Abstract
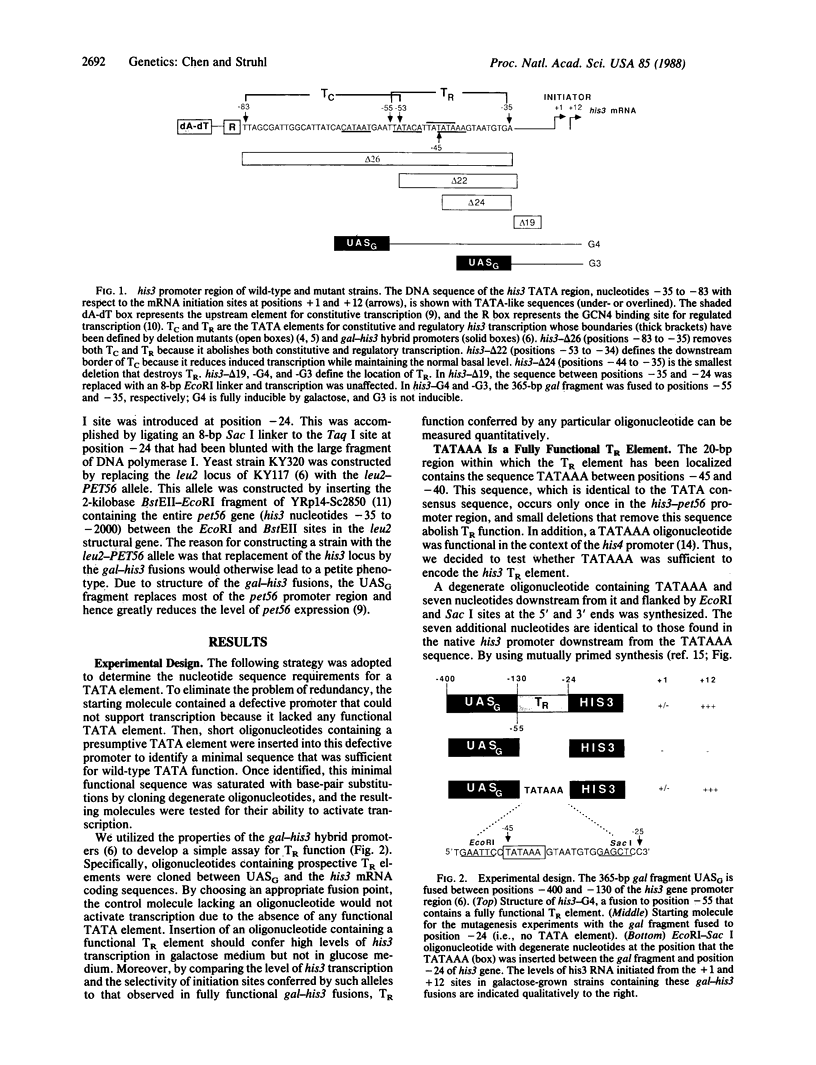
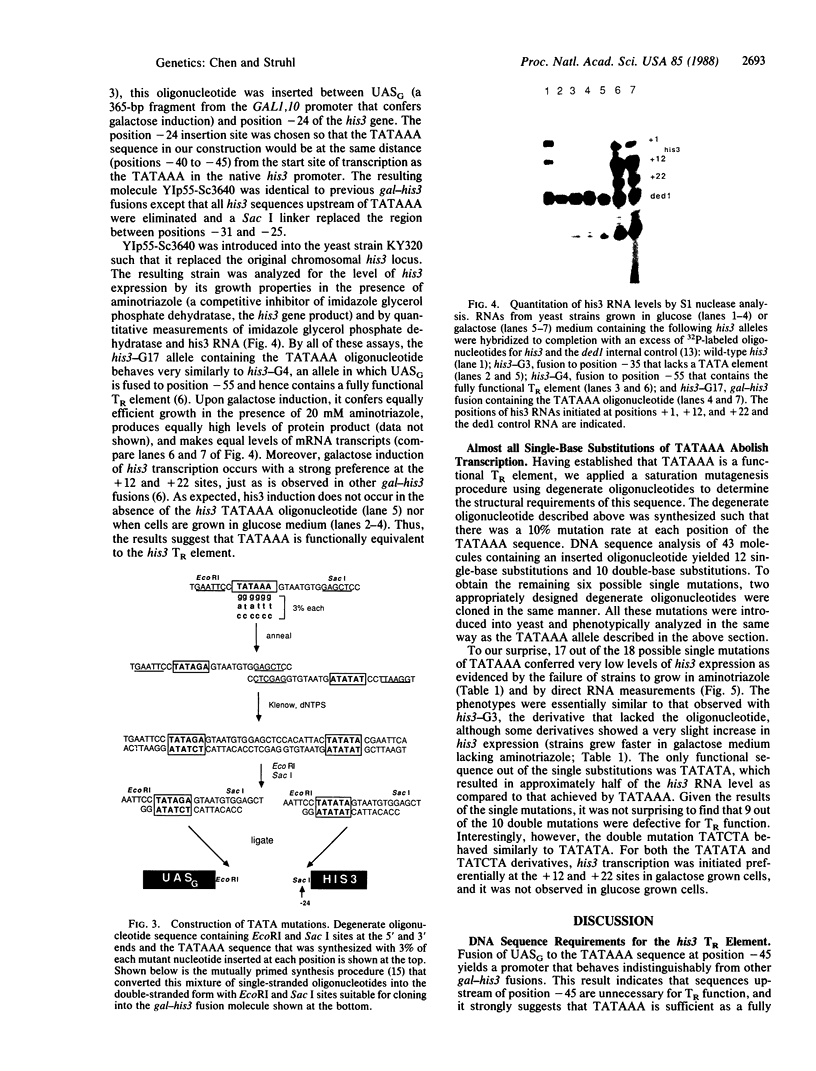
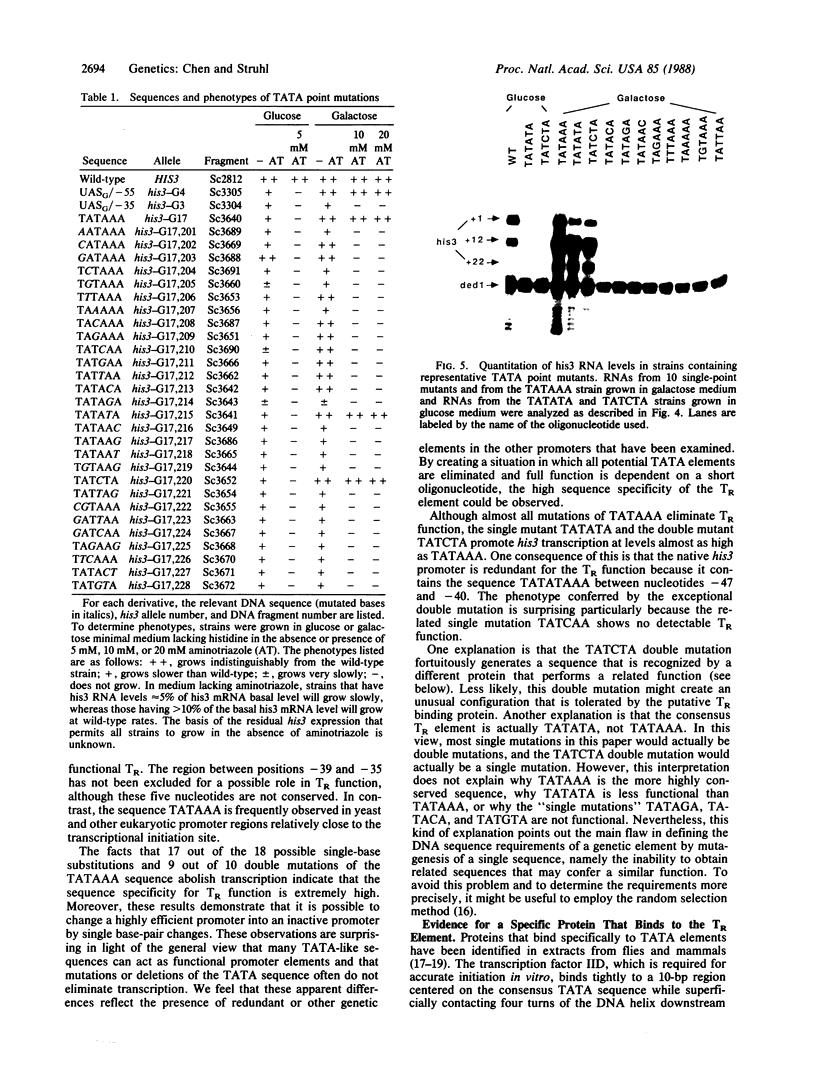
The yeast his3 promoter region contains two distinct classes of "TATA elements," constitutive (TC) and regulatory (TR), that are defined by their interactions with upstream promoter elements, selectivity of initiation sites, and chromatin structure. TC is localized between positions -83 and -53, and TR is localized between positions -55 and -35, regions in which there are several TATA-like sequences. In this study, we used saturation mutagenesis to examine the structural requirements of the his3 TR element necessary for transcriptional induction. To avoid the complications of redundant elements, the phenotypic analysis was carried out by using a gal-his3 hybrid promoter whose function depends on a short oligonucleotide containing the prospective his3 TR element. In this context, an oligonucleotide containing the sequence TATAAA is sufficient for TR function. However, 17 out of the 18 possible single-base substitutions and 9 out of 10 double mutations of this sequence abolish TR function. This strict sequence requirement for TR function strongly suggests that the TR element is a target site for a sequence-specific DNA-binding protein. Further, as the region encoding TC and promoters of certain other yeast genes do not contain a sequence that is compatible with TR function, we suggest that yeast cells contain multiple proteins with distinct sequence specificities that carry out a related "TATA function" and that yeast promoters can be divided into classes based on their downstream promoter elements.
