Abstract
Large volumes of protein sequence and structure data acquired by proteomic studies led to the development of computational bioinformatic techniques that made possible the functional annotation and structural characterization of proteins based on their primary structure. It has become evident from genome-wide analyses that many proteins in eukaryotic cells are either completely disordered or contain long unstructured regions that are crucial for their biological functions. The content of disorder increases with evolution indicating a possibly important role of disorder in the regulation of cellular systems. Transcription factors are no exception and several proteins of this class have recently been characterized as premolten/molten globules. Yet, mammalian cells rely on these proteins to control expression of their 30,000 or so genes. Basic region:leucine zipper (bZIP) DNA-binding proteins constitute a major class of eukaryotic transcriptional regulators. This review discusses how conformational flexibility “built” into the amino acid sequence allows bZIP proteins to interact with a large number of diverse molecular partners and to accomplish their manifold cellular tasks in a strictly regulated and coordinated manner.
Keywords: bZIP proteins, transcription control, intrinsic disorder, molecular recognition motifs, induced protein folding
INTRODUCTION
Sequencing of the whole genomes from diverse specimens together with the application of automated high-throughput molecular biology techniques has allowed an accumulation of a vast systematic data in proteomics, interactomics, transcriptomics, and several other “omics”. The central issue of the ongoing efforts is the identification of function of genes and of the proteins they encode. These studies became possible thanks to powerful techniques for studying protein structure and protein–protein interactions, mapping regulatory elements within gene promoters utilizing gene chips and chromatin precipitation methods, and the development of bioinformatics. This wealth of data, combined with tools for gene-based analysis opened new avenues for systematic discoveries of functionally important features, posed new questions and challenges. The challenge of understanding the organization and dynamics of cellular networks in time and space has shifted the focus of interest from molecular biology toward systems biology. In the case of gene regulatory networks, attempts are being made to delineate all transcription factors (TFs), chromatin regulators and signaling pathways necessary to activate genes in specific tissues and cell types, and to determine when specific gene expression occurs and how it is regulated.
Intrinsically Disordered Proteins
Concomitantly with the widespread employment of genetic methods to identify proteins with specific functions, the application of improved spectroscopic techniques (mainly CD and NMR) to examine macromolecular structures in solution revealed that many proteins lack a well-ordered tertiary structure under physiological conditions. It has been recognized that such proteins (or regions), termed “natively unfolded”, “intrinsically unstructured” or “intrinsically disordered” can exist as dynamic ensembles of interconverting conformers in three disordered structural states: fully unfolded extended random coils, premolten globules that retain some amount of residual secondary structure, and compact but disordered molten globule-like ensembles [1,2]. Disordered regions can be inserted within the single structural domain, usually in the form of long surface loops [3], or serve as flexible linkers between independently folded globular domains. Many examples of such proteins have been described in en excellent review by Dyson and Wright [4]. Surveys of whole genomes showed that the occurrence of long (> 50 residues) unstructured regions is very common in functional eukaryotic proteins [5]. This observation changed the general view on the native structure and its relation to protein function. As accumulated evidence showed the importance of intrinsic disorder (ID) for protein function, in a landmark paper, Wright and Dyson called for reassessing the classic protein structure–function paradigm to include all possible native conformations [6]. Function may arise from any of the conformational states or interconversion between states [5].
In a recent paper, Radivojac et al. [7] proposed to classify a protein as intrinsically disordered if it contains at least one disordered region whose length is sufficient for experimental characterization. Intrinsically disordered proteins (IDPs) have been studied by several methods: NMR (reviewed in [8]), fluorescence, CD and Raman spectroscopy, hydrodynamic and calorimetric methods, and small-angle X-ray scattering, as well as other biophysical techniques [9]. Proteins and protein domains experimentally confirmed to be unstructured fall into several functional categories including effectors, scavengers, assemblers and entropy chains [5,10], and have been implicated in the regulation of cell cycle control, transcription and signal transduction [11]. IDPs were subjects of extensive recent studies, focused reviews [2,4,7,10–15] and commentaries [1,5,6].
Intrinsic Disorder Can be Predicted from the Amino Acid Sequence
The large volume of accumulated sequence data made possible the development of computational techniques that utilize the information contained in amino acid sequences to predict structural disorder. A comparison of sequences from ordered and disordered regions of proteins revealed significant differences in their respective amino acid composition. Several artificial neural network based predictors of natively disordered regions (PONDRs) have been developed based on training of the selected sequence feature (Ref. [2] and references therein). The work of Vucetic and colleagues [16] indicated that the amino acid sequence underlying ID also specifies distinct properties of the polypeptide that correlate with its function. This observation raised the possibility of assigning IDPs to functionally specific classes (flavors) based on their amino acid composition alone. Implementation of a machine learning method for predicting protein function from sequence indeed demonstrated that the inclusion of disordered features improved prediction accuracy for certain functional categories, and made possible the annotation of several orphan human proteins [17]. The manifestations of disorder at the primary structure level include a low sequence complexity, in particular a low content of bulky, hydrophobic residues combined with a high proportion of charged/polar residues, resulting in a large net charge of the protein at neutral pH. Uversky et al. [18] showed that it is possible to predict whether a given amino acid sequence encodes a folded or a natively unfolded protein based on a charge-hydropathy plot. This principle has been employed in development of the protein disorder predictor server FoldIndex [19], to identify local disordered regions on a per residue basis. Other predictors based on specific sequence attributes are PreLink [20] (low hydrophobic cluster content), NORSp [21] (solvent-accessible regions devoid of secondary structure), GlobPlot [22], which uses a scale for propensities for secondary structures and random coils, and based on sequence profiles, DISOPRED2 program. A different approach is employed by the IUPred algorithm, which evaluates the energy resulting from interresidue interactions and has not been trained against a dataset of disordered protein regions. For reviews of the currently available disorder predictors, see [23,24]. Each of these predictors suffers from different limitations and performs better for one type of disorder than for another (e.g., DISOPRED2 and PreLink are able to predict short disordered regions in the context of globally ordered protein, whereas the charge-hydropathy method usually indicates unstructured random coils). Therefore, a reliable prediction of protein disorder has to combine outputs from several predictors based on different physical principles and/or definitions of disorder [23–25]. The current estimate of accuracy for the ab initio methods such as PONDR is about 70% [23]. It is anticipated that further improvement of computational techniques for sequence-based ID prediction could increase the level of accuracy to its upper limit of 85%–90% [7].
Application of these techniques in the whole genomes-wide studies revealed an abundance of proteins containing ID regions, their evolutionary conservation and increasing content in higher organisms, suggesting the importance of protein disorder for regulation of cellular processes [1,17,26–28]. ID is prevalent among proteins involved in cell signaling and DNA/RNA recognition in eukaryotic cells [15,29]. Recently, Romero et al. [30] showed that the majority of alternative spliced peptide fragments were associated with disordered protein regions, and the researchers postulated a relationship between increased ID content and alternative splicing phenomena in multicellular eukaryotes. By affecting mainly natively unstructured regions, alternative splicing may increase functional diversity of the proteome without disrupting the structural integrity of protein components.
The Role of Intrinsic Disorder in Protein Function
The dynamic flexibility associated with the intrinsic lack of structure may confer multiple benefits for protein function, particularly for biomolecular recognition and organization of multicomponent cellular networks. Unstructured regions enable proteins to contact their ligands over a large binding surface [31] and permit binding to multiple targets. Structural plasticity allows a protein to adopt more than one conformation, depending upon the ligand, and to recognize diverse targets (reviewed in [4]).
What are the advantages for molecular recognition between flexible macromolecules compared to rigid systems? In both systems the bimolecular reaction requires diffusional formation of an encounter pair that further evolves into a stable complex in a number of steps. According to the early model of “conformational selection”, a multitude of possibilities for conformational searches offered by the presence of flexible regions in interacting protein(s) result in much better selectivity compared to stably folded molecules. It has been anticipated that binding which involves many isomerization steps is much more efficient in terms of rate and specificity than the “lock–and–key“ type of recognition with a large number of unsuccessful fittings. Furthermore, specific biological function often requires significant conformational reorganization of the interacting proteins. The best example is the regulation of enzymatic activity by the allosteric effect. Small adjustments to the conformation were successfully described by the “induced fit” model. However, there are very many examples of proteins that are partially or completely disordered in the unbound state and undergo function-related disorder–order transitions upon binding to their respective partners [12,18,32]. The most relevant explanation for this phenomenon comes from the requirement for highly specific but weak, readily reversible interactions in very many biological processes. The importance of transient interactions between both proteins and nucleic acids is most evident in transcription, wherein transcription regulatory proteins exchange contacts with a variety of cofactors (see below). In contrast to interactions between rigid molecules, in which stability of the complex usually correlates with specificity, in the binding associated with large structural changes, high specificity (the best fit) is achieved through conformational adjustments, often at the expense of binding affinity. The molecular mechanisms for target-assisted protein folding and the role of flexibility in determining the binding specificity have recently been addressed by a number of experimental and theoretical studies [12,33–39].
Coupling of folding and binding facilitates control and regulation of the binding thermodynamics (reviewed in [4,40]) and increased rates of macromolecular associations [41]. These abilities can be attributed to the large solvated surface area of the polypeptide chain in an extended conformation. It is well documented that the burial of hydrophobic surfaces and the consequent release of hydration waters to the bulk solvent can significantly contribute to the thermodynamics of binding [42]. The ordering of structure, which leads to a decrease in protein surface area and exclusion of a large amount of solvent, will enhance this effect. It has been shown that higher levels of structural ordering are present in specific complexes, when compared to nonspecific complexes [43,44]. Protein folding and protein–ligand interactions often involve entropy–enthalpy compensation mechanism [45]. The entropy change that modulates the free energy of binding depends mainly on the relative contributions from two opposing components, one being solvent release, which increases entropy, and the other being change of conformational entropy. Thus the loss of entropy associated with folding of a flexible domain, which results in the reduction of binding affinity, can be to some extent compensated by a significant entropy gain that arises from ordering of structure upon specific binding. The mechanisms by which folding is coupled to binding are not well understood. It has been suggested that the interplay between structural ordering and desolvation may allow the action of multiple mechanisms of sequential and selective enhancement of interactions [40].
Flexibility in molecular recognition has been analyzed by molecular dynamics simulations initiated from crystal structure of ternary complexes [34], and by simulating the association of various monomers into higher oligomeric complexes [36]. Several models of folding-associated binding were proposed based on the energy landscape theory of protein folding [35]. The process of folding (binding) occurs via searches for energetically favorable intra- (inter-) molecular interactions on an energy landscape shaped like a funnel, with a narrow lower part representing the folded (bound) state. The reaction proceeds downhill along the free energy gradient. Wang and colleagues [35,38] derived the thermodynamic free energy expression for the flexible binding. They showed that inherent hydrophobic interactions and cooperativity between folding and binding lead to dynamic fluctuations between the “non-native” partially folded and closely bound conformational states. To avoid trapping within local energy minima, the binding energy landscape has to be biased (funneled) toward the native binding state. This implies certain distribution of protein apolar residues between the hydrophobic core (for the purpose of folding stability) and surface (for the purpose of binding). The balance between binding affinity and specificity depends on the ratio of the binding transition temperature versus the trapping transition temperature. This criterion implies loss of some affinity to enable the molecules to reach the best fit.
Yet another advantage of coupling binding to folding may be the increased rate of complex formation. The large surface area of IDPs that is available for interactions provides a greater capture radius, compared to a compact globular protein. Using the same methodology, based on analogy between folding and binding, Shoemaker et al. [41] proposed a “fly-casting” mechanism for binding interactions of IDPs. According to this model, the unstructured polypeptide binds weakly over a long distance and then “reels in” the target during the protein folding process [41].
A recently published theory implies that protein disorder optimizes allosteric coupling via ensemble-mediated mechanism [46]. The presence of ID in segments containing one or both of the coupled binding sites generates an ensemble of states that is “optimally poised to respond” to binding. Upon binding to the ligand, the ensemble of states is redistributed. Significantly, such a mechanism depends only on the relative stabilities of the domains and not on the specific structural basis of that stability and can be related to experimentally obtainable values such as stability and binding affinity.
Direct experimental evidence for the coupled binding and folding was provided by Sugase et al. [33], who demonstrated that the binding of the phosphorylated kinase inducible activation domain of CREB (pKID) to the KIX domain of CBP involves the formation of an ensemble of unstructured encounter complexes, which are stabilized by non-specific hydrophobic contacts as well as the formation of partially folded intermediate states. Using NMR titration and 15N relaxation dispersion techniques, the researchers characterized kinetics of binding process, the populations and structure of intermediate states, and showed that the formation of the final fully folded complex with the structure of pKID stabilized by intermolecular interactions occurs without dissociation from KIX.
Interaction-prone regions with the potential to fold upon complexation have been named “molecular recognition features” (MoRFs) [32,47]. The flexibility of binding by IDPs depends on the inherent conformational preferences of disordered regions. In some cases the residual structure of those peptides is not completely random but exhibits local predominance of a particular secondary structure, usually helical, that is later stabilized in the bound state (inherent-structure mechanism). It has been proposed that such nascent, transiently formed structures serve as initial inter-chain contact points and contribute to the reduction of entropic penalty and increased affinity of binding. In an alternative mode of binding (induced-structure mechanism), the intrinsic conformational preferences of disordered regions are suppressed by interactions with the partner. Thus disordered chains with low local structural preferences may adopt multiple conformations upon binding to different partners [47].
In addition to MoRFs, short sequences that form “linear motifs” recognized by globular domains (e.g., phospoSP binding to WW domains, PXXP binding to the SH3 domain), are usually located within unstructured regions [48–50]. These motifs are key mediators of crucial biological interactions. It has been suggested that specific binding occurs via short primary contact site(s) and subsequent adhering of the longer flexible chain to the surface of the binding partner [51]. The extended polypeptide chain often displays several binding motifs, which enable the protein to bind simultaneously to multiple ligands or to form multivalent interactions with one.
Furthermore, IDPs often mediate formation of multi-protein assemblies as well as self-assembly of biological macromolecules [52]. These attributes make ID regions of proteins indispensable in organizing cell signaling and gene regulatory networks. Cellular processes carried out by the extensive protein interaction networks are constructed like electrical and engineering control systems, where most switches and feedback mechanisms rely on assembly and disassembly of multi-component complexes [53]. A majority of the proteins in cellular networks make only one or two connections (date hubs), while, on the other hand relatively few hubs mediate a large number of links (party hubs). Party hub proteins interact simultaneously with many partners, while date hubs interact with different partners at different times [15,54]. The ability of party hub proteins to bind multiple partners with high specificity depends on their conformational plasticity. The study of Singh et al. [55] revealed the significant enrichment of ID in party hub proteins, underscoring the importance of disorder for transient binding interactions. Moreover, a majority of the experimentally determined phosphorylation sites in eukaryotic proteins, as well as sites of proteolytic digestion and other modifications, are located within segments with sequence features indicative of intrinsic disorder [56,57]. Thus, ID promotes binding diversity, renders highly specific but reversible interactions, facilitates tight regulation, rapid turnover, provides a kinetic advantage for macromolecular associations, and ensures quick responses to external signals [6,14].
Intrinsic Disorder and Transcriptional Regulation
Activation of transcription involves complexes of many different proteins that recruit components of the transcriptional machinery to the promoter region and also induce changes in the structure of chromatin. At the DNA level, gene expression is directed by sequence-specific DNA binding TFs, which specifically recognize and bind to cis-regulatory sequences in target genes and subsequently activate or repress their transcription. It was recognized early on into the studies of transcriptional activation [58,59] that critical events regulating the initiation of transcription cannot depend on specific and rigid complementary surfaces of participating proteins. The variety of arrangements and stoichiometries observed in the preinitiation complexes, the undefined position of cis regulatory sequences in respect to the promoter that they activate, as well as the modular nature of specific DNA-binding proteins suggested that regulation of an assembly and activity of transcriptional complexes could not be achieved if every component had an extremely high affinity or specificity for another [60]. Also, lack of sequence similarity among trans-activating domains (TADs), which seemed to consist mainly of “acid blobs” and “negative noodles” with undefined conformation supported this conclusion [59]. Subsequently, many TADs were characterized as mostly unfolded when studied as separate peptide units [61]. Moreover, it has been demonstrated (using NMR and small-angle X-ray scattering) that TADs of p53 and herpes simplex virus VP16 remain unstructured in the full-length protein context [62,63].
Although the principles of gene regulation are the same in case of prokaryotes and eukaryotes, the corresponding transcription regulatory proteins exhibit striking differences in their molecular architecture. Eukaryotic TFs are on average twice as long as their prokaryotic counterparts and contain extended regions of protein disorder [29,64]. Recently, Liu et al. [64] reported that at least 82% of eukaryotic TFs possess extended regions of ID as predicted by the PONDR software and charge-hydropathy plots. In a parallel study, Minezaki et al. [29] compared the content of disorder (assessed by the DISOPRED2 program) in transcriptional activator proteins from different species. They found that up to 49% of the entire sequence of human TF proteins is occupied by ID regions. This is in contrast to their prokaryotic counterparts where long stretches of unstructured region are rare, and for which several 3-dimensional structures of full-length proteins are available in the Protein Data Bank (PDB). This discrepancy probably reflects differences in the complexity of transcriptional apparatus, chromatin structure, and regulatory pathways used to regulate gene transcription in eukaryotes and prokaryotes [29].
A salient difference between transcriptional regulation in the two kingdoms arises from the densely packed structure of chromatin in eukaryotic cells, which restricts access of the basal transcriptional machinery to the promoter region [65]. Furthermore, in contrast to prokaryotic gene regulation whereby transcription can be modulated by a single monomeric protein, transcriptional activation in eukaryotes is generally controlled by the concerted action of many signal-regulated TFs and cofactors, which form multiprotein–DNA complexes and modify chromatin structure in the promoter and enhancer regions. This combinatorial regulation is organized in a highly hierarchical manner and allow for tightly controlled integration of independent signaling pathways in biological responses [66,67]. Clearly, the complexity of the nuclear transcriptional machinery required evolution of more sophisticated regulatory molecules that would be capable to handle tasks of overcoming the repressive effects of chromatin and suppressor proteins, as well as the competitive recruitment of coactivators.
This review focuses on the role of structural disorder in the function of the basic region:leucine zipper (bZIP) transcriptional regulators. bZIP TF proteins constitute the largest and the most conserved [68] superfamily of TF proteins that operate exclusively in eukaryotes. bZIP TFs are involved in vital cellular functions and regulate development, metabolism and responses to environmental changes [69]. This functional versatility arises from their ability to interact with each other and with structurally unrelated TF proteins [70].
