Abstract
Recent experiments showed that the linear double-stranded DNA in bacteriophage capsids is both highly knotted and neatly structured. What is the physical basis of this organization? Here we show evidence from stochastic simulation techniques that suggests that a key element is the tendency of contacting DNA strands to order, as in cholesteric liquid crystals. This interaction favors their preferential juxtaposition at a small twist angle, thus promoting an approximately nematic (and apolar) local order. The ordering effect dramatically impacts the geometry and topology of DNA inside phages. Accounting for this local potential allows us to reproduce the main experimental data on DNA organization in phages, including the cryo-EM observations and detailed features of the spectrum of DNA knots formed inside viral capsids. The DNA knots we observe are strongly delocalized and, intriguingly, this is shown not to interfere with genome ejection out of the phage.
Keywords: DNA packaging, Monte Carlo simulations
How is genomic DNA organized? This simple yet formidable question has not yet been fully answered, not even for bacteriophages, viruses of bacteria which span only a few tens of nanometers in size (1). Thanks to experimental advancements in single-molecule manipulation and imaging techniques (2), we now understand in remarkable detail the process of DNA loading inside the phage capsid, and its subsequent ejection (3–7). Nevertheless, the problem of DNA organization and storage inside the capsid (i.e., after completion of the loading process and before ejection) has proved much more elusive because of the experimental and computational difficulties in probing the dense state of viral DNA molecules, which are several μm long and are packaged inside capsids having their diameter comparable with the DNA persistence length (50 nm).
Here we show that several experimentally accessible features of viral genome packaging can be reproduced and understood via a suitable mesoscopic model of double-stranded DNA which can be studied by computer simulations. In addition to accounting for the well-known bending rigidity and thickness of the DNA, our model incorporates a crucial phenomenological aspect, the fact that contacting DNA segments are chiral and meet with a preferred twist angle. To the best of our knowledge, this interaction has not been previously considered in models for DNA organization inside bacteriophages, although it is known to be ultimately responsible for the appearance of cholesteric phases in dense suspensions of DNA segments (8–11).
There are two main kinds of experimental assays for genome organization in bacteriophages, one probing the geometry, the other the topology of the confined genome. Both give important terms of reference for any study of dsDNA packaging.
On one hand, advanced cryo-EM techniques that do not rely on a priori symmetry assumptions have revealed that close to the capsid internal wall, the DNA of ɛ15 and ϕ29 is arranged in ordered concentric shells (12, 13). An increasingly disordered DNA arrangement, possibly due to the loss of experimental resolution, was instead observed when moving away from the capsid internal surface.
The second type of assay was carried out on specific mutants of the P2 and P4 bacteriophages, whose cohesive DNA ends could meet and anneal inside the capsid. Once the ends anneal, their interaction is so stable that the topology of the molecules is further experimental manipulation (14–16). After extraction from the capsid, these DNAs were subjected to electrophoretic migration in a gel to infer their topological knot type (17–20). The analysis of the knot spectrum proved to be extremely important in other biological contexts (21–24). This analysis also provided several important indications about genome organization within phages. First, the large majority of the molecules were found to be knotted (16, 19). Second, the knot complexity was high because knots with minimal crossing numbers ranging up to 40 were reported (19). Finally, the population of the simplest types of knots was found to be strongly biased towards torus knots and against achiral ones (20). In particular, the populations of the achiral knot 41 and of the twist knot 52 were much smaller than those of the torus knots 31 and 51.
The observed knot spectrum provides an important, if indirect, indication of DNA arrangement, as it cannot be explained by simple models for semiflexible polymers subject to spatial confinement. For example, the extremely high (95%) knotting probability is at odds with predictions from DNA-packaging models where ordered and mostly unknotted spools are obtained as a result of minimization of the bending energy (25). Accounting for thermal disorder in semiflexible polymers confined in a spherical cavity produces, on the other hand, highly entangled configurations (26). Although the resulting disorder reflects in a high fraction of knots and high knot complexity, it fails to account for the observed bias among the simplest types of knots (27, 28). At present, the latter only appears to be qualitatively reproduced in a mildly confined ring polymer sampled with a preferential bias for configurations with high writhe (20). In this regard, it is important to observe that the writhe, which is widely used to characterize open or closed chains (29–31), provides a quantitative measure of global geometric properties. This observation leads to the question of what plausible local DNA interactions are responsible for the observed knot spectrum (possibly, but not necessarily, also resulting in the writhe bias).
Results and Discussion
Our programme in this paper is to set up a mesoscopic model able to explain these two major experimental observations: the DNA surface ordering and the DNA knot spectrum. Like previous work (see, e.g., refs. 20, 26, 28, and 32–35), we build on the notion that viral capsids are permeable to ions in solution (36), which effectively screen electrostatic interactions inside the capsid.
To capture these essential polymer physics considerations, we model dsDNA as a semiflexible chain of beads having a hard core diameter equal to 2.5 nm. The chain bending rigidity is introduced based on the known value (50 nm) of the DNA persistence length (37). The DNA self-interaction associated to steric hindrance and self-repulsion is accounted for by a truncated Lennard–Jones potential and a Debye–Hueckel potential, with the Debye screening length set to ≈0.9 nm, corresponding to a buffer with a (physiological) 0.1-M NaCl concentration.
We note that the model does not account either for DNA torsional rigidity or for desolvation effects. The latter are very important in systems where DNA strands are packed at interaxial distances smaller than 3 nm (38). For the particular systems considered here, model calculations (33, 39) indicate that the expected average separation of the packaged DNA ought to be larger than this spatial threshold and therefore, to mantain the model complexity at a minimal level, no attempt is made to incorporate effects resulting from interaction with the solvent.
