Abstract
Mutations in superoxide dismutase 1 (SOD1, EC 1.15.1.1) cause familial amyotrophic lateral sclerosis (fALS); with aggregated forms of mutant protein accumulating in spinal cord tissues of transgenic mouse models and human patients. Mice over-expressing wild-type human SOD1 (WT hSOD1) do not develop ALS-like disease, but co-expression of WT enzyme at high levels with mutant SOD1 accelerates the onset of motor neuron disease compared to mice expressing mutant hSOD1 alone. Spinal cords of mice expressing both proteins contain aggregated forms of mutant protein and, in some cases, evidence of co-aggregation of WT hSOD1 enzyme. In the present study, we used a cell culture model of mutant SOD1 aggregation to examine how the presence of WT SOD1 affects mutant protein aggregation, finding that co-expression of WT SOD1, human (hSOD1) or mouse (mSOD1), delayed the formation of mutant hSOD1 aggregates; in essence appearing to slow the aggregation rate. In some combinations of WT and mutant hSOD1 co-expression, the aggregates that did eventually form appeared to contain WT hSOD1 protein. However, WT mSOD1 did not co-aggregate with mutant hSOD1 despite displaying a similar ability to slow mutant hSOD1 aggregation. Together, these studies indicate that WT SOD1 (human or mouse), when expressed at levels equivalent to the mutant protein, modulates aggregation of FALS-mutant hSOD1.
Keywords: superoxide, dismutase, aggregation, amyotrophic lateral sclerosis
Introduction
Amyotrophic lateral sclerosis (ALS) is the most common form of motor neuron disease in adults (Bruijn et al. 2004). A small percentage of ALS cases presents a family history of autosomal dominant inheritance, with approximately 20% of all familial ALS (fALS) linked to mutations in superoxide dismutase 1 (SOD1, EC 1.15.1.1) (Rosen et al. 1993). More than 100 mutations in the 153 amino acid SOD1 protein have been identified in familial ALS [for review see (Valentine et al. 2005)]. The majority of these mutations are point mutations, although a few deletion, insertion and frameshift mutations have also been identified. Some of these types of mutations cause early terminations to C-terminally truncate the protein. However, in no case has there been a mutation linked to fALS that would lead to the absence of protein from the mutant allele. Studies of mice lacking SOD1 have demonstrated that the absence of the wild-type (WT) SOD1 protein does not cause motor neuron disease (Reaume et al. 1996). Thus, the disease-linked mutations appear to impart a toxic property to SOD1, which eventually causes the death of motor neurons.
How mutations in SOD1 cause motor neuron death remains uncertain, but several mechanisms have been proposed to explain the gain of toxic property (Valentine et al. 2005). One of the proposed mechanisms is the formation of protein aggregates, which have been proposed to interfere with one or more critical cellular processes (Johnston et al. 2000;Boillee et al. 2006;Okado-Matsumoto and Fridovich 2002;Kunst 2004). SOD1 positive inclusions are found in spinal cord tissue of fALS patients (Shibata et al. 1996;Matsumoto et al. 1996;Kato et al. 1999b;Sasaki et al. 1998;Kato et al. 1999a;Kokubo et al. 1999;Watanabe et al. 2001), and in spinal cords of transgenic mice that express SOD1-linked fALS mutant proteins (Wong et al. 1995;Bruijn et al. 1997;Stieber et al. 2000;Watanabe et al. 2001;Wang et al. 2002a;Wang et al. 2002b;Wang et al. 2003;Sasaki et al. 2005;Wang et al. 2005a;Wang et al. 2005b;Sasaki et al. 2007). In our experience, the detection of mutant SOD1 aggregation is best accomplished biochemically, using a detergent extraction and sedimentation technique (Wang et al. 2002b;Wang et al. 2002a;Wang et al. 2003;Wang et al. 2005b;Wang et al. 2005a;Wang et al. 2006). Detergent-insoluble species of SOD1 are distinguished by the property of forming structures that are not dissociated by non-ionic detergents and are of sufficient size to sediment upon centrifugation at high speed; properties associated with protein aggregation (Wang et al. 2003). For all familial mutant SOD1 proteins studied to date, detergent-insoluble and sedimentable forms of mutant SOD1 can also be generated in cell culture models (Wang et al. 2003;Wang et al. 2006). Thus, to date there has been a strong correlation between the aggregation of mutant SOD1 and toxicity.
In SOD1-linked fALS, mutant SOD1 proteins are co-expressed with WT SOD1 at 1:1 ratios of synthesis (Borchelt et al. 1994). Whether toxicity of mutant SOD1 is modulated by interactions between WT and mutant protein, or by the activity of WT SOD1, has been addressed in several experimental models. Mice over-expressing WT human SOD1 (hSOD1) appear largely normal (Gurney et al. 1994;Wong et al. 1995), although there have been reports of abnormalities in mice that express very high levels of WT hSOD1 (Dal Canto and Gurney 1995;Tu et al. 1996). In a study of mice that express human G85R SOD1, eliminating the expression of normal endogenous WT mouse SOD1 (mSOD1) or over-expressing WT hSOD1 [by crossing to a line of mice produced by Wong and colleagues (Wong et al. 1995)] had no obvious effects on disease onset, progression, or pathology (Bruijn et al. 1998). However, a later study found that mice co-expressing high levels of WT hSOD1 [by crossing to a line of mice produced by Gurney and colleagues (Gurney et al. 1994)] and G93A hSOD1 showed earlier disease onset than mice expressing the G93A mutant alone (Jaarsma et al. 2000). Recently, Deng and colleagues (Deng et al. 2006) reported that crossing the Gurney WT hSOD1 transgenic mice with mice harboring three different fALS mutants (A4V, G93A, and L126Z hSOD1) caused accelerated disease onset, which was accompanied by the appearance of aggregated SOD1. In the case of the A4V mutant mouse model, no evidence of mutant protein aggregation or disease symptoms were detected in the absence of additional WT hSOD1. A second interesting outcome was the observation that WT hSOD1 protein co-purified with the mutant SOD1 aggregates in mice that co-expressed WT and L126Z hSOD1 (Deng et al. 2006). Thus, one explanation for the decrease in age to onset could be that the addition of WT hSOD1 promoted a more rapid aggregation of mutant protein.
In this study we have used a cell culture model of mutant SOD1 aggregation to ask whether WT SOD1 directly promotes the aggregation of mutant SOD1. We found that, in cell culture systems, the co-expression of WT SOD1 (human or mouse) with mutant hSOD1 (A4V, G85R, G93A SOD1) appeared to slow the rate of mutant SOD1 aggregation. However, aggregation of mutant SOD1 is not completely blocked by the co-expression of WT protein; and in some cases we observed clear evidence for the co-purification of WT hSOD1 with the mutant hSOD1 aggregates. Interestingly, we observed that WT mSOD1 also slowed, without completely blocking, the aggregation of mutant hSOD1. However, unlike WT hSOD1, WT mSOD1 was not found in aggregates. These results demonstrate that WT hSOD1 has direct effects on the aggregation of mutant SOD1 with a tendency to slow mutant protein aggregation.
