Abstract
Parkin is a multidomain E3 ligase associated with autosomal recessive Parkinson disease. The N-terminal ubiquitin-like domain (Ubld) of parkin functions with the S5a proteasomal subunit, positioning substrate proteins for degradation. In addition the parkin Ubld recruits the endocytotic protein Eps15, allowing the E3 ligase to ubiquinate Eps15 distal from its parkin-interacting site. The recognition sequences in the S5a subunit and Eps15 for the parkin Ubld are ubiquitin-interacting motifs (UIM). Each protein has two UIM sequences separated by a 50-residue spacer in S5a, but only ∼5 residues in Eps15. In this work we used NMR spectroscopy to determine how the parkin Ubld recognizes the proteasomal subunit S5a compared with Eps15, a substrate for ubiquitination. We show that Eps15 contains two flexible α-helices each encompassing a UIM sequence. The α-helix surrounding UIM II is longer than that for UIM I, a situation that is reversed from S5a. Furthermore, we show the parkin Ubld preferentially binds to UIM I in the S5a subunit. This interaction is strongly diminished in a K48A substitution, found near the center of the S5a interacting surface on the parkin Ubld. In contrast to S5a, parkin recruits Eps15 using both its UIM sequences resulting in a larger interaction surface that includes residues from β1 and β2, not typically known to interact with UIM sequences. These results show that the parkin Ubld uses differential surfaces to recruit UIM regions from the S5a proteasomal subunit compared with Eps15 involved in cell signaling.
Introduction
Modification of proteins by ubiquitin is an essential biochemical process that signals proteins for degradation via the 26 S proteasome and also for non-proteolytic processes such as cell cycle and cell division, protein trafficking, endocytosis, and DNA repair (1–3). Three enzymes in the ubiquitination pathway (E1, E2, and E3) label a targeted protein with ubiquitin. The E3 enzymes are important for mediating the transfer of ubiquitin onto the target protein through their interaction with both the E2 enzyme and substrate and provide the specificity for target protein recognition. Autosomal recessive juvenile parkinsonism (ARJP)2 is an early-onset familial form of the disease that is clinically indistinguishable from the more prevalent idiopathic form of Parkinson disease. Mutations in several genes have been identified in ARJP patients, although the most commonly mutated gene encodes the E3 ubiquitin-protein ligase parkin (4–6). Mutations in parkin account for ∼50% of all ARJP cases. Parkin is a 465-residue multidomain E3 ligase comprising an N-terminal ubiquitin-like domain (Ubld) followed by a unique parkin-specific domain, two RING domains (RING0, RING1), an in-between RING (IBR) domain, and a C-terminal RING domain (RING2) (7, 8). Mutations associated with ARJP are found throughout the parkin protein and have profound affects on the folding and functionality of the protein. For example, missense mutations in the C terminus of parkin have been shown to disrupt its function with E2 ubiquitin-conjugating enzymes UbcH7 (6) and UbcH8 (9) leading to alterations in the ubiquitination levels of substrates such as cyclin E (10), p38 (11), and synphilin (12).
The N-terminal Ubld domain in parkin although not directly involved in E2 interactions has been shown to be essential for the ligase activity of parkin because deletion or mutation of this domain results in impaired E3 ligase activity (6, 13–15). Disease state mutations localized to the Ubld have been shown to cause different effects. For example, a R42P substitution in the Ubld causes complete unfolding of the domain (16), whereas other mutations have much smaller affects on the domain stability (5, 13, 17). Mutations that do not affect the stability of parkin have been suggested to disrupt protein interactions with E2 enzymes, other modulating proteins or substrates. The three-dimensional structure of the parkin Ubld (18, 19) shows it shares a similar three-dimensional fold as ubiquitin, although like other Ublds it does not have the ability to be conjugated to other proteins as observed for ubiquitin. Rather, the function of the Ubld is proposed to act as a protein adapter within multidomain proteins allowing the other domains to carry out a diverse array of processes (20, 21). Representative Ubld-containing members include hPLIC-2 (Dsk2 in yeast) that uses its N-terminal Ubld, collagen-like, and ubiquitin-associated domains for spindle body duplication and hHR23a/b (Rad23 in yeast) involved in nucleotide excision repair, which has at least four distinct domains including an N-terminal Ubld, two ubiquitin-associated domains, and a region identified as the binding site for the repair protein XPC (22–24). Parkin along with HOIL-1 are the only proteins identified to date that possess a Ubld along with multiple RING domains important for ubiquitination. Parkin, as well as hPLIC-1, hPLIC-2, and hHR23a/b have been shown to interact with the 26 S proteasome (17, 18, 25–27) through their Ublds by possibly acting to recruit substrates targeted for degradation to the proteasome machinery. Within the 26 S proteasome, the parkin Ubld has been shown to interact with the S5a subunit of the 19 S regulatory particle. In addition, recent evidence shows that parkin can interact with the endocytic protein Eps15 (18, 28). In this role the Ubld is suggested to position parkin to facilitate monoubiquitination of Eps15 at a distal site. Ubiquitination of Eps15 has been proposed to compete with the parkin Ubld for binding leading to the displacement of the parkin protein allowing for parkin to be recycled for other ubiquitination pathways. The ubiquitination of Eps15, however, precludes Eps15 from interacting with the epidermal growth factor receptor allowing for prolonged signaling through the phosphatidylinositol 3-kinase-Akt pathway, which is important for neuronal survival (28).
The interaction of the parkin Ubld with the 19S regulatory subunit S5a and Eps15 is mediated by ubiquitin-interacting motifs (UIM) (29). These short sequences display alternating patterns of large and small side chain amino acids frequently adjacent to acidic residues (30). In particular the S5a subunit and Eps15 each contain two UIMs near their C termini. Many proteins have multiple copies of the UIM positioned in tandem, although spacing of the UIMs is irregular and the requirement for specific UIMs for binding to a specific protein is not clear. For example, in Eps15 only the second UIM is needed to recruit ubiquitin (31, 32), whereas deletion or substitution in either UIM sequence abrogates the parkin Ubld interaction (28). Similarly, it has been suggested that both UIM sequences from S5a are required for interaction with the Ubld from parkin (18). The interaction of parkin with ubiquitin is unknown. A preference for ubiquitin interaction to the second UIM is present in the S5a subunit (33), whereas the hPLIC-1 Ubld interacts primarily with the S5a first UIM sequence (17). Little information is available showing how parkin differentiates between UIM sequences in the S5a proteasomal subunit and in Eps15, a ubiquitination target.
In this work we have determined that parkin uses two different modes to distinguish between the UIM sequences in S5a and Eps15. In S5a, the Ubld of parkin interacts primarily through UIM I. This interaction is diminished in the K48A substitution in parkin, reported to result from a missense mutation in ARJP (34). For Eps15, we have used NMR spectroscopy to show the UIM-contacting region of Eps15 has a similar structure as S5a (33), comprising two separated α-helices. In contrast to S5a, however, both Eps15 UIM sequences are required for parkin binding that use an enlarged Ubld protein interaction surface not observed previously for Ubld-UIM coordination.
EXPERIMENTAL PROCEDURES
Cloning
The DNA fragments encoding the UIMs from human S5a-(196–309) and human Eps15-(846–896) were cloned into a modified pET21a vector (Novagen) containing a His6 C-terminal tag. The pET21a vector was modified to include a tobacco etch virus protease cleavage site before the C-terminal His6 tag. Ubiquitin from Saccharomyces cerevisiae was expressed from a pET3a vector as previously described (35). The parkin Ubld and UbldK48A proteins were expressed from a pET44a vector as previously described (16). All mutants were created using the QuikChange Site-directed Mutagenesis kit (Stratagene, La Jolla, CA).
