Abstract
ClpB and Hsp104 are members of the AAA+ (ATPases associated with various cellular activities) family of proteins and are molecular machines involved in thermotolerance. They are hexameric proteins containing 12 ATP binding sites with two sites per protomer. ClpB and Hsp104 possess some innate protein remodeling activities; however, they require the collaboration of the DnaK/Hsp70 chaperone system to disaggregate and reactivate insoluble aggregated proteins. We investigated the mechanism by which ClpB couples ATP utilization to protein remodeling with and without the DnaK system. When wild-type ClpB, which is unable to remodel proteins alone in the presence of ATP, was mixed with a ClpB mutant that is unable to hydrolyze ATP, the heterohexamers surprisingly gained protein remodeling activity. Optimal protein remodeling by the heterohexamers in the absence of the DnaK system required approximately three active and three inactive protomers. In addition, the location of the active and inactive ATP binding sites in the hexamer was not important. The results suggest that in the absence of the DnaK system, ClpB acts by a probabilistic mechanism. However, when we measured protein disaggregation by ClpB heterohexamers in conjunction with the DnaK system, incorporation of a single inactive ClpB subunit blocked activity, supporting a sequential mechanism of ATP utilization. Taken together, the results suggest that the mechanism of ATP utilization by ClpB is adaptable and can vary depending on the specific substrate and the presence of the DnaK system.
Keywords: DnaJ, DnaK, GrpE, protein disaggregation
Bacterial ClpB and yeast Hsp104 are ATP-dependent protein remodeling machines that function to disaggregate protein aggregates and reactivate proteins after extreme stress conditions (1–3). In the cell, ClpB acts in conjunction with the DnaK chaperone system and Hsp104 acts with the Hsp70 chaperone system (4, 5). DnaK and Hsp70 are members of another large, ubiquitous family of ATP-dependent molecular chaperones that mediate protein reactivation and remodeling in concert with two cochaperones, DnaJ and GrpE in prokaryotes and Hsp40 and NEF in eukaryotes (6). Alone, neither the DnaK/Hsp70 chaperone system nor ClpB/Hsp104 has the ability to reactivate large insoluble aggregates.
ClpB/Hsp104 exists as a hexameric ring with an axial channel (7–10). Each protomer contains two AAA+ (ATPases associated with various cellular activities) nucleotide-binding domains separated by a hinge region and preceded by an N-terminal domain (1, 7). The two AAA+ domains contain characteristic motifs, including Walker A and B and sensor-1 and -2 motifs, as well as an arginine finger (11, 12). Situated in the first AAA+ domain is a long coiled-coil region, referred to as the middle domain, which is unique to ClpB, Hsp104, and their homologs.
In vitro ClpB/Hsp104 solubilizes and reactivates protein aggregates in ATP-dependent reactions in collaboration with the DnaK/Hsp70 chaperone system (1–3). Although the roles of the two chaperone systems in disaggregation are not fully understood, it is likely that ClpB/Hsp104 is the primary protein disaggregating machine, and DnaK/Hsp70 facilitates the interaction of ClpB/Hsp104 with aggregates (1–3). However, DnaK/Hsp70 may have additional functions.
Evidence suggests that ClpB/Hsp104 acts by forcibly extracting polypeptides from aggregates and translocating the unfolded regions of polypeptides through its axial channel (1–3). This substrate translocation mechanism has been established for ClpA and ClpX, two Clp ATPases that interact with a proteolytic component, ClpP (13). Substrates are unfolded and threaded through the central channels of ClpA and ClpX directly into the chamber of ClpP where degradation occurs. Work from Bukau and colleagues, in which ClpB and Hsp104 were engineered to contain the ClpP interaction loop from ClpA, supports a similar mechanism for ClpB and Hsp104 (14, 15).
The demonstration that ClpB and Hsp104 have an innate ability to unfold and activate proteins in the absence of the DnaK/Hsp70 chaperone system provides additional support for an unfolding and translocation mechanism (16). However, ATP alone is ineffective in promoting these protein remodeling activities. Instead, mixtures of ATP and ATPγS, a slowly hydrolyzed ATP analog, are required for remodeling, suggesting that with the mixture of nucleotides substrates can be held (a function requiring ATP binding and supported by ATPγS) and unfolded and translocated (functions requiring ATP hydrolysis).
To understand how ClpB couples ATP binding and hydrolysis to protein remodeling, we investigated the contribution of the 12 ATP binding sites to the overall chaperone activity of ClpB both alone and in the presence of the DnaK chaperone system.
Results
ClpB Hexamers with a Balance of Active and Inactive Nucleotide Binding Sites Are Required for Optimal Protein Remodeling in the Absence of the DnaK System.
The 12 ATP binding sites of hexameric ClpB are arranged in two rings of six, with each protomer providing one nucleotide-binding site to each ring, referred to here as Ring-1 and Ring-2 (Fig. 1A). Ring-1 of the hexamer is comprised of the N-terminal ATP binding sites, and Ring-2 is comprised of the C-terminal ATP-binding sites.
Fig. 1.
