Abstract
Light signals are fundamental to the growth and development of plants. Red and far-red light are sensed using the phytochrome family of plant photoreceptors. Individual phytochromes display both unique and overlapping roles throughout the life cycle of plants, regulating a range of developmental processes from seed germination to the timing of reproductive development. The evolution of multiple phytochrome photoreceptors has enhanced plant sensitivity to fluctuating light environments, diversifying phytochrome function, and facilitating conditional cross-talk with other signalling systems. The isolation of null mutants, deficient in all individual phytochromes, has greatly advanced understanding of phytochrome functions in the model species, Arabidopsis thaliana. The creation of mutants null for multiple phytochrome combinations has enabled the dissection of redundant interactions between family members, revealing novel regulatory roles for this important photoreceptor family. In this review, current knowledge of phytochrome functions in the light-regulated development of Arabidopsis is summarised.
Keywords: Arabidopsis, light signals, photomorphogenesis, phytochrome
Introduction
The importance of light signals in regulating plant growth has been documented for centuries. Indeed, Darwin himself provides detailed observations of the developmental processes occurring following emergence of a dark-grown (etiolated) seedling into the light, in a book written with his son, Francis, ‘The power of movement in plants’ (Darwin and Darwin, 1880). In this work, the authors record important roles for light throughout plant development, including leaf ’sleep’ movements (epinasty/hyponasty) and the bending of plant stems towards bright lateral light (phototropism). Now collectively termed photomorphogenesis, the regulation of plant growth by light signals is known to involve three main families of information-transducing photoreceptors, the red (R) and far-red (FR) light-absorbing phytochromes and the UV-A/blue light-absorbing cryptochromes and phototropins (Schäfer and Nagy, 2006). The phytochrome (phy) family of photoreceptors are reversibly photochromic biliproteins. These exist as dimers with each monomer comprising an apoprotein covalently attached to a light-absorbing linear tetrapyrrole chromophore, phytochromobilin (Lagarias and Rapoport, 1980). Phytochromes are synthesized in the dark in a biologically inactive R-absorbing (Pr) form. Biological activity is acquired upon photoconversion to the far-red light-absorbing (Pfr) form in a reaction optimized at red wavelengths. Photoconversion of Pfr back to the biologically inactive Pr form is optimized at FR wavelengths, resulting in a dynamic photoequilibrium of Pr and Pfr in natural light conditions. Following conversion to the Pfr form, phytochromes translocate to the nucleus (Sakamoto and Nagatani, 1996; Nagatani, 2004; Kircher et al., 1999, 2002). A primary mechanism of phytochrome signalling involves physical interaction of the phytochrome photoreceptor with a subfamily of bHLH transcription factors, the PHYTOCHROME INTERACTING FACTORS (PIFs) (reviewed in Duek and Fankhauser, 2005; Monte et al., 2007). The PIFs display a diverse array of regulatory functions controlling photomorphogenesis (Ni et al., 1998, 1999; Huq and Quail, 2002; Kim et al., 2003; Huq et al., 2004; Khanna et al., 2004, 2007; Monte et al., 2004; Leivar et al., 2008; Lorrain et al., 2008; Shin et al., 2009; Stephenson et al., 2009). The physical interaction of phytochromes with PIFs leads to the latter's phosphorylation, ubiquitination, and degradation via the 26S proteosome (Bauer et al., 2004; Park et al., 2004; Shen et al., 2005, 2007; Al-Sady et al., 2006). This elegantly simple system enables plants to alter gene expression rapidly in response to fluctuations in the light environment. Phytochrome interaction with PIFs can conversely lead to turnover of the phytochrome photoreceptor, providing a dual mechanism of regulating plant development (Al-Sady et al., 2008).
Plants contain multiple phytochromes. Three discrete apoprotein-encoding genes (PHYA–PHYC) are conserved within angiosperms (Mathews et al., 1995). Additional phytochromes have been identified in dicotyledonous plants and are thought to represent the products of more recent gene duplication events (Mathews et al., 1995; Mathews and Sharrock, 1997). In the model species Arabidopsis thaliana, five genes (PHYA–E) encoding phytochrome apoproteins have been sequenced and characterized (Sharrock and Quail, 1989; Clack et al., 1994). The PHYB protein displays greatest sequence similarity to PHYD (approximately 80%), and forms an evolutionary distinct subgroup with PHYE (Goosey et al., 1997). The PHYA protein is most closely related to PHYC yet the phyA photoreceptor displays unique properties. In contrast to other phytochromes, phyA displays rapid lability in the Pfr form and can signal during rapid photoconversion between Pr and Pfr. Previously termed ‘type I phytochrome’, phyA is the predominant phytochrome in etiolated seedlings but is rapidly degraded to much lower steady-state levels upon transfer to light (Clough and Vierstra, 1997). In this way, phyA serves initially as a highly sensitive light ‘antenna’, enabling the rapid promotion of de-etiolation upon soil emergence. The highly sensitive, non-reversible functions of phyA in response to low quantities of light are termed Very Low Fluence Responses (VLFRs). The proteolysis of phyA displays first order reaction kinetics and is therefore greatest upon transfer to light environments establishing a high proportion of Pfr (e.g. R). Continuous irradiation of wavelengths establishing a low percentage of Pfr (e.g. FR) results in photo-cycling of phyA between the Pr and Pfr forms and signalling via the High Irradiance Response (HIR) mode (Hennig et al., 2000). Photoreversible responses are mediated by ‘type II’ phytochromes displaying relative stability in the Pfr form (phyB–E) and are termed Low Fluence Responses (LFRs). Phytochrome B is the most abundant phytochrome in light-grown plants (Sharrock and Clack, 2002).
The identification of null mutants, deficient in each of the five family members, has enabled the roles of individual phytochromes to be elucidated in the model species Arabidopsis thaliana (Table 1). The first phytochrome-deficient mutants reported in Arabidopsis were a series of phyB alleles in the Landsberg erecta (La-er) background (phyB-1 to phyB-8). These were initially identified as ‘hy3’ elongated hypocotyl mutants in white light (W) (Koornneef et al., 1980). In parallel studies in the Furuya and Quail laboratories, hy3 seedlings were shown to display severely reduced levels of PHYB protein, suggesting that the hy3 phenotype results from a mutation in the PHYB gene (Nagatani et al., 1991; Somers et al., 1991). This was later confirmed by the Chory laboratory through DNA sequence analysis and genetic complementation (Reed et al., 1993). Two further phyB alleles (phyB-9 and phyB-10) were also identified in this study in the Columbia (Col) and La-er backgrounds, respectively. Mutants deficient in phyA were isolated in three separate screens by the Chory, Quail, and Whitelam/Harberd laboratories. In these studies, the authors exploited the signalling behaviour of phyA in the HIR mode to identify mutants displaying an etiolated appearance in continuous FR. Multiple phyA alleles were isolated through these screens; phyA-1 and phyA-2 in La-er (Whitelam et al., 1993), phyA-101–phyA-103 in RLD (Parks and Quail, 1993; Dehesh et al., 1993), phyA-201 and phyA-202 in La-er (Nagatani et al., 1993), phyA-203–phyA-208 in La-er (Reed et al., 1994) and phyA 209–phyA-211 in Col (Reed et al., 1994). Natural genetic variation provided the source of a mutant deficient in phyD. The phyD-1 mutation was identified as a 15 bp deletion in the PHYD gene of the naturally occurring Wassileweskija (Ws) accession by the Sharrock laboratory (Aukerman et al., 1997). A screen of mutagenized phyAphyB double mutants by the Whitelam laboratory later provided the phyE-1 allele in La-er (Devlin et al., 1998). In this work, the authors isolated plants displaying internode growth between rosette leaves and early flowering, ultimately revealing a 1 bp deletion at the PHYE locus. The Arabidopsis phytochrome mutant collection was completed in 2003 with the identification of multiple PHYC alleles by both the Quail and Whitelam laboratories. The PHYC-1 allele was identified via PCR screening of a T-DNA insertion mutant collection in the Ws background (Franklin et al., 2003a; Monte et al., 2003), the PHYC-2 allele was identified as a T-DNA-insertion mutant in the Col background in a screen of the Ecker-Alonzo collection (Monte et al., 2003), whilst a fast neutron deletion approach was used to identify phyC-3 in the Col background from the Maxygen collection (Monte et al., 2003). The isolation of separate null mutants in all five phytochromes has facilitated the construction of multiple, higher-order mutants, deficient in a variety of phytochrome combinations. Analyses of both individual and multiple phytochrome-deficient mutants has subsequently provided a refined picture of phytochrome functions throughout plant development, an overview of which is presented here.
Table 1.
Summary of phy mutant alleles
References refer to manuscripts citing original isolation.
