Abstract
The transition from initiation to elongation of the RNA polymerase (RNAP) is an important stage of transcription that often limits the production of the full-length RNA. Little is known about the RNAP transition kinetics and the steps that dictate the transition rate, because of the challenge in monitoring subpopulations of the transient and heterogeneous transcribing complexes in rapid and real time. Here, we have dissected the complete transcription initiation pathway of T7 RNAP by using kinetic modeling of RNA synthesis and by determining the initiation (IC) to elongation (EC) transition kinetics at each RNA polymerization step using single-molecule and stopped-flow FRET methods. We show that the conversion of IC to EC in T7 RNAP consensus promoter occurs only after 8- to 12-nt synthesis, and the 12-nt synthesis represents a critical juncture in the transcriptional initiation pathway when EC formation is most efficient. We show that the slow steps of transcription initiation, including DNA scrunching/RNAP–promoter rotational changes during 5- to 8-nt synthesis, not the major conformational changes, dictate the overall rate of EC formation in T7 RNAP and represent key steps that regulate the synthesis of full-length RNA.
Keywords: abortive synthesis, FRET, rate-limiting, T7 RNA polymerase, transcription transition
The recruitment of RNA polymerase (RNAP) to the promoter and isomerization to a competent open complex are important regulatory steps in gene transcription (1, 2). Similarly, the transition from abortive initiation to processive elongation is an essential event of transcription that often dictates the rate at which the full-length RNA is made (3–9). During the abortive initiation phase, the RNAP maintains contacts with the promoter and catalyzes RNA polymerization by scrunching the template DNA (10–13). After a certain length of RNA is made, the RNAP switches transcription from promoter-specific to promoter-independent and processive RNA synthesis. Single-subunit and multisubunit RNAPs switch from the initiation complex (IC) to the elongation complex (EC) by undergoing specific structural rearrangements. For example, the single-subunit T7 RNAP protein undergoes major refolding of its N-terminal domain during EC transition that releases promoter contacts and results in the formation of RNA channel (14, 15). The bacterial RNAP makes this transition by releasing the sigma factor from the promoter (7, 16–19), and in eukaryotic RNAP II, this event is marked by the removal of several transcription factors, including TFIIB, TFIIE, TFIIF, and TFIIH (20). Although the initiation stage and the transition from IC to EC are major points of regulation that ultimately control gene expression, the rate-limiting steps governing these multistep processes and the kinetics of transition have not been elucidated.
The challenge in studying the kinetic pathway of initiation and the IC-to-EC transition has been the transient and heterogeneous nature of the initially transcribing complexes. Studies of the transient heterogeneous intermediates necessitate the development of rapid and real-time techniques that can monitor the relevant subpopulations in the time period of RNA synthesis. Although numerous crystal structures of the T7 RNAP complex at various intermediate steps have provided exquisite structural details (13–15, 21, 22), several more of such snapshots will be required to decipher the complete transcription pathway.
To study the kinetics of IC-to-EC transition, we developed single-molecule and stopped-flow FRET assays that monitored the bending and unbending conformational changes in the promoter DNA as the transcription reaction proceeded from initiation to elongation. Structural studies of T7 RNAP have shown that the promoter DNA is highly bent in IC with 2- to 7-nt transcript, whereas the DNA is significantly less bent in the EC (13–15). By placing fluorescent donor and acceptor dyes at critical positions within the promoter DNA (Fig. 1), we were able to structurally resolve the IC and EC species of T7 RNAP by their FRET signatures and quantify the rates of EC formation at each RNA polymerization step. By determining the distributions of IC and EC as a function of reaction time, we measure the kinetics of EC formation in T7 RNAP and show that transition occurs between 8- and 12-nt RNA synthesis. We show that the stage immediately after 12-nt synthesis represents a critical juncture in the transcriptional initiation pathway when EC formation is most efficient. Comparison of the RNA polymerization rates and the EC formation rates reveals that the overall formation of EC is rate-limited by the slow steps of 2-nt synthesis and DNA scrunching/RNAP–promoter rotational conformational changes. Thus, our studies reveal that DNA scrunching during initiation that triggers EC formation in RNAPs represents a key stage that regulates the rate of full-length RNA synthesis.
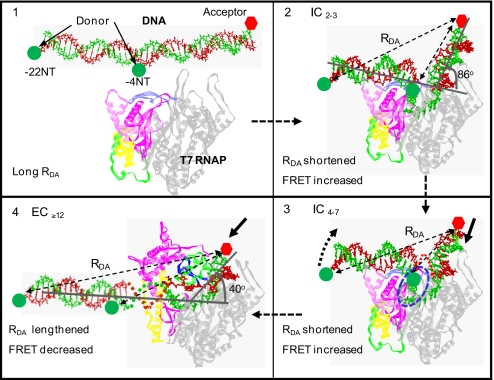
Fig. 1.
Conformational changes in the promoter DNA during the initiation and the transition from initiation to elongation. The distance between the donor and the acceptor in the free promoter DNA is relatively large (A) and is shortened in the initiation complexes IC2–4 because of sharp bending (B). It is further shortened as IC2–4 changes to IC4–7 because of DNA bending, scrunching (in blue dotted region), and rotation (dotted curved arrow) changes (C). The conversion of IC to EC significantly reduces DNA bending that increases the D–A distance (D).
