Abstract
A region between residues 414 and 466 in the cardiac ryanodine receptor (RyR2) harbors more than half of the known NH2-terminal mutations associated with cardiac arrhythmias and sudden death. To gain insight into the structural basis of this NH2-terminal mutation hotspot, we have determined its location in the three-dimensional structure of RyR2. Green fluorescent protein (GFP), used as a structural marker, was inserted into the middle of this mutation hotspot after Ser-437 in the RyR2 sequence. The resultant GFP-RyR2 fusion protein, RyR2S437-GFP, was expressed in HEK293 cells and characterized using Ca2+ release, [3H]ryanodine binding, and single cell Ca2+ imaging studies. These functional analyses revealed that RyR2S437-GFP forms a caffeine-and ryanodine-sensitive Ca2+ release channel that possesses Ca2+- and caffeine-dependence of activation indistinguishable from that of wild type (wt) RyR2. HEK293 cells expressing RyR2S437-GFP displayed a propensity for store-overload induced Ca2+ release similar to that in cells expressing RyR2-wt. The three-dimensional structure of the purified RyR2S437-GFP was reconstructed using cryo-electron microscopy and single particle image processing. Subtraction of the three-dimensional reconstructions of RyR2-wt and RyR2S437-GFP revealed the location of the inserted GFP, and hence the NH2-terminal mutation hotspot, in a region between domains 5 and 9 in the clamp-shaped structure. This location is close to a previously mapped central disease-causing mutation site located in a region between domains 5 and 6. These results, together with findings from previous studies, suggest that the proposed interactions between the NH2-terminal and central regions of RyR2 are likely to take place between domains 5 and 6, and that the clamp-shaped structure, which shows substantial conformational differences between the closed and open states, is highly susceptible to disease-causing mutations.
Cardiac arrhythmia, a leading cause of sudden death, has been linked to a number of genes encoding ion channels, including the cardiac sarcoplasmic recticulum Ca2+ release channel (ryanodine receptor, RyR2) (1–4). To date, more than 40 mutations in RyR2 have been associated with at least two forms of cardiac arrhythmias, catecholaminergic polymorphic ventricular tachycardia (CPVT) and arrhythmogenic right ventricular dysplasia type 2 (ARVD2). Most of these mutations are located in the N-terminal (between aa 164–466), central (2246–2504) and C-terminal (3778–4959) regions of RyR2 (Fig. 1). Interestingly, those mutations in the skeletal muscle RyR (RyR1) that are linked to malignant hyperthermia (MH) and central core disease (CCD) are largely clustered in the corresponding regions of RyR1; some sites even correspond exactly (2, 4) (Fig. 1). These similar distributions suggest that the molecular mechanisms by which these disease-linked RyR1 and RyR2 mutations alter the intrinsic properties of the channel are conserved.
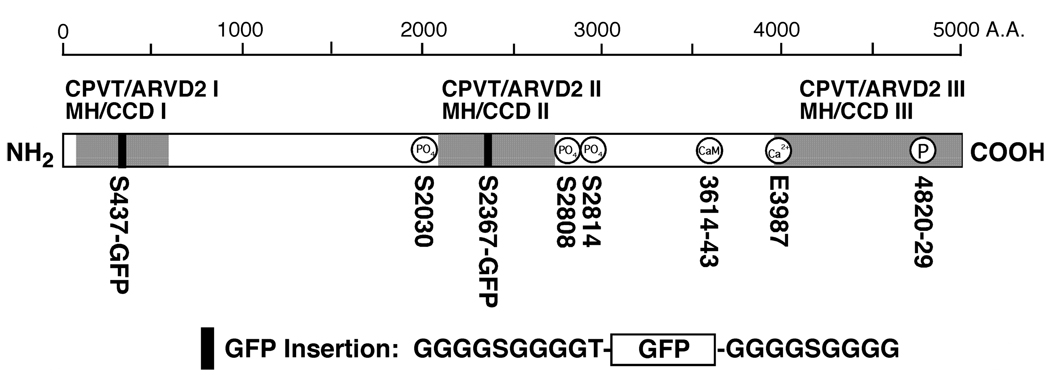
Fig. 1. Insertion of GFP into the RyR2 sequence after residue Ser-437.
The linear sequence of RyR2 is denoted by an open rectangle. The NH2-terminal, central, and COOH-terminal mutation regions, corresponding to CPVT/ARVD2 I and MH/CCD I, CPVT/ARVD2 II and MH/CCD II, and CPVT/ARVD2 III and MH/CCD III, respectively, are indicated by the shaded areas. GFP flanked by two Gly-rich spacers was inserted into the NH2-terminal region (CPVT/ARVD2 I and MH/CCD I) after Ser-437 and into the central region (CPVT/ARVD2 II and MH/CCD II) after residue Ser-2367 of the RyR2 sequence, as indicated by the filled boxes. The phosphorylation sites (S2030, S2808, and S2814), the calmodulin binding site (CaM, 3614–3643), the proposed cytosolic Ca2+ sensor (E3987), and the proposed pore-forming segment (4820–4829) are also shown.
A number of disease-linked RyR2 mutations have been expressed and functionally characterized. A common finding of these studies is that most of the RyR2 mutations enhance the activity of the channel upon stimulation (1–4). This enhancement of function activity is also a consistent feature with most of the RyR1 MH and CCD mutations (5–7). However, the precise mechanism by which channel function is altered by disease-linked mutations is not well defined. Wehrens et al. showed that CPVT RyR2 mutations reduced the binding affinity of a 12.6 kDa FK506 binding protein (FKBP12.6) to RyR2, a protein believed to stabilize the RyR2 channel. When RyR2 is phosphorylated by PKA, FKBP12.6 dissociated from RyR2, leading to faulty calcium release (8). If so, CPVT mutant channels should exhibit an enhanced activity upon PKA phosphorylation. However, George et al. have demonstrated that CPVT RyR2 mutations augmented Ca2+ release in a manner independent of both FKBP12.6 and PKA phosphorylation (9). Consistent with the latter authors, we have demonstrated that the mutations R176Q and L433P in the N-terminal region; S2246L, R2474S, and T2504M in the central region; and N4104K, Q4201R, R4496C, I4867M, and N4895D in the C-terminal region do not affect the RyR2-FKBP12.6 interaction (10). Furthermore, we have found that these mutations increase the sensitivity of RyR2 to activation by luminal Ca2+ and reduce the threshold for spontaneous Ca2+ release during store Ca2+ overload, a process that we have termed as store-overload induced Ca2+ release (SOICR) (10, 11). Since SOICR can lead to delayed after-depolarizations (DADs), which in turn can produce triggered arrhythmias, a reduced threshold for SOICR as a result of enhanced luminal Ca2+ activation of RyR2 would increase the propensity for DADs, cardiac arrhythmias, and sudden death. Hence, an altered luminal Ca2+ response of the channel is a common molecular defect of disease-linked RyR2 mutations. However, how mutations located in different regions of the channel similarly alter luminal Ca2+ activation is not well understood.
Ikemoto and his colleagues have proposed that the NH2-terminal and central regions form two domains that physically interact in the three-dimensional structure of RyR1 and RyR2, and that disease-causing mutations are exposed at the interface of the two domains (4, 12, 13). They further proposed that inter-domain interactions (“zipped” domains) are involved in stabilizing the closed state of the channel, and that disease-causing mutations weaken the interactions (produce domain “unzipping”), thereby destabilizing the channel and making it more susceptible to activation by stimuli (12). Although an increasing body of biochemical and functional evidence supports this domain “unzipping” hypothesis, structural evidence is lacking.