The potential energy terms introduced above are known to be sufficient to account with remarkable detail for the main conformational properties for unconstrained DNA in solutions of high ionic strength (40). As discussed later, these ingredients alone are not sufficient to reproduce either the cryo-EM or the P4 DNA knotting data. To make contact with these experiments, we include another crucial term in our short-ranged potential, which introduces a preferred “twist angle” between DNA segments that are close in space (Fig. 1). This cholesteric twist interaction has a strong phenomenological basis and leads to the onset of cholesteric phases in dense solutions of DNA fragments, typically for DNA densities between about 160 and 380 mg/mL (41). Hence, accounting for the cholesteric interaction appears mandatory for systems where the DNA density is larger than ≈ 150 mg/mL, as in the case for several phages, including P4, where a lower bound for the DNA density is ≈ 200 mg/mL (calculated assuming homogeneous density). Microscopically, the interaction results from an interplay of the helical nature of DNA (which favors the juxtaposition of contacting DNA segments at an angle reflecting the groove “inclination”) and the interaction between the effective electrostatic charges on the double-helical molecule (42).
Fig. 1.
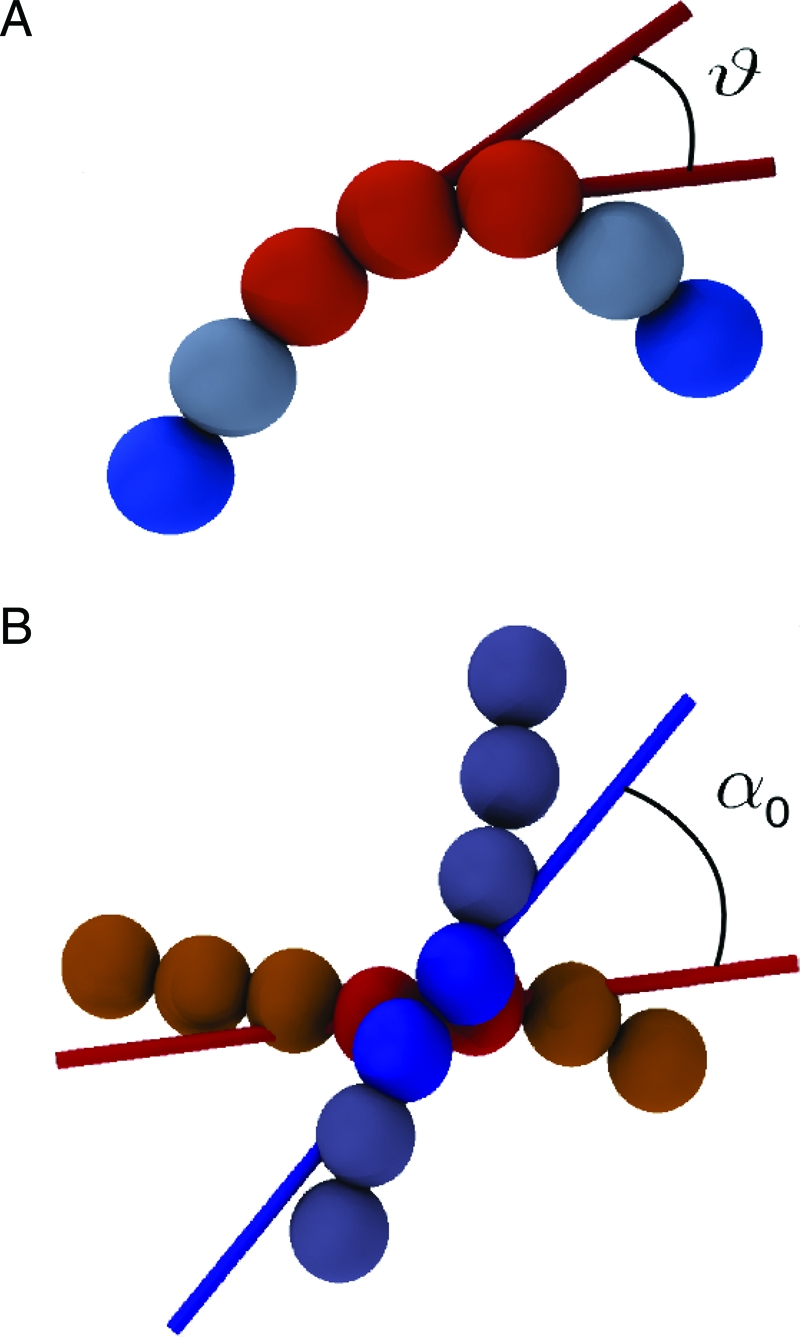
DNA is modeled as a semiflexible chain of beads with diameter equal to 2.5 nm. The flexural rigidity of the DNA molecule is treated by introducing a potential energy term that disfavors bent-chain configurations. As shown in A, the local degree of bending is measured through the angle θ formed by the virtual bonds that join the centers of consecutive beads. As shown in B, a cholesteric interaction between contacting portions of DNA is introduced by favoring a preferential twist angle, α0, between virtual bonds that have a close spatial separation. The figure highlights the pairs of consecutive beads (shown as spheres) bridged by the virtual bonds. The directionality of the latter is shown with colored sticks. The twist angle is calculated accounting for the nonoriented character of dsDNA.