Seedling establishment
Germination
The promotion of seed germination in many species requires light (Casal and Sanchez, 1998). The involvement of a R/FR-reversible photoreceptor in mediating seed germination was first demonstrated in the pioneering ‘flip flop’ experiments of Harry Borthwick and colleagues in 1952. In this work, Grand Rapids lettuce seed were treated with alternating R and FR treatments and germination efficiency analysed. The results were striking. In seeds receiving the R treatment last, almost 100% germination was achieved. A markedly different response was observed in seeds receiving the FR treatment last, with much lower percentages of germination recorded (Borthwick et al, 1952). These experiments provided the first evidence of phytochromes operating reversibly in the LFR mode. The involvement of individual phytochromes in mediating Arabidopsis seed germination has been revealed in multiple mutant studies. Comparisons of mutants deficient in phyA, phyB, and phyA/phyB have shown a predominant role for phyB in regulating germination in R via the LFR mode whilst phyA mediates VLFRs in R and FR (Botto et al., 1996; Shinomura et al., 1996). Phytochrome A can, in addition, promote germination in continuous FR in the HIR mode (Johnson et al., 1994; Reed et al., 1994; Hennig et al., 2002). Observations of R/FR-reversible germination in phyAphyB double mutants (phyAB) suggested the participation of an additional phytochrome in this response (Poppe and Schäfer, 1997). The subsequent isolation of mutants deficient in phyD and phyE enabled the roles of these phytochromes to be examined (Aukerman et al., 1997; Devlin et al., 1998). Whilst only a small role was observed for phyD, deficiency of phytochromes A, B, and E severely impaired light-induced germination and abolished R/FR-reversibility, confirming the redundant interactions between these phytochromes (Hennig et al., 2002). An intriguing observation from this study was the requirement of phyE to promote germination in continuous FR. Germination of monogenic phyE mutants was severely impaired in blue light (B) and abolished in FR, a phenotype similar to that observed in phyA mutants. Immunoblot analyses revealed wild-type (WT) levels of PHYA in phyE mutants, raising the interesting possibility that phyE itself may operate as a FR photoreceptor in the regulation of germination (Hennig et al., 2002). Alternatively, this response may be mediated by phyA/phyE heterodimers, although the absence of obvious phyA heterodimerization with type II phytochromes in vitro would appear to preclude this notion (Sharrock and Clack 2004; Clack et al., 2009).
More recent analyses of mutants deficient in combinations of phyA, phyB, and phyE have shown ambient temperature to modulate the light-regulation of Arabidopsis germination. In this work, phytochrome family members were shown to display altered functional hierarchies at different temperatures (Heschel et al., 2007). At warmer temperatures (>22 °C) phyB adopted a prominent role in promoting germination, followed by phyA and phyE. At cooler temperatures (<16 °C) phyE displayed functional dominance, with phyB displaying an accessory role (Heschel et al., 2007). The increased functional dominance of phyE over phyB at cooler temperatures parallels observations in flowering inhibition (see ‘Reproductive development’), raising the interesting possibility that phyE abundance may exceed phyB levels in these conditions.
De-etiolation
Dark-grown seedlings display a ‘skotomorphogenic’ phenotype. This is characterized by an elongated hypocotyl, unexpanded cotyledons within a protective apical hook, and the absence of chlorophyll. Following germination and/or soil emergence, light signals initiate a variety of de-etiolation responses to promote photoautotrophic survival. These include inhibition of hypocotyl elongation, opening of the apical hook, expansion of cotyledons, and synthesis of chlorophyll. Phytochromes perform a variety of overlapping functions in seedling de-etiolation, in combination with the cryptochrome UV-A/blue light photoreceptors cry1 and cry2.
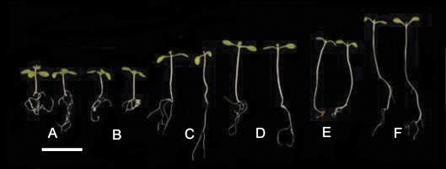
Monogenic phyA mutants display a WT de-etiolation phenotype in W and R (Fig. 1; Dehesh et al., 1993; Nagatani et al., 1993; Parks and Quail, 1993; Whitelam et al., 1993; Reed et al., 1994). The most characteristic phenotype of phyA mutants, and the basis upon which they were identified, is an absence of responsivity in the FR-HIR mode. When grown in continuous FR, phyA mutants display a skotomorphogenic phenotype, confirming the unique role of phyA in mediating de-etiolation in these conditions (Dehesh et al., 1993; Nagatani et al., 1993; Parks and Quail, 1993; Whitelam et al., 1993). Comparative transcriptional profiling of etiolated WT and phyA mutants subject to FR treatment has revealed more than 800 phyA-regulated genes, providing the first insights in to the phyA transcriptional network (Tepperman et al., 2001). Mutants deficient in phyA have also been shown to display elongated hypocotyls in continuous B, presumably because of a lack of phyA activity in the HIR mode (Whitelam et al., 1993; Neff and Chory, 1998). Phytochrome B is the predominant phytochrome regulating de-etiolation in W and R. Mutants deficient in phyB are characterized by elongated hypocotyls, reduced cotyledon expansion, and reduced chlorophyll synthesis (Fig. 1; Koorneef et al., 1980; Nagatani et al., 1991; Somers et al., 1991; Reed et al., 1993). Despite these striking phenotypes, transcription profiles of etiolated phyB mutants transferred to R were shown not to differ dramatically from WT controls (Tepperman et al., 2004). Subsequent mutant analyses revealed a dominant role for phyA in regulating rapid gene expression responses upon R transfer (Tepperman et al., 2006; Quail, 2007). Redundancy between phyA and phyB has been reported in R-mediated hypocotyl inhibition and cotyledon expansion (Neff and Van Volkenburgh, 1994; Reed et al., 1994; Casal and Mazella, 1998; Neff and Chory, 1998). In these studies, phyAB mutants were shown to display modestly longer hypocotyls and smaller cotyledons than phyB monogenic mutants, revealing roles for phyA which are normally masked by the presence of phyB. The stimulation of chlorophyll synthesis by light has been demonstrated to involve the combined actions of phyA, phyB, cry1, and cry2, acting largely through the regulation of the glutamyl tRNA reductase- encoding gene, HEMA1 (Reed et al., 1994; McCormac and Terry, 2002).
Fig. 1.
WT (A), phyA (B), phyB (C), phyBDE (D), phyABE (E), and phyABDE (F) seedlings grown in 8 h photoperiods of white light at 120 μmol m−2 s−1. Scale bar represents 5 mm.
The majority of photomorphogenic analyses are performed at photon irradiances of <50 μmol m−2 s−1. Growth of seedlings at higher photon irradiances of R (>100 μmol m−2 s−1) has revealed photoprotection of phyA from proteolytic degradation and considerable phyA activity (Franklin et al., 2007). Under these conditions, phyB mutants displayed markedly greater hypocotyl inhibition and cotyledon expansion than seedlings grown at lower photon irradiances. The irradiance-dependent enhancement of de-etiolation was largely absent in phyAB mutants, confirming a significant role for phyA in mediating this response. These findings suggest that, in many natural light environments, where photon irradiances are considerably greater than those achievable in laboratory conditions, the contribution of phyA to seedling establishment may be greater than previously considered, even in light conditions which establish a high proportion of Pfr.
Despite high sequence similarity between PHYB and PHYD, the role of phyD in seedling de-etiolation appears minor. Wassilewskija seedlings containing a natural phyD deletion were observed to display only marginally longer hypocotyls in R than plants containing an introgressed PHYD gene (Aukerman et al., 1997). A synergistic relationship was observed between phyB and phyD in R, with phyBD mutants displaying greater hypocotyl elongation than the combined increases in both monogenic mutants (Aukerman et al., 1997). This study also showed phyD to perform minor accessory roles to phyB in both the W-mediated promotion of cotyledon expansion and anthocyanin accumulation. Interestingly, phyD does not appear to perform these roles in La-er, revealing natural genetic variation in phytochrome function (Aukerman et al., 1997). Phytochrome E has a small role in seedling de-etiolation. Monogenic phyE mutants were shown to be indistinguishable from WT controls in a variety of light conditions (Devlin et al., 1998). The combined loss of phyB and phyE has been shown to result in slightly smaller cotyledon size in R- grown seedlings than loss of phyB alone (Franklin et al., 2003b). It is therefore likely that phyE performs a minor redundant role in this response. The isolation of multiple phyC mutants enabled the roles of all five phytochromes to be determined in Arabidopsis seedling de-etiolation (Franklin et al., 2003a; Monte et al., 2003). Mutants deficient in phyC displayed greater hypocotyl elongation than WT controls in R. No additivity was, however, observed in mutants lacking phyB and phyC, suggesting phyC to operate through modulation of phyB function. The recently described obligate heterodimerization of phyC with phyB might explain these observations (Clack et al., 2009). Despite displaying more sequence similarity to PHYA than PHYB, D or E, no role for phyC in mediating seedling de-etiolation in FR was observed (Franklin et al., 2003a; Monte et al., 2003).
An important, frequently overlooked point, is that the phenotypes of the null mutants of all five phytochromes are indistinguishable from WT seedlings when grown in the dark. This observation establishes that the Pr form of the photoreceptor has no detectable intrinsic biological activity, an issue of some debate in earlier years in the photomorphogenesis field.
