Abstract
FGF21 is a hormone produced in liver and fat that dramatically improves peripheral insulin sensitivity and lipid metabolism. We show here that obese mice with genetically reduced levels of a key hepatic transcriptional coactivator, PGC-1α, have improved whole-body insulin sensitivity with increased levels of hepatic and circulating FGF21. Gain- and loss-of-function studies in primary mouse hepatocytes show that hepatic FGF21 levels are regulated by the expression of PGC-1α. Importantly, PGC-1α-mediated reduction of FGF21 expression is dependent on Rev-Erbα and the expression of ALAS-1. ALAS-1 is a PGC-1α target gene and the rate-limiting enzyme in the synthesis of heme, a ligand for Rev-Erbα. Modulation of intracellular heme levels mimics the effect of PGC-1α on FGF21 expression, and inhibition of heme biosynthesis completely abrogates the down-regulation of FGF21 in response to PGC-1α. Thus, PGC-1α can impact hepatic and systemic metabolism by regulating the levels of a nuclear receptor ligand.
Keywords: liver, metabolism, diabetes, coactivator, fasting
Maintenance of hepatic energy homeostasis is complex and under precise molecular control. Adaptive changes in enzyme expression levels are often controlled at the level of transcription by nuclear hormone receptors and other transcription factors (1). Members of the peroxisome proliferator activated receptor (PPAR)-gamma coactivator-1 (PGC-1s) family play an important role in the tight regulation of enzyme expression within the liver. Fasting induces PGC-1α expression, allowing this protein to coactivate several transcription factors including FOXO1, glucocorticoid receptor, nuclear respiratory factor-1 (NRF-1), hepatocyte nuclear factor-4α, retinoid-related orphan receptors (RORs), and PPARα. This leads to increased expression of key enzymes involved in gluconeogenesis, fatty acid oxidation, heme biosynthesis, and the circadian clock (2–4).
The importance of the PGC-1 coactivators in the maintenance of liver metabolism is illustrated in several mouse models. Mice with a tissue-specific loss of one allele of hepatic PGC-1α expression exhibit fasting-induced steatosis and develop hepatic insulin resistance on a high-fat diet (5). Hepatic PGC-1α levels are increased in mouse models of diabetes and obesity (6–9), and are inversely correlated with insulin resistance in humans (10). Although it is clear that the PGC-1s play a key role in regulating the hepatic response to nutritional cues, the molecular pathways are complex.
So far, PGC-1s have been shown only to act as potent positive regulators of transcription, because they promote local chromatin-remodeling events and formation of the preinitiation complex (11). PGC-1s recruit histone acetyltransferases (HAT)-containing protein complexes (through interactions with CBP/p300), and the TRAP/Mediator complex (by interacting with TRAP220/Med1) (12) in response to hormonal or physiological cues (reviewed in refs. 3 and 13). PGC-1α can associate with proteins that negatively affect its coactivator function (e.g., p160/Mib) (14, 15), but there is no evidence that PGC-1s can directly mediate transcriptional repression. On the contrary, PGC-1-containing complexes compete with corepressor binding to initiate transcription of inactive genes (16, 17).
In our current study, we identify PGC-1α as an important negative regulator of fibroblast growth factor-21 (FGF21) expression in the liver. FGF21, a member of the FGF family, is a hepatic hormone that potently regulates peripheral glucose tolerance, torpor, and hepatic lipid metabolism (18–20). The ability of FGF21 to protect against diet-induced obesity, improve insulin sensitivity, stimulate adipose tissue lipolysis, and lower triglyceride levels in diabetic rodents and monkeys makes it a very attractive candidate drug for the treatment of obesity and other metabolic diseases in humans (21–23). Here, we investigate the mechanism by which PGC-1α represses FGF21 gene expression, and suggest a mechanism by which reduction in hepatic PGC-1α expression increases whole-body insulin sensitivity and glucose tolerance. Our data uncover a negative-feedback loop linking PGC-1α-mediated induction of heme biosynthesis to the activity of the transcriptional corepressor Rev-Erbα. This pathway highlights the complexity of metabolic gene regulation and expands the role of Rev-Erbα as a PGC-1α target in hepatic metabolism.
Results
Genetically Reduced Hepatic PGC-1α Improves Whole-Body Glucose Homeostasis.
We have previously shown that chronically reducing levels of hepatic PGC-1α impairs fasting-induced fatty acid oxidation and causes insulin resistance in liver (5). As dysregulation of hepatic lipid metabolism and insulin sensitivity are major contributing factors to the pathogenesis of diabetes, nonalcoholic fatty liver disease (NAFLD), obesity, and atherosclerosis (24, 25), we asked whether chronic reductions in hepatic PGC-1α would contribute to or exacerbate metabolic disease. To test this hypothesis, we induced obesity and insulin resistance in the mice with genetic ablation of one allele of hepatic PGC-1α (liver heterozygous, LH mice) by feeding them a diet high in saturated fat and sucrose.
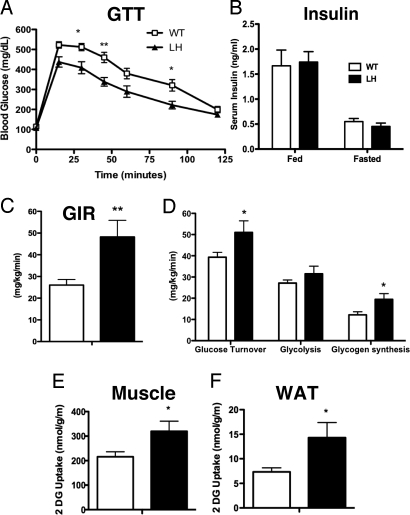
LH mice exhibited no differences in fed or fasted weight, fat mass, or lean mass after 1–4 months of high-fat feeding (Fig. S1 A–C). Paradoxically, when challenged with an i.p. injection of glucose, obese LH mice had significantly improved glucose tolerance compared to littermate, wild-type (WT) mice (Fig. 1A). The improvement in glucose utilization was only evident after high-fat feeding, as WT and LH mice were equally glucose tolerant on regular chow (Fig. S1D). Circulating insulin levels were similar in WT and LH mice (Fig. 1B), prompting us to assess peripheral insulin sensitivity by hyperinsulinemic-euglycemic clamp studies. Consistent with an increase in whole-body insulin sensitivity, the glucose infusion rate required to maintain euglycemia in high-fat fed LH mice was significantly higher than in WT controls (Fig. 1C). Moreover, LH mice had an approximate 30% increase in whole-body glucose turnover and an approximate 60% increase in glycogen synthesis (Fig. 1D). Increased 2-deoxy[14C]glucose uptake was evident in both muscle (Fig. 1E) and adipose (Fig. 1F) tissues from LH mice.