The DNA model (see the Materials and Methods section for further details) is used to study the packaging of the P4 genome. The latter consists of 10 kilo-base pairs (kb) and hence is modeled with chains of 1,360 beads. On the other hand, the P4 capsid is approximated as a nondeformable sphere with a diameter of 45 nm (43). The process of DNA loading inside the capsid is followed via a kink-jump simulation scheme (44), which accounts for thermal fluctuations of the loaded portion of the chain and its stochastic growth inside the capsid.
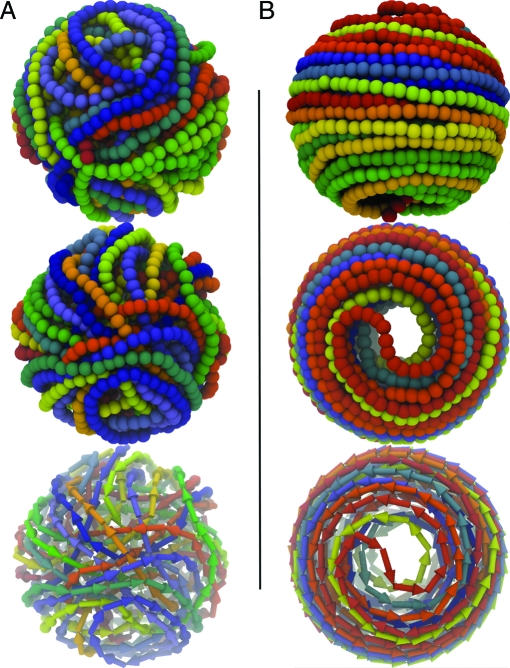
In the absence of the cholesteric term, the configurations generated by packaging the full P4 genome have the typical appearance shown in Fig. 2 A. The highly disordered organization of the DNA at the inner surface of the capsid is apparent, at odds with the cryo-EM experiments.
Fig. 2.
Model conformations of the fully packaged 10 kb-long P4 genome in the absence (A) and presence (B) of the cholesteric potential (of strength k c = 1 K B T range Δ = 3 nm and biasing angle α0 = 1°). In each of the two frames, the first two images present front and top views of DNA arrangement. In both cases, a rainbow-coloring scheme (red → yellow → green → blue) is used to follow the indexing of the chain beads (the red end is the one rooted at the portal motor location). The progression of the DNA chain and its winding directonality are also highlighted in the images at bottom, where an oriented arrow is used to represent each triplet of beads.
Introducing the cholesteric orienting potential, with the interaction strength, range, and amplitude of the angular bias deduced from available data (10, 42, 45), results in a dramatic change of DNA ordering at the surface (see Fig. 2 B). Projection views of the resulting arrangement (see SI Appendix) convey a level of order consistent with cryo-EM observations of various bacteriophages (12, 13). This finding provides a strong indication that the introduction of the cholesteric potential profoundly affects the organization of DNA inside the capsids. Although the arrangement of Fig. 2 B resembles an inverse spool, the coloring scheme reveals both an appreciable degree of interweaving of the layers and the occurrence of hairpin defects. In particular, it is observed that there is (i) no systematic progression of the chain from bottom to top (or vice-versa) of the capsid and (ii) no unique winding directionality. These aspects, particularly the second, reflect the apolar character of the cholesteric interaction because the symmetry of the DNA double-helix implies that the potential bias is insensitive to the directional orientation of interacting DNA segments.
The analysis of the DNA-packaging process reveals that initially the DNA chain tends to form a coaxial spool, with loops having maximum (equatorial) diameter. As more DNA is fed inside, the spool proceeds maintaining the initial winding directionality (which can change from run to run) and moving away from the equatorial region toward the poles. In doing so, it attains tighter radii of curvature and the accompanying increase in bending energy makes it possible for the chain to change winding directionality near the poles producing hairpin defects, like the one shown in Fig. 2 B.
We next turn to the second and key issue, namely the bias in the knot spectrum of circularized DNA molecules. Characterizing the knot spectrum requires the collection of hundreds of configurations where DNA is fully loaded inside the capsid. The duration of each of the packaging simulations depends very strongly on the final filling fraction of the capsid. In fact, the effective rate of DNA loading shows a noticeable reduction upon increasing the fraction of packed genome. For reasons of computational efficiency, we therefore took as a term of reference not the knotting experiments on the full P4 genome but those on the half-P4 genome recently carried by Trigueros and Roca (46). In this study, the knotting of the 4.7kb-long DNA still occurred inside the P4 capsid and produced a spectrum that, except for a decrease in the average level of knot complexity, maintained the same biases of the full-genome case among the simplest types of knots. The shorter genome is still expected to attain local packing densities in the cholesteric range (see SI Appendix) and therefore provides an ideal, computationally viable, test bed for the proposed model.
The same simulation setup described before was used to package chains of 640 beads (corresponding to 4.7 kb-long DNA) inside the P4 model capsid (see Movies S1 and S2 in the SI Appendix). Interestingly, in most of the configurations we generated, the two DNA ends were found at the surface of the confining spherical capsid. This is the simplest nonambiguous situation where the topological state of the molecule can be established by closing the DNA chain outside the capsid (“at infinity”).
Several hundred configurations were generated for various values of the cholesteric potential strength k c (from 0.25 K B T to 3.0K B T), for different values of the interaction range Δ (from ≈3 to 5 nm) and of the preferred twist angle α0 (from 0° to 10°). These values cover the range of available estimates for k c, Δ, and α0 (10, 42, 45). In general, increasing the strength of the cholesteric interaction simplifies the average topological complexity of the generated (closed) configurations. In spite of this, the relative probability of occurrence of the simplest types of knots was found to be only mildly affected by the specific parametrization of the cholesteric potential and was therefore taken as a meaningful term of comparison against available experimental data. As, aside for a difference in the average knot complexity, the key features of the half- and full-genome knot spectrum are consistent with each other, we take as reference the 10 kb case, for which finer experimental data are available (20). As shown later, the best quantitative agreement between the experimental and computed relative probabilities of nontrivial knots was found for k c = 1 K B T, Δ = 5 nm and α0 = 1°. Unless otherwise stated, the results presented hereafter pertain to this choice of parameters.