Results and Discussion
Single-Molecule FRET Monitors Initiation and Elongation Complexes.
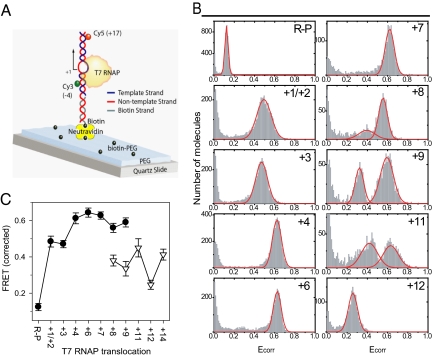
The consensus T7 promoter carrying a single pair of FRET fluorophores at −4 and +17 positions (with respect to the transcription start site at +1) was immobilized to a quartz slide via biotin–neutravidin link (Fig. 2A). After forming the DNA–T7 RNAP complex, an incomplete set of NTPs or NTP plus 3′ dNTP was added to initiate transcription and to halt RNA polymerization at selected positions. The FRET values of the halted transcriptional complexes were determined by analyzing single molecules (23, 24). The single-molecule donor (ID) and acceptor (IA) fluorescence intensities were collected after 1 min of reaction start and were corrected for leakage from donor and acceptor channels and from changes in quantum yields of the donor by using Eq. 1, and the corrected FRETs (Ecorr) were plotted both as histograms (Fig. 2B) and as peak FRET values at various RNAP translocation positions (Fig. 2C).
Fig. 2.
T7 RNAP transcription measured by single-molecule FRET. (A) Promoter DNA with Cy3 (donor) on the −4NT and Cy5 (acceptor) on +17T were surface-immobilized by using avidin-biotin linkage. Quartz surface was precoated with PEG (mixed with biotin-PEG) to achieve specific biotin conjugation and to prevent nonspecific interaction with the glass surface. (B) FRET histograms show Ecorr on the x axis and counts of transcribing molecules on the y axis as T7 RNAP transcription was halted at designated positions by using limiting NTPs with or without 3′ dNTP. (C) Peak FRET efficiencies of R–P and halted transcribing complexes at various positions in IC (filled circles) and EC (open triangles) are shown (SEM came from more than two replicate measurements).
The T7 RNAP–promoter binary complex (R–P) has a relatively low FRET efficiency (Ecorr = 0.12), which indicates a large D–A distance and insignificant promoter DNA bending in R–P (25). Upon adding either 3′ dGTP (a nonpolymerizing GTP analog) or GTP, the RNAP translocates to +1/+2 or +3 when the average FRET efficiency increases to ≈0.45 (Fig. 2 B and C), which is consistent with a shorter D–A distance in these initially transcribing open complexes due to sharp ≈90° bend in the DNA (15, 25). A single population of complexes from +2 to +7 (i.e., making 2- or 7-nt maximum-length RNA) with progressively increasing peak Ecorr was observed (Fig. 2B). The presence of a single population indicates that the halted complexes are predominantly in the “forward translocated” state; i.e., bound to a complete RNA transcript for that position (26), which is consistent with the slow and limiting rate of RNA dissociation from these initiation complexes (27). The progressively increasing FRET value is consistent with progressively increasing DNA bending and scrunching in these initially transcribing complexes (12).
Beyond +7, the FRET histograms showed two peaks: a new low-FRET population with Ecorr ≈ 0.3, along with the high-FRET population with Ecorr ≈ 0.6 (Fig. 2B). The low-FRET population progressively increased in magnitude from +8 to +11, whereas the high-FRET population decreased. By +12 and at further positions, only a single population at the low-FRET value was observed. The new low-FRET species appeared after 8-nt synthesis when the transition from IC to EC was observed from other studies of T7 RNAP (28–31). Therefore, the low-FRET population was assigned as the EC state. The lower FRET value of EC compared with the IC is consistent with the promoter DNA being less bent in EC (13, 15).
Rates of IC-to-EC Transition from Single-Molecule FRET.
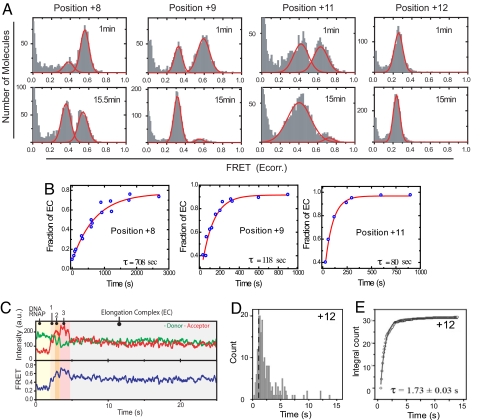
Being able to resolve the IC and EC allowed us to measure the kinetics of EC formation after each RNA polymerization step. Complexes halted at +7 or before displayed stable high FRET for up to 30 min after addition of saturated NTPs, indicating no detectable EC formation at or before +7. On the other hand, complexes stalled to generate 8-, 9-, and 11-nt-long RNA displayed a time-dependent transition from the high- to the low-FRET value population (Fig. 3A and Fig. S1). The kinetics of high-to-low FRET change were fit to an exponential equation to determine the mean lifetimes (τ) of EC formation. The mean τ of EC formation (or the rate constant of EC formation, 1/τ) was ≈12 min at +8 (rate constant ≈0.0014 s−1), ≈2 min at +9 (≈0.0083 s−1), and ≈1.3 min at +11 (0.013 s−1), and it was <1 min at +12 (Fig. 3B).
