Abstract
Telomerase, the essential enzyme that maintains telomere length, contains two core components, TERT and TR. Early studies in yeast and mouse showed that loss of telomerase leads to phenotypes only after several generations, due to telomere shortening. However, recent studies have suggested additional roles for telomerase components in transcription and the response to DNA damage. To examine these potential telomere length maintenance-independent roles of telomerase components, we examined first generation mTR−/− and mTERT−/− mice with long telomeres. We used gene expression profiling and found no genes that were differentially expressed in mTR−/− G1 mice and mTERT−/− G1 mice compared with wild-type mice. We also compared the response to DNA damage in mTR−/−G1 and mTERT−/− G1 mouse embryonic fibroblasts, and found no increase in the response to DNA damage in the absence of either telomerase component compared to wild-type. We conclude that, under physiologic conditions, neither mTR nor mTERT acts as a transcription factor or plays a role in the DNA damage response.
INTRODUCTION
The telomerase enzyme is essential for telomere length maintenance; it catalyzes the addition of telomeric repeats onto telomeres (1). When telomerase is absent or deficient, telomeres shorten with each cell division (2–4). When telomeres become critically short, they induce a DNA damage response and cells senesce or undergo apoptosis (5–7). Thus, telomerase is required for the long-term growth of cells. The core components of telomerase are conserved in all eukaryotes; the reverse transcriptase component TERT is the catalytic protein subunit that carries out telomere repeat addition (8). The essential telomerase RNA component TR is needed for both enzyme activity and to provide the template for the telomeric repeats that are synthesized (9,10). In addition to TERT and TR, different species have additional species-specific telomerase components.
Experiments in both yeast and mouse cells have shown that deletion of telomerase results in loss of cell division capacity only after telomeres become short. In yeast, deletion of either the TERT component, EST2, or the TR component, TLC1, has no phenotype in the first few generations, but results in decreased growth potential after an increased number of cell divisions (11,12). A similar delayed phenotype is seen in mouse; the deletion of either mouse telomerase component, mTERT or mTR, shows no phenotype in the first few generations when telomeres are long (4,13). However, after four to six generations of interbreeding, short telomeres lead to loss of cell division capacity and subsequent loss of tissue renewal in tissues of high turnover (14,15). Similar phenotypic delay in the response to loss of telomerase has been described in Caenorhabditis elegans (16) and Arabidopsis thaliana (17) TERT mutants, again with first-generation telomerase-null mutants being phenotypically normal. Thus, across phyla, the role of telomerase in telomere length maintenance is conserved, as is the effect of short telomeres on cell division capacity.
Although short telomeres limit cell division, recent studies have suggested that, in addition to their role in telomerase activity, TERT and TR may have cellular functions that are independent of telomere elongation. The acute knock-down of TR using either siRNAs or ribozymes has been reported to rapidly reduce cancer cell growth and to induce a set of glycolytic genes (18–20). Acute knock-down of the TERT component with siRNAs was reported to alter histone modification and sensitize human cells to DNA damage (21), and to affect hematopoiesis in zebrafish (22). siRNA studies are known to have off-target effects that result in phenotypes unrelated to the gene targeted (23). In addition to these knock-down studies, a number of groups have concluded from overexpression of TERT that this protein may have functions outside of its role in telomere elongation. TERT overexpression was reported to protect against cell death (24), perhaps by interfering with p53-mediated apoptosis (25). Overexpression of TERT was also reported to rapidly induce growth-promoting genes (26), activate the Myc and Wnt pathways (27) and stimulate hair follicle stem cell proliferation (28). Overexpression studies are one way to approach an understanding of gene function; however, they need to be interpreted in the context of other experiments that take alternative approaches (29). Overexpression, by design, generates a hypermorph that has an excess of the given activity. Further, in some cases, overexpression can result in a neomorph that inadvertently displays a new phenotype not associated with the original gene (30). This may be due to inappropriate processing of high-level overexpressed proteins (31) or inappropriate oligomerization of overexpressed proteins with other cellular proteins (32) that affects downstream targets. Given these caveats with both siRNA and overexpression, we wanted to assess the role of the loss of these two telomerase components in a controlled genetic setting.
As telomerase mutations are associated with human disease (33), we wanted to carefully examine what role telomerase might play independent of telomere length maintenance. We employed a well-defined genetic system, gene deletion in mice, to examine the effects of loss of telomerase components. Mice make an excellent model system for understanding telomeres and telomerase. Telomeres shorten progressively in both human and mouse cells (2,4,34) and the cellular response to short telomeres is conserved (7,35,36). Finally, the disease phenotypes seen in mice with telomere dysfunction faithfully replicate human disease (15,37). We took advantage of the fact that the C57BL/6J mouse strain has long telomeres. Deletion of telomerase components in this genetic background eventually leads to telomere shortening. However, telomeres do not become critically short until after three to four generations of interbreeding the telomerase-null mice. This allowed us to use the first-generation telomerase-null mice, mTR−/− G1, and independently, mTERT−/− G1, to separate the telomere length effects from any effect that is due to the acute loss of the telomerase components. We examined gene expression profiles and compared the mTR−/− G1 mice and mTERT−/− G1 mice with each other and with wild-type mice. Since not all functions of the DNA damage response are mediated through transcriptional changes, we also analyzed the DNA damage response in mTR−/− G1 and mTERT−/− G1 mouse embryonic fibroblasts (MEFs). We found that in these first-generation telomerase-null animals, when telomeres are long, there were no measurable effects in either transcriptional profiles or DNA damage response when compared to wild-type mice.
MATERIALS AND METHODS
Mice
mTR−/− mice were generated as previously described (4). mTERT−/− mice were obtained from Dr. Lea Harrington (13). First-generation (G1) mTR−/− and mTERT−/− mice were derived by intercrossing mTR+/− and mTERT+/− heterozygotes, respectively. Both strains were extensively backcrossed onto the C57BL/6J genetic background and the heterozygotes were maintained by breeding heterozygotes to wild-type to avoid haploinsufficiency causing telomere shortening.
Isolation and growth of MEFs
MEFs were isolated using a modified version of the published protocol (4). Briefly, embryos from day E12.5–E14.5 were isolated from crosses between either mTR+/− or mTERT+/− C57BL/6J mice. The embryos were minced and partially digested in 0.25% trypsin (Gibco) for 30 min at 4°C, followed by a 5-min incubation at 37°C. After centrifugation, cells were plated in 10-cm plates containing DMEM (Gibco) supplemented with 10% FBS (Gibco), 1X PSF (Gibco) and 1X Normocin (InvivoGen). MEFs were incubated at 37°C until confluent, and passaged according to the 3T3 protocol (38). Cells were treated with 5 µM camptothecin (Sigma) for 4 h or 5 Gy gamma rays using a cesium irradiator (Nordion Gammacell 40 Exactor). In all experiments, MEFs at ∼80% confluency were treated at P5 and harvested using 0.05% trypsin (Gibco).
