Abstract
We report the fabrication of devices in which one single-walled carbon nanotube (SWCNT) spans a barrier between two fluid reservoirs, enabling direct electrical measurement of ion transport through the tube. A fraction of the tubes pass anomalously high ionic currents. Electrophoretic transport of small single stranded DNA oligomers through these tubes is marked by large transient increases in ion current and was confirmed by PCR analysis. Each current pulse contains about 107 charges, an enormous amplification of the translocated charge. Carbon nanotubes simplify the construction of nanopores, permit new types of electrical measurements, and may open avenues for control of DNA translocation.
We report the use of single-walled carbon nanotubes (SWCNTs) as nanopores for analyzing molecular transport properties. Nanopores are orifices of molecular diameter that connect two fluid reservoirs. At this length-scale, the passage of even a single molecule generates a detectable change in the flow of ionic current through the pore (1, 2). They can be used as single-molecule Coulter counters, and form the basis of proposed new approaches to DNA sequencing (3). The first nanopore devices were based on pore-proteins (4–7), but more recently pores have been fabricated by drilling (and sometimes partially refilling) solid-state materials (8–12). Nanochannels have been formed by etching silicon nanowires (13) and channels with one nanoscale dimension have been etched into glass (14) or quartz (15).
Carbon nanotubes are obvious candidates for the fabrication of nanopore structures. Pressure driven gas, water and ion transport has been recorded through membranes composed of many multiwalled carbon nanotubes (16) or double-walled carbon nanotubes (17). These experiments showed that the water flow-rate is greatly enhanced inside the tube, an effect confirmed by molecular dynamics simulations (18). DNA has been passed through a 100 nm diameter carbon nanotube (19) and 50 nm wide hydrophilic channels (13). It seems counter-intuitive that hydrophilic DNA would enter the hydrophobic interior of a SWCNT but simulations show that both RNA (20) and DNA (21) will translocate through 1.5 to 2 nm diameter tubes. The simulations were carried out using very large electric fields (tenths of a volt per nm) to generate observable motion on the simulation timescale. This result leaves open the possibility that some measurable translocation might occur at the much smaller fields that could be implemented in the laboratory. Here, we report direct measurement of this translocation.
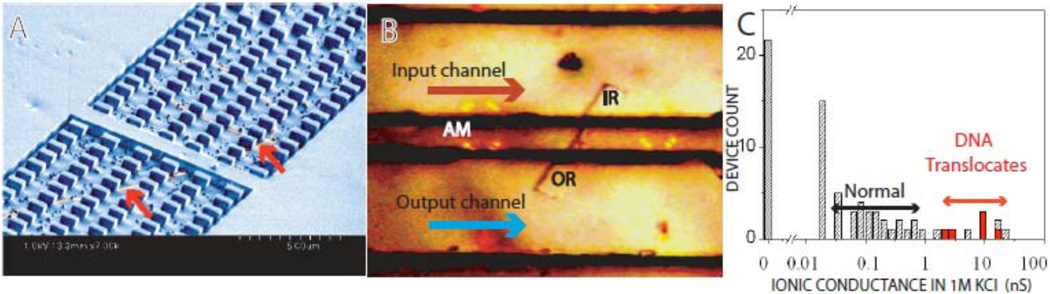
We have made a device in which one SWCNT spans a barrier between two fluid reservoirs [see Fig. 1 and supplementary materials (22)]. Relative to CNT membranes (16, 17) this arrangement makes it possible to detect signals from the translocation of a single molecule and to correlate transport with the properties of individual SWCNTs. We grew well-separated SWCNTs on the surface of oxidized silicon wafers and formed fluid reservoirs along the path of chosen tubes using e-beam lithography. An SEM image of a device at this stage is shown in Fig. 1A, where the SWCNT is just visible on each side of the barrier. An oxygen plasma was used to remove the exposed parts of the SWCNT, leaving the SWCNT under the barrier intact (Fig. S4I) (22,23). The fluidic pathway was completed by placing a poly (dimethyl siloxane) (PDMS) cover on top of the chip (Fig. IB).
Fig. 1.