Fig. 1.
High-fat fed LH mice exhibit improved glucose tolerance and insulin sensitivity. (A) Blood glucose levels in 16-week high fat diet (HFD)-fed WT or LH mice after an overnight fast (time 0) and at the indicated times after i.p. injection of glucose. *, P < 0.05 by two-way ANOVA and Bonferroni posthoc tests at each point (n = 16). (B) Circulating insulin levels in high-fat fed or overnight fasted mice. Glucose infusion rate (GIR), (C) whole body glucose turnover, glycolysis, and glycogen synthesis (D), and glucose uptake in muscle (E) and fat (F) during hyperinsulinemic/euglycemic clamp. Values are means ± SEM, n = 7–11 (*, P < 0.05). White bars, WT mice; black bars, LH mice.
Hepatic PGC-1α Levels Impact FGF21 Expression.
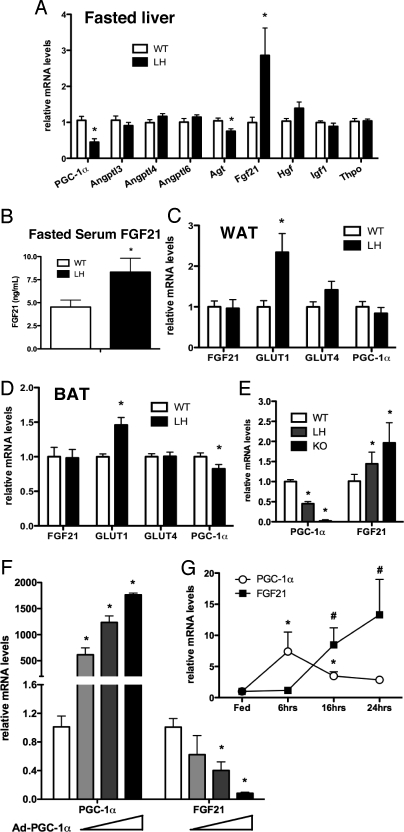
Considering the reduction in PGC-1α expression was limited to the liver, the change in peripheral insulin sensitivity was surprising in light of our earlier results demonstrating hepatic insulin resistance and increased triglyceride levels in LH mice (5). This suggests that hepatic PGC-1α regulates the expression or secretion of a molecule that modulates peripheral glucose metabolism. To identify candidate hormones, we quantified mRNA expression of a number of liver-secreted hormones known to affect peripheral insulin sensitivity or glucose metabolism. Both fasted and fed LH mice had ≈50% lower expression of PGC-1α compared to littermate controls (Fig. 2A and Fig. S1E). Strikingly, hepatic FGF21 mRNA levels were increased approximately 3-fold in LH mice fasted for 24 h compared to fasted controls (Fig. 2A). The mRNA levels of hepatic FGF21 were also increased in overnight (Fig. S2A) and 48-h fasted LH mice fed regular chow (Fig. S2B), demonstrating that the increased levels of FGF21 expression were not diet-dependent. There were no significant differences in FGF21 gene expression levels in fed WT versus LH mice (Fig. S2A).
Fig. 2.
PGC-1α negatively regulates hepatic FGF21 expression. (A) Hepatic mRNA levels in high-fat fed mice fasted for 24 h. (B) Circulating levels of FGF21 in 24-h fasted mice. Bars, means ± SEM. (n = 9), expressed relative to WT. mRNA expression in (C) epididymal WAT, (D) BAT (means ± SEM), and (E) primary hepatocytes from either WT, LH, or PGC-1α knockout mice (KO) (means ± SD). (F) mRNA expression in WT primary hepatocytes infected with increasing amounts of an adenovirus expressing PGC-1α (Ad-PGC-1α). Values are means ± SD of duplicates expressed relative to levels in cells infected with Ad-GFP. (G) Fasting time course of hepatic PGC-1α and FGF21 gene expression in wild-type mice. PGC-1α (*, P < 0.05) and FGF21 #, P < 0.05) mRNA levels were normalized to their corresponding controls (mice fed ad libitum) (mean ± SEM, n = 9).
Hepatic FGF21 expression is induced by long-term fasting or a ketogenic diet and administration of FGF21 to obese mice lowers blood glucose levels and increases insulin sensitivity in muscle, adipose, and heart (18–21). Consistent with the increase in hepatic mRNA, circulating levels of FGF21 were higher (Fig. 2B). Furthermore, the expression of Glut1, a glucose transporter regulated by FGF21 (20), was increased in the white (WAT) and brown adipose tissue (BAT) of LH mice (Fig. 2 C and D). Importantly, the mRNA levels of FGF21 in adipose tissue were similar between WT and LH mice, implicating the liver as the primary source of the increased FGF21. Increases in circulating FGF21 have little effect in lean mice (20, 21) and may explain why increases in glucose tolerance are only observed after high-fat feeding. Interestingly, the levels of PGC-1α were modestly decreased in BAT of high fat-fed LH mice (Fig. 2D). This decrease was diet-dependent, as PGC-1α levels in BAT of chow-fed LH mice were not different (Fig. S2C).
The dysregulation of FGF21 was cell autonomous, as primary hepatocytes isolated from either LH or PGC-1α KO knockout mice had increased basal FGF21 expression (Fig. 2E). Similarly, acute knock-down of endogenous PGC-1α in hepatocytes led to increased FGF21 expression (Fig. S2D). Furthermore, exogenous expression of PGC-1α in primary hepatocytes led to a dose-dependent decrease in FGF21 expression (Fig. 2F).