Protein Expression and Purification
The Ubld, UbldK48A, and the UIM-containing regions from S5a and Eps15 were overexpressed in the Escherichia coli BL21(DE3) Codon Plus strain. The bacteria were grown at 37 °C overnight in 2× YT medium (10 ml) containing the antibiotic carbenecillin (50 μg/ml) and chloramphenicol (34 mg/ml). The culture was diluted 1:100 in 2× YT medium (10 ml in 1 liter) containing the same antibiotics. Expression was induced at an A600 of 0.6–0.7 by the addition of 0.7 mm isopropyl 1-thio-β-d-galactopyranoside and allowed to grow overnight at 15 °C with constant shaking. Ubiquitin was expressed in the E. coli BL21(DE3)pLysS strain. The bacteria were grown at 37 °C overnight in LB medium (10 ml) containing the antibiotic carbenecillin (50 μg/ml) and chloramphenicol (34 mg/ml). The culture was diluted 1:100 in LB medium containing the same antibiotic and induced when an A600 of 0.4 was reached by the addition of 0.4 mm isopropyl 1-thio-β-d-galactopyranoside. The cells were allowed to grow for 4 h at 37 °C with constant shaking. For the production of 15N,13C-labeled proteins, cells were grown in M9 minimal media containing 1.0 g/liter of 15NH4Cl and 2.0 g/liter of [13C]glucose. Ubiquitin and Ubld constructs were purified as previously described (16, 35). The UIMs from S5a and Eps15 were expressed with a His6 tag and purified using a Ni-NTA fast protein liquid chromatography affinity column (GE Healthcare) followed by cleavage of the His6 tag with tobacco etch virus, and then reloaded onto the Ni-NTA column. Further purification was achieved by size exclusion chromatography. The integrity of all proteins was confirmed by electrospray ionization mass spectrometry (UWO Biological Mass Spectrometry Laboratory).
S5a Binding Assays
Purified His6-tagged S5a-(196–309) was mixed with untagged Ubld, UbldK48A, or ubiquitin at a 1:2 molar ratio, respectively, in a total volume of 600 μl and placed on a rotating shaker at 4 °C for 1 h. The mixture was then loaded onto a Ni-NTA spin column (Qiagen) pre-equilibrated in binding buffer (20 mm sodium phosphate, 10 mm imidazole, 300 mm NaCl, pH 8). The column was washed twice with 600 μl of binding buffer and then eluted (20 mm sodium phosphate, 250 mm imidazole, 300 mm NaCl, pH 8). Elution samples were fractionated by electrophoresis in a polyacrylamide gel and stained with Coomassie Brilliant Blue dye. This process was repeated for experiments using His6-tagged S5a-(196–309) and untagged ubiquitin and UbldK48A and also for His6-tagged S5a-(196–309)UIMII-5A and untagged Ubld. Protein concentrations were determined by their extinction coefficient in guanidine hydrochloride (36).
NMR Spectroscopy
NMR experiments were performed on 600 MHz Varian Inova spectrometers equipped with either a 13C-enhanced triple resonance cold probe with Z gradients or a xyz gradient, triple resonance probe. Standard pulse sequences from the Varian BioPack package were used. 1H chemical shifts were referenced directly to internal 4,4-dimethyl-4-silapentane-1 sulfonic acid at 0 ppm, and the 13C and 15N chemical shifts were indirectly referenced from this value (37). HNCA and CBCA(CO)NH (38, 39) experiments were recorded on an 15N,13C-S5a-(196–309)UIMII-5A sample and analyzed to obtain sequence-specific 1HN, 15N, 13Cα, and 13Cβ backbone assignments for the residues that shifted due to the 5-alanine substitution. HNCA, HNCACB, CBCA(CO)NH, and HNCO experiments were recorded on wild-type Eps15-(846–896). The data were analyzed to obtain sequence-specific 1HN, 15N, 13Cα, 13Cβ, and 13C′ backbone assignments. All spectra were processed with NMRPipe (40) software using a 60° shifted cosine-squared function in 1H and 15N to minimize artifacts in the spectra and analyzed by using NMRView (41). Chemical shift index analysis (42) was performed using Hα, Cα, and C′ chemical shifts for Eps15-(846–896).
Titration Experiments
A solution of 15N-labeled parkin Ubld (0.5 mm) was prepared in 10 mm KH2PO4, 1 mm EDTA, 1 mm dithiothreitol, 150 mm NaCl, 30 μm 4,4-dimethyl-4-silapentane-1-sulfonic acid, 10% D2O, pH 7.0. Aliquots from a concentrated solution of unlabeled S5a-(196–309) (3 mm) in the same buffer were added to a maximum 2.5 eq of S5a-(196–309):Ubld. Sensitivity enhanced 1H-15N HSQC spectra (43) were recorded at 25 °C for each S5a-(196–309) addition. Chemical shift perturbations in the Ubld were measured for each residue as a function of addition of S5a-(196–309), and analyzed by non-linear regression and global fitting using Prism 4 software to obtain equilibrium dissociation constants. Experiments were repeated using 15N-labeled S5a-(196–309) (0.5 mm) and unlabeled Ubld (3 mm). Identical experiments were conducted for 15N-labeled Ubld (0.5 mm) and unlabeled Eps15-(846–896) as well as the reciprocal experiment. The change in chemical shift was measured according to the equation ((0.2 × ΔδN2) + ΔδH2))1/2, where ΔδN and ΔδH represent the change in nitrogen and proton chemical shifts (in parts per million) upon protein addition.
Isothermal Titration Calorimetry
Experiments were carried out on a MicroCal VP-ITC isothermal titration calorimeter. All purified protein samples were dialyzed into 10 mm KH2PO4, 1 mm tris(2-carboxyethyl)phosphine, 150 mm NaCl at pH 7.0. For titrations, purified Eps15-(846–896) and Ubld were prepared to 4.98 mm and 269 μm, respectively, using dialysis buffer, and each was degassed prior to titration. Ubld was loaded into the cell (∼1.4 ml) and Eps15-(846–896) was loaded into the syringe. Titration of the UIMs was performed at 25 °C starting with an initial injection (3 μl), followed by 65 larger injections (5 μl), with spacings of 300 s. The sample cell was stirred at 300 rpm throughout the experiment. The buffer blank performed under the same conditions showed negligible heats of binding (<0.010 μcal/s). The dissociation constant (Kd) and stoichiometry of binding (N) were obtained by non-linear least squares fitting of the parkin Ubld versus the UIMs data to a one-site model provided with the data analysis software (Origin 7). Baselines were subtracted automatically using Origin software. Titration of purified Eps15-(846–896) with ubiquitin was done under identical conditions. All protein concentrations were determined in triplicate by amino acid analysis (Advanced Protein Technology Centre, The Hospital for Sick Children) and their extinction coefficient in guanidine hydrochloride (28).
Circular Dichroism Spectropolarimetry
CD spectra were collected on a Jasco J-810 instrument (Biomolecular Interactions and Conformations Facility, University of Western Ontario). Spectra for 10 scans (250–190) were recorded and averaged at 25 °C. A 1-mm path length cell was used and the buffer background was subtracted.
Cross-linking Experiments
Samples of Eps15-(846–896), parkin Ubld, and the cross-linker bis-sulfosuccinimidyl suberate were prepared in 10 mm KH2PO4, 1 mm tris(2-carboxyethyl)phosphine, 150 mm NaCl at pH 7.0. For cross-linking experiments, solutions of Eps15-(846–896) (60 μm) and/or Ubld (60 μm) in the presence and absence of 10-fold excess bis-sulfosuccinimidyl suberate were incubated at room temperature. Samples were removed every 15 min, quenched with 20 mm Tris buffer, and analyzed by SDS-PAGE.