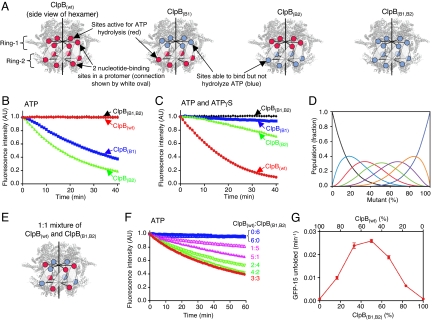
Optimal protein unfolding by ClpB requires that ClpB hexamers have both active and inactive ATP hydrolytic sites. (A) Diagram illustrating the location of the 12 ATP binding sites in hexamers of ClpB(wt), single Walker B mutants in the first, ClpB(B1), or second, ClpB(B2), nucleotide-binding domain and a double Walker B mutant, ClpB(B1,B2). The 12 sites are superimposed on a model of a ClpB hexamer generated from the crystal structure of a ClpB monomer (39). (B and C) Protein unfolding of GFP-15 by ClpB(wt), ClpB(B1), ClpB(B2), and ClpB(B1,B2) in the presence of ATP (B) or ATP and ATPγS in a 1:1 ratio (C) as described in the Methods. (D) Mathematical model, generated as described in ref. 20, depicting the theoretical populations of wild-type and mutant ClpB heterohexamers that contain no (black), one (aqua), two (red), three (green), four (purple), five (orange), or six (blue) mutant subunits as a function of percent mutant. (E) Diagram showing representative locations of wild-type (red) and Walker B mutant ATP binding sites (blue) for a heterohexamer of ClpB(wt) and ClpB(B1,B2) in a 1:1 ratio. (F) GFP-15 unfolding in the presence of ATP using mixtures of ClpB(wt) and ClpB(B1,B2) in various ratios. (G) The rate of GFP-15 unfolding by mixtures of ClpB(wt) and ClpB(B1,B2) plotted as a function of percent ClpB(B1,B2) in the mixture. In B, C, and F, representative experiments of three replicates are shown. In G, data are means ± SEM (n = 3). Some error bars are covered by the plot symbols.
Previous observations demonstrated that ATP hydrolysis at <12 ATP binding sites of the ClpB hexamer is required to elicit the innate protein remodeling activity of ClpB, and ATP hydrolysis at all 12 sites prohibits activity (16). In the work presented here, we used a green fluorescent protein (GFP) fusion protein containing a C-terminal 15 aa peptide, GFP-15, as a substrate for remodeling. We measured protein unfolding by monitoring the decrease in fluorescence with time in the presence of a mutant GroEL (17) that binds unfolded proteins but does not release them. ClpB wild-type, ClpB(wt), was unable to unfold GFP-15 in the presence of ATP (Fig. 1B). However, when mixtures of ATP and ATPγS were used in a 1:1 ratio, there was a large decrease in GFP fluorescence (Fig. 1C). Thus, if some ATP binding sites hydrolyze ATP slowly, ClpB(wt) activity is elicited.
Single Walker B mutants (Fig. 1A), with either an E279A substitution in Ring-1, ClpB(B1), or an E678A substitution in Ring-2, ClpB(B2), unfolded GFP-15 with ATP alone and were inhibited by ATPγS, supporting the conclusion that hydrolysis by <12 ATP binding sites elicits activity (Fig. 1 B and C). These mutants contain six wild-type nucleotide-binding sites per hexamer and six mutant sites that are able to bind but not hydrolyze ATP (18). A double Walker B mutant (Fig. 1A) with both E279A and E678A substitutions, ClpB(B1,B2), was unable to catalyze unfolding of GFP-15 with ATP or mixtures of ATP and ATPγS, since all of its nucleotide-binding sites are defective in ATP hydrolysis (19) (Fig. 1 B and C). Therefore, ClpB can perform protein unfolding when there is only one active nucleotide-binding site per protomer, and the active nucleotide-binding sites are all in Ring-1 or Ring-2 of the hexamer. These results also show that hydrolysis by the two rings does not need to be coupled for protein remodeling by ClpB alone.
To investigate whether the intrinsic remodeling activity of ClpB demands that all six of the active sites reside in Ring-1 or Ring-2 of the hexamer, or if the active sites can be distributed between the two rings, we performed subunit mixing experiments. Recent studies have shown that the ClpB hexamer is a dynamic complex with subunits reshuffling on a time scale of a minute (20, 21). Thus, in subunit mixing experiments, heterohexamers are expected to be represented by a binomial distribution that varies as a function of the molar ratio of each subunit present, assuming subunits have an equal ability to be incorporated into hexamers (Fig. 1D). We mixed the double Walker B mutant, ClpB(B1,B2), with ClpB(wt) (shown schematically in Fig. 1E) and measured GFP-15 unfolding in the presence of ATP as the sole nucleotide. Surprisingly, we saw that the heterohexamers were active, although each protein alone was inactive in GFP-15 unfolding (Fig. 1F). By varying the ratio of ClpB(wt) and ClpB(B1,B2) while holding the total ClpB concentration constant, we observed maximal activity when there were approximately three active sites in each ring of the hexamer [50% ClpB(wt) and 50% ClpB(B1,B2)] (Fig. 1 F and G). Therefore, ClpB is able to catalyze protein remodeling when there are both active and inactive sites in Ring-1 and Ring-2 of the hexamer and the subunits contain either two active or two inactive nucleotide-binding sites.