As both PGC-1α and FGF21 are induced during fasting, PGC-1α-mediated down-regulation of FGF21 seems paradoxical. A detailed time course of hepatic gene expression in fasting mice showed that expression levels of PGC-1α increased rapidly at the initiation of fasting and decreased with prolonged fasting (Fig. 2G). In contrast, the levels of FGF21 displayed an expression pattern opposite to that of PGC-1α, with significant increases in mRNA levels detected only after long-term fasting. Thus, the levels of hepatic FGF21 inversely correlated with the levels of PGC-1α in both cultured cells and in rodents.
Rev-Erbα Negatively Regulates FGF21 Expression.
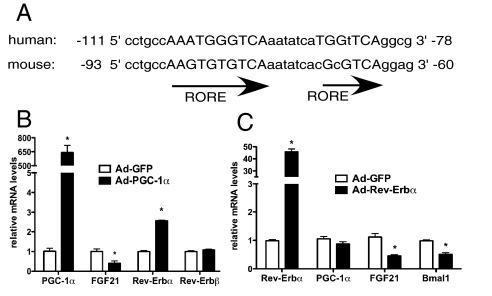
There is no evidence from either structure or function studies indicating that PGC-1α plays a direct role in transcriptional repression (26). However, there are several reports suggesting that increases in PGC-1α levels can negatively impact the expression of some genes (27, 28). Thus, we focus here on delineating the mechanism by which modulation of PGC-1α expression inversely affects FGF21 levels. In silico analysis of the mouse FGF21 promoter revealed a highly conserved RORE consensus half site between −87 and −77 and another putative RORE half site eight base pairs downstream between −69 and −63 (Fig. 3A). Rev-Erbα (NR1D1) is a transcriptional repressor which binds to RORE sites and recruits the nuclear receptor corepressor (NCoR)/histone deacetylase 3 (HDAC3) corepressor complex to potentiate down-regulation of gene expression (29, 30). PGC-1α increases Rev-Erbα/β expression in liver (4). Therefore, PGC-1α-mediated increases in Rev-Erbα and/or Rev-Erbβ expression may mediate the negative effects of PGC-1α on hepatic FGF21.
Fig. 3.
Rev-Erbα decreases FGF21 expression in primary hepatocytes. (A) Alignment of human and mouse FGF21 promoter sequences. Capitalized letters correspond to consensus RORE half sites. mRNA expression levels in primary hepatocytes expressing either (B) PGC-1α, (C) Rev-Erbα, or GFP (control). Values are mean ± SD of triplicates expressed relative to GFP, representative of three individual experiments, *, P < 0.05.
When PGC-1α was expressed in primary hepatocytes to levels that decreased FGF21 expression, we observed a 2-fold induction of Rev-Erbα, but not of Rev-Erbβ (Fig. 3B). To address whether Rev-Erbα regulates FGF21, primary hepatocytes were transduced with an adenovirus expressing FLAG-tagged Rev-Erbα. Indeed, increased Rev-Erbα led to a decrease in both FGF21 and Bmal1, a known Rev-Erbα target, with no significant change in PGC-1α expression (Fig. 3C). Furthermore, Rev-Erbα expression diminished RORα activation of the FGF21 promoter (Fig. S3), although the direct binding sites have yet to be determined.
FGF21 Expression Is Controlled by Intracellular Heme Concentrations.
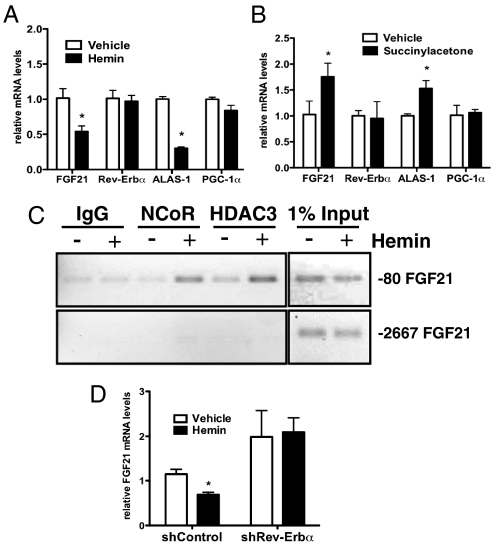
The repressive activity of Rev-Erbα is potentiated by binding of its ligand, heme, stimulating recruitment of the corepressors NCoR and HDAC3 (31, 32). This is of special interest for our study as PGC-1α is an important regulator of heme (33). Upon treatment of primary hepatocytes with hemin, which mimics heme (32), a significant decrease in FGF21 (Fig. 4A) was observed. Heme potently inhibits its own synthesis by decreasing the level of delta-aminolevulinate synthase-1 (ALAS-1), the rate-limiting enzyme in hepatic heme biosynthesis, as part of a negative feedback loop (34). Similarly, depletion of heme by succinylacetone (an irreversible inhibitor of aminolevulinic acid dehydratase) led to a significant increase in both FGF21 and ALAS-1 mRNA expression, confirming a role for endogenous heme in the control of FGF21 (Fig. 4B). Chromatin immunoprecipitation (ChIP) assays showed that heme treatment increased recruitment of NCoR and HDAC3 to the region of the FGF21 promoter containing the predicted RORE site, but not to a more upstream region (Fig. 4C). When Rev-Erbα levels were lowered by shRNA (Fig. S4A), hemin treatment failed to reduce FGF21 (Fig. 4D), consistent with Rev-Erbα playing a role in heme-mediated regulation of hepatic FGF21 expression. Transduction of shRev-Erbα in primary hepatocytes reduced endogenous Rev-Erbα mRNA expression by 50% and had no effect on the expression of the related isoform, Rev-Erbβ (Fig. S4B).
Fig. 4.
Heme negatively regulates FGF21 expression via Rev-Erbα in primary hepatocytes. Gene expression (A and C) or ChIP analysis of the FGF21 promoter (B) in primary hepatocytes treated with either vehicle (media), 30 μM hemin (A and B), or (C) succinylacetone. (D) FGF21 mRNA expression after hemin treatment of primary hepatocytes expressing either shControl or shRev-Erbα. Values are means ± SD of triplicate values, representative of three independent experiments, *, P < 0.05.
