Abstract
Thrombin is the pivotal serine protease enzyme in the blood cascade system. Phe-Pro-Arg-chloromethylketone (PPACK), phosphate and phosphonate ester inhibitors form a covalent bond with the active-site Ser of thrombin. PPACK, a mechanism-based inhibitor, and the phosphate/phosphonate esters form adducts that mimic intermediates formed in reactions catalyzed by thrombin. Therefore, the dependence of the inhibition of human α-thrombin on the concentration of these inhibitors, pH, and temperature was investigated. The second-order rate constant, ki/Ki, and the inhibition constant, Ki, for inhibition of human α-thrombin by PPACK are (1.1 ± 0.2) × 107 M−1 s−1 and (2.4 ± 1.3) × 10−8 M at pH 7.00 in 0.05 M phosphate buffer, 0.15 M NaCl, and 25.0 ± 0.1˚C, and in good agreement with previous reports. The activation parameters at pH 7.00, 0.05M phosphate buffer 0.15 M NaCl, are ΔH‡ = 10.6 ± 0.7 kcal/mol and ΔS‡ = 9 ± 2 cal/mol deg. The pH dependence of the second-order rate constants of inhibition is bell shaped. Values of pKa1 and pKa2 are 7.3 ± 0.2 and 8.8 ± 0.3, respectively, at 25.0 ± 0.1 °C. A phosphate and a phosphonate ester inhibitor gave higher values, 7.8 and 8.0, for pKa1 and 9.3 and 8.6 for pKa2. They inhibit thrombin over six orders of magnitude less efficiently than PPACK does. The deuterium solvent isotope effect for the second-order rate constant at pH 7.0 and 8.3 at 25.0 ± 0.1°C is unity within experimental error in all three cases, indicating the absence of proton transfer in the rate-determining step for the association of thrombin with the inhibitors. But in a 600 MHz 1H NMR spectrum of the inhibition adduct at pH 6.7 and 30 °C, a peak at 18.10 ppm with respect to TSP appears with PPACK, which is absent in the 1H NMR spectrum of a solution of the enzyme between pH 5.3–8.5. The peak at low field is an indication of the presence an SSHB at the active site in the adduct. The deuterium isotope effect on this hydrogen bridge is 2.2 ± 0.2 (ϕ = 0.45). The presence of an SSHB is also established with a signal at 17.34 ppm for a dealkylated phosphate adduct of thrombin.
Keywords: Enzyme mechanisms, blood cascade enzymes, solvent isotope effects, short strong hydrogen bonds, 1H NMR
Thrombin is the pivotal serine protease enzyme in the blood cascade system.(1–6) Thrombin is a highly specific and efficient catalyst of the hydrolysis of one or two peptide bonds in large precursor proteins of blood clotting.(6–11) In fact, thrombin fulfills a dual role: procoagulant and anticoagulant. The two are coordinated in a sophisticated manner. As the control of blood clotting has broad implications in human health, the regulation of human α-thrombin by a broad range of inhibitors has been a main target of investigations and drug design.(12–15) Small-molecule inhibitors, which may not be efficient enough from a medical point of view, serve as great probes of the mechanisms of thrombin action. PPACK is the most effective mechanism-based affinity label of a serine protease. It forms a covalent bond with the active-site Ser of thrombin and cross links with His57 at the active site.(16–19) PPACK forms a tetrahedral adduct with thrombin, which should be a good mimic of intermediates formed in the acylation of thrombin in the reactions it catalyzes. The great potency of PPACK lies in the composition of the peptide portion of the inhibitor, which complements the S1-S3 subsites of thrombin: a critical Arg in the P1 position, a Pro in the P2 position, and a hydrophobic Phe in the P3 position.
The mechanism of inhibition of thrombin by these small-molecule inhibitors begins similarly to the binding of the normal substrate. Thrombin, as a serine protease, contains a catalytic-triad consisting of Ser195, His57 and Asp102.(3;20–24) Ser195 is the nucleophile which is activated by general-base catalysis of proton removal by His57. Asp102 acts in tandem as it holds His57 in place via a hydrogen bond. Nucleophilic attack by Ser195 at the amide carbonyl group of the substrate results in the formation of a tetrahedral intermediate, which is stabilized by main-chain amides in the oxyanion hole for binding the oxyanion. A proton from His57 is then donated to the N of the leaving group in the tetrahedral intermediate, which causes its collapse and release of the first product peptide/protein and the formation the acylenzyme intermediate. The acylenzyme intermediate is attacked by water with general-base assistance from His57 and Asp102 to form another tetrahedral intermediate. The collapse of this tetrahedral intermediate leads to the release of the second product regenerating the starting enzyme. In laboratory experiments, especially enzyme activity assays, chromogenic or fluorogenic activated oligopeptide substrates are used.(23) Upon thrombin-catalyzed hydrolysis, these peptide amides release the leaving group in stoichiometric amount to substrate loss.
It has been proposed that the hydrogen bond donor-acceptor distances across the catalytic triad contract during catalysis, which lowers the activation barrier for the subject reaction.(25–27) Because thrombin is a very efficient catalyst of the breakdown of its natural and analytical substrates, it is likely to employ such structural change to stabilize the transition states for hydrolysis of its substrates. This notion is supported by the observed large solvent deuterium isotope effects in the hydrolysis of many thrombin substrates.(22;28) These isotope effects between 2.5 and 3.5 are most likely primary effects, meaning that protons are transferred in the rate-determining step of the hydrolysis reaction. The origin of this difference between proton and deuteron transfer at the transition state lies in the loss of the difference in zero-point energies existing in the ground state vibration of H/D in bonds to heavy atoms. (25;26;29–32)
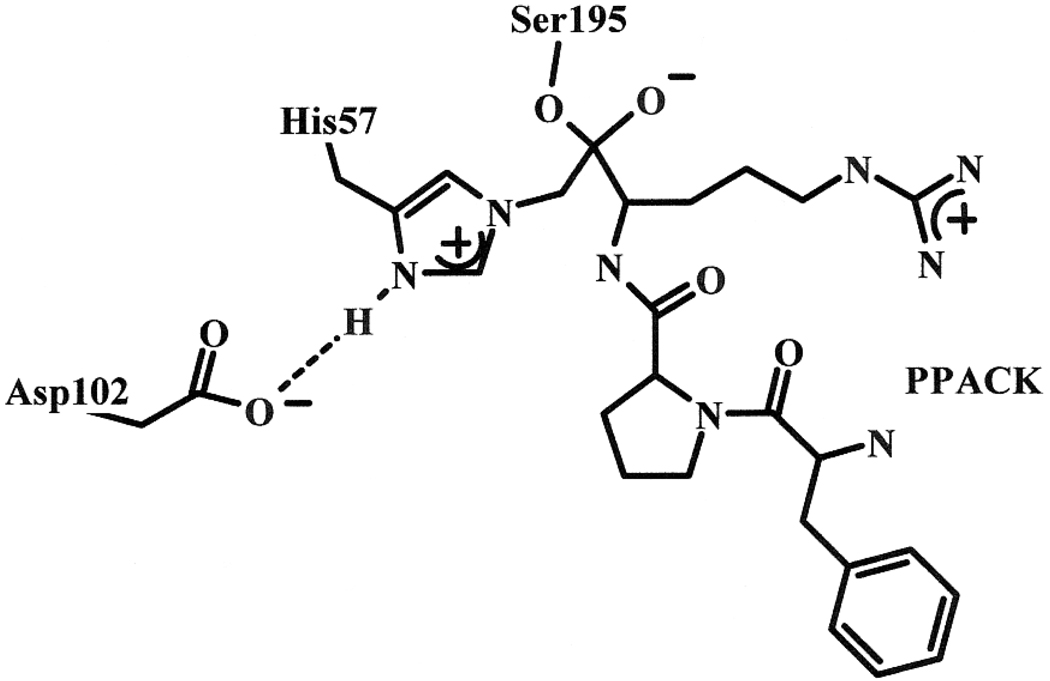
Another probe of hydrogen bridges occurring at the active site of enzymes in transition-state analogs has been used. A unique signal appears in high-resolution 1H NMR spectra at low field between 14 and 21 ppm, in many cases of transition-state-analog adducts of enzymes with inhibitors that form tetrahedral intermediates.(33–45) The low-field signals can also be observed with some native enzymes at pH below 6. The phenomena have been interpreted as the presence of a short-strong-hydrogen bond (SSHB) at the active site of the enzyme. It forms upon protonation of a key base catalyst, which occurs even at pH above 6 when interacting with a modifier. It was shown that the hydrogen bond is most likely one formed between His57δNH and Asp102γO in serine proteases.(46–49) The best affinity label of thrombin is PPACK.(16;18;19) When Ser195 adds to the carbonyl group on PPACK, the tetrahedral intermediate formed freezes as no bond can be cleaved around the central carbonyl C. In fact, due to the vicinity of the nucleophilic His57εN, the methylene C subsequently cross links by alkylating His57 and displacing the chloride ion leaving group, as shown on Figure 1. This results in irreversible inhibition of the enzyme by PPACK. The correspondence between the subsites of the enzyme and the positions of the amino acid residues on the inhibitor enforce tight interactions at the active site. In turn, the H-bonds in the catalytic triad may become compressed, which may be detected by high-resolution 1H NMR measurements.(34)
Figure 1.
