULTS
Endomembrane System Functionality and Protein Targeting Are Altered in mvp1-1
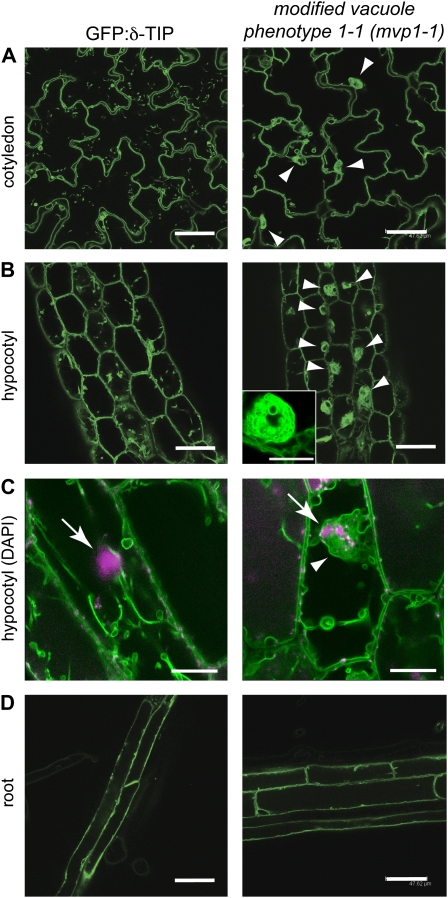
The mvp1-1 mutant was isolated via visually detectable defects in vacuolar protein targeting. mvp1-1 was previously categorized within the aggregates of GFP fluorescence class of mutants, based on static subcellular GFP:δ-TIP-labeled membrane structures. In mvp1-1, aggregates were perinuclear and restricted to aerial tissues (Fig. 1; Avila et al., 2003).
Figure 1.
The mvp1-1 mutant has perinuclear aggregates of GFP:δ-TIP fluorescence in aerial tissues. Confocal images of 7-d-old seedlings are shown. A, In cotyledons, parental plants and mvp1-1 mutants show GFP:δ-TIP fluorescence in the tonoplast, but mutants also accumulate the fusion protein in static aggregates (arrowheads). B, Similar aggregate structures are visible in hypocotyl cells (arrowheads), shown in detail in the inset. C, An overlay of the DNA-specific fluorescent dye 4′,6-diamidino-2-phenylindole (DAPI; magenta; arrows) and GFP:δ-TIP (green; arrowhead) shows aggregates around the nucleus in mvp1-1. D, Aggregate structures were not detected in root tissues in mvp1-1. Bars = 50 μm in A, B, and D and 15 μm in C.
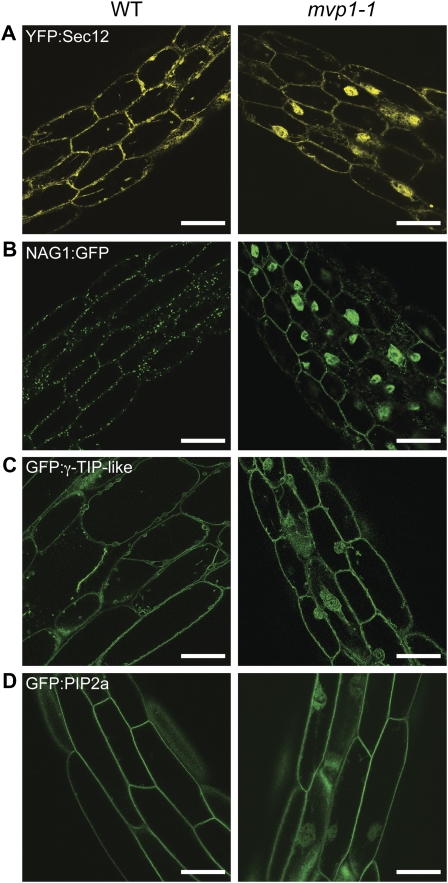
To assess the effects of mvp1-1 on trafficking of other endomembrane markers, it was outcrossed to lines expressing different reporters. Yellow fluorescent protein (YFP):SEC12, an ER marker (Barlowe and Schekman, 1993; G. Drakakaki and N.V. Raikhel, unpublished data) was observed as a web-like network at the periphery of wild-type cells (Fig. 2A), whereas in mvp1-1, YFP:SEC12 was delivered to the ER but the perinuclear aggregates were also visible (Fig. 2A).
Figure 2.
The mvp1-1 mutation disrupts protein targeting to the ER, Golgi, vacuole, and plasma membrane. Confocal images of 7-d-old hypocotyl tissues show that the mvp1-1 mutation leads to partial aggregation of endomembrane fusion proteins. A, YFP:Sec12 in the ER. B, Golgi fusion NAG1:GFP. C, Tonoplast protein GFP:γ-TIP-like. D, Plasma membrane fusion GFP:PIP2a. WT, Wild type. Bars = 50 μm.
NAG1:GFP, a Golgi marker (Burke et al., 1992; Grebe et al., 2003), labeled punctate structures located around the cell periphery (Fig. 2B), whereas in mvp1-1, the marker was also found in the aggregates (Fig. 2B). The tonoplast-localized GFP:γ-TIP-like protein (Cutler et al., 2000) mislocalized to the cellular aggregates in mvp1-1, indicating mistargeting of other tonoplast proteins (Fig. 2C). Plasma membrane intrinsic protein 2A (PIP2A; Cutler et al., 2000; Quigley et al., 2002) localized to the plasma membrane in wild-type and mutant plants but accumulated in mvp1-1 aggregates (Fig. 2D).
We considered the possibility that aggregate formation was caused, in part, by overexpression of fluorescent protein markers, so we examined both mvp1-1 and mvp1-2 (described below) seedlings containing no 35Spro-driven constructs in their genetic backgrounds. Seedlings were stained with the endocytic styryl dye FM4-64 and examined by confocal microscopy (Supplemental Fig. S1). We observed aggregates in both alleles, indicating that aggregate formation was due solely to the mutation in the MVP1 gene.
These data indicate that mvp1-1 widely disrupts endomembrane trafficking and causes a block early in trafficking pathways. Because the plant endomembrane system is essential for an array of physiological pathways, we assessed endomembrane functionality in mvp1-1 by evaluating the endomembrane-associated processes such as growth, gravitropism, carbon utilization, abiotic stress responses, and pathogen defense. All of these processes were negatively affected in the mvp1-1 mutant (Supplemental Fig. S2).
MVP1 Maps to a Gene Encoding a Putative Member of the MyAP Family
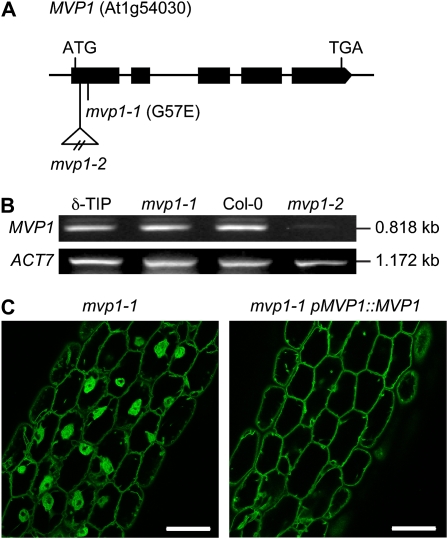
Rough mapping using DNA isolated from F2 progeny of an outcross to the Landsberg erecta accession placed the mvp1-1 mutation between markers ciw1 and nga280 on chromosome 1. This interval was approximately 2,500 kb, or 12.5 centimorgan (Supplemental Fig. S4). We narrowed this interval to 110 kb (0.4 centimorgan) using a fine-mapping population composed of 493 F2 mutant plants. Targeted sequencing of 24 genes in the interval identified a 1-bp change (G→A) in the first exon of At1g54030, whose annotation was based on similarity to oilseed rape (Brassica napus) MyAPs with a GDSL lipase motif. This change in the MVP1 gene resulted in a missense mutation that converted a predicted Gly to a Glu (Fig. 3A). We isolated a second allele, mvp1-2, that had a T-DNA insertion in the first exon of MVP1 (Fig. 3A). This allele had phenotypic defects similar to mvp1-1, including subcellular aggregates as visualized by FM4-64 (Supplemental Fig. S1) and sensitivity to NaCl (Supplemental Fig. S3). The mvp1-1 point mutation did not reduce transcript levels relative to the wild type, but the mvp1-2 insertion allele showed significantly reduced transcript (Fig. 3B). The mvp1-1 line was successfully complemented with a construct that consisted of the native MVP1 gene and approximately 2.0 kb of sequence upstream of the translation start site (Fig. 3C). The combination of complementation results and two independent alleles having similar phenotypes confirmed that At1g54030 is MVP1.
Figure 3.