PGC-1α Regulates FGF21 Expression Through Modulation of Heme Levels.
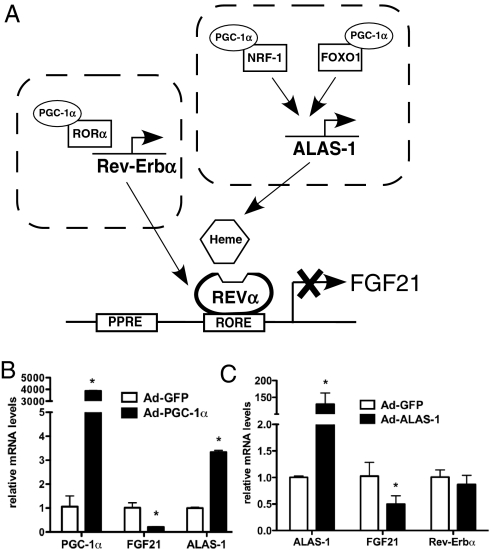
In liver cells, heme levels are tightly regulated. PGC-1α regulates heme levels in liver cells through coactivation of NRF-1 and FOXO1 to increase the expression of ALAS-1, the rate-limiting enzyme in heme biosynthesis (33). Thus, in addition to regulating Rev-Erbα expression, a PGC-1α-mediated increase in intracellular heme concentration may amplify the negative effect of PGC-1α on FGF21 expression (Fig. 5A). Indeed, when PGC-1α levels were increased in primary hepatocytes, the expression of ALAS-1 increased concurrent with a decrease in FGF21 (Fig. 5B). Consistent with a role for heme in the regulation of FGF21, overexpression of ALAS-1 in primary hepatocytes led to a significant decrease in FGF21 expression (Fig. 5C).
Fig. 5.
Increased ALAS-1 decreases FGF21 expression. (A) Schematic representation of the possible role of PGC-1α in the regulation of FGF21 via increased Rev-Erbα expression and/or heme biosynthesis. mRNA expression levels in primary hepatocytes expressing either (B) PGC-1α or (C) HA-ALAS-1. Values are mean ± SD of triplicate values expressed relative to control virus (GFP) and are representative of two independent experiments, *, P < 0.05.
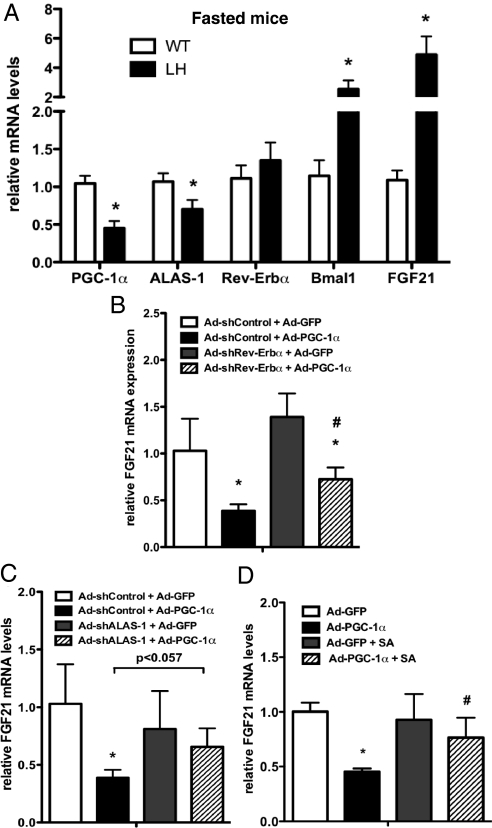
To determine the relative importance of Rev-Erbα and heme in PGC-1α-mediated down-regulation of FGF21, the expression levels of Rev-Erbα and ALAS-1 were examined in LH mice. Interestingly, although FGF21 mRNA levels were significantly higher in obese, fasted LH mice compared to controls, Rev-Erbα levels were similar in both genotypes (Fig. 6A). However, fasted ALAS-1 expression levels were significantly lower in LH mice compared to WT controls. Moreover, although Rev-Erbα levels were unaltered, Bmal1 expression was inappropriately high in LH fasted livers, consistent with a dysregulation in Rev-Erbα activity. It has been shown that the activity of Rev-Erbα as a transcriptional repressor is dependent on ligand/heme binding (31, 32). Our results suggest that rather than direct modulation of Rev-Erbα levels, the availability of heme as a Rev-Erbα ligand may play a more dominant role in PGC-1α-dependent repression of FGF21 expression.
Fig. 6.
Heme biosynthesis is required for PGC-1α-mediated repression of FGF21. (A) Hepatic mRNA levels in high-fat fed mice after a 24-h fast. Bars, means ± SEM (n = 9–11) expressed relative to WT fasted values. mRNA expression in primary hepatocytes coinfected with either Ad-GFP or Ad-PGC-1α, and Ad-shControl or (B) Ad-shRev-Erbα, or (C) Ad-ALAS-1, as indicated. Values are relative to control (Ad-GFP + Ad-shControl). (D) mRNA levels in primary hepatocytes expressing either PGC-1α or GFP and treated with either vehicle or succinylacetone. Values are means ± SD of triplicate values, representative of two independent experiments, *, P < 0.05 compared to Ad-GFP, #, P < 0.05 compared to Ad-PGC-1α.
To test this hypothesis, the ability of expressed PGC-1α to reduce FGF21 expression was examined when either Rev-Erbα or ALAS-1 levels were reduced by shRNA. Upon reduction of endogenous levels of Rev-Erbα in hepatocytes, we observed a trend toward increased basal FGF21 expression not reaching significance (Figs. 4D and 6B). In the presence of overexpressed PGC-1α, cells with decreased levels of Rev-Erbα levels had increased FGF21 expression compared to cells infected with the shControl virus (Fig. 6B). However, the levels of FGF21 were still reduced when compared to GFP infected controls, indicating that reducing Rev-Erbα alone was not sufficient to block the effect of PGC-1α. This may be because high levels of PGC-1α expression, the result of using virus titers sufficient to ensure coinfection of cells with both viruses, led to up-regulation of both Rev-Erbα and Rev-Erbβ mRNA (Fig. S4C). Rev-Erbβ also binds heme and acts as a repressor on RORE sites (32) and may compensate for the loss of Rev-Erbα.