BZIP BASICS
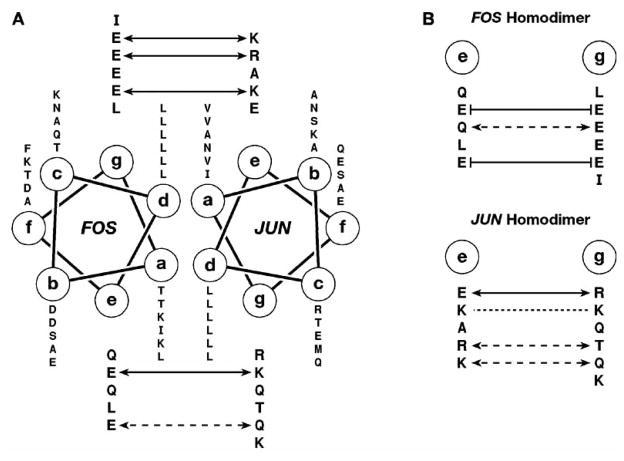
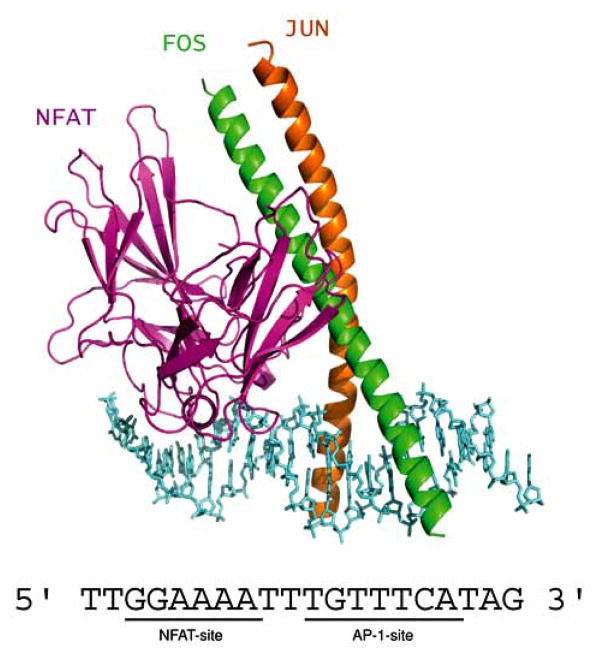
Members of the bZIP superfamily bind to target DNA sites as homodimers or heterodimers and recognize related but distinct palindromic sequences (Fig. 1A). The prototypical bZIP protein, CCAAT/enhancer binding protein α (C/EBPα), was discovered over 20 years ago and was one of the first mammalian transcription factors to be purified and cloned [71]. These studies established that the bZIPs DNA- binding domain (DBD) consists of a positively charged segment, the basic region (BR), linked to a sequence of heptad repeats of leucine residues called the leucine zipper (LZ) [72]. Soon after, many proteins were categorized in this class, including proto-oncogenes Fos and Jun. As shown by a number of crystal structures [73–77], bZIP peptides bind to their cognate DNA duplexes as dimers of uninterrupted α helices which form a chopsticks-like structure (Fig. 1A). Dimerization is mediated by the LZ segments, which form two parallel coiled-coil α-helices wrapped around each other. The two helices, which are positioned nearly perpendicularly to the DNA double helix, diverge smoothly toward their amino termini and each BR segment contacts one-half of a palindromic site in the DNA major groove. This is the simplest known protein–DNA recognition motif.
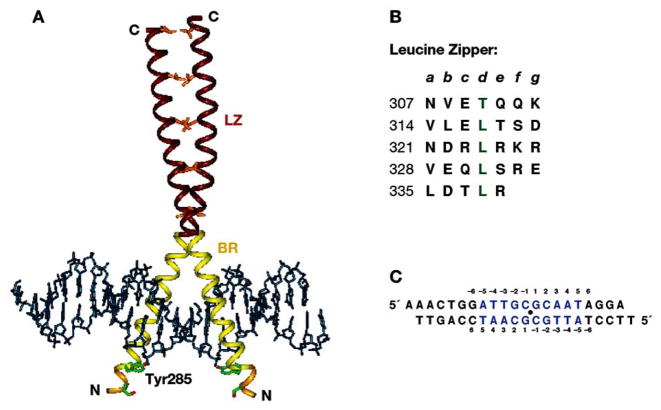
Fig. 1.
(A) Overall structure of the C/EBPα bZIP–DNA complex (PDB code 1NWQ). LZ, leucine zipper; BR, basic region; EBR, extended basic region. (B) The LZ sequence displayed as heptad repeats. Numbering refers to the complete rat C/EBPα protein sequence. Lower-case letters (top line) show positions of leucine zipper residues as they would appear on a standard helical wheel representation of a coiled-coil dimer; “d” corresponds to the position of leucine residues. Coiled-coil structure is stabilized by the van der Waals interactions between residues at a and d positions from both helices and by electrostatic interactions between residues located at g and e positions. (C), The DNA sequence in crystals. Consensus C/EBP recognition site is indicated in blue, and the center of symmetry as a solid circle.
In contrast to many instances of stably folded DNA-binding domains (e.g. GAL4 [78], and p53 tumor suppressor protein [79]), the bipartite DBD of bZIP proteins assumes a well-ordered, stable structure only when bound to a specific DNA site [80,81]. At physiological concentrations, in the absence of DNA, unfolded monomers are in dynamic equilibrium with the folded dimers [82]. A concentration-dependent folding/unfolding transition is characterized by a fast rate of the subunit exchange. Binding to a specific DNA duplex stabilizes the coiled-coil dimer and induces helical folding of the basic region.
Based on their DNA-binding specificities, bZIP proteins have been traditionally arranged into several families, each of which recognizes a unique DNA motif (Fig. 2A). The structural basis of specific DNA recognition has been discussed in several review articles [75,83] and will not be addressed here in detail. Briefly, bZIP TFs recognize their cognate DNA sites through base contacts made by five residues within the basic region motif characteristic for each family. These five positions are highly conserved among bZIP transcription factors and contain invariant Asn and Arg residues. Crystal structures of bZIP domains bound to their cognate DNA duplexes have revealed functional variability of the conserved residues in DNA recognition (Fig. 2B). For that reason, no universal code relating the BR sequence motif to its preferred DNA binding-site sequence has been established; however members of the same family are believed to recognize their cognate sites in the same manner. bZIP proteins show relaxed DNA binding specificity beyond their respective core consensus sequences, and they can recognize similar sequences with lower affinity. Selected bZIP proteins can form stable DNA complexes as cross-family heterodimers [84,85]. Heterotypic dimerization generates an expanded repertoire of specific DNA sites (composed of different half-sites) recognized by bZIP factors. Alternative methods of classification therefore have been proposed based on sequence similarity and dimerization properties [85], and recently on the phylogeny [68] of their BR-LZ domains. In contrast, members of the superfamily differ widely in their amino acid sequence outside the conserved bZIP domain. Only short conserved subregions could be identified, even in the case of proteins belonging to the same family (see Fig. (7)).
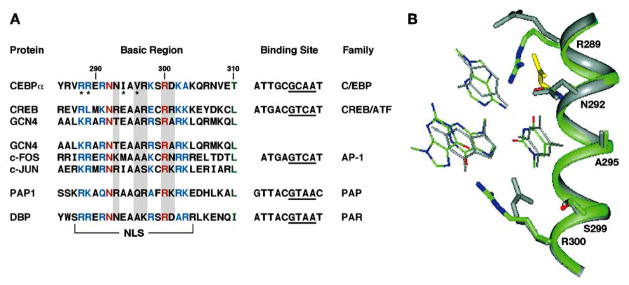
Fig. 2.
DNA recognition. (A) Basic region amino acid sequences and DNA binding sites for representatives of bZIP families. The invariant Asn and Arg are shown in red, conserved basic residues in blue, and the first residue of the leucine zipper is green. Residues that form the KIM motif are marked by asterisks. The cores of the half-sites are underlined. (B), Comparison of conformations of the conserved side chains in the basic regions of C/EBPα (green) and GCN4 (gray). The side chain of the invariant Asn residue from the PAP1 structure is shown in yellow. (Adapted from [77]).
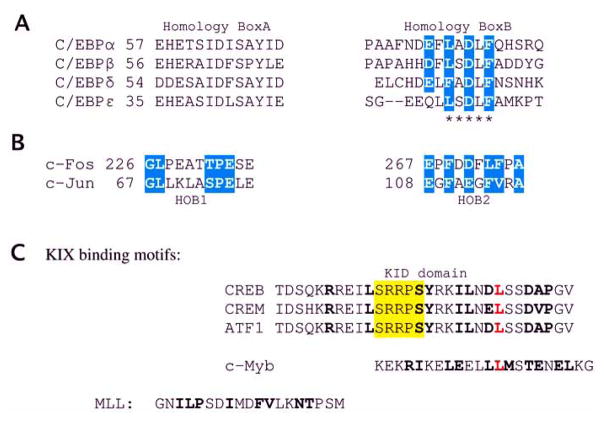
Fig. 7.
Protein recognition motifs identified in TADs. (A) Sequence alignment of Box A and B homologies present in C/EBPs. Residues comprising TAF9 binding motif are marked by asterisks. (B) Homology boxes, HOB1 and HOB2, from c-Jun/c-Fos. Identical residues representing consensus and sequence similarities with Box B are highlighted in cyan. (C) KIX binding sequences. Residues contacting KIX are shown in bold print. The conserved phosphorylation motif in KID domains is highlighted in yellow and the critical Leu (see text) is shown in red.
Mammalian bZIP Proteins
By 1995, 82 distinct DNA-binding bZIP proteins from a variety of eukaryotic sources had been cloned and characterized (reviewed by Hurst [69]). Complete sequencing of several genomes provided opportunities to examine the whole array of bZIP TFs contained in given organisms. Vinson and his colleagues identified and classified bZIP proteins encoded by the Drosophila and human genomes [85,86]. Most recently a database bZIPDB, which contains information on protein interactions and TF-target gene relationships for 49 human bZIP TFs, has been developed [87].
Mammalian bZIPs comprise eight major families, which recognize C/EBP, TRE, CRE, CRE-like and PAR sites (Table 1). Maf bZIPs form a separate class of TFs as they recognize a substantially longer DNA sequence, MARE. Maf proteins contain extended homology region (EHR) preceding BR, which enables them to make a broader contacts with DNA [88]. Three members of the family, referred to as “small Mafs” (MafG, MafF and MafK), serve as obligatory heterodimeric partners for the members of CNC-bZIP family unique for mammals. CNC–small Maf heterodimers recognize the asymmetrical NF-E2 site composed of a half site of TRE or CRE and half site of the MARE [89]. Alignment of BR:LZ sequences of human bZIP TFs and their dimerization properties can be found in [85].
Table 1.
Human bZIP Proteins
| Protein | Length | Preferred Binding Sites for Homodimers or Obligatory Heterodimers | Structural Domainsa | Dimerization Properties | |
|---|---|---|---|---|---|
| BR:LZ | Other | ||||
| C/EBP family; reviewed in [181] | |||||
| C/EBPα | 358 | 280–344 | |||
| C/EBPβ | 345 | C/EBP: GATTGCGCAATC | 269–333 | Homodimers and heterodimers within family, as well as heterodimers with Fos, Jun, ATF2 and ATF4 proteins | |
| C/EBPδ | 269 | 189–253 | |||
| C/EBPε | 281 | 202–266 | |||
| C/EBPγ | 150 | Also CRE and PAR sites | 60–124 | ||
| AP-1 family; reviewed in [96] | |||||
| Jun subfamily: | |||||
| c-Jun | 331 | 250–314 | |||
| JunD | 347 | 266–330 | Jun proteins form heterodimers with Fos and ATF proteins to generate AP-1 TFs. | ||
| JunB | 347 | 266–330 | |||
| Fos subfamily: | |||||
| c-Fos | 380 | TRE (AP-1): ATGAG/CTCAT | 135–199 | In addition, Jun proteins form homodimers and heterodimers with CNC, and Maf proteins; | |
| FosB | 338 | 153–217 | |||
| Fra1 | 271 | 103–167 | |||
| Fra2 | 326 | 122–186 | |||
| BATF | 125 | 24–88 | ATF4 and ATF5 heterodimerize with C/EBPs, and Nrf2; whereas CREB5 with ATF2 | ||
| ATF3 | 181 | Also CRE sites | 84–148 | ||
| ATF2&4 subfamily: | |||||
| ATF2 | 487 | 350–414 | 25–49b | ||
| ATF7 | 494 | 341–405 | 7–31b | ||
| CREB5 | 508 | 373–437 | 16–40b | ||
| ATF4 | 351 | 276–340 | |||
| ATF5 | 282 | 206–270 | |||
| CREB/ATF family; reviewed in [209] | |||||
| CREB | 341 | 280–331 | |||
| CREM | 345 | 287–308 | |||
| CREB-H | 461 | 240–304 | |||
| CREB3 | 395 | CRE: TGACGTCA | 172–236 | Homodimers | |
| OASIS | 519 | 288–352 | |||
| BBF2H7 | 520 | 291–355 | |||
| ATF1 | 271 | 211–268 | |||
| ATF6 family; reviewed in [210] | |||||
| ATF6 | 670 | UPRE: TGACGTGG/A | 304–368 | ||
| CREBL1 | 703 | 323–387 | Homodimers | ||
| CREB4 | 395 | Other CRE-like sites | 215–279 | ||
| Xbp-1 | 261 | 68–132 | |||
| PAR family; ref. [211] and references therein | |||||
| DBP | 325 | 253–317 | |||
| TEF | 303 | PAR: ATTACGTAAT | 231–295 | ||
| HLF | 295 | 223–287 | Homodimers | ||
| E4BP4 | 462 | 71–135 | |||
| MAF family; reviewed in [88] | |||||
| cMAF | 403 | 284–350 | |||
| MAFB | 323 | MARE: TGCTGACTCAGCA, | 234–300 | Large Mafs (cMAF, MAFB, NRL) form heterodimers with Fos or Jun; small Mafs (MafF, -G, -K) form homodimers or heterodimers with CNC or Fos family members | |
| NRL | 237 | 157–221 | 133–172c | ||
| MafF | 164 | 49–113 | 25–62c | ||
| MafG | 162 | TGCTGACGTCAGCA | 46–113 | 25–62c | |
| MafK | 156 | 49–113 | 25–62c | ||
| CNC family; reviewed in [89] | |||||
| BACH1 | 736 | 553–619 | 34–130d | ||
| BACH2 | 841 | NF-ET: TGCTGACTCAT | 646–708 | 37–133d | |
| NF-E2 | 373 | 264–328 | Heterodimers with small Mafs | ||
| NRF1 | 772 | 671–735 | |||
| Nrf2 | 605 | Also ARE, MARE and CRE sites | 495–559 | ||
| Nrf3 | 694 | 576–640 | |||
identified by SMART;
Zinc-finger, ZNF_C2H2;
EHR (see text);
BTB/POZ
STRUCTURAL CHARACTERIZATION AND DOMAIN ORGANIZATION
Early biophysical studies in solution (based mainly on CD and NMR spectroscopy) revealed the lack of ordered structure beyond the DNA-binding domain for several bZIP proteins [90]. Only few members of this superfamily contain regions that are stably folded in the absence of the ligand (Table 1). The three dimensional structure was determined for the zinc finger-like domain identified in the three factors ATF2, ATF7 and CREB5, comprising a subgroup of ATF family [91] (PDB ID code 1BH1), the HER domain of MafG [92] (PDB code 1K1V), and the BTB/POZ domain from BACH1 (PDB code 2IHC).
In general, in order to perform their biological functions, sequence-specific DNA binding TF proteins must translocate to the nucleus, and, once bound to DNA, either activate or repress transcription. The structural basis and molecular mechanisms explaining how these proteins are able to carry on their dual and sometimes opposing functions in the regulation of gene expression, depending on the cellular context, has just begun to be revealed. Transcriptional regulatory proteins are composed of modules that may act independently of each other. The order in which these segments are arranged into functional protein molecules varies between the factors (Fig. 3). The functional domains have been identified by a combination of biochemical and deletion mutagenesis experiments and their borders are usually poorly defined.
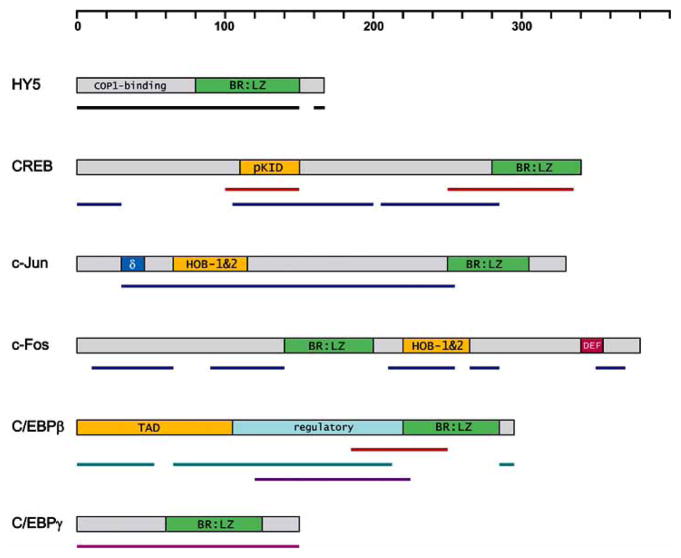
Fig. 3.
Schematic representation of domain organization and predictions of ID regions (referred in various publications) for selected bZIP TFs. Regions predicted as disordered by a variety of prediction programs are marked by colored lines: black, PONDR [97]; dark blue, DISOPRED2 [29]; red, FoldIndex [98]; cyan, Loops/Coil definition; violet, NORSp [100]; magenta, charge-hydrophaty plot [18]. Depicted functionally relevant features are discussed in the text. Boundaries for BR:LZ domain (green) are shown as defined by SMART [102]. Regions necessary for transactivation are shown in orange. δ, and DEF domains contain docking sites for JNK and ERK, respectively. The indicated scale corresponds to the number of amino acids.
Functional domains of bZIP factors other than DBDs may include multiple transactivation domains (TADs), nuclear import/export segments and diverse regulatory elements. Distinct repression domains have also been identified [69,94]. For example, each, c-Fos and c-Jun, has a number of domains which activate transcription and docking sites for several kinases, such as JNK or ERK, which modulate their activities [95,96]. Often, transdominant repressors are truncated bZIP variants, which retain LZ domain but either lack the activation region, or have a modified BR sequence [69]. A characteristic feature of bZIP TFs is that their nuclear localization signal (NLS) is contained within the BR, which performs other functions in addition to sequence-specific DNA recognition (see below).
Dimerization, DNA-binding and transactivation/repression activities are localized to autonomous modules, which have been studied as separate polypeptide chains. Structural and functional characterization of these segments is described in the subsequent sections of this review. The information on structural properties of bZIPs in the full-length protein context or on their putative intramolecular interactions is limited. Biophysical characterization was reported for the cAMP-responsive element binding protein (CREB) [90] and a plant bZIP TF, HY5 [97]. Studies of whole CREB by CD spectroscopy revealed that, when unbound to DNA, the protein contains 20% α-helix, 9% β-strand, and 34% β-turn, whereas 37% of its residues exist in the random coil state. DNA-binding induced a 5% increase of the α-helical content consistent with folding of the BR sequence. However, an isolated N-terminal fragment encompassing the entire region required for transactivation has a significantly lower content of secondary structure than could be predicted by subtracting values measured for the C-terminal bZIP domain. This loss of structure in the truncated version of CREB could indicate that the bZIP domain affects the structure or stability of the rest of the molecule [90]. It was recently demonstrated that sequence-specific DNA binding of CREB induces a global conformational change within the CREB monomer, which affects structure of the kinase inducible domain (KID), rendering it refractory to the action of protein phosphatase 1 [98]. The comprehensive structural and functional characterization of HY5, a plant bZIP TF, by experimental and theoretical methods has been reported by Yoon et al. [97]. Using limited proteolysis in combination with mass spectrometry, CD and NMR spectroscopy, the authors demonstrated that HY5 contains a stable helical LZ domain located at the C-terminus, a molten-globule like BR region, and intrinsically disordered 77 residues on the N-terminal part. The full-length HY5 exhibited a noncooperative CD melting profile characteristic of proteins lacking stable tertiary structure.