We extended these observations with ClpB(B1,B2) and ClpB(wt) heterohexamers by testing two other protein remodeling reactions. In a disaggregation assay, mixtures of ClpB(B1,B2) and ClpB(wt) were able to reactivate heat-inactivated GFP in the presence of ATP alone (Fig. 2A). Maximal reactivation was observed with a 1:1 ratio of the two ClpB proteins. Mixtures of ClpB(B1,B2) and ClpB(wt) were also tested in the RepA activation assay in which inactive RepA dimers are converted to monomers that bind DNA with high affinity (22). In this assay as well, maximal activation was seen with a 1:1 mixture of the two proteins (Fig. 2A). Thus, in additional protein remodeling reactions, heterohexamers of ClpB(B1,B2) and ClpB(wt) are functional, while neither protein separately exhibits significant activity.
Fig. 2.
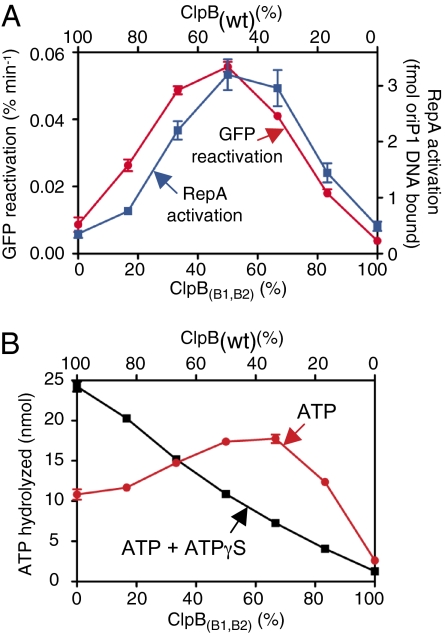
ClpB hexamers containing approximately six active ATP hydrolytic sites are required for optimal chaperone activity. (A) Reactivation of heat-aggregated GFP and activation of RepA was measured as described in the Methods using various ratios of ClpB(wt) to ClpB(B1,B2). GFP reactivation rates (left axis) and DNA binding by RepA (right axis) were plotted as a function of percent ClpB(B1,B2). (B) ATPase activity by mixtures of ClpB(wt) and ClpB(B1,B2) in the presence of ATP or ATP and ATPγS in a 1:1 ratio was measured as described in the Methods. ATPase activity is plotted as a function of percent ClpB(B1,B2). Data are means ± SEM (n = 3). Some error bars are covered by plot symbols.
When ATP hydrolysis was measured, approximately 2-fold higher steady-state hydrolysis was observed with a 1:1 mixture of ClpB(wt) and ClpB(B1,B2) compared to ClpB(wt) alone (Fig. 2B) (19). This suggests that the activity of wild-type subunits within the heterohexamer is stimulated. For example, at a 1:1 ratio of ClpB(wt) to ClpB(B1,B2), the ATPase activity of the wild-type subunits must increase approximately 4-fold to account for the observed increase in ATP hydrolysis. For comparison, when we measured ATP hydrolysis by mixtures of ClpB(wt) and ClpB(B1,B2) in the presence of ATP and ATPγS, hydrolysis decreased linearly as the percent of ClpB(B1,B2) in the mixture was increased (Fig. 2B). ATP hydrolysis by ClpB(wt) alone was approximately 2-fold higher with a 1:1 mixture of ATP and ATPγS than with ATP alone (Fig. 2B) (16). Thus, conditions optimal for protein remodeling are also optimal for ATP hydrolysis.
Our results with the single Walker B mutants and heterohexamers of ClpB(wt) and ClpB(B1,B2) suggest that for remodeling activity by ClpB alone: (i) protomers need not contain a pair of active sites as long as there are a total of six active sites in either Ring-1 or Ring-2 of the hexamer, and (ii) there need not be six functional sites in a single ring as long as there are approximately six active sites in the hexamer. Together these results indicate that the innate remodeling activity of ClpB might simply require a total of approximately six active ATP hydrolytic sites, independent of the location of the sites within the subunit or within the hexamer.
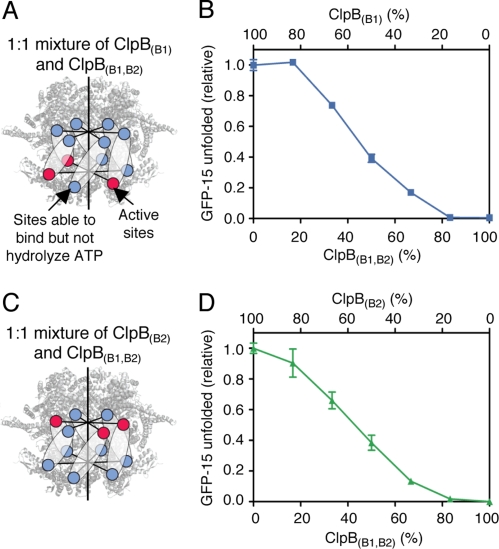
To test this possibility, we measured protein unfolding by heterohexamers of the two single Walker B mutants, ClpB(B1) and ClpB(B2) (Fig. 3A). When ClpB(B1) and ClpB(B2) were mixed in various ratios while keeping the total concentration of ClpB constant, there was no significant gain or loss of function (Fig. 3B). Interpretation of these results depends upon ClpB(B1) and ClpB(B2) forming heterohexamers, which is likely, since each single Walker B mutant is capable of forming heterohexamers with both ClpB(wt) and ClpB(B1,B2) (shown in Fig. 4 B and D and Fig. 5B). The results suggest that ClpB is active in protein remodeling when the hexamer contains six active ATP hydrolytic sites irrespective of the position of the active sites within the hexamer.