Phytochrome co-action with UV-A/blue light photoreceptors
Seedling de-etiolation involves the interaction of phytochrome and cryptochrome signalling. Although the exact nature of co-action remains uncertain, physical interactions between PHYA/CRY1 and PHYB/CRY2 have been reported (Ahmad et al., 1998; Mas et al., 2000). Analysis of mutants deficient in multiple photoreceptors has shown complex interplay between phyA, phyB, and cry1 in seedling de-etiolation. The presence of cry1 was shown to be required for phyB-mediated cotyledon expansion in B and enhanced phyA and phyB-mediated chlorophyll production in R (Neff and Chory, 1998). In a separate study, conditional synergism was observed between cry1 and phyB in hypocotyl inhibition and cotyledon unfolding (Casal and Mazzella, 1998). A functional interaction between cry1 and phyD has also been observed in the W-enhancement of R-mediated hypocotyl inhibition (Hennig et al., 1999). Mutants deficient in phyC have been reported to display hyposensitivity to low photon irradiances of blue light with respect to the regulation of hypocotyl elongation (Franklin et al., 2003a). Under these conditions cry2 function predominates (Lin et al., 1998), suggesting a possible functional interaction between these photoreceptors.
In addition to phytochrome/cryptochrome co-actions, a red light pretreatment, mediated by phytochromes A and B, has been demonstrated to enhance phototropic curvature of Arabidopsis hypocotyls in blue light, suggesting functional interaction between phytochrome and phototropin photoreceptors (Parks et al., 1996; Janoudi et al., 1997a, b).
Gravitropic orientation
Gravity provides a continuous directional signal to plants, ensuring roots grow downwards towards water and nutrients in the soil and shoots grow upwards towards sunlight. When grown in the dark, Arabidopsis hypocotyls display negative gravitropism and grow more or less vertically upwards against the gravitational vector. This strategy presumably enables buried seedlings to reach the soil surface as efficiently as possible. When subject to either continuous or intermittent R treatment, however, randomization of the direction of hypocotyl growth is observed (Liscum and Hangarter, 1993). This strategy enhances plant fitness through facilitating seedling emergence in patchy low light environments (Allen et al., 2006). Mutant analyses have revealed redundant roles for both phyA and phyB in regulating light-mediated randomization. Red light-mediated hypocotyl randomization was observed in both phyA and phyB single mutants, but was absent in mutants lacking both photoreceptors (Poppe et al., 1996; Robson and Smith, 1996). Significant hypocotyl randomization was also observed in WT seedlings following pulses of FR (Poppe et al., 1996). Mutants deficient in phyA displayed negative gravitropism in these conditions, confirming a role for this photoreceptor in mediating gravitropic sensitivity in the VLFR mode (Poppe et al., 1996). Light can also modulate gravitropic responses in roots. Mutants lacking phyB have been shown to display reduced rates of gravitropic curvature, a response which may be attributed to their altered elongation rates (Correll and Kiss, 2005).
Plant architecture
Phytochromes perform many overlapping roles in the regulation of plant architecture. The dissection of individual phytochrome functions has been made possible through the construction of mutants deficient in multiple phytochrome combinations. Arabidopsis phyA mutants display a WT adult phenotype in W and R (Fig. 2; Whitelam et al., 1993; Franklin et al., 2007). By contrast, phyB performs a dominant role in suppressing petiole elongation and apical dominance in light-grown plants. Mutants, deficient in phyB display significantly elongated petioles, reduced leaf area, and increased apical dominance (Fig. 2; Nagatani et al., 1991; Reed et al., 1993). These shoot architectural adaptations are accompanied by increased root hair growth (Reed et al., 1993). Suppression of axes elongation growth in WT plants is facilitated by the redundant actions of phytochromes A, D, and E. When grown in W and R, phyB-deficient mutant combinations display progressively more elongated phenotypes with increasing phytochrome deficiency (Fig. 2; Devlin et al., 1998, 1999; Franklin et al., 2007). Changes in light quality directly manipulate the phyB-, D-, and E-mediated suppression of elongation growth, providing plants with the capacity to alter their architecture in response to the threat of vegetational shading (see ‘Shade avoidance’). Mutants deficient in phyC have also been shown to display increased petiole elongation in W, suggesting an accessory role for this phytochrome in modulating leaf architecture (Franklin et al., 2003a; Monte et al., 2003).
Fig. 2.
WT (A), phyA (B), phyB (C), phyABE (D), and phyABDE (E) plants grown in 8 h photoperiods of white light at 120 μmol m−2 s−1. Scale bar represents 10 mm.
Arabidopsis displays a compact rosette phenotype. Maintenance of this growth habit is regulated by the redundant interactions of phyA, phyB, and phyE. Mutants deficient in these three phytochromes display visible internodes between rosette leaves (Fig. 2D). This phenotype is not visible in single or double mutant combinations and was the basis upon which the phyE mutant was isolated (Devlin et al., 1998). Interestingly, elongated internodes have also been recorded in phyBcry1 double mutants when grown at 22 °C. This phenotype was not observed in plants grown at cooler temperatures, suggesting conditional modulation of photoreceptor interactions (Mazzella et al., 2000).
Shade avoidance
One of the greatest threats to plant survival in natural environments is light limitation through shading by neighbouring vegetation. In shade-intolerant species, phytochromes perform a major role in the detection of neighbouring vegetation and initiation of escape responses before canopy closure. Light transmitted through or reflected from living vegetation is depleted in R and B wavebands, which are absorbed by chlorophyll and carotenoid pigments and used for photosynthesis. Far-red and green wavebands are enriched in reflected/transmitted light, resulting in a reduction in the ratio of R to FR (R:FR). This parameter directly modifies phytochrome activity and can be more precisely defined as follows: photon irradiance between 660 nm and 670 nm/photon irradiance between 725 nm and 735 nm. When growing in close proximity to neighbouring vegetation, plants experience a reduction in R:FR and initiate a suite of developmental responses termed the shade avoidance syndrome (reviewed in Smith and Whitelam, 1997). These include elongation of stems and petioles and increased apical dominance. Such responses serve to elevate leaves within a canopy and enable plants to overtop competitors. In this way, reflected FR signals can initiate adaptive responses before leaves are actually shaded (Ballaré et al., 1990). If light foraging is unsuccessful and the low R:FR signal persists then flowering is accelerated, enabling seed production in unfavourable conditions (Halliday et al., 1994).
When growing under a vegetational canopy, reductions in R:FR are accompanied by reductions in photosynthetically active radiation (PAR), in particular R and B wavebands. Architectural adaptations to canopy shading therefore involve the interaction of phytochrome signalling with UV-A/blue light photoreceptor signalling networks (reviewed in Franklin, 2008). The suppression of shade avoidance responses in high R:FR is mediated predominantly by phyB. The dramatically elongated, early flowering phenotype of phyB mutants is often therefore described as ‘constitutive shade avoidance’ (Nagatani et al., 1991; Somers et al., 1991). Observations of residual shade avoidance responses in phyB mutants of multiple species provided evidence for the involvement of additional phytochromes in R:FR signalling (Whitelam and Smith, 1991; Smith et al., 1992; Robson et al., 1993). Analyses of Arabidopsis mutants, deficient in multiple phytochrome combinations subsequently revealed redundant roles for phyD and E. The roles of phyD and phyE in shade avoidance were investigated using End-Of-Day-FR (EOD-FR) treatments. Plants subject to EOD-FR maintain a low Pfr status throughout the subsequent dark period and mimic plants grown in low R:FR. When grown in W, phyD mutants displayed no obvious morphological phenotype or aberrant response to EOD-FR (Aukerman et al., 1997). The combined deficiency of phyB and phyD, however, was shown to result in longer hypocotyls, longer petioles, and earlier flowering than phyB deficiency alone, suggesting redundancy of the phyB and phyD function in suppressing shade avoidance (Aukerman et al., 1997; Devlin et al., 1999). As with phyD, monogenic mutants deficient in phyE were observed to phenocopy WT plants in multiple light conditions. Double mutants, deficient in both phyB and phyE displayed longer petioles and earlier flowering than phyB monogenic mutants, supporting an additional redundant role for phyE in suppressing these responses (Devlin et al., 1998). Confirmation of the redundant activities of phytochromes B, D, and E in mediating shade avoidance was provided by analysis of triple phyBDE mutants which showed no further response to reductions in R:FR or EOD-FR (Franklin et al., 2003b).
Enrichment of FR wavelengths in reflected/transmitted light can lead to enhanced phyA signalling in the HIR mode. Such activity provides some inhibition of elongation growth, thereby limiting shade avoidance responses. Laboratory analyses have shown phyA mutants to display enhanced elongation responses to low R:FR treatment when compared to WT controls (Johnson et al.,1994; Smith et al., 1997; Salter et al., 2003). The adaptive significance of this phyA-mediated ‘antagonism’ of shade avoidance has been demonstrated in the field by Yanovsky and colleagues. In these experiments, Arabidopsis phyA mutants were grown under the natural shade of densely planted Triticum aestivum plants. Mutants deficient in phyA displayed extreme elongation growth, poor cotyledon development, and a lower survival rate (25%) than WT seedlings (58%) (Yanovsky et al., 1995).