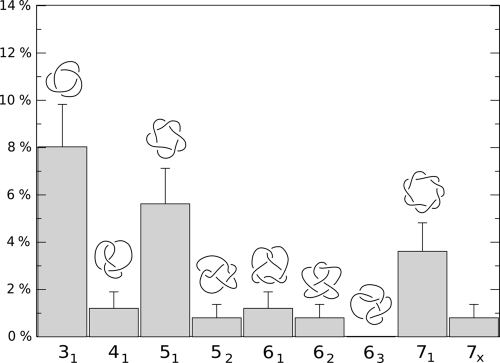
The packaging dynamics were used to generate 250 configurations which, on average, appeared to be highly knotted (with ≈70% of them being nontrivial knots). After all possible geometrical simplifications, ≈20% of the configurations remained too complex to be correctly identified against a lookup table of knots having crossing number smaller than 17*. In particular, the average number of crossings after simplification was 12 (with ≈ 25% of the knots with more than 14 crossings), consistent with experimental findings (46). Among the simplest types of knots, those with up to 7 crossings, the most populated ones were torus knots. As shown in the histogram of Fig. 3 the achiral knot 41 is severely suppressed when compared with both the simpler torus knot 31 and the more complex torus knot 51. The latter is, furthermore, in excess of the 52 counterpart, reversing the trend observed in models of confined self-avoiding flexible polymers (20, 28). Overall, at least 20% of the conformations correspond to the torus knots 31, 51, 71, 819, 91, and 10124 which, in fact, appear to possess an average lower energy than knot types of comparable complexity (see SI Appendix).
Fig. 3.
Knot spectrum. Probability of occurrence of the simplest types of prime knots in 4.7 Kb-long DNA inside the P4 capsid (the cholesteric interaction is taken into account); the error bar is calculated from the Poissonian statistics. Chiral and torus knots prevail, as evident from the predominance of 31, 51, 61, and 71 knots. The index 7x is used for the cumulative set of 72, 73, 74, and 75 knots.
For the purpose of a comparison with the experimental data of ref. 20, we recall that the experimental relative population of 31, 41, 51, and 52 knots were 57%, 3.8%, 39%, and < 1%, respectively. Within estimated uncertanties, the knots populations in Fig. 3 is in good agreement with the above data as the relative probability profile is the following: 51% for 31, 8% for 41, 36% for 51, and 5% for 52. The comparison indicates a level of accord with experiments that was not achieved in previous studies where both equilibrium and dynamical processes were used to sample compact configurations of DNA models of varying complexity (20, 25, 27, 28).
Our results therefore suggest that the knot spectrum can be used to discriminate the viability of several force fields for DNA interactions in confined geometries. Indeed, as expected, simulations with k c = 0, when compared with the experimental data, lack the characteristic bias toward torus knots and provide too complex a knot spectrum. We also performed simulations with self-attractive force fields (coming, e.g., from multivalent counterions in the buffer) which led to better spooling but, once more, produced a knot spectrum qualitatively different from the experimental one.
It is worth pointing out that the bias in favor of torus knots is not accompanied by a detectable bias on the geometrical writhe, which has an average value compatible with 0. A related observation is that no statistically significant difference is observed between the types of handedness of the occurring torus knots. These two aspects reflect the small bias angle α0 = 1° of the cholesteric interaction which, on the length scale given by the capsid size, essentially promotes a nematic, collinear ordering of contacting packaged strands. In fact, the knot spectrum is compatible with the one obtained for a perfectly nematic bias, α0 = 0°. Furthermore, in spite of the apolar character of the interaction, it is found that most pairs of contacting strands are codirectional (assume that orientation is given by chain indexing) so that pronounced differences in the handedness of torus knots are observed upon using larger absolute values of α0. For example, α0 = 10° produces an appreciable majority of right-handed torus knots (see SI Appendix). The results suggest that future experiments where the handedness of torus knots in circularized P4 DNA is probed in dependence of the concentration of polycations (which can modulate the biasing angle) would be valuable to confirm the present findings.
The spatial organization of the DNA in the capsid reproduced by our model can also explain the apparent contradiction between the occurrence of highly knotted DNA configurations inside the mutant phage capsids and the necessity of a highly efficient delivery, by ejection, of the wild-type genome into the cytoplasm of the infected cell (4, 32, 33, 47, 48). In this respect, it is important to recall that, though most of the experiments on the knotting of the P4 DNA have been carried out for the P4 tailless mutant, a nonnegligible fraction of knots (47%) was found also for the wild-type mature P4 phage. In this case, only one of the DNA ends is inside the capsid, and the other is anchored to the phage tail. At variance with the mutant case, the annealing of the two cohesive ends occurs outside the phage after the destruction of the capsid. Considering that a partial DNA relaxation occurs between the extraction of the DNA and its circularization (which “traps” its knotted state), the fact that a substantial fraction of knotted DNA is still found under these conditions is therefore indicative of two facts. First, DNA is highly entangled inside the wild-type virus. As these viruses are infective, the second conclusion is that the entanglement does not hinder the ejection of DNA into the host cell.