Identity of MVP1. A, A schematic drawing of MVP1 shows the mvp1-1 point mutation in the first exon of At1g54030 and a second insertional allele, mvp1-2, also in the first exon. B, RT-PCR analysis of MVP1 and ACTIN7 control transcript levels in parental GFP:δ-TIP (δ-TIP), mvp1-1, wild-type Col-0, and mvp1-2. C, Expression of the wild-type MVP1 gene under control of its native promoter in mvp1-1 mutant plants restores normal vacuolar architecture, as shown here in hypocotyls. Bars = 50 μm.
The MVP1 gene has a 1.97-kb genomic sequence and a 1.25-kb coding sequence, with five exons and four introns. The full-length cDNA (1.43 kb) encodes a 417-amino acid protein with a molecular mass of approximately 46 kD and pI of 6.7. Analysis using the SignalP 3.0 prediction server (Emanuelsson et al., 2007) indicated that MVP1 had a signal peptide from residues 27 to 41 (S score > 0.6, with an 89.7% probability) that facilitates entry into the ER and was cleaved between residues 50 and 51 to yield a protein of approximately 40.5 kD (Supplemental Fig. S5).
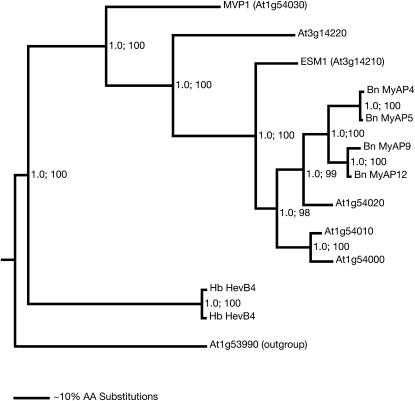
Sequence similarity searches showed that MVP1 was a member of a large GDSL lipase protein family. This family consisted of 907 members from eight different fully sequenced plant genomes, including 118 members from Arabidopsis, and all organisms represented in the UniProt database. Within this superfamily, the sequences with closest identity to MVP1 formed a distinct subclade of putative MyAPs, based on sequence similarities to glycoproteins found in myrosinase complexes of oilseed rape (Falk et al., 1995; Taipalensuu et al., 1996; Fig. 4; Supplemental Fig. S6; Supplemental Table S4). Myrosinases are β-glucosidases that hydrolyze glucosinolates, which are sulfur- and nitrogen-containing secondary metabolites. The glucosinolate-myrosinase system is found almost exclusively in the order Capparales, which includes the Brassicaceae plants oilseed rape and Arabidopsis. Upon tissue disruption, glucosinolates are hydrolyzed by myrosinases. The hydrolysis yields an aglucone intermediate that either spontaneously rearranges into an isothiocyanate or, combined with the actions of specifier and modifier proteins, into epithionitriles, nitriles, or thiocyanates. Arabidopsis contains six myrosinase genes, TGG1 to TGG6. TGG1 and TGG2 are expressed in aerial tissue, albeit with somewhat different cell type expression: TGG1 is found in phloem-associated and stomatal guard cells, whereas TGG2 is found only in phloem-associated cells. The functions of the two proteins appear to be redundant, because glucosinolate breakdown in single null mutants was similar to that in the wild type (Barth and Jander, 2006).
Figure 4.
Phylogram of the nearest neighbors of MVP1. Protein sequence similarity searches with BLASTP were used to assemble the GDSL superfamily. The identified 907 members were organized in a large neighbor-joining tree. MVP1 mapped in this tree to a subclade with 12 members sharing amino acid sequence identities of greater than 40%. These 12 sequences plus one more distant sequence (less than 35% identity) were used to generate the phylogram shown here with the MrBayes software. The distant sequence (At1g53990) served as an outgroup for rooting the tree. The posterior probabilities, generated by the Bayesian analysis, are given by the first value at each branch point. The same sequences were used for a neighbor-joining analysis with the PHYLIP software. This analysis resulted in a tree with an identical branching pattern. The relative bootstrap values derived from 1,000 resampling steps in the neighbor-joining analysis are given as percentages by the second value at each branch point. In the given tree, the most closely related members are five sequences from Arabidopsis and four from oilseed rape (Bn). A rubber tree (Hevea brasiliensis) homolog is more distantly related (Hb). A relative distance scale is given at the bottom. AA, Amino acids.
The clade that includes MVP1 also contained four sequences for oilseed rape MyAPs and six additional Arabidopsis genes (Fig. 4), only one of which, EPITHIOSPECIFIER MODIFIER1 (ESM1), has been characterized. The gene was identified as part of a quantitative trait locus on chromosome 3 that, in combination with the EPITHIOSPECIFIER PROTEIN (ESP) gene, affected the nitrile-to-isothiocyanate ratio in glucosinolate metabolism (Lambrix et al., 2001; Zhang et al., 2006).
The MVP1 expression pattern was examined using the Genevestigator reference expression database and analysis tool (Hruz et al., 2008). The MVP1 transcript was found throughout Arabidopsis tissues (Supplemental Fig. S7), with highest expression in imbibed seed. These data were consistent with our observation of unusual punctate structures visible in mature mvp1 embryos before the formation of the central vacuole (Supplemental Fig. S8). MVP1 was also expressed throughout the seedling, rosette leaves, and inflorescence, but not in pollen. Interestingly, MVP1 was expressed in roots, where we did not observe any aggregate formation. MVP1 transcript was detected at moderate levels during all developmental stages of the Arabidopsis life cycle, with highest mRNA accumulation in germinated seeds (Supplemental Fig. S9). We also carried out reverse transcription (RT)-PCR for the MVP1, TGG2, and ACTIN7 genes using total RNA isolated from 2-week-old seedlings, 4-week-old roots and rosette leaves, inflorescence stems, flowers, and green siliques (Supplemental Fig. S10; Supplemental Table S3). The data from this experiment were consistent with that reported in the Genevestigator database.
Our laboratory has previously identified MVP1 in the vacuole proteome, specifically in the tonoplast fraction (Carter et al., 2004), and a variety of observations suggest that MVP1 may also function in plant defense. First, as discussed above, ESM1 has been shown to be involved in defense against insect herbivory (Lambrix et al., 2001; Zhang et al., 2006), indicating a possible role in defense for this family. Second, microarray data show that MVP1 expression was up-regulated by both pathogen infestation and methyl jasmonate (Supplemental Fig. S11), a well-characterized plant defense hormone (Cheong and Choi, 2003). MVP1 transcript accumulation was also induced by the bacterial pathogens Agrobacterium tumefaciens and Pseudomonas syringae, the fungal necrotrophs Botrytis cinerea and Alternaria brassicicola, and the oomycete Phytophthora infestans (Supplemental Fig. S11).
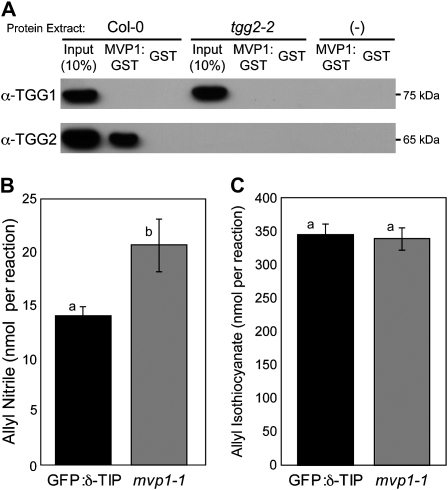
MVP1 Specifically Associates with the Myrosinase Protein TGG2 and Modulates Glucosinolate Hydrolysis
Specifier proteins such as ESM1 do not have direct hydrolytic activity on glucosinolates, but they do influence the type of glucosinolate hydrolysis product formed (Wittstock and Burow, 2007). To elucidate the molecular function of MVP1, we tested its annotation as a putative MyAP by assessing the ability of MVP1:glutathione S-transferase (GST) recombinant protein to interact with TGG1 and TGG2. In a pull-down experiment using crude Arabidopsis extracts, we detected a specific interaction between MVP1:GST and TGG2 that was absent in the tgg2-2 knockout mutant (Fig. 5A). Furthermore, we did not detect a similar interaction between MVP1:GST and TGG1, even in the tgg2-2 mutant, suggesting that MVP1 interacted with TGG2 in a highly specific manner. We also tested whether MVP1 interacted with other known defense-related β-glucosidase enzymes. Neither PYK10 (Nagano et al., 2008) nor PEN2 (Lipka et al., 2005; Bednarek et al., 2009; Clay et al., 2009) interacted with MVP1 in GST pull-down assays (data not shown). We examined the microarray databases to determine the coexpression patterns of MVP1 and TGG2. MVP1 and TGG2 are coexpressed in aerial tissues, where we observed the aggregate formation phenotype (Supplemental Figs. S7 and S9). There was no detectable expression of TGG2 in roots, where we did not observe aggregates. Queries of the developmental expression patterns of the two genes revealed that both were expressed throughout the plant life cycle (Supplemental Figs. S7, S9, and S10).
Figure 5.