Fig. 3.
Protein unfolding by mixtures of ClpB(B1) and ClpB(B2). (A) Diagram showing a heterohexamer containing ClpB(B1) and ClpB(B2) in a 1:1 ratio. (B) GFP-15 unfolding by mixtures of ClpB(B1) and ClpB(B2) was measured as described in the Methods. Rates are expressed as a fraction relative to ClpB(B2) alone (0.060 ± 0.002 min−1) and plotted as a function of percent ClpB(B1) in the mixture. Data are means ± SEM (n ≥ 3). Some error bars are covered by plot symbols.
Fig. 4.
Protein unfolding by mixtures of ClpB(B1,B2) and either ClpB(B1) or ClpB(B2). (A and C) Diagrams showing heterohexamers of ClpB(B1,B2) with ClpB(B1) (A) or ClpB(B2) (C) in a 1:1 ratio. (B and D) GFP-15 unfolding by mixtures of ClpB(B1,B2) and ClpB(B1) (B) or ClpB(B2) (D) as described in the Methods. Unfolding rates are expressed as a fraction relative to ClpB(B1) (0.035 ± 0.001 min−1) (B) or ClpB(B2) (0.060 ± 0.002 min−1) (D) alone and plotted as a function of percent ClpB(B1,B2) in the mixture. Data are means ± SEM (n ≥ 3). Some error bars are hidden by plot symbols.
Fig. 5.
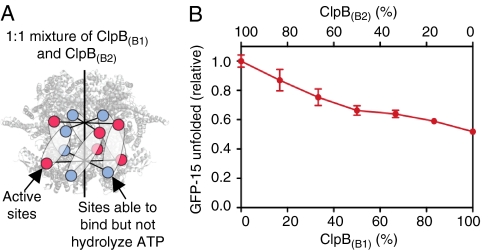
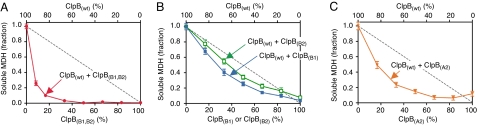
Disaggregation of MDH by mixtures of ClpB(wt) and ClpB mutants in conjunction with DnaK, DnaJ, and GrpE. (A–C) Recovery of soluble MDH from heat-induced aggregates was measured as described in the Methods for mixtures of ClpB(wt) with ClpB(B1,B2) (A), ClpB(B1) (B), ClpB(B2) (B), or ClpB(A2) (C) in the presence of DnaK/DnaJ/GrpE. In A–C, the dashed gray line represents the linear decrease expected for a probabilistic mechanism where the activity of the hexamer is proportional to the number of wild-type subunits. The fraction of soluble MDH recovered in reactions containing 100% ClpB(wt) was set equal to one. With conditions used, ClpB(wt) solubilized 53% ± 1% of the MDH. Data are means ± SEM (n ≥ 3). Some error bars are covered by plot symbols.
Mechanism of ATP Utilization by ClpB in the Absence of the DnaK System.
Our observations that heterohexamers of ClpB(wt) and ClpB(B1,B2) perform remodeling activities demonstrate that protein unfolding and remodeling can be carried out with approximately three active protomers per hexamer (Figs. 1 and 2). These results rule out a concerted mechanism of ATP utilization within a ring, where all six active sites of a ring simultaneously bind ATP, hydrolyze ATP, and release ADP. They also rule out a strictly sequential mechanism, where activity depends upon an endless cycle of ATP utilization around the ring. Instead, the observations point to either a probabilistic mechanism, involving a random order of mutually independent hydrolysis events or a semisequential mechanism, in which the sequence and timing of ATP hydrolysis operates in a sequential manner that proceeds around the ring with hydrolysis pausing occasionally and then resuming at another site in the ring.
To further explore the mechanism of ATP utilization by ClpB, we varied the number of active sites in one ring while maintaining six inactive sites in the other ring. We first decreased the number of active sites in Ring-2 by increasing the percentage of ClpB(B1,B2) in mixtures with ClpB(B1) (Fig. 4A). We observed that incorporation of one or two inactive sites in Ring-2 had little effect on hexamer activity, while incorporation of four or five inactive sites significantly inhibited activity (Fig. 4B). We next changed the number of active sites in Ring-1 by varying the ratio of ClpB(B1,B2) in mixtures with ClpB(B2) (Fig. 4C) and observed similar results (Fig. 4D). Taken together, these experiments show that ClpB is able to function in protein remodeling with one inactive ring and approximately three or more active sites in the other ring of the hexamer (Fig. 4 B and D). Thus, the results best support a probabilistic or a semisequential mechanism of ATP utilization by the ClpB rings. However, six active sites per hexamer are optimal for protein remodeling in the absence of the DnaK system (Figs. 1 F and G and 23–4).
