Abstract
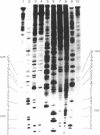
The DNase I cleavage-inhibition analysis shows binding sites of approximately 2 or 3 base pairs--in particular, 5' N-G-C sequences--for the green-colored CoIII and fully oxidized FeIII complexes of bleomycin. The apparent binding constant of the bleomycin-CoIII complex is quite similar for glucosylated and nonglucosylated phage T4 DNAs, whereas poly[d(I-C)] clearly gives a smaller binding constant than does poly[d(G-C)]. In contrast to the covalent attachment of the guanine N-7 with aflatoxin B1, the modification of the guanine 2-amino group with anthramycin remarkable inhibits the DNA cleavages at 5' G-C and 5' G-T sites by the FeIII and CoIII complex systems of bleomycin. These results strongly indicate that metallobleomycin binds in the minor groove of B-DNA and that the 2-amino group of guanine adjacent to the 5' side of the cleaved pyrimidine base is one key element of the specific 5' G-C or G-T recognition by the bleomycin-metal complex. A possible binding mode of metallobleomycin in the DNA helix has been proposed by computer-constructed model building.
Full text
PDF