Structure of PPACK covalently linked to the catalytic triad of thrombin (modified from Brookhaven Protein Data Bank entry 1SHH).(50)
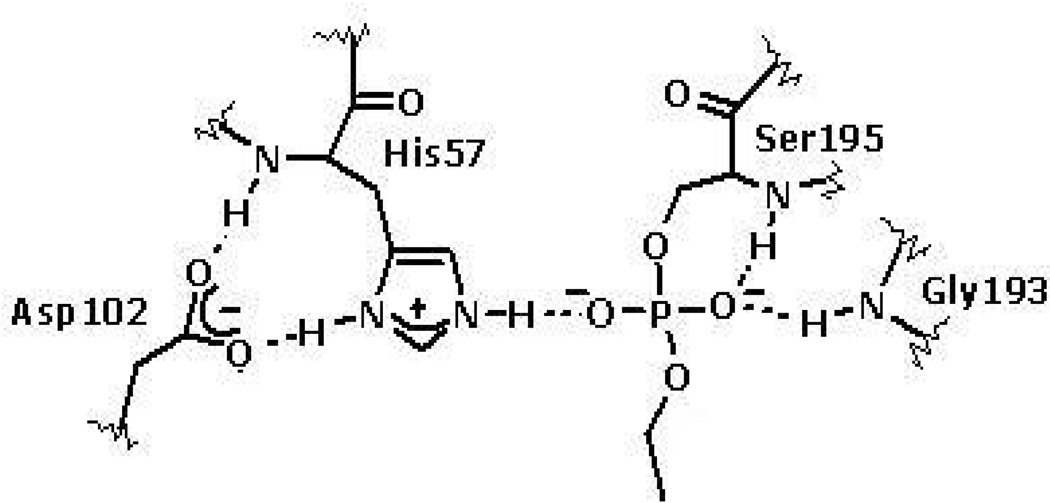
Phosphate and phosphonate esters have been broadly used for the inhibition of serine hydrolases. They also attach covalently to the active site Ser and the resulting ester generally resists nucleophilic attack at P due to the surrounding negative charge density, which is further exacerbated if one of the ligands dealkylates or hydrolyzes from the central P atom shown in Figure 2 for 4-nitrophenyl diethylphosphate (paraoxon).(51) The covalent phosphate or phosphonate adducts resemble the tetrahedral intermediate formed after water attack on the acyl enzyme in the hydrolysis of substrates.(52–59)
Figure 2.
The structure of paraoxon-inhibited thrombin after deethylation (modified using Brookhave Protein Data Bankd structure 1SHH).(50)
In this work, the main goal was to find and characterize SSHBs that α-human thrombin forms with mechanism-based inhibitors that mimic intermediates occurring in its catalytic reaction with its natural substrates. The dependence of the inhibition of human α-thrombin by PPACK on inhibitor concentration, pH, and temperature were investigated first. The pH dependences of inhibition by a phosphate and a phosphonate ester inhibitor were also investigated to determine the pH optimum for kinetic solvent isotope effect studies. One salient result of this study is that kinetic solvent isotope effects are near one on the second-order rate constants for inhibition indicating the absence of proton transfer in the rate-determining step for the association of thrombin with PPACK and with the weaker phospohonate ester inhibitors,. But in a 600 MHz 1H NMR spectrum of the covalently modified thrombin at pH 6.7 and 30 °C, a peak at 18.10 ppm appears with PPACK and one at 17.34 ppm with paraoxon after deethylation, which are absent in the 1H NMR spectrum of a solution of the enzyme between pH 5.3 and 8.5. The peak at low field is typical of a SSHB forming at the active site in the adduct. The deuterium isotope effect on this hydrogen bridge is 2.2 ± 0.2 in the PPACK-modified thrombin. The SSHB is robust as proven in temperature studies of the line width of the resonance.
Materials and Methods
Materials
Anhydrous dimethyl sulfoxide (DMSO), heavy water with 99.9 % deuterium content and anhydrous methanol, were purchased from Aldrich Chemical Co. All buffer salts were reagent grade and were purchased from either Aldrich, Fisher, or Sigma Chemical Co. The proton sponge 1,8-bis(dimethylamino) naphthalene was from Sigma Chemical Co. H-D-Phe-Pip-Arg-4-nitroanilide.2HCl (pNA) (S-2238) 99% (TLC) was purchased from Diapharma Group Inc.. PPACK was purchased from BioMol. Paraoxon was from Aldrich Chemical Co. and the sarin analogue, 4-nitrophenyl-2-propyl methylphosphonate (NPMP) was synthesized in this lab previously.(35;60) Human α-thrombin, 36,500 Da, 3181 NIH u/mg activity (∼98% purity) in pH 6.5, 0.05 M sodium citrate buffer, 0.2 M NaCl, 0.1% PEG-8000 was purchased from Enzyme Research Laboratories.
Instruments
Spectroscopic measurements were performed with a Perkin-Elmer Lambda 6 or Lambda 35 UV-Vis Spectrophotometer connected to a PC. The temperature was monitored using a temperature probe connected to a digital readout device. Either a Neslab RTE-4 or a Lauda 20 circulating water bath was used for temperature control. 1H NMR spectra of inhibited human α-thrombin were obtained by a Varian Inova 600 MHz NMR instrument at Olson Labs, Department of Chemistry, Rutgers University, Newark, NJ.
Solutions
Buffer solutions were prepared from the appropriate analytical grade salts using double distilled deionized water. Buffers were prepared by weight from Tris-base and Tris-HCl at 0.02 M, 0.15 M NaCl, 0.1% PEG4000 at pH 8.0 for enzyme assays. All other buffers were 0.05 M of the respective buffer salt and 0.15 M of NaCl with 0.1% PEG-4000 added. Calculated amounts of HCl or NaOH were used to adjust the buffer pH when needed. All buffers were further filtered using a 0.2 µm Nylon Membrane Filter. Buffers for pH-dependence studies were as follows; phosphate for pH 6.03 – 7.47, citrate for pH 6.52, HEPES for pH 7.49 – 7.79 and barbital for pH 8.07 and 8.54, while 0.05 M Tris or Bis-Tris was used between pH 8.20 and 9.60. For the isotope effect study D2O buffers were made identically to the aqueous buffer with phosphate at pH 7.47, pD 8.25 and with Tris at pH 8.07, pD 8.87. pD values were calculated from the pH electrode reading plus 0.4.(22;29–32) A Delta electronic pH meter was used for pH measurements.
Thrombin Activity Assay
Assays were carried out in the absence or presence of inhibitors in 1.0 mL total volume with 10 µL injection of each an appropriate stock solution of the substrate in DMSO and a thrombin stock solution. Initial rates of hydrolysis of 3–5 × 10−5 M (>10 Km) S-2238, were measured by monitoring the release of 4-nitroaniline at 400 nm for 10–60 sec. Thrombin concentrations, [E], were then calculated from the Vmax values using kcat = 95 ± 20 s−1 in pH 8.0, 0.02 M Tris buffer at 25.0 ± 0.1 °C.(22)
Kinetic Procedures Using Discontinuous Sampling
Inhibition of thrombin with PPACK
The thrombin stock solution was diluted in the appropriate buffer to result in a 20 mL solution of 3 – 5×10−1 M concentration. An aliquot of 980 µL was incubated in a cuvette in the temperature-controlled cell compartment of the spectrophotometer to reach thermal equilibrium. Thrombin inhibition was initiated by injecting 10 µL of a thermally equilibrated 1.1 × 10−7 to 3.65 × 10−7 M PPACK solution and mixing. The reaction mixture was promptly incubated for a time interval between 15 to 200 sec after which 10 µL of 3–5 × 10−3 M solution of S-2238 substrate in DMSO was quickly added to measure the remaining enzyme activity. Pseudo-first-order rate constants were calculated by fitting the exponential function to the remaining enzyme activity versus time of inhibition for ten successive incubation times. Thrombin activity remained constant for the duration of the experiment in the 288–308 K temperature range.
Inactivation of α-thrombin with paraoxon and MPNP
In a typical experiment, 250 µL of a ∼5 × 10−7 M solution of thrombin buffered at the desired pH was prepared An aliquot of 180 µL of the resultant solution was drawn, 10 µL of either 0.014 M paraoxon solution in methanol or 0.0014 M MPNP solution in 10−3 M HCl was added and the reaction mixture was incubated under temperature control. Ten µL aliquots were drawn at appropriate time intervals and the reaction was quenched by dilution into a cuvette containing 980 µL pH 8.00, 0.02M Tris buffer. Ten µL of 3–5 × 10−3 M S-2238 in DMSO was added and the cell inverted several times to initiate the assay reaction. Pseudo-first-order rate constants for inhibition were calculated from the declining activities of thrombin for 4 half-lives. The second-order rate constants were calculated from kobs divided by the concentration of the inhibitor.
Low-field 1H NMR measurements
The samples were prepared by mixing 0.2–0.5 mM thrombin with three-fold excess of PPACK or 10-fold excess of paraoxon or NPMP in pH 6.7, 0.02 M citrate buffer, 0.01 M phosphate buffer, 0.2 M NaCl, 0.1% PEG8000. The D2O content was ∼7% in general and ∼ 45–55% for the isotope effect studies. Thrombin activity was measured before and after inhibition. A 99% loss of activity was achieved with PPACK and 90% loss of activity was obtained with paraoxon and NPMP. The 1H NMR samples were probed for protein aggregation by SDS-PAGE-electrophoresis. Only one band corresponding to the 36,500 Da of thrombin was obtained.
A 5 mm triple resonance probe was used. Water excitation was avoided by using a 1331 pulse sequence and a 90° pulse width of 30 µs was applied for a 512 ms acquisition time including a 2.5 s relaxation delay. One dimensional 1H NMR spectra were acquired for free and covalently modified thrombin at 5 – 40 °C with 1000–4000 transients. The phosphate and phosphonate ester-inhibited thrombin samples were allowed to "age" for 50 hours and were then reexamined. D/H fractionation factors were measured from deshielded resonances at maximal sensitivity by dividing samples identically into two, one in buffered H2O and the other in identically buffered D2O. The proton sponge 1,8-bis(dimethylamino) naphthalene was dissolved in CD3CN then titrated with H2SO4. It was placed in a capillary to serve as an external chemical shift reference for quantitation of deshielded resonances.