The intrinsic structural disorder in HY5 was also implied by the primary structure analysis. The sequence of the whole protein, as well as that of investigated fragments (including the segment corresponding to a bZIP peptide), fell into the disordered region of the charge-hydropathy plot [18] and displayed a bias toward disorder-promoting residues in amino acid composition. The PONDR suite [99] also predicted that, except for 10 residues at the C-terminal region, HY5 is intrinsically disordered. According to a survey performed by Minezaki et al. [29], human bZIP-activating proteins, which do not possess functional domains other than TADs, may contain up to 70% disordered residues. Predictions of ID regions in members of the C/EBP family were reported by Miller [100]. Based on the charge-hydropathy plot [18], C/EBPγ was originally predicted to be completely unfolded, and more recently, 125 out of its 150 residues were predicted to be disordered by PONDR [64]. CREB, MafF and BATF are among the human TFs with the highest levels of predicted disorder, based on PONDR genome-wide analysis, with 87.68, 83.54% and 80.80% of predicted overall disorder respectively [64]. Examples of regions predicted as disordered in several bZIP proteins are depicted in Fig. (3). Despite this abundance of predicted ID regions, bZIP TFs are underrepresented in a database of protein disorder (Dis-Prot) [101], which lists only experimentally verified cases of intrinsically unstructured proteins, such as HY5.
LEUCINE ZIPPER: MUTUALLY INDUCED FOLDING BY SELF-ASSOCIATIONS
Monomeric amphipathic helices are unstable and LZ segments fold into coiled-coil helical structures upon association with another subunit(s). BZIP peptides which cannot homodimerize (e.g., Fos and ATF4) are disordered in solution in the absence of their complementary partner [82,103]. The coil-to-α-helix transition upon dimerization is a reversible, concentration-dependent process. Dimerization constants of bZIP proteins are in the μM range, and the estimated lifetime of dimers at 25 °C was less than 1 sec for the GCN4 homodimer [80] and less than 10 sec for the Fos–Jun heterodimer [82]. Unfolding and reassembly of double-stranded coiled-coil structures facilitates subunit exchange between dimers. Such exchange is thought to provide a general mechanism for selective regulation of gene expression.
Thermodynamic Characterization
In the best studied case, that of yeast TF GCN4, the thermal unfolding transition occurs at 70 °C and 50 °C for concentrations of 5 × 10−4 M and 5 × 10−6 M, respectively [104]. Initial experiments utilizing CD and NMR spectrometry [80,105], calorimetry [104] and stop-flow kinetics [106] indicated that temperature- or denaturant-induced unfolding of a LZ coiled-coil structure is a cooperative, two-state transition. A series of further studies have revealed that it is a much more complicated process [107,108]. Combined optical and differential scanning calorimetry investigations of the thermal denaturation of several LZ peptides related to GCN4 show that the reaction involves several distinct steps [108]. The melting of the molecule starts at 0 °C with the fraying of the N-terminus of the leucine zipper, followed by structural changes that presumably involve repacking of the coiled-coil structure. These two concentration- independent steps are followed, at higher temperatures, by cooperative dissociation and unfolding of the two strands. Equilibrium and kinetic CD experiments demonstrated that both the stability of the dimer and the unfolding and refolding rate constants depend on the helical propensity of LZ sequences [109]. A proposed folding mechanism for GCN4-derived peptides assumes rapid equilibrium between fully unfolded peptides and partially helical (predominantly at the C-terminus) conformers, which are able to form the dimeric transition state populated by partial coiled-coil structures. The presence of nucleating helices is critical for the formation of a transition state. The increased helical propensities of variant peptides lead to the acceleration of the folding reaction, and possibly, to lowering of the transition state free energy. Residual elements of secondary structures displayed by ID regions in their unbound state are thought to play a pivotal role in the thermodynamics of folding-associated binding interactions [110].
Dimerization Specificity
Depending on the sequence of their LZ domain, bZIP proteins can form homodimers and/or form heterodimers with bZIP proteins from their own or different subfamilies. Selective dimerization is critical for bZIP TF’s biological function. Heterodimers within the same family retain their DNA-binding specificity but exhibit different transactivating potential and synergy with other regulatory proteins. Cross-family heterodimerization affects DNA binding activity as well as the stability of the DNA-protein complexes, and is fundamental for the diversification of binding specificities of bZIP TFs. Heterotypic dimerization generates an expanded constellation of dimers with distinct functional properties from a small number of monomers [111]. For example, 53 unique bZIP domains identified in the human genome have the potential to create 1,405 unique dimers [85,112]. This powerful regulatory mechanism depends on the proper assembly of the specific partners. For these reasons, the dimerization properties of LZ-containing TFs have been a subject of extensive investigation. Analysis of available crystal and solution structures [113], molecular modeling [114] and mutagenesis studies [84] has helped to establish sets of rules, which make it possible to predict interaction preferences for bZIP peptides with great precision [85,112,115]
Dimerization selectivity depends on the “complementarity” of the putative partners, which is defined by the precise distribution of hydrophilic and hydrophobic residues in the LZ regions. The dimer interface is formed by the residues located at the a, d, e, and g positions of the (abcdefg)n heptad repeat (see Fig. 1). Residues at a and d positions are usually aliphatic and comprise the hydrophobic core of the duplex. An exception is the a position of the second heptad of many bZIP proteins, which is often occupied by asparagine to prevent higher order oligomerization [116]. The e and g positions are typically occupied by polar amino acids and influence both stability and specificity of the coil-coil. Residues located in the g position of one subunit and residues at the e position of the second subunit, which is situated five steps closer to the carboxyl terminus, are poised to interact in one direction by a “knob to holes” packing arrangement of the two helices, and often form electrostatic inter-helical bridges g↔e′ (‘denotes the second subunit of the dimer). Homodimerization and heterodimerization are prevented when the facing side chains in position e and g create steric or charge repulsion [74]. Analysis of crystal structures of the GCN4 homodimer and Fos/Jun heterodimers bound to their cognate DNA sequences, and mutagenesis studies have established that inter-helical salt bridges are the most decisive determinants of dimerization selectivity [85]. In choosing the dimerization partner, preventing repulsive interactions appears to be more important than increasing the number of attractive interactions. This so-called “i+5” rule explains dimerization preferences for members of the Fos and Jun subfamilies (Fig. 4). A more precise determination of dimerization specificity involves consideration of the coupling energy for g↔e′ pairs, which can be calculated based on a double-mutant thermodynamic cycle [85,105].
Fig. 4.
Mechanism of dimerization specificity for Fos and Jun peptides. (A) A helical wheel diagram of the Fos–Jun coiled-coil. The sequence is read from N to C termini outwards from the wheel. The Fos–Jun heterodimer is stabilized by four electrostatic attractive interactions (solid arrows) and by a hydrogen bond interaction (dashed arrows) between the g and e positions. (B) Potential interactions within homodimer interfaces of Fos and Jun. The formation of Fos homodimer is prevented by four electrostatic repulsive interactions (solid lines). The formation of Jun homodimer is driven by two positive electrostatic interactions and it is additionally stabilized by hydrogen bonds and the interactions between Lys residues (dotted line), wherein the repulsive electrostatic energies between like charges are overcome by the favorable Van der Waals interactions between the methylenes of lysine side chains and hydrophobic amino acids in the a and d positions [85].
Partnering selectivity for 49 human bZIP derived peptides were tested experimentally using coiled-coil arrays [117]. These studies confirmed most of the earlier predictions, but also identified several unexpected interactions. Analysis of fully sequenced proteomes has uncovered many more details that have to be taken into consideration in order to understand the mechanisms of diverse bZIP dimerization patterns [118]. New aspects of regulatory mechanisms underlying biological activities of bZIP TFs that emerged from proteomic studies were reviewed in [112]. As noted by the authors, the current challenge is to understand how the specific dimerization and intermolecular interactions of these proteins are regulated in changing cellular environments.
In vivo, the repertoire of bZIPs capable of associating in a given cell depends on the expression pattern of the respective proteins and on their responses to external stimuli. Furthermore, the reversible nature of the dimerization process facilitates the interconversion between early and late activated complexes, and contributes to time and stimuli -dependent differential activation of specific genes. Such modulation of biological activities by control of dimer composition has been observed in the C/EBP family [119] and in the Fos/Jun family [70]. Through the change of dimeric partner, small Mafs can switch transcriptional activity from repression to activation [89].
THE MANY FACES OF THE BASIC REGION
The short BR performs a remarkable number of functions critical for the biological activity of bZIP proteins: cytoplasm-to-nucleus translocation, specific DNA recognition and protein–protein interactions possibly contributing to the regulation of gene expression. These functions can be regulated by intramolecular interactions of BRs with other regions, their phosphorylation and binding to auxiliary proteins.
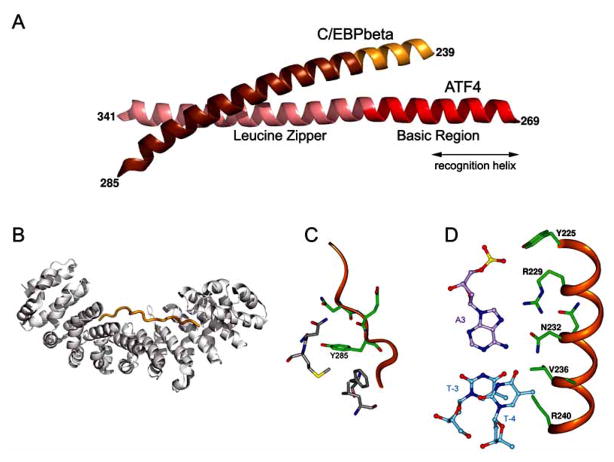
In the unbound state, the BR elements of bZIP proteins exhibit differential, temperature-dependent helical content. It has been shown by CD and NMR spectroscopy that in the absence of DNA, BR of GCN4 and C/EBPβ are largely unfolded in solution [80,120] and have been described as a dynamic ensemble of transiently formed helical structures. The population of transient helical conformations increases with decreasing temperature [121]. The BR of plant TF, HY5, has been characterized as a molten globule [97]. In contrast, in the absence of DNA, the ATF4 BR adopts a stable helical conformation, as revealed by the crystal structure of the ATF4–C/EBPβ heterodimer (see Fig. 5A) [103]. The propensity to form a helical structure is defined by the amino acid sequence. Conserved BR sequences of selected bZIP proteins are preceded by helix-capping residues, which may be responsible for stabilizing their partial α-helical structures [122].
Fig. 5.
Conformational flexibility and binding polymorphism of BR from C/EBPs (orange). (A) Structure of C/EBPβ(224–285)–ATF4(280–341) heterodimer in the absence of DNA; (PDB code 1CI6). Note that the entire region corresponding to the C/EBPβ recognition helix is missing. (B) Structure of the classic, bipartite NLS peptide (orange) bound to armadillo domain of importin α (gray); (PDB code 1EJY). (C) Hypothetical interactions of C/EBPα BR peptide with the BRCT domain of BRCA1. Y285 corresponds to Y225 in C/EBPβ. (D) C/EBPβ recognition helix (residues 225-240) bound to cognate DNA (PDB code 1H8A).
The DNA-dependent folding transition of the basic domain has been documented by CD, fluorescence, and NMR spectroscopic techniques. The requirement of coupling of local folding to site-specific DNA binding has been deduced from theoretical analysis of calorimetric data. Spolar and Record [44] proposed that the significant gain in total association entropy observed in specific protein-DNA interactions is mainly due to changes in water-accessible surface area, and can be explained by the mechanism of protein folding coupled to binding. During such a process, the loss of conformational entropy resulting from structural ordering may be compensated by gain of entropy arising from the burial of large nonpolar surfaces on the complex formation. Non-specific binding, which involves only electrostatic interactions with the DNA phosphodiester backbone, was shown to not produce this ordering effect [43,44]. It has subsequently been suggested that nascent helices transiently formed in the unbound state may significantly reduce the entropy penalty associated with DNA binding by a flexible domain [121], and contribute to the increased affinity of specific binding [122]. Thus, the balance between the specificity and affinity of DNA binding depends on the amino acid sequence and can be regulated by the length of partial helical structures existing in the basic domain prior to complex formation.
The BR can also maintain specific interactions with viral and cellular proteins that function as transcriptional coactivators. The multiple factor-bridging protein 1 (MBF1) stimulates transcription through selective associations with distinct subclasses of bZIP proteins, linking them to the general transcription factor TATA-box binding protein (TBP) [123]. It has been demonstrated that bZIP of Jun (but not Fos) binds directly to the N-terminus of the human TBP-associated factor-1 (hTAF1), causing a de-repression of TFIID-driven transcription [124]. In certain cells, CRE-dependent transcription is enhanced by TORC (transducer of regulated CREB activity) protein, which mediates interactions of CREB bZIP with the TAF4/TAFII130 component of TFIID [125]. Specific BR sequences are also targeted by the human T-cell leukemia virus Tax, whereas the hepatitis B virus pX interacts with a broad range of bZIP proteins [126]. Miotto and Struhl [127] showed that the selective interactions with MBF1 and the related Chameau histone acetylases (or HBO1, the human homolog) are mediated by BR residues that face away from the DNA in the protein-DNA complex. Chameau interacts with AP-1 during Drosophila development via two arginine residues (R221 and R232 in JunD), whereas binding of MBF1 to JunD requires glutamate or glutamine residues located in the fork region, in addition to the two arginines. MBF1 and Chameau compete for binding to the BR of JunD and do not cooperate in its transcriptional activation. However, either MBF1 or Chameau can synergistically coactivate AP-1 genes with pX.
The disordered structure of the BR prior to specific DNA binding facilitates a variety of protein–protein interactions, which regulate bZIP activity. All information necessary to direct bZIP translocation to the nucleus is contained within the BR [69,128]. The NLS motif of bZIPs consists of two clusters of Lys/Arg residues separated by a linker of 10–12 residues (Fig. 2). This type of classic NLS is recognized by a component of transport receptors, importin-α/Kapα [129,130]. The NLS-binding domain of importin-α comprises 10 helical repeats (armadillo motifs), which form a concave surface containing multiple pockets for NLS cargoes. The crystal structure of the NLS peptide–importin-α complex [131] revealed that the bipartite NLS peptide binds to the receptor in an extended conformation (Fig. 5B). The two basic clusters are located in separate specific binding pockets of the receptor, whereas residues from the linker are poorly ordered.
Nuclear import/export as well as DNA-binding of many bZIP proteins, including Jun, C/EBPβ, and several plant factors, is regulated by phosphorylation [132]. For example, phosphorylation of the conserved Ser239 in C/EBPβ impairs its binding to the cognate DNA sequence and induces nuclear export [133]. It has been proposed that the SXEY sequence motif contained in the BR, which is conserved in this family, may be a site of phospho-dependent interactions with BRCA1 [100]. Other functional sites present in this region include the kinase interaction motif (KIM) [100], and a cysteine residue which is a sensor of oxidative stress in c-Jun, JunD, c-Fos, and EB1 [134,135].
The binding polymorphism and conformational plasticity exhibited by BRs underscore the importance of structural disorder for promoting transient interactions with multiple partners. Several of these interactions are mutually exclusive and may be utilized for the sequential ordering of cellular events in response to external signals. Importantly, the binding site for transcriptional coactivators such as MBF1, TORC, and Chameau/HBO1 is created only upon binding of bZIP protein to cognate DNA sequence. This ensures occurrence of these associations in the proper time and location.
ASSEMBLY OF BZIP–DNA COMPLEXES
Dimerization is considered a prerequisite for DNA-binding activity, as LZ mutants that cannot dimerize fail to form stable protein–DNA complexes [72]. However, the sequence of events leading to the formation of a dimeric protein–DNA complex is a matter of debate. In principle, two main pathways are possible: (1) initial formation of a bZIP dimer followed by DNA binding (dimer pathway), or (2) initial formation of a bZIP monomer–DNA complex that subsequently binds a second bZIP monomer (monomer pathway) [136] (see Fig. 6). Several bZIP peptides have been shown to readily dimerize in the absence of DNA, whereas there are only few examples of monomeric bZIP–DNA complexes. On the other hand, the bZIP dimer is relatively unstable, and when not bound to DNA it rapidly dissociates to monomers [82,104]. bZIPs, which cannot form homodimers will be present in solution as monomers in the absence of a complementary partner, as shown for Fos. Wu et al. [137] observed that homodimerizing CREB protein also exists primarily as a monomer in solution, and its dimerization is DNA-dependent. It has further been suggested that the weak association constants of dimerizing TFs ensure that stable protein–DNA complexes can assemble only upon binding to a specific sequence [138]. The rate of assembly via the dimer pathway is expected to be limited by the rate of protein dimerization. In contrast, protein–DNA association is directed by long distance electrostatic interactions and should be much faster. Importantly, if the monomer–DNA intermediate is unstable relative to the dimer–DNA complex, it is less likely to become kinetically trapped at a nonspecific site. The monomer pathway thus offers kinetic advantages and provides a more efficient means for discrimination against nonspecific binding [139]. Schepartz and colleagues provided compelling kinetic and spectroscopic evidence that certain bZIPs (e.g., ATF-2, Fos and Jun) and related helix-loop-helix zipper families (e.g., Max) follow an assembly pathway in which two monomers sequentially bind to DNA and form their dimerization interface while bound [139,140]. The obligatory dimer pathway of DNA recognition has also been challenged by others [141,142].
Fig. 6.
Possible intermediates in the monomer and dimer pathways during assembly of bZIP–DNA complexes.
The shortcoming of these studies is the lack of data on the behavior of full-length bZIP proteins, in which intramolecular interactions may influence dimerization and DNA-binding properties. In many instances DNA binding activity of bZIP proteins (e.g., C/EBPβ), is autorepressed in their inactive state [143]. Also, dimerization is critical for the stability of C/EBP proteins, which in monomeric form undergo rapid ubiquitination-dependent degradation by the 26S proteosome [144]. Moreover, interactions of several TFs with importins have been observed to occur in the dimeric form [128,145]. These considerations indicate that both pathways, and sometimes a combination of two may be utilized in vivo. The mechanism of assembly may depend on many factors such as the helical propensity of the BR and LZ sequences (see above), the expression pattern of suitable partners, and the presence of auxiliary proteins [126] and may vary on a case-by-case basis.
THE ELUSIVE TRANSACTIVATION DOMAIN
Sequence-specific DNA binding TFs (transcriptional activators and repressors) stimulate transcription of their target genes by regulating the assembly and/or activity of transcriptional initiation complexes. They elicit their effect by recruiting members of a diverse family of coactivators, which initiate local opening of chromatin structure and mediate recruitment of the RNA polymerase II complex (Pol II) to the transcriptional start site. Transcription-activating proteins possess specialized distinct domain(s) responsible for transactivation, which interact with the basal transcriptional machinery either directly or via mediator proteins. Activation is a complex, cooperative process that requires dynamic rearrangement of contacts between TADs, general transcription factors, various coactivators, and chromatin remodeling factors.
TADs themselves often have modular structure and may be composed of multiple small transactivating elements (TEs). Some of these elements show little independent activity, but can synergistically activate transcription. At the sequence level, TAD regions are usually enriched in a single amino acid, a property that was the basis for an early classification scheme that divided TADs into groups comprising acidic, basic, glutamine-rich, proline-rich, serine/threonine-rich and isoleucine-rich domains (reviewed in [146]). However, further biochemical studies have indicated that the preponderance of one or two amino acids does not correlate with specific function, which rather seems to depend on short sequences with specific patterns of hydrophobic and aromatic residues [147]. TADs show very little sequence similarity, even among members of the same family, and yet they often compete for a common target protein and may inhibit each other by a mechanism referred to as “squelching” [148].