One of the first genes reported to display reversible regulation of transcript abundance by low R:FR was the homeodomain leucine zipper (HD Zip) transcription factor ATHB2 (formerly HAT4) (Carabelli et al., 1993). Elevated levels of ATHB2 transcript were recorded in phyB mutants, confirming a role for this phytochrome in repressing ATHB2 expression (Carabelli et al., 1996). Further increases in ATHB2 transcripts were, however, still observed in phyB mutants following low R:FR and EOD-FR treatments, suggesting the involvement of additional phytochrome(s) in controlling this response. Studies of mutants deficient in multiple phytochrome combinations have since confirmed that R:FR-ratio-mediated changes in ATHB2 expression are regulated by the redundant actions of phyB and phyE (Franklin et al., 2003b). Microarray analyses have now identified multiple genes displaying altered transcript abundance following low R:FR and simulated shade treatments of light-grown plants (Devlin et al., 2003; Salter et al., 2003; Sessa et al., 2005). As with ATHB2, many of these genes were observed to display enhanced transcript abundance in phyB mutants (Devlin et al., 2003; Salter et al., 2003). One such gene (PIL1), encoding a bHLH transcription factor displaying high sequence similarity to PIF3, displayed an increase in transcript abundance of over 100-fold within 30 min of low R:FR treatment. Triple mutants deficient in phytochromes B, D, and E showed no further increase in PIL1 transcripts following low R:FR treatment, confirming the redundant interactions of these photoreceptors in mediating this response (Salter et al., 2003). Interestingly, a number of genes displaying low R:FR-mediated de-repression in light-grown plants have also been reported to display phytochrome-mediated repression following transfer of etiolated seedlings to both FR and R, including PIL1 (Tepperman et al., 2001, 2004; Quail, 2007; Hwang and Quail, 2008). Together, these data indicate that phytochrome-regulation of gene expression is a key regulatory mechanism underlying photomorphogenesis at multiple stages of development.
Stomatal development
Further roles for phyB in modulating plant architecture have recently been proposed in the regulation of stomatal development. Mutants deficient in phyB were shown to display reduced stomatal index (SI) at higher photon irradiances of both W and R (Boccalandro et al., 2009; Casson et al., 2009). Stomatal index represents the ratio of the number of stomata in a given area, divided by the number of stomata and other epidermal cells in the same area, thereby providing an indication of the extent of stomatal differentiation. Increased photon irradiance is well documented to result in increased SI, an adaptation which likely enhances gas exchange during conditions of high photosynthetic activity (Lake et al., 2001). This response displayed significant attenuation in phyB mutants (Casson et al., 2009). A decreased SI was additionally recorded in WT plants subject to EOD-FR treatment (Boccalandro et al., 2009). The reduced SI observed in phyB mutants was recorded to result in reduced transpiration per unit leaf area which ultimately enhanced water use efficiency (WUE) in these plants (Boccalandro et al., 2009). The authors propose that phyB-mediated increases in stomatal differentiation serve to enhance photosynthesis in high R:FR/high PAR environments, a strategy which is implemented at the expense of WUE.
Reproductive development
A key role of phytochromes in natural light environments involves the monitoring of daylength or photoperiod which, together with temperature, provide plants with important seasonal information. Many plants use seasonal cues to coincide the timing of reproductive development with conditions of optimal climate and/or competitive advantage. Daylength perception requires the integration of light signals with the plant's endogenous oscillators, circadian clocks (Thomas, 2006). Considerable progress has been made in elucidating the molecular components of the Arabidopsis circadian system, which comprises at least three interlocking transcriptional/translational feedback loops (reviewed in Ueda, 2006; McClung, 2006, 2008; Hotta et al., 2007; Pruneda-Paz et al., 2009). The first identified loop (termed ‘the central oscillator’) contains two light-regulated Myb domain transcription factors, CIRCADIAN AND CLOCK ASSOCIATED 1 (CCA1) and LONG HYPOCOTYL (LHY) which, together, regulate abundance of the PSEUDO RESPONSE REGULATOR (PRR) protein, TIMING OF CAB1 (TOC1) and the recently identified CHE, a TCP transcription factor. A second interlocking loop incorporates the PRR proteins PRR7 and PRR9 whilst a third loop involves GI and possibly PRR5. Arabidopsis circadian clocks run with a period of between 22 h and 29 h (Michael et al., 2003) and require daily input or ‘entrainment’ signals from the environment. The ability to co-ordinate photosynthesis and metabolism with day/night cycles has been shown to confer considerable competitive advantage and, ultimately, to enhance plant survival (Dodd et al., 2005).
Entrainment of the circadian clock
Light signals provide important day/night entrainment information to plant circadian clocks and are transduced by multiple photoreceptors. The photoreceptors involved in light input to the circadian clock have been elucidated in Arabidopsis, through analyses of null mutants. To date, roles have been established for phytochromes A, B, D, and E and the cryptochromes 1 and 2 (Somers et al., 1998; Devlin and Kay, 2000). The role of phyC has not been examined. Reporter gene analyses in a variety of mutant backgrounds have shown conditional modulation of circadian period by phytochrome. Increased photon irradiance shortens period length in diurnal organisms such as Arabidopsis (Aschoff, 1979; Millar et al., 1995). Mutants deficient in phyA have been shown to display longer periods of CHLOROPHYLL A/B BINDING PROTEIN::LUCIFERASE (CAB::LUC) expression at lower photon irradiances of R (<2 μmol m−2 s−1), whilst a similar phenotype was observed in phyB mutants at higher photon irradiances (>10 μmol m−2 s−1) (Somers et al., 1998). Mutants deficient in both phytochromes (phyAB) displayed a long period phenotype at all photon irradiances examined (Devlin and Kay, 2000). A further role for phyA was recorded at lower photon irradiances of B, consistent with the role of this phytochrome in B sensing (Somers et al., 1998). Comparative analyses of phyABD and phyABE mutants in R revealed small additional roles for phyD and phyE in regulating CAB::LUC period length, which were masked by the presence of phyB in WT plants. An unusual result from this study was the observation that cry1 mutants displayed deficiencies in the perception of low photon irradiance R, similar to those observed in phyA mutants. As cryptochromes show no absorption peak in R, the authors conclude that cry1 acts as a signal transduction component of phyA in these conditions (Devlin and Kay, 2000).
Flowering
The timing of Arabidopsis floral transition is regulated by environmental stimuli via convergence of multiple signalling cascades. Transcriptional regulators act on floral integrators which, in turn, alter expression of meristem identity genes to promote flowering (reviewed in Turck et al., 2008). Arabidopsis is a characteristic ‘Long-Day-Plant’ (LDP), displaying accelerated flowering in longer photoperiods. This process is determined by the length of the dark period and is regulated via an ‘external co-incidence’ of light with a threshold level of the rhythmically cycling transcriptional activator CONSTANS (CO). This co-incidence leads to elevated expression of the floral integrator FLOWERING TIME (FT) and, ultimately, flowering (Yanovsky and Kay, 2002). Photoreceptor mutant analyses have revealed roles for both phyA and cry2 in perceiving long photoperiods. Photoperiodic sensitivity can be investigated in laboratory experiments by extending short photoperiods with low quantities of incandescent light (thereby not enhancing photosynthetic activity) and by mimicking short day conditions with ‘night break’ light treatments during the dark period. Flowering time studies have shown phyA mutants to display insensitivity to both daylength extensions and night-break treatments, confirming a role for this phytochrome in photoperiodic perception (Johnson et al., 1994; Reed et al., 1994). These data are supported by observations of late flowering in phyA mutants grown in long photoperiods (Johnson et al., 1994; Neff and Chory, 1998). Analyses of multiple photoreceptor-deficient mutants in monochromatic light have also revealed a redundant role for phyA in the B-mediated regulation of flowering. When grown in continuous B, phyA, cry1, and cry2 mutants all flowered at a similar time to WT plants. Pronounced late flowering was, however, recorded in all double mutant combinations confirming the redundant interactions of these three photoreceptors in B-mediated floral promotion (Mockler et al., 2003).
Phytochrome B acts redundantly with phyD and phyE to repress flowering in high R:FR conditions (see ‘Shade avoidance’). Relief of floral repression requires prolonged exposure to low R:FR, ensuring that the precocious transition to reproductive development does not occur in response to transient shading (Halliday et al., 1994). Interestingly, the functional hierarchy of phytochromes in mediating floral repression has been shown to display regulation by ambient growth temperature. When grown at 22 °C, phyB mutants are characterized by their early flowering phenotype (Goto et al., 1991; Whitelam and Smith, 1991; Reed et al., 1993). When grown at 16 °C, however, similar flowering times were recorded in phyB and WT plants (Halliday et al., 2003). As with the promotion of seed germination (see ‘Germination’), phyE appears to adopt a dominant role in cooler conditions (Halliday and Whitelam, 2003). When grown in short photoperiods at 16 °C, phyABD mutants flowered with a similar number of rosette leaves to WT plants. Early flowering was, however, observed in phyABDE mutants, confirming the significance of phyE function in these conditions. Transcript abundance of the floral integrator FLOWERING TIME (FT) displayed a positive correlation with the loss of dominant phytochrome function at 16 °C and 22 °C suggesting that phytochromes inhibit flowering through repression of FT expression (Halliday et al., 2003).
Mutant studies have also revealed roles for phyC in the regulation of Arabidopsis flowering time. Accelerated flowering was observed in phyC-deficient mutants in the Col background when grown in short days (Monte et al., 2003). Comparison of phyAC mutants with monogenic parents also revealed a small redundant role for phyC in the detection of long photoperiods with phyA (Monte et al., 2003). No role for phyC in the regulation of flowering was observed in the Ws background (Franklin et al., 2003a), suggesting the existence of natural variation in phyC function. Quantitative genetic analyses have since suggested that allelic variation at the PHYC locus accounts for considerable latitudinal variation in flowering time, thereby providing support for this notion (Balasubramanian et al., 2006).