Our study provides valuable insight into both of these aspects. Consistent with the first observation, we have verified that both in the wild-type case, where one end of the DNA is kept anchored at the portal motor location, or the mutant case, when both ends are free to reptate and anneal inside the capsid, the majority of the configurations are found to be knotted upon closure at infinity.
The implications of our results for the ejection process are best discussed in relation to the fact that the occurring knots are not tight but highly delocalized. For example, for the most abundant simple knot types, 31 and 51, the knotted portion of the chain (49) is, on average ≈60% (ranging from a minimum of 30% to a maximum of 85%). This result indicates an extremely high degree of knots delocalization, as 31 and 51 knots can be tied very tightly with as little as ≈5% of the chain. Based on this quantitative observations we formulate the proposal that the delocalization of the knots is a key factor for avoiding a plug-like obstruction of the exit pore during DNA ejection, which invariably occurs in the presence of tight knots (50).
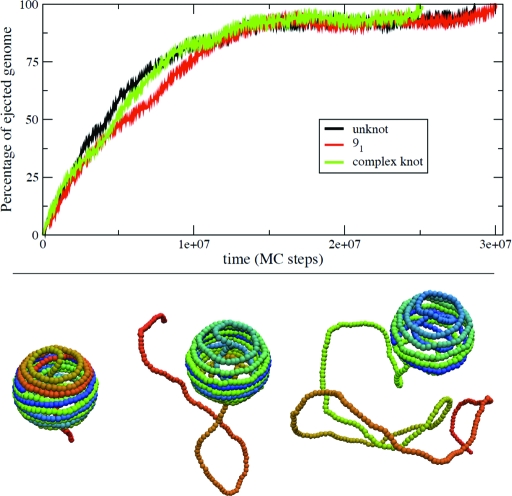
In principle, knot delocalization may not be sufficient to ensure an effective delivery of the genome, as even delocalized knots may be badly entangled geometrically and slow down ejection. We have verified that this is not the case for the knotted configurations generated with our model. To establish this point, we have performed a series of simulations where fully loaded chains with different topological state (upon closure at infinity) were left free to diffuse out of the capsid by taking advantage of thermodynamic forces, which disfavor the confined, packaged, state (51). Strikingly, the ejection kinetics were found to be largely independent of the initial topological state, and all the configurations are ejected without any significant geometrical or topological hindrance (see Fig. 4 and Movies S1 and S2 in the SI Appendix).
Fig. 4.
Percentage of ejected genome as a function of time (in simulation steps, each step corresponding to about 1–5 ns), for three different starting configurations of the fully loaded half-P4 genome. (Upper) The initial configurations correspond to an unknot (black curve), a 91 knot (red curve), and a knot that was too complex to be identified against a lookup table of prime knots of up to 16 crossings (green curve). The ejection proceeds in a fashion that is largely independent of the initial knotted state. A slowing down of the ejection is seen after 70% of the genome has ejected. The ejection speed up for the very last portion of the genome is ascribable to the entropic pulling of the expelled chain (this regime is expected to be sensitive to various details such as the presence of a phage tail). (Lower) Snapshots of the ejection process for the 91 initial knot.
In conclusion, we have presented a theoretical study of several aspects of DNA packaging in viral capsids for which experimental measurements are available and that were unaccounted for by previous theoretical and computational investigations of the problem. The typical mesoscopic description of DNA employed by such studies was that of a self-avoiding semiflexible chain. This treatment is highly successful in reproducing the observed features of unconstrained DNA (40). Our findings show that the known rich phenomenology of densely packed DNA inside phages can be satisfactorily reproduced only when accounting for the known preference of contacting DNA portions to meet with a preferred twist angle.
If this is done, the salient features of the spectrum of knots observed experimentally for P4 DNA are well reproduced. The structures we find are also consistent with the cryo-EM data, which show regular organization close to the surface of the capsid. This organization is at odds with most previous simulation data on bacteriophage DNA, which show much more disordered packing if, as we do here, a buffer with monovalent counterions is considered. The results presented provide not only a consistent explanation of the experimental data on viral DNA organization but are also used to formulate predictions regarding the DNA winding directionality inside capsids and the handedness of torus knots. We hope these can guide the design of future experiments aimed at advancing the current understanding of DNA organization inside bacteriophages.
Materials and Methods
DNA Model.
Double-stranded DNA is modeled as an open chain of N spherical beads of diameter σ = 2.5 nm (each bead therefore comprises slightly less than 8 base pairs). In the following, we shall indicate with the position of the center of the ith bead and with the virtual bond vector connecting beads i and i + 1.
The connectivity of the chain is treated within the finitely extensible nonlinear elastic model (52), and by further requiring that the bond length is never either smaller than 0.7 σ or larger than 1.3 σ.
The bending rigidity of DNA is captured with a standard Kratky Porod potential,
and the bending energy amplitude is set so to reproduce the known persistence length (l p = 50 nm) of unconstrained DNA, κb = K B Tl p/σ, where K B is the Boltzmann constant and T = 300 K is the temperature.
The mesoscopic DNA model includes three types of chain self-interaction, namely Van der Waals, screened electrostatic and cholesteric interactions (38). The Van der Waals interaction is modeled by using a truncated pairwise Lennard–Jones potential,
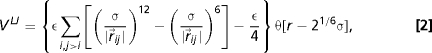 |
where ɛ = k B T, and . Screened electrostatic interactions are accounted for via a Debye–Huckel potential,
where l B is the Bjerrum length (0.7 nm in water), a is the distance between two elementary charges (≈1 nm for B-DNA; see, e.g., refs. 26 and 53), and l D is the Debye length, which we take equal to 0.9 nm (corresponding to 0.1 M solution of monovalent counterions such as NaCl).