Reducing ALAS-1 levels with shRNA (Fig. S5A) had no effect on basal FGF21 expression in primary hepatocytes (Fig. 6C). However, under these conditions exogenous PGC-1α expression failed to significantly reduce FGF21 gene expression, regardless of increased Rev-Erbα/β mRNA (Fig. S5B). To investigate whether this was in fact due to heme depletion and not to a nonspecific effect of ALAS-1 knockdown, we reduced levels of intracellular heme with succinylacetone. Similarly, the PGC-1α-mediated decrease in FGF21 was abrogated in cells treated with succinylacetone (Fig. 6D), confirming the importance of heme in the regulation of FGF21 by PGC-1α.
Discussion
The role of PGC-1α as a potent regulator of gene expression related to energy metabolism has been well documented. However, most studies detailing how PGC-1α affects transcription of target genes have been limited to its role as a direct activator of transcriptional programs (reviewed in refs. 2, 3, and 35). We now show that PGC-1α negatively modulates the expression of FGF21 in fasted liver, independent of direct coactivation, via regulated expression of a nuclear receptor ligand. We show in primary hepatocytes that PGC-1α negatively affects the expression of FGF21 by regulating expression of both Rev-Erbα and its ligand, heme. Furthermore, the data presented here show a direct dependence of FGF21 expression on the concentration of intracellular heme, expanding the current knowledge of how FGF21 expression is regulated in fasted liver.
PGC-1α is a key regulator of the hepatic fasting response (5, 9, 36–40). A major role of PGC-1α in the fasted liver is to promote expression of genes involved in lipid oxidation through coactivation of PPARα (41, 42). It was therefore very surprising that loss of PGC-1α in liver led to increased expression of FGF21, a hormone induced during fasting and starvation in a PPARα-dependent manner (18, 19). This data emphasizes a key distinction in the metabolic demands of liver during fasting versus starvation. During fasting, the liver increases its own energy expenditure to promote glycogenolysis, gluconeogenesis, and fatty acid oxidation, providing nutrients for peripheral tissues. As nutrient deprivation progresses, the body enters a more energy-sparing state and hepatic metabolism switches to the production of ketones. PGC-1α levels are highest in the early stages of fasting and gradually decrease as the length of food deprivation nears starvation (Fig. 2). This correlates well with the notion that positive PGC-1α function is essential during a physiological need to boost hepatic metabolism (26). FGF21 levels increase only after long-term fasting (Fig. 2 and refs. 18 and 19), when it is important for the regulation of adipose tissue catabolism and torpor, a state of decreased activity and metabolism. It was recently shown that FGF21 signals in an autocrine fashion to regulate hepatic carbohydrate and fatty acid metabolism and that some of its actions are PGC-1α-dependent (43). The mechanism of this regulation appears indirect, but illustrates an intimate and important relationship between these two molecules. Thus, the relationship between PGC-1α and FGF21 expression may be a fulcrum for the switch from a normal fasting state to a more profound starvation/energy-sparing state.
The regulation of heme levels further expands the hepatic PGC-1α-regulated gene set to include targets of the Rev-Erbα, a transcriptional repressor important for maintenance of the circadian clock, lipid metabolism, inflammation, and adipogenesis (29, 30). PGC-1α knockout mice have a disrupted circadian rhythm, and PGC-1α regulates circadian gene expression through coactivation of RORα (4). Our data suggests an alternative mechanism by which PGC-1α activity regulates the hepatic circadian clock. In addition to directly modulating the expression levels of transcription factors within the circadian pathway, PGC-1α amplifies the effect of Rev-Erbα by increasing heme levels in response to hormonal cues. Interestingly, hepatic PGC-1α, ALAS-1, and benzofibrate-induced FGF21 expression oscillate according to a circadian rhythm and FGF21 levels inversely correlate to the expression pattern of Rev-Erbα and ALAS-1 in liver (4, 44, 45). Furthermore, heme feeds back to inhibit PGC-1α expression through activation of Rev-Erbα (46). Heme also regulates aspects of the clock independent of Rev-Erbα, such as binding to neuronal PAS 2 (NPAS2) to impact expression of Bmal1 and Period (44, 47). Thus, the rhythmicity of PGC-1α and its target genes in liver is intimately correlated to the circadian clock.
There is mounting evidence that dysregulation of PGC-1α in many tissues leads to imbalances in energy metabolism and exacerbation of diseases including diabetes and obesity (5, 48–50). The data presented here clearly demonstrate that moderate changes in hepatic PGC-1α expression have effects on both the liver and extra-hepatic metabolic pathways to impact peripheral glucose homeostasis and insulin sensitivity. Uncovering the relationship linking PGC-1α and FGF21 provides insight into the cross-talk between liver and other metabolically active tissues and expands the role of this unique transcriptional coactivator in energy homeostasis.
Materials and Methods
Detailed methods are available in SI Text. Primer sequences are listed in Table S1.
Generation of Liver-Specific Heterozygous PGC-1α Mice.
Liver-specific heterozygous mice were generated as described in ref. 5. Male mice were fed a regular chow diet (5008I, PharmaServ) or a high fat diet (58% kcal fat, D12331, Research Diets Inc.), as indicated. All experiments were performed in accordance with the Animal Facility Institutional Animal Care and Use Committee regulations.
Hyperinsulemic/Euglycemic Clamp Studies.
Mice were fed a diet high in fat for 4 weeks before the clamp. Fat and lean body masses were assessed by 1H magnetic resonance spectroscopy (Bruker BioSpin). Hyperinsulemic/euglycemic clamp studies were performed as described in ref. 51.
Glucose Tolerance Test (GTT) and Serum Hormone Levels.