Materials and methods
Expression plasmids
All of the WT and mutant human SOD1 proteins are expressed from expression plasmids based on the pEF-BOS vector (Mizushima and Nagata 1990) containing cDNA SOD1 genes; all SOD1 vectors used in this study have been previously described (Borchelt et al. 1994;Wang et al. 2003;Karch and Borchelt 2008). The GFP cDNA was purchased from Clontech (Mountain View, CA, USA) and inserted into pcDNA3.1(A)-Myc (Invitrogen, Carlsbad, CA, USA).
Transfections and biochemical analysis of SOD1 solubility
Human embryonic kidney HEK293FT cells, which express the SV40 large T-antigen and permit episomal replication of the pEF-BOS plasmid as well as strong enhancement of transcription, were purchased from Invitrogen (Carlsbad, CA, USA). HEK293FT cells were cultured in 60 mm lysine-coated dishes (BD Biosciences Bedford, MA, USA) and transiently transfected, at 90–95% confluency, with either one SOD1 construct (4 µg), equimolar amounts of two SOD1 constructs (~2 µg each) or equimolar amounts of one SOD1 construct and the GFP construct (total 4 µg). Transfections were performed using Lipofectamine 2000, following the manufacturer’s protocol (Invitrogen, Carlsbad, CA, USA). Cells were harvested 24 or 48 hours after transfection by scraping in 1x PBS. The cell pellet was extracted in detergent and mutant SOD1 aggregates were separated by centrifugation using the detergent extraction and centrifugation assay, as previously described (Karch and Borchelt 2008). This procedure generates two fractions termed S1 (detergent-soluble cellular protein) and P2 (detergent-insoluble cellular protein). The latter containing aggregated forms of mutant SOD1.
SDS-PAGE and immunoblotting
Protein concentrations for the detergent-soluble proteins (S1 fraction) and detergent-insoluble proteins (P2 fraction) were obtained using the bicinchoninic acid assay (Pierce Biotechnology, Rockford, IL, USA). Five micrograms of S1 fractions and twenty micrograms of P2 fractions were run in 18% Tris-Glycine polyacrylamide gels (Invitrogen, Carlsbad, CA, USA) and transferred onto nitrocellulose membranes (Optitran BA-S 85, Whatman Inc., New Jersey, USA). Membranes were blocked in 5% nonfat milk in PBS before incubating them with human/mouse SOD1 primary antibody (1:2500) for one hour at room temperature or overnight at 4°C (Pardo et al. 1995). Followed by 3 washes in PBS-T (1x PBS, 0.1% Tween 20) for 10 minutes each, primary antibodies were detected by incubations with goat anti-rabbit IgG, 1:5000 (KPL, Gaithersburgh, MD, USA) for an hour at room temperature. After another 3 washes in PBS-T, secondary chemiluminescence was visualized with a Fujifilm imaging system (FUJIFILM Life Science, Stamford, CT, USA).
Estimation of aggregation propensity and statistical analysis
The aggregation propensity of SOD1 mutants was assessed by comparing the ratio of immunolabeled SOD1 protein in the P2 vs. S1 fractions. Notably, the amount of protein analyzed by immunoblots from these two fractions was not equivalent; in all cases 20 µg of protein was analyzed from the P2 fraction and 5 µg from the S1 fraction. The intensities of the SOD1 immunoreactive bands in the S1 and P2 fractions establish a ratio value for a particular mutant in a particular immunoblot. To normalize the data from different experiments, each immunoblot that was quantified included a positive control (A4V SOD1 for Fig. 1, Fig. 4, Fig. 5 and Supplemental Fig. 2; or G85R SOD1 for Fig. 3), which were used to normalize the data (A4V and G85R show equivalent aggregation propensities and the ratio values for these positive controls was set to 1). Differences in aggregation propensity were assessed by paired Student t-test (GraphPad Prism 4.0, San Diego, CA, USA), and each experiment was repeated at least three times.
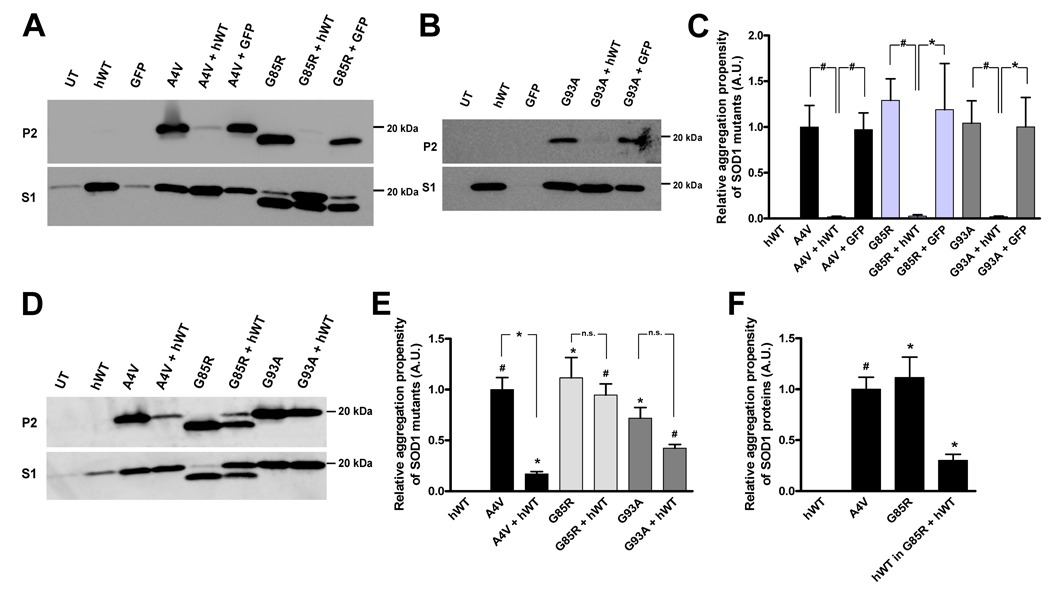
Fig. 1.
Human WT SOD1 modulates the aggregation of mutant SOD1 in cultured cells. A, B & D. Immunoblots of detergent-insoluble (P2 fraction, upper panels) and detergent-soluble fractions (S1 fraction, lower panels) from HEK293FT cells co-transfected with expression plasmids for WT hSOD1 (or GFP) and mutant SOD1 for 24 (A & B) or 48 hours (D). UT: untransfected cells. hWT: cells transfected with WT hSOD1 construct. Mutant SOD1 constructs noted by A4V, G85R, and G93A notation. C & E. Quantification of the relative aggregation potentials of the studied SOD1 mutants when expressed alone and with WT hSOD1. Bars represent mean ± SEM (n = 3–9). Statistical analysis compares the aggregation of WT hSOD1 to each mutant or mutant co-transfected with WT; *p < 0.05, #p < 0.005, n.s. indicates non-significant differences. F. Quantification of the relative aggregation propensity of WT hSOD1 when co-expressed with G85R hSOD1 mutant for 48 hours. The levels of insoluble mutant SOD1 and WT hSOD1 in cells co-transfected with G85R hSOD1 were significantly different from control WT hSOD1 alone *p < 0.05, #p < 0.005.