To understand the structural basis of the mechanism underlying disease-causing mutations, we have recently mapped the site of a mutation in the central region to a spot between domains 5 and 6 in the three-dimensional structure of RyR2 (14). In the present study, to directly test the hypothesis, we attempted to locate the site of a mutation in the NH2-terminal region in the three-dimensional structure of RyR2. To this end, a RyR2-GFP fusion protein, RyR2S437-GFP, in which GFP was inserted into the NH2-terminal region after residue Ser-437, was constructed and expressed in HEK293 cells. Functional studies demonstrated that the GFP insertion does not grossly alter the structure and function of RyR2. RyR2S437-GFP protein was purified and then subjected to cryo-electron microscopy (Cryo-EM) and single-particle image processing. Three-dimensional reconstruction enabled localization of the inserted GFP to a site between domains 5 and 9 that is close to the central mutation site mapped previously, consistent with the zipping/unzipping hypothesis.
EXPERIMENTAL PROCEDURES
Materials
Restriction endonucleases and DNA modifying enzymes were purchased from New England BioLabs Inc (Ipswich, MA). The anti-RyR and anti-GFP antibodies were obtained from Affinity BioReagents (Golden, CO). Soybean phosphatidylcholine was obtained from Avanti Polar Lipids, Inc (Alabaster, AL). ((.3-[(3-Cholamidopropyl)-dimethylammonio]-1-propane sulfonate (CHAPS) and other reagents were purchased from Sigma (St. Louis, MO).
Cell Culture and DNA Transfection
HEK293 cells were maintained in Dulbecco’s modified Eagle’s medium as described previously (15). HEK293 cells grown on 100-mm tissue culture dishes for 18–20 hours after subculture were transfected with 12 µg of wild type RyR2 (RyR2-wt) or RyR2S437-GFP cDNAs using Ca2+ phosphate precipitation (16).
Construction of RyR2S437-GFP
The cloning and construction of the 15-kb full-length cDNA encoding the mouse cardiac RyR2 has been described previously (17). To generate the RyR2S437-GFP cDNA construct, the Cla I (2350)-Not I (14904) fragment was first removed by digesting the full-length RyR2 cDNA with Cla I and Not I, the remaining portion of the RyR2 cDNA was then ligated with the use of a Cla I-Not I adaptor to form the RyR2 (Cla I-Not I)− construct. The Cla I-Not I adaptor was generated by annealing two short primers, 5'-CGATCCGC-3' (forward) and 5'-GGCCGCGGAT-3' (reverse). The cDNA encoding GFP flanked by Gly-rich spacers and an AscI site was obtained by PCR as described previously (18). The Asc I site was introduced into the sequence of the RyR2 (Cla INot I)− construct after Ser-437 by overlap-extension using the polymerase chain reaction (PCR) (19). The “outer” primers used were: forward, 5’-GTGGATGTGAAATCAGCACGA-3’; reverse, 5’-GCTGATCAGCCAGTCGAGGGA-3’. The primers for introducing the Asc I site were: forward, 5’ GATGCTCTCAGCGGGCGCGCCAAGCTGCC CACCATTGAC-3’; reverse, 5’-GGTGGGCTGCTTGGCGCGCCCGCTGAGAG CATCCAGGCC -3’. The BamH I (1153)-EcoR I (1590) PCR fragment containing the Asc I site was used to replace the BamH I (1153)-EcoR I (1590) fragment in the RyR2 (Cla I-Not I)− construct. The missing Cla I (2350)-Not I (14904) fragment was then added back, to yield the full-length RyR2 cDNA with the inserted Asc I site. The Asc I-Asc I fragment containing GFP and the spacers was then subcloned into the full-length RyR2 cDNA at the introduced Asc I site. The sequences of all PCR fragments and the orientation of the inserted GFP cDNA were verified by DNA sequencing analysis.
Cryo-electron Microscopy and Image Processing
The expression and purification of RyR2S437-GFP was carried out as described previously (18). The purified RyR2S437-GFP was diluted 5–10 fold with EM dilution buffer (20 mM Na.PIPES, pH 7.2, 400 mM KCl, 3 mM EGTA, 0.5% CHAPS, 2 mM DTT, and 2 mg/ml leupeptin). Grids were prepared for cryo-EM according to standard methods (20). Micrographs were recorded using low-dose protocols on a Philips EM420, equipped with a low-dose kit and a GATAN 626 cryotransfer holder, at 38,600× ± 2%, as verified by a tobacco mosaic virus standard. Each exposure corresponded to an electron dose of ~10 e−/ Å2. Micrographs were checked for drift, astigmatism, and the presence of Thon rings by optical diffraction. Selected electron micrographs were digitized on a Zeiss/Imaging scanner (Z/I Imaging Corporation, Huntsville, AL) with a step size of 14 µm. Images were processed using the SPIDER/WEB software package (21), and three-dimensional reconstructions were calculated using projection matching procedure (18, 22). The final three-dimensional reconstructions of RyR2-wt and RyR2S437-GFP were computed from 5,434 and 4,948 particles, respectively. Four-fold symmetry was enforced in both reconstructions. The final resolution was estimated to be 34Å by Fourier shell correlation (with a cut-off of 0.5) for RyR2-wt, and 33Å for RyR2S437-GFP. (23). The difference map was calculated by subtraction of the three-dimensional volume of the RyR2-wt from the RyR2S437-GFP volume.
Generation of Stable, Inducible HEK293 Cell Lines Expressing RyR2-wt and RyR2S437-GFP, and Single Cell Ca2+ Imaging
Stable, inducible HEK293 cells expressing RyR2-wt and RyR2S437-GFP were generated using the Flp-In T-Rex Core Kit from Invitrogen as described previously (10). Intracellular Ca2+ transients in HEK293 cells were measured using single-cell Ca2+ imaging and the fluorescent Ca2+ indicator dye fura-2 acetoxymethyl ester (fura-2 AM) as described previously (10).
Ca2+ Release and [3H]Ryanodine Binding Assays
Measurements of free cytosolic Ca2+ concentrations in the transfected HEK293 cells using fluorescent Ca2+ indicator dye fluo-3, and of equilibrium [3H]ryanodine binding to cell lysate were done as described previously (10).
GST-FKBP12.6 Pull Down, Immunoprecipitation, and Immunoblotting Analyses
GST-FKBP12.6 pull-down, immunoprecipitation, and immunoblotting were carried out as described previously (18, 24).
RESULTS
Insertion of Green Fluorescent Protein (GFP) into the NH2-terminal Region of RyR2 after Serine-437
A number of mutations in the NH2-terminal region of RyR2 have been linked to CPVT or ARVD2. Of these NH2-terminal mutations, six are located between residues 414 and 466. We attempted to map the location of this hotspot in the three-dimensional structure of RyR2. Our approach was to use GFP, which contains 238 amino acids and is much smaller than RyR2 (~5000 amino acids), as a structural marker, and to insert it into the region of interest. Three-dimensional reconstructions of the RyR2-wt and GFP-inserted RyR2 were then generated and compared. Barring major conformational rearrangements due to disruption, the primary difference between the two structures would correspond to the inserted GFP. Accordingly, we generated a RyR2-GFP fusion construct (RyR2S437-GFP) in which GFP was inserted into the RyR2 sequence after residue Ser-437, near the middle of a mutation hotspot (aa 414–466) (Fig. 1). To minimize potential perturbations of the folding and function of RyR2, two glycine-rich spacers were added on both side of GFP (25, 26)(Fig.1).
Functional Expression of RyR2S437-GFP
To determine whether the GFP-inserted RyR2 was properly expressed, we transfected HEK293 cells with the RyR2S437-GFP cDNA, and subsequently examined them using fluorescence microscopy. As shown in Fig. 2, green fluorescence in HEK293 cells was readily detected after transfection (Fig. 2A), while no fluorescence was observed in HEK293 cells transfected with RyR2-wt lacking the GFP insertion (data not shown). Since the fluorescence of GFP depends on the proper folding of its own structure and the structure of the inserted region (26, 27), the appearance of green fluorescence in transfected HEK293 cells indicates that the inserted GFP and its neighboring region is likely to have folded properly.