MVP1:GST associates with the myrosinase protein TGG2, and mvp1-1 impacts glucosinolate hydrolysis. A, Crude extracts from Col-0 or tgg2-2 leaves were incubated with MVP1:GST or GST glutathione-Sepharose beads. Eluted proteins were subjected to immunoblot analysis with anti-TGG1 and TGG2 antibodies. B and C, Gas chromatography was used to analyze hydrolysis products of allyl glucosinolate substrate by 4-week-old rosette tissue extracts. mvp1-1 mutants show significantly increased allyl nitrile production (B) but not isothiocyanate production (C). Six biological replicates of each genotype were used for this analysis. Letters indicate statistically similar or different groupings as determined by Student's t test. Error bars represent se.
We next evaluated the capacity of mvp1-1 to hydrolyze glucosinolates. ESM1 was previously characterized by its ability to alter hydrolysis of glucosinolate such that an esm1 mutant showed increased nitrile production (Zhang et al., 2006). We performed similar experiments to assess the effect of the mvp1-1 mutation on glucosinolate hydrolysis. Extracts from rosette leaves of 4-week-old GFP:δ-TIP and mvp1-1 plants were incubated with allyl glucosinolate substrate and analyzed by gas chromatography for the presence of nitrile and isothiocyanate breakdown products. A natural knockout polymorphism in ESP leads to a lack of epithionitrile specifier activity and hence epithionitriles within the Columbia-0 (Col-0) ecotype (Zhang et al., 2006; Burow et al., 2009). However, the mvp1-1 mutation caused an increase in the amount of nitrile formed (Fig. 5B). There was no statistically significant difference in isothiocyanate production (P = 0.53; Fig. 5C). These changes in glucosinolate hydrolysis were statistically significant but not as severe as in the esm1 knockout mutant, which increases nitrile production to almost 10% and leads to a dramatic decrease in the production of isothiocyanate (Zhang et al., 2006).
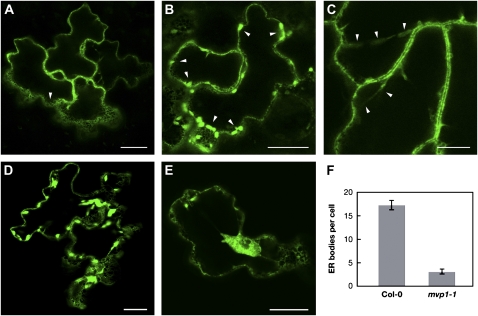
MVP1:GFP Localizes to the ER, ER Bodies, and Tonoplast
We previously identified MVP1 in the tonoplast (Carter et al., 2004). In order to determine whether there are additional sites of localization, we transiently expressed MVP1:GFP in Arabidopsis seedlings (Marion et al., 2008). MVP1 showed an ER-like pattern with a characteristic network of GFP around the cell periphery (Fig. 6A), dotted with spindle-shaped structures that appeared to be ER bodies (Fig. 6B; Matsushima et al., 2003). We also observed GFP fluorescence in transvacuolar strands (Fig. 6C), confirming that MVP1 was localized to the tonoplast.
Figure 6.
MVP1:GFP is found in the ER, ER bodies, and tonoplast, and the mvp1 mutation results in decreased numbers of ER bodies per cotyledon epidermal cell. A, MVP1:GFP localization shows a characteristic ER net-like pattern (arrowhead). Bar = 20 μm. B, Elongated spindle-shaped ER body structures (arrowheads). Bar = 20 μm. C, Transvacuolar strands (arrowheads). Bar = 10 μm. D and E, Col-0 and mvp1-1 seedlings were transiently transformed with a GFP:HDEL binary construct and examined via confocal microscopy. Bars = 20 μm. F, Number of ER bodies per cell. Col-0 = 17.3 ± 1.00 (n = 40 cells); mvp1-1 = 3.14 ± 0.522 (n = 29 cells). Error bars represent se.
ER bodies are ER-derived organelles present in both constitutive and induced forms, where the induced forms are present in wounded tissues (Ogasawara et al., 2009). Electron microscopy has shown that the bodies are localized to the ER lumen (Hawes et al., 2001). Furthermore, the MVP1 gene (referred to as GLL25 in that study) was found to be down-regulated in the ER body formation mutant nai1 (Nagano et al., 2008). Because there was strong localization of MVP1:GFP to the ER bodies, we examined whether there was a connection between MVP1 and ER body formation. A fusion of GFP to the ER retention signal HDEL is known to label ER bodies in stable Arabidopsis transformants (Matsushima et al., 2003). Here, we used transient transformation of Arabidopsis seedlings with the same construct and found a similar pattern in wild-type plants (Fig. 6D). However, mvp1-1 accumulated GFP signal in aggregates and reduced numbers of ER bodies in epidermal cells (Fig. 6, E and F), indicating that MVP1 may play a role in ER body formation.
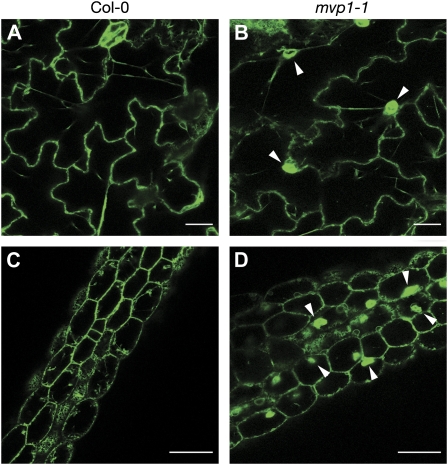
TGG2 Trafficking and Localization Are Altered in mvp1 Mutants
After demonstrating that MVP1 specifically interacts with the myrosinase protein TGG2, we sought to evaluate trafficking of TGG2 in the mvp1 mutant. A TGG2:GFP binary construct was stably transformed into mvp1-1 plants. Resistant lines were isolated, and progeny were examined by confocal microscopy. The majority of the fluorescent reporter was localized to aggregates, with the remainder localized to the ER and, occasionally, to ER bodies (Fig. 7), demonstrating that TGG2 trafficking is impeded in the mvp1-1 mutant. We also examined TGG1 in the mvp1 background. We created a TGG1:GFP binary construct and transiently transformed this construct and the TGG2:GFP binary construct used for stable transformation into Col-0, mvp1-1, and mvp1-2 seedlings. Confocal microscopy of transformed seedlings revealed that TGG2:GFP localization in wild-type and mutant seedlings was consistent with that observed for stably transformed seedlings, and the marker was localized primarily to the aggregates. The localization of TGG1:GFP was identical to that of TGG2:GFP in wild-type and mutant backgrounds (Supplemental Fig. S12). These results reinforce our hypothesis that the mvp1 mutations cause global trafficking blocks early in the endomembrane trafficking system.
Figure 7.
The myrosinase TGG2 has a similar localization as MVP1 and is mislocalized to the aggregates in mvp1-1 mutants. TGG2:GFP is found in the ER, ER bodies, and transvacuolar strands in epidermal cells of stably transformed wild-type Arabidopsis seedlings. TGG2:GFP additionally localizes to aggregates in mvp1-1. A and B, Epidermal cells of cotyledons. Bars = 20 μm. C and D, Epidermal cells of hypocotyls. Arrowheads point to aggregates. Bars = 50 μm.
Given the association of MVP1 and TGG2, and the effects of the mvp1-1 mutation on nitrile levels, we asked if there were trafficking defects in other known glucosinolate metabolism mutants. Using FM4-64, we showed that neither esm1 nor esp knockout lines formed aggregate structures as observed in the mvp1-1 alleles (Supplemental Fig. S1). We did not observe any other obvious malformations in membrane structures of the esm1 and esp mutants. We also tested the gravitropic responses of the esm1 and esp mutants and did not detect any differences compared with the wild-type response (data not shown), suggesting that neither of these proteins has trafficking functions.
MVP1 Is Not a Lipase
Although MVP1 was annotated as a lipase, its putative lipolytic sequence was divergent from the canonical GDSL motif, which in wild-type Col-0 plants is GDGL. The modified catalytic sequence raised questions about whether MVP1 was actually a lipase. We fused both wild-type and mutant proteins to GST tags to facilitate purification after expression in Escherichia coli. Recombinant MVP1 protein was isolated and purified along with the secreted GDSL lipase protein GLIP1 (Oh et al., 2005), which was used as a positive control. The glip1 mutant protein and GST were also expressed and used as negative controls (Supplemental Fig. S13A). Hydrolysis of the substrate p-nitrophenyl acetate was measured over time to determine lipase activity. GLIP1 hydrolyzed the substrate in a linear fashion over time, whereas the glip1 mutant protein was deficient in esterase activity. Although we used a 5-fold higher amount of MVP1 recombinant protein compared with the positive control (5 versus 1 μg), MVP1 did not hydrolyze p-nitrophenyl acetate (Supplemental Fig. S13B). The mvp1-1 mutant protein showed a similar lack of activity. We concluded that MVP1 did not act as an esterase for this particular substrate in vitro, and the mvp1-1 mutation had no effect on this lack of activity.