Fig. 3.
Kinetics of IC-to-EC transition measured by single-molecule FRET. (A) Distribution of FRET values of the halted complexes 1 min and 15 min after reaction start. Data for +8, +9, and +11 were fitted to two Gaussian distributions (smoothed curves), and data for +12 were fitted to a single distribution to quantify IC and EC fractions. (B) The fraction of EC increases with single-exponential kinetics, with the time of reaction providing the indicated mean τ. (C) Single-molecule FRET time trajectory of transcription reaction halted at +12 measured by using the DNA SM-NT2. (Upper) Time course of donor and acceptor intensities. (Lower) The apparent FRET, Eapp, calculated without the γ correction factor in Eq. 1. (D) Dwell times in the high-FRET state (State 3 in C) were determined from >200 molecules. (E) Integrated frequency counts from the selected portion of the dwell time (right half of the dashed line in D) fit well to a single-exponential decay function to yield a mean τ of 1.7 s for EC formation at +12.
The relatively fast rate of EC formation after 12-nt synthesis was monitored more accurately by analyzing the real-time FRET trajectories of transcription reactions under single-molecule conditions. In the example trace shown in Fig. 3C, soon after adding NTPs to R–P, the apparent FRET value increased from ≈0.3 to ≈0.5 (state 1) and to ≈0.6 (state 2) until it reached a maximal value of ≈0.7 (state 3). After spending a certain amount of time in state 3, the apparent FRET at ≈0.7 dropped to ≈0.45 abruptly, and it remained such until photobleaching of the acceptor after ≈30 s. We propose that the sudden decrease in FRET, which is faster than the time resolution of the measurement, represents the unbending of the promoter due to EC formation. Based on the apparent FRET values of the halted complexes in Fig. S2, we assigned state 1 as IC2/3; state 2 as IC4/5; and state 3 as IC5/6 to 8/12. The mean τ of state 3, therefore, sets the lower limit to how fast EC formation can occur after 12-nt RNA synthesis (Fig. 3D). The dwell time distribution of state 3 was measured, and the slow phase was fit to a single exponential decay function to yield a mean τ of 1.7 s, or the EC formation rate constant of 0.6 s−1 (Fig. 3 D and E). Similar values of mean τ were obtained for transcription halted at +14 (τ = 2.2 s) or +15 (τ = 2.3 s; Fig. S3). The results indicate that although the switch from IC to EC starts at +8, the observed rate constant of EC formation is slow during 8- to 11-nt synthesis and increases by ≈400-fold when the RNA reaches a length of 12 nt.
Kinetics of IC-to-EC Transition by Stopped-Flow FRET Measurements.
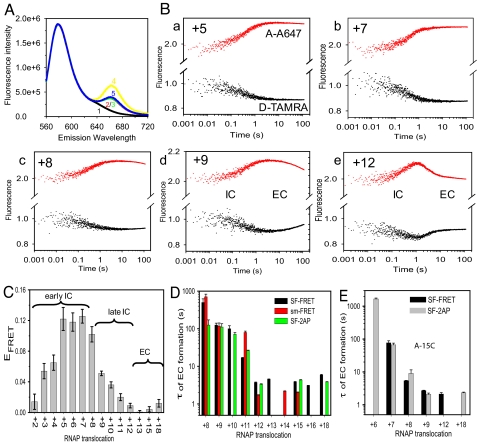
The IC-to-EC transition kinetics were measured under ensemble reaction conditions by using the stopped-flow method. The consensus T7 promoter was labeled with the donor–acceptor (D–A) fluorophore pair at opposing ends of the DNA on −22 nontemplate (NT) and +18 template (T), respectively, to report on global changes in the promoter structure in actively transcribing DNA–RNAP complexes (Fig. 1). Free DNA or DNA–RNAP has little FRET because of the large end-to-end distance of ≈140 Å in the linear 40-bp DNA (Fig. 4A). The transcriptional complex halted at +6 showed a FRET value of ≈0.15, indicating a significantly shortened D–A distance of ≈86 Å. At +15, the FRET value decreased, indicating movement of the DNA ends away from each other.
Fig. 4.
T7 RNAP transcription measured by stopped-flow FRET. (A) Fluorescence emission spectra after excitation at 550 nm for the free DNA seq1 (100 nM) singly labeled with TAMRA at −22NT (curve 1) or doubly labeled with TAMRA (at −22NT) and A647 (at +18T) (curve 2), DNA in the binary complex with 150 nM T7 RNAP (curve 3), DNA in +6 halted complex (with 1 mM GTP plus 0.4 M ATP, curve 4), or +15 halted complex (with 1 mM GTP plus 0.4 mM ATP plus 0.4 mM CTP, curve 5). Donor fluorescence was normalized to that of D-only free DNA to display the sensitized acceptor fluorescence. (B) Representative real-time traces of donor (black) and acceptor (red) fluorescence intensity changes monitoring DNA conformational changes during transcription by T7 RNAP (a–e). (C) FRET values of complexes halted at various positions after 120 s of reaction start. (D) Mean τ values of EC formation from stopped-flow experiments (black bars; see B and Fig. S4), single-molecule experiments (red bars; see Fig. 3B), or 2AP fluorescence changes (green bars; see Fig. S6). Error bars show SD based on at least three independent measurements. (E) Mean τ values of EC formation on the A-15C promoter from stopped-flow FRET (black bars; see Fig. S7) and 2AP fluorescence (gray bars).