When expressed as recombinant proteins, TADs appear to be disordered. Structural disorder was first experimentally documented in the TAD of the Herpes Simplex Virus trans-activating protein VP16 [149]. Among bZIPs, the TAD region of CREB [90], the C- terminal part of the ATF-2 TAD [91], the C-terminal TAD of Fos [150], and most recently TAD of the plant bZIP TF HY5 [97] have been experimentally characterized as disordered. These results suggested that TAD regions may adopt ordered structures only when bound to their molecular partners. The observed low-sequence similarity, combined with the lack of well-defined structural features of these modules has made it difficult to understand their apparent ability to maintain common, specific interactions with multiple coregulators. Furthermore, in many cases the domain boundaries were poorly defined [151]. Characterization of the conformational propensities of different TADs and their specific contacts with coactivators have been further complicated by their involvement in complex interdependent networks of intermolecular interactions regulated by phosphorylation and other posttranscriptional modifications (see below).
It is now well established that the majority of TADs rely on relatively short segments with specific patterns of hydrophobic residues to recognize their target proteins. The commonly accepted model is that of folding-associated recognition. Increased binding specificity at the expense of thermodynamic stability, which characterizes binding coupled to folding of ID regions (see above), facilitates transient interactions with a range of different proteins with major structural and functional differences. For example, depending on cellular cues, the disordered C-terminal TAD of c-Fos may interact with TBP, with the general coactivator CREB-binding protein (CBP), or with the regulatory factor Smad3 [150,152]. Similarly, c-Myc transactivating activity is regulated by binding to variety of factors including TBP, MM-1 and p21 (Ref. [151] and references therein). In this way, the flexibility of cellular response is a direct consequence of the conformational plasticity of TADs.
The versatility of binding and the affinity toward target proteins depend on the presence of specific binding motifs, and the structural and dynamic properties of the polypeptide. Both direct induction of secondary structure and wrapped folding-on-binding onto the target protein surface have been observed [4,12,33]. The functional recognition motifs are often flanked by clusters of charged amino acids. Hermann et al. [153] proposed a two-step model for the association of such TADs with their partners. In the first step, initial low-affinity complex formation is driven by ionic interactions, and is followed by slow interconversion into a more stable form stabilized by hydrophobic interactions. The second step is accompanied by folding of the TAD into a structure that closely fits the molecular surface of the ligand. Recent work of Sugase et al. [33] unraveled the mechanism by which the phosphorylated TAD of CREB (pKID) folds on the surface of KIX domain of CBP (see below). Initially, the disordered pKID forms an ensemble of transient encounter complexes at multiple sites on the KIX surface, which is followed by the formation of an intermediate complex, in which the pKID is only partially folded. In this case, transient encounter complexes are stabilized predominantly by non-specific hydrophobic contacts.
TE segments containing short protein recognition motifs often display residual secondary structure features in their unbound states. These partially formed structures can facilitate formation of complexes and contribute to a decrease in the entropic penalty of binding, in the same manner as discussed earlier in the case of BR-LZ domains [47,110]. Hydrophobic-solvent inducible amphipathic-helical segments have been observed in several acidic TADs [154] and could be predicted by sequence analysis.
Examples of common protein recognition motifs identified within TADs of various TFs are shown in Fig. (7). The TADs regions of Jun subfamily contain two adjacent motifs, HOB1 and HOB2, which serve as cooperating activation modules. Related sequences are also present in c-Fos, but not in other subfamily members. Similarly, activating C/EBP proteins have conserved regions named Box A and Box B embedded in their common acidic TADs [155]. Segments corresponding to HOB2 from Fos/Jun and Box B from C/EBP proteins share weak sequence similarity. The entire fragment encompassing Box A and B is required for interactions of C/EBPs with the TAZ2 domain of CBP/p300 [155,156] and possibly the same segment also mediates interaction with TBP [155]. Homology BOX B comprises the L/FXXLF motif and corresponds to a “signature helix” found in TADs of many transcriptional activators, including the tumor suppressor protein, p53 [157]. The L/FS/ADLF sequence, conserved among C/EBP family members, has been found in other TFs (e.g., ALL1, NFAT1) and has been identified as a critical component of the TAF9 binding motif [158].
Regulation by Phosphorylation
Accumulated evidence indicates that the transactivating activity of many TFs is regulated by phosphorylation. Phosphorylation of TADs may modulate direct binding to protein ligands, as well as intramolecular interactions. All the serine residues present in the p53 TAD were found to be phosphorylated by several distinct kinases. Depending on the phosphorylation status, p53 may interact with MDM2, TAF9, and/or the TAZ2 domain of p300 (reviewed in [159]). The transactivation capacity of Jun, JunD, and ATF-2 is stimulated by the Jun N-terminal kinase (JNK), which specifically phosphorylates Ser63 and Ser73 within the HOB1 motif (see Fig. (7)), whereas the analogous Thr232 in HOB1 of Fos is phosphorylated by ERK [160], reviewed by Wagner [96]. Activation of CREB transcriptional activity depends on the recruitment of the transcriptional coactivator CBP. Association of CREB with the KIX domain of CBP is induced by protein kinase A (PKA) mediated phosphorylation of Ser133, which is located in the kinase-inducible domain (KID) of CREB [161]. In response to ionizing radiation, ATM-dependent phosphorylation of CREB on Ser121 inhibits CREB–CBP interactions [162]. A conserved serine in homology BOX A of C/EBPβ has been shown to be phosphorylated by CDK1 in a cell cycle-dependent manner [163]. Furthermore, the primary phosphorylation often generates new phosphodependent protein–protein interaction motifs. Fos phosphorylated by ERK on multiple residues within its C-terminal TAD becomes a target of prolyl isomerase Pin1 activity. It is thought that conformational changes induced by isomerization of the peptidyl-prolyl bond lead to further enhancement of Fos transcriptional activity [164]. It was noted [100] that phosphorylation of the serine residue within BOX A that is conserved among activating members of the C/EBP family would generate the pSXXI/L (pS denotes phosphorylated serine) motif, which could be recognized by a pair of BRCT repeats from the C-terminus of the PAX-transactivation-domain-interacting protein (PTIP) [165]. Conformational flexibility enables these TADs to interact with both modifying enzymes and their cognate coactivators, thus allowing for context-dependent recognition of TFs by signaling and transcriptional machineries, respectively [166].
Constitutive and Inducible Recognition of Activators by the KIX Domain of CBP/p300
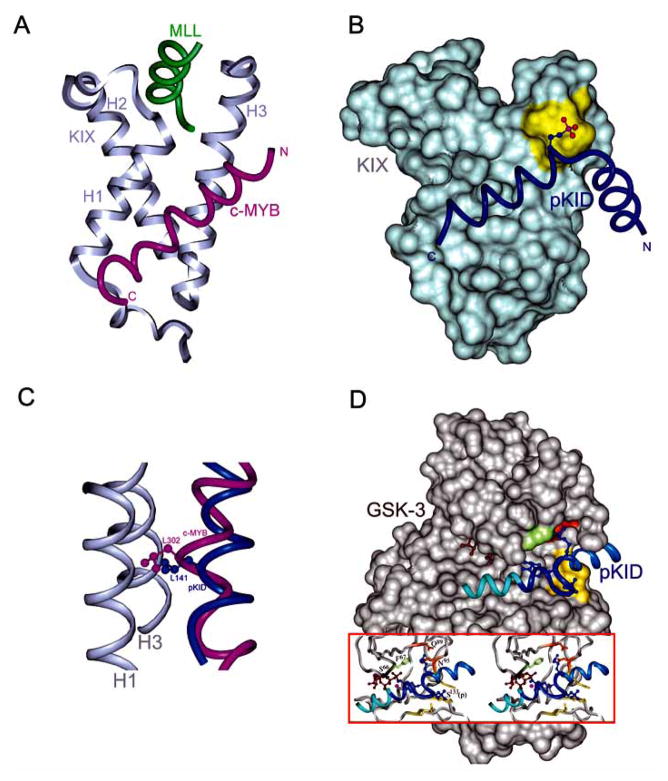
Structural and mechanistic insights into folding of unstructured peptides induced by binding have been revealed by solution structures of complexes of distinct CBP/p300 domains (see below) with several classes of TADs [167–170]. Particularly well characterized by structural and thermodynamic methods are interactions involving the KIX domain, which can recognize TADs of many different TFs. These studies reveal the role of the propensity to form helical structures of unbound peptides in constitutive binding and a structural basis for phosphorylation-regulated inducible binding of activators to KIX. Solution structures of KIX in complex with different peptides are shown in Fig. (8). The KIX domain adopts a compact helical bundle structure composed of three α helices (H1, H2 and H3) and two short 310 helices held together by an extensive hydrophobic core. Two patches of hydrophobic residues are located on the opposite sides of the KIX surface, which serve as binding sites for distinct classes of TADs. The KIX domain of CBP can participate in both, phosphorylation-dependent and phosphorylation- independent, interactions with TFs. Association of CREB with the KIX domain of CBP requires phosphorylation of Ser133 located in the kinase-inducible domain (KID) of CREB [161]. The solution structure of pKID bound to the KIX domain [171] is shown in Fig. (8B). As demonstrated by NMR studies [171], free phosphorylated KID (pKID) is mostly unstructured in solution. Upon binding to KIX, the pKID peptide folds into two separate helices (N- and C-terminal) joined by a loop and positioned almost perpendicularly to each other. Both helices are stabilized by packing against the surface of KIX. The C-terminal helix is situated in the shallow hydrophobic groove of the KIX domain formed by the H1 and H3 helices and contributes most of the interactions with KIX. The phosphorylated Ser133 is located at the N terminus of the C-terminal helix of pKID in the vicinity of Tyr658 and Lys662 of KIX. Mutagenesis studies indicated that hydrogen bonding interaction between pSer133 and Tyr658 plays decisive role in stabilization of the pKID–KIX complex. Basal affinity of unphosphorylated KID to KIX is two orders of magnitude lower than the affinity of KID in the phosphorylated state.
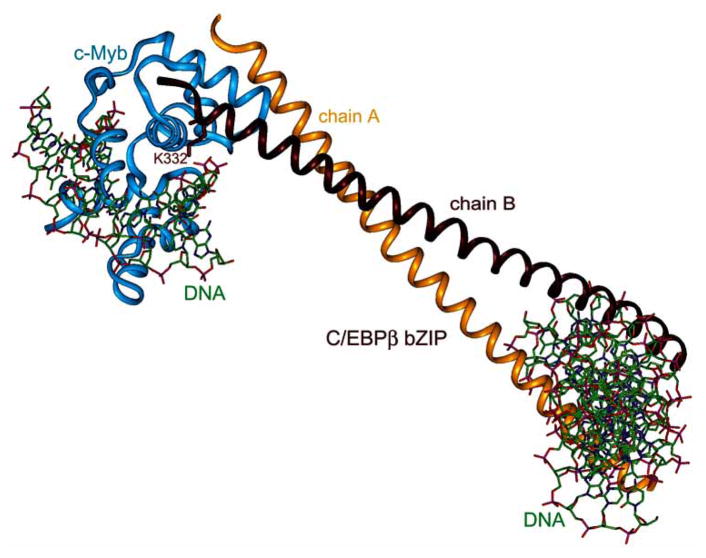
Fig. 8.
Binding commonality and binding diversity displayed by pKID domain of CREB. (A) Synergistic binding of c-MYB (purple) and MLL (green) to KIX (gray); (PDB code 2AGH). (B) pKID (navy) bound to KIX; (PDB code 1KDX). Solvent accessible surface of KIX is shown in gray, Tyr658 in yellow, side chain of phosphorylated Ser133 is represented as balls-and-sticks. (C) Superposition of pKID and c-MYB peptides bound to the same site on KIX (competitive binding). The bend of the α helix of c-Myb facilitates optimal interactions with KIX and enables a critical Leu side chain to penetrate much more deeply than the equivalent Leu from pKID does into the hydrophobic pocket of KIX. (D) Model structure of pKID bound to GSK-3β (beige). The phosphate binding site is depicted in yellow; (adapted from [174]).
On the other hand, c-Myb peptide interacts with KIX via the same binding surface as pKID in a constitutive fashion. Upon binding to KIX, c-Myb peptide folds into a single 16 residue-long α helix that bends to optimize interactions with KIX. Nearly half of the interactions between the two proteins are provided by a critical leucine residue from c-Myb (Leu302) located at the kink of the helix, which inserts its side chain deeply into the hydrophobic pocket of KIX. c-Myb binds to KIX with an affinity sevenfold higher than unphosphorylated KID, and 20–50 fold lower than phosphorylated KID. These differences enable c-Myb to be a constitutive activator, whereas upon phosphorylation, CREB competes efficiently with c-Myb and other constitutive activators for binding to a limited amount of CBP [172]. Structured-based sequence alignment (see Fig. 7C) shows the lack of sequence similarity between KID domains of CREB/ATF proteins and the c-Myb peptide. It is believed that the ability of the activator to form an amphipathic helix is necessary to form a constitutive low affinity complex with KIX. Analysis of interactions by isothermal calorimetry (ITC) is consistent with enthalpy driven complex formation between pKID and KIX, and with binding of c-Myb to KIX depending on both enthalpy and entropy components. These different modes of binding arise from the differences in the length and distribution of nascent helical structures present in unbound peptides.
There is another deep hydrophobic groove created by side chains of three helices located on the opposite side of KIX, which can bind mixed lineage leukemia protein (MLL) and possibly Jun, HTLV-1 Tax, and HIV Tat-1. Recent studies showed that KIX is able to bind simultaneously to two different factors in a cooperative manner (Fig. 8A) [170]. ITC experiments demonstrated that binding of MLL increases the affinity of KIX for either pKID or c-Myb; conversely MLL binds with a twofold higher affinity to the KIX–c-Myb binary complex than to KIX alone. The structure of the KIX–c-Myb–MLL ternary complex revealed allosteric changes in the MLL binding site of KIX, which resulted in the formation of favorable electrostatic interactions between KIX and c-Myb that may account for the synergy between two factors participating in the regulation of the same gene.
Phosphorylation of CREB Ser133 creates a potential phosphorylation site for glycogen synthase kinase 3 (GSK-3β) at CREB Ser129. GSK-3β recognizes and modifies substrates that are phosphorylated at position +3 with respect to the target phosphorylation site (consensus SXXXpS, where pS denotes phosphorylated serine residue) [173]. Molecular modeling studies [174] indicate that, in the ternary GSK-3, ATP, pKID complex, two helices of pKID are almost collinear. pKID peptide interacts with GSK-3β residues: Phe67, Gln89, and Asn95, whereas the binding pocket for phosphorylated Ser133 consists of side chains of Arg96, Arg180 and Lys205 of GSK-3β (Fig. 8D).
FLEXIBLE REGULATORY REGIONS
The functional domains containing a residual structure are usually separated by stretches of disordered regions lacking any elements of the regular secondary structure. In contrast to regions with the potential to fold into the helix upon binding to the proper ligand, these regions remain unstructured, irrespective of their environment. Their role can be as simple as spacers or they may provide the large conformational freedom required for the activation and/or repression domains of the DNA bound TFs to reach other components of transcriptional apparatus. Very often, however, these unstructured linkers harbor important functional sites and play critical role in the activation and regulation of biological functions of numerous proteins [57]. For example, two regulatory segments termed RD1 and RD2, which regulate DNA binding and transactivating activity of C/EBPβ [143] and C/EBPε [175], as well as the δ domain, which control TAD of c-Jun, were predicted to be disordered by a variety of prediction methods [100] (see Fig. 3).
Polypeptide chains in extended coil form are characterized by lower sequence complexity and higher net charge than other types of disorder and often have a high content of proline residues [16]. According to recent analysis, most significant amino acid patterns associated with protein disorder are clusters of three or four glutamic acids and those containing prolines [176]. Consistent with this type of sequence bias, solvent-accessible, unstructured regions are sensitive to proteolysis [177] and are often enriched in PEST sequences (e.g., regions rich in proline, glutamic acid, serine and threonine residues) that confer multiple protein degradation signals and phosphorylation targets [178]. Significantly, in several cases unstructured regions were directly recognized and cleaved by the 20S proteosome [14]. Structureless regulatory regions also serve as platforms for docking sites and recognition motifs facilitating interactions with specific modifying enzymes.
Transcriptional regulatory proteins are end points of many signal transduction cascades and their activities are subjected to multiple modes of regulation [166,179]. In response to external stimuli TFs have to be inactivated as quickly as they are induced. A number of short-lived factors such as NF-κB, AP-1 and C/EBPs, undergoes selective degradation by ubiquitin proteasome pathway [144,180]. Activity of TFs is also controlled by phosphorylation, sumoylation and acetylation on lysine residues and methylation on argin-ine and lysine residues. Flexible regulatory regions of C/EBP proteins are targeted for sumoylation [175] in addition to phosphorylation and contain putative phospho-Ser/Thr recognition sites for WW and Polo-box modules of Pin1 and Polo-like kinases, respectively [100]. Majority of bZIP proteins are downstream targets for mitogen-activated protein (MAP) kinases (e.g., the components of RAS pathway) [181], which often act in concerted manner with Ser/Thr phosphatases to regulate TFs in a graded fashion (reviewed in [166]). MAP kinases phosphorylate very similar motifs containing minimal consensus sequence Ser/Thr-Pro and recognize specific substrates through binding surfaces (docking grooves) located outside the catalytic active site. These docking grooves bind to short peptide motifs (docking sites) residing in target proteins. Depending on the number and sequence of these recognition motifs, substrates are phosphorylated by specific subsets of MAPKs (reviewed in [182]). For example, a D docking site located within the δ domain of c-Jun (Fig. 3) mediates phosphorylation of c-Jun by JNK, whereas a DEF motif is required for ERK phosphorylation of c-Fos. On the other hand, JunD, which contains both D and DEF motifs, undergoes phosphorylation either by ERK or JNK. Certain proteins (e.g., JunB) contain the docking site but lack the phospho-acceptor motif and act as scaffolds for assembly of complexes containing components of MAP kinase cascades. Such a mode of specific recognition requires large area of interactions between the kinase and its substrate that can not be achieved by interactions of two globular proteins and depends on the ability of the unstructured region of one to wrap around the other.
MULTIPROTEIN-DNA COMPLEXES IN COMBINATORIAL GENE REGULATION
Transcriptional regulation of the target gene requires the synergetic action of multiple TFs bound to cis-regulatory elements, and cooperation of diverse families of coregulators [183,184]. Activator-binding sites are often clustered into enhancers that function as separate regulatory modules. Specificity in transcription is achieved by cooperative recruitment of several sequence-specific DNA-binding TFs and formation of multiprotein-enhancer DNA complexes termed enhanceosomes [185]. The gene-specific architecture of such a complex is dependent on the spatial arrangement of cis-regulatory elements and the correct array of bound activators which together generate a network of protein–protein and protein–DNA interactions unique to a given enhancer. TFs bound to composite regulatory sites modulate each other’s activity, thus, a particular factor can assume different functions when bound to different enhancers. Moreover, cooperative interactions of TFs increase stability of the mul-tiprotein–DNA complex, enhance the recognition specificity, and further extend the combinatorial potential of transcription regulation. Importantly, concerted action of numerous signal-activated TFs allows for integration of independent signaling pathways at a specific promoter.