The assay described above tested activity on a short-chain acyl substrate (esterase assay); thus, we also conducted plate assays to determine whether MVP1 could hydrolyze different lipid substrates. BL21 (DE3) E. coli carrying various expression constructs was spotted on the plates containing one of three substrates: tributyrin (for esterase activity), Tween 20 (lipase activity), and Tween 80 (lipase activity). All plates were spotted with E. coli cells expressing MVP1:GST, mvp1-1:GST, or an empty vector; E. coli expressing a GDSL lipase from the tropical tree Jacaranda mimosifolia was used as a positive control (Kram et al., 2008). Both the MVP1 and mvp1-1 constructs were negative for the formation of halos and precipitate, further demonstrating that MVP1 does not act as a lipase or esterase (data not shown).
DISCUSSION
Trafficking and Physiological Defects in mvp1
In an effort to identify unknown components of the cellular protein trafficking machinery, we initiated an ethyl methanesulfonate mutagenesis screen using a seed population carrying a GFP:δ-TIP fusion (Avila et al., 2003) and screened for lines that mislocalized the reporter protein. In this study, we describe the cloning and characterization of one of these mutant genes, mvp1.
The mvp1 mutant exhibited a single perinuclear aggregate per cell in aerial tissues. We did not observe any mislocalization of endomembrane system markers in root tissue. This is consistent with our previous observation that the root and shoot endomembrane systems are uncoupled and that vacuolar biogenesis has tissue-specific components (Avila et al., 2003). Interestingly, although this defect was observed only in aerial tissues of Arabidopsis seedlings, RT-PCR and microarray data indicated that the MVP1 gene was actively transcribed in roots. There were, however, other root-associated phenotypes: roots from mutant plants were shorter than their wild-type counterparts and more sensitive to NaCl. It is possible that while MVP1 is expressed in roots and does appear to have some functions in that tissue, they are not the same as those in aerial tissues. Alternatively, MVP1 may have similar functions in both root and aerial tissues, but the different tissue phenotypes may arise from the fact that TGG2, which may be dependent upon MVP1 for proper subcellular localization, is not present in root tissue; thus, MVP1 may have different binding partners that do not cause an aggregation phenotype.
Although mvp1-1 was isolated based on defects in targeting of GFP:δ-TIP, studies with additional endomembrane system markers indicated a global disruption of the endomembrane system, including the ER, Golgi, and plasma membrane. Altered targeting of the ER protein fusion YFP:Sec12 demonstrated a role for MVP1 early in the endomembrane system, which may have caused a backup of proteins destined for other compartments into subcellular aggregates. Because the ER is the first of several sequential steps that eventually lead to the vacuole and plasma membrane, it is likely that the ER disruption caused downstream defects in targeting to other compartments. We also observed unusual punctate structures and an altered pattern of GFP fluorescence in mature Arabidopsis embryos; furthermore, the perinuclear aggregates were formed as soon as 2 d after germination. This gives further support to the importance of MVP1 in organization of the endomembrane system.
Phenotypic observations of reduced growth, altered gravitropic response, poor utilization of stored carbon, decreased salt tolerance, and increased pathogen sensitivity provided further evidence that endomembrane trafficking and vacuole functionality were severely impacted by mvp1-1. While it is possible that MVP1 may play a direct role in any one of these processes, it seems more likely that the observed endomembrane defects lead to structural defects or to altered targeting of endomembrane proteins such as pumps or transporters that are required for vacuole functionality. We also noted a specific sensitivity of mvp1-1 mutants to the fungal necrotroph A. brassicicola. Interestingly, the defect that leads to susceptibility to A. brassicicola does not impair the mutant's response to the oomycete Hyaloperonospora parasitica or the bacterium P. syringae, despite the fact that MVP1 is induced by these pathogens. The mechanism of MVP1-mediated defense against A. brassicicola may thus provide some interesting insight into both the trafficking pathway mediated by MVP1 and the defense molecules that are specific for this pathogen.
The multiorganelle aggregates in mvp1-1 were similar to structures observed in two katamari mutants, kam1/mur3 (Tamura et al., 2005) and kam2/grv2 (Tamura et al., 2007; Silady et al., 2008). The kam mutants accumulate aggregates of a vacuolar protein fusion, GFP:2SC, in a manner similar to mvp1. KAM2/GRV2 encodes a homolog of the animal protein RME-8, which is involved in endocytic trafficking from the plasma membrane. It localizes to punctate structures that do not colocalize with any of the Golgi-, trans-Golgi network-, endosome, or prevauoloar compartment-specific fluorescent markers that were examined. The kam1/mur3 mutants also feature perinuclear aggregates, but the wild-type protein is localized to the Golgi. Therefore, although phenotypically similar, these proteins had no sequence identity to MVP1 or other glucosinolate-related genes, indicating that perinuclear aggregate formation could be the result of several distinct disruptions of the endomembrane system. It may be possible that the lesions in all these genes prevent interaction with other trafficking proteins, thus causing accumulation at the site of the failed interaction.
Cloning and Identification of MVP1 as a MyAP
Positional cloning experiments showed that the mutation that causes the mvp1 phenotype was in At1g54030. At1g54030 was annotated as a MyAP/GDSL lipase and is part of a large family of plant lipases. Significantly, it was most closely related to MyAPs from both Arabidopsis and oilseed rape.
It has been shown that some myrosinases can form complexes with two types of proteins, myrosinase-binding proteins (MBPs) and MyAPs. The first characterized MyAP was described as a 40-kD glycoprotein that copurified with the myrosinase complex from oilseed rape seeds (Falk et al., 1995). It is not known whether these complexes are present in the plant or form upon tissue damage. The Arabidopsis annotations are based on sequence identity to the oilseed rape MyAPs and not on experimental data. The only MyAP characterized in Arabidopsis is ESM1. ESM1 is found in the same clade as MVP1, and together with MVP1 and five other Arabidopsis and four oilseed rape proteins, it forms a vacuole-localized clade. It has not yet been determined whether ESM1 directly interacts with myrosinase or ESP. The MyAPs feature GDSL-like lipase motifs, but no lipase activity has yet been definitively established for any of the MyAPs. A number of proteomic studies have detected ESM1 in the vacuole (Carter et al., 2004; Shimaoka et al., 2004; Huttlin et al., 2007). Five other putative MyAPs were detected in vacuoles (Carter et al., 2004). Three of these were also identified in a study of the Arabidopsis tonoplast (Shimaoka et al., 2004). In another proteomics study, Huttlin et al. (2007) identified peptides for ESM1 in the organellar fraction but other putative MyAPs in both the organellar and microsomal fractions.
MVP1 specifically interacted with TGG2 and not TGG1. Given this finding, we examined glucosinolate hydrolysis capability of the mvp1-1 mutant. Extracts of mvp1-1 rosette leaves incubated with an allyl glucosinolate substrate had a significant increase in nitrile output. MVP1 and its closest Arabidopsis relative, ESM1, share 41% protein sequence identity and have been shown here to have similar effects on hydrolysis of allyl glucosinolate in vitro; however, it is unclear whether this activity serves as the sole cellular function for either protein. ESM1 is the most abundant protein in the Arabidopsis (Col-0 accession) vacuole (Carter et al., 2004); the difference in abundance of ESM1 and MVP1 may account for the disparity between their respective phenotypes. Furthermore, the increase in nitrile output may be due primarily to TGG1 activity; it may be that in the absence of MVP1, there is more substrate available for TGG1. It is also worth noting that we examined the esm1 and esp knockout mutants for trafficking defects to determine whether it plays a role in the endomembrane system similar to mvp1-1. The esm1 and esp mutants displayed no such differences, and gravitropism assays also did not uncover any divergence from the wild-type response. These data indicate that neither of these proteins has trafficking-related functions. Thus, while MVP1 and ESM1 both alter glucosinolate hydrolysis away from nitrile formation, they differ in their effects on the endomembrane system. These divergent functions may be explained by the presence of an intact GDSL motif in ESM1 that is altered in MVP1. It could follow that the observed glucosinolate hydrolysis changes in mvp1 may be a downstream result of the primary trafficking defect.
MVP1 is part of a larger protein family classified as GDSL lipases. The wild-type MVP1 GDGL motif was mutated to EDGL in the mvp1 mutant. The Ser residue contained in the GDSL motif is crucial for enzymatic activity (Akoh et al., 2004), and multiple site-directed mutagenesis studies of lipases/esterases from Aeromonas hydrophila (Hilton and Buckley, 1991), Lactobacillus helveticus (Fenster et al., 2000), and Vibrio mimicus (Shaw et al., 1994) have demonstrated that mutagenesis of the Ser residue in the GDSL motif completely abolishes enzymatic activity. We found that MVP1 did in fact lack lipase activity.