RNA isolation, microarray hybridization and analysis
Livers from age- (1.5–3 months) and sex-matched (male) wild-type, mTR−/− G1 and mTERT−/− G1 C57BL/6J mice were homogenized in 15 ml TRIzol reagent (Invitrogen) using a dounce homogenizor. RNA was extracted following the TRIzol protocol. In brief, the liver homogenates were extracted with chloroform, followed by 2-propanol precipitation and a 75% ethanol wash. Pellets were resuspended and incubated in a DNase I (Promega)/RNase inhibitor (New England BioLabs) cocktail, followed by a final phenol/chloroform extraction. RNA was precipitated with 100% ethanol, resuspended in DEPC water and cleaned-up using the RNeasy kit (Qiagen). For RNA isolation from MEFs a modified protocol was used. Following the 75% ethanol wash, an in-column DNase digestion using the RNeasy kit (Qiagen) was performed prior to the clean-up step. MEFs were derived from embryos from day E12.5–14.5. RNA samples (1 µg) from three biological replicates were sent for hybridization on Mouse Gene 1.0 ST Arrays (Affymetrix) at the Johns Hopkins HiT Center Microarray Facility. To avoid any genetic background effect, knock-out mice and MEFs were matched to wild-type mice and MEFs derived from the same parents (i.e. mTR−/− G1 and mTR WT, mTERT−/− G1 and mTERT WT). Due to mouse availability, the liver samples were processed in two batches. The first batch contained one biological replicate for a wild-type/mTERT−/− G1 and wild-type/mTR−/− G1 pair. The remaining samples were processed and hybridized in a second batch. The expression signals were adjusted for batch effect with the R package ComBat, using the non-parametric empirical Bayes method [http://statistics.byu.edu/johnson/ComBat/; (39)]. Gene expression was analyzed using volcano plots and heat maps, which were generated and analyzed with the Spotfire software (TIBCO). Raw data have been deposited at the NCBI Gene Expression Omnibus (GEO) repository (GSE16731).
Quantitative RT-PCR
Total RNA (2 µg) from mTERT−/− G1, mTR−/− G1 and wild-type MEFs was reverse transcribed using random hexamers and Superscript III reverse transcriptase (Invitrogen), following the manufacturer’s instructions. Quantitative RT-PCR was performed for mTERT and mTR using a CFX96 thermocycler (Bio-Rad). Each qRT-PCR reaction contained 1× SYBR Green Supermix and 5 µM of each primer. Roughly, 5 ng cDNA were amplified per reaction. The expression in each sample was normalized to HPRT. The cycling conditions for mTR were as follows: 5 min at 95°C; 15 s at 95°C, 30 s at 68°C, 45 s at 72°C, 10 s at 82°C (35 cycles); 3 min at 72°C. For each cycle, fluorescence readings were performed at the 82°C step, to avoid generation of primer dimers. For mTERT, we followed the cycling conditions and primer sequences described elsewhere (40). Primer sequences for mTR were the following: RT_mTR_F 5′-TGTGGGTTCTGGTCTTTTGTTCTCCG-3′, RT_mTR_R 5′-GTTTTTGAGGCTCGGGAACGCG-3′, HPRT_F 5′-TGATCAGTCAACGGGGGACA-3′, HPRT_R 5′-TTCGAGAGGTCCTTTTCACCA-3′. To assess for genomic DNA contamination, in each run we included a control in which no reverse transcriptase was added (− RT). All samples did not have significant DNA contamination, as fluorescent intensities were not above background levels. Three different samples for each genotype were run in triplicate and the normalized average was reported.
Western blot
For whole-cell lysates (WCLs), MEFs were harvested after treatment and washed twice in cold PBS. Pellets were resuspended in 1× SDS sample buffer (1× Tris–Cl/SDS pH 6.8 (0.5 M Tris–Cl, 0.4% SDS), 0.05% glycerol, 0.04% SDS, 0.002% bromophenol blue, 0.5% 2-mercaptoethanol), boiled at 100°C for 5 min and chilled on ice for 3 min. After centrifugation, WCLs (15 µl) were fractionated on sodium dodecyl sulfate polyacrylamide gel electrophoresis (SDS–PAGE; 4–15% Tris–HCl, Bio-Rad), and transferred onto 0.2-µm PVDF membranes (Millipore). Blocking (2 h at 4°C), and multiplexed primary antibody incubation (overnight at 4°C) were performed in 1% casein/0.1% Tween-20. Primary antibodies were as follows: rabbit phospho-p53 (Ser 15) (Cell Signaling), rabbit anti-γ-H2AX (Ser 139) (Abcam) and mouse anti-β-actin (Abcam). After incubation with secondary antibodies conjugated to near-infrared dyes (IRDye® 680 anti-mouse and 800 anti-rabbit, LI-COR), blots were scanned on a two-channel near-infrared Odyssey scanner (LI-COR). Band intensities were quantified using the Odyssey software (LI-COR), normalized to β-actin levels and expressed as arbitrary units. Experiments were performed in three individual replicates.
Immunofluorescence
MEFs at ∼80% confluence were cultured, treated and processed on two-well chamber slides (Nunc Lab-Tek II). Briefly, cells were washed twice with PBS and fixed in 4% formaldehyde. Fixed cells were blocked (2 h at room temperature) and incubated with primary antibody (overnight at 4°C) using 10% normal goat serum/0.1% Triton X-100. Primary antibodies were as follows: rabbit anti-γ-H2AX (Ser 139) (Abcam), and rabbit anti-phospho-53BP1 (Ser 1778) (Cell Signaling). Secondary antibody incubation was performed for 2 h at room temperature using an Alexa fluor 488-conjugated secondary antibody (Invitrogen) and 1× DAPI nucleic-acid stain (Invitrogen). Slides were analyzed on an Axioskop fluorescence microscope (Zeiss), at 25× magnification. For quantitation, 100 cells were scored and classified into 0, <10 and >10 categories, according to the number of foci. Cells with >10 nuclear foci were considered to be undergoing a strong DNA damage response.
Cell proliferation assay
Cell proliferation was determined using the Click-iT™ EdU cell proliferation assay kit (Invitrogen), which relies on a copper-catalyzed reaction between the alkyne-containing nucleoside analog EdU and the Alexa Fluor 488-conjugated azide. Briefly, MEFs were plated and treated in two-well chamber slides. EdU was added immediately after treatment and cells were incubated for 16 h at 37°C, to allow cells to undergo a full DNA replication cycle. Cells were fixed in 3.7% formaldehyde and permeabilized with 0.5% Trixon X-100. Cells were washed with 3% BSA and S-phase cells were detected with the Click-iT™ reaction cocktail for 30 min. Slides were washed, stained with Hoechst nucleic-acid stain and analyzed on an Axioskop fluorescence microscope (Zeiss). For each slide, 100 MEFs were scored and classified as EdU-positive or -negative.
Statistical analysis
For the microarray analysis, the Spotfire software (TIBCO) was used to calculate P-values using a two-way analysis of variance (ANOVA) test. Statistical significance for qRT-PCR and the functional DNA damage experiments was calculated with Microsoft Excel’s statistical analysis package, using a paired Student’s t-test. A P-value below a threshold of 0.05 was considered statistically significant.
RESULTS
Gene expression profiles of mTERT–/– and mTR–/– G1 mice are similar to wild-type
To test the consequences of the loss of each of the telomerase components, mTERT and mTR, we examined transcriptional profiles from wild-type, mTERT–/– G1 and mTR–/– G1 mice. As mentioned above, first-generation C57BL/6J mTR–/– G1 and mTERT–/– G1 mice have long telomeres, and no phenotypes are seen in these telomerase-null mice until telomeres become short after interbreeding for three to six generations (4,13,14). By comparing wild-type with mTERT–/– G1 and mTR–/– G1 mice, we can separate a telomere effect from any telomere length maintenance-independent effects of mTERT or mTR deletion. We analyzed samples from three independent mice for each genotype. To control for potential genetic variation, for each matched set of mTR–/– and wild-type or mTERT–/– and wild-type samples, the wild-type mice were progeny from the same parents as the knock-outs.