Mechanism of ATP Utilization by ClpB When Disaggregating Substrates That Require the DnaK System.
To test whether the mechanism of ATP utilization by ClpB is the same when ClpB works on aggregates with the DnaK system as when it works alone, we monitored disaggregation by mixtures of ClpB(wt) and ClpB(B1,B2) in conjunction with DnaK, DnaJ, and GrpE. We used aggregated malate dehydrogenase (MDH) as a substrate, since previous studies showed that MDH disaggregation by ClpB in combination with the DnaK system was approximately 20-fold greater than disaggregation by either chaperone alone (23, 24). As the percent of ClpB(B1,B2) in mixtures with ClpB(wt) was increased, we observed a rapid exponential decrease in MDH disaggregation, compared to the linear decrease expected if a probabilistic mechanism was used (Fig. 5A). The loss of activity when the percentage of ClpB(B1,B2) is low indicates that incorporation of approximately one protomer with two hydrolytically defective ATP binding sites blocks disaggregation of MDH. Our results indicate a striking departure from the protein remodeling and ATP-ase activities seen in the absence of the DnaK system (Figs. 1 F and G and 2 A and B) and from the ATPase data (Fig. 2B). They suggest that for disaggregation of protein aggregates in collaboration with the DnaK system, cooperative interactions between all six ClpB subunits are required. Similar inter-subunit coupling during remodeling has been observed for Thermus thermophilus ClpB using another substrate, aggregated α-glucosidase (20).
We next measured the effects of single nucleotide binding site mutants in mixtures with ClpB(wt). We used the two single Walker B mutants discussed above, ClpB(B1) and ClpB(B2). In addition, we tested a ClpB Walker A mutant, ClpB(A2), with a K611T substitution in Ring-2 that renders it unable to bind ATP in Ring-2.* All three mutants are defective in disaggregation of MDH in combination with the DnaK system (23, 25) (Fig. 5 B and C). As the percentage of ClpB(B1), ClpB(B2), or ClpB(A2) in mixtures with ClpB(wt) was increased, disaggregation of MDH decreased more rapidly than the linear decrease expected for a probabilistic mechanism of action (Fig. 5 B and C). However, incorporating approximately two inactive sites in the same ring inhibited disaggregation less than incorporating approximately one protomer with two inactive sites (Fig. 5 A and B). These observations suggest that for ClpB to work on substrates that require collaboration with the DnaK system, both inter- and intra-ring communication between ClpB subunits is important. Together the data support a sequential or semisequential mechanism of ATP utilization within each ring and within the hexamer. Moreover, they imply that the mechanism of ATP utilization by ClpB can vary depending on the substrate and the requirement for the DnaK system.
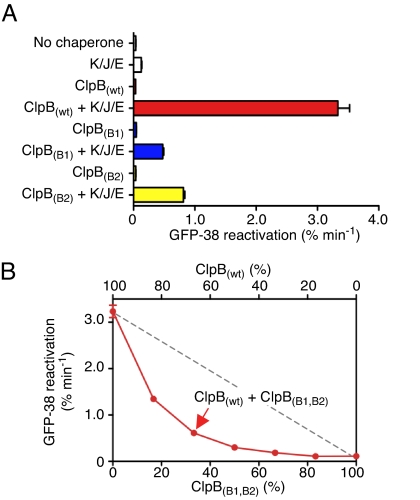
To address the question of whether the specific aggregate influences ATP utilization requirements, we compared disaggregation of aggregated MDH to that of heat-aggregated GFP-38, a GFP fusion protein containing a C-terminal 38 aa peptide. Aggregated GFP-38, like MDH, required the combination of ClpB(wt) and the DnaK system for reactivation, as measured by the regain of GFP fluorescence with time (Fig. 6A). ClpB(B1) and ClpB(B2) were each able to reactivate GFP-38 at a low rate in the presence of the DnaK system, although neither mutant was able to detectably disaggregate MDH (Figs. 5B and 6A). These results suggest that with some aggregates, six active ATP binding sites are sufficient to carry out limited disaggregation in conjunction with the DnaK system.
Fig. 6.
Disaggregation of GFP-38 by mixtures of ClpB(wt) and ClpB mutants in conjunction with DnaK, DnaJ, and GrpE. (A) The rate of reactivation of heat-inactivated GFP-38 by ClpB(wt) or ClpB Walker B mutants was measured with and without DnaK/DnaJ/GrpE as described in the Methods. (B) The rate of GFP-38 reactivation by mixtures of ClpB(wt) and ClpB(B1,B2) with DnaK/DnaJ/GrpE was measured as described in the Methods and plotted as a function of percent ClpB(B1,B2). The dashed gray line represents the linear decrease expected for a probabilistic mechanism where the activity of the hexamer is proportional to the number of wild-type subunits. Data are means ± SEM (n = 3). Some error bars are hidden by plot symbols.
To extend the comparison of the two aggregates, we measured disaggregation of GFP-38 by mixtures of ClpB(B1,B2) and ClpB(wt). Disaggregation activity decreased exponentially as the ratio of ClpB(B1,B2) to ClpB(wt) in the mixture increased (Fig. 6B). The data provide further support that cooperative interactions between subunits are important and that ATP utilization by ClpB is likely through a sequential or semisequential mechanism when ClpB acts in protein disaggregation in conjunction with the DnaK system.