Blood glucose was measured in tail blood by using a standard glucometer. Serum insulin concentrations were determined by ELISA (Assay Core, Joslin Diabetes Center) and serum levels of FGF21 were determined by RIA (Phoenix Pharmaceuticals). For glucose tolerance tests, animals were fasted overnight before i.p. injection of 2 g/kg D-glucose. Glucose was measured in tail vein blood at intervals after glucose injection.
RNA Isolation and Quantitative RT-PCR.
RNA from frozen tissue or cultured cells was reverse transcribed and quantified with the Applied Biosystems Real-Time PCR System, Sybr-green PCR master mix, and the ΔΔct threshold cycle method. Gene expression levels were normalized to TBP mRNA and expressed relative to control levels.
Creation of cDNA Expression Plasmids, shRNA Constructs, and Adenoviruses.
The short hairpin sequences targeting mouse Rev-Erbα (NR1D1) and mouse ALAS-1 are: shControl: 5′-ACAACAGCCACAACGTCTATA-3′, shRev-Erbα: 5′-GCGCTTTGCATCGTTGTTCAA-3′, and shALAS-1: 5′-CCAAGATAGTAGCATTTGAAA-3′. The shPGC-1α adenovirus (5′- GGTGGATTGAAGTGGTGTAGA-3′) was a gift from Marc Montminy (La Jolla, CA). Adenoviruses for human FLAG-Rev-Erbα and HA-ALAS-1 were created by using the pAd-Track-CMV/Ad-Easy adenoviral vector system (Stratagene), according to manufacturer's instructions. Adenoviruses expressing GFP or PGC-1α were created as described in ref. 9. For shRNA adenoviruses, each short hairpin, preceded by the human U6 promoter, was inserted into pAd-Track vector lacking the CMV promoter.
Primary Hepatocyte Isolation.
Primary mouse hepatocytes from 10- to 12-week-old male mice were isolated by collagen perfusion and percoll gradient purification. Cells were seeded on collagen-coated plates and maintained in DMEM supplemented with 0.2% BSA, 4.5 g/L glucose, 2 mM sodium pyruvate, 0.1 μM dexamethasone, and 1 nM insulin (maintenance media).
Cell Culture and Treatment.
To access basal expression, primary hepatocytes were harvested 48 h after plating. For overexpression and knock-down studies, cells were transduced 1 day after isolation with the indicated adenovirus diluted in maintenance media. Eight to 16 hours post-infection, cells were incubated in media lacking insulin and dexamethasone for an additional 24 h. A 1 mM stock solution of hemin (Sigma) was prepared fresh in 0.1 M NaOH (pH adjusted to 8). Cells were treated for 24 h with 30 μM hemin to increase intracellular heme or 5 mM succinylacetone (SA) (Sigma) to inhibit heme biosynthesis. For inhibition of PGC-1α-induced heme biosynthesis, SA was added to media 16 h post-infection and cells were incubated for an additional 32 h.
Protein Isolation and Western Blot Analysis.
Protein samples, solubilized in RIPA buffer, were resolved by SDS/PAGE, blotted, and incubated with antibodies toward Rev-Erbα (Cell Signaling), HA (Roche), or Hsp90 (Cell Signaling).
ChIP Assay.
Primary hepatocytes were infected as described. Cells were treated with 30 μM hemin for 3 h before fixation with 1% formaldehyde for 30 min. ChIP assays were performed as described in ref. 52. Immunoprecipitation was preformed with antibodies against NCoR (Affinity Bioreagents), HDAC3 (Abcam), or IgG (as a negative control).
Statistical Analysis.
Statistical significance (*, P < 0.05) was assessed by ANOVA or Student's t test by using Prism 5.0 software (GraphPad Software).
Supplementary Material
Acknowledgments.
We thank Dr. Marc Montminy for the shPGC-1α adenovirus, Dr. Mitch Lazar and Nan Wu for the human FLAG-Rev-Erbα construct, and Drs. Folliott Martin Fisher and Srikripa Devarakonda for helpful discussion. This work was supported by the H.L. Holmes Award (National Research Council of Canada) and a postdoctoral fellowship from the Canadian Institutes of Health Research (J.L.E.), and National Institutes of Health Grants R01 DK-061562 (to B.M.S.), and R01 DK-40936, P30 DK-45735, and U24 DK-59635 (to G.I.S.) support the work in this manuscript.
Footnotes
The authors declare no conflict of interest.
This article contains supporting information online at www.pnas.org/cgi/content/full/0912533106/DCSupplemental.