In the stopped-flow experiments, R–P was mixed with limiting set of NTPs or NTPs plus 3′ dNTP mixture, and donor and acceptor fluorescence changes were measured in real time. At the start, in all of the reactions, a rapid increase in FRET—i.e., donor fluorescence decrease and corresponding increase in acceptor fluorescence—was observed (Fig. 4B and Fig. S4). This early-phase FRET increase is consistent with DNA bending during initiation. Starting at +8 (Fig. 4Bc), a time-dependent decrease in FRET was observed after 1 s of initiating the reactions. This late-phase FRET decrease due to EC formation became more prominent after 9-nt synthesis (Fig. 4Bd), and most efficient in terms of rate and magnitude after 12-nt synthesis (Fig. 4Be).
The stopped-flow kinetic traces were fit to Eqs. 2–5 (SI Text) to determine the rate and magnitude of FRET changes during ongoing transcription and after reactions were halted at various positions. The rate constant of the early-phase FRET increase observed in all of the reactions is consistent with the measured rate constant of transcription initiation (Fig. S5). The early-phase FRET amplitude, however, was low in the RNAP–DNA complex, but it increased progressively to ≈0.13 at +5 (Fig. S5), consistent with progressive bending of the DNA during initiation. FRET remained at the high value even after 2 min in the +5 to +7 reactions, but starting at +8, it showed a time-dependent decrease, reaching its minimum value in +12 (Fig. 4C). The kinetics of the late-phase FRET decrease were fit to an exponential function to obtain the following EC formation rate constants: ≈1 × 10−3 s−1 at +8 (τ = 16 min), ≈0.01 s−1 at +9 (τ = 1.7 min) and +10, ≈0.05 s−1 at +11 (τ = 20 s), and ≈0.3 s−1 at ≥+12 (τ = 3 s). Thus, the mean τ of IC-to-EC transition from single molecule analysis, using internally labeled DNA constructs (Fig. 4D, red bars), and from the ensemble methods, using terminally labeled DNA constructs (Fig. 4D, black bars), are in excellent agreement. These results indicate that the observed rate constant of EC formation increases progressively as the transcript RNA length increases from 8 nt to 12 nt. There is a ≈10-fold increase in rate from 8 to 9, ≈5-fold increase from 9 to 10, ≈7-fold increase from 11 to 12, and a cumulative 300- to 600-fold increase in the transition rate as the RNA extends from 8 to 12 nt.
During the IC-to-EC transition, the initially opened transcription bubble from −4 to +2 of the promoter DNA recloses to regenerate the duplex region (initial bubble collapse) (28–30). The kinetics of initial bubble reannealing were monitored by following the decrease in the fluorescence of 2-aminopurine (2AP) placed at −4NT (28). The 2AP fluorescence decrease (Fig. 4D, green bars) occurred at the same rate constants as the FRET decrease (Fig. S6). Based on these results, we conclude that there is tight coupling between global and local DNA conformational changes during the IC-to-EC transition (25, 32).
To determine whether changes in the promoter sequence alter the efficiency and/or the location of the IC-to-EC transition, the stopped-flow FRET studies were carried out with an altered T7 promoter with base pair mutation A-15C in the upstream promoter region (28, 33). The late-phase FRET decrease indicative of the IC-to-EC transition was detectable in the A-15C promoter as early as 7-nt synthesis and was most efficient after 9-nt synthesis (Fig. 4E and Fig. S7) in contrast to after 12 nt in the consensus promoter. The observed rate of EC formation in the A-15C promoter, however, was not faster than the consensus promoter (Fig. 4E). Similar observations of promoter release and bubble reannealing at earlier locations have been reported with the C-9A mutant promoter (28). These results indicate that alterations in the promoter sequence (such as A-15C) can change the location of the IC-to-EC transition (8–9 nt in A-15C vs. 12 nt in consensus promoter), but in this case without affecting the overall rate of IC-to-EC transition (Fig. 4 D and E).
Kinetics of RNA Synthesis.