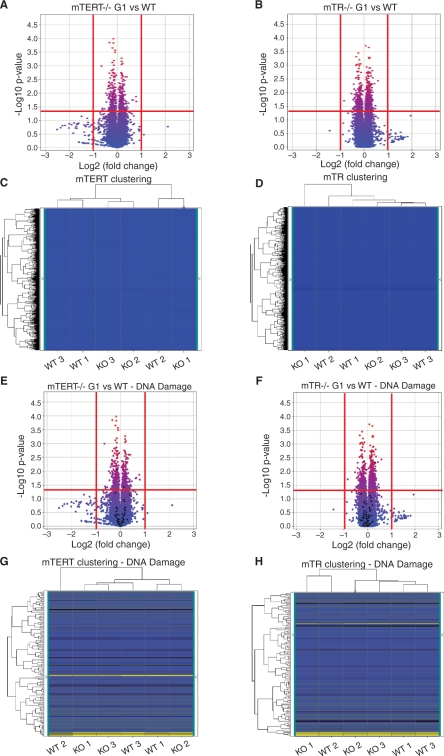
We isolated liver RNA from age- and sex-matched wild-type, mTERT–/– G1 and mTR–/– G1 mice, and hybridized it to Mouse Gene 1.0 ST microarrays from Affymetrix. We compared whole-genome transcriptional profiles in the mTERT–/– G1 and wild-type, and the mTR–/– G1 and wild-type data sets separately, using volcano plots and heat maps (Figure 1). Volcano plots represent gene expression levels according to the fold change and statistical significance. The horizontal axis represents the impact of telomerase absence on transcript abundance, expressed as the fold change between the two experimental groups, on a Log2 scale. The vertical axis represents the statistical reliability of the fold change, expressed as a P-value between samples, on a negative Log10 scale. Of the genes that showed significant levels of expression (P < 0.05) that we could analyze, we found none whose levels were either increased or decreased at least 2-fold when comparing either mTR–/– G1 and wild-type or mTERT–/– G1 and wild-type mice (Figure 1A and B). We next used heat maps generated by hierarchical clustering of gene expression levels to determine if the knock-out mice were different from the wild-type mice. We found that knock-out and wild-type samples clustered randomly, indicating that gene expression levels were similar in all mice (Figure 1C and D). These experiments indicate that mTERT–/– G1 and mTR–/– G1 mice have similar transcriptional profiles compared to their wild-type counterparts.
Figure 1.
Whole genome and DNA damage response-specific transcriptional profiles of mTERT–/– G1 and mTR–/– G1 mice are similar to wild-type. RNA expression in three different mice for each genotype was examined and transcriptional profiles were compared to wild-type. Volcano plots show the P-value for correlation of the three replicates on the y-axis and the fold change between genotypes on the x-axis. Values above the horizontal red line at Y = 1.3 are considered statistically significant (P < 0.05). The vertical red lines mark the threshold for 2-fold up- or down-regulation. Genes to the right and left of these lines are considered to be up- or down-regulated, respectively. Heat maps represent the level of gene expression across samples and the hierarchical clustering of these. (A and B) Volcano plots of whole-genome transcriptional profiles of mTERT–/– G1 and mTR–/– G1 mice compared to wild-type. (C and D) Heat maps and clustering analysis of mTERT–/– G1 and mTR–/– G1 mice compared to wild-type. (E and F) Volcano plots of DNA damage response-specific transcriptional profiles of mTERT–/– G1 and mTR–/– G1 mice compared to wild-type. DNA damage response genes are highlighted in black against whole genome volcano plots. (G and H) Heat maps and clustering analysis of DNA damage response gene expression of mTERT–/– G1 and mTR–/– G1 mice compared to wild-type. Blue represents low signal values, yellow represents high signal values and black represents the mid-range.
As described in the ‘Introduction’ section, recent studies have concluded that mTERT may play telomere length maintenance-independent roles in cell proliferation, DNA damage response and in developmental pathways, such as the Wnt pathway (26–28,41). To examine these pathways specifically, we analyzed the RNA expression levels of genes involved in the DNA damage response, apoptosis, cell cycle and the proliferation-promoting Wnt, Erbb (EGF) and MAPK pathways. We analyzed each pathway independently by selecting all genes involved in the pathway and highlighting them against our whole-genome data using both volcano plots and heat maps. We found no significant differences in the level of gene expression for any of these pathways in mTERT–/– G1 versus wild-type or mTR–/– G1 versus wild-type mice (Figure 1E–H and Supplementary Figure S2; data not shown). Finally, we compared whole-genome and pathway-specific expression patterns between mTERT–/– G1 and mTR–/– G1 mice, as well as between the corresponding wild-type samples. Again, we found no significant up- or down-regulation in the gene expression in either of these comparisons (Supplementary Figure S3, data not shown).
It has been suggested that the failure to observe phenotypic consequences of telomerase loss in first-generation mice may be due to a compensatory pathway masking the effects of the loss of telomerase components in the mTR–/– and mTERT–/– mice (27,41). Genetic compensation is a well-established phenomenon in which one or a set of genes can compensate for the loss of a gene, so that no phenotype is seen when that gene is not functional. The presence of paralogs and redundant genes in organisms is thought to exemplify this need for potential compensation (42). In well-established cases of genetic compensation, transcriptional upregulation of the genes that are compensating is seen (43). To determine whether such compensation might occur in early cell divisions after the loss of TR or TERT, we examined gene expression profiles in MEFs derived from heterozygous mTR+/– and mTERT+/– crosses. These MEFs represent cells from early embryonic stages that have just experienced the loss of mTR or mTERT, and as such would allow us to see any transcriptional upregulation that may compensate for the recent loss of these telomerase components. We found no significant differences in the transcriptional profiles between mTR–/– G1 and wild-type or mTERT–/– G1 and wild-type MEFs for any genes in the Wnt, DNA damage or apoptosis pathways (Supplementary Figures S4 and S5, and data not shown). Taken together, our liver and MEF data are consistent and support the conclusion that mTERT and mTR do not regulate gene expression levels.
We also examined the array data specifically for proteins that are associated with telomerase. We did not find a change in the levels of mTERT or the accessory components Nop10, Nhp2 or dyskerin when one component was knocked out (Supplementary Figure S1 and Table S1; data not shown). As telomerase RNA might not be amplified under conditions set to find mRNAs, to examine the level of mTR and mTERT in the two different knock-outs, we used quantitative RT-PCR to directly measure changes in both mTERT and mTR in wild-type and knock-out MEFs. We found that mTR was not detected in the mTR–/– G1 MEFs and mTERT was not detected in the mTERT–/– G1 MEFs, as expected. mTERT was present at comparable levels both in the wild-type and mTR–/– G1 samples. We observed a slight (∼1.5-fold, P = 0.028) decrease in mTR transcript in the mTERT–/– G1 samples, suggesting that the absence of mTERT might affect the stability of the telomerase RNA (Supplementary Figure S1 and Table S1).
mTERT–/– and mTR–/– cells do not show a difference in DNA damage response induction compared to wild-type
Changes in gene expression levels might be expected if telomerase components play a direct role in transcription; however, some phenotypes might be due to functional changes that do not involve transcriptional changes. To further examine a potential telomere length maintenance-independent role of mTR and mTERT in DNA damage, we used a sensitive functional assay using mTR–/– G1, mTERT–/– G1 and wild-type MEFs. Three independent early-passage MEF cultures of each genotype were treated with two different DNA-damaging agents, camptothecin and gamma rays, to induce a DNA damage response. Camptothecin is a topoisomerase I inhibitor that generates double-strand breaks (DSBs) during DNA replication (44). Gamma rays act directly on DNA, inducing DSBs in the sugar-phosphate backbone. In each cell line, we monitored p53 and H2AX (γ-H2AX) phosphorylation in response to DNA damage as examples of well-characterized damage responses (45,46). The phosphorylation levels were monitored with phospho-specific antibodies on western blots, using β-actin as an internal loading control. We also measured the accumulation of γ-H2AX and phospho-53BP1 (47) in DNA damage-induced foci using immunofluorescence. Finally, we monitored the rate of cell proliferation in response to both camptothecin and gamma irradiation.