In summary the results presented here suggest that the mechanism of ATP utilization by the two rings of ClpB can change dependent upon the specific substrate and the requirement for the DnaK chaperone system.
Discussion
The surprising result that the heterohexamer of ClpB(wt) and ClpB(B1,B2) is active under conditions where neither homohexamer is active shows that modulation of the hydrolytic cycle of ClpB elicits protein remodeling. Moreover, approximately six active nucleotide-binding sites are required for maximal protein remodeling activity by ClpB alone (Figs. 1 and 2). These observations are consistent with our previous findings that ClpB and Hsp104 perform protein remodeling alone in the presence of mixtures of ATP and ATPγS (16). Together the data suggest that remodeling activity is elicited when some sites hydrolyze ATP and other sites bind ATP, but hydrolyze it slowly or not at all. One interpretation is that the sites that bind but do not hydrolyze ATP help stabilize interactions between ClpB and the substrate as well as interactions among ClpB protomers in the hexamer, while the sites that hydrolyze ATP are necessary for substrate unfolding and translocation. The balance between the two types of sites influences the efficiency of protein remodeling. However, modulating the ATP hydrolytic cycle is not sufficient to elicit the full activity of ClpB, since in vivo, mutants defective in ATP hydrolysis in either ring are also defective for thermotolerance (18). One possibility is that the substrate and the DnaK system regulate ATP utilization, consistent with our previous results showing that ATP hydrolysis is stimulated by the combined presence of ClpB, the DnaK system, and aggregated substrate (23).
Interestingly, our results suggest that the number of hydrolytically active sites sufficient to perform remodeling activity and the mechanism of ATP utilization can vary depending on the substrate and the involvement of the DnaK system. When ClpB acts in the absence of the DnaK system, the mechanism of ATP utilization by the two rings is likely probabilistic, although a semisequential mechanism is also possible. This is based on experiments showing that approximately three active protomers per hexamer are optimal for remodeling activity. In addition, approximately three active ATP hydrolytic sites in a ring are sufficient for protein remodeling when the other ring contains six sites defective in ATP hydrolysis. In contrast, when ClpB acts on aggregated substrates that require the DnaK system, ClpB protomers must work together in a cooperative fashion, supporting a sequential mechanism of ATP utilization. However, the observations that heterohexamers can tolerate protomers with one active and one inactive ATP binding site provide evidence for a semisequential mechanism of ATP utilization within the separate rings.
Probabilistic, semisequential, and sequential modes of action have been proposed for other AAA+ proteins (10, 26, 27). For example, another hexameric Clp protein, ClpX, has been shown to function in a probabilistic manner by Martin, Baker, and Sauer (26). In contrast, Saibil, Lindquist, and colleagues recently proposed that Hsp104, the yeast homolog of ClpB, uses a sequential mechanism, based on electron microscopic data showing asymmetry in the hexameric model (10). Crystal structures showing asymmetric conformations have been observed for several AAA+ helicases, suggesting that those proteins act by a sequential mechanism (27). Additionally, subunit mixing studies provided evidence for a semisequential mechanism of action by MCM and RuvB and a sequential mechanism for T7gp4 (28–32).
While further biochemical and structural analyses of ClpB and its interaction with the DnaK system are needed, the mechanistic insights revealed by this study of ClpB may extend to Hsp104 and other ClpB homologs.
Methods
Proteins and DNA.
P1 RepA (22), GFP (33), GFP-15 (34), ClpB and ClpB mutants (35), GroELtrap (17), DnaK (36), DnaJ (36), GrpE (36), and [3H]oriP1 DNA (22) (4,475 cpm/fmol) were prepared as described. pET-GFP-38 was created by inserting GAT and GAC codons before the multicloning site stop codon of pET-GFP-X30-H6 (37) by QuikChange (Stratagene) mutagenesis. GFP-38 was purified as described for GFP-X30-H6 (37). MDH (Roche) was labeled with 3H as described in ref. 38. Protein concentrations given are for monomeric GFP, GFP-15, GFP-38, MDH, and DnaK; dimeric RepA, DnaJ, and GrpE; hexameric ClpB; and tetradecameric GroELtrap.
Assays.
GFP-fusion protein unfolding (16), GFP reactivation (16), MDH disaggregation (23), RepA activation (16), and ATPase (16) assays were performed as described with slight modifications described in the SI Methods. GFP-38 reactivation was performed as described in the SI Methods. For subunit mixing experiments, control experiments were conducted to demonstrate that the activity of ClpB(wt), ClpB(B1), and ClpB(B2) decreased linearly upon dilution over the range of concentrations used.
Supplementary Material
Acknowledgments.
We thank Danielle Johnston, Jodi Camberg, Marika Miot, and Olivier Genest for critical reading of the manuscript and helpful discussions. We thank Michal Zolkiewski (Kansas State University) for the plasmid pET20b-K611T and BK Lee (National Cancer Institute) for generating mathematical models. This work was supported by the Intramural Research Program of the National Institutes of Health (NIH) National Cancer Institute Center for Cancer Research.
Footnotes
The authors declare no conflict of interest.
This article contains supporting information online at www.pnas.org/cgi/content/full/0911937106/DCSupplemental.