RESULTS
The three-dimensional structure of a fragment of the S5a proteasomal subunit that contains two UIM sequences (residues 196–306; S5a-(196–309)) has been previously determined by NMR spectroscopy (33). The structure comprises a non-compact arrangement of three well structured α-helical regions connected by flexible linker regions (Fig. 1). The first helix spans residues Pro214–Glu245, and contains the first UIM motif 216LALAL220 (UIM I). The third helix spans residues Leu278–Gln296, and contains the second UIM motif 287IAYAM291 (UIM II). Acidic residues surround both UIMs (30) and are involved in binding interactions (Fig. 1A). The ubiquitin-like domains from hPLIC-1 and hHR23a/b interact differently with the S5a-(196–309) region. For example, hHR23a binds with high affinity (12 μm) to only UIM II (33, 44), whereas hPLIC-1 binds selectively to UIM I (17). It has been suggested that the preference for binding is a result of the UIM amino acid composition that best accommodates the individual Ubld (33). Unlike S5a, where more than 50 residues separate the UIM sequences, the UIM regions in Eps15 are very close together (Fig. 1). There is little structural information about this region in Eps15 or how the Ubld in parkin recognizes the UIM sequences in Eps15 or the S5a proteasomal subunit. In this work the secondary structure for the UIM-containing region of Eps15 was determined and the differences in the interaction between Eps15 and S5a with the parkin Ubld identified. This data may help distinguish how the parkin Ubld targets a protein for degradation via the S5a subunit in the 26 S proteasome compared with an adapter role assisting in the ubiquitination of a substrate (Eps15).
FIGURE 1.
UIM regions in the proteasomal subunit S5a and the endocytotic protein Eps15. A, sequence alignment of the UIM containing regions from Homo sapiens S5a and Eps15 (30). The sequence of S5a, containing a 5-alanine substitution in the second UIM S5a-(196–309)UIMII-5A is also shown. The UIM motif identified by Hofmann and Falquet (30) is shown below, where Φ is a large hydrophobic residue, e/d denotes a preference for an acidic residue, and X is any amino acid. The constructs used in this work extend from residues 196–309 in S5a (S5a-(196–309)) and 846–896 in Eps15 (Eps15-(846–896)). A box with inset numbers indicates the number of residues in the inter-UIM linker for S5a and Eps15. B, ribbon drawing of the three-dimensional structure of S5a-(196–309) (Protein Data Bank code 1YX4) (33) highlighting the regions (red) for the central hydrophobic regions of UIM I and II. This figure was produced using the program PyMOL (52).
The Parkin Ubld Interacts with UIM I from S5a
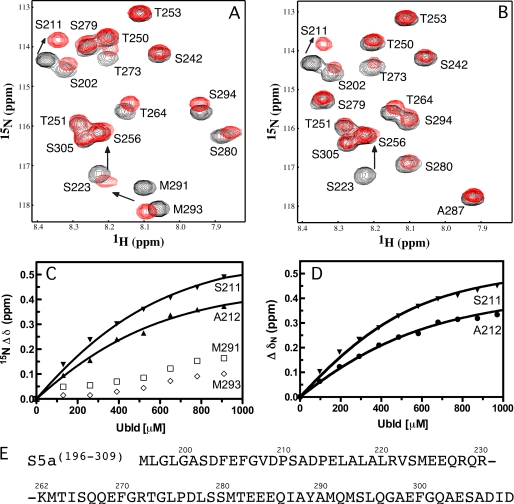
To determine whether the parkin Ubld interacts with one or both UIMs in the S5a subunit we used NMR spectroscopy to characterize the interactions between the Ubld and an S5a fragment (S5a-(196–309)) containing both UIM sequences. All of the resonances from the 1H-15N HSQC of S5a-(196–309) could be accounted for based on previously published data for this protein (44). We incrementally titrated a solution of 15N-labeled S5a-(196–309) with unlabeled parkin Ubld, and acquired standard 1H-15N HSQC experiments at each titration point (Fig. 2). The chemical shift changes were plotted and perturbations were identified for UIMs I and II. From this data it was clear that residues in UIM I were perturbed to a much greater extent than those in UIM II. For example, residues Ser211 and Ala212 in UIM I shifted >0.3 ppm in the 15N dimension compared with <0.1 ppm for Met291 and Met293 in UIM II (Figs. 2 and supplemental S1). Furthermore, the magnitude of the chemical shift changes observed for UIM II is much smaller than observed for its binding to hHR23a (26). The change in nitrogen chemical shift for residues around UIM I (Gly207, Ser211, Ala212, and Glu215) could be fit to a one-site binding equation with a Kd of 217 ± 51 μm (Fig. 2 and Table 1). This dissociation constant is very similar for other UIMs interacting with Ublds, which are generally low affinity and in the range of 10–500 μm (39). In contrast, attempts to fit the residues near UIM II (Met291 and Met293) yielded a much poorer binding curve with an apparent Kd ≫ 2000 μm. To confirm that only UIM I was required for the interaction with the Ubld of parkin, we introduced a five-alanine substitution in UIM II (287IAYAM291 substituted to 287AAAAA291; S5a-(196–309)UIMII-5A). The alanine substitution was selected to maintain the α-helical structure of the UIM while abolishing binding of the UIM similar to that previously reported for S5a-(196–309) when binding to ubiquitin (45). The 1H-15N HSQC of S5a-(196–309)UIMII-5A showed no gross structural changes from the substitutions in UIM II as only the substituted residues and their neighbors experienced chemical shift perturbations (Fig. 2). A solution of purified Ubld was then titrated into a 15N-labeled S5a-(196–309)UIMII-5A sample and an 1H-15N HSQC was recorded for each addition. Little change in the chemical shifts for the alanines or surrounding residues in UIM II were noted, whereas the chemical shift changes in the UIM I remained similar to the wild-type S5a-(196–309) (Figs. 2D and supplemental S1). Using residues near UIM I (Gly207, Ser211, Ala212, and Glu215) a Kd of 121 ± 24 μm was determined for S5a-(196–309)UIMII-5A binding to the parkin Ubld (Fig. 2, Table 1). The increased affinity of UIM I for the Ubld when the second UIM is removed suggests that the presence of UIM II could be interfering with the binding of UIM I to the Ubld at the high protein concentrations used. A similar experiment using a UIM I mutant, S5a-(196–309)UIMI-5A was monitored by NMR spectroscopy using 15N-labeled S5a-(196–309)UIMI-5A with increasing amounts of parkin Ubld (data not shown). We noted some residues such as Ser202, Thr273, and Thr251 experienced line broadening perhaps suggestive of a weaker interaction, although residues Ser211 and Ala212, which shift in the wild-type S5a, do not change significantly. However, little difference was noted for residues near UIM II similar to observations for both wild-type S5a-(196–309) or S5a-(196–309)UIMII-5A. These experiments re-affirm that the S5a UIM I region is the major site of interaction with the parkin Ubld but that replacement of the three central hydrophobic residues with alanine may not be sufficient to completely abolish binding.
FIGURE 2.