The Subcellular Localization of MVP1 and Its Role in TGG2 Localization
In order to better understand the molecular function of MVP1, we carried out subcellular localization studies. Our results showed that MVP1 is localized to the ER and ER bodies and, to a lesser extent, the tonoplast. In comparison, the closest relative to MVP1, ESM1, has been found in the vacuole and leaf peroxisomes (Carter et al., 2004; Shimaoka et al., 2004; Huttlin et al., 2007; Reumann et al., 2007), and sequence analysis suggests that it could be localized to the ER (Zhang et al., 2006). The different subcellular distributions of these proteins could also explain their differing functions in Arabidopsis.
Localization of MVP1 to the ER bodies may explain, in part, the altered pathogen response in the mvp1 mutant. Using an ER body-specific GFP fusion marker, we detected significantly fewer ER bodies in mvp1 plants compared with wild-type plants, where most of the label in mutant plants was localized to the aggregate. It is not clear at this point whether the mvp1 mutation affects ER body formation per se, but MVP1 transcript is reduced in the ER body formation mutant nai1 (Nagano et al., 2008), so the loss or reduction of functional MVP1 protein may affect ER body formation.
The subcellular localization of the TGG2:GFP reporter fusion was essentially identical to that of MVP1:GFP in both wild-type and mutant backgrounds, in agreement with our in vitro data that the two proteins physically associate. The fact that we were able to observe TGG1:GFP, with which TGG2 does not interact, in the perinuclear aggregates of mvp1-1 and mvp1-2 further demonstrates that the mvp1 mutations cause global trafficking defects.
Previously published studies suggest that MBPs, and not MyAPs, promote the formation of higher order myrosinase complexes (Eriksson et al., 2002). Additionally, in Arabidopsis, PYK10, a β-glucosidase and one of the major protein constituents of ER bodies, has been found in active and inactive forms. The active version of PYK10 forms higher order complexes that contain additional β-glucosidases, jacalin-related lectins, and a GDSL-lipase/myrosinase-associated protein (At1g54000/GLL22). It is thought that two types of jacalin-related proteins regulate the size of the PYK10 complex and that they do so in an antagonistic manner, one promoting complex formation and the other inhibiting the formation of new complexes. PYK10-BINDING PROTEIN1 is an inhibitory binding protein. Therefore, although existing data favor MBPs as a determining factor in myrosinase complex formation, we cannot yet rule out a role for MVP1 in this process.
By extension, it is also not yet clear whether the observed downstream vacuole-related defects in the mutant alleles are due to additional interactions of MVP1 with other vacuole proteins or are simply a secondary effect of damaging ER function or, alternatively, allowing TGG2 to act in an inappropriate manner. Two recent studies have established a role for glucosinolates and the PEN2 atypical myrosinase in fungal defense (Bednarek et al., 2009; Clay et al., 2009). It was further hypothesized that the PEN2-dependent antifungal pathway arose as a means of detoxification for toxic compounds generated in the course of glucosinolate hydrolysis (Bednarek et al., 2009). Thus, by controlling the localization of TGG2, MVP1 may control hydrolysis of glucosinolates and the subsequent production of toxic compounds in the plant.
It is clear, based on the observed phenotypes of the fluorescent protein marker lines and the ER localization of MVP1, that the trafficking defect in the mvp1 mutants occurs early in the endomembrane transport pathway, most likely in the ER. It may be possible that MVP1 facilitates exit from the ER or that overaccumulation of unchaperoned TGG2 in mvp1 results in blocked transport from the ER. Important issues to address will be when and where TGG2 and MVP1 may interact in planta and what other proteins may reside in this putative complex. Another important issue to resolve is whether MVP1 has any additional interacting partners besides TGG2 or other proteins in a putative TGG2 complex. It will also be important to determine whether MVP1 interacts with either of the active root myrosinases, TGG4 or TGG5, although the absence of aggregates in root tissue and the fact that the TGG4 and TGG5 genes are part of different myrosinase subfamilies may argue against such an association (Andersson et al., 2009; Wang et al., 2009).
An outstanding question at this point is whether MVP1 has unique functions among the Arabidopsis MyAPs. Our examination of endomembrane structure in the esm1 mutant would suggest so, as might the fact that the other Arabidopsis MyAPs have retained the catalytic Ser that is universally conserved in GDSL lipases. A systematic evaluation of lipase activity and endomembrane system integrity in the corresponding mutants of the other family members may help better define the functions of MVP1 and its relatives and lead to a better understanding of glucosinolate metabolism specificity.
CONCLUSION
We demonstrated that MVP1 affected trafficking from the ER and downstream vacuole-related processes, such as growth, gravitropism, and pathogen defense. MVP1 specifically interacted with TGG2 in aerial tissues, and the glucosinolate hydrolysis profile of the mutant was altered compared with that of the wild-type parent. It is not clear at this point whether MVP1 actually acts as a direct modifier protein in specifying glucosinolate hydrolysis products or, rather, acts in a chaperone-like manner by ensuring the proper trafficking and localization of TGG2. Further proteomic and colocalization analyses and biochemical characterization of MVP1 will be instrumental in resolving these questions. It is clear, however, that our original screening strategy is robust and has allowed us to identify a heretofore uncharacterized trafficking component.
MATERIALS AND METHODS
Plant Material and Growth Conditions
The mvp1-1 mutant was isolated from a population of Arabidopsis (Arabidopsis thaliana) 35S:GFP:δ-TIP plants (Col-0 ecotype; Cutler et al., 2000) treated with ethyl methanesulfonate (Avila et al., 2003). mvp1-1 was backcrossed to GFP:δ-TIP twice. mvp1-1 plants were outcrossed to wild-type Col-0 to remove the GFP:δ-TIP transgene. Homozygous mvp1-1 (−GFP) plants were crossed with plants expressing fluorescent fusions to endomembrane proteins specific for the ER (YFP:Sec12; G. Drakakaki and N.V. Raikhel, unpublished data), Golgi (35S:NAG1:GFP; Grebe et al., 2003), vacuole (35S:GFP:γ-TIP-like; Cutler et al., 2000), and plasma membrane (35S:GFP:PIP2a; Cutler et al., 2000). mvp1-2 mutants were obtained from the SALK T-DNA collection (SALK_030621).
Seeds were surface sterilized in 50% bleach and 0.1% Triton X-100 solution and grown on plates containing 0.5× Murashige and Skoog medium + MES buffer (PlantMedia) and 1% Suc. Plants were incubated at 22°C under long-day conditions (16 h of light/8 h of dark), except for the gravitropism, carbon limitation, and pathogen sensitivity assays, conditions for which are described in Supplemental Materials and Methods S1.
Confocal Laser Scanning Microscopy
Confocal images were collected with a Leica TCS SP2/UV laser scanning confocal microscope system, and high-throughput visual inspection of mapping populations was carried-out with the BD Pathway HT system (BD Biosciences; Agee and Carter, 2009). Endomembrane structures were visualized after incubation in a solution of liquid Murashige and Skoog growth medium supplemented with 10 μm FM4-64 (Molecular Probes, Invitrogen) for the described time periods. To visualize DNA, seedlings were incubated with 10 μg mL−1 4′,6-diamidino-2-phenylindole in phosphate-buffered saline for 40 to 60 min.
Positional Cloning and Complementation
mvp1-1 mutants (Col-0 background expressing 35S:GFP:δ-TIP) were outcrossed to Landsberg erecta to generate a mapping population. Forty-eight F2 recombinant mvp1-1 homozygotes were analyzed with 22 simple sequence length polymorphism markers spanning the five Arabidopsis chromosomes (Lukowitz et al., 2000). Markers were developed from the Monsanto (Cereon) Arabidopsis Polymorphism Collection (Jander et al., 2002) for fine-scale mapping of 493 F2 recombinant mvp1-1 mutants (for primer sequences, see Supplemental Table S1). Twenty-four genes were sequenced in the 110-kb fine-map region.
A 4.0-kb fragment containing upstream sequence, the full-length MVP1 gene, and downstream sequences was amplified from Col-0 genomic DNA using the following primers: At1g54030 L (5′-GGGGACAAGTTTGTACAAAAAAGCAGGC-3′) and At1g54030 R (5′-GGGGACCACTTTGTACAAGAAAGCTGGG-3′). The PCR product was cloned into the pDONR207 vector and then cloned into the pMDC100 binary vector using previously published methods (Curtis and Grossniklaus, 2003). DNA was transformed into Agrobacterium tumefaciens strain GV2101 for introduction into homozygous mvp1-1 plants by floral dip (Clough and Bent, 1998). T1 seeds were selected on agar growth medium supplemented with 50 mg mL−1 kanamycin and examined by laser scanning confocal microscopy.
Transcript levels for MVP1 were analyzed with RT-PCR for 25 cycles using gene-specific primers 5′-CTTCCCTCTTCTTCTTCTCCA-3′ and 5′-CGTTGATAGAGCCAGTTCCA-3′. Primers for the ACTIN7 gene were 5′-AAAATGGCCGATGGTGAGG-3′ and 5′-ACTCACCACCACGAACCAG-3′.