References
- 1.Doyle SM, Wickner S. Hsp104 and ClpB: Protein disaggregating machines. Trends Biochem Sci. 2009;34:40–48. doi: 10.1016/j.tibs.2008.09.010. [DOI] [PubMed] [Google Scholar]
- 2.Mogk A, Haslberger T, Tessarz P, Bukau B. Common and specific mechanisms of AAA+ proteins involved in protein quality control. Biochem Soc Trans. 2008;36:120–125. doi: 10.1042/BST0360120. [DOI] [PubMed] [Google Scholar]
- 3.Zolkiewski M. A camel passes through the eye of a needle: Protein unfolding activity of Clp ATPases. Mol Microbiol. 2006;61:1094–1100. doi: 10.1111/j.1365-2958.2006.05309.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Parsell DA, Kowal AS, Singer MA, Lindquist S. Protein disaggregation mediated by heat-shock protein Hsp104. Nature. 1994;372:475–478. doi: 10.1038/372475a0. [DOI] [PubMed] [Google Scholar]
- 5.Mogk A, et al. Identification of thermolabile Escherichia coli proteins: Prevention and reversion of aggregation by DnaK and ClpB. EMBO J. 1999;18:6934–6949. doi: 10.1093/emboj/18.24.6934. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Bukau B, Horwich AL. The Hsp70 and Hsp60 chaperone machines. Cell. 1998;92:351–366. doi: 10.1016/s0092-8674(00)80928-9. [DOI] [PubMed] [Google Scholar]
- 7.Lee S, et al. The structure of ClpB: A molecular chaperone that rescues proteins from an aggregated state. Cell. 2003;115:229–240. doi: 10.1016/s0092-8674(03)00807-9. [DOI] [PubMed] [Google Scholar]
- 8.Lee S, Choi JM, Tsai FT. Visualizing the ATPase cycle in a protein disaggregating machine: Structural basis for substrate binding by ClpB. Mol Cell. 2007;25:261–271. doi: 10.1016/j.molcel.2007.01.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Wendler P, et al. Atypical AAA+ subunit packing creates an expanded cavity for disaggregation by the protein-remodeling factor Hsp104. Cell. 2007;131:1366–1377. doi: 10.1016/j.cell.2007.10.047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Wendler P, et al. Motor mechanism for protein threading through Hsp104. Mol Cell. 2009;34:81–92. doi: 10.1016/j.molcel.2009.02.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Hanson PI, Whiteheart SW. AAA+ proteins: Have engine, will work. Nat Rev Mol Cell Biol. 2005;6:519–529. doi: 10.1038/nrm1684. [DOI] [PubMed] [Google Scholar]
- 12.Ogura T, Whiteheart SW, Wilkinson AJ. Conserved arginine residues implicated in ATP hydrolysis, nucleotide-sensing, and inter-subunit interactions in AAA and AAA+ ATPases. J Struct Biol. 2004;146:106–112. doi: 10.1016/j.jsb.2003.11.008. [DOI] [PubMed] [Google Scholar]
- 13.Sauer RT, et al. Sculpting the proteome with AAA(+) proteases and disassembly machines. Cell. 2004;119:9–18. doi: 10.1016/j.cell.2004.09.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Tessarz P, Mogk A, Bukau B. Substrate threading through the central pore of the Hsp104 chaperone as a common mechanism for protein disaggregation and prion propagation. Mol Microbiol. 2008;68:87–97. doi: 10.1111/j.1365-2958.2008.06135.x. [DOI] [PubMed] [Google Scholar]
- 15.Weibezahn J, et al. Thermotolerance requires refolding of aggregated proteins by substrate translocation through the central pore of ClpB. Cell. 2004;119:653–665. doi: 10.1016/j.cell.2004.11.027. [DOI] [PubMed] [Google Scholar]
- 16.Doyle SM, et al. Asymmetric deceleration of ClpB or Hsp104 ATPase activity unleashes protein-remodeling activity. Nat Struct Mol Biol. 2007;14:114–122. doi: 10.1038/nsmb1198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Weber-Ban EU, Reid BG, Miranker AD, Horwich AL. Global unfolding of a substrate protein by the Hsp100 chaperone ClpA. Nature. 1999;401:90–93. doi: 10.1038/43481. [DOI] [PubMed] [Google Scholar]
- 18.Mogk A, et al. Roles of individual domains and conserved motifs of the AAA+ chaperone ClpB in oligomerization, ATP hydrolysis, and chaperone activity. J Biol Chem. 2003;278:17615–17624. doi: 10.1074/jbc.M209686200. [DOI] [PubMed] [Google Scholar]
- 19.Weibezahn J, Schlieker C, Bukau B, Mogk A. Characterization of a trap mutant of the AAA+ chaperone ClpB. J Biol Chem. 2003;278:32608–32617. doi: 10.1074/jbc.M303653200. [DOI] [PubMed] [Google Scholar]
- 20.Werbeck ND, Schlee S, Reinstein J. Coupling and dynamics of subunits in the hexameric AAA+ chaperone ClpB. J Mol Biol. 2008;378:178–190. doi: 10.1016/j.jmb.2008.02.026. [DOI] [PubMed] [Google Scholar]