Data Analysis
The irreversible inhibition of thrombin by the covalent inhibitors (I) can be modeled by the following equation:(18;61)
| Equation 1 |
In this model Ki is an equilibrium constant, kd/ka, where ka is the rate constant for the association of the enzyme and the inhibitor to form the enzyme inhibitor complex (EI) while kd is the rate constant for the dissociation of the enzyme and inhibitor from the EI complex. After the EI complex is formed, ki is the first-order rate constant for bond formation between the enzyme and inhibitor to result in EI*, the covalently modified thrombin. This is a minimalistic approach especially for thrombin inhibition with a chloromethyl ketone as PPACK, which forms a hemiketal anion ensued by cross alkylation.(62) However, we have not employed the requisite mechanistic probes for an elucidation of the relative rates of individual steps of inhibition leading to loss of enzyme activity.
The rate constant observed in the experiments has a hyperbolic dependence on [I] according to the Michaelis-Menten formalism shown in equation 2:(23)
| Equation 2 |
Alternatively, as the concentration of the inhibitors were typically well below Ki, the observed rate constant was modeled by equation 3:(18)
| Equation 3 |
Activation parameters were calculated by nonlinear fitting of the Eyring equation (Eq 4) to the temperature dependence of the second-order rate constants, measured with 1.47 × 10−9 M PPACK at pH 7.0.
| Equation 4 |
where kB is the Boltzman constant, h is Planck’s constant, R is the gas constant, T is the temperature, ΔS‡ is the activation entropy and ΔH‡ is the activation enthalpy to reach the transition state.(63) The data were plotted according to the linearized Eyring equation:
| Equation 5 |
The pH dependence of the pseudo-first-order rate constant of inhibition by PPACK, paraoxon and MPNP was plotted and evaluated according to a single pKa and to a two-pKa model (Eq 6).(23) The latter gave more consistent and precise results.
| Equation 6 |
All fitting was at the 95% confidence level using simple robust or explicit error propagation (from the inverse of the errors in the dependent variable) using GraFit 5.(64) Structures were drawn using program VMD, Visual Molecular Dynamics.(65)
NMR data analysis was performed with the Mestrelab Research software or using SpinWorks. Fractionation factors (ϕ)were calculated from the integrated signals in 7% and 55% D2O buffers as follows:
I = [Imax(X)] ϕ/(1-X)+X], where X = mole fraction of H2O; I = observed intensity and Imax is maximal intensity at X = 1.0.
Results
Kinetic Experiments
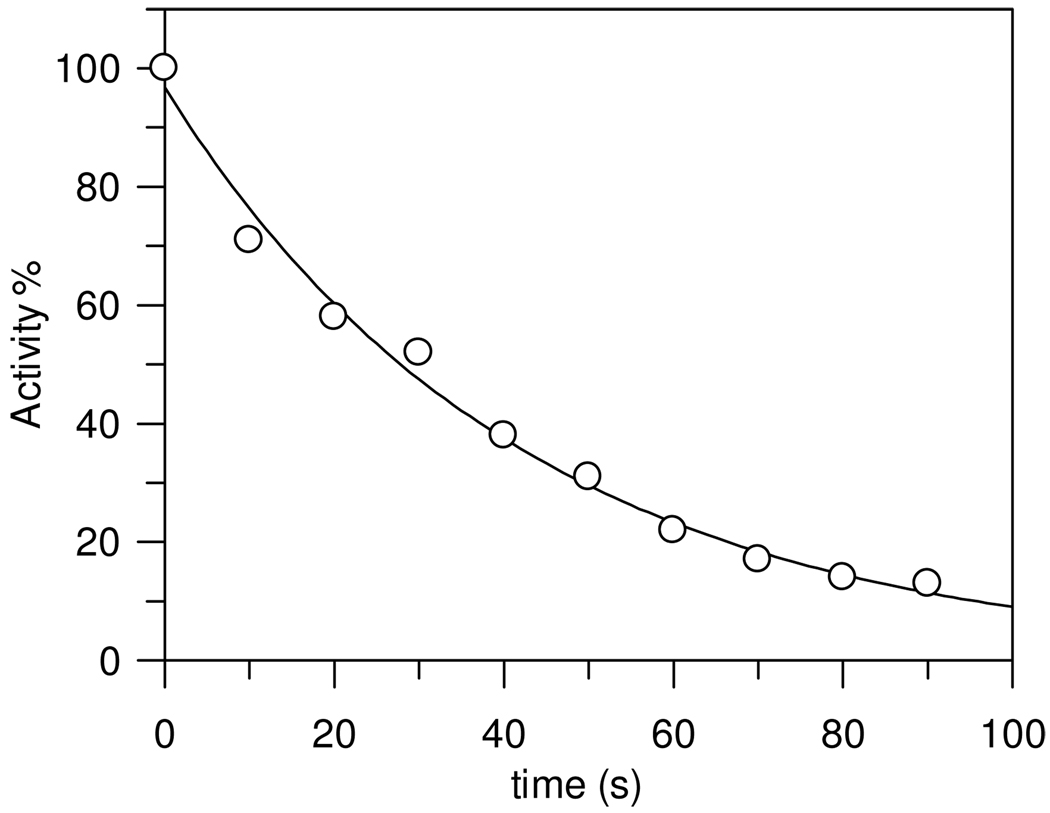
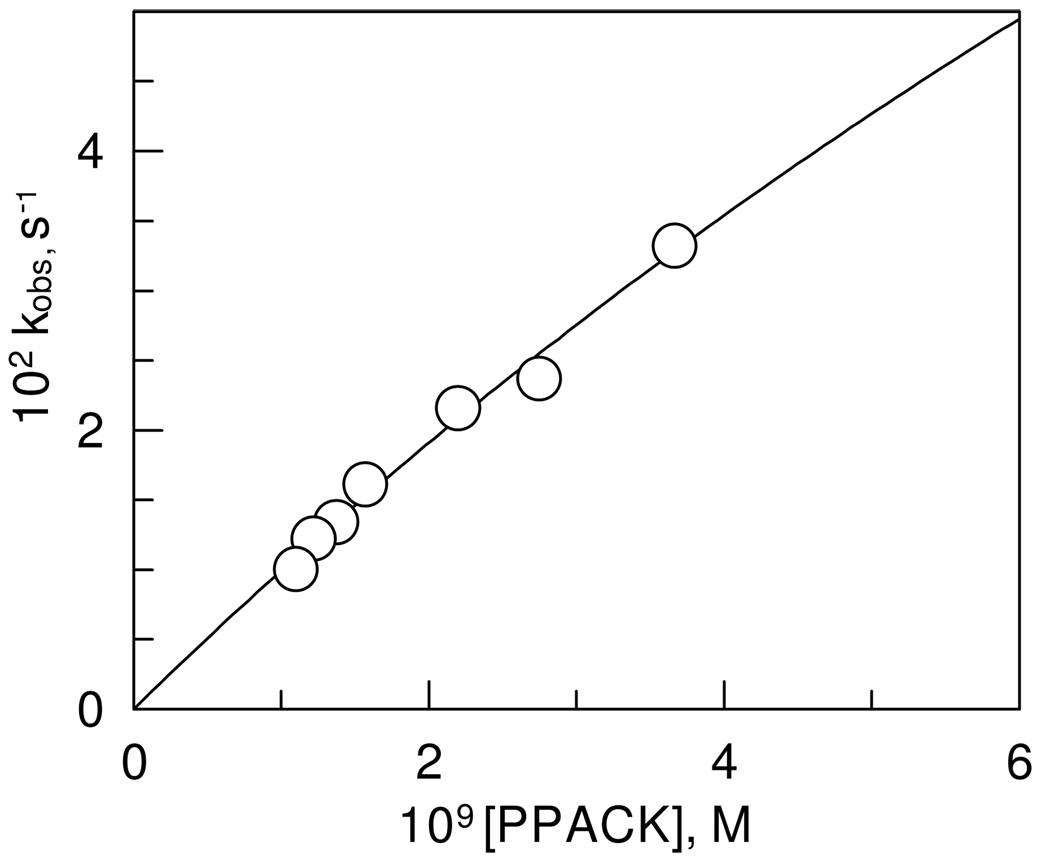
The time dependence of decreasing thrombin activity in the presence of PPACK in the concentration rage of 1.1 × 10−9 M to 3.7 × 10−9 M was exponential approaching first order as shown in Figure 3. The second-order rate constant calculated from the observed rate constant is ki/Ki = 1.1 ± 0.2 × 107 M−1 s−1 at pH 7.00, 0.05 M phosphate buffer, 25.0 ± 0.1 °C. This is in very good agreement with an estimate of Kettner and Shaw, ki/Ki of 1.15 × 107 M−1 s−1, at pH 7.0, 0.05 M PIPES buffer and 25 °C for bovine thrombin.(16;18) More recent studies of the inhibition of human α-thrombin with PPACK using different methods gave similar results.(19) However, the observed first-order rate constants showed a non-linear dependence on PPACK concentration and equation 2 was fitted to the data to obtain values of ki = 0.25 ± 0.12 s−1 and Ki = (2.4 ± 1.3) × 10−8 M (Figure 4). The large errors in these parameters are due to the difficulty in obtaining rate constants by conventional methods at concentrations of PPACK above the range on Figure 4, which would reach saturation of the enzyme with PPACK.
Figure 3.
Time-dependent decline in human α-thrombin activity in the presence of 2.2 × 10−9 M PPACK at pH 7.47, 0.05 M phosphate buffer NaCl 0.15 M, and 25.0 ± 0.1 °C. One standard deviation in the data is indicated by the diameter of the circle
Figure 4.
Dependence of the observed rate constants on the concentration of PPACK at pH 7.00, 0.05 M phosphate buffer, NaCl 0.15 M, and 25.0 ± 0.1 °C. One standard deviation in the data is indicated by the diameter of the circle.