Freezing tolerance
Further cross-talk between light and temperature signalling pathways has been identified in the regulation of Arabidopsis freezing tolerance. In order to survive sub-zero temperatures, many plants require a period of low temperature (<4 °C), termed cold acclimation. Exposure to low temperature elevates expression of a number of genes encoding proteins which protect plants against freezing damage through membrane stabilization and the accumulation of compatible solutes (Chinnusumay et al., 2007). Phytochromes have been shown to perform a role in the regulation of one such regulon, the C-repeat-Binding-Factor (CBF) regulon (Franklin and Whitelam, 2007). Low R:FR treatment at 16 °C was shown to elevate expression of both CBF transcription factors and their downstream targets, the COLD-REGULATED (COR) genes, leading to enhanced freezing tolerance. Intriguingly, the linkage of CBFs to downstream target genes was uncoupled at 22 °C. Mutant analyses revealed repression of the CBF regulon to be mediated by phyB and phyD in a non-redundant manner (Franklin and Whitelam, 2007). The authors speculate that the cooler temperatures and prolonged twilight reductions in low R:FR experienced by plants growing at Northern latitudes may confer some seasonal protection against sudden freezing snaps during warmer than average autumn months.
Future perspectives
Phytochromes perform a diverse array of regulatory functions throughout plant development (Table 2). The isolation and construction of Arabidopsis mutants deficient in both single and multiple phytochrome combinations has provided an invaluable set of tools to explore the conditional and redundant interactions of individual family members. Observations of heterodimerization between phytochromes B–E may suggest further functional diversification of these photoreceptors (Sharrock and Clack, 2004). A more recent study has suggested obligate heterodimerization of phyC and phyE with other Type II phytochromes (Clack et al., 2009), although the de-etiolation and survival of phyABDE mutants through to flowering in R would appear to suggest some activity of phyC, either as homodimers or monomers in planta (Franklin et al., 2003b, 2007). Co-action between phytochromes and UV-A/blue light photoreceptors enables plants to precisely monitor changes in light quantity and quality throughout development. Moreover, early studies of plant photomorphogenesis revealed that protocols combining red and blue light treatments are more effective than either waveband in isolation (reviewed in Mohr,1994). These observations led to the provocative suggestion that the sole function of UV-A/blue light photoreceptors was to enhance the activity of phytochrome, which acted as a ‘single effector’ to mediate photomorphogenic responses (Mohr, 1994). Despite the fundamental importance of this issue to our understanding of plant photobiology, an unequivocal answer as to whether UV-A/blue light photoreceptors can operate independently of phytochrome action has yet to be provided. The construction of quadruple phytochrome-deficient mutants has enabled investigation into the functional capabilities of individual phytochromes in the absence of other family members. To date, phyABDE and phyBCDE plants have been created, containing only phyC and phyA, respectively. The germination, de-etiolation and survival through to flowering of these mutants in both W and R shows the diverse functional capabilities of individual phytochromes; roles which are usually masked by the actions of other family members in WT plants (Fig. 1; Franklin et al., 2003b, 2007). The existence of Arabidopsis quadruple mutants, deficient in four family members, should now enable the implementation of crossing strategies designed to produce a totally phytochrome null plant. The successful construction of a phyABCDE quintuple mutant will, ultimately, address a long unresolved question in photomorphogenesis research, whether phytochromes are an obligate requirement for the germination, growth and reproduction of flowering plants.
Table 2.
Summary of phytochrome functions, elucidated through analysis of Arabidopsis mutants
Dedication
This review is dedicated to the memory of Professor Garry Whitelam (1955–2008). Garry was an internationally respected figure in plant photomorphogenesis research and made numerous important contributions to the field. In addition to leading many advances in plant shade avoidance research, Garry was at the forefront of phytochrome mutant isolation and the elucidation of redundant phytochrome functions through multiple mutant analyses. He will be greatly missed.
Acknowledgments
Work in the laboratory of PHQ was supported by National Institutes of Health Grant GM-47475, Department of Energy Grant DEFG03-87ER13742, and USDA Agricultural Research Service Current Research Information System Grant 5335-21000-027-00D. KAF is a Royal Society University Research Fellow.
References
- Ahmad M, Jarillo JA, Smirnova O, Cashmore AR. The CRY1 blue light photoreceptor of Arabidopsis interacts with phytochrome A in vitro. Molecular Cell. 1998;1:939–948. doi: 10.1016/s1097-2765(00)80094-5. [DOI] [PubMed] [Google Scholar]
- Al-Sady B, Kikis EA, Monte E, Quail PH. Mechanistic duality of transcription factor function in phytochrome signalling. Proceedings of the National Academy of Sciences, USA. 2008;105:2232–2237. doi: 10.1073/pnas.0711675105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Allen T, Ingles PJ, Praekelt U, Smith H, Whitelam GC. Phytochrome-mediated agravitropism in Arabidopsis hypocotyls requires GIL1 and confers a fitness advantage. The Plant Journal. 2006;46:641–648. doi: 10.1111/j.1365-313X.2006.02727.x. [DOI] [PubMed] [Google Scholar]
- Al-Sady B, Ni W, Kircher S, Scháfer E, Quail PH. Photoactivated phytochrome induces rapid PIF3 phosphorylation as a prelude to proteosome-mediated degradation. Molecular Cell. 2006;23:439–446. doi: 10.1016/j.molcel.2006.06.011. [DOI] [PubMed] [Google Scholar]
- Aschoff J. Circadian rhythms: influences of internal and external factors on the period measured in constant conditions. Zeitschrift für Tierpsychologie. 1979;49:225–249. doi: 10.1111/j.1439-0310.1979.tb00290.x. [DOI] [PubMed] [Google Scholar]
- Aukerman MJ, Hirschfeld M, Wester L, Weaver M, Clack T, Amasino RM, Sharrock RA. A deletion in the PHYD gene of the Arabidopsis Wassilewskija ecotype defines a role for phytochrome D in red/far-red light sensing. The Plant Cell. 1997;9:1317–1326. doi: 10.1105/tpc.9.8.1317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Balasubramanian S, Sureshkumar S, Agrawal M, Michael TP, Wessinger C, Maloof JN, Clark R, Warthmann N, Chory J, Weigel D. The PHYTOCHROME C photoreceptor gene mediates natural variation in flowering and growth responses of Arabidopsis thaliana. Nature Genetics. 2006;38:711–715. doi: 10.1038/ng1818. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ballaré CL, Scopel AL, Sánchez RA. Far-red radiation reflected from adjacent leaves: an early signal of competition in plant canopies. Science. 1990;247:329–332. doi: 10.1126/science.247.4940.329. [DOI] [PubMed] [Google Scholar]
- Bauer D, Viczián A, Kircher S, et al. Constitutive Photomorphogenesis 1 and multiple photoreceptors control degradation of Phytochrome Interacting Factor 3, a transcription factor required for light signalling in Arabidopsis. The Plant Cell. 2004;16:1433–1445. doi: 10.1105/tpc.021568. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boccalandro HE, Rugnone ML, Moreno JE, Ploschuk EL, Serna L, Yanovsky MJ, Casal JJ. Phytochrome B enhances photosynthesis at the expense of water-use-efficiency in Arabidopsis. Plant Physiology. 2009;150:1083–1092. doi: 10.1104/pp.109.135509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Borthwick HA, Hendricks SB, Parker MW, Toole EH, Toole VK. A reversible photoreaction controlling seed germination. Proceedings of the National Academy of Sciences, USA. 1952;38:662–666. doi: 10.1073/pnas.38.8.662. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Botto JF, Sanchez RA, Whitelam GC, Casal JJ. Phytochrome A mediates the promotion of seed germination by very low fluences of light and canopy shade light in Arabidopsis. Plant Physiology. 1996;110:439–444. doi: 10.1104/pp.110.2.439. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carabelli M, Morelli G, Whitelam GC, Ruberti I. Twilight-zone and canopy shade induction of the ATHB-2 homeobox gene in green plants. Proceedings of the National Academy of Sciences, USA. 1996;93:3530–3535. doi: 10.1073/pnas.93.8.3530. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carabelli M, Sessa G, Ruberti I, Morelli G. The Arabidopsis ATHB-2 and -4 genes are strongly induced by far-red-rich light. The Plant Journal. 1993;4:469–479. doi: 10.1046/j.1365-313x.1993.04030469.x. [DOI] [PubMed] [Google Scholar]