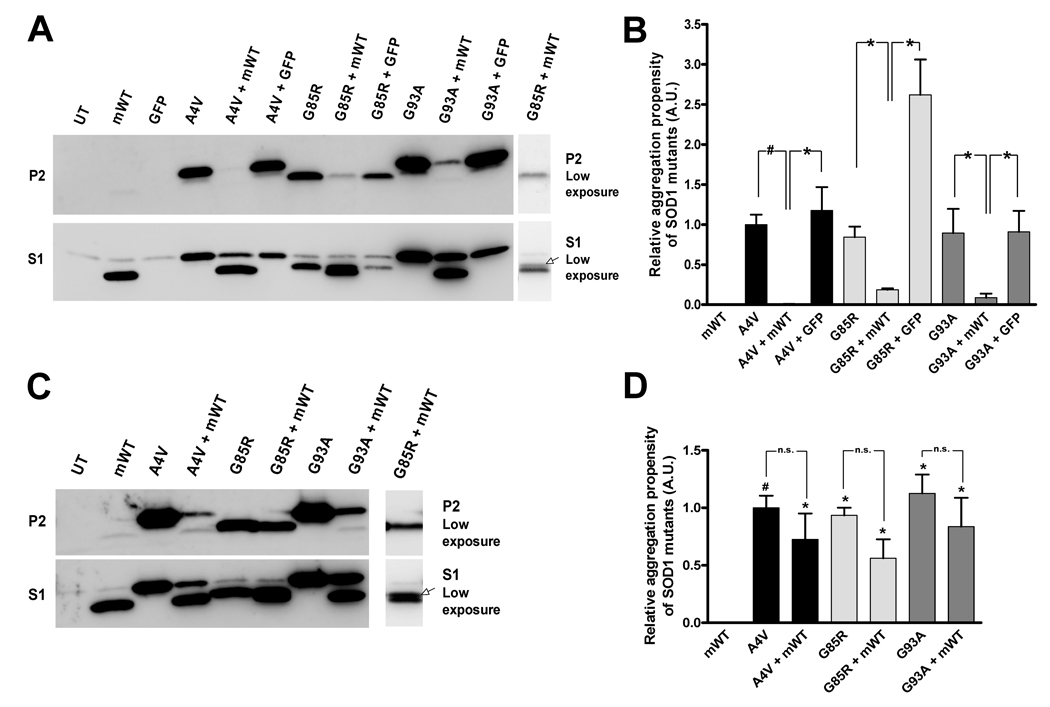
Fig. 4.
Mouse WT SOD1 modulates the aggregation of mutant hSOD1 in cultured cells without evidence of co-aggregation. A & C. Immunoblots of detergent-insoluble (P2 fraction, upper panels) and detergent-soluble fractions (S1 fraction, lower panels) from 24 (A) and 48 hour (C) incubations of HEK293FT cells co-transfected with WT mSOD1 (or GFP in A) and mutant hSOD1. UT: untransfected cells. mWT: cells transfected with WT mSOD1 construct. Mutant SOD1 constructs noted as indicated in Fig. 1. B & D. Quantification of the relative aggregation potentials of the studied SOD1 mutants when expressed alone and with WT mSOD1. Bars represent mean ± SEM (n = 3–6). Statistical analysis compares the aggregation of WT mSOD1 to each mutant, or mutant co-transfected with WT mSOD1; *p < 0.05, #p < 0.005, n.s. indicates non-significant differences.
Fig. 5.
Cysteine 111 is not required for the co-aggregation of WT hSOD1 with G85R hSOD1. A. Immunoblots of detergent extracted cells that co-expressed C111S and G85R hSOD1 proteins for 48h. UT: untransfected cells. hWT: cells transfected with WT hSOD1 construct. Mutant SOD1 constructs noted by G85R, and C111S notation. B. Quantification of the elative aggregation propensity of mutant hSOD1 proteins. Bars represent mean ± SEM. N=4. Statistical analysis compares the aggregation of WT hSOD1 to each mutant as indicated on the figure (A4V, G85R, or C111S), *p < 0.05, #p < 0.005. Comparison is also made between C111S alone to C111S in cells co-transfected with G85R, *p < 0.05.
Fig. 3.
SOD1 mutants with high propensity to aggregate (A4V and G93A SOD1) do not interfere with aggregation of G85R SOD1. A. Immunoblot of P2 (upper panel) and S1 (lower panel) fractions of singly and doubly transfected HEK293FT cells. UT: untransfected cells. WT and mutant SOD1 notations are the same as in Fig 1. B. Quantification of the relative aggregation potentials of the G85R SOD1 when expressed alone and with other SOD1 constructs. Bars represent mean ± SEM (n = 4–8). Statistical analysis compares the aggregation of WT hSOD1 to G85R or G85R co-transfected with another construct *p < 0.05, #p < 0.005. Only G85R + WT was significantly different from G85R alone *p < 0.05; n.s. indicates non-significant differences.
Analysis of detergent-soluble and insoluble fractions by hybrid linear ion-trap Fourier-transform ion cyclotron resonance mass spectrometry (FTMS)
For mass spectrometry analysis, six 60 mm culture dishes were co-transfected with WT and G93A hSOD1 constructs, then combined and extracted in detergent as described above. Ultimately, the S1 and P2 fractions were combined into final volumes of 600µl and 100µl respectively. Portions of these fractions were chromatographed by HPLC as previously described (Shaw et al. 2008). SOD1 containing fractions from HPLC chromatography were quickly thawed and 7 µL was loaded into nano-electrospray emitters (Proxeon) for immediate analysis using a nano-electrospray source equipped mass spectrometer (LTQ-FT Ultra, Thermo, San Jose). Samples were analyzed in positive ion mode with 1.8 kV typically required for stable nanospray performance. Full mass spectra were recorded over a mass range 600–2000 (m/z, Da) with resolution set at 100,000 at m/z = 400. Typically, 50 transients were averaged prior to recording a single MS spectrum. FTMS analyses were repeated twice.
Results
Differential detergent extraction and centrifugation techniques have been demonstrated as an approach to separate mutant SOD1 complexes of high molecular weight (presumed aggregates and defined as such here) that are produced in both transgenic mouse tissue and cultured cells (Johnston et al. 2000;Wang et al. 2002a;Wang et al. 2002b;Wang et al. 2003;Wang et al. 2005a;Wang et al. 2005b;Wang et al. 2006). In mutant mice, detergent-insoluble SOD1 aggregates accumulate as the animals age and disease-associated symptoms worsen (Wang et al. 2003). The formation of SOD1 aggregates can be modeled by high level expression of mutant SOD1 in human HEK293FT cells (Wang et al. 2003;Karch and Borchelt 2008). Using this model system we sought to examine whether WT SOD1 modulates the aggregation of mutant SOD1.
Co-expression of WT and mutant hSOD1 in cultured HEK293FT cells reduced the level of detergent-insoluble mutant SOD1 proteins that accumulate in 24 hours (Fig. 1). HEK293FT cells transfected with A4V, G85R or G93A SOD1 alone formed detergent-insoluble aggregates that sedimented upon ultracentrifugation, whereas cells expressing both WT and mutant hSOD1 produced little or no detergent-insoluble SOD1 protein (Figs. 1A and B, upper panel). Instead, both WT and mutant hSOD1 proteins were found only in soluble fractions (Figs. 1A and B, lower panel). To control for non-specific effects of co-transfection, such as reduced mutant protein expression that may have caused a reduction in aggregation, we co-expressed the mutant SOD1 constructs (A4V, G85R and G93A SOD1) with an expression plasmid for GFP and performed the detergent extraction and centrifugation assay. In each case, the expression of GFP did not affect the aggregation of mutant SOD1 (Figs. 1A and B, upper panel). In these cell culture experiments, a large fraction of the mutant SOD1 remains fully soluble in detergent (Figs. 1A and B, lower panels). The levels of soluble SOD1 protein provide a good indication of protein expression, indicating that all mutant proteins were expressed at high levels relative to non-transfected control cells. Immunoblot data from at least four experiments for each set of WT and mutant hSOD1 co-transfections was quantified and analyzed statistically (Fig. 1C; see Methods - Estimation of aggregation propensity and statistical analysis for an explanation of the methodology used to quantify relative aggregation propensities), providing clear evidence that in our cell culture model the co-expression of WT SOD1 modulates mutant hSOD1 aggregation.