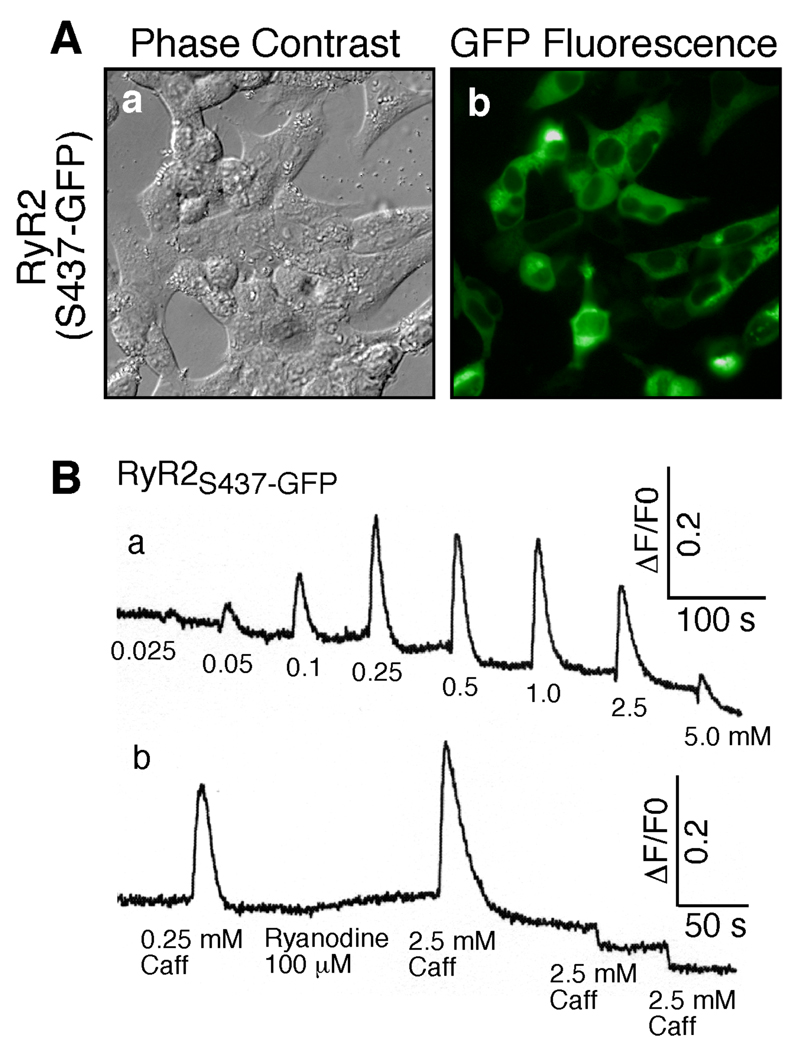
Fig. 2. Expression and functional characterization of RyR2S437-GFP.
(A) HEK293 cells grown on glass coverslips were transfected with RyR2S437-GFP. Phase-contrast images (a) and GFP-fluorescence (b) of transfected cells were recorded under the fluorescence microscope. (B) The fluorescent intensity of the fluo-3-loaded HEK293 cells transfected with RyR2S437-GFP cDNA was monitored continuously before and after addition of various concentrations of caffeine (a), or before and after the sequential addition of 0.25 mM caffeine, 100 µM ryanodine, and three successive doses of 2.5 mM caffeine (b). The sharp decreases in fluorescence intensity (b) immediately after the third and forth doses of caffeine were due to fluorescence quenching by caffeine. Similar results were obtained from three separate experiments.
To assess whether the expressed RyR2S437-GFP was functional, we measured intracellular Ca2+ release in transfected HEK293 cells, using the fluorescent Ca2+ indicator dye fluo-3. As shown in Fig. 2B, HEK293 cells transfected with the RyR2S437-GFP cDNA displayed multiple Ca2+ release events in response to repeated stimulation by caffeine (Fig. 2Ba), an agonist of RyRs. However, transfected cells pretreated with ryanodine responded only to the first, but not the second or third, caffeine stimulation due to the modification of RyR2 by ryanodine (Fig. 2Bb). The same responses to caffeine and ryanodine were observed in HEK293 cells transfected with RyR2-wt (28), indicating that RyR2S437-GFP, like RyR2-wt, is able to form a caffeine- and ryanodine-sensitive Ca2+ release channel in HEK293 cells. The immediate drop in fluorescence after the second and third caffeine stimulations (Fig. 2Bb) was caused by fluorescence quenching by caffeine (28). It should also be noted that no caffeine- or ryanodine-induced Ca2+ release was detected in non-transfected HEK293 cells, or in cells transfected with pCDNA3 vector DNA (data not shown).
Responses of RyR2S437-GFP to Ca2+, Caffeine, and Ca2+ Overload
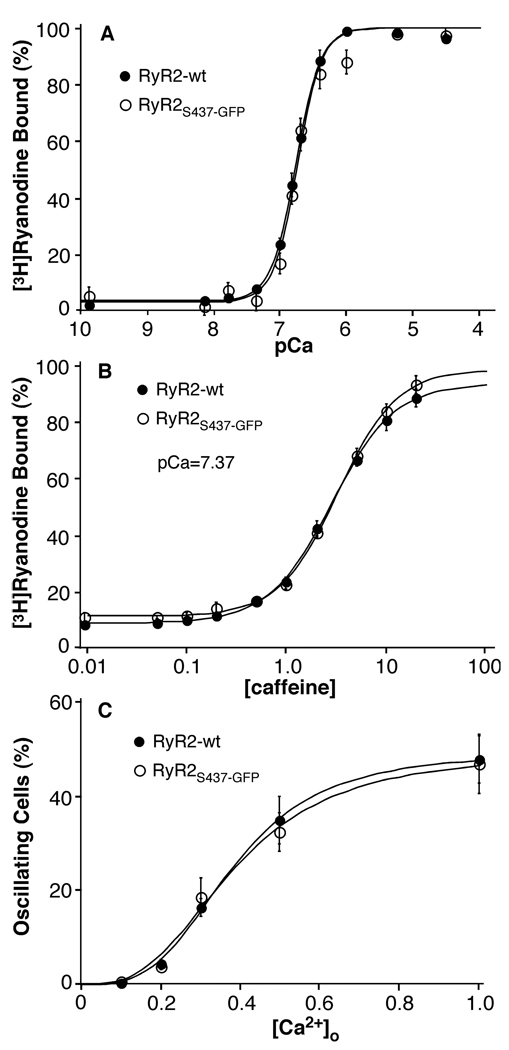
We have recently shown that the CPVT RyR2 mutation L433P does not markedly alter the Ca2+ dependence of [3H]ryanodine binding, but it does increase the sensitivity of the channel to activation by caffeine, and it does enhance the propensity for spontaneous Ca2+ release during store Ca2+ overload, or SOICR (10). The fact that GFP was inserted into the RyR2 sequence after S437, a mere four residues away, raised the possibility that the GFP insertion could alter the response of the channel to activation by various stimuli. To test this, we first assessed the Ca2+ dependence of [3H]ryanodine binding to RyR2S437-GFP. [3H]ryanodine binding has been widely used as a functional assay for RyR channel activity, since ryanodine binds only to the open state of the channel (29). Fig. 3A shows [3H]ryanodine binding to RyR2S437-GFP in the presence of a wide range of Ca2+ concentrations. Analysis of the Ca2+ dependence of [3H]ryanodine binding yielded an EC50 of 0.19 ± 0.01 µM (mean ± SEM, n=6), a value similar to EC50 of RyR2-wt (0.17 ± 0.14 µM, n=4). We then measured [3H]ryanodine binding to RyR2-wt and RyR2S437-GFP in the presence of various concentrations of caffeine. As shown in Fig.3B, RyR2-wt and RyR2S437-GFP displayed very similar responses to caffeine activation. The EC50 values for caffeine activation of [3H]ryanodine binding were 2.85 ± 0.21mM (n=5) for RyR2-wt and 3.32 ± 0.34mM (n=5) for RyR2S437-GFP. These values are not significantly different (P = 0.27, Student’s t-test).