- Casal JJ, Mazella MA. Conditional synergism between cryptochrome 1 and phytochrome B is shown by the analysis of phyA, phyB and hy4 simple, double and triple mutants in Arabidopsis. Plant Physiology. 1998;118:19–25. doi: 10.1104/pp.118.1.19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Casal JJ, Sánchez RA. Phytochromes and seed germination. Seed Science Research. 1998;8:317–329. [Google Scholar]
- Casson SA, Franklin KA, Gray JE, Grierson CS, Whitelam GC, Hetherington AM. Phytochrome B and PIF4 regulate stomatal development in response to light quantity. Current Biology. 2009;19:229–234. doi: 10.1016/j.cub.2008.12.046. [DOI] [PubMed] [Google Scholar]
- Chinnusumay V, Zhu J, Zhu JK. Cold stress regulation of gene expression in plants. Trends in Plant Science. 2007;12:444–451. doi: 10.1016/j.tplants.2007.07.002. [DOI] [PubMed] [Google Scholar]
- Clack T, Mathews S, Sharrock RA. The phytochrome apoprotein family in Arabidopsis is encoded by five genes: the sequences and expression of PHYD and PHYE. Plant Molecular Biology. 1994;25:413–427. doi: 10.1007/BF00043870. [DOI] [PubMed] [Google Scholar]
- Clack T, Shokry A, Moffet M, Liu P, Faul M, Sharrock RA. Obligate heterodimerization of Arabidopsis phytochromes C and E and interaction with the PIF3 basic helix-loop-helix transcription factor. The Plant Cell. 2009;21:786–799. doi: 10.1105/tpc.108.065227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clough RC, Vierstra RD. Phytochrome degradation. Plant, Cell and Environment. 1997;20:713–721. [Google Scholar]
- Correll MJ, Kiss JZ. The roles of phytochromes in elongation and gravitropism of roots. Plant and Cell Physiology. 2005;46:317–323. doi: 10.1093/pcp/pci038. [DOI] [PubMed] [Google Scholar]
- Darwin C, Darwin F. The power of movement in plants. London: John Murray; 1880. [Google Scholar]
- Dehesh K, Franci C, Parks BM, Seeley KA, Short TW, Tepperman JM, Quail PH. Arabidopsis HY8 locus encodes phytochrome A. The Plant Cell. 1993;5:1081–1088. doi: 10.1105/tpc.5.9.1081. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Devlin PF, Kay SA. Cryptochromes are required for phytochrome signaling to the circadian clock but not for rhythmicity. The Plant Cell. 2000;12:2499–2509. doi: 10.1105/tpc.12.12.2499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Devlin PF, Yanovsky MJ, Kay SA. A genomic analysis of the shade avoidance response in Arabidopsis. Plant Physiology. 2003;133:1617–1629. doi: 10.1104/pp.103.034397. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Devlin PF, Patel SR, Whitelam GC. Phytochrome E influences internode elongation and flowering time in Arabidopsis. The Plant Cell. 1998;10:1479–1487. doi: 10.1105/tpc.10.9.1479. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Devlin PF, Robson PRH, Patel SR, Goosey L, Sharrock RA, Whitelam GC. Phytochrome D acts in the shade-avoidance syndrome in Arabidopsis by controlling elongation and flowering time. Plant Physiology. 1999;119:909–915. doi: 10.1104/pp.119.3.909. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dodd AN, Salanthia N, Hall A, Kévei E, Tóoth R, Nagy F, Hibberd JM, Millar AJ, Webb AAR. Plant circadian clocks increase photosynthesis, growth, survival and competitive advantage. Science. 2005;309:630–633. doi: 10.1126/science.1115581. [DOI] [PubMed] [Google Scholar]
- Duek PD, Fankhauser C. bHLH class transcription factors take centre stage in phytochrome signaling. Trends in Plant Science. 2005;10:51–54. doi: 10.1016/j.tplants.2004.12.005. [DOI] [PubMed] [Google Scholar]
- Franklin KA. Shade avoidance. New Phytologist. 2008;179:930–944. doi: 10.1111/j.1469-8137.2008.02507.x. [DOI] [PubMed] [Google Scholar]
- Franklin KA, Allen T, Whitelam GC. Phytochrome A is an irradiance-dependent red light sensor. The Plant Journal. 2007;50:108–117. doi: 10.1111/j.1365-313X.2007.03036.x. [DOI] [PubMed] [Google Scholar]
- Franklin KA, Davis SJ, Stoddart WM, Vierstra RD, Whitelam GC. Mutant analyses define multiple roles for phytochrome C in Arabidopsis thaliana photomorphogenesis. The Plant Cell. 2003a;15:1981–1989. doi: 10.1105/tpc.015164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Franklin KA, Praekelt U, Stoddart WM, Billingham OE, Halliday KJ, Whitelam GC. Phytochromes B, D and E act redundantly to control multiple physiological responses in Arabidopsis. Plant Physiology. 2003b;131:1340–1346. doi: 10.1104/pp.102.015487. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Franklin KA, Whitelam GC. Light-quality regulation of freezing tolerance in Arabidopsis thaliana. Nature Genetics. 2007;39:1410–1413. doi: 10.1038/ng.2007.3. [DOI] [PubMed] [Google Scholar]
- Goosey L, Palecanda L, Sharrock RA. Differential patterns of expression of the Arabidopsis PHYB, PHYD and PHYE phytochrome genes. Plant Physiology. 1997;115:959–969. doi: 10.1104/pp.115.3.959. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goto N, Kumagai T, Koornneef M. Flowering responses to light-breaks in photomorphogenic mutants of Arabidopsis thaliana, a long day plant. Physiologia Plantarum. 1991;83:209–215. [Google Scholar]
- Halliday KJ, Koornneef M, Whitelam GC. Phytochrome B and at least one other phytochrome mediate the accelerated flowering response of Arabidopsis thaliana L. to low red/far-red ratio. Plant Physiology. 1994;104:1311–1315. doi: 10.1104/pp.104.4.1311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Halliday KJ, Salter MG, Thingnaes E, Whitelam GC. Phytochrome control of flowering is temperature sensitive and correlates with expression of the floral integrator FT. The Plant Journal. 2003;33:875–885. doi: 10.1046/j.1365-313x.2003.01674.x. [DOI] [PubMed] [Google Scholar]
- Halliday KJ, Whitelam GC. Changes in photoperiod or temperature reveal roles for phyD and phyE. Plant Physiology. 2003;131:1913–1920. doi: 10.1104/pp.102.018135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hennig L, Büche C, Schäfer E. Degradation of phytochrome A and the high irradiance response in Arabidopsis: a kinetic analysis. Plant, Cell and Environment. 2000;23:727–734. [Google Scholar]
- Hennig L, Funk M, Whitelam GC, Schäfer E. Functional interaction of cryptochrome 1 and phytochrome D. The Plant Journal. 1999;20:289–294. doi: 10.1046/j.1365-313x.1999.t01-1-00599.x. [DOI] [PubMed] [Google Scholar]
- Hennig L, Stoddart WM, Dieterle M, Whitelam GC, Schäfer E. Phytochrome E controls light-induced germination of Arabidopsis. Plant Physiology. 2002;128:194–200. [PMC free article] [PubMed] [Google Scholar]
- Heschel MS, Selby J, Butler C, Whitelam GC, Sharrock RA, Donohue K. A new role for phytochromes in temperature-dependent germination. New Phytologist. 2007;174:735–741. doi: 10.1111/j.1469-8137.2007.02044.x. [DOI] [PubMed] [Google Scholar]
- Hotta CT, Gardner MJ, Hubbard KE, Baek SJ, Dalchau N, Suhita D, Dodd AN, Webb AAR. Modulation of environmental responses of plants by circadian clocks. Plant, Cell and Environment. 2007;30:333–349. doi: 10.1111/j.1365-3040.2006.01627.x. [DOI] [PubMed] [Google Scholar]
- Huq E, Al-Sady B, Hudson ME, Kim C, Apel K, Quail PH. PHYTOCHROME-INTERACTING FACTOR 1 is a critical bHLH regulator of chlorophyll biosynthesis. Science. 2004;305:1937–1941. doi: 10.1126/science.1099728. [DOI] [PubMed] [Google Scholar]
- Huq E, Quail PH. PIF4, a phytochrome-interacting bHLH factor functions as a negative regulator of phytochrome B signaling in Arabidopsis. EMBO Journal. 2002;21:2441–2450. doi: 10.1093/emboj/21.10.2441. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hwang Y-S, Quail PH. Phytochrome-regulated PIL1 derepression is developmentally modulated. Plant and Cell Physiology. 2008;49:501–511. doi: 10.1093/pcp/pcn024. [DOI] [PubMed] [Google Scholar]
- Janoudi AK, Gordon WR, Wagner D, Quail P, Poff KL. Multiple phytochromes are involved in red-light induced enhancement of first positive phototropism in Arabidopsis thaliana. Plant Physiology. 1997a;113:975–979. doi: 10.1104/pp.113.3.975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Janoudi AK, Konjevic R, Whitelam GC, Gordon W, Poff KL. Both phytochrome A and phytochrome B are required for the normal expression of phototropism in Arabidopsis thaliana seedlings. Physiologia Plantarum. 1997b;101:278–282. [Google Scholar]