To examine the effects of WT hSOD1 on mutant SOD1 aggregation over time, we extended the interval between transfection and harvest to 48 hours. Interestingly we found that WT hSOD1 differentially affected the aggregation of the different SOD1 mutants (A4V, G85R and G93A SOD1; Fig. 1D, E). As compared to cells expressing A4V SOD1 alone, cells co-transfected with vectors for WT and A4V hSOD1 continued to accumulate less detergent-insoluble mutant protein (Fig. 1D, upper panel). However, aggregation was not blocked as these cells contained significantly more detergent-insoluble SOD1 than cells transfected with WT hSOD1 (Fig. 1E). The levels of detergent-soluble SOD1 protein in extracts from cells co-transfected with WT and A4V hSOD1 indicated relatively high levels of expressed protein (Fig 1D, lower panel). However, because the WT and A4V hSOD1 proteins could not be distinguished by SDS-PAGE and the low amount of detergent-insoluble SOD1 in the co-transfected cells, we could not determine whether the insoluble hSOD1 is limited to mutant protein. When WT hSOD1 was co-expressed with G85R hSOD1, it was possible to differentiate the WT and mutant hSOD1 proteins by SDS-PAGE; the G85R variant migrates anomalously in SDS-PAGE, running slightly faster than the expected size (Hayward et al. 2002;Wang et al. 2003;Wang et al. 2006). In cells co-expressing WT with G85R hSOD1, we observed significant accumulation of detergent-insoluble mutant protein at 48 hours. More interestingly, WT hSOD1 was clearly detected in the detergent-insoluble fraction (Fig. 1D upper panel, and F). We estimate the relative ratio of G85R to WT hSOD1 in the insoluble fraction to be about 4 to 1. In cells co-expressing WT and G93A hSOD1 for 48 hours, a significant amount of SOD1 was detected in the detergent-insoluble fraction (Fig. 1D and E). To determine whether WT hSOD1 was present in the detergent-insoluble fraction, we analyzed these fractions by hybrid linear ion-trap Fourier-transform ion cyclotron resonance mass spectrometry (FTMS). FTMS analysis revealed the presence of both WT and G93A hSOD1 in both the detergent-insoluble and -soluble fractions (Fig. 2). In the detergent-insoluble fractions, however, the amount of WT hSOD1 was about 10 fold less than G93A hSOD1 (Fig. 2A P2,); while in the soluble fractions, the levels of WT and G93A hSOD1 were similar (Fig. 2B S1,). Overall, these results are consistent with the discoveries of detergent-insoluble WT hSOD1 in spinal cords of transgenic mice co-expressing WT and mutant hSOD1 (Deng et al. 2006). We interpret our data in cell culture as evidence that WT hSOD1 primarily slows the rate of mutant SOD1 aggregation, but ultimately when aggregates form, the WT protein may inefficiently co-aggregate with some mutant SOD1 variants.
Fig. 2.
Comparison of SOD1 molecular mass profiles from soluble (S1) and insoluble (P2) extracts of HEK293FT cells co-expressing human WT and G93A hSOD1. SOD1 was recovered from spinal cord extracts, solubilized and purified by reverse-phase chromatography for electrospray-ionization mass spectrometry (see methods). An appropriate chromatographic fraction containing the co-eluting WT and mutant hSOD1 proteins was analyzed by nano-electrospray using a hybrid linear ion-trap Fourier-transform mass spectrometer (LTQ-FT; Thermo Fisher) operating with resolution 100,000 at m/z=400. Zero charge molecular mass profiles were deconvoluted from raw Fourier-transform mass spectra of SOD1 recovered from pellet (A, P2 top) and supernatant (B, S1 bottom) fractions using Xtract software (Thermo Fisher). The bars represent the individual 13C isotopomers with the most intense approximating the average mass of the protein. Minor sodiated adducts are typical in these experiments and are seen with both WT and mutant hSOD1 proteins equally.
To control for the effects of co-transfection and for the possibility that SOD1 proteins of differing sequences might interfere with aggregation, we also co-transfected G85R SOD1 with WT, A4V, and G93A hSOD1 constructs. In these experiments we took advantage of the anomalous migration of G85R hSOD1 to examine how the co-expression of two different SOD1 mutants might affect their aggregation. Cells co-expressing G85R SOD1 with either A4V or G93A SOD1 produced detergent-insoluble forms of each SOD1 mutant (Fig. 3A, upper panel). As described above, the presence of mutant hSOD1 protein in the S1 fraction for each transfection indicated that only a portion of the total protein adopted the insoluble conformation (Fig. 3A, lower panel). Quantification of multiple independent experiments demonstrated that whether expressed alone or in combination with the A4V or G93A SOD1 variants, the propensity of G85R SOD1 to aggregate was not significantly altered (Fig. 3B). These data suggest that the apparent reduction in aggregation caused by the co-expression of WT with mutant hSOD1 is not due to some non-specific effect of co-transfection or some non-specific effect of interactions between two different SOD1 subunits, but rather appears to be due to a specific property of the WT hSOD1 protein.
The human and mouse WT SOD1 protein share 83.6% identity at the level of amino acid sequence (Supplemental Fig. 1); 25 amino acid differences in the 153 residue protein. Thus we next sought to investigate whether these differences in protein sequence would affect the ability of WT SOD1 to modulate the aggregation rate of mutant SOD1. HEK293FT cells co-transfected with WT mouse SOD1 (mSOD1) and mutant hSOD1 proteins (A4V, G85R and G93A SOD1) showed a significant reduction in the amount of detergent-insoluble SOD1 aggregates produced in 24 hours (Fig. 4A, B). Interpretation of the immunoblots of cells co-transfected with WT mSOD1 and G85R hSOD1 was complicated by the fact that these proteins migrated to very near the same position in SDS-PAGE. However, these proteins could be resolved in gels exposed for short intervals, allowing for the detection of both WT mSOD1 and G85R hSOD1 in the detergent-soluble protein fraction (Fig. 4A, right lower panel). Despite a significant reduction in the amount of insoluble G85R hSOD1 in these co-transfected cells, aggregation was not blocked and it was possible to demonstrate that the detergent-insoluble fraction contained only G85R hSOD1 (Fig. 4A, right upper panel). Quantification of the relative aggregation propensity of the hSOD1 mutants in cells co-transfected with WT mSOD1 revealed a significant reduction in the amount of detergent-insoluble mutant hSOD1 protein that accumulated in 24 hours (Fig 4B). Thus, WT mSOD1 has the same capacity as WT hSOD1 to slow the rate of mutant hSOD1 to aggregate.