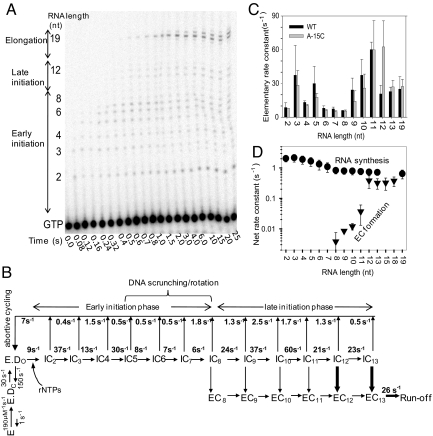
The kinetics analysis of IC-to-EC transition provides the macroscopic rate constant of EC formation that includes the steps of abortive RNA synthesis up to the maximum length of RNA made in the reaction. To determine whether the step/s of initiation or the transition itself dictates the observed rate of EC formation, we dissected the initiation pathway and determined the elementary rate constants of each RNA synthesis and RNA dissociation steps and compared them to the observed rate constant of EC formation. The time course of RNA elongation starting from the individual NTPs was measured by using rapid-mixing methods with the consensus T7 promoter. T7 RNAP undergoes abortive cycling both during the early initiation phase (2–8 nt) and the late initiation phase (9 to 12/13; Fig. 5A). After RNAs of lengths 12–13 nt are made, transcription becomes processive until 19-nt runoff formation. The individual RNA products from 2 nt and 19 nt made as a function of time were quantified, and their formation, decay, and accumulation were globally fit to the transcription model (Fig. 5B and Fig. S8). In the transcription model, we assumed that promoter binding and opening are fast steps relative to the 2-nt synthesis step (25, 27, 32, 34, 35). The fittings provided the elementary rate constants of RNA synthesis and RNA dissociation (Fig. 5C), which revealed that the 2-nt RNA was made with a rate constant of ≈8 s−1, which is consistent with previous reports (27, 34). Interestingly, RNA synthesis rate constants increased from ≈8 s−1 for the 2GTP→2-nt step to ≈30 s−1 for 4→5. But during the stage when DNA scrunching and RNAP–promoter rotation conformational changes occur in T7 RNAP from 5–8 nt (12), RNA synthesis steps are slow between 6 and 8 s−1. Curiously, after 8-nt, the RNA synthesis rate constant increased suddenly to ≈24 s−1 for the 8→9 step and remained high between 20 and 60 s−1 for the rest of the steps until runoff synthesis (Fig. 5C).
Fig. 5.
Kinetics of RNA synthesis. (A) Sequencing gel shows the time course of RNA synthesis (2- to 19-nt runoff) on the consensus T7 promoter (DNA sequence P-22 from refs. 8 and 44). (B) The complete transcription initiation pathway of T7 RNAP with the experimentally determined rate constants for each of the elementary steps. The rate constants for RNAP binding to the T7 consensus promoter and open complex formation were obtained from previous studies (32, 35). The rate constants of RNA synthesis and RNA dissociation were derived from global fit of the RNA-elongation kinetics in A to the model in B (Fig. S8). Efficient EC formation from IC on 12/13-nt RNA is shown in bold arrows. (C) Elementary rate constants of 2- to 19-nt RNA synthesis steps for the consensus promoter (black bars) and the A-15C promoter (gray bars). Error bars represent the range of data from two or more measurements. (D) Net rate constants of making RNA of 2- to 19-nt lengths (circles) and EC formation (triangles) at those translocation steps on the consensus promoter. The net rate constants of EC formation were obtained from the data in Fig. 4D. The plotted values are means ± SE of measurements of three independent measurements.
The sudden increase in RNA polymerization rate after 8-nt synthesis coincides with the stage when the specificity loop loses its interactions with the promoter (28, 36). Loss-of-specificity loop interactions could increase the rate of RNA synthesis by removing the barrier for the rotational conformational change, facilitating further elongation of the RNA from 8 nt until EC formation. Thus, the dissection of the initiation pathway of transcription shows that DNA scrunching and RNAP–promoter rotation steps from 5- to 8-nt RNA synthesis are the bottleneck during transcription initiation, responsible for the rate-limiting synthesis of 9- to 12-nt RNAs. The kinetic pathway of A-15C promoter was similarly dissected (Fig. S8A), which showed that even in this altered promoter, the RNA synthesis steps during DNA scrunching and RNAP–promoter rotation stages are the slowest.
To compare the kinetics of RNA synthesis to EC formation, we calculated the net rate constants of RNA synthesis. The synthesis of 2- to 19-nt RNA occurs by a multistep process, and therefore a net rate constant calculated by using Eq. 2 indicates how fast a particular length of RNA is made. The net rate constant of making a 2-nt RNA is ≈2 s−1, but it decreases progressively as the RNA gets longer, reaching a value of 0.8 s−1 for the 8 nt and remaining at that value until runoff synthesis (Fig. 5D). These net rate constants of RNA synthesis were compared to the EC formation rate constants at each RNA synthesis step to determine what limits EC formation. The 8-nt RNA is made with a net rate constant of 0.8 s−1, whereas the observed rate constant of EC formation at this position is 0.004 s−1 (Figs. 3B and 4D). Thus, EC formation at +8 is 200-fold slower than making 8-nt RNA. Similar comparison shows that 9- to 11-nt RNAs are made faster than EC formation at those positions. Hence, EC formation, even though observed between 8 and 11 nt synthesis, is relatively slow and inefficient. On the other hand, EC is formed at +12 (0.3 to 0.6 s−1) as fast as the 12-nt RNA is made (0.7 s−1; Fig. 5D). These results indicate that IC12 readily converts to EC12, whereas IC8–11 species do not.