Fig. 3. Ca2+- and caffeine-dependent activation and response to Ca2+ overload of RyR2S437-GFP.
(A) [3H]ryanodine binding to cell lysates prepared from HEK293 cells transfected with RyR2-wt (filled circles) or RyR2S437-GFP (open circles) was carried out at various concentrations of Ca2+ in the presence of 5 nM [3H]ryanodine. (B) [3H]ryanodine binding to cell lysates prepared from HEK293 cells transfected with RyR2-wt or RyR2S437-GFP was carried out at various concentrations of caffeine in the presence of ~43 nM (pCa=7.37) Ca2+ and 5 nM [3H]ryanodine. (C) Stable, inducible HEK293 cells expressing RyR2-wt or RyR2S437-GFP were grown on glass coverslips. The cells were induced with tetracycline for 24 hours and loaded with 5 mM fura-2-AM in KRH buffer for 20 min at room temperature. The cells were continuously perfused with KRH buffer containing 0.1, 0.2, 0.3, 0.5, or 1.0 mM CaCl2. The fluorescent intensity of individual cells was continuously monitored using single cell Ca2+ imaging. The fractions of RyR2-wt and RyR2S437-GFP cells that displayed Ca2+ oscillations at various [Ca2+]o are shown. Data shown are mean ± SEM from 3–6 separate experiments.
Furthermore, to determine whether the insertion of GFP into the NH2-terminal region of the channel altered SOICR, we generated a stable, tetracycline-inducible HEK293 cell line expressing RyR2S437-GFP. Store Ca2+ overload was induced by elevating the external Ca2+ concentration ([Ca2+]o), and the occurrence of SOICR was monitored via single cell Ca2+ imaging. As shown in Fig. 3C, the fraction of cells that displayed Ca2+ oscillations at each [Ca2+]o in HEK293 cells expressing RyR2S437-GFP was very similar to that in cells expressing RyR2-wt. These observations indicate that the insertion of GFP after Ser-437 did not grossly alter the sensitivity of RyR2 to Ca2+-or caffeine-activation, or the propensity for SOICR to occur.
Purification of RyR2S437-GFP
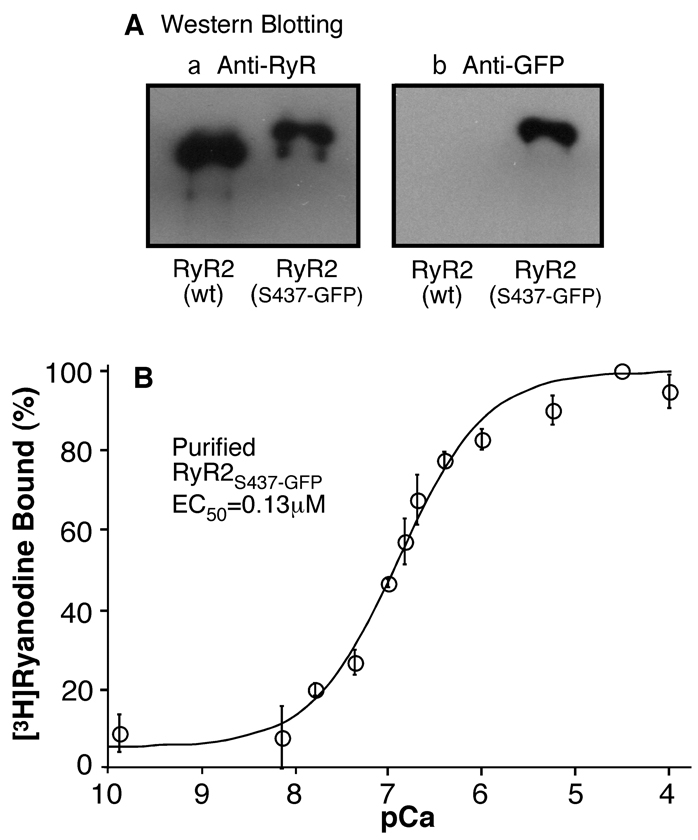
HEK293 cells transfected with RyR2S437-GFP cDNA were solubilized by the detergent CHAPS, and the RyR2S437-GFP protein was purified in a single step using GST-FKBP12.6, which binds to RyR2 with high affinity. The purified RyR2S437-GFP protein was analyzed by SDS-PAGE and immunoblotting. A single high molecular weight band, which migrated at a slightly slower rate than did the RyR2-wt band, was detected in the purified sample of RyR2S437-GFP (Fig. 4Aa), consistent with the addition of GFP. To further confirm the insertion of GFP into RyR2, the purified samples were immunoblotted with an anti-GFP antibody. As shown in Fig. 4Ab, the anti-GFP antibody recognized the purified RyR2S437-GFP sample, but not the purified RyR2-wt sample.
Fig. 4. Immuno-blotting of and [3H]ryanodine binding to the purified RyR2S437-GFP.
(A) The RyR2S437-GFP protein was purified from cell lysate by affinity chromatography using GST-FKBP12.6 as the affinity ligand. The purified RyR2S437-GFP and RyR2-wt proteins were solubilized, separated in 6% SDS-PAGE, and transferred to nitrocellulose membranes. The membrane was probed either with an anti-RyR antibody or an anti-GFP antibody for Western blotting. Note that the RyR2S437-GFP protein migrated at a slightly slower rate in SDS-PAGE than did the RyR2-wt, due to the insertion of GFP. Also note that the anti-GFP antibody reacted only with RyR2S437-GFP, but not with RyR2-wt. (B) [3H]ryanodine binding to purified RyR2S437-GFP protein (open circles) was carried out at various concentrations of Ca2+ in the presence of 5 nM [3H]ryanodine. Data shown are mean ± SEM from 3 separate experiments.
[3H]ryanodine binding was then employed to assess whether the purified RyR2S437-GFP protein remained functional. Fig. 4B shows [3H]ryanodine binding to the purified RyR2S437-GFP protein in the presence of a wide range of Ca2+ concentrations. Analysis of the Ca2+ dependence of [3H]ryanodine binding with the Hill equation yielded an EC50 of 0.13 ± 0.02 µM (mean ± SEM, n=3), indicating that the purified RyR2S437-GFP channel is capable of binding [3H]ryanodine with a Ca2+ dependence of activation similar to that of RyR2-wt (18).
Three-dimensional Reconstruction of RyR2S437-GFP
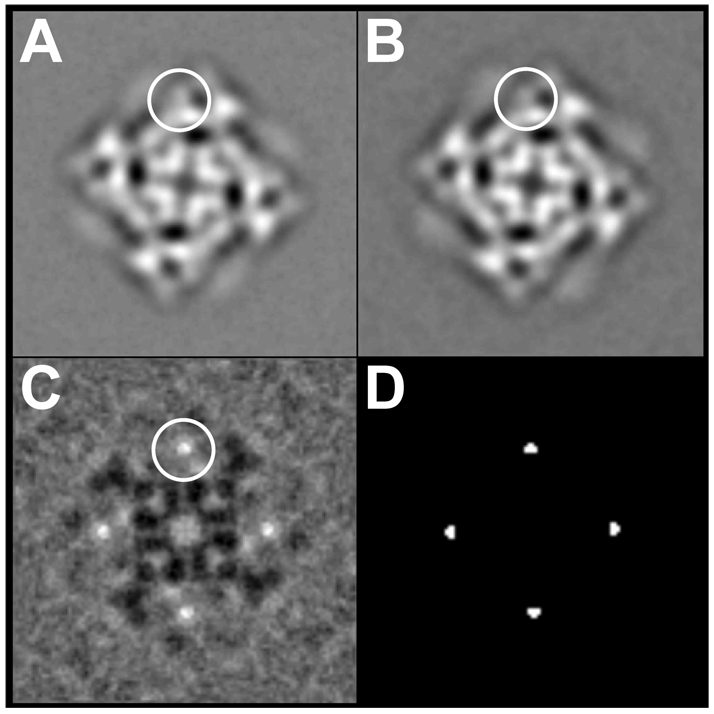
We next imaged the purified RyR2S437-GFP proteins using Cryo-EM. The purified proteins were diluted in a lipid- and sucrose-free buffer and applied to carbon-coated EM grids, which were rapidly frozen in liquid ethane. The frozen samples were then subjected to cryo-EM. Fig. 5 shows a typical electron micrograph of frozen-hydrated RyR2S437-GFP. Individual particles with apparently intact structure and assuming multiple orientations were readily detected. Particles that were oriented with their 4-fold symmetry axes perpendicular to the carbon support film were selected, and their images were aligned using cross-correlation methods (Fig. 6). The overall two-dimensional projection of RyR2S437-GFP (Fig. 6A) was very similar to that of RyR2-wt reported previously (Fig. 6B). To identify subtle localized differences between these two projected structures, a difference map was generated by subtracting the two-dimensional average of RyR2-wt from that of RyR2S437-GFP (Fig. 6C). The brightest areas in the difference map correspond to the most significant positive densities, which represent extra protein mass present in RyR2S437-GFP, but absent from RyR2-wt. These areas were located within the "clamp" structures that form the corners of the square-shaped receptor (Fig. 6C) (30, 31). The differences, which are highly significant, with a confidence level of >99.9% (Fig. 6D) (32), correspond to the additional mass contributed by the GFP insertion in RyR2S437-GFP.