- 21.Haslberger T, et al. Protein disaggregation by the AAA+ chaperone ClpB involves partial threading of looped polypeptide segments. Nat Struct Mol Biol. 2008;15:641–650. doi: 10.1038/nsmb.1425. [DOI] [PubMed] [Google Scholar]
- 22.Wickner S, Hoskins J, McKenney K. Function of DnaJ and DnaK as chaperones in origin-specific DNA binding by RepA. Nature. 1991;350:165–167. doi: 10.1038/350165a0. [DOI] [PubMed] [Google Scholar]
- 23.Doyle SM, Hoskins JR, Wickner S. Collaboration between the ClpB AAA+ remodeling protein and the DnaK chaperone system. Proc Natl Acad Sci USA. 2007;104:11138–11144. doi: 10.1073/pnas.0703980104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Schlieker C, Tews I, Bukau B, Mogk A. Solubilization of aggregated proteins by ClpB/DnaK relies on the continuous extraction of unfolded polypeptides. FEBS Lett. 2004;578:351–356. doi: 10.1016/j.febslet.2004.11.051. [DOI] [PubMed] [Google Scholar]
- 25.Mogk A, et al. Refolding of substrates bound to small Hsps relies on a disaggregation reaction mediated most efficiently by ClpB/DnaK. J Biol Chem. 2003;278:31033–31042. doi: 10.1074/jbc.M303587200. [DOI] [PubMed] [Google Scholar]
- 26.Martin A, Baker TA, Sauer RT. Rebuilt AAA+ motors reveal operating principles for ATP-fuelled machines. Nature. 2005;437:1115–1120. doi: 10.1038/nature04031. [DOI] [PubMed] [Google Scholar]
- 27.Enemark EJ, Joshua-Tor L. On helicases and other motor proteins. Curr Opin Struct Biol. 2008;18:243–257. doi: 10.1016/j.sbi.2008.01.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Mezard C, Davies AA, Stasiak A, West SC. Biochemical properties of RuvBD113N: A mutation in helicase motif II of the RuvB hexamer affects DNA binding and ATPase activities. J Mol Biol. 1997;271:704–717. doi: 10.1006/jmbi.1997.1225. [DOI] [PubMed] [Google Scholar]
- 29.Hishida T, Iwasaki H, Han YW, Ohnishi T, Shinagawa H. Uncoupling of the ATPase activity from the branch migration activity of RuvAB protein complexes containing both wild-type and ATPase-defective RuvB proteins. Genes Cells. 2003;8:721–730. doi: 10.1046/j.1365-2443.2003.00670.x. [DOI] [PubMed] [Google Scholar]
- 30.Moreau MJ, McGeoch AT, Lowe AR, Itzhaki LS, Bell SD. ATPase site architecture and helicase mechanism of an archaeal MCM. Mol Cell. 2007;28:304–314. doi: 10.1016/j.molcel.2007.08.013. [DOI] [PubMed] [Google Scholar]
- 31.Crampton DJ, Mukherjee S, Richardson CC. DNA-induced switch from independent to sequential dTTP hydrolysis in the bacteriophage T7 DNA helicase. Mol Cell. 2006;21:165–174. doi: 10.1016/j.molcel.2005.11.027. [DOI] [PubMed] [Google Scholar]
- 32.Singleton MR, Sawaya MR, Ellenberger T, Wigley DB. Crystal structure of T7 gene 4 ring helicase indicates a mechanism for sequential hydrolysis of nucleotides. Cell. 2000;101:589–600. doi: 10.1016/s0092-8674(00)80871-5. [DOI] [PubMed] [Google Scholar]
- 33.Hoskins JR, Kim SY, Wickner S. Substrate recognition by the ClpA chaperone component of ClpAP protease. J Biol Chem. 2000;275:35361–35367. doi: 10.1074/jbc.M006288200. [DOI] [PubMed] [Google Scholar]
- 34.Hoskins JR, Yanagihara K, Mizuuchi K, Wickner S. ClpAP and ClpXP degrade proteins with tags located in the interior of the primary sequence. Proc Natl Acad Sci USA. 2002;99:11037–11042. doi: 10.1073/pnas.172378899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Zolkiewski M, Kessel M, Ginsburg A, Maurizi MR. Nucleotide-dependent oligomerization of ClpB from Escherichia coli. Protein Sci. 1999;8:1899–1903. doi: 10.1110/ps.8.9.1899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Skowyra D, Wickner S. The interplay of the GrpE heat shock protein and Mg2+ in RepA monomerization by DnaJ and DnaK. J Biol Chem. 1993;268:25296–25301. [PubMed] [Google Scholar]
- 37.Hoskins JR, Wickner S. Two peptide sequences can function cooperatively to facilitate binding and unfolding by ClpA and degradation by ClpAP. Proc Natl Acad Sci USA. 2006;103:909–914. doi: 10.1073/pnas.0509154103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Hoskins JR, Pak M, Maurizi MR, Wickner S. The role of the ClpA chaperone in proteolysis by ClpAP. Proc Natl Acad Sci USA. 1998;95:12135–12140. doi: 10.1073/pnas.95.21.12135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Diemand AV, Lupas AN. Modeling AAA+ ring complexes from monomeric structures. J Struct Biol. 2006;156:230–243. doi: 10.1016/j.jsb.2006.04.011. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.