When we extended the interval between transfection and harvest to 48 hours, we observed that A4V, G85R and G93A hSOD1, when expressed with WT mSOD1, were able to form detectable amounts of detergent-insoluble SOD1 aggregates (Fig. 4C). We observed that in measures of aggregation propensity, which compensates for any changes in the expression of mutant hSOD1 that may occur when co-transfected with WT mSOD1, the presence of WT mSOD1 had no significant effect on aggregation of mutant hSOD1 (Fig. 4D). Interestingly, WT mSOD1, unlike WT hSOD1, did not seem to co-aggregate with any of the mutants even after the longer 48 hour interval (Fig. 4C, upper panel). In co-transfections of A4V or G93A hSOD1 mutants with WT mSOD1, the amount of mSOD1 detected in the insoluble fraction was not different from that of cells transfected with mSOD1 alone (Fig. 4C; p > 0.05, n = 4 independent replications). Moreover, we did not detect mSOD1 in detergent-insoluble fractions of cells co-expressing WT mSOD1 and G85R hSOD1 at a level greater than that of cells expressing mSOD1 alone (p > 0.20, n = 3 independent replications). Thus, although WT mSOD1 possesses an ability that is similar to WT hSOD1 in modulating the aggregation of mutant proteins, it lacks a feature that allows for co-sedimentation with mutant hSOD1.
The differing ability of WT mSOD1 and hSOD1 to co-aggregate with mutant hSOD1 is a finding that appears to be consistent with a recent report suggesting that a specific cysteine residue in hSOD1 may mediate the co-aggregation of WT and mutant hSOD1 (Cozzolino et al. 2008). The cysteine residue at position 111 of hSOD1 has been identified as a potential mediator of disulfide cross-linking between mutant and WT hSOD1 (Cozzolino et al. 2008). Mouse SOD1 encodes serine at position 111, and thus could not generate such disulfide linkages. To directly test this hypothesis, we used a previously described cDNA hSOD1 that encodes serine as position 111 (C111S hSOD1) (Karch and Borchelt 2008) in co-transfection with G85R hSOD1. The mutant C111S hSOD1 is not an ALS mutation; in most species the position equivalent to 111 encodes serine. Previous studies have established that this mutant does not spontaneously aggregate (Cozzolino et al. 2008;Karch and Borchelt 2008). In C111S and G85R hSOD1 co-transfections, cells were harvested after 48 hours, which was the interval needed to observe WT and G85R hSOD1 co-aggregation. Consistent with previous studies, all C111S hSOD1 fractionated to the detergent-soluble fraction (Fig. 5A, compare upper and lower panels). However, in cells co-transfected with C111S and G85R hSOD1 and harvested 48 hours later, we found both proteins in the detergent-insoluble fraction Quantification of the aggregation propensity of each SOD1 protein was performed as previously described, showing significant accumulation of aggregated C111S hSOD1 in the co-transfected cells (Fig. 5B). This finding suggests that the co-aggregation of WT hSOD1 with mutant protein is not dependent upon a disulfide linkage between cysteine 111 of WT protein and cysteine residues of mutant hSOD1.
Discussion
In the present study, we examined the potential for WT SOD1 to influence the aggregation of mutant SOD1 as defined by the formation of structures that are insoluble in non-ionic detergent and sediment upon ultracentrifugation. In a cell culture model of mutant SOD1 aggregation, we found that the presence of WT hSOD1 or mSOD1 significantly lowered the amount of mutant hSOD1 (A4V, G85R and G93A) in aggregates after 24 hours. Upon longer incubation (48 hours), we observed significant aggregation of G85R and G93A hSOD1, but continued attenuation of A4V aggregate levels. We also observed that WT hSOD1 can co-sediment with mutant G85R and G93A SOD1 aggregates; however, the predominant species of SOD1 in these aggregates was mutant protein. Importantly, these effects of WT hSOD1 or mSOD1 on the aggregation of mutant protein were specific to WT protein. Co-expression of G85R hSOD1 with either A4V or G93A hSOD1 showed no evidence of slowed aggregation rates; detergent-insoluble forms of both mutant proteins were readily detectable in 24 hours. From these findings, we conclude that WT SOD1 possess a capacity to modulate the aggregation of the mutant protein, with the primary effect being to slow aggregation rates.
Human but not mouse WT SOD1 can co-aggregate with mutant SOD1
One mechanism by which WT protein could slow aggregation of mutant protein, but then ultimately become a component of such aggregates, is via direct protein-protein interactions between the WT and mutant proteins at the level of nucleation, or growth, of the aggregate. In many aggregate structures, the stacking of peptide chains of identical sequence is crucial to the formation of stable oligomeric structures (Petty and Decatur 2005;Shorter and Lindquist 2005). Such stacking forces have been proposed in prion protein conformational changes and it is well established that the presence of two prion proteins with single amino acid substitutions can slow aggregation (Petty et al. 2005;Hsiao et al. 1994). With this notion in mind, we tested whether WT mSOD1 could produce the same effects as WT hSOD1 on the aggregation of mutant hSOD1. The WT mSOD1 protein contains 25 amino acid differences from the human protein (see Supplemental Fig. 1). Despite these numerous sequence differences, WT mSOD1 retains the ability to slow aggregation of mutant hSOD1, presumably through direct protein-protein interactions. However, WT mSOD1 does not co-sediment with the mutant hSOD1 aggregates. This latter outcome could indicate that the numerous sequence differences between human and mouse SOD1 disrupt the types of close protein-protein interactions that would be required in the assembly of SOD1 aggregates.
WT and mutant hSOD1 protein-protein interaction: role of disulfide bonding and cysteine 111
Our observation that WT mSOD1 does not co-aggregate with mutant hSOD1 is consistent with a recently a proposed mechanism of WT and mutant SOD1 co-aggregation that suggested a role for inter-subunit disulfide crosslinking between cysteine residues at position 111 (Cozzolino et al. 2008). In a heterodimer of WT and mutant SOD1 subunits, these cysteines would be in close proximity near the dimer interface and thus could mediate an inter-subunit bridge. Mouse SOD1 encodes serine at position 111 and would be incapable of forming such a disulfide bridge. To test the role of disulfide linkages between cysteine 111 residues in the co-aggregation of WT and mutant hSOD1, we mutated cysteine 111 of WT hSOD1 to serine and then co-transfected this construct with G85R hSOD1; finding that we could still detect co-aggregation of this modified WT hSOD1 with mutant protein. We cannot rule out the possibility that cysteine 111 of the mutant hSOD1 mediates a disulfide linkage with another cysteine in C111S hSOD1 (cysteines at positions 6, 57, or 146); however, it is clear that the linkage cannot be between cysteine 111 of the two proteins. Additionally, we have noted that we can supplement the buffers used in our detergent extraction protocols with as much as 30% β-mercaptoethanol (β-ME, a strong reducing agent) without noting significant reductions in the amount of mutant SOD1 that fractionates to detergent-insoluble fractions (Supplemental Fig. 2). These data provide compelling evidence that disulfide cross-linking is not a primary mechanism by which the structure of aggregates are maintained [also see (Karch and Borchelt 2008)]; and we think it unlikely that disulfide-linkages are responsible for the co-sedimentation of WT hSOD1 with mutant hSOD1. Rather, we suggest that the co-sedimentation of WT hSOD1 with mutant hSOD1 is likely to involve more intimate protein-protein interactions.