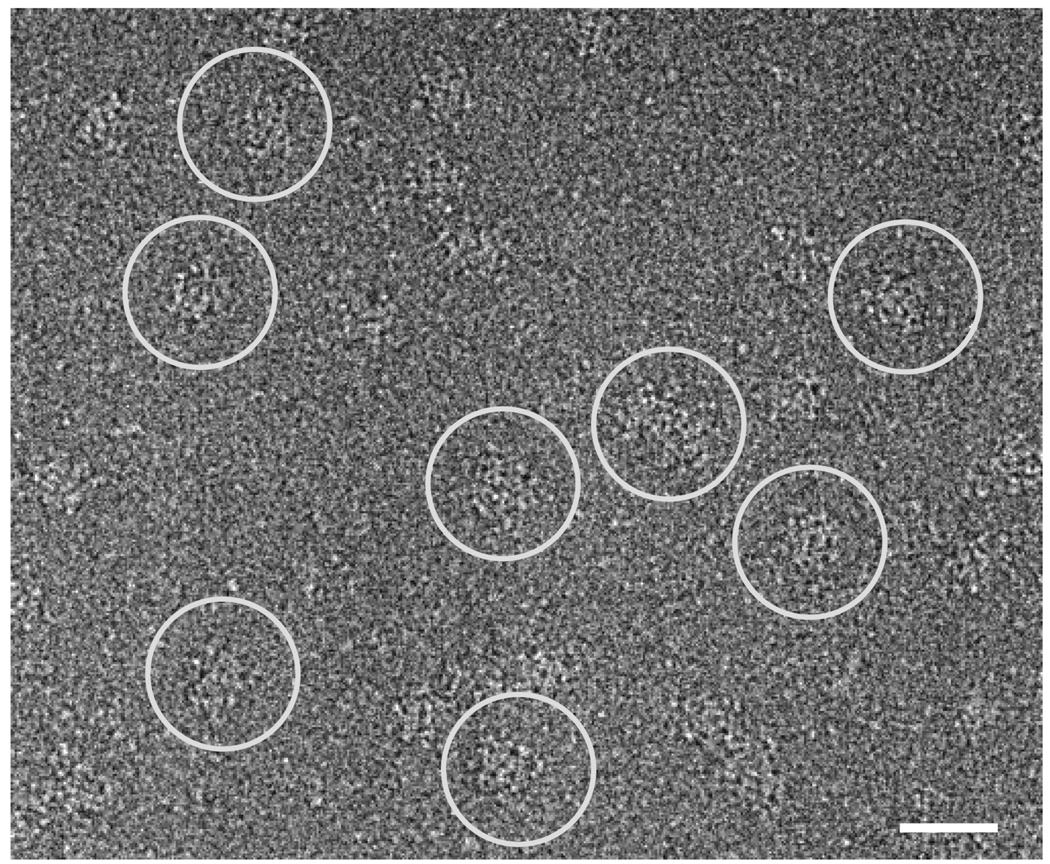
Fig. 5. Cryo-electron microscopy of RyR2S437-GFP.
A portion of a cryo-EM micrograph of the purified RyR2S437-GFP proteins embedded in a thin layer of vitreous ice is shown. Several individual RyR2T1874-GFP particles are marked with white circles. The scale bar represents 500Å.
Fig. 6. Two-dimensional averages of RyR2S437-GFP and RyR2-wt.
(A) Two-dimensional average of RyR2S437-GFP (n=263 particle images) “top” view; (B) The top view of the two-dimensional average of RyR2-wt (n=269 particle images); (C) Difference map obtained by subtracting (B) from (A). The top view represents the projection of the channel as seen from the cytoplasmic side. The largest differences shown in (C), corresponding to the additional masses due to the GFP insertion, are seen as bright white areas, one of which is circled, in the clamp-shaped domains. (D) Map of statistically significant regions of difference obtained by t-test; map is displayed at >99.9% confidence level. The width of each frame is 544 Å.
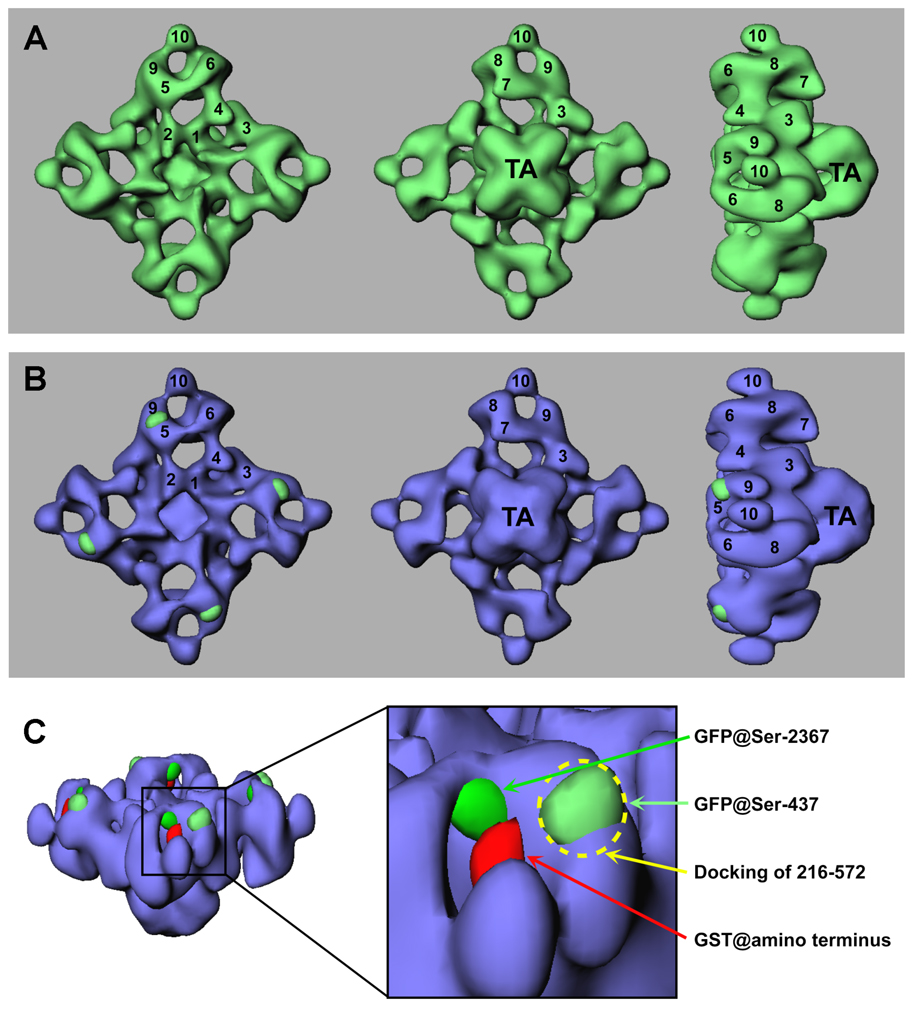
To obtain a more detailed structure of RyR2S437-GFP and to more precisely map the location of the inserted GFP, three-dimensional volumes are computed and compared. Fig. 7A shows a surface representation of the resulting three-dimensional reconstruction of RyR2S437-GFP in three orientations (top, bottom, and side views). The reconstructed structure of RyR2S437-GFP resembles a mushroom shape, which consists of two major components: a large square cytoplasmic assembly (290×290×130Å) encompassing 10 distinct domains (labeled by numerals in Fig. 7), and a smaller transmembrane assembly (120×120×70Å, labeled “TA”). The final resolution of the reconstructed RyR2S437-GFP was estimated to be 33 Å. Although the overall three-dimensional reconstruction of RyR2S437-GFP is very similar to that of RyR2-wt, some small differences exist. The most noticeable difference was found in the region between domains 5 and 9 in RyR2S437-GFP, which appears to be larger than the corresponding region in RyR2-wt. To further analyze these differences, a three-dimensional difference map was generated by subtracting the three-dimensional volume of RyR2-wt from that of RyR2S437-GFP. The difference regions were displayed in green and superimposed on the three-dimensional reconstruction of RyR2-wt (shown in blue) (Fig. 7B). This difference map clearly shows four significant differences located between each pair of domains 5 and 9 in the cytoplasmic assembly. It should be noted first, that 4-fold symmetry was imposed, and second, that these are the only significant differences that appear when the three-dimensional difference map is displayed at a density threshold similar to the one used to image the RyR2S437-GFP and RyR2-wt structures. Since the GFP tag was inserted into each RyR2 monomer, and since RyR2 is a homotetramer composed of four identical monomers, the difference would be expected to repeat four times in the three-dimensional difference map. Consistent with the notion that these four structural repeated differences are a result of GFP insertion, the calculated volume of each of these four areas showing the most difference corresponds to a molecular mass of 28 kDa, assuming a protein density of 1.37 g/cm3 (33). This value agrees well with the molecular mass of GFP. Furthermore, the locations of the major differences observed in the three-dimensional reconstruction (Fig. 7) are consistent with those observed in the averaged two-dimensional projections (Fig. 6). Based on these observations, we conclude that the NH2 mutation region, as indicated by the inserted GFP at Ser-437, is located between domains 5 and 9 in each of the clamp-shaped structures of RyR2.
Fig. 7. Three-dimensional surface representation of RyR2S437-GFP and difference map.
(A) The three-dimensional reconstruction of RyR2S437-GFP is shown in green. (B) Difference map (RyR2S437-GFP – RyR2-wt) shown in green is superimposed on the three-dimensional reconstruction of the RyR2-wt (in blue). The three-dimensional reconstructions are shown in three views: left, top views from the cytoplasmic surface, which in situ would face the transverse-tubule; middle, views towards the bottom of the channel (i.e., as it would appear if viewed from the lumen of the sarcoplasmic reticulum); right, side views. The numerals on the cytoplasmic assembly indicate the distinguishable domains. (C) A detailed view of the clamp-shaped structures, showing the relative positions of the inserted GFPs at Ser-437 and Ser-2367, the inserted GST at the amino terminus (in red), and the docking of a domain formed by residues 216–572 in RyR1, as indicated by a dash circle.
DISCUSSION