Parkin Ubld interacts with UIM I in S5a. Selected regions of the 600 MHz 1H-15N HSQC spectra showing the interaction of the parkin Ubld with 15N-labeled (A) S5a-(196–309) or (B) S5a-(196–309)UIMII-5A. In both figures the S5a-(196–309) (or S5a-(196–309)UIMII-5A) is shown in the absence of parkin Ubld (black contours) and presence of 2 eq of parkin Ubld (red contours). The spectra were collected using 500 μm S5a protein in 10 mm KH2PO4, 1 mm EDTA, 1 mm dithiothreitol, 150 mm NaCl, pH 7, at 25 °C. The spectrum of S5a-(196–309) was assigned according to triple resonance experiments and published work (44). The spectrum of S5a-(196–309)UIMII-5A was assigned by triple resonance NMR methods. Spectra are presented at different contour levels to account for line broadening in the S5a proteins upon Ubld addition. Binding curves based on NMR titration data are shown in C and D. In C the S5a-(196–309) titration data is plotted and fit using peaks near UIM I (Ser211 and Ala212) and UIM II (Met291 and Met293) In D, the S5a-(196–309)UIMII-5A titration data are plotted using residues from UIM I only (Ser211 and Ala212). The sequence of the S5a UIM regions is shown in E with the portion of the linker region between Arg230 and Lys262 omitted for clarity.
TABLE 1.
Binding affinities of the parkin Ubld with S5a and Eps15 proteins
| Protein (cell)a | Protein (titrant)b | Kd | Technique |
|---|---|---|---|
| μm | |||
| Ubld | S5a-(196–309) | 233 ± 60 | NMR |
| S5a-(196–309) | Ubld | 217 ± 51c | NMR |
| S5a-(196–309)UIMII-5A | Ubld | 121 ± 24c | NMR |
| Ubld | Eps15-(846–896) | 139 ± 19 | NMR |
| Ubld | Eps15-(846–896) | 193 ± 30 | ITC |
a Protein (cell) denotes the protein in cell (ITC) or 15N-labeled protein (NMR).
b Protein (titrant) denotes the protein in the syringe (ITC) or unlabeled (NMR).
c Residues from UIM I (Gly207, Ser211, Ala212, and Glu215) were used.
To determine the interacting surface on the parkin Ubld for the S5a subunit we monitored the chemical shift changes for Ubld residues using a 15N-labeled Ubld sample titrated with unlabeled S5a (Fig. 3A). The peaks were easily tracked throughout the titration to their final position after the addition of 2.5 eq of S5a-(196–309). Several residues had chemical shift perturbations that were more than double the average perturbation including Phe4 and Phe13 at the N terminus, Arg42–Ile44 in β3, Ala46, Lys48, Glu49, and Arg51 in β4, Leu61 in the α2-β5 linker, and Ile66–Val67 in β5, and Arg75 at the C terminus. One of the most perturbed residues was Lys48, which had a chemical shift change of nearly 2 ppm in the 15N dimension. The largest chemical shift changes were mapped onto the surface of the parkin Ubld to obtain the interaction surface for S5a (Fig. 3B). Most residues were localized primarily on the five-strand β-sheet face of the Ubld with little perturbation of the long α-helix positioned on the backside of the domain. A contiguous surface containing residues Phe13, Arg42–Ile44, Ala46, Lys48, Glu49, Arg51, Leu61, Ile66–Val67, Gln71, and Arg75 is formed on the parkin Ubld used for the interaction of UIM I in S5a.
FIGURE 3.
The parkin Ubld binding surface for UIM I of S5a-(196–309). A, selected regions of the 600 MHz 1H-15N showing the interaction of S5a-(196–309) with 15N-labeled parkin Ubld. The spectra show the parkin Ubld in the absence of S5a-(196–309) (black contours) and presence of 2 eq of S5a-(196–309) (red contours). Spectra are presented at different contour levels to account for line broadening in the Ubld protein upon S5a-(196–309) addition. B, ribbon and surface drawing of the parkin Ubld showing the interacting surface for UIM I in S5a. The surface is colored according to the magnitudes of the chemical shift changes ((0.2 × ΔδN2) + ΔδH2))1/2 observed in NMR experiments resulting from the interaction with the S5a-(196–309). Residues that shifted more than 0.5 S.D. above the average chemical shift change (0.024 ppm) are indicated in red. This figure was produced using the program PyMOL (52) using the human parkin Ubld structure (Protein Data Bank 1IYF) (18).
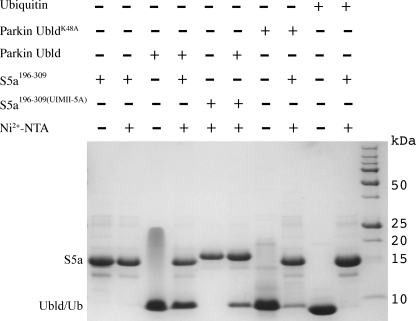
The observation that the UIM I region of S5a and the region surrounding Lys48 in the Ubld are necessary for interaction was assessed by affinity pulldown experiments. Fig. 4 shows that purified His6-tagged S5a-(196–309) bound to Ni-NTA successfully pulled down untagged parkin Ubld (lane 4). Furthermore, the His6-tagged S5a-(196–309)UIMII-5A pulled down similar amounts of the Ubld compared with the wild-type S5a-(196–309) (lane 6). In contrast, negative control experiments using His6-tagged S5a-(196–309) and ubiquitin showed no detectable binding (lane 10) similar to results obtained by Wang and co-workers (33). Due to the large perturbation of Lys48 in the Ubld of parkin during NMR experiments, we tested the requirement for this residue in the interaction with S5a-(196–309). As shown in Fig. 4, the Ubld containing the single alanine substitution (K48A) interacted poorly with S5a-(196–309) (lane 8). Because UbldK48A retains a similar structure and stability as the wild-type protein (16) this experiment shows that Lys48 in the parkin Ubld is an important residue for S5a interaction.
FIGURE 4.
A substitution in the Ubld binding surface impairs S5a interaction. His6-S5a-(196–309) or His6-S5a-(196–309)UIMII-5A were incubated with 2 molar eq of the parkin Ubld, UbldK48A, or ubiquitin for 1 h and then loaded onto a Ni-NTA spin column. Following elution from the column the samples were fractionated on a Tricine gel and then stained with Coomassie dye. The figure shows the purified proteins His-S5a-(196–309) (lane 1), parkin Ubld (lane 3), parkin UbldK48A (lane 7), and ubiquitin (lane 9); purified His6-S5a-(196–309) (lane 2) and His6-S5a-(196–309)UIMII-5A (lane 5) after elution from the Ni-NTA column; and bound parkin Ubld with His6-S5a-(196–309) (lane 4) and His6-S5a-(196–309)UIMII-5A (lane 6), His6-S5a-(196–309) with parkin UbldK48A (lane 8), and His6-S5a-(196–309) with ubiquitin (lane 10). Molecular weight markers are shown at the right of the gel.
Eps15 Contains Two α-Helical UIM Regions
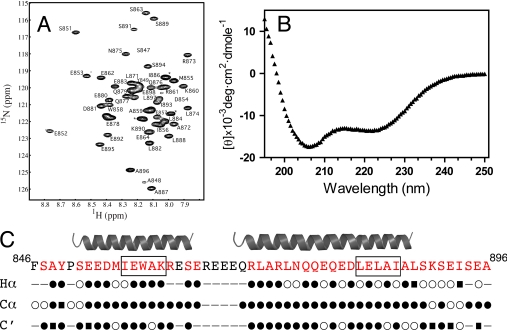
Because little structural information is available for the UIM-containing region of Eps15, a 50-amino acid construct of this protein containing both UIM regions (residues 846–896; Eps15–846-896)) was created and analyzed by NMR spectroscopy and circular dichroism spectropolarimetry. The 1H-15N HSQC spectrum of Eps15-(846–896) showed a dispersed series of resonances indicative of a folded protein and allowed the backbone assignment for Eps15-(846–896) to be completed (Fig. 5). Circular dichroism spectropolarimetry of purified Eps15-(846–896) showed the presence of well defined minimums at 206 and 225 nm indicative of a protein with about 40% α-helical structure (Fig. 5). The α-helical regions for Eps15-(846–896) were identified using NMR assignments for the Cα, Hα, and C′ atoms and the chemical shift index (42). This analysis (Fig. 5C) showed two distinct α-helical regions in Eps15-(846–896) encompassing UIM I (Glu852–Ser863) and UIM II (Arg870–Ala887). Both helices cover the UIM regions and several residues toward their N termini. The shorter α-helix includes UIM I, whereas a significantly longer α-helix surrounds UIM II. This observation is opposite that found for S5a where a longer helix encompassing UIM I and UIM II is found in the shorter helix. Furthermore, the linker region between the helices in Eps15 is about 40 residues shorter compared with S5a. On the surface, these two distinctions suggest that interactions between the parkin Ubld with Eps15 should differ from those of S5a.