Sequence Family Analysis
Related protein sequences were identified by BLASTP searches with an E value cutoff of 1e−10 against the protein sequences from nine databases: Arabidopsis (The Arabidopsis Information Resource annotation 7), Oryza sativa (The Institute for Genomic Research annotation 5), Populus trichocarpa (Joint Genome Institute [JGI] version 1.1), Chlamydomonas reinhardtii (JGI version 3.1), Ostreococcus tauri (JGI version 2.0), Phaeodactylum tricornutum (JGI version 2.0), Physcomitrella patens (JGI version 1.1), Selaginella moellendorffii (JGI version 1.0), and the UniProt database (February 2008). Duplicated sequences within and between databases were removed from the data set. Multiple sequence alignments were generated with the T-Coffee program (Notredame et al., 2000), and motif searches were performed with custom scripts.
An initial global multiple alignment and guide tree was constructed for all 907 sequences. The guide tree was calculated with the PHYLIP package (Felsenstein, 1997) using the PROTDIST program for calculating the distance matrix, the neighbor-joining method for tree construction, and the midpoint method in RETREE for defining the root of the tree. A subclade in the guide tree with 12 members including MVP1 was selected for the final phylogenetic analysis. These 12 protein sequences plus one outgroup (At1g53990) sequence were used to compute a high-quality multiple alignment. The final phylogenetic analysis was performed with two different tree construction methods: the Bayesian phylogenic inference implemented in the program MrBayes (Ronquist and Huelsenbeck, 2003) and the neighbor-joining method from the PHYLIP package (see above). The Markov chain Monte Carlo analysis with MrBayes was performed according to the recommendations of the program manual (version 3.1). The mixed amino acid substitution model was chosen and the simulation was set to 2,000,000 generations. The final tree from Figure 4 was plotted with the APE package from the statistical R environment using the At1g53990 sequence as outgroup for rooting the tree (Paradis et al., 2004; R Development Core Team, 2009). The neighbor-joining analysis with PHYLIP was performed as described above. Bootstrap values for this analysis were computed from 1,000 alignment resampling steps.
Purification of MVP1:GST and Lipase Activity Assays
Cloning and protein expression were performed as described previously (Oh et al., 2005) using PCR primers with appended sequences for BamHI and XhoI cloning sites (lowercase and underlined): 5′-gagaggatccATGCTTTTGATACCTTC-3′ (forward) and 5′-gagactcgagTTATATCATAAAGGAG-3′ (reverse). The mvp1-1 (G57E) mutant clone was generated by mutagenic PCR using the following primers with one mismatch (lowercase): 5′-GACGCTCTTTGTGTTCGaAGATGGTCTTTACGACGCC-3′ (forward) and 5′-GTCGTAAAGACCATCTtCGAACACAAAGAGCGTCTG-3′ (reverse). Protein expression was induced and lipase activity was measured as described using GLIP1 and glip1 recombinant proteins as positive and negative controls, respectively (Oh et al., 2005). To assay for MVP1 lipase activity in E. coli, we performed assays as described previously (Sommer et al., 1997). Briefly, isolated E. coli BL21 (DE3) colonies transformed with the MVP1:GST or mvp1:GST expression construct, or empty vector, were dotted on substrate plates containing 100 mg mL−1 ampicillin, 1 mm isopropylthio-β-galactoside, 1 mm CaCl2, and one of the following lipid substrates (1% [v/v]): Tween 20, Tween 80, or tributyrin. On plates with tributyrin as a lipid substrate, the presence of clear halos around the bacterial colonies was indicative of esterase activity. Correspondingly, the formation of a precipitate around bacterial colonies on Tween 20 or Tween 80 plates demonstrated lipase activity. E. coli heterologously expressing a GDSL lipase, JNP1, from Jacaranda mimosifolia was used as a positive control (Kram et al., 2008).
Myrosinase Association Assay
Partially purified MVP1:GST or GST proteins were bound to glutathione-Sepharose 4B (Amersham Biosciences) and incubated with protein extracts from Arabidopsis Col-0 or tgg2-2 (SAIL_237_G11; Barth and Jander, 2006) in pull-down buffer and incubated at 4°C for 1 h with shaking (Song et al., 2006). Beads were washed four times with pull-down buffer, and bound proteins were eluted and separated by 12% SDS-PAGE followed by immunoblot analysis with anti-TGG1 or anti-TGG2 antibodies, which were kindly provided by Ikuko Hara-Nishimura (Ueda et al., 2006).
Glucosinolate Structural Specificity
The structural outcome of glucosinolate activation was assayed using a modified version of a previously published protocol (Lambrix et al., 2001). Briefly, 100 mg of 4-week-old rosette tissue was homogenized in 1 mL of MES acid buffer (pH 6.0) and 0.4 μmol of allyl glucosinolate and incubated for 5 min. Glucosinolate activation products were extracted with dichloromethane for analysis by gas chromatography as described using a gas chromatography-mass spectral detector (Agilent HP 6890 with an Agilent 5973N mass spectral detector), and peak identities for nitrile and isothiocyanate were established by comparison with published mass spectra (Spencere and Daxenbichler, 1980; Lambrix et al., 2001). Differences between the genotypes were tested using Student's t tests.
Preparation of Binary Constructs and Transformations
MVP1, TGG1, and TGG2 were amplified using the primers shown in Supplemental Table S2. The forward primers contained the attB1 sites. The reverse primers contained attB2 sites and lacked termination codons in order to create C-terminal GFP fusions.
The PCR products were cloned into the pDONR207 vector (Invitrogen) and sequenced. We then carried out an LR recombination reaction using LR clonase (Invitrogen) and the Gateway-compatible binary vector pGWB5 (from Tsuyoshi Nakagawa, Research Institute of Molecular Genetics, Shimane University).
mvp1-1 plants were stably transformed using the floral dip method (Clough and Bent, 1998). Col-0, mvp1-1, and mvp1-2 plants were transiently transformed using a vacuum infiltration procedure (Marion et al., 2008).
The Arabidopsis Genome Initiative locus identifier for the MVP1 gene is At1g54030. ACTIN7 is At5g09810, GLIP1 is At5g40990, and SALT OVERLY SENSITIVE2 (SOS2) is At5g35410. TGG1 is At5g26000 and TGG2 is At5g25980.
Supplemental Data
The following materials are available in the online version of this article.
Supplemental Figure S1. mvp1, esm, and esp mutants stained with FM4-64.
Supplemental Figure S2. Phenotypic analysis of mvp1-1.
Supplemental Figure S3. NaCl treatment of mvp1 mutants.
Supplemental Figure S4. Positional cloning of mvp1-1.
Supplemental Figure S5. MVP1 contains a putative signal peptide for entry into the ER.
Supplemental Figure S6. Sequence alignment of putative myrosinase-associated proteins from Arabidopsis and oilseed rape.
Supplemental Figure S7. Spatial expression of MVP1, Arabidopsis MyAPs, and TGG2.
Supplemental Figure S8. GFP aggregates in early mvp1 seedling development.
Supplemental Figure S9. Developmental expression patterns of MVP1, Arabidopsis MyAPs, and TGG2.
Supplemental Figure S10. RT-PCR of MVP1 and TGG2.
Supplemental Figure S11. MVP1 expression is induced by pathogens and methyl jasmonate.
Supplemental Figure S12. Vacuolar delivery of TGG2:GFP and TGG1:GFP is impeded in mvp1 alleles.
Supplemental Figure S13. MVP1 does not act as a GDSL lipase.
Supplemental Table S1. Primers used for fine mapping of mvp1-1.
Supplemental Table S2. Primers used to create GFP fusion constructs.
Supplemental Table S3. Primers used for RT-PCR.
Supplemental Table S4. Percent sequence identity among putative myrosinase-associated proteins.
Supplemental Materials and Methods S1.
Supplementary Material
Acknowledgments
We thank Georgia Drakakaki for YFP:Sec12 seeds, Ben Scheres for NAG1:GFP, Chris Somerville for GFP:δ-TIP, GFP:γ-TIP-like, and GFP:PIP2a, Jian-Kang Zhu and Rebecca Stevenson for sos2-1, Georg Jander for tgg mutant seeds, and Ikuko Hara-Nishimura for the TGG1 and TGG2 antibodies. We thank Tsuyoshi Nakagawa for the pGWB5 vector. Colleen Knoth and Thomas Eulgem provided H. parasitica isolates and graciously performed sensitivity tests. We also thank Dayoung Kang and So Youn Won for technical assistance.
This work was supported by the U.S. Department of Energy/Energy Biosciences (grant no. DE–FG02–02ER15295 to N.V.R.), by the Plant Signaling Network Research Center funded by the Korea Science and Engineering Foundation (grant to O.K.P.), by the National Science Foundation (Career Award no. MCB–0642843 to H.J.), and by a Postdoctoral Fulbright Fellowship Award (grant no. FU–2006–0248) to A.R.