A. A single nanopore device was fabricated by growing SWCNTs at low densities on an oxidized Si wafer. We used cobalt catalyst particles with ethanol vapor as the carbon source in conditions most likely to produce high-quality SWCNTs with an outside diameter of 1 to 2 nm (30). A 700 nm layer of PMMA resist is spun on and reservoirs opened over selected tubes with electron beam lithography. The exposed regions of SWCNTs were removed by O2 plasma etch. The SEM image of the device shows a 2 µm barrier prior to removal of the exposed SWCNT (pointed to by arrows). Pillars in the reservoir support the PDMS cover. B. Optical micrograph taken through a PDMS cover. The reservoirs (“IR” = input, “OR” = output) span the barrier between PDMS channels at an angle of about 60°. “AM” marks the location of one set of alignment markers. C. Current flows through the single SWCNTs and not a leakage path. With the SWCNT bridging the gap and opened, most tubes pass currents in the expected range (“Normal”), but 20% pass unexpectedly large currents. Some of these (marked in red) also passed DNA oligomers. These data are limited to the subset of devices exposed to short plasma etches for which control experiments show no leakage (supplementary information).
Each chip also contained control devices lacking the bridging SWCNT (supplementary materials) to check the integrity of barrier, including devices with unopened SWCNTs. We used a mild plasma treatment such that 100% of the devices lacking CNTs did not leak (Fig. S5), although this approach resulted in a large fraction of tubes that were not opened (20 %), as determined by SEM imaging (Figure S4F). The fluid reservoirs were filled with 1M KCl, and Ag/AgCl electrodes (BASI MF-2078) were used to measure the conductance across the reservoirs connected by the SWCNT. The devices passed current if, and only if they were spanned by a SWCNT that was opened (Fig. 1C), so the interface between the tube and the PMMA does not appear to provide a leakage path. This conclusion was verified by chemically tethering PEG molecules to one, or both ends of the CNTs. The current was reduced in one direction of bias when the tube was modified at one end, and in both directions of bias when the tube was modified at both ends (Fig. S6).
The ionic conductance of a tube of electrolyte should be given by G=6.02 × 1026(µK + µCl)cKCleπD2/4L Where µk=7.62 × 10−8 m2/V s, µcl= 7.91 × 10−8 m2/V s, ckcl is the KCl concentration in mole/l, e the electronic charge, D the tube diameter, and L the tube length. Table 1 shows that there is no correlation between the tube diameter and ionic conductance. The ionic conductance spans nearly four orders of magnitude (Fig. 1C) with only the lowest conductances (the range marked “normal” in Figure 1C) being consistent with the classical formula for cKCl = 1M, 1 nm < D < 5 nm (Fig. S7) and L = 2µm. We also measured the electronic properties of some of the tubes (Table 1) using both their response as FETs and Raman scattering (Figs. S8–S10).The SWCNTs with the highest ionic conductance are all metallic.
Table 1.
Relation of ionic conductance with electrical properties (VT is the threshold voltage for semiconducting tubes). These measurements do not discriminate between metallic SWCNTs and bundles containing a metallic tube, but most of the tubes are single walled (30).
| FET Device ID | Ionic Current | Diameter (nm) | Electrical Property |
|---|---|---|---|
| HL_4_1_41 AP3 | 10.7 ± 0.05 nA/0.4V | 2.0 | Metallic |
| HL_4_1_10AB6 | 3.4 ± 0.04nA/0.5V | 1.7(1.5*) | Metallic† |
| HL_4_1_39 P6 | 2.5 ± 0.07 nA/0.4Va | 4.2 | Metallic |
| HL_4_1_41 AZ3 | 1.91 ± 0.05 nA/0.4V | - | Metallic |
| HL_4_1_41 N2 | 0.98 ± 0.04 nA/0.4V (VT ~10V) |
0.9 | Semiconducting |
| HL_4_1_37 AB20 | 0.46± 0.03 nA/0.5V (VT ~ 10V) |
1.3* | Semiconducting† |
| HL_4_1_41 AS3 | 0.07 ± 0.02 nA/0.4V (VT ~ 25V) |
1.8 | Semiconducting |
| HL_4_1_37 Z22 | 0.1n ± 0.03 nA/0.5V (VT ~10V) |
1.1* | Semiconducting† |
| HL_4_1_41 M8 | <10pA/0.4V (VT ~ 25V) |
3.4 | Semiconducting |
Raman scattering was used to determine diameters marked * and confirm electronic properties marked †.