Full text
PDF

Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Chen E. Y., Seeburg P. H. Supercoil sequencing: a fast and simple method for sequencing plasmid DNA. DNA. 1985 Apr;4(2):165–170. doi: 10.1089/dna.1985.4.165. [DOI] [PubMed] [Google Scholar]
- Chen W., Tabor S., Struhl K. Distinguishing between mechanisms of eukaryotic transcriptional activation with bacteriophage T7 RNA polymerase. Cell. 1987 Sep 25;50(7):1047–1055. doi: 10.1016/0092-8674(87)90171-1. [DOI] [PubMed] [Google Scholar]
- Davison B. L., Egly J. M., Mulvihill E. R., Chambon P. Formation of stable preinitiation complexes between eukaryotic class B transcription factors and promoter sequences. Nature. 1983 Feb 24;301(5902):680–686. doi: 10.1038/301680a0. [DOI] [PubMed] [Google Scholar]
- Guarente L. Regulatory proteins in yeast. Annu Rev Genet. 1987;21:425–452. doi: 10.1146/annurev.ge.21.120187.002233. [DOI] [PubMed] [Google Scholar]
- Hope I. A., Struhl K. GCN4 protein, synthesized in vitro, binds HIS3 regulatory sequences: implications for general control of amino acid biosynthetic genes in yeast. Cell. 1985 Nov;43(1):177–188. doi: 10.1016/0092-8674(85)90022-4. [DOI] [PubMed] [Google Scholar]
- Losick R., Pero J. Cascades of Sigma factors. Cell. 1981 Sep;25(3):582–584. doi: 10.1016/0092-8674(81)90164-1. [DOI] [PubMed] [Google Scholar]
- McKnight S., Tjian R. Transcriptional selectivity of viral genes in mammalian cells. Cell. 1986 Sep 12;46(6):795–805. doi: 10.1016/0092-8674(86)90061-9. [DOI] [PubMed] [Google Scholar]