Finally, the cholesteric twist interaction potential is
where
where is the distance between the centers of mass of the two bonds and Δ is the spatial range of the interaction. The twist angle, α, formed by two bonds, and is defined by
With this definition of α the cholesteric interaction is apolar, i.e., insensitive to the reversal of any of the two bonds and . The apolarity is required in consideration of the symmetry of the DNA double helix. The preferential twist angle is indicated as α0. A comparison with the case of polar interaction is provided as supporting information.
The above potential arguably represents the simplest phenomenological way to account for the preferential twist angle formed by contacting DNA segments, in a chain-of-beads DNA model. This interaction, which is responsible for the well-documented phenomenology of cholesteric phases of DNA (41), results from the complex interplay of various atomic interactions, which only in recent years are being systematically addressed (11, 45).
Viable values for the phenomenological parameters k c and α0 can be obtained from available estimates of k t and k 22, measured and computed for dsDNA left-handed cholesteric phases. Specifically, it is found that α0 should be of the order of 1° (45), though larger values could be obtained if the capsid is permeated by polycations. In addition, considering the energy associated with the introduction of a cholesteric twist in a hexagonal arrangement of parallel DNA strands (11), one has k c ≈ 1 K B T at T = 300K. Finally, the interaction range Δ is expected to be ≈3–5 nm (11, 42).
Kink-Jump Dynamics.
The simulations were performed by using the constant-temperature kink-jump stochastic dynamics scheme (44). This algorithm consists of the Metropolis acceptance/rejection of attempted local polymer deformations. The elementary move entails a displacement of randomly picked bead by a fraction of its diameter. The time step associated with each kink-jump move can be mapped with the time span of 1–5 ns, over which an isolated bead would diffuse over a length comparable with such a fraction. At all stages of the loading process, one end of the DNA is held fixed at a specific “portal motor” position on the surface of the spherical capsid (45 nm diameter) confining the beads. The progressive insertion of the genome performed by the motor is described by the occasionally attempted addition of beads at the free end of DNA until the desired length of the chain inside the capsid is reached. The half-P4 genome packaging process lasts several ms, consistent with the time span covered by other DNA-packaging simulations (35, 54). The chain growth process is expected to capture the realistic aspects of the DNA insertion inside the capsid, which is expected to mostly progress by reptation (48).
Simulations of spontaneous (free) ejection are carried out within the kink-jump scheme by modifying the potential field so that there is a hole (radius 5 nm) on the surface of the capsid from which DNA can exit. To model the presence of a collar in the phage, beads close to the opening are subjected to a harmonic potential toward the center of the opening.
DNA Circularization.
Packaging simulations are performed by using an open chain, which eventually must be closed in order to identify its topological (knotted) state. The closure procedure can be delicate, as different ways of connecting the two ends produce different knots. The standard procedure used in this work is to connect the ends after prolonging them radially out of the capsid. This scheme is equivalent to a closure at infinity. Virtually all configurations produced with this scheme had their ends at the surface; hence, for those configuration the radial closure unambiguously identifies the knot.
Supplementary Material
Acknowledgments.
We acknowledge financial support from Italian Ministry of Education Grant PRIN and from Swiss National Science Foundation Grant 3100A0-116275. This work has made use of the resources provided by the Edinburgh Compute and Data Facility.
Note Added in Proof.
Other works of relevance to the present article include the study of Katzav et al. (55), who employed a mean-field approach to characterize ordering effects in packaged DNA, and the one of Arsuaga et al. (56), in which the formation of torus and chiral knots in confined ring polymers was promoted by favoring coaxial spooling with a constant winding direction.
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
This article contains supporting information online at www.pnas.org/cgi/content/full/0907524106/DCSupplemental.
The KnotScape software package created by J. Hoste and M. Thistlethwaite was used to identify prime and composite knots of up to 16 crossings.
References
- 1.Earnshaw WC, Harrison SC. DNA arrangement in isometric phage heads. Nature. 1977;268:598–602. doi: 10.1038/268598a0. [DOI] [PubMed] [Google Scholar]
- 2.Smith DE, et al. The bacteriophage straight phi29 portal motor can package DNA against a large internal force. Nature. 2001;413:748–752. doi: 10.1038/35099581. [DOI] [PubMed] [Google Scholar]
- 3.Leforestier A, et al. Bacteriophage T5 DNA ejection under pressure. J Mol Biol. 2008;384:730–739. doi: 10.1016/j.jmb.2008.09.035. [DOI] [PubMed] [Google Scholar]
- 4.Gelbart WM, Knobler CM. Virology. Pressurized viruses. Science. 2009;323:1682–1683. doi: 10.1126/science.1170645. [DOI] [PubMed] [Google Scholar]