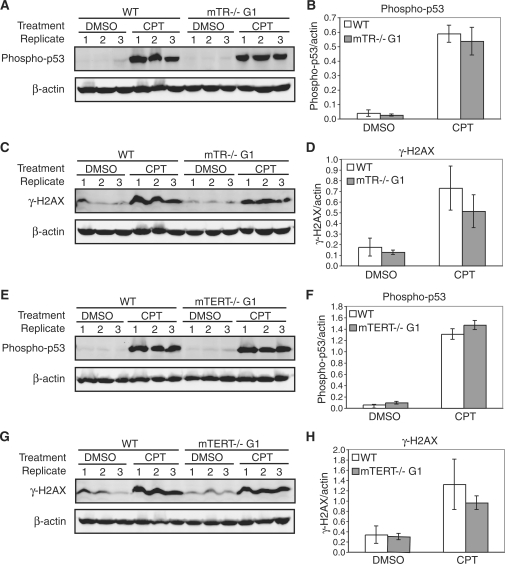
Upon DNA damage induction with 5 µM camptothecin, the level of phospho-p53 and γ-H2AX increased significantly in wild-type, mTERT–/– G1 and mTR–/– G1 MEFs, in all three replicate cell lines (Figure 2, see figure legend for P-values). However, there was no significant difference in the phosphorylation levels between wild-type and mTERT–/– G1 or mTR–/– G1 MEFs (Figure 2), under both untreated [dimethyl sulfoxide (DMSO)] and treated [camptothecin (CPT)] conditions. As one exception, we did observe a small change in the phosphorylation levels of H2AX in mTR–/– G1 MEFs (Figure 2D). Interestingly, the levels of γ-H2AX were slightly lower in the knock-out cells, opposite of what has been reported (21).
Figure 2.
Camptothecin induces a similar DNA damage response in wild-type, mTR–/– G1 and mTERT–/– G1 MEFs. The phosphorylation of p53 and H2AX in untreated (DMSO) and cells treated with 5 µM camptothecin (CPT) was measured by western blot using phospho-specific antibodies. Quantitative graphs show the arithmetic mean and SEM of the expression levels for three replicate experiments. (A) Western blots of cells treated with camptothecin were probed with a phospho-p53 specific antibody in wild-type and mTR–/– G1 MEFs. (B) Normalized phospho-p53 expression levels: wild-type DMSO compared to mTR–/– G1 DMSO, P = 0.28; wild-type CPT compared to mTR–/– G1 CPT, P = 0.21; wild-type DMSO compared to wild-type CPT, P = 0.005; mTR–/– G1 DMSO compared to mTR–/– G1 CPT, P = 0.01. (C) Western blots of cells treated with camptothecin were probed with a phospho-H2AX (γ-H2AX) specific antibody in wild-type and mTR–/– G1 MEFs. (D) Normalized γ-H2AX expression levels: wild-type DMSO compared to mTR–/– G1 DMSO, P = 0.32; wild-type CPT compared to mTR–/– G1 CPT, P = 0.03; wild-type DMSO compared to wild-type CPT, P = 0.02; mTR–/– G1 DMSO compared to mTR–/– G1 CPT, P = 0.04. (E) Western blots of cells treated with camptothecin were probed with a phospho-p53 specific antibody in wild-type and mTERT–/– G1 MEFs. (F) Normalized phospho-p53 expression levels: wild-type DMSO compared to mTERT–/– G1 DMSO, P = 0.19; wild-type CPT compared to mTERT–/– G1 CPT, P = 0.16; wild-type DMSO compared to wild-type CPT, P = 0.002; mTERT–/– G1 DMSO compared to mTERT–/– G1 CPT, P = 0.001. (G) Western blots of cells treated with camptothecin were probed with a phospho-H2AX (γ-H2AX) specific antibody in wild-type and mTERT–/– G1 MEFs. (H) Normalized γ-H2AX expression levels: wild-type DMSO compared to mTERT–/– G1 DMSO, P = 0.69; wild-type CPT compared to mTERT–/– G1 CPT, P = 0.08; wild-type DMSO compared to wild-type CPT, P = 0.037; mTERT–/– G1 DMSO compared to mTERT–/– G1 CPT, P = 0.005.
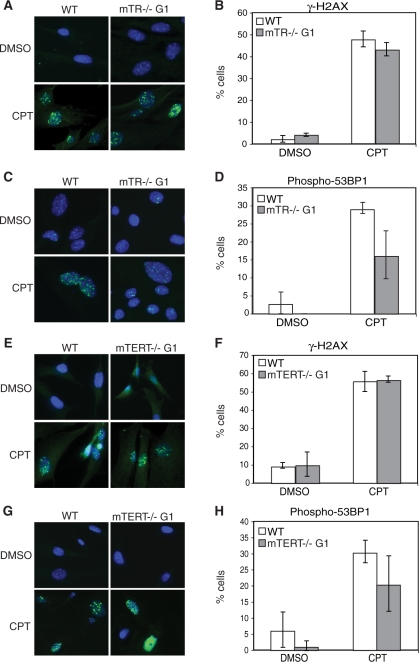
To further examine the DNA damage response, we examined damage-induced foci that accumulate in cells repairing DNA damage (48). We monitored the induction of γ-H2AX and phospho-53BP1 foci by immunofluorescence. Treatment with camptothecin generated a significant accumulation of γ-H2AX- and phospho-53BP1 foci in wild-type, mTERT–/– G1 and mTR–/– G1 MEFs (Figure 3). We found no significant difference in the number of phospho-53BP1 foci between wild-type and mTR–/– G1 MEFs and in the number of γ-H2AX foci between wild-type and mTERT–/– G1 MEFs, under untreated and treated conditions (Figure 3). There was a small decrease in γ-H2AX foci number in mTR–/– G1 MEFs compared to wild-type (Figure 3B) and in phospho-53BP1 foci number in mTERT–/– G1 MEFs compared to wild-type (Figure 3H), which may be due to experimental fluctuation. We monitored cell proliferation by EdU incorporation in treated and untreated samples. Proliferation was decreased by camptothecin treatment in the three MEF cultures. Both mTR–/– G1 and mTERT–/– G1 MEFs were affected to a similar level compared to wild-type MEFs (Supplementary Figure S6A and B). A similar finding for mTERT–/– MEFs using etoposide as a damaging agent was recently reported (49).
Figure 3.
Camptothecin induces accumulation of γ-H2AX and phospho-53BP1 foci in wild-type, mTR–/– G1 and mTERT–/– G1 MEFs. γ-H2AX and phospho-53BP1 foci in untreated (DMSO) and cells treated with 5 µM camptothecin (CPT) were visualized using immunofluorescence. Representative images of each protein and cell line are shown. Quantitative graphs show the fraction of cells that accumulated >10 foci. Graphs show the arithmetic mean and SEM for three replicate experiments. (A) Immunofluorescence detects camptothecin-induced γ-H2AX foci in wild-type and mTR–/– G1 MEFs. (B) Quantification of cells with >10 γ-H2AX foci: wild-type DMSO compared to mTR–/– G1 DMSO, P = 0.18; wild-type CPT compared to mTR–/– G1 CPT, P = 0.034; wild-type DMSO compared to wild-type CPT, P = 0.003; mTR–/– G1 DMSO compared to mTR–/– G1 CPT, P = 0.002. (C) Immunofluorescence detects camptothecin-induced phospho-53BP1 foci in wild-type and mTR–/– G1 MEFs. (D) Quantification of cells with >10 phospho-53BP1 foci: wild-type DMSO compared to mTR–/– G1 DMSO, P = 0.23; wild-type CPT compared to mTR–/– G1 CPT, P = 0.08; wild-type DMSO compared to wild-type CPT, P = 0.001; mTR–/– G1 DMSO compared to mTR–/– G1 CPT, P = 0.05. (E) Immunofluorescence detects camptothecin-induced γ-H2AX foci in wild-type and mTERT–/– G1 MEFs. (F) Quantification of cells with >10 γ-H2AX foci: wild-type DMSO compared to mTERT–/– G1 DMSO, P = 0.86; wild-type CPT compared to mTERT–/– G1 CPT, P = 0.75; wild-type DMSO compared to wild-type CPT, P = 0.003; mTERT–/– G1 DMSO compared to mTERT–/– G1 CPT, P = 0.009. (G) Immunofluorescence detects camptothecin-induced phospho-53BP1 foci in wild-type and mTERT–/– G1 MEFs. (H) Quantification of cells with >10 phospho-53BP1 foci: wild-type DMSO compared to mTERT–/– G1 DMSO, P = 0.2; wild-type CPT compared to mTERT–/– G1 CPT, P = 0.02; wild-type DMSO compared to wild-type CPT, P = 0.006; mTERT–/– G1 DMSO compared to mTERT–/– G1 CPT, P = 0.07.