Localization of an NH2-terminal Mutation Hotspot within the Clamp-shaped Structure
The NH2-terminal region is one of three major regions in the RyR sequence that harbor disease-causing mutations (2, 4). To understand the structural basis of these mutations, a considerable effort has been made to localize the NH2-terminal domain in the three-dimensional structure of RyRs. The first structural localization of the NH2- terminal region was achieved by fusing glutathione-S-transferase (GST) to the NH2-terminus of the type 3 ryanodine receptor (RyR3) and then comparing three-dimensional reconstructions of the RyR3-wt and the GST-tagged RyR3 (34). That study mapped the GST and, thus, the NH2-terminus, to the middle of the region defined by domains 7–10 within the clamp-shaped structures located at the corners of the cytoplasmic assembly of RyR3 (34). The localization of the NH2-terminus within the clamp-shaped structure was confirmed in subsequent bioinformatics and cryo-EM studies. Baker et al. identified an oxidoreductase-like domain formed by residues 41–420 in RyR1, corresponding to 41–435 in RyR2, and they computationally localized this domain to a region largely defined by domain 5 within the clamp-shaped structures in the cytoplasmic region of RyR1 (35). Similarly, using a sequence based fold recognition method, Serysheva et al. predicted a domain fold for residues 216–572 of RyR1, corresponding to 230–583 in RyR2, based on the crystal structure of the IP3-binding core region of the type 1 IP3 receptor, and they computationally docked this domain fold to a region defined by domains 5 and 9 in the clamp structures (36). Consistent with this computational localization, an antibody raised against a synthetic peptide corresponding to residues 416–434 in RyR1, equivalent to 431–449 in RyR2, was mapped in two-dimensional projection to two major sites within the clamp structures and two minor sites in the central portion of the channel (35). However, the three-dimensional localization of this antibody was not done and so the identity of domains involved could not be unambiguously assessed.
In the present study, we engineered a GFP-RyR2 fusion protein in which GFP was inserted into the RyR2 sequence after residue Ser-437 and then mapped the inserted GFP to a surface-exposed site between domains 5 and 9 within the clamp-shaped region in the three-dimensional structure of RyR2. As shown in Fig. 7C, the three-dimensional location of GFPS437 is largely superimposed on the predicted docking of residues 216–572 of RyR1 (corresponding to 230–583 of RyR2), and it also overlaps with the predicted localization of the C-terminus of the oxidoreductase-like domain formed by residues 41–423 of RyR1 (corresponding 41–435 of RyR2). Hence, the results of the present study confirm and extend those of previous studies, which consistently predicted that the mutation hotspot in the NH2-terminal region around Ser-437 would be located in the vicinity of domains 5 and 9 within the clamp-shaped structure of RyRs.
The Clamp-shaped Structure of RyR is a Crucial Part of the Channel That is Highly Susceptible to Disease-causing Mutations
The clamp-shaped structures at the corners of the cytoplasmic assembly are the regions in the three-dimensional structure of RyRs that display major differences in conformations between the closed and open states (30, 31). Specifically, domains 9 and 10, which are fused in the closed state, are separated in the open conformation. Similarly, domains 5 and 6 are further apart in the open state than in the closed state. Furthermore, the size and shape of each of domains 5, 6, 9, and 10 themselves exhibit some changes when RyR switches from the closed to the open state. Although no major changes were observed between domains 5 and 9, where the GFPS437 was located, at the current level of resolution, this region is likely to undergo some conformational changes as a result of major changes between domains 5 and 6, between domains 9 and 10, and within domains 5 and 9. Considering its involvement in channel gating, the clamp-shaped structure is likely to be susceptible to disease-causing mutations. In support of this view, we have mapped a mutation in the central region (GFPS2367) to a site between domains 5 and 6 in a previous study (14), and we have mapped a mutation site in the NH2-terminal region into a spot between domains 5 and 9 in the present study (Fig. 7C). Based on these findings and on the previous localizations of the NH2-terminus, GFPT1366, and GFPT1874, all of which are within the clamp structure (34, 37, 38), we predict that other disease-causing mutations in the NH2-terminal region and the central region will be found to lie within the clamp-shaped structure.