- Nagawa F., Fink G. R. The relationship between the "TATA" sequence and transcription initiation sites at the HIS4 gene of Saccharomyces cerevisiae. Proc Natl Acad Sci U S A. 1985 Dec;82(24):8557–8561. doi: 10.1073/pnas.82.24.8557. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oettinger M. A., Struhl K. Suppressors of Saccharomyces cerevisiae his3 promoter mutations lacking the upstream element. Mol Cell Biol. 1985 Aug;5(8):1901–1909. doi: 10.1128/mcb.5.8.1901. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oliphant A. R., Nussbaum A. L., Struhl K. Cloning of random-sequence oligodeoxynucleotides. Gene. 1986;44(2-3):177–183. doi: 10.1016/0378-1119(86)90180-0. [DOI] [PubMed] [Google Scholar]
- Oliphant A. R., Struhl K. The use of random-sequence oligonucleotides for determining consensus sequences. Methods Enzymol. 1987;155:568–582. doi: 10.1016/0076-6879(87)55037-6. [DOI] [PubMed] [Google Scholar]
- Parker C. S., Topol J. A Drosophila RNA polymerase II transcription factor contains a promoter-region-specific DNA-binding activity. Cell. 1984 Feb;36(2):357–369. doi: 10.1016/0092-8674(84)90229-0. [DOI] [PubMed] [Google Scholar]
- Sawadogo M., Roeder R. G. Interaction of a gene-specific transcription factor with the adenovirus major late promoter upstream of the TATA box region. Cell. 1985 Nov;43(1):165–175. doi: 10.1016/0092-8674(85)90021-2. [DOI] [PubMed] [Google Scholar]
- Selleck S. B., Majors J. Photofootprinting in vivo detects transcription-dependent changes in yeast TATA boxes. Nature. 1987 Jan 8;325(7000):173–177. doi: 10.1038/325173a0. [DOI] [PubMed] [Google Scholar]