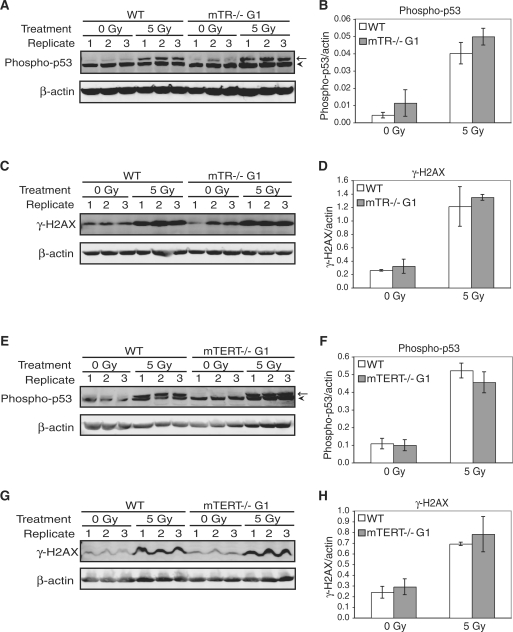
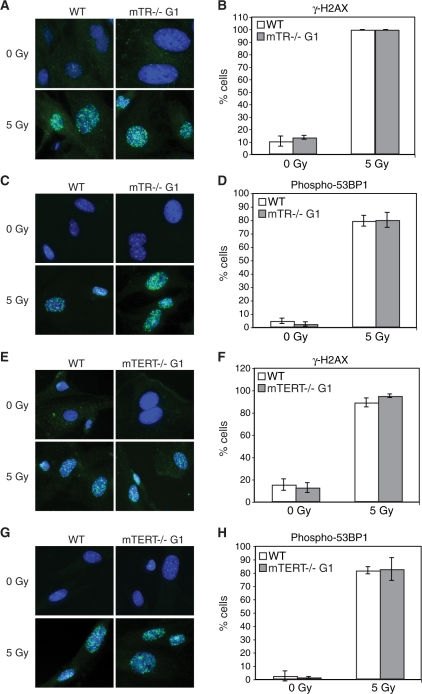
Different types of genotoxic stress activate different DNA damage response pathways (50). To determine whether the different type of genotoxic stress might show a difference in wild-type, mTERT–/– G1 or mTR–/– G1 MEFs, we examined the effect of gamma irradiation on these cells. We first examined the level of damage-induced protein phosphorylation using western analysis as described above. When cells were damaged with 5 Gy gamma rays, the phosphorylation of both p53 and H2AX (γ-H2AX) was significantly increased in all three cell cultures (Figure 4). We found no significant difference in the phosphorylation levels between wild-type, mTERT–/– G1 or mTR–/– G1 MEFs under both untreated (0 Gy) and treated (5 Gy) conditions (Figure 4). We next examined damage-induced foci in response to 5-Gy gamma irradiation. We found a significant accumulation of foci recognized by γ-H2AX- and phospho-53BP1 antibodies in wild-type, mTERT–/– G1 and mTR–/– G1 MEFs (Figure 5). We found no significant difference in the induction of damage-induced foci between wild-type, mTERT–/– G1 and mTR–/– G1 MEFs under both untreated and treated conditions. As described above for camptothecin, cell proliferation was affected by gamma irradiation to a similar extent in wild-type, mTERT–/– G1 and mTR–/– G1 MEFs (Supplementary Figure S6C and D). As further evidence that the mTR–/– and mTERT–/– G1 telomerase-null mice have long functional telomeres, no telomere damage-induced foci (TIFs) (51) were seen in any of the untreated samples. If any telomeres in these mice were critically short, we would have seen TIFs present at telomeres in untreated cells. Overall, these results indicate that when telomeres are not yet short in the first generation, the absence of either mTR or mTERT does not alter the cellular response to DNA damage.
Figure 4.
Gamma irradiation induces a similar DNA damage response in wild-type, mTR–/– G1 and mTERT–/– G1 MEFs. The phosphorylation of p53 and H2AX in untreated (0 Gy) and cells treated with 5 Gy gamma rays (5 Gy) was measured by western blot using phospho-specific antibodies. Quantitative graphs show the arithmetic mean and SEM of the expression levels for three replicate experiments. (A) Western blots of cells treated with gamma rays were probed with a phospho-p53 specific antibody (arrow) in wild-type and mTR–/– G1 MEFs. A cross-reacting band was picked up with this antibody (arrowhead). (B) Normalized phospho-p53 expression levels: wild-type 0 Gy compared to mTR–/– G1 0 Gy, P = 0.23; wild-type 5 Gy compared to mTR–/– G1 5 Gy, P = 0.03; wild-type 0 Gy compared to wild-type 5 Gy, P = 0.006; mTR–/– G1 0 Gy compared to mTR–/– G1 5 Gy, P = 0.003. (C) Western blots of cells treated with gamma rays were probed with a phospho-H2AX (γ-H2AX) specific antibody in wild-type and mTR–/– G1 MEFs. (D) Normalized γ-H2AX expression levels: wild-type 0 Gy compared to mTR–/– G1 0 Gy, P = 0.45; wild-type 5 Gy compared to mTR–/– G1 5 Gy, P = 0.54; wild-type 0 Gy compared to wild-type 5 Gy, P = 0.03; mTR–/– G1 0 Gy compared to mTR–/– G1 5 Gy, P = 0.007. (E) Western blots of cells treated with gamma rays were probed with a phospho-p53 specific antibody (arrow) in wild-type and mTERT–/– G1 MEFs. (F) Normalized phospho-p53 expression levels: wild-type 0 Gy compared to mTERT–/– G1 0 Gy, P = 0.81; wild-type 5 Gy compared to mTERT–/– G1 5 Gy, P = 0.37; wild-type 0 Gy compared to wild-type 5 Gy, P = 0.009; mTERT–/– G1 0 Gy compared to mTERT–/– G1 5 Gy, P = 0.021. (G) Western blots of cells treated with gamma rays were probed with a phospho-H2AX (γ-H2AX) specific antibody in wild-type and mTERT–/– G1 MEFs. (H) Normalized γ-H2AX expression levels: wild-type 0 Gy compared to mTERT–/– G1 0 Gy, P = 0.23; wild-type 5 Gy compared to mTERT–/– G1 5 Gy, P = 0.58; wild-type 0 Gy compared to wild-type 5 Gy, P = 0.025; mTERT–/– G1 0 Gy compared to mTERT–/– G1 5 Gy, P = 0.035.
Figure 5.