Direct Protein–Protein Interactions of bZIPs with Other TF Proteins at Composite Sites on Promoters
Many recognition sequences in natural promoter and enhancer regions deviate from the optimal binding sites for regulatory proteins. The weak binding affinities to these suboptimal sites impose a requirement for interactions with others TFs. The inherent flexibility of bZIP regions facilitates direct association of bZIP proteins with other structurally unrelated TFs (Fig. 9) such as NF-κB/Rel family members, NFAT, cMyb, and SMADs bound to a nearby regulatory element.
Fig. 9.
The NFAT–Fos–Jun–DNA quaternary complex (PDB code 1A02). The ARRE2 DNA sequence in the crystal is shown below the figure. Note the deviation from the consensus sequence (TGAGTCA) recognized by AP-1 proteins. Protein–protein interactions between NFAT and the LZ of AP-1 enable the two TFs to bind DNA cooperatively, and regulate coordinately the IL-2 promoter.
Synergistic regulation of transcription specificity by multiple factors is best illustrated by studies of regulatory complexes containing Jun/Fos proteins [70]. Nonconsensus AP-1 binding sites are present downstream of most NFAT sites in the promoter region of several genes in immune system cells. Both AP-1 and NFAT show only weak independent binding at such composite sites and the productive response to NFAT requires concomitant activation of AP-1 proteins. The activation of NFAT proteins is regulated by calcium and calcineurin, whereas that of AP-1 proteins by RAS/MAP kinase and PKC, thus composite sites, such as NFAT:AP-1, integrate signals from distinct signal transduction pathways. The cocrystal structure of Fos–Jun–NFAT DBDs bound to the DNA fragment containing composite site ARRE2 from the IL-2 gene promoter provided the structural basis for transcriptional cooperativity of these TFs [186]. NFAT and Fos–Jun heterodimer bind on the same face of the DNA to adjacent binding sites (Fig. 9). Interactions of AP-1 with NFAT require substantial bending of both the coiled-coil and the DNA duplex. The majority of the protein–protein interactions are between NFAT and the Fos subunit of AP-1. These contacts help to orient Fos–Jun heterodimer in a unique orientation with respect to DNA, so that Jun binds to the half of the asymmetric AP-1 site close to NFAT. In contrast, both orientations were observed in the crystal of the Fos–Jun–DNA ternary complex [74]. Of note, protein–protein interactions are not critical for cooperative binding of c-Jun–ATF2 heterodimer and IRF-3 to the interferon β enhancer. In this case, the sites for adjacent proteins overlap and the cooperativity of binding arises from sequence-dependent conformability of the DNA [187].
Another well studied example of transcriptional synergy is the cooperation of c-Myb with C/EBP family members in induction of the mim-1 gene expression during myeloid cell differentiation. The binding sites for c-Myb and C/EBP proteins on the mim-1 gene promoter are separated by a sequence of 80 base pairs. As demonstrated by atomic force microscopy, their cooperative interaction involves DNA loop formation. X-ray crystallography provided a high resolution view of c-Myb–C/EBPβ interactions and the mechanism of DNA looping stabilization [188]. In the crystal of the c-Myb–C/EBPβ–DNA complex, C-terminal portions of C/EBPβ chains A and B interact with a subdomain of c-Myb (bound to another DNA fragment) to form a four-helix bundle structure (Fig. 10). The side chain of Lys332 from C/EBPβ chain B makes a salt bridge with the phosphate of c-Myb-bound DNA molecule, presumably contributing to DNA loop formation. The C-terminal LZ extensions of C/EBPβ, which interact with c-Myb are unstructured without c-Myb and assume helical conformation upon binding to c-Myb. Reciprocally, C/EBPβ chain A stabilizes the conformation of a c-Myb loop that interacts with the DNA backbone.
Fig. 10.
The intercomplex interactions between c-Myb bound to one DNA fragment and the C-terminal portions of C/EBPβ homodimer bound to another DNA fragment observed in the crystal structure of the c-Myb–C/EBPβ–DNA complex [188]; PDB code 1H89. The helical fold of C-terminal extensions of C/EBPβ LZ is induced by interaction with c-Myb.
Assembly of TFs on the enhancer may as well be affected by non-DNA binding proteins. For example, binding of C/EBPβ to suboptimal site located on the C-reactive protein gene promoter is enhanced by protein–protein interaction with c-Rel, which is not itself bound to DNA [189]. Also, several viral proteins are known to associate with bZIPs to gain access to the promoter (see above). X-ray crystallographic studies of multiprotein complexes bound to DNA were reviewed in [190] and [191].
Architectural TFs
Long-range interactions between cis-regulatory modules (enhancers, silencers and promoters) are mediated by architectural proteins, which facilitate interactions among proteins bound to separate recognition sites by binding to multiple sequences within enhancers and inducing changes in DNA structure. For example, BACH1 helicase forms heterodimers with small Maf proteins that bind to MARE recognition elements. Oligomerization of BACH1–Maf heterodimer via the N-terminus BTB/POZ domain of BACH1 generates multimeric and multivalent DNA binding complexes that are able to mediate interactions between distant, multiple MAREs and to induce DNA loop structures [93].
Critical to the assembly of many enhanceosomes are the high mobility group A proteins, HGMA (previously known as HMGI(Y)). Members of the HGMA family are non-histone, multifunctional proteins that participate in a variety of cellular processes, including the regulation of chromatin structure (reviewed in [192,193]). The striking feature of these proteins is their enormous structural flexibility and their ability to bind to both DNA and a wide range of proteins from different classes, including many TFs. Each HGMA protein contains three repeats of peptides (AT-hooks), which enable them to bind to the minor groove of short stretches of AT-rich DNA and to induce structural alterations in bound DNA, such as looping, bending, and/or unwinding of linear DNA particles. It has been proposed that HGMA proteins recognize gene-specific arrangements of AT-rich sequences within enhancer region and coordinate incorporation of other nuclear factors to the enhanceosome [192]. Direct interactions of HGMA with ATF-2 and c-Jun were reported in the context of the IFN-β gene enhancer. HGMA also interacts with AP-1 (Il-2 promoter), Jun-B/Fra-2 (HPV18 enhancer), C/EBPβ (adipocyte-specific gene promoters), and C/EBPα on the IR promoter (Ref. [193] and references therein).
Interactions with Coregulators
Enhancer-bound TFs recruit multi-subunit transcriptional coactivator complexes [194], which facilitate the binding and function of Pol II at the core promoter. The interaction of multiple coregulators with TFs in different temporal and spatial contexts provides another possible level of regulation of gene expression. The accessibility of DNA template is modulated by SWI/SNF chromatin remodeling complexes, histone acetyltransferase (HAT) and deacetylase (HDAC) complexes, histone methyltransferases (HMTs) and histone kinases. Recruitment of HATs (e.g., p300 or its ortholog CBP, GCN5/PCAF, MBF1) and HMTs by activators is crucial for activation of many classes of genes, whereas deacetylation of the histone tails is required for repression. Histone lysine methylation is involved in both gene activation and repression, depending upon the specific lysine residue that gets methylated. The possibility of interaction with distinct coregulator complexes underlies the capability of certain TFs to perform tissue and cell type dependent dual functions. Thus C/EBP proteins, which interact with the general coactivator, CBP/p300 and usually act as transcriptional activators, were shown to inhibit the expression of PPARβ gene in mouse keratinocytes through the recruitment of a transcriptional repressor complex containing HDAC-1 [195]. The essential role in the regulation of transcription in eukaryotes is played by the multi-subunit Mediator complex, which activates distinct expression programs via interactions with gene-specific TFs [196]. Other examples of factors found in the regulatory complexes are BRCA1 adapter protein [197] and TORC (see above). These cofactor molecules bridge the sequence-specific DNA-binding proteins to the components of general transcription machinery. On the other hand, a range of TFs binds directly to TBP and its associated factors TAFs [194]. Surprisingly, it was recently found that the terminal kinases of signal transduction cascades which phosphorylates TFs and other components of chromatin, also play a structural role and are stably associated with the genes they activate [198]. As Pokholak et al. [199] demonstrated by genome-wide ChIP-Chip analyses in yeast, activated MAPK and PKA kinases can bind at target genes to factors within transcription complexes.
Examples of bZIP proteins’ interactions with diverse coregulators are presented in Table 2. The individual interactions are too weak to activate target genes and the stable recruitment of cofactors to the promoter requires cooperation between regulatory TFs [200] and/or multivalent activator-coactivator interactions. For example, synergistic binding of two TADs from Nrf2 to CBP is necessary for induction of reporter gene expression, whereas activation of transcription by CREB requires cooperativity between pKID (which recruits CBP) and the glutamine-rich Q2 domain (which recruits TFIID) as well as coordination with other gene-specific TFs [98,200]. The synergy between TFs is facilitated by the CBP or p300 and other scaffolding proteins that are capable of forming simultaneous interactions with multiple TFs. The p300 protein binds to TAD of c-Myb via KIX domain and to TADs of C/EBP proteins via TAZ2 domain, and further stimulates the synergy between c-Myb and C/EBPs in expression of mim-1. CBP/p300 is also capable to link factors bound to different cognate regulatory modules, and is thought to act as a parser of regulatory information [95].
Table 2.
Association of bZIP Proteins with Transcription Coregulator Complexes
| Cofactor(domain) | bZIP TF | References |
|---|---|---|
| CBP/p300 (KIX) | CREB, CREM, ATF1, ATF4, c-Jun | |
| CBP/p300 (PHD/CH2) | ATF2 | Revied in [212,213] |
| CBP/p300 (TAZ2/CH3) | C/EBPs (except -γ), JunB, c-Fos, Nrf2 | |
| CBP/p300 | Par proteins | [214] |
| SWI/SNF (Brg1) | C/EBPα, C/EBPβ | Revied in [215] |
| Mediator (MED23) | C/EBPβ | [196] |
| HDAC-1 complex | C/EBPβ | [195] |
| TORC | CREB (bZIP) | [125] |
| MBF1 | JunD (bZIP) | [123] |
| BRCA1 (BRCTdomain) | BACH1 | [216] |
| BRCA1 | JunB, JunD | [217] |
| TFIID (TBP) | C/EBPα, c-Fos, FosB, ATF4 | [89,150,155] |
| TFIID (TAF4) | CREB (Q2 domain) | [98] |
| TFIID (TAF9) | C/EBPβ | [158] |
| TFIID (TAF1) | c-Jun (bZIP) | [124] |
| TFIIB | C/EBPα, ATF4 | [155,209] |
| TFIIF (RAP30) | ATF4 | [209] |
The cooperative recruitment of TFs, coactivators and chromatin remodeling factors to promoters produces regulatory complexes specific to tissue, cell type and external stimulus. The functions of individual components of such complexes are interdependent and require the concerted action of all the protein in the complex [66,67]. Depending on the cell type and external stimulus, distinct sets of regulatory proteins can assemble at the same promoter. An additional layer of complexity in transcriptional complexes is introduced by enzymatic activities exerted by participating proteins toward each other. HATs such as CBP/p300 and PCAF acetylate variety of TFs (e.g., p53, c-Jun, C/EBPβ, mafG, and CREB) as well as HGMA proteins, altering their DNA-binding ability and transactivation potential [201,202]. Reciprocally, CBP/p300 bound C/EBP proteins facilitate its massive phosphorylation [156,203]. Accumulated evidence implies that BRCA1–BARD1 heterodimer that exhibits the E3-ubiquitin ligase activity may specifically ubiquitylate proteins involved in transcription [204]. Another example is the histone methyltransferase CARM1 that methylates CBP/p300 and disrupts its interaction with CREB, thus inactivating CREB-mediated transcription [205].
A recurring theme in the regulation of assembly and function of transcriptional complexes is structural disorder of participating proteins. Results from CD and NMR studies imply that HGMA proteins are almost entirely unfolded in solution and undergo disorder-order structural transitions upon binding to diverse ligands [192]. CBP/p300, which associates with myriads of TFs and other protein partners, has more than 50% of its residues in predicted intrinsically disordered regions [4]. These regions are located between folded domains and function as flexible linkers. The central two-thirds of BRCA1, which separates the N-terminal RING domain responsible for interaction with BARD1 and the C-terminal BRCT domain, has been predicted to be unstructured [206].
Numerous theories have been proposed to explain the prevalence of unstructured proteins in transcription control. (1) Disordered regions may be considered a sort of “molecular glue”, which is needed to connect together all the components of multiprotein-DNA complexes [29,192]. (2) The ability of IDPs to form transient interactions characterized by high specificity [4,11,55,64] is critical for gene regulatory networks. (3) Rapid clearance and degradation of IDPs provide an additional level of control for turning on and off transcription responses to intracellular signaling [6,13]. (4) Formation of multimolecular complexes and protein–DNA/RNA interactions requires large intermolecular interfaces [207]. ID regions provide for large interfaces without causing cellular crowding or increasing the size of complexes and cells [31]. (5) The “fly-casting” mechanism for binding interactions [41] predicts an increased rate of binding and it may be particularly important in transcriptional processes when the concentrations of regulatory proteins and their targets are low [12]. (6) Conformational adaptability to environment of IDPs enables different modes of regulation [64]. In particular, ID regions are amenable for phosphorylation and other covalent modifications, which modulate activities of TFs as well as the assembly and disassembly of transcription regulatory complexes [4,64]. (7) The existence of a disordered state prior to specific binding may prevent the occurrence of spontaneous interactions with certain partners at inappropriate times or location [64] (e.g., interaction of MBF1 with BR of DNA bound AP-1 proteins). (8) ID optimizes allosteric coupling, thereby facilitate site-to site communication and signal propagation [46]. Taken together, ID underlies a variety of critical aspects of spatial and temporal organization of interaction networks that govern the gene expression program.
SUMMARY
Eukaryotic TF proteins are central components of dynamic supramolecular complexes that control transcription in a combinatorial manner, and all seem to require a considerable level of structural disorder to perform their functions. This notion is particularly relevant to the bZIP class of activators, which do not possess preformed DBD domains. With the exception of few proteins with specialized functional domains other than TADs, the native structure of a typical bZIP protein is a premolten globule. bZIPs are composed of regions with the potential to fold upon complexation and regions that retain irregular conformations independently of their environment. Conformational flexibility is necessary for the formation of sequential, transient intermolecular interactions that regulate bZIPs’ cellular compartmentalization, DNA-binding and transactivating activities and eventually their degradation. bZIP TFs rely on disorder–order transitions of their DBDs for specific dimerization and DNA-binding. Such a mechanism of DNA recognition insures a fast and reliable choice of a specific site and timely binding of coregulators. The ability of bZIP monomers to associate selectively with many different dimerization partners in regulatable manner enables the formation of multiple easily interconverted dimers with distinct DNA and protein binding properties, thus greatly broadening the range of transcriptional control exerted by these factors. Specific dimerization also underlies the dominant repressive capability of naturally occurring truncated variants of these proteins, which retain the bZIP domain, but lack other functional domains. The inherent flexibility of the LZ dimeric coiled-coil domains facilitates physical associations with other factors. The mostly unfolded regions that are responsible for transactivation contain protein recognition motifs and are able to form highly specific but reversible interactions with wide range of protein targets, interactions that can be easily modulated by phosphorylation/dephosphorylation events. Binding diversity and binding commonality with other cofactors are essential for the dynamic and competitive exchange of contacts with multiple components of transcriptional regulatory complexes. Solvent-exposed, unstructured regions serve as flexible linkers between TADs and DNA binding elements. These segments display sites for post-translational modifications, as well as docking domains and recognition motifs for specific modifying enzymes, which regulate the activities of TF proteins in a stimulus-dependent fashion. Structural disorder thus enables multiple modes of regulation of bZIP proteins’ activities and girds their ability to effectively control the cellular patterns of gene expression.
Acknowledgments
I thank my colleagues from MCL for help and discussions, and Michael Gribskov for many useful suggestions and thoughtful comments on the manuscript. This work was supported by the Intramural Research Program of NIH, National Cancer Institute, Center for Cancer Research.