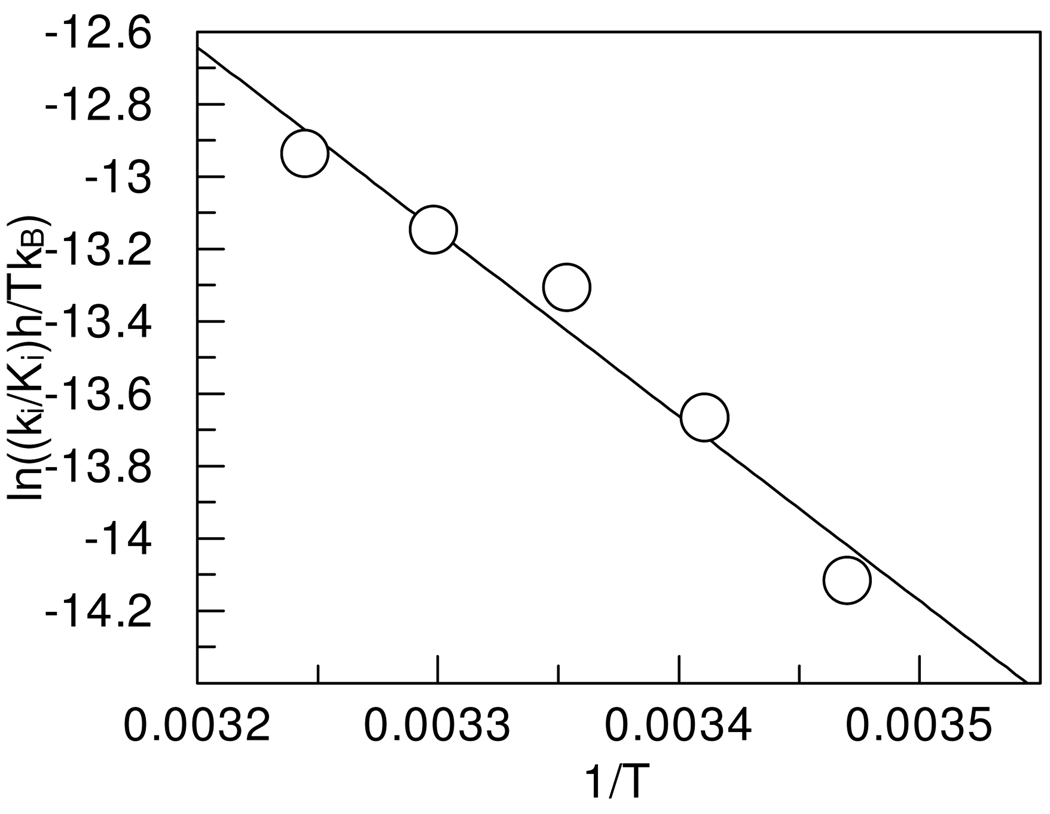
The temperature dependence of the inhibition of thrombin with PPACK at pH 7.00 was evaluated from the Eyring equation and is shown in linear form in Figure 5. The values of ΔH‡ and ΔS‡ were calculated to be 10.6 ± 0.7 kcal/mol and 9 ± 2 cal/mol K, respectively.
Figure 5.
Temperature dependence of the second-order rate constants for the inhibition of thrombin by PPACK at pH 7.00, 0.05 M phosphate buffer NaCl 0.15 M. One standard deviation in the data is indicated by the diameter of the circle.
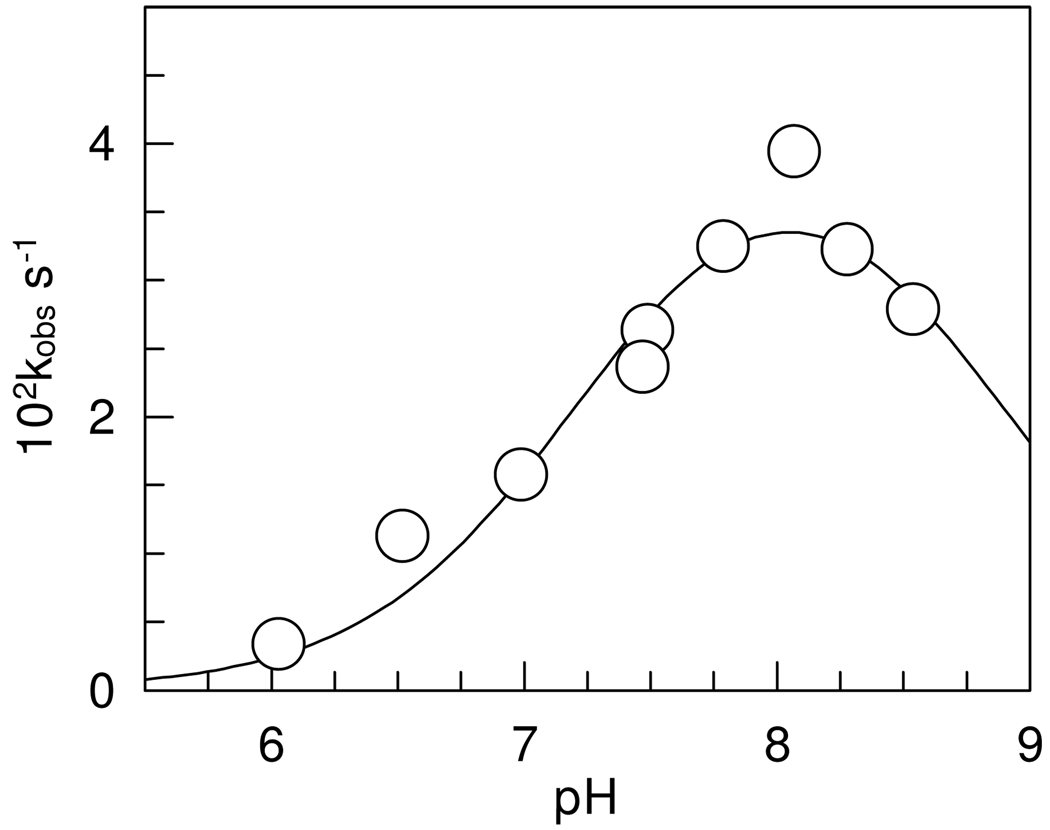
The pH dependence of the first-order inhibition rate constants was bell shaped and gave a maximum of 4.7 ± 0.8 × 10−2 s−1, corresponding to 2.15 × 10 7 M−1 s−1 at pH 8.1, as shown on Figure 6. The calculated pKas are 7.3 ± 0.2 and 8.8 ± 0.3. Fitting a single pKa model to the data gave a 0.2 unit smaller pKa but the parameters had greater errors than those calculated with the two pKa model.
Figure 6.
pH dependence of the pseudo first-order rate constants for the inhibition of human α-thrombin by PPACK at NaCl 0.15 M and 25.0 ± 0.1°C. One standard deviation in the data is indicated by the diameter of the circle.
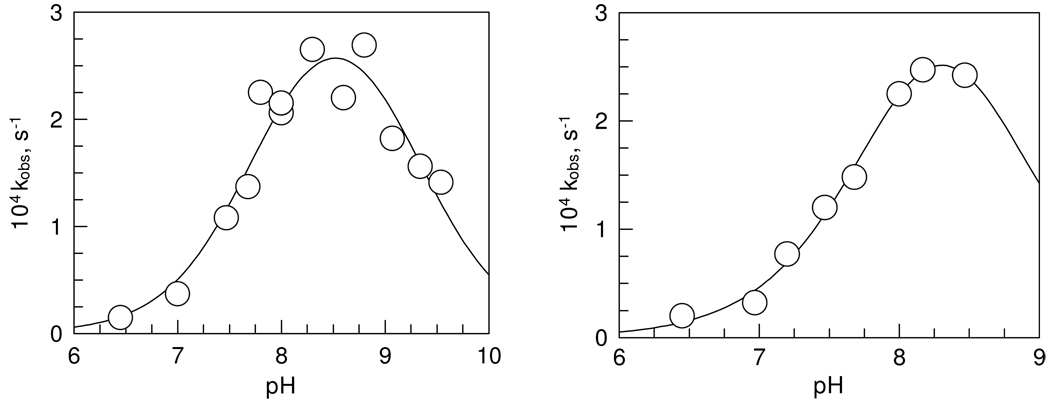
Figure 7 shows similar pH dependence of the inhibition of human α-thrombin by paraoxon and MPNP. The maximal second-order rate constants are 0.47 ± 0.05 M−1 s−1 for paraoxon inhibition, and 6.2 ± 1.0 M−1 s−1 for MPNP inhibition of thrombin at pH 8.3. The two pKa-s calculated from the data are 7.8 ± 0.1 and 9.3 ± 0.2 for paraoxon inhibition and 8.0 ± 0.1 and 8.6 ± 0.2 for MPNP inhibition. Again, the single-pKa model for MPNP gave larger errors than did the double-pKa model and pKa was 0.2 unit lower than the first pKa calculated with the double pKa model..
Figure 7.
pH dependence of the pseudo-first-order rate constant for the inhibition of thrombin with paraoxon (left) and MPNP (right) at NaCl 0.15 M and 25.0 ± 0.1°C. One standard deviation in the data is indicated by the diameter of the circle.