Gamma irradiation induces accumulation of γ-H2AX and phospho-53BP1 foci in wild-type, mTR–/– G1 and mTERT–/– G1 MEFs. The accumulation of γ-H2AX and phospho-53BP1 foci in untreated (0 Gy) and cells treated with 5 Gy gamma rays (5 Gy) was visualized using immunofluorescence. Representative images of each protein and cell line are shown. Quantitative graphs show the fraction of cells that accumulated >10 foci. Graphs show the arithmetic mean and SEM for three replicate experiments. (A) Immunofluorescence detects gamma ray-induced γ-H2AX foci in wild-type and mTR–/– G1 MEFs. (B) Quantification of cells with >10 γ-H2AX foci: wild-type 0 Gy compared to mTR–/– G1 0 Gy, P = 0.43; wild-type 5 Gy compared to mTR–/– G1 5 Gy, P = 1; wild-type 0 Gy compared to wild-type 5 Gy, P = 0.0006; mTR–/– G1 0 Gy compared to mTR–/– G1 5 Gy, P = 0.0007. (C) Immunofluorescence detects gamma ray-induced phospho-53BP1 foci in wild-type and mTR–/– G1 MEFs. (D) Quantification of cells with >10 phospho-53BP1 foci: wild-type 0 Gy compared to mTR–/– G1 0 Gy, P = 0.12; wild-type 5 Gy compared to mTR–/– G1 5 Gy, P = 0.9; wild-type 0 Gy compared to wild-type 5 Gy, P = 0.002; mTR–/– G1 0 Gy compared to mTR–/– G1 5 Gy, P = 0.003. (E) Immunofluorescence detects gamma ray-induced γ-H2AX foci in wild-type and mTERT–/– G1 MEFs. (F) Quantification of cells with >10 γ-H2AX foci: wild-type 0 Gy compared to mTERT–/– G1 0 Gy, P = 0.58; wild-type 5 Gy compared to mTERT–/– G1 5 Gy, P = 0.15; wild-type 0 Gy compared to wild-type 5 Gy, P = 0.005; mTERT–/– G1 0 Gy compared to mTERT–/– G1 5 Gy, P = 0.002. (G) Immunofluorescence detects gamma ray-induced phospho-53BP1 foci in wild-type and mTERT–/– G1 MEFs. (H) Quantification of cells with >10 phospho-53BP1 foci: wild-type 0 Gy compared to mTERT–/– G1 0 Gy, P = 0.67; wild-type 5 Gy compared to mTERT–/– G1 5 Gy, P = 0.89; wild-type 0 Gy compared to wild-type 5 Gy, P = 0.002; mTERT–/– G1 0 Gy compared to mTERT–/– G1 5 Gy, P = 0.04.
DISCUSSION
Telomeres and telomerase play pivotal roles in cell proliferation, tissue maintenance and the growth of cancer cells. To fully understand the consequences that telomerase mutations may have, we set out to examine whether the absence of mTR or mTERT have an effect on transcriptional profiles or the DNA damage response. In our experiments, we found no evidence that, in the setting of wild-type telomere lengths, telomerase regulates gene expression or the DNA damage response.
In our expression analysis we used a 2-fold change as our cut-off for changes in gene expression. We chose a 2-fold difference as a threshold in an attempt to bias in favor of seeing changes in as many differentially expressed genes as possible. Still, we did not find any significant changes. In contrast, previous groups found up to a 30-fold difference in over 600 genes when TERT was overexpressed (26,27). In our functional assays, using two different types of DNA damage we found that the DNA damage response was not altered when either mTR or mTERT were deleted. In a recent paper, similar results were seen using mTERT–/– MEFs and ES cells treated with etoposide and gamma irradiation as a source of genotoxic damage (49).
Our results stand in contrast to recent reports suggesting that telomerase has telomere length maintenance-independent roles. Heterologous overexpression of TERT has been suggested to result in enhancement of cell proliferation (26), hair follicle stem cell proliferation (27,28,52), cell survival (24,53) and protection against apoptosis (24). It has also been suggested that many of these phenotypes are due to transcriptional induction of the Wnt pathway (27,41). As described in the ‘Introduction’ section, overexpression of proteins at high levels can result in neomorphs that have a function which the original protein does not have (30–32). Transgenic mice that overexpress specific proteins are subject to caveats in the interpretation of the results, in part due to the creation of neomorphs. The Tet transcriptional activator used to regulate transgene expression is known to have functional consequences of its own, and the site of integration and number of transgenes present can alter the phenotype (29). Finally, it is known that the Wnt pathway can be inappropriately activated under a variety of experimental conditions and interpretation of activation is subject to specific caveats (54).
In addition to finding no changes when mTERT was deleted, we also found no effects when mTR was deleted. Previous experiments suggested transcriptional defects from knock-down of mTR (18–20). siRNA knock-downs may have off target effects (23,55–57) and in addition, our results may differ from those previously reported, due in part to the use of knock-out animals instead of cancer cell lines. Cancer cell lines are heterogeneous and often have very short telomeres (58). In examining the effects of telomerase component deletion in cell lines, the possibility of short dysfunctional telomeres should be examined, as only a few short telomeres can induce a DNA damage response (59).
In the experiments presented here we found no evidence for a role for mTERT or mTR in transcriptional regulation or the DNA damage response. While in our studies we examined knock-out mice, the results are directly relevant to human disease. Moreover, genetic anticipation is observed in mice null for telomerase components (4,14,15) as well as in families that carry mutant telomerase genes (60,61). The study of the telomerase knock-out mouse thus continues to have relevance for exploring the role of telomeres and telomerase in disease.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online.
FUNDING
The National Institutes of Health (PO1CA16519 project 4 to C.W.G.). Funding for open access charge: National Institutes of Health Grant (PO1CA16519 project 4).
Conflict of interest statement. None declared.
Supplementary Material
ACKNOWLEDGEMENTS
We thank Margaret Strong for breeding the mTR and mTERT mice and for the cell proliferation studies, and Conover Talbot Jr. at the JHU HiT center for assistance with microarray data analysis. We also thank Dr. Brendan Cormack, Dr. Jeremy Nathans, Dr. Mary Armanios and members of the Greider lab for helpful comments on the manuscript.