- 5.Evilevitch A, Lavelle L, Knobler CM, Raspaud E, Gelbart WM. Osmotic pressure inhibition of DNA ejection from phage. Proc Natl Acad Sci USA. 2003;100:9292–9295. doi: 10.1073/pnas.1233721100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Castelnovo M, Evilevitch A. DNA ejection from bacteriophage: Towards a general behavior for osmotic-suppression experiments. Eur Phys J E Soft Matter. 2007;24:9–18. doi: 10.1140/epje/i2007-10205-5. [DOI] [PubMed] [Google Scholar]
- 7.Löf D, Schillén K, Jönsson B, Evilevitch A. Forces controlling the rate of DNA ejection from phage lambda. J Mol Biol. 2007;368:55–65. doi: 10.1016/j.jmb.2007.01.076. [DOI] [PubMed] [Google Scholar]
- 8.Robinson C. The cholesteric phase in polypeptide solutions and biological structures. Mol Cryst. 1966;1:467–494. [Google Scholar]
- 9.Bouligand Y, Livolant F. The organization of cholesteric spherulites. J Physique. 1984;45:1899–1923. [Google Scholar]
- 10.Stanley CB, Hong H, Strey HH. DNA cholesteric pitch as a function of density and ionic strength. Biophys J. 2005;89:2552–2557. doi: 10.1529/biophysj.105.064550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Kornyshev A A, Leikin S, Malinin SV. Chiral electrostatic interaction and cholesteric liquid crystals of DNA. Eur Phys J E. 2002;7:83–93. [Google Scholar]
- 12.Jiang W, et al. Structure of epsilon 15 bacteriophage reveals genome organization and DNA packaging/injection apparatus. Nature. 2006;439:612–616. doi: 10.1038/nature04487. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Comolli LR, et al. Three-dimensional architecture of the bacteriophage phi29 packaged genome and elucidation of its packaging process. Virology. 2008;371:267–277. doi: 10.1016/j.virol.2007.07.035. [DOI] [PubMed] [Google Scholar]
- 14.Wang JC, Martin KV, Calendar R. On the sequence similarity of the cohesive ends of coliphage P4, P2, and 186 deoxyribonucleic acid. Biochemistry. 1973;12:2119–2123. doi: 10.1021/bi00735a016. [DOI] [PubMed] [Google Scholar]
- 15.Liu LF, Perkocha L, Calendar R, Wang JC. Knotted DNA from bacteriophage capsids. Proc Natl Acad Sci USA. 1981;78:5498–5502. doi: 10.1073/pnas.78.9.5498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Liu LF, Davis JL, Calendar R. Novel topologically knotted DNA from bacteriophage P4 capsids: studies with DNA topoisomerases. Nucleic Acids Res. 1981;9:3979–3989. doi: 10.1093/nar/9.16.3979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Shaw SY, Wang JC. Knotting of a DNA chain during ring closure. Science. 1993;260:533–536. doi: 10.1126/science.8475384. [DOI] [PubMed] [Google Scholar]
- 18.Stasiak A, Katritch V, Bednar J, Michoud D, Dubochet J. Electrophoretic mobility of DNA knots. Nature. 1996;384:122–122. doi: 10.1038/384122a0. [DOI] [PubMed] [Google Scholar]
- 19.Arsuaga J, Vázquez M, Trigueros S, Sumners D, Roca J. Knotting probability of DNA molecules confined in restricted volumes: DNA knotting in phage capsids. Proc Natl Acad Sci USA. 2002;99:5373–5377. doi: 10.1073/pnas.032095099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Arsuaga J, et al. DNA knots reveal a chiral organization of DNA in phage capsids. Proc Natl Acad Sci USA. 2005;102:9165–9169. doi: 10.1073/pnas.0409323102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Wasserman SA, Dungan JM, Cozzarelli NR. Discovery of a predicted DNA knot substantiates a model for site-specific recombination. Science. 1985;229:171–174. doi: 10.1126/science.2990045. [DOI] [PubMed] [Google Scholar]
- 22.Spengler SJ, Stasiak A, Cozzarelli NR. The stereostructure of knots and catenanes produced by phage lambda integrative recombination: Implications for mechanism and DNA structure. Cell. 1985;42:325–334. doi: 10.1016/s0092-8674(85)80128-8. [DOI] [PubMed] [Google Scholar]
- 23.Ernst C, Sumners DW. A calculus for rational tangles: Applications to DNA recombination. Math Proc Camb Phil Soc. 1990;108:489–515. [Google Scholar]
- 24.Postow L, Crisona NJ, Peter BJ, Hardy CD, Cozzarelli NR. Topological challenges to DNA replication: conformations at the fork. Proc Natl Acad Sci USA. 2001;98:8219–8226. doi: 10.1073/pnas.111006998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Arsuaga J, Tan RKZ, Vazquez M, Sumners DW, Harvey SC. Investigation of viral DNA packaging using molecular mechanics models. Biophys Chem. 2002;101–102:475–484. doi: 10.1016/s0301-4622(02)00197-7. [DOI] [PubMed] [Google Scholar]
- 26.Forrey C, Muthukumar M. Langevin dynamics simulations of genome packing in bacteriophage. Biophys J. 2006;91:25–41. doi: 10.1529/biophysj.105.073429. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Micheletti C, Marenduzzo D, Orlandini E, Sumners DW. Knotting of random ring polymers in confined spaces. J Chem Phys. 2006;124:64903–64903. doi: 10.1063/1.2162886. [DOI] [PubMed] [Google Scholar]