- Struhl K. Constitutive and inducible Saccharomyces cerevisiae promoters: evidence for two distinct molecular mechanisms. Mol Cell Biol. 1986 Nov;6(11):3847–3853. doi: 10.1128/mcb.6.11.3847. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Struhl K. Direct selection for gene replacement events in yeast. Gene. 1983 Dec;26(2-3):231–241. doi: 10.1016/0378-1119(83)90193-2. [DOI] [PubMed] [Google Scholar]
- Struhl K. Genetic properties and chromatin structure of the yeast gal regulatory element: an enhancer-like sequence. Proc Natl Acad Sci U S A. 1984 Dec;81(24):7865–7869. doi: 10.1073/pnas.81.24.7865. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Struhl K. Naturally occurring poly(dA-dT) sequences are upstream promoter elements for constitutive transcription in yeast. Proc Natl Acad Sci U S A. 1985 Dec;82(24):8419–8423. doi: 10.1073/pnas.82.24.8419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Struhl K. Nucleotide sequence and transcriptional mapping of the yeast pet56-his3-ded1 gene region. Nucleic Acids Res. 1985 Dec 9;13(23):8587–8601. doi: 10.1093/nar/13.23.8587. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Struhl K. Promoters, activator proteins, and the mechanism of transcriptional initiation in yeast. Cell. 1987 May 8;49(3):295–297. doi: 10.1016/0092-8674(87)90277-7. [DOI] [PubMed] [Google Scholar]
- Struhl K. The yeast his3 promoter contains at least two distinct elements. Proc Natl Acad Sci U S A. 1982 Dec;79(23):7385–7389. doi: 10.1073/pnas.79.23.7385. [DOI] [PMC free article] [PubMed] [Google Scholar]