The tube marked a translocated DNA.
We considered whether the excess current be accounted for by electrochemical currents stemming from reduction and oxidation reactions at the end of metallic tubes. A conducting tube suspended in a potential gradient in an electrolyte acts as a bipolar electrode (24) but enormous fields are required to drive electrochemical processes at the ends of a bipolar carbon nanotube electrode (25). Measurements with an electrode contacting the SWCNT directly revealed that electrochemical currents were negligible for the potentials used here (Fig. S11).
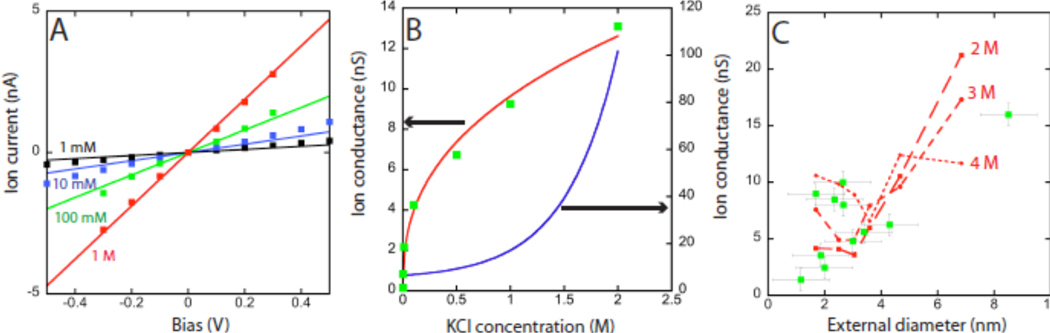
To look for clues to a mechanism for the large ionic currents, we used molecular dynamics simulations coupled with solutions of the Poisson-Nernst-Planck equation for transport in the SWCNT and the outside reservoirs (supporting online materials). The flow rate of water is greatly enhanced inside SWCNTs (17) but the MD simulations showed that the electrophoretic mobility of ions is similar to that in the bulk electrolyte. However, the selective filtering of anions or cations owing to charged end groups (26) can result in a net excess concentration, n, of one charge inside the tube. This charge will, in turn, drive an electroosmotic current. Molecular dynamics simulations further showed that both water and ions flow with an electroosmotic velocity, v, given by v ∝ n0.74 for a (10,10) SWCNT. Both anions and cations are driven in the same direction by an extremely large electroosmotic flow, but only the charge imbalance inside the tube results in a net ionic current proportional to nv, i.e. ∝n1.74. The mechanism of charge accumulation is complex and involves both charged end groups and the electronic properties of the SWCNT, and we have not yet developed a quantitative model for it (see Fig. S12 for further evidence of the role of charged end groups). However, current-voltage curves obtained at different salt concentrations in the reservoirs, c, can be fitted if n = 3.31 c0.22 M (Fig. 2A). This result is equivalent to an ionic conductance that varies as c0.39, shown by the red curve passing through the measured data in Fig. 2B. This dependence on concentration is quite different from the linear dependence expected for a tube of electrolyte, or the saturation at low salt observed for a planar nanopore carrying a surface charge (27).
Fig. 2.
Ion transport in the subset of SWCNTS with high ionic conductance. A. Current vs. voltage applied to Ag/AgCl reference electrodes for a 2 µ long, 1.7 nm diameter SWCNT for various concentrations of KCl electrolyte as marked. The solid lines are simulated as described in the text. B. Ionic conductance as a function of salt concentration. The red line is a fit to the cm dependence suggested by MD simulations. We found 0.33 < m < 0.4 in three different tubes. The blue line shows the salt dependence of conductance measured in a planar nanopore (26).C. In this subset of tubes, current at 1M KCl is better related to diameter (green squares). The red dashed lines show simulations for excess charge densities of 2, 3, and 4 M.
In contrast to the full set of devices, the subset with anomalously highly conductance does show some relation between conductance and tube diameter (green squares, Figure 2C). The red dashed lines show simulated values of ionic conductance as a function of diameter for n = 2, 3 and 4M. The measured data can be accounted for by assuming that variability in the charge of end groups leads to some variability in n.