FIGURE 5.
The UIM regions in Eps15 are found in two distinct α-helices. A, 600 MHz 1H-15N HSQC spectrum showing the assignment of Eps15-(846–896). Peaks that could not be assigned are labeled (*). B, CD spectrum of Eps15-(846–896) at 25 °C shows the minimums at 206 and 222 nm indicative of the α-helical structure. C, secondary structure of Eps15-(846–896) determined from chemical shift index analysis (35) of NMR assignment data for Hα, Cα, and C′ resonances. Symbols are used to denote α-helical (●), β-sheet (■), or random coil (○) structures. The consensus α-helical structure identified is shown above the Eps15 sequence.
The Parkin Ubld Interacts with Both UIMs from Eps15- (846–896)
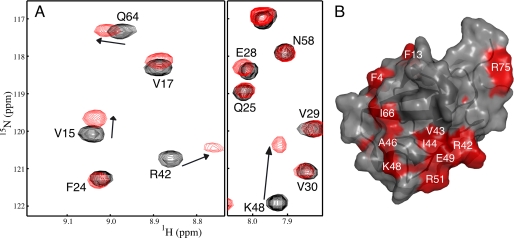
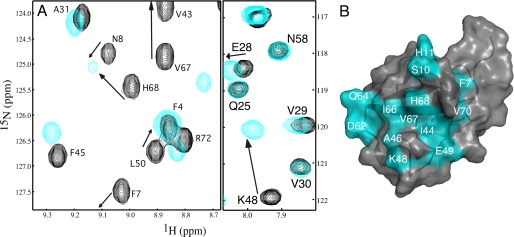
To identify the binding surface for Eps15-(846–896) with the parkin Ubld, a solution of purified 15N-labeled Ubld was incrementally titrated with a solution of purified unlabeled Eps15-(846–896) and monitored by 1H-15N HSQC spectroscopy (Fig. 6). As with the S5a interaction the most perturbed residues were Ala46 and Lys48 in the Ubld whose chemical shifts were changed by more than 1.8 ppm in the nitrogen dimension. Other residues had chemical shift changes that were more than double the average perturbation (0.050) including Phe4, Val5, Phe7, Ser10, His11, Val43, Ile44, Ala46, Lys48–Glu49, Leu61–Gln64, Ile66–His68, and Val70. Within this group were several residues that did not undergo significant shifts upon S5a binding including Val5, Phe7, Ser10, His11, Asp62–Ser65, His68, and Val70 (Fig. 7). In addition many peaks underwent much larger shifts in the Ubld in the presence of Eps15 compared with S5a. When these residues were mapped to the surface of the parkin Ubld (Fig. 6) it was clear that a much larger contiguous binding region was presented compared with that observed for S5a. This included the C-terminal region of β1 through the linker and into β2, regions not utilized by the S5a proteosomal subunit. The Eps15 interaction with the Ubld of parkin was determined by globally fitting four residues (Gly47, Ile44, Phe45, and Gln64), whose chemical shifts could be tracked throughout the titration to a Kd of 139 ± 19 μm. This was confirmed using isothermal titration calorimetry (ITC) experiments that showed a Kd of 193 ± 30 μm (see Table 1).
FIGURE 6.
The Eps15-(846–896) binding surface on the parkin Ubld. A, selected regions of 600 MHz 1H-15N spectra showing the interaction of Eps15-(846–896) with 15N-labeled parkin Ubld. The spectra show the parkin Ubld in the absence of Eps15-(846–896) (black contours) and presence of 2 eq of Eps15-(846–896) (red contours). Spectra are presented at different contour levels to account for line broadening in the Ubld protein upon Eps15-(846–896) addition. B, ribbon and surface drawing of the parkin Ubld showing the interacting surface for Eps15-(846–896). The surface is colored according to the magnitudes of the chemical shift changes ((0.2 × ΔδN2) + ΔδH2))1/2 observed in NMR experiments resulting from the interaction with Eps15-(846–896). Residues that shifted more than 0.5 S.D. above the average chemical shift change (0.050 ppm) are colored in cyan. This figure was produced using the program PyMOL (52) for the structure of human parkin Ubld structure (Protein Data Bank code 1IYF) (18).
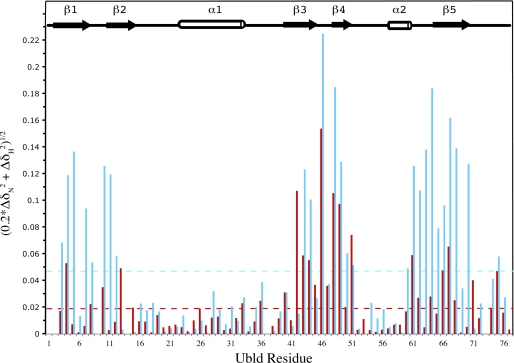
FIGURE 7.
NMR chemical shift perturbation data showing the differences in the parkin Ubld interaction with the UIMs from S5a (red) and Eps15 (cyan). The data are displayed for each residue according to the equation ((0.2 × ΔδN2) + ΔδH2)1/2, where ΔδN and ΔδH represent the changes in nitrogen and proton chemical shift (ppm) upon Ubld binding addition. The dotted lines represent the average chemical shift change for S5a-(196–309) (red) or Eps15-(846–896) (cyan) binding.