Role of WT and mutant SOD1 interactions in disease
In a study by Deng and colleagues (Deng et al. 2006), the co-expression of WT and mutant hSOD1 in transgenic animals, produced by mating two distinct lines of mice, showed earlier onset of disease and earlier age to paralysis; with the symptomatic mice showing high levels of detergent-insoluble forms of both WT and mutant protein. If aggregation of mutant SOD1 were one of the driving forces in age to disease onset, then increasing the concentration of total SOD1, through the addition of WT hSOD1 protein, could potentially decrease the “nucleation” phase of protein aggregation; which is well established to be highly concentration-dependent (Jarrett and Lansbury, Jr. 1992). However, in our cell culture model, we find that the presence of WT hSOD1 slows the aggregation of mutant protein. The most informative data in our experiment is a comparison of mutant SOD1 aggregate loads in cells co-transfected with mutant SOD1 constructs and constructs for GFP to cells co-transfected with mutant and WT SOD1 constructs. As compared to GFP, WT SOD1 co-expression reduced overall amounts of aggregated mutant SOD1 that accumulated in 24 hours. We interpret this finding as evidence that WT hSOD1 does not provide a concentration-dependent enhancement of mutant SOD1 aggregation. Whether the effect of WT SOD1 on mutant SOD1 aggregation occurs at the level of aggregate nucleation is difficult to address in our cell culture system. It is possible that WT SOD1 interferes with the growth phase of aggregation in which small oligomers of protein assemble into larger sedimentable aggregates.
If our cell culture studies accurately model events that occur in vivo, then our data would argue that the basis for accelerated disease onset in the mouse studies of Deng and colleagues (Deng et al. 2006) is not attributable to accelerated rates of mutant SOD1 aggregation. However, the foregoing study demonstrated that mice expressing low levels of A4V hSOD1 never develop disease and do not develop SOD1 aggregates, whereas mice that co-express high levels of WT hSOD1 with low levels of A4V hSOD1 develop disease with spinal cords that contain aggregated SOD1 protein (undetermined whether WT, A4V, or both) (Deng et al. 2006). This latter outcome suggests a direct involvement of SOD1 aggregation in disease pathogenesis.
However, other recent studies have demonstrated that aggregation of mutant SOD1 may be dissociable from the toxic events that drive disease onset. Co-expression of high levels of the copper chaperone for SOD1 (CCS) with G93A SOD1 greatly accelerates the onset of disease while reducing the level of G93A SOD1 aggregation (Son et al. 2007). Moreover, we have recently determined that the accumulation of the larger sedimentable aggregates of mutant SOD1 in fALS mouse models occurs largely after disease onset (Karch and Borchelt, manuscript in preparation). These recent findings indicate that disease onset may not be governed by the rate of mutant protein aggregation. Whether other aspects of disease, such as progression, are related to the rates of mutant protein aggregation is a subject of study.
Although the cell model we use here has high utility in assessing the aggregation propensity of mutant SOD1, it is not well suited for studies of toxicity. The advantage of the model is that aggregation occurs without an exogenous stimulus, such as inhibition of proteasomes or other toxic insult. However, the levels of expression achieved are admittedly well above physiologic levels and thus we are hesitant to conclude that any toxicity observed in this cell model over a 24 or 48 hour period would equate to events occurring over a much longer time frame in either mouse models or humans. Deciphering the mechanism by which WT hSOD1 overexpression heightens the toxicity of mutant SOD1 will require development of more physiologically relevant cell models, or innovative approaches to studying molecular events in animal models.
Conclusions
In a cell culture model of mutant SOD1 aggregation, we find evidence that WT SOD1 is a direct modulator of mutant hSOD1 aggregation, with the predominant effect being to slow aggregation rates. More than 100 mutations in SOD1 have been associated with fALS; given the variability in the biophysical properties of these mutants, we think it is highly likely that the magnitude of the effect of WT protein on the aggregation rate of mutant hSOD1 will vary. Indeed in our small sample of mutants in the present study, we find that that the effect of WT SOD1 on the aggregation of A4V hSOD1 appears to be distinct from that of the G85R or G93A variants. In human SOD1-linked fALS, disease occurs in a setting of equivalent expression of WT and mutant SOD1 subunits. We propose that the modulation of mutant hSOD1 aggregation by WT enzyme may introduce another factor that influences the age to onset, or rate of progression, of disease in humans.
Supplementary Material
Acknowledegments
We thank Ms. Celeste Karch for thoughtful discussions and help in preparation of this manuscript. We also thank Ms. Hilda Brown for her help in generating mutant SOD1 cDNA vectors. We are grateful to Professor Joseph A. Loo for providing access to the LTQ-FT Ultra high resolution mass spectrometer. This work was funded by a grant from the National Institutes of Neurologic Disease and Stroke (P01 NS049134 – Program Project award to Drs. Joan S. Valentine, P. John Hart, D.R. Borchelt, and J.P. Whitelegge - we thank our colleagues for thoughtful discussions) and by a grant from the National Center for Research Resources (S10 RR023045 to Julian Whitelegge).
Abbreviations used
- fALS
familial amyotrophic lateral sclerosis
- SOD1
superoxide dismutase 1
- hSOD1
human SOD1
- mSOD1
mouse SOD1
- WT
wild-type
- β-ME
β-mercaptoethanol.