The author responsible for distribution of materials integral to the findings presented in this article in accordance with the policy described in the Instructions for Authors (www.plantphysiol.org) is: Natasha V. Raikhel (natasha.raikhel@ucr.edu).
The online version of this article contains Web-only data.
Open Access articles can be viewed online without a subscription.
References
- Agee A, Carter D (2009) Whole-organism screening: plants. Methods Mol Biol 486 77–95 [DOI] [PubMed] [Google Scholar]
- Akoh CC, Lee GC, Liaw YC, Huang TH, Shaw JF (2004) GDSL family of serine esterases/lipases. Prog Lipid Res 43 534–552 [DOI] [PubMed] [Google Scholar]
- Andersson D, Chakrabarty R, Bejai S, Zhang J, Rask L, Meijer J (2009) Myrosinases from root and leaves of Arabidopsis thaliana have different catalytic properties. Phytochemistry 70 1345–1354 [DOI] [PubMed] [Google Scholar]
- Avila EL, Zouhar J, Agee AE, Carter DG, Chary SN, Raikhel NV (2003) Tools to study plant organelle biogenesis. Point mutation lines with disrupted vacuoles and high-speed confocal screening of green fluorescent protein-tagged organelles. Plant Physiol 133 1673–1676 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barlowe C, Schekman R (1993) SEC12 encodes a guanine-nucleotide-exchange factor essential for transport vesicle budding from the ER. Nature 365 347–349 [DOI] [PubMed] [Google Scholar]
- Barth C, Jander G (2006) Arabidopsis myrosinases TGG1 and TGG2 have redundant function in glucosinolate breakdown and insect defense. Plant J 46 549–562 [DOI] [PubMed] [Google Scholar]
- Bassham DC (2007) Plant autophagy: more than a starvation response. Curr Opin Plant Biol 10 587–593 [DOI] [PubMed] [Google Scholar]
- Bednarek P, Pislewska-Bednarek M, Svatos A, Schneider B, Doubsky J, Mansurova M, Humphry M, Consonni C, Panstruga R, Sanchez-Vallet A, et al (2009) A glucosinolate metabolism pathway in living plant cells mediates broad-spectrum antifungal defense. Science 323 101–106 [DOI] [PubMed] [Google Scholar]
- Burke J, Pettitt JM, Schachter H, Sarkar M, Gleeson PA (1992) The transmembrane and flanking sequences of β 1,2-n-acetylglucosaminyltransferase I specify medial-Golgi localization. J Biol Chem 267 24433–24440 [PubMed] [Google Scholar]
- Burow M, Losansky A, Müller R, Plock A, Kliebenstein DJ, Wittstock U (2009) The genetic basis of constitutive and herbivore-induced ESP-independent nitrile formation in Arabidopsis. Plant Physiol 149 561–574 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carter C, Pan S, Zouhar J, Avila EL, Girke T, Raikhel NV (2004) The vegetative vacuole proteome of Arabidopsis thaliana reveals predicted and unexpected proteins. Plant Cell 16 3285–3303 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chary SN, Hicks GR, Choi YG, Carter D, Raikhel NV (2008) Trehalose-6-phosphate synthase/phosphatase regulates cell shape and plant architecture in Arabidopsis. Plant Physiol 146 97–107 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheong JJ, Choi YD (2003) Methyl jasmonate as a vital substance in plants. Trends Genet 19 409–413 [DOI] [PubMed] [Google Scholar]
- Clay NK, Adio AM, Denoux C, Jander G, Ausubel FM (2009) Glucosinolate metabolites required for an Arabidopsis innate immune response. Science 323 95–101 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clough SJ, Bent AF (1998) Floral dip: a simplified method for Agrobacterium-mediated transformation of Arabidopsis thaliana. Plant J 16 735–743 [DOI] [PubMed] [Google Scholar]
- Curtis MD, Grossniklaus U (2003) A Gateway cloning vector set for high-throughput functional analysis of genes in planta. Plant Physiol 133 462–469 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cutler SR, Ehrhardt DW, Griffitts JS, Somerville CR (2000) Random GFP:cDNA fusions enable visualization of subcellular structures in cells of Arabidopsis at a high frequency. Proc Natl Acad Sci USA 97 3718–3723 [DOI] [PMC free article] [PubMed] [Google Scholar]
- De DN (2000) Plant Cell Vacuoles: An Introduction. CSIRO Publishing, Collingwood, Australia
- Emanuelsson O, Brunak S, von Heijne G, Nielsen H (2007) Locating proteins in the cell using TargetP, SignalP and related tools. Nat Protoc 2 953–971 [DOI] [PubMed] [Google Scholar]
- Eriksson S, Andréasson E, Ekbom B, Granér G, Pontoppidan B, Taipalensuu J, Zhang J, Rask L, Meijer J (2002) Complex formation of myrosinase isoenzymes in oilseed rape seeds are dependent on the presence of myrosinase-binding proteins. Plant Physiol 129 1592–1599 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Falk A, Ek B, Rask L (1995) Characterization of a new myrosinase in Brassica napus. Plant Mol Biol 27 863–874 [DOI] [PubMed] [Google Scholar]
- Felsenstein J (1997) An alternating least squares approach to inferring phylogenies from pairwise distances. Syst Biol 46 101–111 [DOI] [PubMed] [Google Scholar]
- Fenster KM, Parkin KL, Steele JL (2000) Characterization of an arylesterase from Lactobacillus helveticus CNRZ32. J Appl Microbiol 88 572–583 [DOI] [PubMed] [Google Scholar]
- Grebe M, Xu J, Möbius W, Ueda T, Nakano A, Geuze HJ, Rook MB, Scheres B (2003) Arabidopsis sterol endocytosis involves actin-mediated trafficking via ARA6-positive early endosomes. Curr Biol 13 1378–1387 [DOI] [PubMed] [Google Scholar]
- Hawes C, Saint-Jore C, Martin B, Zheng HQ (2001) ER confirmed as the location of mystery organelles in Arabidopsis plants expressing GFP! Trends Plant Sci 6 245–246 [DOI] [PubMed] [Google Scholar]
- Hilton S, Buckley JT (1991) Studies on the reaction mechanism of a microbial lipase/acyltransferase using chemical modification and site-directed mutagenesis. J Biol Chem 266 997–1000 [PubMed] [Google Scholar]
- Hruz T, Laule O, Szabo G, Wessendorp F, Bleuler S, Oertle L, Widmayer P, Gruissem W, Zimmermann P (2008) Genevestigator V3: a reference expression database for the meta-analysis of transcriptomes. Adv Bioinform 2008: Article 420747 [DOI] [PMC free article] [PubMed]
- Huttlin EL, Hegeman AD, Harms AC, Sussman MR (2007) Comparison of full versus partial metabolic labeling for quantitative proteomics analysis in Arabidopsis thaliana. Mol Cell Proteomics 6 860–881 [DOI] [PubMed] [Google Scholar]
- Jander G, Norris SR, Rounsley SD, Bush DF, Levin IM, Last RL (2002) Arabidopsis map-based cloning in the post-genome era. Plant Physiol 129 440–450 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kato T, Morita MT, Fukaki H, Yamauchi Y, Uehara M, Niihama M, Tasaka M (2002) SGR2, a phospholipase-like protein, and ZIG/SGR4, a SNARE, are involved in the shoot gravitropism of Arabidopsis. Plant Cell 14 33–46 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kram BW, Bainbridge EA, Perera MA, Carter C (2008) Identification, cloning and characterization of a GDSL lipase secreted into the nectar of Jacaranda mimosifolia. Plant Mol Biol 68 173–183 [DOI] [PubMed] [Google Scholar]
- Lambrix V, Reichelt M, Mitchell-Olds T, Kliebenstein DJ, Gershenzon J (2001) The Arabidopsis epithiospecifier protein promotes the hydrolysis of glucosinolates to nitriles and influences Trichoplusia ni herbivory. Plant Cell 13 2793–2807 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lipka V, Dittgen J, Bednarek P, Bhat R, Wiermer M, Stein M, Landtag J, Brandt W, Rosahl S, Scheel D, et al (2005) Pre- and postinvasion defenses both contribute to nonhost resistance in Arabidopsis. Science 310 1180–1183 [DOI] [PubMed] [Google Scholar]
- Lukowitz W, Gillmor CS, Scheible WR (2000) Positional cloning in Arabidopsis: why it feels good to have a genome initiative working for you. Plant Physiol 123 795–805 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lukowitz W, Mayer U, Jürgens G (1996) Cytokinesis in the Arabidopsis embryo involves the syntaxin-related KNOLLE gene product. Cell 84 61–71 [DOI] [PubMed] [Google Scholar]
- Marion J, Bach L, Bellec Y, Meyer C, Gissot L, Faure JD (2008) Systematic analysis of protein subcellular localization and interaction using high-throughput transient transformation of Arabidopsis seedlings. Plant J 56 169–179 [DOI] [PubMed] [Google Scholar]