The net rate constants of RNA synthesis on the A-15C promoter were comparable to the consensus promoter (Fig. S8B). Although EC formation was observed earlier in the A-15C promoter, the rate constant of EC formation at +7 (≈0.01 s−1) or +8 (≈0.15 s−1) was slower than that of 7- or 8-nt synthesis. Only at +9 did the EC formation rate constant (0.4–0.5 s−1) becomes comparable to that of RNA synthesis (Fig. S8B). The results indicate that during active transcription, EC formation primarily occurs at +12 (on the consensus promoter) and at +9 (on the A-15C promoter). Examination of the initiation pathway reveals that EC formation is rate-limited by the slow steps of DNA scrunching and RNAP–promoter rotational steps that bring about the synthesis of 8-nt RNA. We also noted that the overall synthesis rate constant of 12-nt RNA was essentially the same as that of the runoff product, and this implies that the drastic conformational changes associated with transition to EC must be faster than the RNA synthesis rate, or these changes are progressively realized such that they are not rate-limiting.
The conversion from IC to EC is a critical as well as a complex process for all RNAPs. Surprisingly, in T7 RNAP, it is not the rate-limiting step for full-length RNA synthesis. Instead, our studies indicate that full-length RNA production is limited by the efficiency of early-phase initiation and escape from frequent abortive release before the RNA transcript enters the EC-formation phase. Triggering by DNA scrunching has been suggested as the major mechanism behind transition to EC in various model RNAPs (11, 12, 37). We show here that DNA scrunching and rotation of the promoter-interacting domains and the bound upstream promoter also dictate the observed rate of elongation complex formation in T7 RNAP. These steps would serve as prime targets for differential regulation of transcription and gene expression. Indeed, T7 lysozyme, the negative regulator of T7 transcription, targets the steps of initiation rather than elongation in a promoter strength-dependent manner (38–41).
Materials and Methods
Protein and Oligonucleotides.
T7 RNAP was purified as described previously (42, 43). Oligodeoxynucleotides (Integrated DNA Technologies) (Table S1) were gel-purified and labeled with the fluorophores via an aminohexyl linker (C6) as described previously (44).
Single-Molecule FRET Measurements.
The experiments were carried out by using a wide-filed total internal reflection (TIR) microscopy setup (12) and DNA constructs (SM-NT1, SM-NT2, and a tethering B strand; Table S1), with Cy3 and Cy5 being labeled at −4NT and +17T, respectively. The FRET efficiency, Ecorr, was calculated from apparent donor and acceptor signals (ID, IA) by using Eq. 1,
where γ is the ratio of change in average acceptor intensity (ΔIA) to change in average donor intensity (ΔID) before and after acceptor photobleaching (23) (for details, see SI Text).
Stopped-Flow FRET Measurements.
Real-time simultaneous measurements of donor and acceptor fluorescence intensities were carried out on a T-scheme KinTek stopped-flow instrument (Model 2001) (32) (for details, see SI Text).
Transient-State Kinetics of RNA Polymerization and Data Analysis.
The kinetics of de novo RNA synthesis were measured at 25 °C by using a rapid chemical-quench flow instrument equipped with a temperature-controlled water bath (KinTek) according to established procedures (44). The time-dependent formation and decay of each RNA product was fit to the transcription model (Fig. 5B) by using the KinTek Explorer program (KinTek) (45).
To determine the net rate of i-nt RNA synthesis (i ≥ 2), we summed all ≥i-nt products and fit the kinetics to an incomplete gamma function by using a MATLAB-based (Mathworks) global analysis program (gfit; http://gfit.sourceforge.net/) (Eq. 2).
where Y is the amount of i-nt RNA products summed from all ≥i-nt bands, A is the amplitude of each phase from the fitting, k is the stepping rate, t is reaction time, and n represents the number of steps required by the RNAP to synthesize the RNA product since adding the first nucleotide. The averaged overall rate constant (k = 1/τ) for the polymerase to synthesize i-nt RNA is expressed by 1/τ = k/n.
Supplementary Material
Acknowledgments.
This work was supported by National Institutes of Health Grants GM51966 (to S.S.P.) and GM065367 (to T.H.).
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
This article contains supporting information online at www.pnas.org/cgi/content/full/0906979106/DCSupplemental.