- Johnson E, Bradley JM, Harberd NP, Whitelam GC. Photoresponses of light-grown phyA mutants of Arabidopsis: phytochrome A is required for the perception of daylength extensions. Plant Physiology. 1994;105:141–149. doi: 10.1104/pp.105.1.141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Khanna R, Huq E, Kikis EA, Al-Sady B, Lanzatella C, Quail PH. A novel molecular recognition motif necessary for targeting photoactivated phytochrome signalling to specific basic helix-loop-helix transcription factors. The Plant Cell. 2004;16:3033–3044. doi: 10.1105/tpc.104.025643. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Khanna R, Shen Y, Mario CM, Tsuchisaka A, Theologis A, Scháfer E, Quail PH. The basic helix-loop-helix transcription factor PIF5 acts on ethylene biosynthesis and phytochrome signaling by distinct mechanisms. The Plant Cell. 2007;19:3915–3929. doi: 10.1105/tpc.107.051508. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim J, Yi H, Choi G, Shin B, Song P-S, Choi G. Functional characterization of phytochrome interacting factor 3 in phytochrome-mediated light signal transduction. The Plant Cell. 2003;15:2399–2407. doi: 10.1105/tpc.014498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kircher S, Kozma-Bognar L, Kim L, Adam E, Harter K, Scháfer E, Nagy F. Light quality-dependent nuclear import of the plant photoreceptors phytochrome A and B. The Plant Cell. 1999;11:1445–1456. doi: 10.1105/tpc.11.8.1445. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kircher S, Kozma-Bognar L, Fejes E, Speth V, Husselstein-Muller T, Bauer D, Adam E, Scháfer E, Nagy F. Nucleo-cytoplasmic partitioning of plant photoreceptors phytochromes A, B, C, D, and E is differentially regulated by light and exhibits a diurnal rhythm. The Plant Cell. 2002;14:1514–1544. doi: 10.1105/tpc.001156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koornneef M, Rolff E, Spruit CJP. Genetic control of light-inhibited hypocotyl elongation in Arabidopsis thaliana (L.) Heynh. Zeitschrift für Pflanzenphysiologie. 1980;100:147–160. [Google Scholar]
- Lake JA, Quick WP, Beerling DJ, Woodward FI. Plant development. Signals from mature to new leaves. Nature. 2001;411:154. doi: 10.1038/35075660. [DOI] [PubMed] [Google Scholar]
- Lagarias JC, Rapoport H. Chromopeptides from phytochrome: the structure and linkage of the Pfr form of the phytochrome chromophore. Journal of the American Chemical Society. 1980;102:4821–4828. [Google Scholar]
- Leivar P, Monte E, Al-Sady B, Carle C, Storer A, Alonso JM, Ecker JR, Quail PH. The Arabidopsis Phytochrome-Interacting Factor PIF7, together with PIF3 and PIF4, regulates responses to prolonged red light by modulating phyB levels. The Plant Cell. 2008;20:337–352. doi: 10.1105/tpc.107.052142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin C, Yang H, Guo H, Mockler T, Chen J, Cashmore AR. Enhancement of blue light sensitivity of Arabidopsis seedlings by a blue light receptor cryptochrome 2. Proceedings of the National Academy of Sciences, USA. 1998;95:7686–7699. doi: 10.1073/pnas.95.5.2686. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liscum E, Hangarter RP. Genetic evidence that the Pr form of phytochrome B plays a role in Arabidopsis thaliana gravitropism. Plant Physiology. 1993;103:15–19. doi: 10.1104/pp.103.1.15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lorrain S, Allen T, Duek PD, Whitelam GC, Fankhauser C. Phytochrome-mediated inhibition of shade avoidance involves degradation of growth-promoting bHLH transcription factors. The Plant Journal. 2008;53:312–323. doi: 10.1111/j.1365-313X.2007.03341.x. [DOI] [PubMed] [Google Scholar]
- Más P, Devlin PF, Panda S, Kay SA. Functional interaction of phytochrome B and cryptochrome 2. Nature. 2000;408:207–211. doi: 10.1038/35041583. [DOI] [PubMed] [Google Scholar]
- Mathews S, Lavin M, Sharrock RA. Evolution of the phytochrome gene family and its utility for phylogenetic analyses of angiosperms. Annals of the Missouri Botanic Garden. 1995;82:296–321. [Google Scholar]
- Mathews S, Sharrock RA. Phytochrome gene diversity. Plant, Cell and Environment. 1997;20:666–671. [Google Scholar]
- Mazzella MA, Bertero D, Casal JJ. Temperature-dependent internode elongation in vegetative plants of Arabidopsis thaliana lacking phytochrome B and cryptochrome 1. Planta. 2000;210:497–501. doi: 10.1007/PL00008157. [DOI] [PubMed] [Google Scholar]
- McClung CR. Comes a time. Current Opinion in Plant Biology. 2008;11:514–520. doi: 10.1016/j.pbi.2008.06.010. [DOI] [PubMed] [Google Scholar]
- McClung CR. Plant circadian rhythms. The Plant Cell. 2006;18:792–803. doi: 10.1105/tpc.106.040980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McCormac AC, Terry MJ. Light-signalling pathways leading to the co-ordinated expression of HEMA1 and Lhcb during chloroplast development in Arabidopsis thaliana. The Plant Journal. 2002;32:549–559. doi: 10.1046/j.1365-313x.2002.01443.x. [DOI] [PubMed] [Google Scholar]
- Michael TP, Salome PA, Yu HJ, Spencer TR, Sharp EL, McPeek MA, Alonso JM, Ecker JR, McClung CR. Enhanced fitness conferred by naturally occurring variation in the circadian clock. Science. 2003;302:1049–1053. doi: 10.1126/science.1082971. [DOI] [PubMed] [Google Scholar]
- Millar AJ, Straume M, Chory J, Chua N-H, Kay SA. The regulation of circadian photoperiod by phototransduction pathways in Arabidopsis. Science. 1995;267:1163–1166. doi: 10.1126/science.7855596. [DOI] [PubMed] [Google Scholar]
- Mockler T, Yang H, Yu X, Parikh D, Cheng Y-C, Dolan S, Lin C. Regulation of photoperiodic flowering by Arabidopsis photoreceptors. Proceedings of the National Academy of Sciences, USA. 2003;100:2140–2145. doi: 10.1073/pnas.0437826100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mohr H. Coaction between pigment systems. In: Kendrick RE, Kronenberg GHM, editors. Photomorphogenesis in plants. Dordrecht, The Netherlands: Kluwer; 1994. [Google Scholar]
- Monte E, Alonso JM, Ecker JR, Zhang Y, Li X, Young J, Austin-Phillips S, Quail PH. Isolation and characterization of phyC mutants in Arabidopsis reveals complex cross-talk between phytochrome signalling pathways. The Plant Cell. 2003;15:1962–1980. doi: 10.1105/tpc.012971. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Monte E, Al-Sady B, Leivar P, Quail PH. Out of the dark: how the PIFs are unmasking a dual temporal mechanism of phytochrome signalling. Journal of Experimental Botany. 2007;58:3125–3133. doi: 10.1093/jxb/erm186. [DOI] [PubMed] [Google Scholar]
- Monte E, Tepperman JM, Al-Sady B, Kaczorowski KA, Alonso JM, Ecker JR, Li X, Zhang Y, Quail PH. The phytochrome-interacting transcription factor, PIF3, acts early, selectively and positively in light-induced chloroplast development. Proceedings of the National Academy of Sciences, USA. 2004;101:16091–16098. doi: 10.1073/pnas.0407107101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nagatani A. Light-regulated nuclear localization of phytochromes. Current Opinion in Plant Biology. 2004;7:1–4. doi: 10.1016/j.pbi.2004.09.010. [DOI] [PubMed] [Google Scholar]
- Nagatani A, Chory J, Furuya M. Phytochrome B is not detectable in the hy3 mutant of Arabidopsis, which is deficient in responding to end-of-day far-red light treatments. Plant and Cell Physiology. 1991;32:1119–1112. [Google Scholar]
- Nagatani A, Reed JW, Chory J. Isolation and initial characterization of Arabidopsis mutants that are deficient in functional phytochrome A. Plant Physiology. 1993;102:269–277. doi: 10.1104/pp.102.1.269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Neff MM, Chory J. Genetic interaction between phytochrome A, phytochrome B and cryptochrome 1 during Arabidopsis development. Plant Physiology. 1998;118:27–36. doi: 10.1104/pp.118.1.27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Neff MM, Van Volkenburgh E. Light-stimulated cotyledon expansion in Arabidopsis seedlings: the role of phytochrome B. Plant Physiology. 1994;104:1027–1032. doi: 10.1104/pp.104.3.1027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ni M, Tepperman JM, Quail PH. PIF3, a phytochrome-interacting factor necessary for normal photoinduced signal transduction, is a novel basic helix-loop-helix protein. Cell. 1998;95:657–667. doi: 10.1016/s0092-8674(00)81636-0. [DOI] [PubMed] [Google Scholar]
- Ni M, Tepperman JM, Quail PH. Binding of phytochrome B to its nuclear signaling partner PIF3 is reversibly induced by light. Nature. 1999;400:462–466. doi: 10.1038/23500. [DOI] [PubMed] [Google Scholar]
- Park E, Kim J, Lee Y, Shin J, Oh E, Chung WI, Liu JR, Choi G. Degradation of phytochromeinteracting factor 3 in phytochrome-mediated signaling. Plant and Cell Physiology. 2004;45:968–975. doi: 10.1093/pcp/pch125. [DOI] [PubMed] [Google Scholar]