While it might be instructive to study the translocation of simpler polyelectrolytes as a prelude to the study of DNA, methods such as dye-labeling are much less sensitive than the polymerase chain reaction for detecting and counting small numbers of molecules.
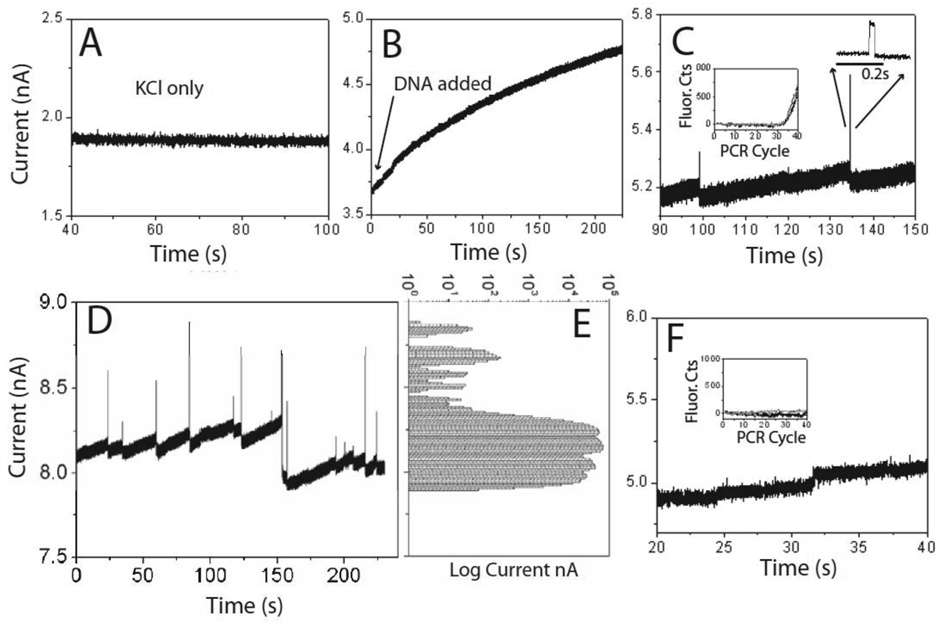
To test for DNA translocation of SWCNTs, we used 60 nt and 120 nt DNA oligomers with sequences that were predicted to be relatively free of secondary structure, with forward and reverse primers chosen to have high melting temperatures to minimize primer dimers and false priming (supporting online materials).Devices were characterized by measuring current flow with 1 or 2 M KCl alone, and then a DNA solution (1 or 2 M KCl, 1mM phosphate buffer, pH7) was flowed into the input reservoir side. A control aliquot was collected from the output reservoir to check for DNA contamination, and a positive bias was then applied to the output side of the device. In the subset of high current tubes, we first observed a slow increase in the background current (Fig. 3, A and B - data are for 0.1 nM DNA). After a time, which varied from a few to tens of minutes, depending on the DNA concentration in the input reservoir, large transient increases in current were observed. These “spikes” were accompanied by large fluctuations in the background current (Fig. 3C). The spikes disappeared when the polarity of the bias across the tube was reversed, and re-appeared when the original bias (positive on the output side) was restored. Quantitative polymerase chain reaction (q-PCR - supporting online materials) showed that DNA was translocated in devices manifesting these large spikes. Translocation occurred only in those tubes with conductances (prior to DNA addition) of > 2 nS (Fig. 1C). Some devices that showed instabilities in the background but no large current spikes (Fig. 3F) gave negative PCR results. We also tested for translocation in “failed” control devices (i.e., lacking the CNT and deliberately over-etched) that displayed leakage current. A few, with very large leakage current showed evidence of DNA in the output well, but none displayed spikes, regardless of the magnitude of the leakage current. Thus, the spikes signal translocation of DNA through the SWCNTs.
Fig. 3.