REFERENCES
- 1.Greider CW, Blackburn EH. Identification of a specific telomere terminal transferase activity in Tetrahymena extracts. Cell. 1985;43:405–413. doi: 10.1016/0092-8674(85)90170-9. [DOI] [PubMed] [Google Scholar]
- 2.Harley CB, Futcher AB, Greider CW. Telomeres shorten during ageing of human fibroblasts. Nature. 1990;345:458–460. doi: 10.1038/345458a0. [DOI] [PubMed] [Google Scholar]
- 3.Dokal I. Dyskeratosis congenita. A disease of premature ageing. Lancet. 2001;358(Suppl):S27. doi: 10.1016/s0140-6736(01)07040-4. [DOI] [PubMed] [Google Scholar]
- 4.Blasco MA, Lee H-W, Hande PM, Samper E, Lansdorp PM, DePinho RA, Greider CW. Telomere shortening and tumor formation by mouse cells lacking telomerase RNA. Cell. 1997;91:25–34. doi: 10.1016/s0092-8674(01)80006-4. [DOI] [PubMed] [Google Scholar]
- 5.Lundblad V, Szostak JW. A mutant with a defect in telomere elongation leads to senescence in yeast. Cell. 1989;57:633–643. doi: 10.1016/0092-8674(89)90132-3. [DOI] [PubMed] [Google Scholar]
- 6.IJpma A, Greider CW. Short telomeres induce a DNA damage response in Saccharomyces cerevisiae. Mol. Biol. Cell. 2003;14:987–1001. doi: 10.1091/mbc.02-04-0057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.d'A;dda di Fagagna F, Reaper PM, Clay-Farrace L, Fiegler H, Carr P, Von Zglinicki T, Saretzki G, Carter NP, Jackson SP. A DNA damage checkpoint response in telomere-initiated senescence. Nature. 2003;426:194–198. doi: 10.1038/nature02118. [DOI] [PubMed] [Google Scholar]
- 8.Lingner J, Hughes TR, Shevchenko A, Mann M, Lundblad V, Cech TR. Reverse transcriptase motifs in the catalytic subunit of telomerase. Science. 1997;276:561–567. doi: 10.1126/science.276.5312.561. [DOI] [PubMed] [Google Scholar]
- 9.Greider CW, Blackburn EH. A telomeric sequence in the RNA of Tetrahymena telomerase required for telomere repeat synthesis. Nature. 1989;337:331–337. doi: 10.1038/337331a0. [DOI] [PubMed] [Google Scholar]
- 10.Chen JL, Opperman KK, Greider CW. A critical stem-loop structure in the CR4-CR5 domain of mammalian telomerase RNA. Nucleic Acids Res. 2002;30:592–597. doi: 10.1093/nar/30.2.592. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Singer MS, Gottschling DE. TLC1: template RNA component of Saccharomyces cerevisiae telomerase. Science. 1994;266:404–409. doi: 10.1126/science.7545955. [DOI] [PubMed] [Google Scholar]
- 12.Lendvay TS, Morris DK, Sah J, Balasubramanian B, Lundblad V. Senescence mutants of Saccharomyces cerevisiae with a defect in telomere replication identify three additional EST genes. Genetics. 1996;144:1399–1412. doi: 10.1093/genetics/144.4.1399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Liu Y, Snow BE, Hande MP, Yeung D, Erdmann NJ, Wakeham A, Itie A, Siderovski DP, Lansdorp PM, Robinson MO, et al. The telomerase reverse transcriptase is limiting and necessary for telomerase function in vivo. Curr. Biol. 2000;10:1459–1462. doi: 10.1016/s0960-9822(00)00805-8. [DOI] [PubMed] [Google Scholar]
- 14.Lee HW, Blasco MA, Gottlieb GJ, Horner JW, Greider CW, DePinho RA. Essential role of mouse telomerase in highly proliferative organs. Nature. 1998;392:569–574. doi: 10.1038/33345. [DOI] [PubMed] [Google Scholar]
- 15.Hao LY, Armanios M, Strong MA, Karim B, Feldser DM, Huso D, Greider CW. Short telomeres, even in the presence of telomerase, limit tissue renewal capacity. Cell. 2005;123:1121–1131. doi: 10.1016/j.cell.2005.11.020. [DOI] [PubMed] [Google Scholar]
- 16.Cheung I, Schertzer M, Rose A, Lansdorp PM. High incidence of rapid telomere loss in telomerase-deficient Caenorhabditis elegans. Nucleic Acids Res. 2006;34:96–103. doi: 10.1093/nar/gkj417. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Fitzgerald MS, Riha K, Gao F, Ren S, McKnight TD, Shippen DE. Disruption of the telomerase catalytic subunit gene from Arabidopsis inactivates telomerase and leads to a slow loss of telomeric DNA. Proc. Natl Acad. Sci. USA. 1999;96:14813–14818. doi: 10.1073/pnas.96.26.14813. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Li S, Rosenberg JE, Donjacour AA, Botchkina IL, Hom YK, Cunha GR, Blackburn EH. Rapid inhibition of cancer cell growth induced by lentiviral delivery and expression of mutant-template telomerase RNA and anti-telomerase short-interfering RNA. Cancer Res. 2004;64:4833–4840. doi: 10.1158/0008-5472.CAN-04-0953. [DOI] [PubMed] [Google Scholar]
- 19.Li S, Crothers J, Haqq CM, Blackburn EH. Cellular and gene expression responses involved in the rapid growth inhibition of human cancer cells by RNA interference-mediated depletion of telomerase RNA. J. Biol. Chem. 2005;280:23709–23717. doi: 10.1074/jbc.M502782200. [DOI] [PubMed] [Google Scholar]
- 20.Bagheri S, Nosrati M, Li S, Fong S, Torabian S, Rangel J, Moore DH, Federman S, Laposa RR, Baehner FL, et al. Genes and pathways downstream of telomerase in melanoma metastasis. Proc. Natl Acad. Sci. USA. 2006;103:11306–11311. doi: 10.1073/pnas.0510085103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Masutomi K, Possemato R, Wong JM, Currier JL, Tothova Z, Manola JB, Ganesan S, Lansdorp PM, Collins K, Hahn WC. The telomerase reverse transcriptase regulates chromatin state and DNA damage responses. Proc. Natl Acad. Sci. USA. 2005;102:8222–8227. doi: 10.1073/pnas.0503095102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Imamura S, Uchiyama J, Koshimizu E, Hanai J, Raftopoulou C, Murphey RD, Bayliss PE, Imai Y, Burns CE, Masutomi K, et al. A non-canonical function of zebrafish telomerase reverse transcriptase is required for developmental hematopoiesis. PLoS ONE. 2008;3:e3364. doi: 10.1371/journal.pone.0003364. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Ma Y, Creanga A, Lum L, Beachy PA. Prevalence of off-target effects in Drosophila RNA interference screens. Nature. 2006;443:359–363. doi: 10.1038/nature05179. [DOI] [PubMed] [Google Scholar]
- 24.Lu C, Fu W, Mattson MP. Telomerase protects developing neurons against DNA damage-induced cell death. Brain Res. Dev. Brain Res. 2001;131:167–171. doi: 10.1016/s0165-3806(01)00237-1. [DOI] [PubMed] [Google Scholar]
- 25.Rahman R, Latonen L, Wiman KG. hTERT antagonizes p53-induced apoptosis independently of telomerase activity. Oncogene. 2005;24:1320–1327. doi: 10.1038/sj.onc.1208232. [DOI] [PubMed] [Google Scholar]
- 26.Smith LL, Coller HA, Roberts JM. Telomerase modulates expression of growth-controlling genes and enhances cell proliferation. Nat. Cell Biol. 2003;5:474–479. doi: 10.1038/ncb985. [DOI] [PubMed] [Google Scholar]
- 27.Choi J, Southworth LK, Sarin KY, Venteicher AS, Ma W, Chang W, Cheung P, Jun S, Artandi MK, Shah N, et al. TERT promotes epithelial proliferation through transcriptional control of a Myc- and Wnt-related developmental program. PLoS Genet. 2008;4:e10. doi: 10.1371/journal.pgen.0040010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Sarin KY, Cheung P, Gilison D, Lee E, Tennen RI, Wang E, Artandi MK, Oro AE, Artandi SE. Conditional telomerase induction causes proliferation of hair follicle stem cells. Nature. 2005;436:1048–1052. doi: 10.1038/nature03836. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Matthaei KI. Genetically manipulated mice: a powerful tool with unsuspected caveats. J. Physiol. 2007;582:481–488. doi: 10.1113/jphysiol.2007.134908. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Zhang JZ. Overexpression analysis of plant transcription factors. Curr. Opin. Plant Biol. 2003;6:430–440. doi: 10.1016/s1369-5266(03)00081-5. [DOI] [PubMed] [Google Scholar]