Low-field 1H NMR spectra
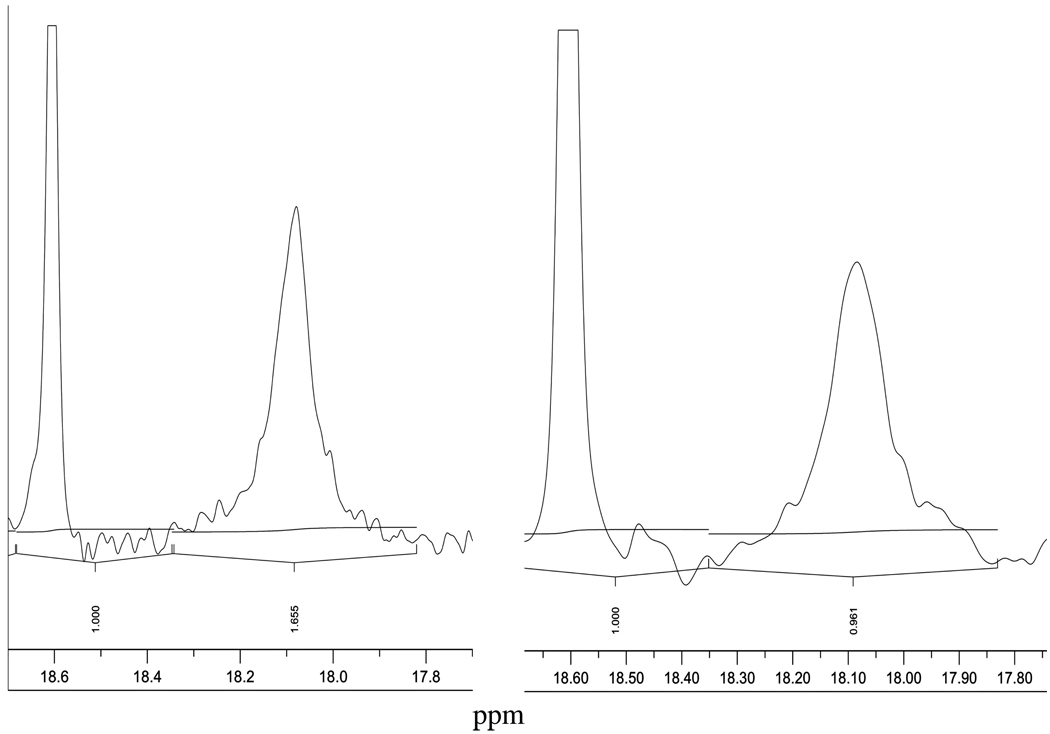
A high-resolution 1H NMR spectrum obtained from the PPACK-inhibited human α-thrombin is shown in Figure 8. The spectrum is focused on the SSHB formed at the active site of thrombin when the covalently bound and cross-linked adduct with PPACK is formed. The peak for the adduct is at 18.10 ± 0.05 ppm in 7% D2O and in 55% D2O, while the peak is positioned at 18.60 ± 0.05 ppm is for the integration standard proton sponge in acetonitrile-d3. Scans identical to the one shown were obtained in three repeats with PPACK-inhibited thrombin.
Figure 8.
Low-field sections of 3000 scans of 600 MHz 1H NMR spectra of the covalently modified human α-thrombin by PPACK at pH 6.7, 0.02 M citrate buffer, 0.01 M phosphate buffer, 0.2 M NaCl, and 0.01% PEG800 at 30.0 ± 0.1°C and; 7% D2O on the left side and 55% D2O on the right side.
The fractionation factor for PPACK-inhibited human α-thrombin is 0.45 ± 0.09 calculated from two values of the integrated resonance at 18.10 ppm at 30.0 ± 0.1°C. The estimated precision in the integration is 20%
The H-bridge in the adduct of thrombin with PPACK was further tested for robustness. The temperature dependence of the line width of the 18.10 ppm signal for PPACK-inhibited thrombin in comparison with the line width of the proton sponge signal, showed sharper signals with rising temperature, presumably due to faster exchange.
The 1H NMR signal with paraoxon-inhibited thrombin at 17.34 ppm in 7% D2O emerged slowly and was maximal after 50 hours. This signal is associated with the dealkyklated adduct which has additional negative charge accumulation on the oxygen in P after the departure of one ethyl group. Control samples of thrombin at several pH in the range 5.3–8.5 did not reveal a signal between 14 and 21 ppm after several thousand scans.
Discussion
Two types of intermediate analogs occurring in thrombin-catalyzed reactions have been studied here with special focus on the H-bridges formed accompanying covalent modification of thrombin. Inhibition of thrombin with PPACK results in a tetrahedral hemiketal anion which forms without the departure of a fragment from the central carbonyl C and thus resembles the tetrahedral intermediate formed after nucleophilic attack on a substrate of thrombin. The other intermediate mimic is of the tetrahedral intermediate formed in deacylation in the substrate reaction sequence after water attack on the acyl enzyme. The analogs of this intermediate are the phosphate and phosphonate ester adducts of thrombin especially after dealkylation (aging).(51–53;55;58;59;66) Significant negative charge accumulation at the oxygen ligands of the central P is the hallmark of these adducts, which is consistent with the charge distribution at the intermediate for deacylation in a natural reaction of thrombin.(67)
Results from Kinetic Studies
Table 1 summarizes the parameters calculated from the data obtained with kinetic measurements.
Table 1.
Summary of Kinetic Data for the Covalent Inhibition of Human α-Thrombin at 25.0 ± 0.1 °C.
| Inhibitor | pKa 1 | pKa2 | ki/Ki M−1 s−1 max | ki, s−1 pH 7.00 | Ki, M pH 7.00 | ΔH‡ kcal/mol | ΔS‡ cal/mol,K |
|---|---|---|---|---|---|---|---|
| PPACK | 7.3 ± 0.2 | 8.8 ± 0.3 | (2.2 ± 0.3) × 107 | 0.24 ± 0.12 | (2.4 ± 1.3) × 10−8 | 10.6 ± 0.7 | 9 ± 2 |
| Paraoxon | 7.8 ± 0.2 | 9.3 ± 0.2 | 0.47 ± 0.05 | > 10−5 | |||
| MPNP | 8.0 ± 0.1 | 8.6 ± 0.2 | 6.2 ± 0.1 | > 10−5 |
The maximal second-order rate constant, ki/Ki, for the inhibition of human α-thrombin with PPACK was determined to be 2.15 × 107 M−1 s−1 at pH 8.1 and 1.07 × 107 M−1 s−1 at pH 7.00 and 25.0 ± 0.1 °C. The value of ΔH‡ = 10.6 ± 0.7 kcal/mol and ΔS‡ = 9 ± 2 cal/mol K are for the activation barrier for the enzyme-catalyzed inhibition reaction. From the second-order rate constant at pH 7.00 and 25.0 ± 0.1 °C, ΔG‡ = 7.85 kcal/mol can be calculated using the Eyring equation. The value of TΔS‡, the difference between ΔG‡ and ΔH‡ is 2,753 cal/mol, which gives ΔS‡ = 9 cal/mol K in full agreement with the value obtained from the nonlinear fit of the temperature dependence of the second-order rate constant. The transition state of the rate-determining step for ki/Ki has more favorable entropy than the free enzyme and 1 M inhibitor have. This result seems consistent with a favorable electrostatic environment for the transition state associated with formation of the hemiketal anion rather than with a transition state for a simple noncovalent association of the enzyme and the inhibitor. The value of ΔH‡ between 10.21 and 11.14 kcal/mol was determined for the acylation rate constant (k2) for the thrombin-catalyzed hydrolysis of S-2238, the structural substrate analog of PPACK, under similar conditions.(68) It seems reasonable to propose that the transitions state for the two reactions have similarity, because they both have a quasi-tetrahedral character with similar charge distribution.
From the pH dependence of thrombin inhibition by PPACK, pKa1 and pKa2 of the enzyme are 7.3 ± 0.2 and 8.8 ± 0.3, respectively. A pKa of 7.3 is consistent with catalysis by His57, as the unprotonated His base is needed to promote the reaction. The second pKa of 8.8, is consistent with the pKa of the amino terminal Ile16 residue, which is engaged in a salt bridge with Asp194 thereby keeping the oxyanion hole in the correct conformation for catalysis. The participation of Ile16 has been reported in other thrombin-catalyzed reactions. (7,22,69)
The Ki value for PPACK inhibition of thrombin is significantly, over 100 fold, smaller than the Km value for the best chromogenic substrate of thrombin, S-2238 at pH 8.(70–71) The tripeptide segment of the two structures are nearly identical as Pip is a Pro analog. Although Km is a complicated constant with contributions from rate constants for elementary chemical steps, its numerical value is identical to the dissociation constant for the reaction of S-2238 with thrombin. The implication is then that Ki for thrombin inhibition by PPACK includes terms that pertain to events ensuing the binding step (Eq.7). It is almost certain that formation of the hemiketal anion, kh, precedes alkylation of His57 by the methylene group and the departure of Cl− ( kalk).
| Equation 7 |
Stein and Trainor(62) have carried out decisive solvent isotope effect and proton inventory measurements on the kinetics of inhibition of human leukocyte elastase by MeOSuc-Ala-Ala- Pro-Val-chloromethyl ketone. On the basis of their results, they ascertain that formation of the hemiketal determines the rate of elastase inhibition by the peptidyl choloromethyl ketone at [I] < Ki and ki represents the alkylation step, kalk, at saturating levels of [I].