Ion current signals of DNA translation: A. Current (2M KCl, 1mM PBS pH 7) prior to DNA addition. B. After DNA addition, current slowly increases. C. 5 minutes after addition of 0.1 nM 60 nt DNA, large positive current spikes appear. These spikes are followed by a drop in baseline over a period of a second or so, and then by a gradual rise leading to the next spike. D. Representative data from another tube (also 60 nt DNA) with the distribution of currents shown in E. The DNA causes large changes in baseline in addition to the spikes. F. Data from a tube that showed both a current increase on DNA addition and baseline fluctuations but no spikes. No translocation was detected by PCR. The insets in C and F show the fluorescence signal from dsDNA dye labels as a function of the PCR cycle number for samples collected from these particular runs.
Quantitative PCR also provides a measure of the number of molecules collected. We collected small aliquots of fluid from the output reservoir by flushing the system through with excess buffer, and concentrated the solution using a Microcon YM-10 centrifugal filter so that we could redilute with PCR buffer. The filter losses were found to be highly variable, more so at low DNA concentrations, and account for much of the stated uncertainty in our results. We calibrated the PCR reaction with known amounts of DNA and, for two data points, calibrated filter losses by adding a known amount of a second sequence (with orthogonal primers) and carrying out a PCR analysis of both the target and calibration samples. The final molecule count was corrected for filter losses and dilution during the sample collection. The various errors in these steps tend to underestimate the amount of DNA that translocated so the final results are probably lower limits. PCR was limited to the first use of a device, and we rejected samples from chips that showed contamination in the control samples collected.
We were able to carry out PCR on samples collected from 12 devices that had a conductance > 2 nS. Of these, four had DNA contamination in control samples, leaving the eight devices listed in Table 2. Two of these showed no spikes and yielded no PCR signal. The remaining six all appeared to pass more than one molecule per spike. In particular, tubes HL-4-1-36 and A136, for which the filter recovery was directly measured with a control sample, passed at least 30 to 40 molecules for each spike. It is possible that the tube fills entirely with DNA, the spike signaling the cooperative emptying (or possibly filling) of the tube. The uncertainties in the PCR measurement are too large to reveal any significant difference between the number of molecules per spike for the 60 nt sample (23, 13, 34, 41) and the 120 nt sample (88, 16) though the spike frequency was much lower in the two 120 nt runs, and the spike duration significantly longer (Fig. S13,S14).
Table 2.
Results of q-PCR tests for translocation in tubes with conductance > 2 nS that gave uncontaminated control signals (data from four other devices that showed contamination in the control sample were rejected).
| Tube | Tube Conductance nS (1M KCI) |
DNA Sample (nt) |
Number of Spikes | Number of Molecules | Molecules per spike |
|---|---|---|---|---|---|
| ADI | 9.7 | 60 | 350±50 | 8000±2000 | 23 ±10 |
| AD2 | 9.5 | 60 | 30±10 | 400±200 | 13 (+17, −13) |
| AA Newl | 19.6 | 120 | 64±10 | 8500±3100 | 88 (+126, −88) |
| AA New2 | 2.7 | 120 | 1500±200 | 24400±5700 | 16 ±7 |
| HL-4-1-36 | 9.6 | 60 | 36±4 | 1224±774* | 34±21* |
| A136 | 1.6 | 60 | 46±5 | 1900±200* | 41±10* |
| HL-4-1-41 | 4.8 | 60 | 0 | 0 | - |
| HL-4-1-40 08 | 2.7 | 60 | 0 | 0 | - |
Errors in spike count reflect the consequences of different cut-off criteria for selecting spikes. Errors in the molecule count were dominated by uncertainties in the filter recovery efficiency, except for the data labeled * which were calibrated with a second oligomer.