Reference List
- Boillee S, Vande VC, Cleveland DW. ALS: a disease of motor neurons and their nonneuronal neighbors. Neuron. 2006;52:39–59. doi: 10.1016/j.neuron.2006.09.018. [DOI] [PubMed] [Google Scholar]
- Borchelt DR, Lee MK, Slunt HS, Guarnieri M, Xu ZS, Wong PC, Brown RH, Jr, Price DL, Sisodia SS, Cleveland DW. Superoxide dismutase 1 with mutations linked to familial amyotrophic lateral sclerosis possesses significant activity. Proc. Natl. Acad. Sci. U. S. A. 1994;91:8292–8296. doi: 10.1073/pnas.91.17.8292. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bruijn LI, Becher MW, Lee MK, Anderson KL, Jenkins NA, Copeland NG, Sisodia SS, Rothstein JD, Borchelt DR, Price DL, Cleveland DW. ALS-linked SOD1 mutant G85R mediates damage to astrocytes and promotes rapidly progressive disease with SOD1-containing inclusions. Neuron. 1997;18:327–338. doi: 10.1016/s0896-6273(00)80272-x. [DOI] [PubMed] [Google Scholar]
- Bruijn LI, Houseweart MK, Kato S, Anderson KL, Anderson SD, Ohama E, Reaume AG, Scott RW, Cleveland DW. Aggregation and motor neuron toxicity of an ALS-linked SOD1 mutant independent from wild-type SOD1. Science. 1998;281:1851–1854. doi: 10.1126/science.281.5384.1851. [DOI] [PubMed] [Google Scholar]
- Bruijn LI, Miller TM, Cleveland DW. Unraveling the mechanisms involved in motor neuron degeneration in ALS. Annu. Rev. Neurosci. 2004;27:732–749. doi: 10.1146/annurev.neuro.27.070203.144244. [DOI] [PubMed] [Google Scholar]
- Cozzolino M, Amori I, Pesaresi MG, Ferri A, Nencini M, Carri MT. Cysteine 111 Affects Aggregation and Cytotoxicity of Mutant Cu,Zn-superoxide Dismutase Associated with Familial Amyotrophic Lateral Sclerosis. J. Biol. Chem. 2008;283:866–874. doi: 10.1074/jbc.M705657200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dal Canto MC, Gurney ME. Neuropathological changes in two lines of mice carrying a transgene for mutant human Cu,ZN SOD, and in mice overexpressing wild type human SOD: a model of familial amyotrophic lateral sclerosis (FALS) Brain Res. 1995;676:25–40. doi: 10.1016/0006-8993(95)00063-v. [DOI] [PubMed] [Google Scholar]
- Deng HX, Shi Y, Furukawa Y, Zhai H, Fu R, Liu E, Gorrie GH, Khan MS, Hung WY, Bigio EH, Lukas T, Dal Canto MC, O'Halloran TV, Siddique T. Conversion to the amyotrophic lateral sclerosis phenotype is associated with intermolecular linked insoluble aggregates of SOD1 in mitochondria. Proc. Natl. Acad. Sci. U. S. A. 2006;103:7142–7147. doi: 10.1073/pnas.0602046103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gurney ME, Pu H, Chiu AY, Dal Canto MC, Polchow CY, Alexander DD, Caliendo J, Hentati A, Kwon YW, Deng H-X, Chen W, Zhai P, Sufit RL, Siddique T. Motor neuron degeneration in mice that express a human Cu,Zn superoxide dismutase mutation. Science. 1994;264:1772–1775. doi: 10.1126/science.8209258. [DOI] [PubMed] [Google Scholar]
- Hayward LJ, Rodriguez JA, Kim JW, Tiwari A, Goto JJ, Cabelli DE, Valentine JS, Brown RH., Jr Decreased Metallation and Activity in Subsets of Mutant Superoxide Dismutases Associated with Familial Amyotrophic Lateral Sclerosis. J. Biol. Chem. 2002;277:15923–15931. doi: 10.1074/jbc.M112087200. [DOI] [PubMed] [Google Scholar]
- Hsiao KK, Groth D, Scott M, Yang SL, Serban H, Rapp D, Foster D, Torchia M, DeArmond SJ, Prusiner SB. Serial transmission in rodents of neurodegeneration from transgenic mice expressing mutant prion protein. Proc. Natl. Acad. Sci. U. S. A. 1994;91:9126–9130. doi: 10.1073/pnas.91.19.9126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jaarsma D, Haasdijk ED, Grashorn JA, Hawkins R, van DW, Verspaget HW, London J, Holstege JC. Human Cu/Zn superoxide dismutase (SOD1) overexpression in mice causes mitochondrial vacuolization, axonal degeneration, and premature motoneuron death and accelerates motoneuron disease in mice expressing a familial amyotrophic lateral sclerosis mutant SOD1. Neurobiol. Dis. 2000;7:623–643. doi: 10.1006/nbdi.2000.0299. [DOI] [PubMed] [Google Scholar]
- Jarrett JT, Lansbury PT., Jr Amyloid fibril formation requires a chemically discriminating nucleation event: studies of an amyloidogenic sequence from the bacterial protein OsmB. Biochemistry. 1992;31:12345–12352. doi: 10.1021/bi00164a008. [DOI] [PubMed] [Google Scholar]
- Johnston JA, Dalton MJ, Gurney ME, Kopito RR. Formation of high molecular weight complexes of mutant Cu, Zn-superoxide dismutase in a mouse model for familial amyotrophic lateral sclerosis. Proc. Natl. Acad. Sci. USA. 2000;97:12751–12576. doi: 10.1073/pnas.220417997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karch CM, Borchelt DR. A Limited Role for Disulfide Cross-linking in the Aggregation of Mutant SOD1 Linked to Familial Amyotrophic Lateral Sclerosis. J. Biol. Chem. 2008;283:13528–13537. doi: 10.1074/jbc.M800564200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kato S, Horiuchi S, Nakashima K, Hirano A, Shibata N, Nakano I, Saito M, Kato M, Asayama K, Ohama E. Astrocytic hyaline inclusions contain advanced glycation endproducts in familial amyotrophic lateral sclerosis with superoxide dismutase 1 gene mutation: immunohistochemical and immunoelectron microscopical analyses. Acta Neuropathol. (Berl) 1999a;97:260–266. doi: 10.1007/s004010050983. [DOI] [PubMed] [Google Scholar]
- Kato S, Saito M, Hirano A, Ohama E. Recent advances in research on neuropathological aspects of familial amyotrophic lateral sclerosis with superoxide dismutase 1 gene mutations: neuronal Lewy body-like hyaline inclusions and astrocytic hyaline inclusions. Histol. Histopathol. 1999b;14:973–989. doi: 10.14670/HH-14.973. [DOI] [PubMed] [Google Scholar]
- Kokubo Y, Kuzuhara S, Narita Y, Kikugawa K, Nakano R, Inuzuka T, Tsuji S, Watanabe M, Miyazaki T, Murayama S, Ihara Y. Accumulation of neurofilaments and SOD1-immunoreactive products in a patient with familial amyotrophic lateral sclerosis with I113T SOD1 mutation. Arch. Neurol. 1999;56:1506–1508. doi: 10.1001/archneur.56.12.1506. [DOI] [PubMed] [Google Scholar]
- Kunst CB. Complex genetics of amyotrophic lateral sclerosis. Am. J. Hum. Genet. 2004;75:933–947. doi: 10.1086/426001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matsumoto S, Kusaka H, Ito H, Shibata N, Asayama T, Imai T. Sporadic amyotrophic lateral sclerosis with dementia and Cu/Zn superoxide dismutase-positive Lewy body-like inclusions. Clin. Neuropathol. 1996;15:41–46. [PubMed] [Google Scholar]
- Mizushima S, Nagata S. pEF-BOS, a powerful mammalian expression vector. Nucleic Acids Res. 1990;18:5322. doi: 10.1093/nar/18.17.5322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Okado-Matsumoto A, Fridovich I. Amyotrophic lateral sclerosis: a proposed mechanism. Proc. Natl. Acad. Sci U. S. A. 2002;99:9010–9014. doi: 10.1073/pnas.132260399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pardo CA, Xu Z, Borchelt DR, Price DL, Sisodia SS, Cleveland DW. Superoxide dismutase is an abundant component in cell bodies, dendrites, and axons of motor neurons and in a subset of other neurons. Proc. Natl. Acad. Sci. U. S. A. 1995;92:954–958. doi: 10.1073/pnas.92.4.954. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Petty SA, Adalsteinsson T, Decatur SM. Correlations among morphology, beta-sheet stability, and molecular structure in prion peptide aggregates. Biochemistry. 2005;44:4720–4726. doi: 10.1021/bi047445a. [DOI] [PubMed] [Google Scholar]