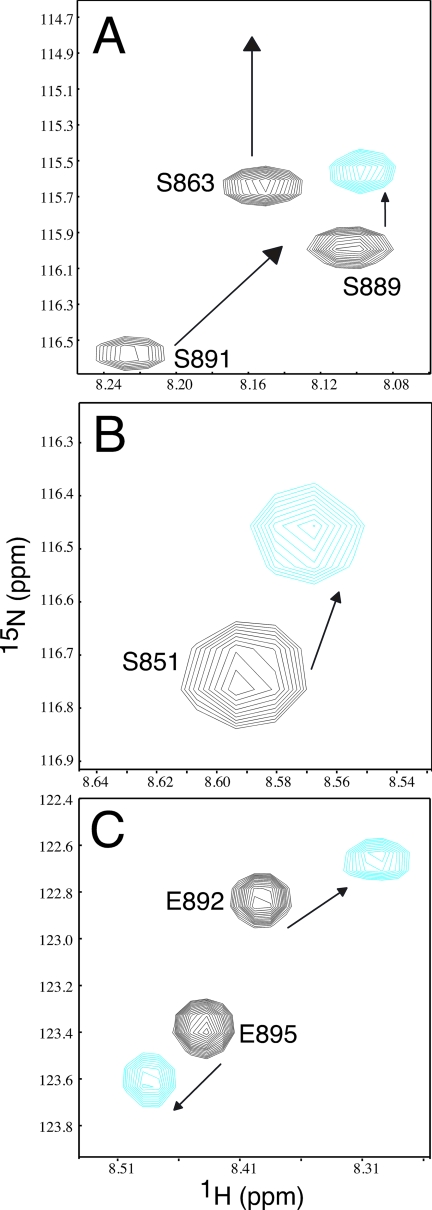
1H-15N HSQC titration experiments using 15N-labeled Eps15-(846–896) and unlabeled parkin Ubld showed that large chemical shift changes were noted in and near both UIM I and II of Eps15 (Fig. 8). For example, residues Ser851, Trp858, and Ser863 near UIM I and Ser889, Ser891, Glu892, and Glu895 in UIM II exhibited obvious changes upon Ubld binding. This observation is in contrast to the S5a interaction with the parkin Ubld where binding was noted preferentially to UIM I. In Eps15 several residues also experienced line broadening caused by the increased rotational correlation time of the complex, likely reflecting residues directly at the Eps15-Ubld interface. The residues included Ser863, Arg873, Ala887, and Ser891, all of which experienced significant chemical shift perturbations ranging in the nitrogen dimension from 0.23 ppm for Arg873 to 1.02 ppm for Ala887 and in the proton dimension from 0.01 for Ser863 to 0.08 for Ser891 (Fig. 8). The observation that both UIM sequences in S5a exhibited chemical shift changes raised the possibility that Eps15-(846–896) was interacting with multiple Ubld proteins akin to the mechanism proposed by Sims and Cohen (46) for the interaction of the UIMs in Rap80 with multiple ubiquitin molecules. To test this, we performed chemical cross-linking experiments with the parkin Ubld and Eps15-(846–896) (supplemental Fig. S2). These experiments revealed that Eps15-(846–896) had a high propensity to form 1:1 complexes with the parkin Ubld with little evidence for a 2:1 (Ubld-Eps15) complex evident.
FIGURE 8.
Eps15-(846–896) uses both UIM sequences to interact with the parkin Ubld. A–C, three different regions of the 600 MHz 1H-15N HSQC spectra showing 15N-labeled Eps15-(846–896) in the absence (black contours) and presence of 1.5 eq of parkin Ubld (cyan contours). The figure shows large changes in chemical shifts and broadening for residues in and near UIM I (Ser863 and Arg873) and UIM II (Ala887 and Ser891) of Eps15. Spectra are presented at different contour levels to account for line broadening in the Eps15-(846–896) protein upon Ubld addition.
DISCUSSION
The UIM-containing region from the endocytotic protein Eps15 was determined to have two α-helical segments separated by a 7-residue linker. A shorter α-helix (Glu852–Ser863) encompasses UIM I, whereas a longer helix (Arg870–Ala887) contains UIM II. The α-helical nature of the individual UIM regions is similar to that observed for other known UIM sequences from Vps27, hHR23, Hrs, and S5a (26, 37, 41–43). It is also interesting that Eps15 displays one α-helix that is clearly longer (UIM II) than the other, a common characteristic observed in the three-dimensional structures of the tandem UIM sequences from the proteolytic subunit S5a (33) and the endosome-sorting protein Vps15 (47). However, the shorter linker between the two helices in Eps15 makes it distinct from these other proteins where the two UIM-containing α-helices are separated by ∼30 and ∼40 residues, respectively.
Eps15 Has a Unique Arrangement of UIM Motifs
Our results show that both UIM I and II in Eps15 are involved in interaction with the parkin Ubld. This finding agrees well with experiments that show mutation to either UIM in Eps15 abolishes the interaction with parkin and reduces its ability to ubiquitinate Eps15 at a distal site (28). The use of both UIM regions by Eps15 is unique compared with other known Ubld-UIM interactions. For example, UIM II from the S5a subunit has been shown to preferentially bind to hHR23a using a series of deletion constructs (25). Furthermore, the ubiquitin-like domain from hPLIC-1 (ubiquilin-1) has been shown to interact with UIM I from Eps15 (48) and UIM I from yeast Rpn10a (17), a homolog of human S5a. The interaction of hPLIC-2 with the S5a subunit has also been observed (27). Although this study did not identify whether UIM I or II was utilized, the high similarity of the Ubld regions for hPLIC-2 and hPLIC-1 (95% identity) suggests a preference for UIM I is likely.
The structure of the hHR23a-S5a complex has been determined using NMR spectroscopy (26) and shows the major sites of interaction with the S5a UIM II arise from residues along β5 flanked by the C termini of β1 and β3 where the largest changes in chemical shift were also noted in hHR23a. Using chemical shift mapping the residues in the parkin Ubld most sensitive to Eps15 binding comprise similar regions but also include β2, the linker preceding β5 and nearly all of β1. This leads to a more extensive surface on the Ubld of parkin compared with that of hHR23a. In addition, the parkin Ubld interaction surface is also more extensive that that identified in this work for the interaction of UIM I from S5a. It is likely that close spacing of the two UIM-containing helices in Eps15 is responsible for this differential interaction with the Ubld of parkin that may correspond to its unique function. Parkin is capable of ubiquitinating both Eps15 and S5a (28, 49). However, these previous experiments show that ubiquitination of Eps15 requires its recruitment via both UIM sequences by the Ubld of parkin. It is possible this mechanism is used to distinguish trafficking substrates such as Eps15 via monoubiquitination from substrates destined for degradation requiring a polyubiquitin signal (28, 31).
Rationale for the Preference of Parkin for S5a UIM I
Our experiments show the parkin Ubld interacts preferentially with UIM I from S5a. In contrast, previous experiments using the individual UIM regions from Rpn10 (S5a) showed no detectable binding to parkin using NMR spectroscopy (18). This result may suggest that the lengths of the individual UIM constructs used may contribute to the strength of the interaction. In our case we utilized substitutions in UIM II rather than deletions to confirm the interaction. Alternatively there could be a secondary area outside the central hydrophobic regions of UIM I (LALAL) and II (IAYAM) that is important for interaction with the Ubld of parkin. Indeed many of the largest chemical shift changes in and around UIM I are observed for residues prior to this region including Gly207, Ala211, Ala212, and Glu215. It is also clear that the type of protein expression tag on the parkin Ubld influences the observed interaction with S5a sequences. For example, co-immunoprecipitation experiments using glutathione S-transferase-tagged parkin Ubld and S5a fail to show an interaction (18, 28), a similar result to that obtained by our group.3 However, the current work shows the interaction is easily observable with smaller, non-dimeric His6-tagged S5a constructs suggesting the glutathione S-transferase moiety may mask the weak interaction. In support of this idea is the interaction of untagged S5a-(196–309) with the parkin Ubld is easily observed using NMR or ITC experiments in this and previous work (18).