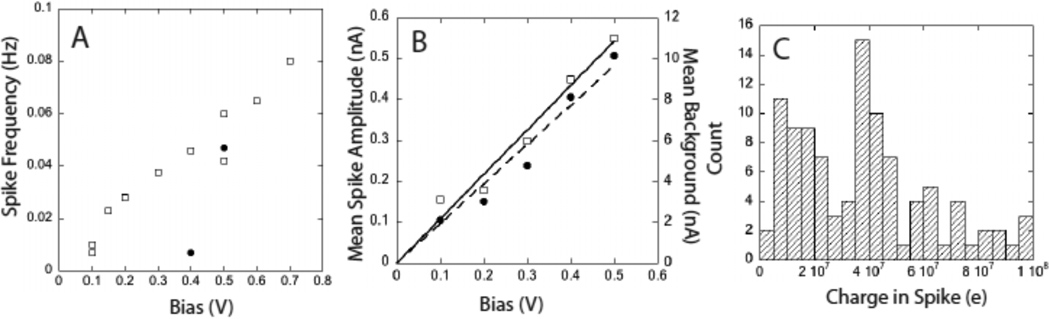
Figure 4A shows data for the spiking-rate as a function of bias for two different tubes passing 60 nt DNA. The spike rate increased with applied bias, and the two tubes showed different threshold biases for the onset of spikes (and hence translocation). For the 60 nt DNA, the spike amplitudes are about 5% of the baseline current (Fig. 4B) and their duration is between 3 and 100 ms, independent of applied bias, so long as it is above the threshold for translocation. The product of the spike duration and amplitude yields the charge contained in each spike (Fig. 4C). This is remarkably large, at about 1 pC or 107 electrons in each spike. Fan et al. explained positive charge spikes observed in nanochannels as a consequence of additional mobile ions brought into the channel by DNA molecules (13). Filling the tubes (2 µm long) with 100 (20 nm long) 60 nt DNA oligomers, each carrying 60 excess electronic charges would account for only 1 part in 10,000 of the observed charge in each spike. The spikes must originate with large changes in the polarization outside the tubes, much as observed in junctions between micro- and nano-channels (28). The charge accumulation caused by the asymmetrical current in the SWCNT might be the source of this polarization, but further modeling is required to shed light on this unusual signal.
Fig. 4.
Characteristics of the translocation signals for 60 nt DNA: A. Spike rate increases with bias after a threshold that depend upon the particular CNT-the two devices here show spike signals above 0.1 (squares) and 0.4 V (circles). B. Spike amplitude (squares) increases linearly with bias, and is about 5% of the background current (circles). C. Distribution of the charge in each spike for the SWCNT in units of the electronic charge, e.
The excess ionic conductance appears to be a characteristic of metallic tubes, and we have proposed a mechanism based on electroosmotic flow resulting from trapped charge. Tubes with high ionic conductance will transport DNA molecules, giving a distinctive and unexpectedly large electrical signal of translocation. This kind of nanopore combines a long channel (in which translocation speed might be slowed) with an “integrated” electrode that might prove useful in new schemes for sequencing DNA by tunneling (3). The ability to select metallic SWCNTs of a desired diameter (29) may open the way for production of devices with particular pore sizes.
Supplementary Material
Footnotes
One sentence summary: Translocation of single-stranded DNA through single-walled carbon nanotubes is accompanied by giant current pulses.
This manuscript has been accepted for publication in Science. This version has not undergone final editing. Please refer to the complete version of record at http://www.sciencemag.org/. The manuscript may not be reproduced or used in any manner that does not fall within the fair use provisions of the Copyright Act without the prior, written permission of AAAS.
References
- 1.Rhee M, Burns MA. Trends in Biotechnology. 2007;25:174. doi: 10.1016/j.tibtech.2007.02.008. [DOI] [PubMed] [Google Scholar]
- 2.Dekker C. Nature Nanotechnology. 2007;2:208. doi: 10.1038/nnano.2007.27. [DOI] [PubMed] [Google Scholar]
- 3.Branton B, et al. Nature Biotechnology. 2008;26:1146. doi: 10.1038/nbt.1495. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Kasianowicz JJ, Brandin E, Branton D, Deamer DW. Proc. Nat. Acad. Sci. 1996;93:13770–13773. doi: 10.1073/pnas.93.24.13770. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Akeson M, Branton D, Kasianowicz JJ, Brandin E. D. W. D. Biophys J. 1999;77:3227. doi: 10.1016/S0006-3495(99)77153-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Kasianowicz J, Henrickson S, Weetall H, Robertson B. Anal. Chem. 2001;73:2268. doi: 10.1021/ac000958c. [DOI] [PubMed] [Google Scholar]