ABBREVIATONS
- TF
Transcription factor
- ID
Intrinsic disorder
- IDP
Intrinsically disordered protein
- DBD
DNA-binding domain
- BR
Basic region
- LZ
Leucine zipper
- TAD
Transactivation domain
- NLS
Nuclear localization signal
Footnotes
USEFUL WEB RESOURCES:
PFAM: Protein families’ database [208]; (http://www.sanger.ac.uk/Software/Pfam/)
DisProt: Database of Protein Disorder [101]; (http://www.disprot.org)
SMART: Simple Modular Architecture Research Tool [102]; (http://smart.embl.de)
bZIPDB: A database of regulatory information for human bZIP transcription factors [87]; (http://biosoft.kaist.ac.kr/bzipdb}
References
- 1.Dunker AK, Obradovic Z. The protein trinity–linking function and disorder. Nat Biotechnol. 2001;19:805–806. doi: 10.1038/nbt0901-805. [DOI] [PubMed] [Google Scholar]
- 2.Uversky VN. Natively unfolded proteins: a point where biology waits for physics. Protein Sci. 2002;11:739–756. doi: 10.1110/ps.4210102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Fukuchi S, Homma K, Minezaki Y, Nishikawa K. Intrinsically disordered loops inserted into the structural domains of human proteins. J Mol Biol. 2006;355:845–857. doi: 10.1016/j.jmb.2005.10.037. [DOI] [PubMed] [Google Scholar]
- 4.Dyson HJ, Wright PE. Intrinsically unstructured proteins and their functions. Nat Rev Mol Cell Biol. 2005;6:197–208. doi: 10.1038/nrm1589. [DOI] [PubMed] [Google Scholar]
- 5.Dunker AK, Brown CJ, Lawson JD, Iakoucheva LM, Obradovic Z. Intrinsic disorder and protein function. Biochemistry. 2002;41:6573–6582. doi: 10.1021/bi012159+. [DOI] [PubMed] [Google Scholar]
- 6.Wright PE, Dyson HJ. Intrinsically unstructured proteins: reassessing the protein structure-function paradigm. J Mol Biol. 1999;293:321–331. doi: 10.1006/jmbi.1999.3110. [DOI] [PubMed] [Google Scholar]
- 7.Radivojac P, Iakoucheva LM, Oldfield CJ, Obradovic Z, Uversky VN, Dunker AK. Intrinsic disorder and functional proteomics. Biophys J. 2007;92:1439–1456. doi: 10.1529/biophysj.106.094045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Dyson HJ, Wright PE. Unfolded proteins and protein folding studied by NMR. Chem Rev. 2004;104:3607–3622. doi: 10.1021/cr030403s. [DOI] [PubMed] [Google Scholar]
- 9.Receveur-Brechot V, Bourhis JM, Uversky VN, Canard B, Longhi S. Assessing protein disorder and induced folding. Proteins. 2006;62:24–45. doi: 10.1002/prot.20750. [DOI] [PubMed] [Google Scholar]
- 10.Tompa P. Intrinsically unstructured proteins. Trends Biochem Sci. 2002;27:527–533. doi: 10.1016/s0968-0004(02)02169-2. [DOI] [PubMed] [Google Scholar]
- 11.Uversky VN, Oldfield CJ, Dunker AK. Showing your ID: intrinsic disorder as an ID for recognition, regulation and cell signaling. J Mol Recognit. 2005;18:343–384. doi: 10.1002/jmr.747. [DOI] [PubMed] [Google Scholar]
- 12.Dyson HJ, Wright PE. Coupling of folding and binding for unstructured proteins. Curr Opin Struct Biol. 2002;12:54–60. doi: 10.1016/s0959-440x(02)00289-0. [DOI] [PubMed] [Google Scholar]
- 13.Fink AL. Natively unfolded proteins. Curr Opin Struct Biol. 2005;15:35–41. doi: 10.1016/j.sbi.2005.01.002. [DOI] [PubMed] [Google Scholar]
- 14.Tompa P. The interplay between structure and function in intrinsically unstructured proteins. FEBS Lett. 2005;579:3346–3354. doi: 10.1016/j.febslet.2005.03.072. [DOI] [PubMed] [Google Scholar]
- 15.Dunker AK, Cortese MS, Romero P, Iakoucheva LM, Uversky VN. Flexible nets. The roles of intrinsic disorder in protein interaction networks. FEBS J. 2005;272:5129–5148. doi: 10.1111/j.1742-4658.2005.04948.x. [DOI] [PubMed] [Google Scholar]
- 16.Vucetic S, Brown CJ, Dunker AK, Obradovic Z. Flavors of protein disorder. Proteins. 2003;52:573–584. doi: 10.1002/prot.10437. [DOI] [PubMed] [Google Scholar]
- 17.Lobley A, Swindells MB, Orengo CA, Jones DT. Inferring function using patterns of native disorder in proteins. PLoS Comput Biol. 2007;3:e162. doi: 10.1371/journal.pcbi.0030162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Uversky VN, Gillespie JR, Fink AL. Why are “natively unfolded” proteins unstructured under physiologic conditions? Proteins. 2000;41:415–427. doi: 10.1002/1097-0134(20001115)41:3<415::aid-prot130>3.0.co;2-7. [DOI] [PubMed] [Google Scholar]
- 19.Prilusky J, Felder CE, Zeev-Ben-Mordehai T, Rydberg EH, Man O, Beckmann JS, Silman I, Sussman JL. FoldIndex: a simple tool to predict whether a given protein sequence is intrinsically unfolded. Bioinformatics. 2005;21:3435–3438. doi: 10.1093/bioinformatics/bti537. [DOI] [PubMed] [Google Scholar]
- 20.Coeytaux K, Poupon A. Prediction of unfolded segments in a protein sequence based on amino acid composition. Bioinformatics. 2005;21:1891–1900. doi: 10.1093/bioinformatics/bti266. [DOI] [PubMed] [Google Scholar]
- 21.Liu J, Rost B. NORSp: Predictions of long regions without regular secondary structure. Nucleic Acids Res. 2003;31:3833–3835. doi: 10.1093/nar/gkg515. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Linding R, Russell RB, Neduva V, Gibson TJ. GlobPlot: Exploring protein sequences for globularity and disorder. Nucleic Acids Res. 2003;31:3701–3708. doi: 10.1093/nar/gkg519. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Ferron F, Longhi S, Canard B, Karlin D. A practical overview of protein disorder prediction methods. Proteins. 2006;65:1–14. doi: 10.1002/prot.21075. [DOI] [PubMed] [Google Scholar]
- 24.Bourhis JM, Canard B, Longhi S. Predicting protein disorder and induced folding: from theoretical principles to practical applications. Curr Protein Pept Sci. 2007;8:135–149. doi: 10.2174/138920307780363451. [DOI] [PubMed] [Google Scholar]
- 25.Oldfield CJ, Cheng Y, Cortese MS, Brown CJ, Uversky VN, Dunker AK. Comparing and combining predictors of mostly disordered proteins. Biochemistry. 2005;44:1989–2000. doi: 10.1021/bi047993o. [DOI] [PubMed] [Google Scholar]
- 26.Liu J, Rost B. Comparing function and structure between entire proteomes. Protein Sci. 2001;10:1970–1979. doi: 10.1110/ps.10101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Ward JJ, Sodhi JS, McGuffin LJ, Buxton BF, Jones DT. Prediction and functional analysis of native disorder in proteins from the three kingdoms of life. J Mol Biol. 2004;337:635–645. doi: 10.1016/j.jmb.2004.02.002. [DOI] [PubMed] [Google Scholar]
- 28.Chen JW, Romero P, Uversky VN, Dunker AK. Conservation of intrinsic disorder in protein domains and families: II. functions of conserved disorder. J Proteome Res. 2006;5:888–898. doi: 10.1021/pr060049p. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Minezaki Y, Homma K, Kinjo AR, Nishikawa K. Human Transcription Factors Contain a High Fraction of Intrinsically Disordered Regions Essential for Transcriptional Regulation. J Mol Biol. 2006;359:1137–1149. doi: 10.1016/j.jmb.2006.04.016. [DOI] [PubMed] [Google Scholar]
- 30.Romero PR, Zaidi S, Fang YY, Uversky VN, Radivojac P, Oldfield CJ, Cortese MS, Sickmeier M, LeGall T, Obradovic Z, Dunker AK. Alternative splicing in concert with protein intrinsic disorder enables increased functional diversity in multicellular organisms. Proc Natl Acad Sci USA. 2006;103:8390–8395. doi: 10.1073/pnas.0507916103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Gunasekaran K, Tsai CJ, Kumar S, Zanuy D, Nussinov R. Extended disordered proteins: targeting function with less scaffold. Trends Biochem Sci. 2003;28:81–85. doi: 10.1016/S0968-0004(03)00003-3. [DOI] [PubMed] [Google Scholar]
- 32.Oldfield CJ, Cheng Y, Cortese MS, Romero P, Uversky VN, Dunker AK. Coupled folding and binding with alpha-helix-forming molecular recognition elements. Biochemistry. 2005;44:12454–12470. doi: 10.1021/bi050736e. [DOI] [PubMed] [Google Scholar]
- 33.Sugase K, Dyson HJ, Wright PE. Mechanism of coupled folding and binding of an intrinsically disordered protein. Nature. 2007;447:1021–1025. doi: 10.1038/nature05858. [DOI] [PubMed] [Google Scholar]
- 34.Verkhivker GM, Bouzida D, Gehlhaar DK, Rejto PA, Freer ST, Rose PW. Simulating disorder-order transitions in molecular recognition of unstructured proteins: where folding meets binding. Proc Natl Acad Sci USA. 2003;100:5148–5153. doi: 10.1073/pnas.0531373100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Wang J, Verkhivker GM. Energy landscape theory, funnels, specificity, and optimal criterion of biomolecular binding. Phys Rev Lett. 2003;90:188101. doi: 10.1103/PhysRevLett.90.188101. [DOI] [PubMed] [Google Scholar]
- 36.Levy Y, Cho SS, Onuchic JN, Wolynes PG. A survey of flexible protein binding mechanisms and their transition states using native topology based energy landscapes. J Mol Biol. 2005;346:1121–1145. doi: 10.1016/j.jmb.2004.12.021. [DOI] [PubMed] [Google Scholar]
- 37.Wang J, Lu Q, Lu HP. Single-molecule dynamics reveals cooperative binding-folding in protein recognition. PLoS Comput Biol. 2006;2:e78. doi: 10.1371/journal.pcbi.0020078. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Wang J, Xu L, Wang E. Optimal specificity and function for flexible biomolecular recognition. Biophys J. 2007;92:L109–L111. doi: 10.1529/biophysj.107.105551. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Lu Q, Lu HP, Wang J. Exploring the mechanism of flexible biomolecular recognition with single molecule dynamics. Phys Rev Lett. 2007;98:128105. doi: 10.1103/PhysRevLett.98.128105. [DOI] [PubMed] [Google Scholar]
- 40.Demchenko AP. Recognition between flexible protein molecules: induced and assisted folding. J Mol Recognit. 2001;14:42–61. doi: 10.1002/1099-1352(200101/02)14:1<42::AID-JMR518>3.0.CO;2-8. [DOI] [PubMed] [Google Scholar]
- 41.Shoemaker BA, Portman JJ, Wolynes PG. Speeding molecular recognition by using the folding funnel: the fly-casting mechanism. Proc Natl Acad Sci USA. 2000;97:8868–8873. doi: 10.1073/pnas.160259697. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Sturtevant JM. Heat capacity and entropy changes in processes involving proteins. Proc Natl Acad Sci USA. 1977;74:2236–2240. doi: 10.1073/pnas.74.6.2236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Duan Y, Wilkosz P, Rosenberg JM. Dynamic contributions to the DNA binding entropy of the EcoRI and EcoRV restriction endonucleases. J Mol Biol. 1996;264:546–555. doi: 10.1006/jmbi.1996.0660. [DOI] [PubMed] [Google Scholar]
- 44.Spolar RS, Record MT., Jr Coupling of local folding to site-specific binding of proteins to DNA. Science. 1994;263:777–784. doi: 10.1126/science.8303294. [DOI] [PubMed] [Google Scholar]
- 45.Dunitz JD. Win some, lose some: enthalpy-entropy compensation in weak intermolecular interactions. Chem Biol. 1995;2:709–712. doi: 10.1016/1074-5521(95)90097-7. [DOI] [PubMed] [Google Scholar]
- 46.Hilser VJ, Thompson EB. Intrinsic disorder as a mechanism to optimize allosteric coupling in proteins. Proc Natl Acad Sci USA. 2007;104:8311–8315. doi: 10.1073/pnas.0700329104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Mohan A, Oldfield CJ, Radivojac P, Vacic V, Cortese MS, Dunker AK, Uversky VN. Analysis of molecular recognition features (MoRFs) J Mol Biol. 2006;362:1043–1059. doi: 10.1016/j.jmb.2006.07.087. [DOI] [PubMed] [Google Scholar]
- 48.Puntervoll P, Linding R, Gemünd C, Chabanis-Davidson S, Mattingsdal M, Cameron S, Martin DMA, Ausiello G, Brannetti B, Costantini A, Ferre F, Maselli V, Via A, Cesareni G, Diella F, Superti-Furga G, Wyrwicz L, Ramu C, McGuigan C, Gudavalli R, Letunic I, Bork P, Rychlewski L, Küster B, Helmer-Citterich M, Hunter WN, Aasland R, Gibson TJ. ELM server: A new resource for investigating short functional sites in modular eukaryotic proteins. Nucleic Acids Res. 2003;31:3625–3630. doi: 10.1093/nar/gkg545. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Neduva V, Linding R, Su-Angrand I, Stark A, de Masi F, Gibson TJ, Lewis J, Serrano L, Russell RB. Systematic discovery of new recognition peptides mediating protein interaction networks. PLoS Biol. 2005;3:e405. doi: 10.1371/journal.pbio.0030405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Apic G, Russell RB. A shortcut to peptides to modulate platelets. Nat Chem Biol. 2007;3:83–84. doi: 10.1038/nchembio0207-83. [DOI] [PubMed] [Google Scholar]
- 51.Csizmok V, Bokor M, Banki P, Klement E, Medzihradszky KF, Friedrich P, Tompa K, Tompa P. Primary contact sites in intrinsically unstructured proteins: the case of calpastatin and microtubule-associated protein 2. Biochemistry. 2005;44:3955–3964. doi: 10.1021/bi047817f. [DOI] [PubMed] [Google Scholar]
- 52.Namba K. Roles of partly unfolded conformations in macromolecular self-assembly. Genes Cells. 2001;6:1–12. doi: 10.1046/j.1365-2443.2001.00384.x. [DOI] [PubMed] [Google Scholar]
- 53.Blundell TL, Fernandez-Recio J. Cell biology: brief encounters bolster contacts. Nature. 2006;444:279–280. doi: 10.1038/nature05306. [DOI] [PubMed] [Google Scholar]
- 54.Han JD, Bertin N, Hao T, Goldberg DS, Berriz GF, Zhang LV, Dupuy D, Walhout AJ, Cusick ME, Roth FP, Vidal M. Evidence for dynamically organized modularity in the yeast protein-protein interaction network. Nature. 2004;430:88–93. doi: 10.1038/nature02555. [DOI] [PubMed] [Google Scholar]
- 55.Singh GP, Ganapathi M, Dash D. Role of intrinsic disorder in transient interactions of hub proteins. Proteins: Struct Funct Bioinformatics. 2007;66:761–765. doi: 10.1002/prot.21281. [DOI] [PubMed] [Google Scholar]
- 56.Iakoucheva LM, Radivojac P, Brown CJ, O’Connor TR, Sikes JG, Obradovic Z, Dunker AK. The importance of intrinsic disorder for protein phosphorylation. Nucleic Acids Res. 2004;32:1037–1049. doi: 10.1093/nar/gkh253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Eisenhaber B, Eisenhaber F. Posttranslational modifications and subcellular localization signals: indicators of sequence regions without inherent 3D structure? Curr Protein Pept Sci. 2007;8:197–203. doi: 10.2174/138920307780363424. [DOI] [PubMed] [Google Scholar]
- 58.Ptashne M, Gann A. Transcriptional activation by recruitment. Nature. 1997;386:569–577. doi: 10.1038/386569a0. [DOI] [PubMed] [Google Scholar]
- 59.Sigler PB. Transcriptional activation. Acid blobs and negative noodles. Nature. 1988;333:210–212. doi: 10.1038/333210a0. [DOI] [PubMed] [Google Scholar]
- 60.Frankel AD, Kim PS. Modular structure of transcription factors: implications for gene regulation. Cell. 1991;65:717–719. doi: 10.1016/0092-8674(91)90378-c. [DOI] [PubMed] [Google Scholar]
- 61.Uesugi M, Nyanguile O, Lu H, Levine AJ, Verdine GL. Induced alpha helix in the VP16 activation domain upon binding to a human TAF. Science. 1997;277:1310–1313. doi: 10.1126/science.277.5330.1310. [DOI] [PubMed] [Google Scholar]
- 62.Ayed A, Mulder FA, Yi GS, Lu Y, Kay LE, Arrowsmith CH. Latent and active p53 are identical in conformation. Nat Struct Biol. 2001;8:756–760. doi: 10.1038/nsb0901-756. [DOI] [PubMed] [Google Scholar]
- 63.Grossmann JG, Sharff AJ, O’Hare P, Luisi B. Molecular shapes of transcription factors TFIIB and VP16 in solution: implications for recognition. Biochemistry. 2001;40:6267–6274. doi: 10.1021/bi0028946. [DOI] [PubMed] [Google Scholar]
- 64.Liu J, Perumal NB, Oldfield CJ, Su EW, Uversky VN, Dunker AK. Intrinsic disorder in transcription factors. Biochemistry. 2006;45:6873–6888. doi: 10.1021/bi0602718. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Struhl K. Fundamentally different logic of gene regulation in eukaryotes and prokaryotes. Cell. 1999;98:1–4. doi: 10.1016/S0092-8674(00)80599-1. [DOI] [PubMed] [Google Scholar]
- 66.Levine M, Tjian R. Transcription regulation and animal diversity. Nature. 2003;424:147–151. doi: 10.1038/nature01763. [DOI] [PubMed] [Google Scholar]
- 67.Michelson AM. Deciphering genetic regulatory codes: a challenge for functional genomics. Proc Natl Acad Sci USA. 2002;99:546–548. doi: 10.1073/pnas.032685999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Amoutzias G, Veron A, Weiner J, III, Robinson-Rechavi M, Bornberg-Bauer E, Oliver S, Robertson D. One Billion Years of bZIP Transcription Factor Evolution: Conservation and Change in Dimerization and DNA-Binding Site Specificity. Mol Biol Evol. 2007;24:827–835. doi: 10.1093/molbev/msl211. [DOI] [PubMed] [Google Scholar]
- 69.Hurst HC. Transcription factors 1: bZIP proteins. Protein Profile. 1995;2:101–168. [PubMed] [Google Scholar]
- 70.Chinenov Y, Kerppola TK. Close encounters of many kinds: Fos-Jun interactions that mediate transcription regulatory specificity. Oncogene. 2001;20:2438–2452. doi: 10.1038/sj.onc.1204385. [DOI] [PubMed] [Google Scholar]
- 71.Landschulz WH, Johnson PF, Mcknight SL. The leucine zipper: a hypothetical structure common to a new class of DNA binding proteins. Science. 1988;240:1759–1764. doi: 10.1126/science.3289117. [DOI] [PubMed] [Google Scholar]
- 72.Landschulz WH, Johnson PF, Mcknight SL. The DNA binding domain of the rat liver nuclear protein C/EBP is bipartite. Science. 1989;243:1681–1688. doi: 10.1126/science.2494700. [DOI] [PubMed] [Google Scholar]
- 73.Ellenberger TE, Brandl CJ, Struhl K, Harrison SC. The GCN4 basic region leucine zipper binds DNA as a dimer of uninterrupted alpha helices: crystal structure of the protein-DNA complex. Cell. 1992;71:1223–1237. doi: 10.1016/s0092-8674(05)80070-4. [DOI] [PubMed] [Google Scholar]
- 74.Glover JN, Harrison SC. Crystal structure of the heterodimeric bZIP transcription factor c-Fos- c-Jun bound to DNA. Nature. 1995;373:257–261. doi: 10.1038/373257a0. [DOI] [PubMed] [Google Scholar]