The second-order rate constant of thrombin inhibition by PPACK is ∼ 630 fold greater than the one for elastase inhibition by the peptidyl chloromethyl ketone of Stein and Trainor. Moreover, kcat/Km for thrombin-catalyzed hydrolysis of S-2238 at pH 8.0 and 25 °C is 2.75 × 107 M−1 s−1, (70–71) and nearly identical to ki/Ki = 2.2 × 107 M−1 s−1 for thrombin inhibition by PPACK under identical conditions. The encounter between thrombin and S-2238 is also known, it is k1 = 1.0 × 108 M−1 s−1, which may include a conformational change. The value of k1 is modified by the ratio k2/(k−1 + k2) = 101/(101+280) = 0.27 to 2.75 × 107 M−1 s−1.(70–71) A crude estimate of the partitioning ratio of hemiketal formation over return to the enzyme-inhibitor complex plus proceeding to alkylation gives 115 for PPACK inhibition of thrombin. Judging from the rate constant for nucleophilic attack on S-2238 by Ser195 of thrombin, 101 s−1 at 25 °C, kh for hemiketal formation is probably < 100 s−1. The inhibition of thrombin by PPACK is so efficient that its rate under saturation of the enzyme with PPACK cannot be obtained accurately using conventional methods. Nevertheless, the ki value in this work is at least 10 times greater than the value reported for elastase inhibition by the peptidyl chloromethyl ketone. The relative order of magnitude of rates between reversal of the hemiketal anion to ketone and alkylation cannot be foretold. Additional insight will have to wait for a more extensive elucidation of the mechanism of inhibition of thrombin by PPACK, which is a future goal of this project.
Paraoxon and MPNP react with thrombin seven and six orders of magnitude slower than PPACK does. The pH dependence of these reactions are quite different. The lower pKa for inhibition by the phosphate and phosphonate esters is a half unit above the lower pKa found for PPACK inhibition. The pKa of, presumably His57, approaches 8.0 indicating the effect of the negative charge accumulating in the phosphonate oxygen of the ester adduct near the catalytic His in the reactant state. The higher pKa for the inhibition of thrombin by paraoxon and MPNP are 9.3 and 8.6, respectively, bracketing the value for PPACK inhibition.
The second-order rate constants are nearly identical in H2O and D2O in all three inhibition experiments indicating no isotope effect on the second-order rate constant. Because the isotope effect is near unity, the rate-limiting step for the association of thrombin with PPACK and the phosphate and phosphonate esters does not include a direct proton transfer. The solvent isotope effect is also 1.0 ± 0.2 for kcat/Km, for the thrombin-catalyzed hydrolysis of S-2238, consequently the fractionation factor for the transition state of the association of thrombin with S-2238 or that of the ensuing conformational change is one.(22) The fractionation factors for the encounter between thrombin and PPACK is 1.0 at pH 7.45 and 0.93 at pH 8.00 within ∼ 10% experimental error. These results with thrombin deviate from the solvent isotope effect obtained for elastase inhibition by the peptidyl chlormethyl ketone, which is 0.65 at [I] <Ki.(62). However, Stein and Trainor leave the possibility open to a transition-state fractionation factor of 1.0 for their system, which is then compensated by terms from the contribution of solvent reorganization in elastase inhibition by the petidyl cholormethyl ketone.
SSHB in covalent adducts
The presence of an SSHB in the covalent adducts of thrombin has been established by the observation of highly deshielded 1H NMR signals at 18.10 and 17.34 ppm respectively for the analogs of the tetrahedral intermediate for acylation and the intermediate for deacylation.
Notably, the resonances for the two cases are nearly identical to those observed in the corresponding analogs of tetrahedral intermediates in serine proteases(37–42) and in the double displacement mechanism of ester hydrolysis catalyzed by cholinesterases.(34–36) In most cases a signal was also observed with the native enzymes below pH 6, which can be assigned to the protonated His. In contrast, we have not found a signal with the native enzyme between pH 5.3 and 8.5. The active site of native thrombin appears to be more restricted than that of other serine proteases, yet proton exchange seems to be faster. This may be due to the vicinity of residues around the proton bridge between the His-Asp pair, which can serve as catalysts of proton transfer in the pH range studied.
One critical measure of SSHBs is a resonance signal for 1H below 14 ppm. The resonances measured here were used for calculating bond length using an empirical relationship between chemical shifts and N-H-O bond distances in small crystals(34–36) to give 2.62–2.64 Å for the SSHB in the PPACK and paraoxon adducts of thrombin, respectively. The distances compare very well with the crystallographic data for the donor acceptor distance in His57δNH and Asp102γO or Asp102γO and Ser214γOH in PPACK-inhibited thrombin.(50) The precision of the recent protein X-ray data is now comparable to the precision in the correlation of chemical shifts with distances measured by X-ray crystallography for small molecules.
An isotope effect of 2.2 ± 0.2 associated with the formation of a proton bridge is calculated from signal intensities of the 18.10 ppm resonance of the inhibition complex in different isotopic mixtures of buffered water. This isotope effect again denotes the presence of an SSHB. The D/H fractionation factor (ϕ) for a H-bridge is essentially an equilibrium constant for the exchange of H/D between a site on a protein and the protic solvent, i.e. L2O (L=H,D). In this case, the active site is where covalent modification occurs resulting in the formation of proton bridging. The equilibrium constant for the process can be defined as follows;
ϕ = [Active site-D][Hsolvent]/[Active site-H][Dsolvent], indicating the preference of the active site for D over H in reference to the solvent, i.e. an inverse deuterium solvent isotope effect. The shorter and stronger the H-bond, the smaller the value of ϕ. The distance between H-bond acceptor and donor atoms may be estimated from the value of ϕ by assuming a double well model for potential energy of the proton vibration in the H-bridge, in which the minima in the wells are centered at one covalent bond length from the donor and acceptor heavy atoms. As the H-bond gets shorter the wells approach each other and become broader and shallower. The broadening of the potential well decreases the zero point vibrational energy of the H-bonded proton. Because of the 2-fold greater mass of a deuteron than a proton, its zero point vibrational energy decreases (2)1/2 –fold less, resulting in a decreased preference for deuterium over protium (ϕ) as the H-bond shortens. The minima are typically separated by distances between 0.4 and 0.69 Å.(45) This is empirically correlated to a third-order polynomial fit of ϕ to the distances between the minima of the two vibrational potential wells.(34;45;72;73) As the covalent bond length in O-H and N-H bonds are close to 1.00 Å, two covalent bond length ( 2.00 Å) are added to the distance between the minima of the wells to yield the distance between donor and acceptor of an H-bond. The precision of this estimate is similar to the one mentioned above and it gives the same bond length of 2.62 Å as estimated from the correlation for chemical shifts for the PPACK- inhibited thrombin.
The small-molecule modifiers of the thrombin active site lack critical remote interactions of natural substrates at exosites. The exosites, especially the fibrinogen binding site, determine the substrate selection, which is in turn regulated by Na+ binding at an adjacent location.(50) The regulation is mediated by water channels, which presumably involves H-bridges and may exhibit solvent isotope effects of two or greater when thrombin performs its role of hydrolyzing its natural substrates. This was found for the first step of hydrolysis of fibrinogen to fibrinopeptide A.(28) On the basis of our findings, the transition state for binding the small inhibitors doesn't show changes in the status of the H-bridges, but the stable adducts of the inhibitors with thrombin show one unique SSHB at the active site. These SSHBs presumably also occur in tetrahedral intermediates on the reaction path catalyzed by thrombin. Proton bridges with SSHBs appear to contribute to the remarkable catalytic prowess of thrombin and other enzymes that employ acid-base catalysis.
Acknowledgment
The contributions of Dr. Renata Kwiecien, Laboratory of Isotopic and Electrochemical Analysis of Metabolism, CNRS UMR6006, University of Nantes, BP 99208, 44322 Nantes, France, is gratefully acknowledged.
Abbreviations
- KSIE
kinetic solvent isotope effect;
- NPMP
4-nitrophenyl 2-propyl methylphosphonate
- OD
optical density
- paraoxon
4-nitrophenyl-diethylphosphonate
- pNA
para-nitroaniline
- RS
reactant state
- Pip
pipecolyl
- PPACK
Phe-Pro-Arg-Chloromethylketone
- S-2238
H-D-Phe-Pip-Arg-pNA.2HCl
- SSHB
short-strong hydrogen bond
- TLC
thin-layer chromatography
- TS
transition state
- TSP
3-(trimethylsilyl)propionate-2,2,3,3-d6
Footnotes
This work was supported in part by the US National Institutes of Health, Grant No 1 R15 HL067754-02.