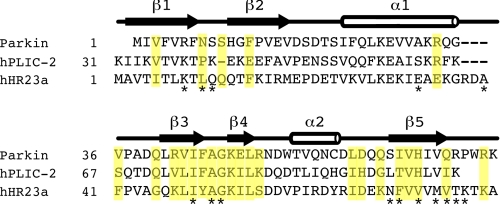
The observation that UIM I from S5a interacts with parkin Ubld is similar to that observed for interaction for the hPLIC-1 Ubld (17) and likely hPLIC-2 based on sequence identity, but in contrast to hHR23a that targets UIM II. Sequence comparison and analysis of these three Ublds (Fig. 9) in combination with observed chemical shift changes upon UIM interaction and the hHR23a-S5a structure (26) sheds some light into the differences in UIM selectivity. For example, both parkin and hHR23a undergo many large chemical shift perturbations in the β3–β4 region including Arg42–Arg51 in parkin (Lys47–Ser56 in hHR23a). Within this region Lys48 in parkin is clearly an integral residue because substitution to an alanine strongly modulates its interaction with S5a. This is a region of high identity between parkin, hHR23a, and the hPLIC proteins with the only significant difference being Arg51 in parkin (Lys82 in hPLIC-1) replaced by Ser56 in hHR23a (Fig. 9), a site more remote from the UIM binding surface. Larger differences exist in the Ubld sequences of parkin, hPLIC-1/2, and hHR23a when examining interacting residues near β1–β2, in β5, and in the α1–β3 linker. For example, in parkin the largest chemical shift changes are noted for Phe4, Ser10, and Phe13 (β1–β2), and Leu61, Ile66, Val67, His68, Gln71, and Arg75 (β5) upon UIM I binding. In hHR23a the largest change observed is Leu10, which is a key anchoring residue packing between Pro275, Leu278, Tye289, and Met293 in UIM II. This position is occupied by Asn8 in parkin (Pro40 in hPLIC-1/2) suggesting a different role for this residue in the UIM I interaction. Furthermore, the larger change in shift for Phe4 (Thr6 in hHR23a) perhaps indicates this residue plays a more important role in the S5a UIM I interaction. In parkin and hPLIC-1/2 residues in β5 are among the most divergent in the Ubld sequence (Fig. 9) from hHR23a. Three key positions, exposed in the structure of the parkin Ubld structure (18) appear to be His68, Val70, and Arg72, occupied by similar residues in hPLIC-1/2 (His102, Leu104, and Lys106), but alternate residues in hHHR23a (Val73, Met75, and Thr77). In hHR23a this section of the Ubld interacts with the IAYAM region, indicating that sequence variation in parkin and hPLIC-1/2 might contribute to a more favored interaction with the LALAL sequence in S5a UIM I. In addition, it has been noted that hHR23a has a 3-residue extension within the α1–β3 linker that is absent in other Ublds (Fig. 9) (26) including parkin and hPLIC-1/2. In hHR23a this region contacts Pro275, Leu278, and Gln286 near the N terminus of UIM II and approaches Leu10 in the β1–β2 loop. In the parkin and hPLIC-2 structures (18, 27), a larger gap exists between the α1–β3 linker and β1–β2 loop indicating similar Ubld-UIM interactions as observed in hHR23a-S5a likely do not occur in parkin·hPLIC-UIM complexes. Together, these three sections are likely the most responsible for the selectivity of the S5a UIM I by the parkin Ubld.
FIGURE 9.
Sequence alignment of the Ubld from parkin, hPLIC-2, and hHR23a. The alignment is based on previously reported secondary structure elements (18, 27) shown above the sequences. Highlighted in yellow are residues that exhibited the largest chemical shifts in parkin upon interaction with UIM I from S5a. Residues that become buried or partly buried in the hHR23a Ubld upon binding to UIM II from S5a are indicated below the sequences (*). The sequence of hPLIC-1 is identical to hPLIC-2 with the exception of Met33, Glu60, His68, and Ser88.
Interaction between the Ubld-containing protein parkin with Eps15 is important because mutation of the UIM sequence results in impaired activity in vivo (31). Nevertheless, these interactions are weak (∼100–200 μm) based on experiments from this work. It has been suggested that the weak Ubld-UIM interactions allow for easy reversibility and in the case of an abundant protein like ubiquitin (10 μm in a mammalian cells (50) offset the need for a strong affinity (51). This hypothesis is less clear for less abundant signaling proteins such as Eps15. The work presented here is the first to show how parkin uses two different methods to recruit UIM sequences, a single UIM with the S5a subunit and two UIM sequences with Eps15. The unique coordination of both UIM sequences in Eps15 by parkin may act as a signal to assist in the trafficking of this protein and distinguish this process from UIM recognition in the degradation pathway.
Supplementary Material
Acknowledgments
We thank Kathryn Barber for excellent technical assistance, Qin Liu for maintenance of the NMR spectrometers in the UWO Biomolecular NMR Facility, and Lee-Ann Briere for training and assistance with the isothermal titration calorimetry in the UWO Biomolecular and Conformations Facility. We also thank Dr. Ted Fon (McGill University) for the cDNA encoding the Eps15 UIM regions used in this work.
This work was supported by research and maintenance grants from the Canadian Institutes of Health Research (to G. S. S.), an award from the Canada Research Chairs Program (to G. S. S.), and a Canadian Institutes of Health Research Doctoral Scholarship (to S. S. S.).

The on-line version of this article (available at http://www.jbc.org) contains supplemental Figs. S1 and S2.
S. S. Safadi and G. S. Shaw, unpublished results.
- ARJP
- autosomal recessive juvenile parkinsonism
- ITC
- isothermal titration calorimetry
- Ubld
- ubiquitin-like domain
- UIM
- ubiquitin-interacting motif
- Ni-NTA
- nickel-nitrilotriacetic acid
- Tricine
- N-[2-hydroxy-1,1-bis(hydroxymethyl)ethyl]glycine
- HSQC
- heteronuclear single quantum coherence.
REFERENCES
- 1.Hershko A., Ciechanover A. (1998) Annu. Rev. Biochem. 67, 425–479 [DOI] [PubMed] [Google Scholar]
- 2.Glickman M. H., Ciechanover A. (2002) Physiol. Rev. 82, 373–428 [DOI] [PubMed] [Google Scholar]
- 3.Pickart C. M. (1997) FASEB J. 11, 1055–1066 [DOI] [PubMed] [Google Scholar]
- 4.Kitada T., Asakawa S., Hattori N., Matsumine H., Yamamura Y., Minoshima S., Yokochi M., Mizuno Y., Shimizu N. (1998) Nature 392, 605–608 [DOI] [PubMed] [Google Scholar]
- 5.Matsumine H., Saito M., Shimoda-Matsubayashi S., Tanaka H., Ishikawa A., Nakagawa-Hattori Y., Yokochi M., Kobayashi T., Igarashi S., Takano H., Sanpei K., Koike R., Mori H., Kondo T., Mizutani Y., Schäffer A. A., Yamamura Y., Nakamura S., Kuzuhara S., Tsuji S., Mizuno Y. (1997) Am. J. Hum. Genet. 60, 588–596 [PMC free article] [PubMed] [Google Scholar]