- 28.Micheletti C, Marenduzzo D, Orlandini E, Sumners DW. Simulations of knotting in confined circular DNA. Biophys J. 2008;95:3591–3599. doi: 10.1529/biophysj.108.137653. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Fuller FB. The writhing number of a space curve. Proc Natl Acad Sci USA. 1971;68:815–819. doi: 10.1073/pnas.68.4.815. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Podtelezhnikov AA, Cozzarelli NR, Vologodskii AV. Equilibrium distributions of topological states in circular DNA: Interplay of supercoiling and knotting. Proc Natl Acad Sci USA. 1999;96:12974–12979. doi: 10.1073/pnas.96.23.12974. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Cerf C, Stasiak A. A topological invariant to predict the three-dimensional writhe of ideal configurations of knots and links. Proc Natl Acad Sci USA. 2000;97:3795–3798. doi: 10.1073/pnas.97.8.3795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Kindt J, Tzlil S, Ben-Shaul A, Gelbart WM. DNA packaging and ejection forces in bacteriophage. Proc Natl Acad Sci USA. 2001;98:13671–13674. doi: 10.1073/pnas.241486298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Purohit PK, Kondev J, Phillips R. Mechanics of DNA packaging in viruses. Proc Natl Acad Sci USA. 2003;100:3173–3178. doi: 10.1073/pnas.0737893100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Marenduzzo D, Micheletti C. Thermodynamics of DNA packaging inside a viral capsid: The role of DNA intrinsic thickness. J Mol Biol. 2003;330:485–492. doi: 10.1016/s0022-2836(03)00584-9. [DOI] [PubMed] [Google Scholar]
- 35.Petrov AS, Harvey SC. Packaging double-helical DNA into viral capsids: Structures, forces, and energetics. Biophys J. 2008;95:497–502. doi: 10.1529/biophysj.108.131797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Leforestier A, Livolant F. Structure of toroidal DNA collapsed inside the phage capsid. Proc Natl Acad Sci USA. 2009;106:9157–9162. doi: 10.1073/pnas.0901240106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Bustamante C, Marko JF, Siggia ED, Smith S. Entropic elasticity of lambda-phage DNA. Science. 1994;265:1599–1600. doi: 10.1126/science.8079175. [DOI] [PubMed] [Google Scholar]
- 38.Strey HH, Podgornik R, Rau DC, Parsegian VA. DNA–DNA interactions. Curr Opin Struct Biol. 1998;8:309–313. doi: 10.1016/s0959-440x(98)80063-8. [DOI] [PubMed] [Google Scholar]
- 39.Inamdar M M, Gelbart M W, Phillips R. Dynamics of DNA ejection from bacteriophage. Biophys J. 2006;91:411–420. doi: 10.1529/biophysj.105.070532. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Rybenkov VV, Cozzarelli NR, Vologodskii AV. Probability of DNA knotting and the effective diameter of the DNA double helix. Proc Natl Acad Sci USA. 1993;90:5307–5311. doi: 10.1073/pnas.90.11.5307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Livolant F, Leforestier F. Condensed phases of DNA: structures and phase transitions. Prog PolymSci. 1996;21:1115–1164. [Google Scholar]
- 42.Cherstvy AG. DNA cholesteric phases: The role of DNA molecular chirality and DNA–DNA electrostatic interactions. J Phys Chem B. 2008;112:12585–12595. doi: 10.1021/jp801220p. [DOI] [PubMed] [Google Scholar]
- 43.Dokland T, Lindqvist BH, Fuller SD. Image reconstruction from cryo-electron micrographs reveals the morphopoietic mechanism in the P2-P4 bacteriophage system. EMBO J. 1992;11:839–846. doi: 10.1002/j.1460-2075.1992.tb05121.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Marenduzzo D, Orlandini E. Dynamics of fibers growing inside soft vesicles. Europhys Lett. 2007;80:48004. [Google Scholar]
- 45.Tombolato F, Ferrarini A. From the double-stranded helix to the chiral nematic phase of B-DNA: A molecular model. J Chem Phys. 2005;122:54908–54908. doi: 10.1063/1.1839859. [DOI] [PubMed] [Google Scholar]
- 46.Trigueros S, Roca J. Production of highly knotted DNA by means of cosmid circularization inside phage capsids. BMC Biotechnol. 2007;7:94–94. doi: 10.1186/1472-6750-7-94. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Siber A, Dragar M, Parsegian VA, Podgornik R. Packing nanomechanics of viral genomes. Eur Phys J E Soft Matter. 2008;26:317–325. doi: 10.1140/epje/i2008-10330-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Gabashvili IS, Grosberg AY. Dynamics of double stranded DNA reptation from bacteriophage. J Biomol Struct Dyn. 1992;9:911–920. doi: 10.1080/07391102.1992.10507966. [DOI] [PubMed] [Google Scholar]
- 49.Marcone B, Orlandini E, Stella AL, Zonta F. What is the length of a knot in a polymer? J Phys A. 2005;38:L15–L21. [Google Scholar]
- 50.Matthews R, Louis AA, Yeomans JM. Knot-controlled ejection of a polymer from a virus capsid. Phys Rev Lett. 2009;102:088101–088101. doi: 10.1103/PhysRevLett.102.088101. [DOI] [PubMed] [Google Scholar]
- 51.Muthukumar M. Translocation of a confined polymer through a hole. Phys Rev Lett. 2001;86:3188–3191. doi: 10.1103/PhysRevLett.86.3188. [DOI] [PubMed] [Google Scholar]
- 52.Kremer K, Grest G. Dynamics of entangled linear polymer melts: A molecular-dynamics simulation. J Chem Phys. 1990;92:5057–5086. [Google Scholar]
- 53.Podgornik R, Hansen PL, Parsegian VA. Elastic moduli renormalization in self-interacting stretchable polyelectrolytes. J Chem Phys. 2000;113:9343–9350. [Google Scholar]
- 54.Rollins GC, Petrov AS, Harvey SC. The role of DNA twist in the packaging of viral genomes. Biophys J. 2008;94:38–40. doi: 10.1529/biophysj.107.126698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Katzav E, Adda–Bedia M, Boudaoud A. A statistical approach to close packing of elastic rods and to DNA packaging in viral capsids. Proc Natl Acad Sci USA. 2006;103:18900–18904. doi: 10.1073/pnas.0609079103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Arsuaga J, Diao Y. DNA knotting in spooling like conformations in Bacteriophages. J Comp Math Meth Med. 2008;9:303–316. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.