- 31.Yang DS, Tandon A, Chen F, Yu G, Yu H, Arawaka S, Hasegawa H, Duthie M, Schmidt SD, Ramabhadran TV, et al. Mature glycosylation and trafficking of nicastrin modulate its binding to presenilins. J. Biol. Chem. 2002;277:28135–28142. doi: 10.1074/jbc.M110871200. [DOI] [PubMed] [Google Scholar]
- 32.Jacobs C, Pirson I. Pitfalls in the use of transfected overexpression systems to study membrane proteins function: the case of TSH receptor and PRA1. Mol. Cell Endocrinol. 2003;209:71–75. doi: 10.1016/j.mce.2003.07.002. [DOI] [PubMed] [Google Scholar]
- 33.Kirwan M, Dokal I. Dyskeratosis congenita: a genetic disorder of many faces. Clin. Genet. 2008;73:103–112. doi: 10.1111/j.1399-0004.2007.00923.x. [DOI] [PubMed] [Google Scholar]
- 34.Prowse KR, Greider CW. Developmental and tissue specific regulation of mouse telomerase and telomere length. Proc. Natl Acad. Sci. USA. 1995;92:4818–4822. doi: 10.1073/pnas.92.11.4818. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Hemann MT, Rudolph L, Strong M, DePinho RA, Chin L, Greider CW. Telomere dysfunction triggers developmentally regulated germ cell apoptosis. Mol. Biol. Cell. 2001;12:2023–2030. doi: 10.1091/mbc.12.7.2023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Wong KK, Chang S, Weiler SR, Ganesan S, Chaudhuri J, Zhu C, Artandi SE, Rudolph KL, Gottlieb GJ, Chin L, et al. Telomere dysfunction impairs DNA repair and enhances sensitivity to ionizing radiation. Nat. Genet. 2000;26:85–88. doi: 10.1038/79232. [DOI] [PubMed] [Google Scholar]
- 37.Armanios M. Syndromes of telomere shortening. Annu. Rev. Genomics Hum. Genet. 2009;10:45–61. doi: 10.1146/annurev-genom-082908-150046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Todaro GJ, Green H. Quantitative studies of the growth of mouse embryo cells in culture and their development into established lines. J. Cell Biol. 1963;17:299–313. doi: 10.1083/jcb.17.2.299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Johnson WE, Li C, Rabinovic A. Adjusting batch effects in microarray expression data using empirical Bayes methods. Biostatistics. 2007;8:118–127. doi: 10.1093/biostatistics/kxj037. [DOI] [PubMed] [Google Scholar]
- 40.Gonzalez-Suarez E, Geserick C, Flores JM, Blasco MA. Antagonistic effects of telomerase on cancer and aging in K5-mTert transgenic mice. Oncogene. 2005;24:2256–2270. doi: 10.1038/sj.onc.1208413. [DOI] [PubMed] [Google Scholar]
- 41.Park JI, Venteicher AS, Hong JY, Choi J, Jun S, Shkreli M, Chang W, Meng Z, Cheung P, Ji H, et al. Telomerase modulates Wnt signalling by association with target gene chromatin. Nature. 2009;460:66–72. doi: 10.1038/nature08137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Nowak MA, Boerlijst MC, Cooke J, Smith JM. Evolution of genetic redundancy. Nature. 1997;388:167–171. doi: 10.1038/40618. [DOI] [PubMed] [Google Scholar]
- 43.Terashima H, Yabuki N, Arisawa M, Hamada K, Kitada K. Up-regulation of genes encoding glycosylphosphatidylinositol (GPI)-attached proteins in response to cell wall damage caused by disruption of FKS1 in Saccharomyces cerevisiae. Mol. Gen. Genet. 2000;264:64–74. doi: 10.1007/s004380000285. [DOI] [PubMed] [Google Scholar]
- 44.Liu LF, Desai SD, Li TK, Mao Y, Sun M, Sim SP. Mechanism of action of camptothecin. Ann. NY Acad. Sci. 2000;922:1–10. doi: 10.1111/j.1749-6632.2000.tb07020.x. [DOI] [PubMed] [Google Scholar]
- 45.Shieh SY, Ikeda M, Taya Y, Prives C. DNA damage-induced phosphorylation of p53 alleviates inhibition by MDM2. Cell. 1997;91:325–334. doi: 10.1016/s0092-8674(00)80416-x. [DOI] [PubMed] [Google Scholar]
- 46.Rogakou EP, Boon C, Redon C, Bonner WM. Megabase chromatin domains involved in DNA double-strand breaks in vivo. J. Cell Biol. 1999;146:905–916. doi: 10.1083/jcb.146.5.905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Schultz LB, Chehab NH, Malikzay A, Halazonetis TD. p53 binding protein 1 (53BP1) is an early participant in the cellular response to DNA double-strand breaks. J. Cell Biol. 2000;151:1381–1390. doi: 10.1083/jcb.151.7.1381. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Ward IM, Minn K, Jorda KG, Chen J. Accumulation of checkpoint protein 53BP1 at DNA breaks involves its binding to phosphorylated histone H2AX. J. Biol. Chem. 2003;278:19579–19582. doi: 10.1074/jbc.C300117200. [DOI] [PubMed] [Google Scholar]
- 49.Erdmann N, Harrington LA. No attenuation of the ATM-dependent DNA damage response in murine telomerase-deficient cells. DNA Repair (Amst) 2009;8:347–353. doi: 10.1016/j.dnarep.2008.11.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Harper JW, Elledge SJ. The DNA damage response: ten years after. Mol. Cell. 2007;28:739–745. doi: 10.1016/j.molcel.2007.11.015. [DOI] [PubMed] [Google Scholar]
- 51.Takai H, Smogorzewska A, de Lange T. DNA damage foci at dysfunctional telomeres. Curr. Biol. 2003;13:1549–1556. doi: 10.1016/s0960-9822(03)00542-6. [DOI] [PubMed] [Google Scholar]
- 52.Flores I, Cayuela ML, Blasco MA. Effects of telomerase and telomere length on epidermal stem cell behavior. Science. 2005;309:1253–1256. doi: 10.1126/science.1115025. [DOI] [PubMed] [Google Scholar]
- 53.Lee J, Sung YH, Cheong C, Choi YS, Jeon HK, Sun W, Hahn WC, Ishikawa F, Lee HW. TERT promotes cellular and organismal survival independently of telomerase activity. Oncogene. 2008;27:3754–3760. doi: 10.1038/sj.onc.1211037. [DOI] [PubMed] [Google Scholar]
- 54.Barolo S. Transgenic Wnt/TCF pathway reporters: all you need is Lef? Oncogene. 2006;25:7505–7511. doi: 10.1038/sj.onc.1210057. [DOI] [PubMed] [Google Scholar]
- 55.Jackson AL, Linsley PS. Noise amidst the silence: off-target effects of siRNAs? Trends Genet. 2004;20:521–524. doi: 10.1016/j.tig.2004.08.006. [DOI] [PubMed] [Google Scholar]
- 56.Snove O, Jr, Holen T. Many commonly used siRNAs risk off-target activity. Biochem. Biophys. Res. Commun. 2004;319:256–263. doi: 10.1016/j.bbrc.2004.04.175. [DOI] [PubMed] [Google Scholar]
- 57.Cullen BR. Enhancing and confirming the specificity of RNAi experiments. Nat. Methods. 2006;3:677–681. doi: 10.1038/nmeth913. [DOI] [PubMed] [Google Scholar]
- 58.Xu L, Blackburn EH. Human cancer cells harbor T-stumps, a distinct class of extremely short telomeres. Mol. Cell. 2007;28:315–327. doi: 10.1016/j.molcel.2007.10.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Hemann MT, Strong M, Hao LY, Greider CW. The shortest telomere, not average telomere length, is critical for cell viability and chromosome stability. Cell. 2001;107:67–77. doi: 10.1016/s0092-8674(01)00504-9. [DOI] [PubMed] [Google Scholar]
- 60.Vulliamy T, Marrone A, Goldman F, Dearlove A, Bessler M, Mason PJ, Dokal I. The RNA component of telomerase is mutated in autosomal dominant dyskeratosis congenita. Nature. 2001;413:432–435. doi: 10.1038/35096585. [DOI] [PubMed] [Google Scholar]
- 61.Armanios M, Chen JL, Chang YP, Brodsky RA, Hawkins A, Griffin CA, Eshleman JR, Cohen AR, Chakravarti A, Hamosh A, et al. Haploinsufficiency of telomerase reverse transcriptase leads to anticipation in autosomal dominant dyskeratosis congenita. Proc. Natl Acad. Sci. USA. 2005;102:15960–15964. doi: 10.1073/pnas.0508124102. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.