References
- 1.Staudinger JL, Lichti K. Cell signaling and nuclear receptors: New opportunities for molecular pharmaceuticals in liver disease. Mol Pharm. 2008;5:17–34. doi: 10.1021/mp700098c. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Finck BN, Kelly DP. PGC-1 coactivators: Inducible regulators of energy metabolism in health and disease. J Clin Invest. 2006;116:615–622. doi: 10.1172/JCI27794. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Lin J, Handschin C, Spiegelman BM. Metabolic control through the PGC-1 family of transcription coactivators. Cell Metab. 2005;1:361–370. doi: 10.1016/j.cmet.2005.05.004. [DOI] [PubMed] [Google Scholar]
- 4.Liu C, Li S, Liu T, Borjigin J, Lin JD. Transcriptional coactivator PGC-1α integrates the mammalian clock and energy metabolism. Nature. 2007;447:477–481. doi: 10.1038/nature05767. [DOI] [PubMed] [Google Scholar]
- 5.Estall JL, et al. Sensitivity of lipid metabolism and insulin signaling to genetic alterations in hepatic PGC-1α expression. Diabetes. 2009 doi: 10.2337/db08-1571. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Koo SH, et al. PGC-1 promotes insulin resistance in liver through PPAR-α-dependent induction of TRB-3. Nat Med. 2004;10:530–534. doi: 10.1038/nm1044. [DOI] [PubMed] [Google Scholar]
- 7.Biddinger SB, et al. Hepatic insulin resistance is sufficient to produce dyslipidemia and susceptibility to atherosclerosis. Cell Metab. 2008;7:125–134. doi: 10.1016/j.cmet.2007.11.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Biddinger SB, et al. Effects of diet and genetic background on sterol regulatory element-binding protein-1c, stearoyl-CoA desaturase 1, and the development of the metabolic syndrome. Diabetes. 2005;54:1314–1323. doi: 10.2337/diabetes.54.5.1314. [DOI] [PubMed] [Google Scholar]
- 9.Yoon JC, et al. Control of hepatic gluconeogenesis through the transcriptional coactivator PGC-1. Nature. 2001;413:131–138. doi: 10.1038/35093050. [DOI] [PubMed] [Google Scholar]
- 10.Croce MA, et al. Hepatic lipin 1beta expression is diminished in insulin-resistant obese subjects and is reactivated by marked weight loss. Diabetes. 2007;56:2395–2399. doi: 10.2337/db07-0480. [DOI] [PubMed] [Google Scholar]
- 11.Puigserver P, et al. Activation of PPARgamma coactivator-1 through transcription factor docking. Science. 1999;286:1368–1371. doi: 10.1126/science.286.5443.1368. [DOI] [PubMed] [Google Scholar]
- 12.Wallberg AE, Yamamura S, Malik S, Spiegelman BM, Roeder RG. Coordination of p300-mediated chromatin remodeling and TRAP/mediator function through coactivator PGC-1α. Mol Cell. 2003;12:1137–1149. doi: 10.1016/s1097-2765(03)00391-5. [DOI] [PubMed] [Google Scholar]
- 13.Feige JN, Auwerx J. Transcriptional coregulators in the control of energy homeostasis. Trends Cell Biol. 2007;17:292–301. doi: 10.1016/j.tcb.2007.04.001. [DOI] [PubMed] [Google Scholar]
- 14.Knutti D, Kressler D, Kralli A. Regulation of the transcriptional coactivator PGC-1 via MAPK-sensitive interaction with a repressor. Proc Natl Acad Sci USA. 2001;98:9713–9718. doi: 10.1073/pnas.171184698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Fan M, et al. Suppression of mitochondrial respiration through recruitment of p160 myb binding protein to PGC-1α: Modulation by p38 MAPK. Genes Dev. 2004;18:278–289. doi: 10.1101/gad.1152204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Borgius LJ, Steffensen KR, Gustafsson JA, Treuter E. Glucocorticoid signaling is perturbed by the atypical orphan receptor and corepressor SHP. J Biol Chem. 2002;277:49761–49766. doi: 10.1074/jbc.M205641200. [DOI] [PubMed] [Google Scholar]
- 17.Guan HP, Ishizuka T, Chui PC, Lehrke M, Lazar MA. Corepressors selectively control the transcriptional activity of PPARγ in adipocytes. Genes Dev. 2005;19:453–461. doi: 10.1101/gad.1263305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Badman MK, et al. Hepatic fibroblast growth factor 21 is regulated by PPARα and is a key mediator of hepatic lipid metabolism in ketotic states. Cell Metab. 2007;5:426–437. doi: 10.1016/j.cmet.2007.05.002. [DOI] [PubMed] [Google Scholar]
- 19.Inagaki T, et al. Endocrine regulation of the fasting response by PPARα-mediated induction of fibroblast growth factor 21. Cell Metab. 2007;5:415–425. doi: 10.1016/j.cmet.2007.05.003. [DOI] [PubMed] [Google Scholar]
- 20.Kharitonenkov A, et al. FGF-21 as a novel metabolic regulator. J Clin Invest. 2005;115:1627–1635. doi: 10.1172/JCI23606. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Xu J, et al. Fibroblast growth factor 21 reverses hepatic steatosis, increases energy expenditure, and improves insulin sensitivity in diet-induced obese mice. Diabetes. 2009;58:250–259. doi: 10.2337/db08-0392. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Kharitonenkov A, Shanafelt AB. FGF21: A novel prospect for the treatment of metabolic diseases. Curr Opin Investig Drugs. 2009;10:359–364. [PubMed] [Google Scholar]
- 23.Chavez AO, et al. Circulating fibroblast growth factor-21 (FGF-21) is elevated in impaired glucose tolerance and type 2 diabetes and correlates with muscle and hepatic insulin resistance. Diabetes Care. 2009;32:1542–1546. doi: 10.2337/dc09-0684. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Watanabe S, Yaginuma R, Ikejima K, Miyazaki A. Liver diseases and metabolic syndrome. J Gastroenterol. 2008;43:509–518. doi: 10.1007/s00535-008-2193-6. [DOI] [PubMed] [Google Scholar]
- 25.Haas JT, Biddinger SB. Dissecting the role of insulin resistance in the metabolic syndrome. Curr Opin Lipidol. 2009;20:206–210. doi: 10.1097/MOL.0b013e32832b2024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Spiegelman BM, Heinrich R. Biological control through regulated transcriptional coactivators. Cell. 2004;119:157–167. doi: 10.1016/j.cell.2004.09.037. [DOI] [PubMed] [Google Scholar]