- 7.Meller A, Branton D. Electrophoresis. 2002;23:2583. doi: 10.1002/1522-2683(200208)23:16<2583::AID-ELPS2583>3.0.CO;2-H. [DOI] [PubMed] [Google Scholar]
- 8.Li J, et al. Nature. 2001;412:166. doi: 10.1038/35084037. [DOI] [PubMed] [Google Scholar]
- 9.Storm A, Chen J, Ling X, Zandbergen H, Dekker C. Nature Mat. 2003;2:537. doi: 10.1038/nmat941. [DOI] [PubMed] [Google Scholar]
- 10.Chang H, et al. App. Phys. Lett. 2006;88:103109. [Google Scholar]
- 11.Kim MJ, Wanunu M, Bell DC, Meller A. Advanced Materials. 2006;18:3149. [Google Scholar]
- 12.Chen P, et al. Nano Lett. 2004;4:1333. doi: 10.1021/nl0494001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Fan R, et al. Nano Lett. 2005;5:1633. doi: 10.1021/nl0509677. [DOI] [PubMed] [Google Scholar]
- 14.Stein D, Kruithof M, Dekker C. Phys. Rev. Lett. 2004;93:035901. doi: 10.1103/PhysRevLett.93.035901. [DOI] [PubMed] [Google Scholar]
- 15.Liang X, Chou SY. Nano Lett. 2008;8:1472. doi: 10.1021/nl080473k. [DOI] [PubMed] [Google Scholar]
- 16.Hinds BJ, et al. Science. 2004;303:62. doi: 10.1126/science.1092048. [DOI] [PubMed] [Google Scholar]
- 17.Holt JK, et al. Science. 2004;312:1034. doi: 10.1126/science.1126298. [DOI] [PubMed] [Google Scholar]
- 18.Joseph S, Alum NR. Nano Lett. 2008;8:452. doi: 10.1021/nl072385q. [DOI] [PubMed] [Google Scholar]
- 19.Ito T, Sun L, Crooks RM. Chemical Communications. 2003;1482 doi: 10.1039/b302511j. [DOI] [PubMed] [Google Scholar]
- 20.Yeh I-C, Hummer G. Proc. Natl. Acad. Sci.(USA) 2004;101:12177. doi: 10.1073/pnas.0402699101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Xie Y, Kong Y, Soh AK, Gao H. J. Chem. Phys. 2007;127:225101. doi: 10.1063/1.2799989. [DOI] [PubMed] [Google Scholar]
- 22.Supporting Online Material Fabrication of devices, Measurements on multiwalled tubes, Plasma etching and device leakage, Blocking of tubes by chemical modification, AFM diameter measurements, Raman Measurements, Measurement of electrical properties, Measurement of electrochemical current, Theoretical modeling and simulation, pH dependence of current, DNA sequences and primers, Translocation data for the 120 nt oligomer and distributions of spike widths, Q-PCR.
- 23.We also made some devices using multiwalled tubes but found that these were much more difficult to open (supporting online materials, Fig. S3)(22).
- 24.Chow K-F, Mavre F, Crooks RM. J. Am Chem Soc. 2008;130:7544. doi: 10.1021/ja802013q. [DOI] [PubMed] [Google Scholar]
- 25.Warakulwit C, et al. Nano Lett. 2008;8:500. doi: 10.1021/nl072652s. [DOI] [PubMed] [Google Scholar]
- 26.Joseph S, Mashl RJ, Jakkobsson E, Alum NR. Nano Letters. 2003;3:1399. [Google Scholar]
- 27.Smeets RMM, et al. Nano Lett. 2006;6:89. doi: 10.1021/nl052107w. [DOI] [PubMed] [Google Scholar]
- 28.Jin X, Joseph S, Gatimu E, Bohn P, Alum NR. Langmuir. 2007;23:13209. doi: 10.1021/la702326v. [DOI] [PubMed] [Google Scholar]
- 29.Tu X, Manohar S, Jagota A, Zheng M. Nature. 2009;460:250. doi: 10.1038/nature08116. [DOI] [PubMed] [Google Scholar]
- 30.Guo XF, et al. Science. 2006;311:356. [Google Scholar]
- 31.We acknowledge valuable discussions with Guigen Zhang, Otto Sankey, Dick Crooks and Murugappan Muthukumar. Xiadong Cui provided us with samples of larger-diameter SWCNTs, Tao Liu assisted us with AFM measurements. This work was supported by grants from the DNA sequencing technology program of the NHGRI, Arizona Technology Enterprises and the Biodesign Institute.
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.