- Parks BM, Quail PH, Hangarter RP. Phytochrome A regulates red light induction of phototropic enhancement in Arabidopsis. Plant Physiology. 1996;110:155–162. doi: 10.1104/pp.110.1.155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parks BM, Quail PH. hy8, a new class of Arabidopsis long hypocotyl mutants deficient in functional phytochrome A. The Plant Cell. 1993;3:39–48. doi: 10.1105/tpc.5.1.39. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pruneda-Paz JL, Breton G, Para A, Kay SA. A functional genomics approach reveals CHE as a component of the Arabidopsis circadian clock. Science. 2009;323:1481–1485. doi: 10.1126/science.1167206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Poppe C, Hangarter RP, Sarrock RA, Nagy F, Schäfer E. The light-induced reduction of the gravitropic growth-orientation of seedlings of Arabidopsis thaliana (L.) Heynh. is a photomorphogenic response mediated synergistically by the far-red absorbing forms of phytochromes A and B. Planta. 1996;119:511–514. doi: 10.1007/BF00195180. [DOI] [PubMed] [Google Scholar]
- Poppe C, Schäfer E. Seed germination of Arabidopsis thaliana phyA/phyB double mutants is under phytochrome control. Plant Physiology. 1997;114:1487–1492. doi: 10.1104/pp.114.4.1487. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Quail PH. Phytochrome-regulated gene expression. Journal of Integrative Plant Biology. 2007;49:11–20. [Google Scholar]
- Reed JW, Nagatani A, Elich T, Fagan M, Chory J. Phytochrome A and phytochrome B have overlapping but distinct functions in Arabidopsis development. Plant Physiology. 1994;104:1139–1149. doi: 10.1104/pp.104.4.1139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reed JW, Nagpal P, Poole DS, Furuya M, Chory J. Mutations in the gene for red/far-red light receptor phytochrome B alter cell elongation and physiological responses throughout Arabidopsis development. The Plant Cell. 1993;5:147–157. doi: 10.1105/tpc.5.2.147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robson PRH, Smith H. Genetic and transgenic evidence that phytochromes and B act to modulate the gravitropic orientation of Arabidopsis thaliana hypocotyls. Plant Physiology. 1996;110:211–216. doi: 10.1104/pp.110.1.211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robson PRH, Whitelam GC, Smith H. Selected components of the shade-avoidance syndrome are displayed in a normal manner in mutants of Arabidopsis thaliana and Brassica rapa deficient in phytochrome B. Plant Physiology. 1993;102:1179–1184. doi: 10.1104/pp.102.4.1179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sakamoto K, Nagatani A. Nuclear localization activity of phytochrome B. The Plant Journal. 1996;10:859–868. doi: 10.1046/j.1365-313x.1996.10050859.x. [DOI] [PubMed] [Google Scholar]
- Salter MG, Franklin KA, Whitelam GC. Gating of the rapid shade avoidance response by the circadian clock in plants. Nature. 2003;426:680–683. doi: 10.1038/nature02174. [DOI] [PubMed] [Google Scholar]
- Schäfer E, Nagy F, editors. Photomorphogenesis in plants and bacteria. 3rd edn. Dordrecht, The Netherlands: Springer; 2006. [Google Scholar]
- Sessa G, Carabelli M, Sassi M, Ciolfi A, Possenti M, Mittempergher F, Becker J, Morelli G, Ruberti I. A dynamic balance between gene activation and repression regulates the shade avoidance response in Arabidopsis. Genes and Development. 2005;19:2811–2815. doi: 10.1101/gad.364005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sharrock RA, Clack T. Patterns of expression and normalized levels of the five Arabidopsis phytochromes. Plant Physiology. 2002;130:442–456. doi: 10.1104/pp.005389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sharrock RA, Clack T. Heterodimerization of type II phytochromes in Arabidopsis. Proceedings of the National Academy of Sciences, USA. 2004;101:11500–11505. doi: 10.1073/pnas.0404286101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sharrock RA, Quail PH. Novel phytochrome sequences in Arabidopsis thaliana: structure, evolution and differential expression of a plant photoreceptor family. Genes and Development. 1989;3:1745–1757. doi: 10.1101/gad.3.11.1745. [DOI] [PubMed] [Google Scholar]
- Shen H, Moon J, Huq E. PIF1 is regulated by light-mediated degradation through the ubiquitin-26S proteosome pathway to optimize photomorphogenesis of seedlings in Arabidopsis. The Plant Journal. 2005;44:1023–1035. doi: 10.1111/j.1365-313X.2005.02606.x. [DOI] [PubMed] [Google Scholar]
- Shen Y, Khanna R, Carle CM, Quail PH. Phytochrome induces rapid PIF5 phosphorylation and degradation in response to red-light activation. Plant Physiology. 2007;145:1043–1051. doi: 10.1104/pp.107.105601. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shin J, Kim K, Kang H, Zulfugarov IS, Bae G, Lee C-H, Lee D, Choi G. Phytochromes promote seedling light responses by inhibiting four negatively-acting phytochrome-interacting factors. Proceedings of the National Academy of Sciences, USA. 2009;106:7660–7665. doi: 10.1073/pnas.0812219106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shinomura T, Nagatani A, Hanzawa H, Kubota M, Watanabe M, Furuya M. Action spectra for phytochrome A- and B-specific photoinduction of seed germination in Arabidopsis thaliana. Proceedings of the National Academy of Sciences, USA. 1996;93:8129–8133. doi: 10.1073/pnas.93.15.8129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith H, Turnbull M, Kendrick RE. Light-grown plants of the cucumber long hypocotyl mutant exhibit both long-term and rapid growth responses to irradiation with supplementary far-red light. Photochemistry and Photobiology. 1992;56:607–610. [Google Scholar]
- Smith H, Whitelam GC. The shade avoidance syndrome: multiple responses mediated by multiple phytochromes. Plant, Cell and Environment. 1997;20:840–844. [Google Scholar]
- Smith H, Xu Y, Quail PH. Antagonistic but complementary actions of phytochromes A and B allow optimum seedling de-etiolation. Plant Physiology. 1997;114:637–641. doi: 10.1104/pp.114.2.637. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Somers DE, Devlin PF, Kay SA. Phytochromes and cryptochromes in the entrainment of the Arabidopsis circadian clock. Science. 1998;282:1488–1490. doi: 10.1126/science.282.5393.1488. [DOI] [PubMed] [Google Scholar]
- Somers DE, Sharrock RA, Tepperman JM, Quail PH. The hy3 long hypocotyl mutant of Arabidopsis is deficient in phytochrome B. The Plant Cell. 1991;3:1263–1274. doi: 10.1105/tpc.3.12.1263. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stephenson PG, Fankhauser C, Terry MJ. PIF3 is a repressor of chloroplast development. Proceedings of the National Academy of Sciences, USA. 2009;106:7654–7659. doi: 10.1073/pnas.0811684106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tepperman JM, Hudson ME, Khanna R, Zhu T, Chang SH, Wang X, Quail PH. Expression profiling of phyB mutant demonstrates substantial contribution of other phytochromes to red-light-regulated gene expression during seedling de-etiolation. The Plant Journal. 2004;38:725–739. doi: 10.1111/j.1365-313X.2004.02084.x. [DOI] [PubMed] [Google Scholar]
- Tepperman JM, Hwang Y-S, Quail PH. phyA dominates in transduction of red-light signals to rapidly-responding genes at the initiation of Arabidopsis seedling deetiolation. The Plant Journal. 2006;5:728–742. doi: 10.1111/j.1365-313X.2006.02914.x. [DOI] [PubMed] [Google Scholar]
- Tepperman JM, Zhu T, Chang HS, Wang X, Quail PH. Multiple transcription-factor genes are early targets of phytochrome A signaling. Proceedings of the National Academy of Sciences, USA. 2001;98:9437–9442. doi: 10.1073/pnas.161300998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thomas B. Light signals and flowering. Journal of Experimental Botany. 2006;57:3387–3393. doi: 10.1093/jxb/erl071. [DOI] [PubMed] [Google Scholar]
- Turck F, Fornara F, Coupland G. Regulation and identity of Florigen: FLOWERING LOCUS T moves to center stage. Annual Reviews in Plant Biology. 2008;59:573–594. doi: 10.1146/annurev.arplant.59.032607.092755. [DOI] [PubMed] [Google Scholar]
- Ueda HR. Systems biology flowering in the plant clock field. Molecular Systems Biology. 2006 doi: 10.1038/msb4100105. doi:10.1038/msb4100105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Whitelam GC, Johnson E, Peng J, Carol P, Anderson MC, Cowl JS, Harberd NP. Phytochrome A null mutants of Arabidopsis display a wild-type phenotype in white light. The Plant Cell. 1993;5:757–768. doi: 10.1105/tpc.5.7.757. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Whitelam GC, Smith H. Retention of phytochrome-mediated shade avoidance responses in phytochrome-deficient mutants of Arabidopsis, cucumber and tomato. Journal of Plant Physiology. 1991;39:119–125. [Google Scholar]
- Yanovsky MJ, Casal JJ, Whitelam GC. Phytochrome A, phytochrome B and HY4 are involved in hypocotyl growth responses to natural radiation in Arabidopsis: weak de-etiolation of the phyA mutant under dense canopies. Plant. Cell and Environment. 1995;18:788–794. [Google Scholar]
- Yanovsky MJ, Kay SA. Molecular basis of seasonal time measurement in Arabidopsis. Nature. 2002;419:308–312. doi: 10.1038/nature00996. [DOI] [PubMed] [Google Scholar]