- Petty SA, Decatur SM. Intersheet rearrangement of polypeptides during nucleation of {beta}-sheet aggregates. Proc. Natl. Acad. Sci. U. S. A. 2005;102:14272–14277. doi: 10.1073/pnas.0502804102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reaume AG, Elliott JL, Hoffman EK, Kowall NW, Ferrante RJ, Siwek DF, Wilcox HM, Flood DG, Beal MF, Brown RH, Jr, Scott RW, Snider WD. Motor neurons in Cu/Zn superoxide dismutase-deficient mice develop normally but exhibit enhanced cell death after axonal injury. Nat. Genet. 1996;13:43–47. doi: 10.1038/ng0596-43. [DOI] [PubMed] [Google Scholar]
- Rosen DR, Siddique T, Patterson D, Figlewicz DA, Sapp P, Hentati A, Donaldson D, Goto J, O'Regan JP, Deng HX. Mutations in Cu/Zn superoxide dismutase gene are associated with familial amyotrophic lateral sclerosis. Nature. 1993;362:59–62. doi: 10.1038/362059a0. [DOI] [PubMed] [Google Scholar]
- Sasaki S, Nagai M, Aoki M, Komori T, Itoyama Y, Iwata M. Motor neuron disease in transgenic mice with an H46R mutant SOD1 gene. J. Neuropathol. Exp. Neurol. 2007;66:517–524. doi: 10.1097/01.jnen.0000263868.84188.3b. [DOI] [PubMed] [Google Scholar]
- Sasaki S, Ohsawa Y, Yamane K, Sakuma H, Shibata N, Nakano R, Kikugawa K, Mizutani T, Tsuji S, Iwata M. Familial amyotrophic lateral sclerosis with widespread vacuolation and hyaline inclusions. Neurology. 1998;51:871–873. doi: 10.1212/wnl.51.3.871. [DOI] [PubMed] [Google Scholar]
- Sasaki S, Warita H, Murakami T, Shibata N, Komori T, Abe K, Kobayashi M, Iwata M. Ultrastructural study of aggregates in the spinal cord of transgenic mice with a G93A mutant SOD1 gene. Acta Neuropathol. (Berl) 2005;109:247–255. doi: 10.1007/s00401-004-0939-7. [DOI] [PubMed] [Google Scholar]
- Shaw BF, Lelie HL, Durazo A, Nersissian AM, Xu G, Chan PK, Gralla EB, Tiwari A, Hayward LJ, Borchelt DR, Valentine JS, Whitelegge JP. Detergent-insoluble aggregates associated with amyotrophic lateral sclerosis in transgenic mice contain primarily full-length, unmodified superoxide dismutase-1. J. Biol. Chem. 2008;283:8340–8350. doi: 10.1074/jbc.M707751200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shibata N, Hirano A, Kobayashi M, Siddique T, Deng HX, Hung WY, Kato T, Asayama K. Intense superoxide dismutase-1 immunoreactivity in intracytoplasmic hyaline inclusions of familial amyotrophic lateral sclerosis with posterior column involvement. J. Neuropathol. Exp. Neurol. 1996;55:481–490. doi: 10.1097/00005072-199604000-00011. [DOI] [PubMed] [Google Scholar]
- Shorter J, Lindquist S. Prions as adaptive conduits of memory and inheritance. Nat. Rev. Genet. 2005;6:435–450. doi: 10.1038/nrg1616. [DOI] [PubMed] [Google Scholar]
- Son M, Puttaparthi K, Kawamata H, Rajendran B, Boyer PJ, Manfredi G, Elliott JL. Overexpression of CCS in G93A-SOD1 mice leads to accelerated neurological deficits with severe mitochondrial pathology. Proc. Natl. Acad. Sci. U. S. A. 2007;104:6072–6077. doi: 10.1073/pnas.0610923104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stieber A, Gonatas JO, Gonatas NK. Aggregates of mutant protein appear progressively in dendrites, in periaxonal processes of oligodendrocytes, and in neuronal and astrocytic perikarya of mice expressing the SOD1(G93A) mutation of familial amyotrophic lateral sclerosis. J. Neurol. Sci. 2000;177:114–123. doi: 10.1016/s0022-510x(00)00351-8. [DOI] [PubMed] [Google Scholar]
- Tu P-H, Raju P, Robinson KA, Gurney ME, Trojanowski JQ, Lee VMY. Transgenic mice carrying a human mutant superoxide dismutase transgene develop neuronal cytoskeletal pathology resembling human amyotrophic lateral sclerosis lesions. Proc. Natl. Acad. Sci. USA. 1996;93:3155–3160. doi: 10.1073/pnas.93.7.3155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Valentine JS, Doucette PA, Potter SZ. Copper-Zinc Superoxide Dismutase and Amyotrophic Lateral Sclerosis. 2005 doi: 10.1146/annurev.biochem.72.121801.161647. [DOI] [PubMed] [Google Scholar]
- Wang J, Slunt H, Gonzales V, Fromholt D, Coonfield M, Copeland NG, Jenkins NA, Borchelt DR. Copper-binding-site-null SOD1 causes ALS in transgenic mice: aggregates of non-native SOD1 delineate a common feature. Hum. Mol. Genet. 2003;12:2753–2764. doi: 10.1093/hmg/ddg312. [DOI] [PubMed] [Google Scholar]
- Wang J, Xu G, Borchelt DR. High molecular weight complexes of mutant superoxide dismutase 1: age-dependent and tissue-specific accumulation. Neurobiol. Dis. 2002a;9:139–148. doi: 10.1006/nbdi.2001.0471. [DOI] [PubMed] [Google Scholar]
- Wang J, Xu G, Borchelt DR. Mapping superoxide dismutase 1 domains of nonnative interaction: roles of intra- and intermolecular disulfide bonding in aggregation. J. Neurochem. 2006;96:1277–1288. doi: 10.1111/j.1471-4159.2005.03642.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang J, Xu G, Gonzales V, Coonfield M, Fromholt D, Copeland NG, Jenkins NA, Borchelt DR. Fibrillar inclusions and motor neuron degeneration in transgenic mice expressing superoxide dismutase 1 with a disrupted copper-binding site. Neurobiol. Dis. 2002b;10:128–138. doi: 10.1006/nbdi.2002.0498. [DOI] [PubMed] [Google Scholar]
- Wang J, Xu G, Li H, Gonzales V, Fromholt D, Karch C, Copeland NG, Jenkins NA, Borchelt DR. Somatodendritic accumulation of misfolded SOD1-L126Z in motor neurons mediates degeneration: alphaB-crystallin modulates aggregation. Hum. Mol. Genet. 2005a;14:2335–2347. doi: 10.1093/hmg/ddi236. [DOI] [PubMed] [Google Scholar]
- Wang J, Xu G, Slunt HH, Gonzales V, Coonfield M, Fromholt D, Copeland NG, Jenkins NA, Borchelt DR. Coincident thresholds of mutant protein for paralytic disease and protein aggregation caused by restrictively expressed superoxide dismutase cDNA. Neurobiol. Dis. 2005b;20:943–952. doi: 10.1016/j.nbd.2005.06.005. [DOI] [PubMed] [Google Scholar]
- Watanabe M, Dykes-Hoberg M, Culotta VC, Price DL, Wong PC, Rothstein JD. Histological evidence of protein aggregation in mutant SOD1 transgenic mice and in amyotrophic lateral sclerosis neural tissues. Neurobiol. Dis. 2001;8:933–941. doi: 10.1006/nbdi.2001.0443. [DOI] [PubMed] [Google Scholar]
- Wong PC, Pardo CA, Borchelt DR, Lee MK, Copeland NG, Jenkins NA, Sisodia SS, Cleveland DW, Price DL. An adverse property of a familial ALS-linked SOD1 mutation causes motor neuron disease characterized by vacuolar degeneration of mitochondria. Neuron. 1995;14:1105–1116. doi: 10.1016/0896-6273(95)90259-7. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.