References
- 1.McClure WR. Rate-limiting steps in RNA chain initiation. Proc Natl Acad Sci USA. 1980;77:5634–5638. doi: 10.1073/pnas.77.10.5634. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Ptashne M, Gann A. Transcriptional activation by recruitment. Nature. 1997;386:569–577. doi: 10.1038/386569a0. [DOI] [PubMed] [Google Scholar]
- 3.Carpousis AJ, Gralla JD. Cycling of ribonucleic acid polymerase to produce oligonucleotides during initiation in vitro at the lac UV5 promoter. Biochemistry. 1980;19:3245–3253. doi: 10.1021/bi00555a023. [DOI] [PubMed] [Google Scholar]
- 4.Rougvie AE, Lis JT. The RNA polymerase II molecule at the 5′ end of the uninduced hsp70 gene of D. melanogaster is transcriptionally engaged. Cell. 1988;54:795–804. doi: 10.1016/s0092-8674(88)91087-2. [DOI] [PubMed] [Google Scholar]
- 5.Panov KI, Friedrich JK, Zomerdijk JC. A step subsequent to preinitiation complex assembly at the ribosomal RNA gene promoter is rate limiting for human RNA polymerase I-dependent transcription. Mol Cell Biol. 2001;21:2641–2649. doi: 10.1128/MCB.21.8.2641-2649.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Hsu LM. Promoter clearance and escape in prokaryotes. Biochim Biophys Acta. 2002;1577:191–207. doi: 10.1016/s0167-4781(02)00452-9. [DOI] [PubMed] [Google Scholar]
- 7.Reppas NB, Wade JT, Church GM, Struhl K. The transition between transcriptional initiation and elongation in E. coli is highly variable and often rate limiting. Mol Cell. 2006;24:747–757. doi: 10.1016/j.molcel.2006.10.030. [DOI] [PubMed] [Google Scholar]
- 8.Muse GW, et al. RNA polymerase is poised for activation across the genome. Nat Genet. 2007;39:1507–1511. doi: 10.1038/ng.2007.21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Hatoum A, Roberts J. Prevalence of RNA polymerase stalling at Escherichia coli promoters after open complex formation. Mol Microbiol. 2008;68:17–28. doi: 10.1111/j.1365-2958.2008.06138.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Ikeda RA, Richardson CC. Interactions of the RNA polymerase of bacteriophage T7 with its promoter during binding and initiation of transcription. Proc Natl Acad Sci USA. 1986;83:3614–3618. doi: 10.1073/pnas.83.11.3614. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Kapanidis AN, et al. Initial transcription by RNA polymerase proceeds through a DNA-scrunching mechanism. Science. 2006;314:1144–1147. doi: 10.1126/science.1131399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Tang GQ, Roy R, Ha T, Patel SS. Transcription initiation in a single-subunit RNA polymerase proceeds through DNA scrunching and rotation of the N-terminal subdomains. Mol Cell. 2008;30:567–577. doi: 10.1016/j.molcel.2008.04.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Durniak KJ, Bailey S, Steitz TA. The structure of a transcribing t7 RNA polymerase in transition from initiation to elongation. Science. 2008;322:553–557. doi: 10.1126/science.1163433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Tahirov TH, et al. Structure of a T7 RNA polymerase elongation complex at 2.9 A resolution. Nature. 2002;420:43–50. doi: 10.1038/nature01129. [DOI] [PubMed] [Google Scholar]
- 15.Yin YW, Steitz TA. Structural basis for the transition from initiation to elongation transcription in T7 RNA polymerase. Science. 2002;298:1387–1395. doi: 10.1126/science.1077464. [DOI] [PubMed] [Google Scholar]
- 16.Travers AA, Burgess RR. Cyclic re-use of the RNA polymerase sigma factor. Nature. 1969;222:537–540. doi: 10.1038/222537a0. [DOI] [PubMed] [Google Scholar]
- 17.Hansen UM, McClure WR. Role of the sigma subunit of Escherichia coli RNA polymerase in initiation. II. Release of sigma from ternary complexes. J Biol Chem. 1980;255:9564–9570. [PubMed] [Google Scholar]
- 18.Gill SC, Weitzel SE, von Hippel PH. Escherichia coli sigma 70 and NusA proteins. I. Binding interactions with core RNA polymerase in solution and within the transcription complex. J Mol Biol. 1991;220:307–324. doi: 10.1016/0022-2836(91)90015-x. [DOI] [PubMed] [Google Scholar]
- 19.Mooney RA, Darst SA, Landick R. Sigma and RNA polymerase: An on-again, off-again relationship? Mol Cell. 2005;20:335–345. doi: 10.1016/j.molcel.2005.10.015. [DOI] [PubMed] [Google Scholar]
- 20.Zawel L, Kumar KP, Reinberg D. Recycling of the general transcription factors during RNA polymerase II transcription. Genes Dev. 1995;9:1479–1490. doi: 10.1101/gad.9.12.1479. [DOI] [PubMed] [Google Scholar]
- 21.Cheetham GM, Steitz TA. Structure of a transcribing T7 RNA polymerase initiation complex. Science. 1999;286:2305–2309. doi: 10.1126/science.286.5448.2305. [DOI] [PubMed] [Google Scholar]
- 22.Cheetham GM, Jeruzalmi D, Steitz TA. Structural basis for initiation of transcription from an RNA polymerase-promoter complex. Nature. 1999;399:80–83. doi: 10.1038/19999. [DOI] [PubMed] [Google Scholar]