- 75.Fujii Y, Shimizu T, Toda T, Yanagida M, Hakoshima T. Structural basis for the diversity of DNA recognition by bZIP transcription factors. Nat Struct Biol. 2000;7:889–893. doi: 10.1038/82822. [DOI] [PubMed] [Google Scholar]
- 76.Schumacher MA, Goodman RH, Brennan RG. The structure of a CREB bZIP. somatostatin CRE complex reveals the basis for selective dimerization and divalent cation-enhanced DNA binding. J Biol Chem. 2000;275:35242–35247. doi: 10.1074/jbc.M007293200. [DOI] [PubMed] [Google Scholar]
- 77.Miller M, Shuman JD, Sebastian T, Dauter Z, Johnson PF. Structural basis for DNA recognition by the basic region leucine zipper transcription factor CCAAT/enhancer-binding protein alpha. J Biol Chem. 2003;278:15178–15184. doi: 10.1074/jbc.M300417200. [DOI] [PubMed] [Google Scholar]
- 78.Wolberger C. Transcription factor structure and DNA binding. Curr Opin Struct Biol. 1993;3:3–10. [Google Scholar]
- 79.Cho Y, Gorina S, Jeffrey PD, Pavletich NP. Crystal structure of a p53 tumor suppressor-DNA complex: understanding tumorigenic mutations. Science. 1994;265:346–355. doi: 10.1126/science.8023157. [DOI] [PubMed] [Google Scholar]
- 80.Weiss MA, Ellenberger T, Wobbe CR, Lee JP, Harrison SC, Struhl K. Folding transition in the DNA-binding domain of GCN4 on specific binding to DNA. Nature. 1990;347:575–578. doi: 10.1038/347575a0. [DOI] [PubMed] [Google Scholar]
- 81.Krylov D, Olive M, Vinson C. Extending dimerization interfaces: the bZIP basic region can form a coiled coil. EMBO J. 1995;14:5329–5337. doi: 10.1002/j.1460-2075.1995.tb00217.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Patel LR, Curran T, Kerppola TK. Energy transfer analysis of Fos-Jun dimerization and DNA binding. Proc Natl Acad Sci USA. 1994;91:7360–7364. doi: 10.1073/pnas.91.15.7360. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Garvie CW, Wolberger C. Recognition of specific DNA sequences. Mol Cell. 2001;8:937–946. doi: 10.1016/s1097-2765(01)00392-6. [DOI] [PubMed] [Google Scholar]
- 84.Vinson CR, Hai T, Boyd SM. Dimerization specificity of the leucine zipper-containing bZIP motif on DNA binding: prediction and rational design. Genes Dev. 1993;7:1047–1058. doi: 10.1101/gad.7.6.1047. [DOI] [PubMed] [Google Scholar]
- 85.Vinson C, Myakishev M, Acharya A, Mir AA, Moll JR, Bonovich M. Classification of human B-ZIP proteins based on dimerization properties. Mol Cell Biol. 2002;22:6321–6335. doi: 10.1128/MCB.22.18.6321-6335.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Fassler J, Landsman D, Acharya A, Moll JR, Bonovich M, Vinson C. B-ZIP proteins encoded by the Drosophila genome: evaluation of potential dimerization partners. Genome Res. 2002;12:1190–1200. doi: 10.1101/gr.67902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Ryu T, Jung J, Lee S, Nam HJ, Hong SW, Yoo JW, Lee DK, Lee D. bZIPDB: a database of regulatory information for human bZIP transcription factors. BMC Genomics. 2007;8:136. doi: 10.1186/1471-2164-8-136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Blank V, Andrews NC. The Maf transcription factors: regulators of differentiation. Trends Biochem Sci. 1997;22:437–441. doi: 10.1016/s0968-0004(97)01105-5. [DOI] [PubMed] [Google Scholar]
- 89.Motohashi H, O’Connor T, Katsuoka F, Engel JD, Yamamoto M. Integration and diversity of the regulatory network composed of Maf and CNC families of transcription factors. Gene. 2002;294:1–12. doi: 10.1016/s0378-1119(02)00788-6. [DOI] [PubMed] [Google Scholar]
- 90.Richards JP, Bachinger HP, Goodman RH, Brennan RG. Analysis of the structural properties of cAMP-responsive element-binding protein (CREB) and phosphorylated CREB. J Biol Chem. 1996;271:13716–13723. doi: 10.1074/jbc.271.23.13716. [DOI] [PubMed] [Google Scholar]
- 91.Nagadoi A, Nakazawa K, Uda H, Okuno K, Maekawa T, Ishii S, Nishimura Y. Solution structure of the transactivation domain of ATF-2 comprising a zinc finger-like subdomain and a flexible subdomain. J Mol Biol. 1999;287:593–607. doi: 10.1006/jmbi.1999.2620. [DOI] [PubMed] [Google Scholar]
- 92.Kusunoki H, Motohashi H, Katsuoka F, Morohashi A, Yamamoto M, Tanaka T. Solution structure of the DNA-binding domain of MafG. Nat Struct Biol. 2002;9:252–256. doi: 10.1038/nsb771. [DOI] [PubMed] [Google Scholar]
- 93.Yoshida C, Tokumasu F, Hohmura KI, Bungert J, Hayashi N, Nagasawa T, Engel JD, Yamamoto M, Takeyasu K, Igarashi K. Long range interaction of cis-DNA elements mediated by architectural transcription factor Bach1. Genes Cells. 1999;4:643–655. doi: 10.1046/j.1365-2443.1999.00291.x. [DOI] [PubMed] [Google Scholar]
- 94.Williamson EA, Xu HN, Gombart AF, Verbeek W, Chumakov AM, Friedman AD, Koeffler HP. Identification of transcriptional activation and repression domains in human CCAAT/enhancer-binding protein epsilon. J Biol Chem. 1998;273:14796–14804. doi: 10.1074/jbc.273.24.14796. [DOI] [PubMed] [Google Scholar]
- 95.Smith JL, Freebern WJ, Collins I, De Siervi A, Montano I, Haggerty CM, McNutt MC, Butscher WG, Dzekunova I, Petersen DW, Kawasaki E, Merchant JL, Gardner K. Kinetic profiles of p300 occupancy in vivo predict common features of promoter structure and coactivator recruitment. Proc Natl Acad Sci USA. 2004;101:11554–11559. doi: 10.1073/pnas.0402156101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Wagner EF. AP-1--Introductory remarks. Oncogene. 2001;20:2334–2335. doi: 10.1038/sj.onc.1204416. [DOI] [PubMed] [Google Scholar]
- 97.Yoon MK, Shin J, Choi G, Choi BS. Intrinsically unstructured N-terminal domain of bZIP transcription factor HY5. Proteins. 2006;65:856–866. doi: 10.1002/prot.21089. [DOI] [PubMed] [Google Scholar]
- 98.Sharma N, Lopez DI, Nyborg JK. DNA binding and phosphorylation induce conformational alterations in the kinase-inducible domain of CREB. Implications for the mechanism of transcription function. J Biol Chem. 2007;282:19872–19883. doi: 10.1074/jbc.M701435200. [DOI] [PubMed] [Google Scholar]
- 99.Romero P, Obradovic Z, Li X, Garner EC, Brown CJ, Dunker AK. Sequence complexity of disordered protein. Proteins. 2001;42:38–48. doi: 10.1002/1097-0134(20010101)42:1<38::aid-prot50>3.0.co;2-3. [DOI] [PubMed] [Google Scholar]
- 100.Miller M. Phospho-Dependent Protein Recognition Motifs Contained in C/EBP Family of Transcription Factors: in Silico Studies. Cell Cycle. 2006;5:2501–2508. doi: 10.4161/cc.5.21.3421. [DOI] [PubMed] [Google Scholar]
- 101.Vucetic S, Obradovic Z, Vacic V, Radivojac P, Peng K, Iakoucheva LM, Cortese MS, Lawson JD, Brown CJ, Sikes JG, Newton CD, Dunker AK. DisProt: a database of protein disorder. Bioinformatics. 2005;21:137–140. doi: 10.1093/bioinformatics/bth476. [DOI] [PubMed] [Google Scholar]
- 102.Letunic I, Goodstadt L, Dickens NJ, Doerks T, Schultz J, Mott R, Ciccarelli F, Copley RR, Ponting CP, Bork P. Recent improvements to the SMART domain-based sequence annotation resource. Nucleic Acids Res. 2002;30:242–244. doi: 10.1093/nar/30.1.242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Podust LM, Krezel AM, Kim Y. Crystal structure of the CCAAT box/enhancer-binding protein beta activating transcription factor-4 basic leucine zipper heterodimer in the absence of DNA. J Biol Chem. 2001;276:505–513. doi: 10.1074/jbc.M005594200. [DOI] [PubMed] [Google Scholar]
- 104.Thompson KS, Vinson CR, Freire E. Thermodynamic characterization of the structural stability of the coiled-coil region of the bZIP transcription factor GCN4. Biochemistry. 1993;32:5491–5496. doi: 10.1021/bi00072a001. [DOI] [PubMed] [Google Scholar]
- 105.Krylov D, Mikhailenko I, Vinson C. A thermodynamic scale for leucine zipper stability and dimerization specificity: e and g inter-helical interactions. EMBO J. 1994;13:2849–2861. doi: 10.1002/j.1460-2075.1994.tb06579.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Sosnick TR, Jackson S, Wilk RR, Englander SW, DeGrado WF. The role of helix formation in the folding of a fully alpha-helical coiled coil. Proteins. 1996;24:427–432. doi: 10.1002/(SICI)1097-0134(199604)24:4<427::AID-PROT2>3.0.CO;2-B. [DOI] [PubMed] [Google Scholar]
- 107.d’Avignon DA, Bretthorst GL, Holtzer ME, Holtzer A. Site-specific thermodynamics and kinetics of a coiled-coil transition by spin inversion transfer NMR. Biophys J. 1998;74:3190–3197. doi: 10.1016/S0006-3495(98)78025-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Dragan AI, Privalov PL. Unfolding of a leucine zipper is not a simple two-state transition. J Mol Biol. 2002;321:891–908. doi: 10.1016/s0022-2836(02)00699-x. [DOI] [PubMed] [Google Scholar]
- 109.Zitzewitz JA, Ibarra-Molero B, Fishel DR, Terry KL, Matthews CR. Preformed secondary structure drives the association reaction of GCN4-p1, a model coiled-coil system. J Mol Biol. 2000;296:1105–1116. doi: 10.1006/jmbi.2000.3507. [DOI] [PubMed] [Google Scholar]
- 110.Fuxreiter M, Simon I, Friedrich P, Tompa P. Preformed structural elements feature in partner recognition by intrinsically unstructured proteins. J Mol Biol. 2004;338:1015–1026. doi: 10.1016/j.jmb.2004.03.017. [DOI] [PubMed] [Google Scholar]
- 111.Lamb P, Mcknight SL. Diversity and specificity in transcriptional regulation: the benefits of heterotypic dimerization. Trends Biochem Sci. 1991;16:417–422. doi: 10.1016/0968-0004(91)90167-t. [DOI] [PubMed] [Google Scholar]
- 112.Vinson C, Acharya A, Taparowsky EJ. Deciphering B-ZIP transcription factor interactions in vitro and in vivo. Biochim Biophys Acta. 2006;1759:4–12. doi: 10.1016/j.bbaexp.2005.12.005. [DOI] [PubMed] [Google Scholar]
- 113.Alber T. Structure of the leucine zipper. Curr Opin Genet Dev. 1992;2:205–210. doi: 10.1016/s0959-437x(05)80275-8. [DOI] [PubMed] [Google Scholar]
- 114.Grigoryan G, Keating AE. Structure-based prediction of bZIP partnering specificity. J Mol Biol. 2006;355:1125–1142. doi: 10.1016/j.jmb.2005.11.036. [DOI] [PubMed] [Google Scholar]
- 115.Fong JH, Keating AE, Singh M. Predicting specificity in bZIP coiled-coil protein interactions. Genome Biol. 2004;5:R11. doi: 10.1186/gb-2004-5-2-r11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Harbury PB, Zhang T, Kim PS, Alber T. A switch between two-, three-, and four-stranded coiled coils in GCN4 leucine zipper mutants. Science. 1993;262:1401–1407. doi: 10.1126/science.8248779. [DOI] [PubMed] [Google Scholar]
- 117.Newman JR, Keating AE. Comprehensive identification of human bZIP interactions with coiled-coil arrays. Science. 2003;300:2097–2101. doi: 10.1126/science.1084648. [DOI] [PubMed] [Google Scholar]
- 118.Deppmann CD, Acharya A, Rishi V, Wobbes B, Smeekens S, Taparowsky EJ, Vinson C. Dimerization specificity of all 67 B-ZIP motifs in Arabidopsis thaliana: a comparison to Homo sapiens B-ZIP motifs. Nucleic Acids Res. 2004;32:3435–3445. doi: 10.1093/nar/gkh653. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.Parkin SE, Baer M, Copeland TD, Schwartz RC, Johnson PF. Regulation of CCAAT/enhancer-binding protein (C/EBP) activator proteins by heterodimerization with C/EBPgamma (Ig/EBP) J Biol Chem. 2002;277:23563–23572. doi: 10.1074/jbc.M202184200. [DOI] [PubMed] [Google Scholar]
- 120.O’Neil KT, Shuman JD, Ampe C, DeGrado WF. DNA-induced increase in the alpha-helical content of C/EBP and GCN4. Biochemistry. 1991;30:9030–9034. doi: 10.1021/bi00101a017. [DOI] [PubMed] [Google Scholar]
- 121.Bracken C, Carr PA, Cavanagh J, Palmer AG., III Temperature dependence of intramolecular dynamics of the basic leucine zipper of GCN4: implications for the entropy of association with DNA. J Mol Biol. 1999;285:2133–2146. doi: 10.1006/jmbi.1998.2429. [DOI] [PubMed] [Google Scholar]
- 122.Hollenbeck JJ, McClain DL, Oakley MG. The role of helix stabilizing residues in GCN4 basic region folding and DNA binding. Protein Sci. 2002;11:2740–2747. doi: 10.1110/ps.0211102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123.Kabe Y, Goto M, Shima D, Imai T, Wada T, Morohashi K, Shirakawa M, Hirose S, Handa H. The role of human MBF1 as a transcriptional coactivator. J Biol Chem. 1999;274:34196–34202. doi: 10.1074/jbc.274.48.34196. [DOI] [PubMed] [Google Scholar]
- 124.Lively TN, Nguyen TN, Galasinski SK, Goodrich JA. The basic leucine zipper domain of c-Jun functions in transcriptional activation through interaction with the N terminus of human TATA-binding protein-associated factor-1 (human TAF(II)250) J Biol Chem. 2004;279:26257–26265. doi: 10.1074/jbc.M400892200. [DOI] [PubMed] [Google Scholar]
- 125.Conkright MD, Canettieri G, Screaton R, Guzman E, Miraglia L, Hogenesch JB, Montminy M. TORCs: transducers of regulated CREB activity. Mol Cell. 2003;12:413–423. doi: 10.1016/j.molcel.2003.08.013. [DOI] [PubMed] [Google Scholar]
- 126.Baranger AM. Accessory factor-bZIP-DNA interactions. Curr Opin Chem Biol. 1998;2:18–23. doi: 10.1016/s1367-5931(98)80031-8. [DOI] [PubMed] [Google Scholar]
- 127.Miotto B, Struhl K. Differential gene regulation by selective association of transcriptional coactivators and bZIP DNA-binding domains. Mol Cell Biol. 2006;26:5969–5982. doi: 10.1128/MCB.00696-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 128.Williams SC, Angerer ND, Johnson PF. C/EBP proteins contain nuclear localization signals imbedded in their basic regions. Gene Expr. 1997;6:371–385. [PMC free article] [PubMed] [Google Scholar]
- 129.Weis K. Importins and exportins: how to get in and out of the nucleus. Trends Biochem Sci. 1998;23:185–189. doi: 10.1016/s0968-0004(98)01204-3. [DOI] [PubMed] [Google Scholar]
- 130.Conti E, Izaurralde E. Nucleocytoplasmic transport enters the atomic age. Curr Opin Cell Biol. 2001;13:310–319. doi: 10.1016/s0955-0674(00)00213-1. [DOI] [PubMed] [Google Scholar]
- 131.Fontes MR, Teh T, Kobe B. Structural basis of recognition of monopartite and bipartite nuclear localization sequences by mammalian importin-alpha. J Mol Biol. 2000;297:1183–1194. doi: 10.1006/jmbi.2000.3642. [DOI] [PubMed] [Google Scholar]
- 132.Harreman MT, Kline TM, Milford HG, Harben MB, Hodel AE, Corbett AH. Regulation of nuclear import by phosphorylation adjacent to nuclear localization signals. J Biol Chem. 2004;279:20613–20621. doi: 10.1074/jbc.M401720200. [DOI] [PubMed] [Google Scholar]
- 133.Buck M, Zhang L, Halasz NA, Hunter T, Chojkier M. Nuclear export of phosphorylated C/EBPbeta mediates the inhibition of albumin expression by TNF-alpha. EMBO J. 2001;20:6712–6723. doi: 10.1093/emboj/20.23.6712. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 134.Bannister AJ, Cook A, Kouzarides T. In vitro DNA binding activity of Fos/Jun and BZLF1 but not C/EBP is affected by redox changes. Oncogene. 1991;6:1243–1250. [PubMed] [Google Scholar]
- 135.Jindra M, Gaziova I, Uhlirova M, Okabe M, Hiromi Y, Hirose S. Coactivator MBF1 preserves the redox-dependent AP-1 activity during oxidative stress in Drosophila. EMBO J. 2004;23:3538–3547. doi: 10.1038/sj.emboj.7600356. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 136.Metallo SJ, Schepartz A. Certain bZIP peptides bind DNA sequentially as monomers and dimerize on the DNA. Nat Struct Biol. 1997;4:115–117. doi: 10.1038/nsb0297-115. [DOI] [PubMed] [Google Scholar]
- 137.Wu X, Spiro C, Owen WG, McMurray CT. cAMP response element-binding protein monomers cooperatively assemble to form dimers on DNA. J Biol Chem. 1998;273:20820–20827. doi: 10.1074/jbc.273.33.20820. [DOI] [PubMed] [Google Scholar]
- 138.Pomerantz JL, Wolfe SA, Pabo CO. Structure-based design of a dimeric zinc finger protein. Biochemistry. 1998;37:965–970. doi: 10.1021/bi972464o. [DOI] [PubMed] [Google Scholar]
- 139.Kohler JJ, Schepartz A. Kinetic studies of Fos. Jun. DNA complex formation: DNA binding prior to dimerization. Biochemistry. 2001;40:130–142. doi: 10.1021/bi001881p. [DOI] [PubMed] [Google Scholar]
- 140.Kohler JJ, Metallo SJ, Schneider TL, Schepartz A. DNA specificity enhanced by sequential binding of protein monomers. Proc Natl Acad Sci USA. 1999;96:11735–11739. doi: 10.1073/pnas.96.21.11735. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 141.Berger C, Piubelli L, Haditsch U, Bosshard HR. Diffusion-controlled DNA recognition by an unfolded, monomeric bZIP transcription factor. FEBS Lett. 1998;425:14–18. doi: 10.1016/s0014-5793(98)00156-2. [DOI] [PubMed] [Google Scholar]
- 142.Kim B, Little JW. Dimerization of a specific DNA-binding protein on the DNA. Science. 1992;255:203–206. doi: 10.1126/science.1553548. [DOI] [PubMed] [Google Scholar]
- 143.Williams SC, Baer M, Dillner AJ, Johnson PF. CRP2 (C/EBP beta) contains a bipartite regulatory domain that controls transcriptional activation, DNA binding and cell specificity. EMBO J. 1995;14:3170–3183. doi: 10.1002/j.1460-2075.1995.tb07319.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 144.Hattori T, Ohoka N, Inoue Y, Hayashi H, Onozaki K. C/EBP family transcription factors are degraded by the proteasome but stabilized by forming dimer. Oncogene. 2003;22:1273–1280. doi: 10.1038/sj.onc.1206204. [DOI] [PubMed] [Google Scholar]
- 145.Lee SJ, Sekimoto T, Yamashita E, Nagoshi E, Nakagawa A, Imamoto N, Yoshimura M, Sakai H, Chong KT, Tsukihara T, Yoneda Y. The structure of importin-beta bound to SREBP-2: nuclear import of a transcription factor. Science. 2003;302:1571–1575. doi: 10.1126/science.1088372. [DOI] [PubMed] [Google Scholar]