- Matsushima R, Kondo M, Nishimura M, Hara-Nishimura I (2003) A novel ER-derived compartment, the ER body, selectively accumulates a β-glucosidase with an ER-retention signal in Arabidopsis. Plant J 33 493–502 [DOI] [PubMed] [Google Scholar]
- Nagano AJ, Fukao Y, Fujiwara M, Nishimura M, Hara-Nishimura I (2008) Antagonistic jacalin-related lectins regulate the size of ER body-type beta-glucosidase complexes in Arabidopsis thaliana. Plant Cell Physiol 49 969–980 [DOI] [PubMed] [Google Scholar]
- Notredame C, Higgins DG, Heringa J (2000) T-Coffee: a novel method for fast and accurate multiple sequence alignment. J Mol Biol 302 205–217 [DOI] [PubMed] [Google Scholar]
- Ogasawara K, Yamada K, Christeller JT, Kondo M, Hatsugai N, Hara-Nishimura I, Nishimura M (2009) Constitutive and inducible ER bodies of Arabidopsis thaliana accumulate distinct β-glucosidases. Plant Cell Physiol 50 480–488 [DOI] [PubMed] [Google Scholar]
- Oh IS, Park AR, Bae MS, Kwon SJ, Kim YS, Lee JE, Kang NY, Lee S, Cheong H, Park OK (2005) Secretome analysis reveals an Arabidopsis lipase involved in defense against Alternaria brassicicola. Plant Cell 17 2832–2847 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paradis E, Claude J, Strimmer K (2004) APE: analyses of phylogenetics and evolution in R language. Bioinformatics 20 289–290 [DOI] [PubMed] [Google Scholar]
- Quigley F, Rosenberg JM, Shachar-Hill Y, Bohnert HJ (2002) From genome to function: the Arabidopsis aquaporins. Genome Biol 3: RESEARCH0001 [DOI] [PMC free article] [PubMed]
- R Development Core Team (2009) R: A Language and Environment for Statistical Computing. R Foundation for Statistical Computing, Vienna (http://www.R-project.org)
- Reumann S, Babujee L, Ma C, Wienkoop S, Siemsen T, Antonicelli GE, Rasche N, Lüder F, Weckwerth W, Jahn O (2007) Proteome analysis of Arabidopsis leaf peroxisomes reveals novel targeting peptides, metabolic pathways, and defense mechanisms. Plant Cell 19 3170–3193 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robatzek S (2007) Vesicle trafficking in plant immune responses. Cell Microbiol 9 1–8 [DOI] [PubMed] [Google Scholar]
- Robert S, Chary SN, Drakakaki G, Li S, Yang Z, Raikhel NV, Hicks GR (2008) Endosidin1 defines a compartment involved in endocytosis of the brassinosteroid receptor BRI1 and the auxin transporters PIN2 and AUX1. Proc Natl Acad Sci USA 105 8464–8469 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rojo E, Gillmor CS, Kovaleva V, Somerville CR, Raikhel NV (2001) VACUOLELESS1 is an essential gene required for vacuole formation and morphogenesis in Arabidopsis. Dev Cell 1 303–310 [DOI] [PubMed] [Google Scholar]
- Ronquist F, Huelsenbeck JP (2003) MrBayes 3: Bayesian phylogenetic inference under mixed models. Bioinformatics 19 1572–1574 [DOI] [PubMed] [Google Scholar]
- Sanderfoot AA, Pilgrim M, Adam L, Raikhel NV (2001) Disruption of individual members of Arabidopsis syntaxin gene families indicates each has essential functions. Plant Cell 13 659–666 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shaw JF, Chang RC, Chuang KH, Yen YT, Wang YJ, Wang FG (1994) Nucleotide sequence of a novel arylesterase gene from Vibrio mimicus and characterization of the enzyme expressed in Escherichia coli. Biochem J 298 675–680 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shimaoka T, Ohnishi M, Sazuka T, Mitsuhashi N, Hara-Nishimura I, Shimazaki K, Maeshima M, Yokota A, Tomizawa K, Mimura T (2004) Isolation of intact vacuoles and proteomic analysis of tonoplast from suspension-cultured cells of Arabidopsis thaliana. Plant Cell Physiol 45 672–683 [DOI] [PubMed] [Google Scholar]
- Silady RA, Ehrhardt DW, Jackson K, Faulkner C, Oparka K, Somerville CR (2008) The GRV2/RME-8 protein of Arabidopsis functions in the late endocytic pathway and is required for vacuolar membrane flow. Plant J 53 29–41 [DOI] [PubMed] [Google Scholar]
- Sohn EJ, Rojas-Pierce M, Pan S, Carter C, Serrano-Mislata A, Madueño F, Rojo E, Surpin M, Raikhel NV (2007) The shoot meristem identity gene TFL1 is involved in flower development and trafficking to the protein storage vacuole. Proc Natl Acad Sci USA 104 18801–18806 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sommer P, Bormann C, Götz F (1997) Genetic and biochemical characterization of a new extracellular lipase from Streptomyces cinnamomeus. Appl Environ Microbiol 63 3553–3560 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Song J, Lee MH, Lee GJ, Yoo CM, Hwang I (2006) Arabidopsis EPSIN1 plays an important role in vacuolar trafficking of soluble cargo proteins in plant cells via interactions with clathrin, AP-1, VTI11, and VSR1. Plant Cell 18 2258–2274 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spencere GF, Daxenbichler ME (1980) Gas chromatography-mass spectrometry of nitriles, isothiocyanates and oxazolidinethiones derived from cruciferous glucosinolates. J Sci Food Agric 31 359–367 [Google Scholar]
- Surpin M, Raikhel N (2004) Traffic jams affect plant development and signal transduction. Nat Rev Mol Cell Biol 5 100–109 [DOI] [PubMed] [Google Scholar]
- Surpin M, Rojas-Pierce M, Carter C, Hicks GR, Vasquez J, Raikhel NV (2005) The power of chemical genomics to study the link between endomembrane system components and the gravitropic response. Proc Natl Acad Sci USA 102 4902–4907 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Surpin M, Zheng H, Morita MT, Saito C, Avila E, Blakeslee JJ, Bandyopadhyay A, Kovaleva V, Carter D, Murphy A, et al (2003) The VTI family of SNARE proteins is necessary for plant viability and mediates different protein transport pathways. Plant Cell 15 2885–2899 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taipalensuu J, Falk A, Rask L (1996) A wound- and methyl jasmonate-inducible transcript coding for a myrosinase-associated protein with similarities to an early nodulin. Plant Physiol 110 483–491 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tamura K, Shimada T, Kondo M, Nishimura M, Hara-Nishimura I (2005) KATAMARI1/MURUS3 is a novel Golgi membrane protein that is required for endomembrane organization in Arabidopsis. Plant Cell 17 1764–1776 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tamura K, Takahashi H, Kunieda T, Fuji K, Shimada T, Hara-Nishimura I (2007) Arabidopsis KAM2/GRV2 is required for proper endosome formation and functions in vacuolar sorting and determination of the embryo growth axis. Plant Cell 19 320–332 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ueda H, Nishiyama C, Shimada T, Koumoto Y, Hayashi Y, Kondo M, Takahashi T, Ohtomo I, Nishimura M, Hara-Nishimura I (2006) AtVAM3 is required for normal specification of idioblasts, myrosin cells. Plant Cell Physiol 47 164–175 [DOI] [PubMed] [Google Scholar]
- Wang M, Sun X, Tan D, Gong S, Meijer J, Zhang J (2009) The two non-functional myrosinase genes TGG3 and TGG6 in Arabidopsis are expressed predominantly in pollen. Plant Sci 177 371–375 [Google Scholar]
- Wittstock U, Burow M (2007) Tipping the scales: specifier proteins in glucosinolate hydrolysis. IUBMB Life 59 744–751 [DOI] [PubMed] [Google Scholar]
- Yano D, Sato M, Saito C, Sato MH, Morita MT, Tasaka M (2003) A SNARE complex containing SGR3/atvam3 and ZIG/VTI11 in gravity-sensing cells is important for Arabidopsis shoot gravitropism. Proc Natl Acad Sci USA 100 8589–8594 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Z, Ober JA, Kliebenstein DJ (2006) The gene controlling the quantitative trait locus EPITHIOSPECIFIER MODIFIER1 alters glucosinolate hydrolysis and insect resistance in Arabidopsis. Plant Cell 18 1524–1536 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu J, Gong Z, Zhang C, Song CP, Damsz B, Inan G, Koiwa H, Zhu JK, Hasegawa PM, Bressan RA (2002) OSM1/SYP61: A syntaxin protein in Arabidopsis controls abscisic acid-mediated and non-abscisic acid-mediated responses to abiotic stress. Plant Cell 14 3009–3028 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zouhar J, Hicks GR, Raikhel NV (2004) Sorting inhibitors (sortins): chemical compounds to study vacuolar sorting in Arabidopsis. Proc Natl Acad Sci USA 101 9497–9501 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.