Reference List
- 1.Furie B, Furie BC. The Molecular Basis of Blood Coagulation. Cell. 1988;53:505–518. doi: 10.1016/0092-8674(88)90567-3. [DOI] [PubMed] [Google Scholar]
- 2.Davie EW, Fujikawa K, Kisiel W. The Coagulation Cascade: Initiation, Maintenance, and Regulation. Biochemistry. 1991;29:10363–10370. doi: 10.1021/bi00107a001. [DOI] [PubMed] [Google Scholar]
- 3.Berliner LJ. Thrombin: Structure and Function. New York: Plenum Press; 1992. [Google Scholar]
- 4.Mann KG, Lorand L. Introduction: Blood Coagulation. Meth. Enzymology. 1993;222:1–10. doi: 10.1016/0076-6879(93)22003-x. [DOI] [PubMed] [Google Scholar]
- 5.Patthy L. Modular Design of Proteases of Coagulation, Fibrinolysis, and Complement Activation: Implications for Protein Engineering and Structure-Function Studies. Meth. Enzymology. 1993;222:10–22. doi: 10.1016/0076-6879(93)22004-y. [DOI] [PubMed] [Google Scholar]
- 6.Dang QD, Vindigni A, Di Cera E. An Allosteric Switch Controls the Procoagulant and Anticoagulant Activities of Thrombin. Proc. Natl. Acad. Sci. USA. 1995;92:5977–5981. doi: 10.1073/pnas.92.13.5977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Stone SR, Betz A, Hofsteenge J. Mechanistic Studies on Thrombin Catalysis. Biochemistry. 1991;30:9841–9848. doi: 10.1021/bi00105a005. [DOI] [PubMed] [Google Scholar]
- 8.Vindigni A, Di Cera E. Release of Fibrinopeptides by the Slow and Fast Forms of Thrombin. Biochemistry. 1996;35:4417–4426. doi: 10.1021/bi952834d. [DOI] [PubMed] [Google Scholar]
- 9.Di Cera E, Dang QD, Ayala Y, Vindigni A. Linkage at Steady State: Allosteric Transitions of Thrombin. Meth. Enzymology. 1995;259:127–144. doi: 10.1016/0076-6879(95)59041-2. [DOI] [PubMed] [Google Scholar]
- 10.Di Cera E, Dang QD, Ayala YM. Molecular mechanisms of thrombin function. Cell Mol. Life Sci. 1997;53:701–730. doi: 10.1007/s000180050091. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Pineda AO, Savvides SN, Waksman G, Di Cera E. Crystal Structure of the Anticoagulant Slow Form of Thrombin. J. Biol. Chem. 2002;277:40177–40180. doi: 10.1074/jbc.C200465200. [DOI] [PubMed] [Google Scholar]
- 12.Vertstraete M, Zoldhelyi P. Novel Antithrombotic Drugs in Development. Drugs. 1995;49:856–884. doi: 10.2165/00003495-199549060-00002. [DOI] [PubMed] [Google Scholar]
- 13.Das J, Kimball DS. Thrombin Active Site Inhibitors. Bioorg. Med. Chem. 1995;3:990–1007. doi: 10.1016/0968-0896(95)00104-o. [DOI] [PubMed] [Google Scholar]
- 14.Jetten M, Peters CAM, Visser A, Grootenhuis PDJ, van Nispen JW, Ottenheijm HCJ. Peptide-derived Transition State Analogue Inhibitors of Thrombin; Synthesis, Activity and Selectivity. Bioorg. Med. Chem. 1995;3:1099–1114. doi: 10.1016/0968-0896(95)00102-m. [DOI] [PubMed] [Google Scholar]
- 15.Lombardi A, De Simone G, Galdiero S, Nastri F, Pavone V. From Natural to Synthetic Multisite Thrombin Inhibitors. Biopolymers. 1999;51:19–39. doi: 10.1002/(SICI)1097-0282(1999)51:1<19::AID-BIP4>3.0.CO;2-G. [DOI] [PubMed] [Google Scholar]
- 16.Kettner C, Shaw E. D-Phe-Pro-ArgCH2Cl-A Selective Affinity Label For Thrombin. Thrombosis Research. 1979;14:969–973. doi: 10.1016/0049-3848(79)90014-8. [DOI] [PubMed] [Google Scholar]
- 17.Nienaber VL, Mersinger LJ, Kettner CA. Structure-Based Understanding of Ligand Affinity Using Human Thrombin as a Model System. Biochemistry. 1996;35:9690–9699. doi: 10.1021/bi952164b. [DOI] [PubMed] [Google Scholar]
- 18.Kettner C, Shaw E. Inactivation of Trypsin-Like Enzymes with Peptides of Arginine Chloromethyl Ketone. Meth. Enzymology. 1981;90:826–842. doi: 10.1016/s0076-6879(81)80065-1. [DOI] [PubMed] [Google Scholar]
- 19.Bock PE. Active Site Selective Labeling of Blood Coagulation Proeinases with Fluorescence Probes by the Use of Thioester Peptide Chloromethyl Ketones I. Specificity of Thrombin Labeling. J. Biol. Chem. 1992;267:14963–14973. [PubMed] [Google Scholar]
- 20.Bode W, Turk D, Karshikov A. The Refined 1.9 A X-ray Crystal Structure of D-Phe-Pro-Arg Chloromethylketone-inhibited Human α-Thrombin: Structure Analysis, Overall Structure, Electrostatic Properties, Detailed Active-Site Geometry, and Structure-Function Relationships. Protein Sci. 1992;1:426–471. doi: 10.1002/pro.5560010402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Bode W, Mayr I, Baumann U, Huber R, Stone SR, Hofsteenge J. The Refined 1.9 A Crystal Structure of Human α-Thrombin: Interaction with D-Phe-Pro-Arg Chloromethylketone and Significance of the Tyr-Pro-Pro-Trp Insertion Segment. EMBO J. 1989;8:3467–3475. doi: 10.1002/j.1460-2075.1989.tb08511.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Enyedy EJ, Kovach IM. Proton Inventory Studies of Thrombin-Catalyzed Reactions of Substrates with Selected P and P' Sites. J. Am. Chem. Soc. 2004;126:6017–6024. doi: 10.1021/ja0320166. [DOI] [PubMed] [Google Scholar]
- 23.Fersht A. Structure and Mechanism in Protein Science. New York, NY: W. H. Freeman and Co; 1999. [Google Scholar]
- 24.Hedstrom L. Serine protease mechanism and specificity. Chem. Rev. 2002;102:4501–4524. doi: 10.1021/cr000033x. [DOI] [PubMed] [Google Scholar]
- 25.Schowen RL. Structural and Energetic Aspects of Protolytic Catalysis by Enzymes: Charge - Relay Catalysis in the Function of Serine Proteases. In: Liebman JF, Greenberg A, editors. Mechanistic Principles of Enzyme Activity. vol. 9. New York: VCH Publishers; 1988. pp. 119–168. [Google Scholar]
- 26.Schowen KB, Limbach HH, Denisov GS, Schowen RL. Hydrogen bonds and proton transfer in general-catalytic transition state stabilization in enzyme catalysis. Biochem. Biophys. Acta. 2000;1458:43–62. doi: 10.1016/s0005-2728(00)00059-1. [DOI] [PubMed] [Google Scholar]
- 27.Schowen RL, Klinman JP, Hynes JT, Limbach HH. Hydrogen Transfer Reactions. Weinheim: Wiley-VCH; 2007. [Google Scholar]
- 28.Zhang D, Kovach IM. Full and Partial Deuterium Solvent Isotope Effect Studies of α-Thrombin-catalyzed Reactions of Natural Substrates. J. Am. Chem. Soc. 2005;127:3760–3766. doi: 10.1021/ja043258o. [DOI] [PubMed] [Google Scholar]
- 29.Alvarez FJ, Schowen RL. Mechanistic Deductions from Solvent Isotope Effects. In: Buncel E, Lee CC, editors. Isotopes in Organic Chemistry. vol. 7. Amsterdam: Elsevier; 1987. pp. 1–60. [Google Scholar]
- 30.Kresge AJ, More O, Powell MF. Solvent Isotopes Effects, Fractionation Factors and Mechanisms of Proton Transfer Reactions. In: Buncel E, Lee CC, editors. Isotopes in Organic Chemistry. vol. 7. Amsterdam: Elsevier; 1987. pp. 177–273. [Google Scholar]
- 31.Venkatasubban KS, Schowen RL. The Proton Inventory Technique. CRC Crit. Rev. Biochem. 1985;17:1–44. doi: 10.3109/10409238409110268. [DOI] [PubMed] [Google Scholar]
- 32.Quinn DM, Sutton LD. Theoretical Basis and Mechanistic Utility of Solvent Isotope Effects. In: Cook PF, editor. Enzyme Mechanism from Isotope Effects. Boston, MA: CRC Press; 1991. pp. 73–126. [Google Scholar]
- 33.Robillard G, Shulman RG. High Resolution Nuclear Magnetic Resonance Studies of the Active Site of Chymotrypsin II. Polarization of Histidine 57 by Substrate Analogs and Competitive Inhibitors. J. Mol. Biol. 1974;86:541–558. doi: 10.1016/0022-2836(74)90179-x. [DOI] [PubMed] [Google Scholar]
- 34.Mildvan AS, Massiah MA, Harris TK, Marks GT, Harrison DHT, Viragh C, Reddy PM, Kovach IM. Short Strong Hydrogen Bonds on Enzymes: NMR and Mechanistic Studies. J. Mol. Structure. 2002;215:163–175. [Google Scholar]
- 35.Massiah MA, Viragh C, Reddy PM, Kovach IM, Johnson J, Rosenberry TL, Mildvan AS. Short, Strong Hydrogen Bonds at the Active Site of Human Acetylcholinesterase: Proton NMR Studies. Biochemistry. 2001;40:5682–5690. doi: 10.1021/bi010243j. [DOI] [PubMed] [Google Scholar]
- 36.Viragh C, Harris TKRPM, Massiah MA, Mildvan AS, Kovach IM. NMR Evidence for a Short, Strong Hydrogen Bond at the Active Site of a Cholinesterase. Biochemistry. 2000;39:16200–16205. doi: 10.1021/bi0022644. [DOI] [PubMed] [Google Scholar]
- 37.Frey PA, Whitt SA, Tobin JB. A Low-Barrier Hydrogen Bond in the Catalytic Triad of Serine Proteases. Science. 1994;264:1927–1930. doi: 10.1126/science.7661899. [DOI] [PubMed] [Google Scholar]
- 38.Tobin JB, Whitt SA, Cassidy CS, Frey PA. Low-Barrier Hydrogen Bonding in Molecular Complexes Analogous to Histidine and Aspartate in the Catalytic Triad of Serine Proteases. Biochemistry. 1995;34:6919–6924. doi: 10.1021/bi00021a002. [DOI] [PubMed] [Google Scholar]