- 23.Ha T, et al. Single-molecule fluorescence spectroscopy of enzyme conformational dynamics and cleavage mechanism. Proc Natl Acad Sci USA. 1999;96:893–898. doi: 10.1073/pnas.96.3.893. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Roy R, Hohng S, Ha T. A practical guide to single-molecule FRET. Nat Methods. 2008;5:507–516. doi: 10.1038/nmeth.1208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Tang GQ, Patel SS. T7 RNA polymerase-induced bending of promoter DNA is coupled to DNA opening. Biochemistry. 2006;45:4936–4946. doi: 10.1021/bi0522910. [DOI] [PubMed] [Google Scholar]
- 26.Margeat E, et al. Direct observation of abortive initiation and promoter escape within single immobilized transcription complexes. Biophys J. 2006;90:1419–1431. doi: 10.1529/biophysj.105.069252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Jia Y, Patel SS. Kinetic mechanism of GTP binding and RNA synthesis during transcription initiation by bacteriophage T7 RNA polymerase. J Biol Chem. 1997;272:30147–30153. doi: 10.1074/jbc.272.48.30147. [DOI] [PubMed] [Google Scholar]
- 28.Bandwar RP, Tang GQ, Patel SS. Sequential release of promoter contacts during transcription initiation to elongation transition. J Mol Biol. 2006;360:466–483. doi: 10.1016/j.jmb.2006.05.029. [DOI] [PubMed] [Google Scholar]
- 29.Gong P, Esposito EA, Martin CT. Initial bubble collapse plays a key role in the transition to elongation in T7 RNA polymerase. J Biol Chem. 2004;279:44277–44285. doi: 10.1074/jbc.M409118200. [DOI] [PubMed] [Google Scholar]
- 30.Liu C, Martin CT. Promoter clearance by T7 RNA polymerase. Initial bubble collapse and transcript dissociation monitored by base analog fluorescence. J Biol Chem. 2002;277:2725–2731. doi: 10.1074/jbc.M108856200. [DOI] [PubMed] [Google Scholar]
- 31.Guo Q, Nayak D, Brieba LG, Sousa R. Major conformational changes during T7RNAP transcription initiation coincide with, and are required for, promoter release. J Mol Biol. 2005;353:256–270. doi: 10.1016/j.jmb.2005.08.016. [DOI] [PubMed] [Google Scholar]
- 32.Tang GQ, Patel SS. Rapid binding of T7 RNA polymerase is followed by simultaneous bending and opening of the promoter DNA. Biochemistry. 2006;45:4947–4956. doi: 10.1021/bi052292s. [DOI] [PubMed] [Google Scholar]
- 33.Guo Q, Sousa R. Weakening of the T7 promoter-polymerase interaction facilitates promoter release. J Biol Chem. 2005;280:14956–14961. doi: 10.1074/jbc.M500518200. [DOI] [PubMed] [Google Scholar]
- 34.Jia Y, Patel SS. Kinetic mechanism of transcription initiation by bacteriophage T7 RNA polymerase. Biochemistry. 1997;36:4223–4232. doi: 10.1021/bi9630467. [DOI] [PubMed] [Google Scholar]
- 35.Stano NM, Levin MK, Patel SS. The +2 NTP binding drives open complex formation in T7 RNA polymerase. J Biol Chem. 2002;277:37292–37300. doi: 10.1074/jbc.M201600200. [DOI] [PubMed] [Google Scholar]
- 36.Place C, Oddos J, Buc H, McAllister WT, Buckle M. Studies of contacts between T7 RNA polymerase and its promoter reveal features in common with multisubunit RNA polymerases. Biochemistry. 1999;38:4948–4957. doi: 10.1021/bi982689e. [DOI] [PubMed] [Google Scholar]
- 37.Brieba LG, Sousa R. T7 promoter release mediated by DNA scrunching. EMBO J. 2001;20:6826–6835. doi: 10.1093/emboj/20.23.6826. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Moffatt BA, Studier FW. T7 lysozyme inhibits transcription by T7 RNA polymerase. Cell. 1987;49:221–227. doi: 10.1016/0092-8674(87)90563-0. [DOI] [PubMed] [Google Scholar]
- 39.Zhang X, Studier FW. Mechanism of inhibition of bacteriophage T7 RNA polymerase by T7 lysozyme. J Mol Biol. 1997;269:10–27. doi: 10.1006/jmbi.1997.1016. [DOI] [PubMed] [Google Scholar]
- 40.Huang J, Villemain J, Padilla R, Sousa R. Mechanisms by which T7 lysozyme specifically regulates T7 RNA polymerase during different phases of transcription. J Mol Biol. 1999;293:457–475. doi: 10.1006/jmbi.1999.3135. [DOI] [PubMed] [Google Scholar]
- 41.Stano NM, Patel SS. T7 lysozyme represses T7 RNA polymerase transcription by destabilizing the open complex during initiation. J Biol Chem. 2004;279:16136–16143. doi: 10.1074/jbc.M400139200. [DOI] [PubMed] [Google Scholar]
- 42.Davanloo P, Rosenberg AH, Dunn JJ, Studier FW. Cloning and expression of the gene for bacteriophage T7 RNA polymerase. Proc Natl Acad Sci USA. 1984;81:2035–2039. doi: 10.1073/pnas.81.7.2035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Jia Y, Kumar A, Patel SS. Equilibrium and stopped-flow kinetic studies of interaction between T7 RNA polymerase and its promoters measured by protein and 2-aminopurine fluorescence changes. J Biol Chem. 1996;271:30451–30458. doi: 10.1074/jbc.271.48.30451. [DOI] [PubMed] [Google Scholar]
- 44.Tang GQ, Bandwar RP, Patel SS. Extended upstream A-T sequence increases T7 promoter strength. J Biol Chem. 2005;280:40707–40713. doi: 10.1074/jbc.M508013200. [DOI] [PubMed] [Google Scholar]
- 45.Johnson KA, Simpson ZB, Blom T. Global Kinetic Explorer: A new computer program for dynamic simulation and fitting of kinetic data. Anal Biochem. 2009;387:20–29. doi: 10.1016/j.ab.2008.12.024. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.