- 6.Shimura H., Hattori N., Kubo S., Mizuno Y., Asakawa S., Minoshima S., Shimizu N., Iwai K., Chiba T., Tanaka K., Suzuki T. (2000) Nat. Genet. 25, 302–305 [DOI] [PubMed] [Google Scholar]
- 7.Hristova V. A., Beasley S. A., Rylett R. J., Shaw G. S. (2009) J. Biol. Chem. 284, 14978–14986 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Morett E., Bork P. (1999) Trends Biochem. Sci. 24, 229–231 [DOI] [PubMed] [Google Scholar]
- 9.Zhang Y., Gao J., Chung K. K., Huang H., Dawson V. L., Dawson T. M. (2000) Proc. Natl. Acad. Sci. U.S.A. 97, 13354–13359 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Staropoli J. F., McDermott C., Martinat C., Schulman B., Demireva E., Abeliovich A. (2003) Neuron 37, 735–749 [DOI] [PubMed] [Google Scholar]
- 11.Corti O., Hampe C., Koutnikova H., Darios F., Jacquier S., Prigent A., Robinson J. C., Pradier L., Ruberg M., Mirande M., Hirsch E., Rooney T., Fournier A., Brice A. (2003) Hum. Mol. Genet. 12, 1427–1437 [DOI] [PubMed] [Google Scholar]
- 12.Chung K. K., Zhang Y., Lim K. L., Tanaka Y., Huang H., Gao J., Ross C. A., Dawson V. L., Dawson T. M. (2001) Nat. Med. 7, 1144–1150 [DOI] [PubMed] [Google Scholar]
- 13.Henn I. H., Gostner J. M., Lackner P., Tatzelt J., Winklhofer K. F. (2005) J. Neurochem. 92, 114–122 [DOI] [PubMed] [Google Scholar]
- 14.Shimura H., Schlossmacher M. G., Hattori N., Frosch M. P., Trockenbacher A., Schneider R., Mizuno Y., Kosik K. S., Selkoe D. J. (2001) Science 293, 263–269 [DOI] [PubMed] [Google Scholar]
- 15.Sato S., Chiba T., Sakata E., Kato K., Mizuno Y., Hattori N., Tanaka K. (2006) EMBO J. 25, 211–221 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Safadi S. S., Shaw G. S. (2007) Biochemistry 46, 14162–14169 [DOI] [PubMed] [Google Scholar]
- 17.Ko H. S., Uehara T., Tsuruma K., Nomura Y. (2004) FEBS Lett. 566, 110–114 [DOI] [PubMed] [Google Scholar]
- 18.Sakata E., Yamaguchi Y., Kurimoto E., Kikuchi J., Yokoyama S., Yamada S., Kawahara H., Yokosawa H., Hattori N., Mizuno Y., Tanaka K., Kato K. (2003) EMBO Rep. 4, 301–306 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Tashiro M., Okubo S., Shimotakahara S., Hatanaka H., Yasuda H., Kainosho M., Yokoyama S., Shindo H. (2003) J. Biomol. NMR 25, 153–156 [DOI] [PubMed] [Google Scholar]
- 20.Hartmann-Petersen R., Gordon C. (2004) Semin. Cell Dev. Biol. 15, 247–259 [DOI] [PubMed] [Google Scholar]
- 21.Imai Y., Soda M., Inoue H., Hattori N., Mizuno Y., Takahashi R. (2001) Cell 105, 891–902 [DOI] [PubMed] [Google Scholar]
- 22.Biggins S., Ivanovska I., Rose M. D. (1996) J. Cell Biol. 133, 1331–1346 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Hofmann K., Bucher P. (1996) Trends Biochem. Sci. 21, 172–173 [PubMed] [Google Scholar]
- 24.Masutani C., Araki M., Sugasawa K., van der Spek P. J., Yamada A., Uchida A., Maekawa T., Bootsma D., Hoeijmakers J. H., Hanaoka F. (1997) Mol. Cell. Biol. 17, 6915–6923 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Hiyama H., Yokoi M., Masutani C., Sugasawa K., Maekawa T., Tanaka K., Hoeijmakers J. H., Hanaoka F. (1999) J. Biol. Chem. 274, 28019–28025 [DOI] [PubMed] [Google Scholar]
- 26.Mueller T. D., Feigon J. (2003) EMBO J. 22, 4634–4645 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Walters K. J., Kleijnen M. F., Goh A. M., Wagner G., Howley P. M. (2002) Biochemistry 41, 1767–1777 [DOI] [PubMed] [Google Scholar]
- 28.Fallon L., Bélanger C. M., Corera A. T., Kontogiannea M., Regan-Klapisz E., Moreau F., Voortman J., Haber M., Rouleau G., Thorarinsdottir T., Brice A., van Bergen En Henegouwen P. M., Fon E. A. (2006) Nat. Cell Biol. 8, 834–842 [DOI] [PubMed] [Google Scholar]
- 29.Hurley J. H., Lee S., Prag G. (2006) Biochem. J. 399, 361–372 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Hofmann K., Falquet L. (2001) Trends Biochem. Sci. 26, 347–350 [DOI] [PubMed] [Google Scholar]
- 31.Polo S., Sigismund S., Faretta M., Guidi M., Capua M. R., Bossi G., Chen H., De Camilli P., Di Fiore P. P. (2002) Nature 416, 451–455 [DOI] [PubMed] [Google Scholar]
- 32.Fisher R. D., Wang B., Alam S. L., Higginson D. S., Robinson H., Sundquist W. I., Hill C. P. (2003) J. Biol. Chem. 278, 28976–28984 [DOI] [PubMed] [Google Scholar]
- 33.Wang Q., Young P., Walters K. J. (2005) J. Mol. Biol. 348, 727–739 [DOI] [PubMed] [Google Scholar]
- 34.Dev K. K., van der Putten H., Sommer B., Rovelli G. (2003) Neuropharmacology 45, 1–13 [DOI] [PubMed] [Google Scholar]
- 35.Hodgins R., Gwozd C., Arnason T., Cummings M., Ellison M. J. (1996) J. Biol. Chem. 271, 28766–28771 [DOI] [PubMed] [Google Scholar]
- 36.Stoscheck C. M. (1990) Methods Enzymol. 182, 50–68 [DOI] [PubMed] [Google Scholar]
- 37.Harris R. K., Becker E. D., Cabral de Menezes S. M., Goodfellow R., Granger P. (2002) Solid State Nucl. Magn. Reson. 22, 458–483 [DOI] [PubMed] [Google Scholar]
- 38.Kay L. E., Ikura M., Tschudin R., Bax A. (1990) J. Magn. Reson. 89, 496–514 [DOI] [PubMed] [Google Scholar]
- 39.Grzesiek S., Bax A. (1992) J. Am. Chem. Soc. 114, 6291–6293 [Google Scholar]
- 40.Delaglio F., Grzesiek S., Vuister G. W., Zhu G., Pfeifer J., Bax A. (1995) J. Biomol. NMR 6, 277–293 [DOI] [PubMed] [Google Scholar]
- 41.Johnson B. A. (2004) Methods Mol. Biol. 278, 313–352 [DOI] [PubMed] [Google Scholar]
- 42.Wishart D. S., Sykes B. D., Richards F. M. (1992) Biochemistry 31, 1647–1651 [DOI] [PubMed] [Google Scholar]
- 43.Kay L. E., Keifer P., Saarinen T. (1992) J. Am. Chem. Soc. 114, 10663–10665 [Google Scholar]
- 44.Wang Q., Walters K. J. (2004) J. Biomol. NMR 30, 231–232 [DOI] [PubMed] [Google Scholar]
- 45.Young P., Deveraux Q., Beal R. E., Pickart C. M., Rechsteiner M. (1998) J. Biol. Chem. 273, 5461–5467 [DOI] [PubMed] [Google Scholar]
- 46.Sims J. J., Cohen R. E. (2009) Mol. Cell 33, 775–783 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Swanson K. A., Kang R. S., Stamenova S. D., Hicke L., Radhakrishnan I. (2003) EMBO J. 22, 4597–4606 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Regan-Klapisz E., Sorokina I., Voortman J., de Keizer P., Roovers R. C., Verheesen P., Urbé S., Fallon L., Fon E. A., Verkleij A., Benmerah A., van Bergen en Henegouwen P. M. (2005) J. Cell Sci. 118, 4437–4450 [DOI] [PubMed] [Google Scholar]
- 49.Uchiki T., Kim H. T., Zhai B., Gygi S. P., Johnston J. A., O'Bryan J. P., Goldberg A. L. (2009) J. Biol. Chem. 284, 12622–12632 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Haas A. L., Bright P. M. (1985) J. Biol. Chem. 260, 12464–12473 [PubMed] [Google Scholar]
- 51.Hicke L., Schubert H. L., Hill C. P. (2005) Nat. Rev. Mol. Cell Biol. 6, 610–621 [DOI] [PubMed] [Google Scholar]
- 52.DeLano W. L. (2002) The PyMOL Molecular Graphics System, DeLano Scientific, Palo Alto, CA [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.