- 39.Cassidy CS, Lin J, Frey PA. A New Concept for the Mechanism of Action of Chymotrypsin: The Role of the Low-Barrier Hydrogen Bond. Biochemistry. 1997;36:4576–4584. doi: 10.1021/bi962013o. [DOI] [PubMed] [Google Scholar]
- 40.Lin J, Westler WM, Cleland WW, Markley JL, Frey PA. Fractionation factors and activation energies for exchange of the low barrier hydrogen bonding proton in peptidyl trifluoromethyl ketone complexes of chymotrypsin. Proc. Natl. Acad. Sci. 1998;95:14664–14668. doi: 10.1073/pnas.95.25.14664. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Lin J, Cassidy CS, Frey PA. Correlations of the Basicity of His57 with Transition State Analogue Binding, Substrate Reactivity, and the Strength of the Low-Barrier Hydrogen Bond in Chymotrypsin. Biochemistry. 1998;37:11940–11948. doi: 10.1021/bi980278s. [DOI] [PubMed] [Google Scholar]
- 42.Halkides CJ, Wu YQ, Murray CJ. A Low-Barrier Hydrogen Bond in Subtilisin: 1H and 15N NMR Studies with Peptidyl Trifluoromethyl Ketones. Biochemistry. 1996;35:15941–15948. doi: 10.1021/bi961805f. [DOI] [PubMed] [Google Scholar]
- 43.Ash EL, Sudmeier JL, De Fabo EC, Bachovchin WW. A Low-barrier Hydrogen Bond in the Catalytic Triad for Serine Proteases? Theory Versus Experiment. Science. 1997;278:1128–1132. doi: 10.1126/science.278.5340.1128. [DOI] [PubMed] [Google Scholar]
- 44.Kahayaoglu A, Haghjoo K, Guo F, Jordan F, Kettner C, Felfoldi F, Polgar L. Low Barrier Hydrogen Bond is Absent in the Catalytic Triads in the Ground State but is Present in a Transition-state Complex in the Prolyl Oligopeptidase Family of Serine Proteases. J. Biol. Chem. 1997;272:25547–25554. doi: 10.1074/jbc.272.41.25547. [DOI] [PubMed] [Google Scholar]
- 45.Bao D, Huskey PW, Kettner CA, Jordan F. Hydrogen Bonding to Active-Site Histidine in Peptidyl Boronic Acid Inhibitor Complexes of Chymotrypsin and Subtilisin: Proton Magnetic Resonance Assignments and H/D Fractionation. J. Am. Chem. Soc. 1999;121:4684–4689. [Google Scholar]
- 46.Bachovchin WW. Confirmation of the Assignment of the Low-field Proton Resonance of Serine Proteases by Using Specifically Nitrogen-15 Labeled Enzyme. Proc Natl Acad Sci U S A. 1985;82:7948–7951. doi: 10.1073/pnas.82.23.7948. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Tsilikounas E, Rao T, Gutheil WG, Bachovchin WW. 15N and 1H NMR Spectroscopy of the Catalytic Histidine in Chloromethyl Ketone-inhibited Complexes of Serine Proteases. Biochemistry. 1996;20:2437–2444. doi: 10.1021/bi9513968. [DOI] [PubMed] [Google Scholar]
- 48.Bachovchin WW, Wong WY, Farr-JonesS S, Shenvi AB, Kettner CA. Nitrogen-15 NMR Spectroscopy of the Catalytic-triad Histidine of a Serine Protease in Peptide Boronic Acid Inhibitor Complexes. Biochemistry. 1988;27:7689–7697. doi: 10.1021/bi00420a018. [DOI] [PubMed] [Google Scholar]
- 49.Bachovchin WW. 15N NMR Spectroscopy of Hydrogen-bonding Interactions in the Active Site of Serine Proteases: Evidence for a Moving Histidine Mechanism. Biochemistry. 1986;25:7751–7759. doi: 10.1021/bi00371a070. [DOI] [PubMed] [Google Scholar]
- 50.Pineda AO, Carrell CJ, Bush LA, Prasad S, Caccia S, Chen ZW, Mathews FS, Di Cera E. Molecular Dissection of Na+ Binding to Thrombin. J. Biol. Chem. 2004;279:31842–31853. doi: 10.1074/jbc.M401756200. [DOI] [PubMed] [Google Scholar]
- 51.Kovach IM. Stereochemistry and secondary reactions in the irreversible inhibition of serine hydrolases by organophosphorus compounds. J. Phys. Org. Chem. 2004;17:602–614. [Google Scholar]
- 52.Bencsura A, Enyedy I, Viragh C, Akhmetshin R, Kovach IM. Phosphonate Ester Active Site Probes of Acetylcholinesterase, Trypsin, and Chymotrypsin. In: Quinn DM, Balasubramanian AS, Doctor BP, Taylor P, editors. Enzymes of The Cholinesterase Family. New York: Plenum Press; 1995. pp. 155–162. [Google Scholar]
- 53.Bencsura A, Enyedy I, Kovach IM. Origins and Diversity of the Aging Reaction in Phosphonate Adducts of Serine Hydrolase Enzymes: What Characteristics of the Active Site Do They Probe. Biochemistry. 1995;34:8989–8999. doi: 10.1021/bi00028a007. [DOI] [PubMed] [Google Scholar]
- 54.Enyedy EJ, Kovach IM. Modulation of the Activity of Human α-Thrombin with Phosphonate Ester Inhibitors. Bioorg. Med. Chem. 1997;5:1531–1541. doi: 10.1016/s0968-0896(97)00099-0. [DOI] [PubMed] [Google Scholar]
- 55.Enyedy I, Bencsura A, Kovach IM. Interactions in Tetravalent and Pentavalent Phosphonate Esters of Ser at the Active Site of Serine Enzymes. Phosphorus, Sulfur, and Silicon. 1996;109–110:249–252. [Google Scholar]
- 56.Kovach IM. Structure and Dynamics of Serine Hydrolase-Organophosphate Adducts. J. Enzyme Inhib. 1988;2:199–208. doi: 10.3109/14756368809040726. [DOI] [PubMed] [Google Scholar]
- 57.Kovach IM, Larson M, Schowen RL. Catalytic Recruitment in the Inactivation of Serine Proteases by Phosphonate Esters. Recruitment of Acid-Base Catalysis. J. Am. Chem. Soc. 1986;108:5490–5495. [Google Scholar]
- 58.Kovach IM, McKay L, Vander Velde D. Diastereomeric Phosphonate ester Adducts of Chymotrypsin: 31P- NMR Measurements. Chirality. 1993;5:143–149. doi: 10.1002/chir.530050307. [DOI] [PubMed] [Google Scholar]
- 59.Kovach IM, Enyedy EJ. Active-Site-Dependent Elimination of 4-Nitrophenol from 4-Nitrophenyl Alkylphosphonyl Serine Protease Adducts. J. Am. Chem. Soc. 1998;120:258–263. [Google Scholar]
- 60.Bennet A, Kovach IM, Schowen RL. Rate-Limiting P-O Fission in the Self-Stimulated Inactivation of Acetylcholinesterase by 4-Nitrophenyl 2-Propyl Methylphosphonate. J. Am. Chem. Soc. 1988;110:7892–7893. [Google Scholar]
- 61.Kitz R, Wilson IB. Esters of Methanesulfonic Acid as Irreversible Inhibitors of Acetylcholinesterase. J. Biol. Chem. 1962;237:3245–3249. [PubMed] [Google Scholar]
- 62.Stein RL, Trainor DA. Mechanism of Inactivation of Human Leukocyte Elastase by a Chlorromethyl Ketone: Kinetic and Solvent Isotope Effect Studies. Biochemistry. 1986;25:5414–5419. doi: 10.1021/bi00367a011. [DOI] [PubMed] [Google Scholar]
- 63.Schowen RL. Catalytic Power and Transition-State Stabilization. In: Gandour RD, Schowen RL, editors. Transition States of Biochemical Proceses. New York: Plenum; 1978. pp. 77–114. [Google Scholar]
- 64.Leatherbarrow RJ. Staines, U.K.: Ertihacus Sofware Ltd; 1992. GraFit User's Guide. [Google Scholar]
- 65.Humphrey W, Dalke A, Schulten K. Visual Molecular Dynamics. J. Molec. Graphics. 1996;14:33–38. doi: 10.1016/0263-7855(96)00018-5. [DOI] [PubMed] [Google Scholar]
- 66.Kovach IM. Ligand and Active-Site Dependent P-O Versus C-O Bond Cleavage in Organophosphorus Adducts of Serine Hydrolases. Phosphorus, Sulfur and Silicon. 1999;144–146:537–540. [Google Scholar]
- 67.Kovach IM, Huhta D. Comparative Study of the Charge Distribution in Tetravalent Carbonyl Transients and Organophosphorus Adducts of Trypsin. Theochem. 1991;79:335–342. [Google Scholar]
- 68.DiCera E, De Cristofaro R, Albright DJ, Fenton JW. Linkage between Proton Binding and Amidase Activity in Human α-Thrombin: Effect of Ions and Temperature. Biochemistry. 1991;30:7913–7924. doi: 10.1021/bi00246a007. [DOI] [PubMed] [Google Scholar]
- 69.Lottenberg R, Hall JA, Blinder M, Binder EP, Jackson CM. The Action of Thrombin on Peptide p-Nitroanilide Substrates. Substrate Selectivity and Examination of Hydrolysis under Different Reaction Conditions. Biochem. Biophys. Acta. 1983;742:539–557. doi: 10.1016/0167-4838(83)90272-8. [DOI] [PubMed] [Google Scholar]
- 70.Wells CM, DiCera E. Thrombin is a Na+-Activated Enzyme. Biochemistry. 1992;31:11721–11730. doi: 10.1021/bi00162a008. [DOI] [PubMed] [Google Scholar]
- 71.Zhang D, Kovach IM, Sheehy JP. Locating the rate-determining step(s) for three-step hydrolase-catalyzed reactions with dynafit. BBA, Proteins and Proteomics. 2008;1784:827–833. doi: 10.1016/j.bbapap.2008.02.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Cleland WW, Kreevoy MM. Low Barrier Hydrogen Bonds and Enzymic Catalysis. Science. 1994;264:1887–1890. doi: 10.1126/science.8009219. [DOI] [PubMed] [Google Scholar]
- 73.Smirnov SN, Benedict H, Golubev NS, Denisov GS, Kreevoy MM, Schowen RL, Limbach HH. Exploring Zero-point energies and hydrogen bond geometries along proton transfer pathways by low-temperature NMR. Can. J. Chem. 1999;77:943–949. [Google Scholar]