- 27.Zhang Y, Castellani LW, Sinal CJ, Gonzalez FJ, Edwards PA. Peroxisome proliferator-activated receptor-gamma coactivator 1α (PGC-1α) regulates triglyceride metabolism by activation of the nuclear receptor FXR. Genes Dev. 2004;18:157–169. doi: 10.1101/gad.1138104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Jeong JH, Cho S, Pak YK. Sterol-independent repression of low density lipoprotein receptor promoter by peroxisome proliferator activated receptor gamma coactivator-1α (PGC-1α) Exp Mol Med. 2009;41:406–416. doi: 10.3858/emm.2009.41.6.046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Preitner N, et al. The orphan nuclear receptor REV-ERBα controls circadian transcription within the positive limb of the mammalian circadian oscillator. Cell. 2002;110:251–260. doi: 10.1016/s0092-8674(02)00825-5. [DOI] [PubMed] [Google Scholar]
- 30.Yin L, Lazar MA. The orphan nuclear receptor Rev-erbα recruits the N-CoR/histone deacetylase 3 corepressor to regulate the circadian Bmal1 gene. Mol Endocrinol. 2005;19:1452–1459. doi: 10.1210/me.2005-0057. [DOI] [PubMed] [Google Scholar]
- 31.Yin L, et al. Rev-erbα, a heme sensor that coordinates metabolic and circadian pathways. Science. 2007;318:1786–1789. doi: 10.1126/science.1150179. [DOI] [PubMed] [Google Scholar]
- 32.Raghuram S, et al. Identification of heme as the ligand for the orphan nuclear receptors REV-ERBα and REV-ERBβ. Nat Struct Mol Biol. 2007;14:1207–1213. doi: 10.1038/nsmb1344. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Handschin C, et al. Nutritional regulation of hepatic heme biosynthesis and porphyria through PGC-1α. Cell. 2005;122:505–515. doi: 10.1016/j.cell.2005.06.040. [DOI] [PubMed] [Google Scholar]
- 34.Zheng J, Shan Y, Lambrecht RW, Donohue SE, Bonkovsky HL. Differential regulation of human ALAS1 mRNA and protein levels by heme and cobalt protoporphyrin. Mol Cell Biochem. 2008;319:153–161. doi: 10.1007/s11010-008-9888-0. [DOI] [PubMed] [Google Scholar]
- 35.Handschin C, Spiegelman BM. Peroxisome proliferator-activated receptor gamma coactivator 1 coactivators, energy homeostasis, and metabolism. Endocr Rev. 2006;27:728–735. doi: 10.1210/er.2006-0037. [DOI] [PubMed] [Google Scholar]
- 36.Herzig S, et al. CREB regulates hepatic gluconeogenesis through the coactivator PGC-1. Nature. 2001;413:179–183. doi: 10.1038/35093131. [DOI] [PubMed] [Google Scholar]
- 37.Puigserver P, et al. Insulin-regulated hepatic gluconeogenesis through FOXO1-PGC-1α interaction. Nature. 2003;423:550–555. doi: 10.1038/nature01667. [DOI] [PubMed] [Google Scholar]
- 38.Rhee J, et al. Regulation of hepatic fasting response by PPARγ coactivator-1α (PGC-1): Requirement for hepatocyte nuclear factor 4α in gluconeogenesis. Proc Natl Acad Sci USA. 2003;100:4012–4017. doi: 10.1073/pnas.0730870100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Burgess SC, et al. Diminished hepatic gluconeogenesis via defects in tricarboxylic acid cycle flux in peroxisome proliferator-activated receptor gamma coactivator-1α (PGC-1α)-deficient mice. J Biol Chem. 2006;281:19000–19008. doi: 10.1074/jbc.M600050200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Estall JL, Arany Z. Regulation of hepatic metabolism by PGC-1 transcriptional coactivators. Cellscience. 2007;4:225–236. [Google Scholar]
- 41.Vega RB, Huss JM, Kelly DP. The coactivator PGC-1 cooperates with peroxisome proliferator-activated receptor alpha in transcriptional control of nuclear genes encoding mitochondrial fatty acid oxidation enzymes. Mol Cell Biol. 2000;20:1868–1876. doi: 10.1128/mcb.20.5.1868-1876.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Finck BN, et al. Lipin 1 is an inducible amplifier of the hepatic PGC-1α/PPARα regulatory pathway. Cell Metab. 2006;4:199–210. doi: 10.1016/j.cmet.2006.08.005. [DOI] [PubMed] [Google Scholar]
- 43.Potthoff MJ, et al. FGF21 induces PGC-1α and regulates carbohydrate and fatty acid metabolism during the adaptive starvation response. Proc Natl Acad Sci USA. 2009;106:10853–10858. doi: 10.1073/pnas.0904187106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Kaasik K, Lee CC. Reciprocal regulation of haem biosynthesis and the circadian clock in mammals. Nature. 2004;430:467–471. doi: 10.1038/nature02724. [DOI] [PubMed] [Google Scholar]
- 45.Oishi K, Uchida D, Ishida N. Circadian expression of FGF21 is induced by PPARα activation in the mouse liver. FEBS Lett. 2008;582:3639–3642. doi: 10.1016/j.febslet.2008.09.046. [DOI] [PubMed] [Google Scholar]
- 46.Wu N, Yin L, Hanniman EA, Joshi S, Lazar MA. Negative feedback maintenance of heme homeostasis by its receptor, Rev-erbα. Genes Dev. 2009;23:2201–2209. doi: 10.1101/gad.1825809. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Dioum EM, et al. NPAS2: A gas-responsive transcription factor. Science. 2002;298:2385–2387. doi: 10.1126/science.1078456. [DOI] [PubMed] [Google Scholar]
- 48.Patti ME, et al. Coordinated reduction of genes of oxidative metabolism in humans with insulin resistance and diabetes: Potential role of PGC1 and NRF1. Proc Natl Acad Sci USA. 2003;100:8466–8471. doi: 10.1073/pnas.1032913100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Pihlajamaki J, et al. Haplotypes of PPARGC1A are associated with glucose tolerance, body mass index and insulin sensitivity in offspring of patients with type 2 diabetes. Diabetologia. 2005;48:1331–1334. doi: 10.1007/s00125-005-1800-9. [DOI] [PubMed] [Google Scholar]
- 50.Handschin C, et al. Abnormal glucose homeostasis in skeletal muscle-specific PGC-1α knockout mice reveals skeletal muscle-pancreatic beta cell crosstalk. J Clin Invest. 2007;117:3463–3474. doi: 10.1172/JCI31785. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Choi CS, et al. Continuous fat oxidation in acetyl-CoA carboxylase 2 knockout mice increases total energy expenditure, reduces fat mass, and improves insulin sensitivity. Proc Natl Acad Sci USA. 2007;104:16480–16485. doi: 10.1073/pnas.0706794104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Holmquist-Mengelbier L, et al. Recruitment of HIF-1α and HIF-2α to common target genes is differentially regulated in neuroblastoma: HIF-2α promotes an aggressive phenotype. Cancer Cell. 2006;10:413–423. doi: 10.1016/j.ccr.2006.08.026. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.