- 146.Johnson PF, Sterneck E, Williams SC. Activation domains of transcriptional regulatory proteins. J Nutr Biochem. 1993;4:386–398. [Google Scholar]
- 147.Triezenberg SJ. Structure and function of transcriptional activation domains. Curr Opin Genet Dev. 1995;5:190–196. doi: 10.1016/0959-437x(95)80007-7. [DOI] [PubMed] [Google Scholar]
- 148.Gill G, Ptashne M. Negative effect of the transcriptional activator GAL4. Nature. 1988;334:721–724. doi: 10.1038/334721a0. [DOI] [PubMed] [Google Scholar]
- 149.O’Hare P, Williams G. Structural studies of the acidic transactivation domain of the Vmw65 protein of herpes simplex virus using 1H NMR. Biochemistry. 1992;31:4150–4156. doi: 10.1021/bi00131a035. [DOI] [PubMed] [Google Scholar]
- 150.Campbell KM, Terrell AR, Laybourn PJ, Lumb KJ. Intrinsic structural disorder of the C-terminal activation domain from the bZIP transcription factor Fos. Biochemistry. 2000;39:2708–2713. doi: 10.1021/bi9923555. [DOI] [PubMed] [Google Scholar]
- 151.Fladvad M, Zhou K, Moshref A, Pursglove S, Safsten P, Sunnerhagen M. N and C-terminal sub-regions in the c-Myc trans-activation region and their joint role in creating versatility in folding and binding. J Mol Biol. 2005;346:175–189. doi: 10.1016/j.jmb.2004.11.029. [DOI] [PubMed] [Google Scholar]
- 152.Metz R, Bannister AJ, Sutherland JA, Hagemeier C, O’Rourke EC, Cook A, Bravo R, Kouzarides T. c-Fos-induced activation of a TATA-box-containing promoter involves direct contact with TATA-box-binding protein. Mol Cell Biol. 1994;14:6021–6029. doi: 10.1128/mcb.14.9.6021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 153.Hermann S, Berndt KD, Wright AP. How transcriptional activators bind target proteins. J Biol Chem. 2001;276:40127–40132. doi: 10.1074/jbc.M103793200. [DOI] [PubMed] [Google Scholar]
- 154.Schmitz ML, dos Santos Silva MA, Altmann H, Czisch M, Holak TA, Baeuerle PA. Structural and functional analysis of the NF-kappa B p65 C terminus. An acidic and modular transactivation domain with the potential to adopt an alpha-helical conformation. J Biol Chem. 1994;269:25613–25620. [PubMed] [Google Scholar]
- 155.Nerlov C, Ziff EB. CCAAT/enhancer binding protein-alpha amino acid motifs with dual TBP and TFIIB binding ability cooperate to activate transcription in both yeast and mammalian cells. EMBO J. 1995;14:4318–4328. doi: 10.1002/j.1460-2075.1995.tb00106.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 156.Kovacs KA, Steinmann M, Magistretti PJ, Halfon O, Cardinaux JR. CCAAT/enhancer-binding protein family members recruit the coactivator CREB-binding protein and trigger its phosphorylation. J Biol Chem. 2003;278:36959–36965. doi: 10.1074/jbc.M303147200. [DOI] [PubMed] [Google Scholar]
- 157.Kussie PH, Gorina S, Marechal V, Elenbaas B, Moreau J, Levine AJ, Pavletich NP. Structure of the MDM2 oncoprotein bound to the p53 tumor suppressor transactivation domain. Science. 1996;274:948–953. doi: 10.1126/science.274.5289.948. [DOI] [PubMed] [Google Scholar]
- 158.Choi Y, Asada S, Uesugi M. Divergent hTAFII31-binding motifs hidden in activation domains. J Biol Chem. 2000;275:15912–15916. doi: 10.1074/jbc.275.21.15912. [DOI] [PubMed] [Google Scholar]
- 159.Anderson CW, Appella E. The p53 Tumor Suppressor Pathway and Cancer. In: Zanbetti GP, editor. Posttranslational modificaitons of p53: Upstream signaling. Springer Science+Business Media, Inc; New York: 2005. pp. 95–114. [Google Scholar]
- 160.Bannister AJ, Brown HJ, Sutherland JA, Kouzarides T. Phosphorylation of the c-Fos and c-Jun HOB1 motif stimulates its activation capacity. Nucleic Acids Res. 1994;22:5173–5176. doi: 10.1093/nar/22.24.5173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 161.Du K, Montminy M. CREB is a regulatory target for the protein kinase Akt/PKB. J Biol Chem. 1998;273:32377–32379. doi: 10.1074/jbc.273.49.32377. [DOI] [PubMed] [Google Scholar]
- 162.Shanware NP, Trinh AT, Williams LM, Tibbetts RS. Coregulated ataxia telangiectasia-mutated and casein kinase sites modulate cAMP-response element-binding protein-coactivator interactions in response to DNA damage. J Biol Chem. 2007;282:6283–6291. doi: 10.1074/jbc.M610674200. [DOI] [PubMed] [Google Scholar]
- 163.Shuman JD, Sebastian T, Kaldis P, Copeland TD, Zhu S, Smart RC, Johnson PF. Cell cycle-dependent phosphorylation of C/EBPβ mediates oncogenic cooperativity between C/EBPβ and H-RasV12. Mol Cell Biol. 2004;24:7380–7391. doi: 10.1128/MCB.24.17.7380-7391.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 164.Monje P, Hernandez-Losa J, Lyons RJ, Castellone MD, Gutkind JS. Regulation of the transcriptional activity of c-Fos by ERK. A novel role for the prolyl isomerase PIN1. J Biol Chem. 2005;280:35081–35084. doi: 10.1074/jbc.C500353200. [DOI] [PubMed] [Google Scholar]
- 165.Manke IA, Lowery DM, Nguyen A, Yaffe MB. BRCT repeats as phosphopeptide-binding modules involved in protein targeting. Science. 2003;302:636–639. doi: 10.1126/science.1088877. [DOI] [PubMed] [Google Scholar]
- 166.Gardner KH, Montminy M. Can you hear me now? Regulating transcriptional activators by phosphorylation. Sci STKE. 2005;2005:e44. doi: 10.1126/stke.3012005pe44. [DOI] [PubMed] [Google Scholar]
- 167.Kwong PD, Wyatt R, Majeed S, Robinson J, Sweet RW, Sodroski J, Hendrickson WA. Structures of HIV-1 gp120 envelope glycoproteins from laboratory-adapted and primary isolates. Structure Fold Des. 2000;8:1329–1339. doi: 10.1016/s0969-2126(00)00547-5. [DOI] [PubMed] [Google Scholar]
- 168.Freedman SJ, Sun ZY, Poy F, Kung AL, Livingston DM, Wagner G, Eck MJ. Structural basis for recruitment of CBP/p300 by hypoxia-inducible factor-1 alpha. Proc Natl Acad Sci USA. 2002;99:5367–5372. doi: 10.1073/pnas.082117899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 169.Campbell KM, Lumb KJ. Structurally distinct modes of recognition of the KIX domain of CBP by Jun and CREB. Biochemistry. 2002;41:13956–13964. doi: 10.1021/bi026222m. [DOI] [PubMed] [Google Scholar]
- 170.De Guzman RN, Goto NK, Dyson HJ, Wright PE. Structural basis for cooperative transcription factor binding to the CBP coactivator. J Mol Biol. 2006;355:1005–1013. doi: 10.1016/j.jmb.2005.09.059. [DOI] [PubMed] [Google Scholar]
- 171.Radhakrishnan I, Perez-Alvarado GC, Parker D, Dyson HJ, Montminy MR, Wright PE. Solution structure of the KIX domain of CBP bound to the transactivation domain of CREB: a model for activator:coactivator interactions. Cell. 1997;91:741–752. doi: 10.1016/s0092-8674(00)80463-8. [DOI] [PubMed] [Google Scholar]
- 172.Zor T, De Guzman RN, Dyson HJ, Wright PE. Solution structure of the KIX domain of CBP bound to the transactivation domain of c-Myb. J Mol Biol. 2004;337:521–534. doi: 10.1016/j.jmb.2004.01.038. [DOI] [PubMed] [Google Scholar]
- 173.Roach PJ. Multisite and hierarchal protein phosphorylation. J Biol Chem. 1991;266:14139–14142. [PubMed] [Google Scholar]
- 174.Ilouz R, Kowalsman N, Eisenstein M, Eldar-Finkelman H. Identification of novel glycogen synthase kinase-3beta substrate-interacting residues suggests a common mechanism for substrate recognition. J Biol Chem. 2006;281:30621–30630. doi: 10.1074/jbc.M604633200. [DOI] [PubMed] [Google Scholar]
- 175.Kim J, Sharma S, Li Y, Cobos E, Palvimo JJ, Williams SC. Repression and coactivation of CCAAT/enhancer-binding protein epsilon by sumoylation and protein inhibitor of activated STATx proteins. J Biol Chem. 2005;280:12246–12254. doi: 10.1074/jbc.M413771200. [DOI] [PubMed] [Google Scholar]
- 176.Lise S, Jones DT. Sequence patterns associated with disordered regions in proteins. Proteins. 2005;58:144–150. doi: 10.1002/prot.20279. [DOI] [PubMed] [Google Scholar]
- 177.Fontana A, Polverino dL, De FV, Scaramella E, Zambonin M. Probing the partly folded states of proteins by limited proteolysis. Fold Des. 1997;2:R17–R26. doi: 10.1016/S1359-0278(97)00010-2. [DOI] [PubMed] [Google Scholar]
- 178.Singh GP, Ganapathi M, Sandhu KS, Dash D. Intrinsic un-structuredness and abundance of PEST motifs in eukaryotic proteomes. Proteins. 2006;62:309–315. doi: 10.1002/prot.20746. [DOI] [PubMed] [Google Scholar]
- 179.Hill CS, Treisman R. Transcriptional regulation by extracellular signals: mechanisms and specificity. Cell. 1995;80:199–211. doi: 10.1016/0092-8674(95)90403-4. [DOI] [PubMed] [Google Scholar]
- 180.Ciechanover A, DiGiuseppe JA, Bercovich B, Orian A, Richter JD, Schwartz AL, Brodeur GM. Degradation of nuclear oncoproteins by the ubiquitin system in vitro. Proc Natl Acad Sci USA. 1991;88:139–143. doi: 10.1073/pnas.88.1.139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 181.Ramji DP, Foka P. CCAAT/enhancer-binding proteins: structure, function and regulation. Biochem J. 2002;365:561–575. doi: 10.1042/BJ20020508. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 182.Sharrocks AD, Yang SH, Galanis A. Docking domains and substrate-specificity determination for MAP kinases. Trends Biochem Sci. 2000;25:448–453. doi: 10.1016/s0968-0004(00)01627-3. [DOI] [PubMed] [Google Scholar]
- 183.Narlikar GJ, Fan HY, Kingston RE. Cooperation between complexes that regulate chromatin structure and transcription. Cell. 2002;108:475–487. doi: 10.1016/s0092-8674(02)00654-2. [DOI] [PubMed] [Google Scholar]
- 184.Orphanides G, Reinberg D. A unified theory of gene expression. Cell. 2002;108:439–451. doi: 10.1016/s0092-8674(02)00655-4. [DOI] [PubMed] [Google Scholar]
- 185.Merika M, Thanos D. Enhanceosomes. Curr Opin Genet Dev. 2001;11:205–208. doi: 10.1016/s0959-437x(00)00180-5. [DOI] [PubMed] [Google Scholar]
- 186.Chen L, Glover JN, Hogan PG, Rao A, Harrison SC. Structure of the DNA-binding domains from NFAT, Fos and Jun bound specifically to DNA. Nature. 1998;392:42–48. doi: 10.1038/32100. [DOI] [PubMed] [Google Scholar]
- 187.Panne D, Maniatis T, Harrison SC. Crystal structure of ATF-2/c-Jun and IRF-3 bound to the interferon-beta enhancer. EMBO J. 2004;23:4384–4393. doi: 10.1038/sj.emboj.7600453. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 188.Tahirov TH, Sato K, Ichikawa-Iwata E, Sasaki M, Inoue-Bungo T, Shiina M, Kimura K, Takata S, Fujikawa A, Morii H, Kumasaka T, Yamamoto M, Ishii S, Ogata K. Mechanism of c-Myb-C/EBPß cooperation from separated sites on a promoter. Cell. 2002;108:57–70. doi: 10.1016/s0092-8674(01)00636-5. [DOI] [PubMed] [Google Scholar]
- 189.Cha-Molstad H, Young DP, Kushner I, Samols D. The interaction of C-Rel with C/EBPbeta enhances C/EBPbeta binding to the C-reactive protein gene promoter. Mol Immunol. 2007;44:2933–2942. doi: 10.1016/j.molimm.2007.01.015. [DOI] [PubMed] [Google Scholar]
- 190.Wolberger C. Multiprotein-DNA complexes in transcriptional regulation. Annu Rev Biophys Biomol Struct. 1999;28:29–56. doi: 10.1146/annurev.biophys.28.1.29. [DOI] [PubMed] [Google Scholar]
- 191.Ogata K, Sato K, Tahirov TH. Eukaryotic transcriptional regulatory complexes: cooperativity from near and afar. Curr Opin Struct Biol. 2003;13:40–48. doi: 10.1016/s0959-440x(03)00012-5. [DOI] [PubMed] [Google Scholar]
- 192.Reeves R, Beckerbauer L. HMGI/Y proteins: flexible regulators of transcription and chromatin structure. Biochim Biophys Acta. 2001;1519:13–29. doi: 10.1016/s0167-4781(01)00215-9. [DOI] [PubMed] [Google Scholar]
- 193.Sgarra R, ;Tessari MA, Di Bernardo J, Rustighi A, Zago P, Liberatori S, Armini A, Bini L, Giancotti V, Manfioletti G. Discovering high mobility group A molecular partners in tumour cells. Proteomics. 2005;5:1494–1506. doi: 10.1002/pmic.200401028. [DOI] [PubMed] [Google Scholar]
- 194.Naar AM, Lemon BD, Tjian R. Transcriptional coactivator complexes. Annu Rev Biochem. 2001;70:475–501. doi: 10.1146/annurev.biochem.70.1.475. [DOI] [PubMed] [Google Scholar]
- 195.Di Poi N, Desvergne B, Michalik L, Wahli W. Transcriptional repression of peroxisome proliferator-activated receptor beta/delta in murine keratinocytes by CCAAT/enhancer-binding proteins. J Biol Chem. 2005;280:38700–38710. doi: 10.1074/jbc.M507782200. [DOI] [PubMed] [Google Scholar]
- 196.Conaway RC, Sato S, Tomomori-Sato C, Yao T, Conaway JW. The mammalian Mediator complex and its role in transcriptional regulation. Trends Biochem Sci. 2005;30:250–255. doi: 10.1016/j.tibs.2005.03.002. [DOI] [PubMed] [Google Scholar]
- 197.Starita LM, Parvin JD. The multiple nuclear functions of BRCA1: transcription, ubiquitination and DNA repair. Curr Opin Cell Biol. 2003;15:345–350. doi: 10.1016/s0955-0674(03)00042-5. [DOI] [PubMed] [Google Scholar]
- 198.Edmunds JW, Mahadevan LC. Cell signaling. Protein kinases seek close encounters with active genes. Science. 2006;313:449–451. doi: 10.1126/science.1131158. [DOI] [PubMed] [Google Scholar]
- 199.Pokholok DK, Zeitlinger J, Hannett NM, Reynolds DB, Young RA. Activated signal transduction kinases frequently occupy target genes. Science. 2006;313:533–536. doi: 10.1126/science.1127677. [DOI] [PubMed] [Google Scholar]
- 200.Zhang X, Odom DT, Koo SH, Conkright MD, Canettieri G, Best J, Chen H, Jenner R, Herbolsheimer E, Jacobsen E, Kadam S, Ecker JR, Emerson B, Hogenesch JB, Unterman T, Young RA, Montminy M. Genome-wide analysis of cAMP-response element binding protein occupancy, phosphorylation, and target gene activation in human tissues. Proc Natl Acad Sci USA. 2005;102:4459–4464. doi: 10.1073/pnas.0501076102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 201.Vries RG, Prudenziati M, Zwartjes C, Verlaan M, Kalkhoven E, Zantema A. A specific lysine in c-Jun is required for transcriptional repression by E1A and is acetylated by p300. EMBO J. 2001;20:6095–6103. doi: 10.1093/emboj/20.21.6095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 202.Cesena TI, Cardinaux JR, Kwok R, Schwartz J. CCAAT/enhancer-binding protein (C/EBP) beta is acetylated at multiple lysines: acetylation of C/EBPbeta at lysine 39 modulates its ability to activate transcription. J Biol Chem. 2007;282:956–967. doi: 10.1074/jbc.M511451200. [DOI] [PubMed] [Google Scholar]
- 203.Schwartz C, Beck K, Mink S, Schmolke M, Budde B, Wenning D, Klempnauer KH. Recruitment of p300 by C/EBPbeta triggers phosphorylation of p300 and modulates coactivator activity. EMBO J. 2003;22:882–892. doi: 10.1093/emboj/cdg076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 204.Boulton SJ. BRCA1-Mediated Ubiquitylation. Cell Cycle. 2006;5:1481–1486. doi: 10.4161/cc.5.14.2930. [DOI] [PubMed] [Google Scholar]
- 205.Xu W, Chen H, Du K, Asahara H, Tini M, Emerson BM, Montminy M, Evans RM. A transcriptional switch mediated by cofactor methylation. Science. 2001;294:2507–2511. doi: 10.1126/science.1065961. [DOI] [PubMed] [Google Scholar]
- 206.Mark WY, Liao JC, Lu Y, Ayed A, Laister R, Szymczyna B, Chakrabartty A, Arrowsmith CH. Characterization of segments from the central region of BRCA1: an intrinsically disordered scaffold for multiple protein-protein and protein-DNA interactions? J Mol Biol. 2005;345:275–287. doi: 10.1016/j.jmb.2004.10.045. [DOI] [PubMed] [Google Scholar]
- 207.Lo CL, Chothia C, Janin J. The atomic structure of protein-protein recognition sites. J Mol Biol. 1999;285:2177–2198. doi: 10.1006/jmbi.1998.2439. [DOI] [PubMed] [Google Scholar]
- 208.Bateman A, Birney E, Cerruti L, Durbin R, Etwiller L, Eddy SR, Griffiths-Jones S, Howe KL, Marshall M, Sonnhammer EL. The Pfam protein families database. Nucleic Acids Res. 2002;30:276–280. doi: 10.1093/nar/30.1.276. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 209.Hai T, Hartman MG. The molecular biology and nomenclature of the activating transcription factor/cAMP responsive element binding family of transcription factors: activating transcription factor proteins and homeostasis. Gene. 2001;273:1–11. doi: 10.1016/s0378-1119(01)00551-0. [DOI] [PubMed] [Google Scholar]
- 210.Bailey D, O’Hare P. Transmembrane bZIP Transcription Factors in ER Stress Signaling and the Unfolded Protein Response. Antioxid Redox Signal. 2007:9. doi: 10.1089/ars.2007.1796. [DOI] [PubMed] [Google Scholar]
- 211.Hunger SP, Li S, Fall MZ, Naumovski L, Cleary ML. The proto-oncogene HLF and the related basic leucine zipper protein TEF display highly similar DNA-binding and transcriptional regulatory properties. Blood. 1996;87:4607–4617. [PubMed] [Google Scholar]
- 212.Goodman RH, Smolik S. CBP/p300 in cell growth, transformation, and development. Genes Dev. 2000;14:1553–1577. [PubMed] [Google Scholar]
- 213.McManus KJ, Hendzel MJ. CBP, a transcriptional coactivator and acetyltransferase. Biochem Cell Biol. 2001;79:253–266. [PubMed] [Google Scholar]
- 214.Lamprecht C, Mueller CR. D-site binding protein transactivation requires the proline- and acid-rich domain and involves the coactivator p300. J Biol Chem. 1999;274:17643–17648. doi: 10.1074/jbc.274.25.17643. [DOI] [PubMed] [Google Scholar]
- 215.Nerlov C. The C/EBP family of transcription factors: a paradigm for interaction between gene expression and proliferation control. Trends Cell Biol. 2007;17:318–324. doi: 10.1016/j.tcb.2007.07.004. [DOI] [PubMed] [Google Scholar]
- 216.Yu X, Chini CC, He M, Mer G, Chen J. The BRCT domain is a phosphoprotein binding domain. Science. 2003;302:639–642. doi: 10.1126/science.1088753. [DOI] [PubMed] [Google Scholar]
- 217.Hu YF, Li R. JunB potentiates function of BRCA1 activation domain 1 (AD1) through a coiled-coil-mediated interaction. Genes Dev. 2002;16:1509–1517. doi: 10.1101/gad.995502. [DOI] [PMC free article] [PubMed] [Google Scholar]