RyRs are regulated by a number of endogenous and exogenous ligands and protein effectors, including Ca2+, Mg2+, ATP, FKBPs, calmodulin, DHPR, PKA, CaMKII, caffeine, and procaine (39, 40). The location of the FKBP12.0/FKBP12.6 binding site was mapped to a region between domains 9 and 3, adjacent to the clamp-shaped structure (41, 42), while the binding site for calmodulin was located in a region between domains 3 and 4 (43). The binding sites for other ligands and modulators are unknown. Given the fact that the clamp-shaped structure is associated with disease-causing mutations and conformational changes involved in channel gating, it would not be surprising to find that the clamp-shaped structure is also a target of channel modulators. In this regard, it is of interest to note that the NH2-terminal sequence of RyR exhibits a high degree of homology to the IP3-binding core of the IP3 receptor (36), suggesting that the NH2-terminal region of RyR may contain binding sites for endogenous regulators. It is also interesting that the computationally predicted location of the C-terminal part of a domain formed by residues 216–572 of RyR1 (corresponding to 230–583 of RyR2) is very close to the FKBP12 binding site in the three-dimensional structure of RyR1 (36), suggesting that the NH2-terminal region is involved in FKBP-RyR interaction. Consistent with this possibility, we have demonstrated that the deletion of the first 305 NH2-terminal residues abolishes FKBP12.6 binding (24).
Implication for Disease Mechanisms Associated with the NH2-terminal and Central Mutations in RyRs
We have recently shown that disease-causing mutations in the N-terminal, central, and C-terminal regions of the RyR2 sequence enhance the sensitivity of the channel to activation by luminal Ca2+ (10, 11). The C-terminal region is folded within trans-membrane assembly in three-dimensional structure and encompasses the channel conduction pathway. It contains regions that are accessible to luminal Ca2+ and likely involved in the activation of the channel by luminal Ca2+. However, it is more difficult to imagine how mutations in the NH2-terminal and central regions would affect luminal Ca2+ sensing, which is presumably controlled by the C-terminal region. Ikemoto and his colleagues have proposed that although they are far apart in the primary sequence of RyR2, the NH2-terminal and central regions of RyR are located in close proximity in the three-dimensional structure of the channel (12). Those authors further hypothesize that disease-causing mutations are located on the surface of the NH2-terminal and central domains. Mutations weaken normal inter-domain interactions, thereby promoting channel opening upon stimulation via long-range (>100 Ǻ) conformational alterations that link the NH2-terminal and central regions to the COOH-terminal region (12). A test of this domain “zipping-unzipping” hypothesis would be to locate the NH2-terminal and central mutation regions in the three-dimensional structure. To this end, we have mapped a central mutation site to an area bridging domains 5 and 6, and an NH2- terminal mutation hotspot to a site between domains 5 and 9. The two sites are separated by about 30Å. Hence, our data support the idea that the NH2-teminal and central mutation regions are closely neighboring in the three-dimensional structure of RyR2. However, given the moderate resolution of our three-dimensional reconstructions, it is impossible to pinpoint the areas of interaction between the NH2-terminal and central mutation regions. Since the location of GFPS437 was about 30Å away from that of GFPS2367, it is unlikely that the NH2-teminal mutations near Ser-437 directly interacts with the central mutation site around Ser-2367. On the other hand, it has been proposed that the region containing another mutation R614 directly interacts with the central mutation region (13). Thus, it will be of great interest to map residue R614 onto the three-dimensional structure of RyR2.
Further work is needed to define how many and precisely which domains are involved in the cardiac and skeletal muscle diseases that arise from mutations in the NH2-terminal and central-regions of RyR’s. Based upon the results discussed so far, it seems that domain 5 is certainly involved and that domains 6 and/or possibly domain 9 are as well. The simplest interpretation of our structural findings that is consistent with Ikemoto’s domain-domain interaction hypothesis is that the NH2-terminal mutations are located in domain 5 and the central mutations are in domain 6, and that alteration of interactions between these two domains is responsible for the observed phenotypes. However, we cannot yet exclude the other domains in the clamp-shaped structure (7, 8, 10 in Fig. 7) until the boundaries of the mutation hotspots are better defined.
An interesting observation from the present study was that the insertion of GFP into an NH2-terminal mutation hotspot after residue Ser-437 did not significantly alter the functional properties of RyR2, despite the fact that mutations around the GFP-insertion site (Ser-437) alter channel function. HEK293 cells expressing RyR2S437-GFP displayed responses to caffeine, ryanodine, and Ca2+ overload similar to those of HEK293 cells expressing RyR2-wt. The Ca2+- and caffeine-dependent activations of [3H]ryanodine binding to RyR2-wt and RyR2S437-GFP are indistinguishable. The reason for the lack of functional impact of the inserted GFP is not clear, but is likely related to the position of insertion, and the structure of the inserted GFP. The inserted GFP was located in the middle of a small stretch of 12 amino acid residues that are highly divergent among RyR isoforms. Disease-causing RyR mutations are located on either sides of this region. It is possible that this stretch of amino acids forms a surface-exposed loop, and that insertion of GFP into this loop does not alter the nearby structures. Furthermore, the inserted GFP was flanked by 9–10 amino acid-spacers containing eight glycine residues, which should provide a high degree of flexibility and minimize steric hindrance. Since the GFP-inserted region is highly divergent among RyR isoforms, the impact of the GFP-insertion on the function of the RyR channels may be isoform-dependent. Investigations are currently underway to determine whether the insertion of GFP into the corresponding region of RyR1 alters the functional properties of the RyR1 channel.
In summary, despite an increasing body of biochemical and functional evidence suggesting that disease-causing RyR mutations are located along the domain interface between the NH2-terminal and central regions of RyRs and weaken inter-domain interactions (the domain “unzipping” hypothesis), there has been little structural evidence for these inter-domain interactions. To test this domain unzipping hypothesis, we have started to map different mutation regions in the three-dimensional structure of RyR2. Our data indicate that although they are ~2000 residues apart in the primary sequence of RyR2, the NH2-terminal and central mutation regions are closely located in the three-dimensional structure of RyR2.
Acknowledgments
This work was supported by research grants from the Canadian Institutes of Health Research and the Heart and Stroke Foundation of Canada, Alberta to S.R.W.C., the American Heart Association grant 0430076N to Z.L., and the National Institutes of Health grant AR40615 to T.W. We would like to thank the Wadsworth Center’s DNA Sequencing Core and Electron Microscopy Core Facilities, and the Resource for Visualization of Biological Complexity (NIH Biotechnological Resource Grant RR01219).
REFERENCES
- 1.Scoote M, Williams AJ. Biochem. Biophys. Res. Commun. 2004;322:1286–1309. doi: 10.1016/j.bbrc.2004.08.034. [DOI] [PubMed] [Google Scholar]
- 2.Priori SG, Napolitano C. J. Clin. Invest. 2005;115:2033–2038. doi: 10.1172/JCI25664. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Kontula K, Laitinen PJ, Lehtonen A, Toivonen L, Viitasalo M, Swan H. Cardiovasc. Res. 2005;67:379–387. doi: 10.1016/j.cardiores.2005.04.027. [DOI] [PubMed] [Google Scholar]
- 4.Yano M, Yamamoto T, Ikeda Y, Matsuzaki M. Nat. Clin. Pract. Cardiovasc. Med. 2006;3:43–52. doi: 10.1038/ncpcardio0419. [DOI] [PubMed] [Google Scholar]
- 5.Loke J, MacLennan DH. Am. J. Med. 1998;104:470–486. doi: 10.1016/s0002-9343(98)00108-9. [DOI] [PubMed] [Google Scholar]
- 6.Lyfenko AD, Goonasekera SA, Dirksen RT. Biochem. Biophys. Res. Commun. 2004;322:1256–1266. doi: 10.1016/j.bbrc.2004.08.031. [DOI] [PubMed] [Google Scholar]
- 7.Treves S, Anderson AA, Ducreux S, Divet A, Bleunven C, Grasso C, Paesante S, Zorzato F. Neuromuscul. Disord. 2005;15:577–587. doi: 10.1016/j.nmd.2005.06.008. [DOI] [PubMed] [Google Scholar]
- 8.Wehrens X, Lehnart S, Huang F, et al. Cell. 2003;113:829–840. doi: 10.1016/s0092-8674(03)00434-3. [DOI] [PubMed] [Google Scholar]
- 9.George CH, Higgs GV, Lai FA. Circ. Res. 2003;93:531–540. doi: 10.1161/01.RES.0000091335.07574.86. [DOI] [PubMed] [Google Scholar]
- 10.Jiang D, Wang R, Xiao B, Kong H, Hunt DJ, Choi P, Zhang L, Chen SRW. Circ. Res. 2005;97:1173–1181. doi: 10.1161/01.RES.0000192146.85173.4b. [DOI] [PubMed] [Google Scholar]
- 11.Jiang D, Xiao B, Yang D, Wang R, Choi P, Zhang L, Cheng H, Chen SRW. Proc. Nat. Acad. Sci., USA. 2004;101:13062–13067. doi: 10.1073/pnas.0402388101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Ikemoto N, Yamamoto T. Front. Biosci. 2002;7:d671–d683. doi: 10.2741/A803. [DOI] [PubMed] [Google Scholar]
- 13.Murayama T, Oba T, Hara H, Wakebe K, Ikemoto N, Ogawa Y. Biochem. J. 2006 doi: 10.1042/BJ20061040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Liu Z, Wang R, Zhang J, Chen SR, Wagenknecht T. J. Biol. Chem. 2005;280:37941–37947. doi: 10.1074/jbc.M505714200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Chen SRW, Li X, Ebisawa K, Zhang L. J. Biol. Chem. 1997;272:24234–24246. doi: 10.1074/jbc.272.39.24234. [DOI] [PubMed] [Google Scholar]
- 16.Sambrook J, Fritsch EF, Maniatis T. 1989 [Google Scholar]
- 17.Zhao M, Li P, Li X, Zhang L, Winkfein RJ, Chen SR. J. Biol. Chem. 1999;274:25971–25974. doi: 10.1074/jbc.274.37.25971. [DOI] [PubMed] [Google Scholar]
- 18.Liu Z, Zhang J, Li P, Chen SR, Wagenknecht T. J. Biol. Chem. 2002;240:24. doi: 10.1074/jbc.M208124200. [DOI] [PubMed] [Google Scholar]
- 19.Ho SN, Hunt HD, Horton RM, Pullen JK, Pease LR. Gene. 1989;77:51–59. doi: 10.1016/0378-1119(89)90358-2. [DOI] [PubMed] [Google Scholar]
- 20.Wagenknecht T, Frank J, Boublik M, Nurse K, Ofengand J. J. Mol. Biol. 1988;203:753–760. doi: 10.1016/0022-2836(88)90207-0. [DOI] [PubMed] [Google Scholar]
- 21.Frank J. Three-dimensional electron microscopy of macromolecular assemblies. 2nd ed. New York: Oxford University Press; 2006. pp. 71–142. [Google Scholar]
- 22.Penczek PA, Grassucci RA, Frank J. Ultramicroscopy. 1994;53:251–270. doi: 10.1016/0304-3991(94)90038-8. [DOI] [PubMed] [Google Scholar]
- 23.Malhotra A, Penczek P, Agrawal RK, Gabashvili IS, Grassucci RA, Junemann R, Burkhardt N, Nierhaus KH, Frank J. J. Mol. Biol. 1998;280:103–116. doi: 10.1006/jmbi.1998.1859. [DOI] [PubMed] [Google Scholar]
- 24.Masumiya H, Wang R, Zhang J, Xiao B, Chen SRW. J. Biol. Chem. 2003;278:3786–3792. doi: 10.1074/jbc.M210962200. [DOI] [PubMed] [Google Scholar]
- 25.Doi N, Yanagawa H. Methods Mol. Biol. 2002;183:49–55. doi: 10.1385/1-59259-280-5:049. [DOI] [PubMed] [Google Scholar]
- 26.Niwa H, Inouye S, Hirano T, Matsuno T, Kojima S, Kubota M, Ohashi M, Tsuji FI. Proc. Natl. Acad. Sci. U. S. A. 1996;93 doi: 10.1073/pnas.93.24.13617. 13617-1322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Waldo GS, Standish BM, Berendzen J, Terwilliger TC. Nat. Biotechnol. 1999;17 doi: 10.1038/10904. 691-65. [DOI] [PubMed] [Google Scholar]
- 28.Chen SR, Li P, Zhao M, Li X, Zhang L. Biophys. J. 2002;82 doi: 10.1016/s0006-3495(02)75587-2. 2436-247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Tanna B, Welch W, Ruest L, Sutko JL, Williams AJ. J. Gen. Physiol. 1998;112:55–69. doi: 10.1085/jgp.112.1.55. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Orlova EV, Serysheva I, van Heel M, Hamilton SL, Chiu W. Nat. Struct. Biol. 1996;3:547–552. doi: 10.1038/nsb0696-547. [DOI] [PubMed] [Google Scholar]
- 31.Serysheva I, Schatz M, van Heel M, Chiu W, Hamilton SL. Biophys. J. 1999;77:1936–1944. doi: 10.1016/S0006-3495(99)77035-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Liu Z, Zhang J, Sharma MR, Li P, Chen SR, Wagenknecht T. Proc. Natl. Acad. Sci. U. S. A. 2001;98:6104–6619. doi: 10.1073/pnas.111382798. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Radermacher M, Rao V, Grassucci R, Frank J, Timerman AP, Fleischer S, Wagenknecht T. J. Cell Biol. 1994;127:411–423. doi: 10.1083/jcb.127.2.411. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Liu Z, Zhang J, Sharma MR, Li P, Chen SR, Wagenknecht T. Proc. Natl. Acad. Sci. U. S. A. 2001;98:6104–6619. doi: 10.1073/pnas.111382798. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Baker ML, Serysheva II, Sencer S, Wu Y, Ludtke SJ, Jiang W, Hamilton SL, Chiu W. Proc. Natl. Acad. Sci. U. S. A. 2002;99:12155–12160. doi: 10.1073/pnas.182058899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Serysheva II, Hamilton SL, Chiu W, Ludtke SJ. J. Mol. Biol. 2005;345:427–431. doi: 10.1016/j.jmb.2004.10.073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Zhang J, Liu Z, Masumiya H, Wang R, Jiang D, Li F, Wagenknecht T, Chen SRW. J. Biol. Chem. 2003;278:14211–14218. doi: 10.1074/jbc.M213164200. [DOI] [PubMed] [Google Scholar]
- 38.Liu Z, Zhang J, Wang R, Wayne Chen SR, Wagenknecht T. J. Mol. Biol. 2004;338:533–545. doi: 10.1016/j.jmb.2004.03.011. [DOI] [PubMed] [Google Scholar]
- 39.Meissner G. Annu. Rev. Physiol. 1994;56:485–508. doi: 10.1146/annurev.ph.56.030194.002413. [DOI] [PubMed] [Google Scholar]
- 40.Hamilton SL. Cell Calcium. 2005;38:253–260. doi: 10.1016/j.ceca.2005.06.037. [DOI] [PubMed] [Google Scholar]
- 41.Wagenknecht T, Grassucci R, Berkowitz J, Wiederrecht GJ, Xin HB, Fleischer S. Biophys. J. 1996;70:1709–1715. doi: 10.1016/S0006-3495(96)79733-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Sharma MR, Jeyakumar LH, Fleischer S, Wagenknecht T. Biophys. J. 2002;82:644a. doi: 10.1529/biophysj.105.063503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Wagenknecht T, Berkowitz J, Grassucci R, Timerman AP, Fleischer S. Biophys. J. 1994;67:2286–2295. doi: 10.1016/S0006-3495(94)80714-3. [DOI] [PMC free article] [PubMed] [Google Scholar]