I. INTRODUCTION
Several bunyaviruses including Rift Valley fever virus (RVFV), Crimean-Congo hemorrhagic fever and Hantaan viruses are responsible for potentially lethal hemorrhagic fevers and therefore are of significant medical and public health importance. Unfortunately, there are no FDA-approved vaccines available for any of these viruses. The real threat posed by RVFV, coupled with the fact that there currently is no effective licensed vaccine for human use, clearly illustrates the need for more RVFV vaccine research and development. A better understanding of bunyavirus-associated pathogenesis remains critical to the development of effective vaccines and antiviral compounds to combat hemorrhagic fevers. Many vaccine candidates and therapeutics developed through traditional methods either fail to provide protection or result in unacceptable adverse effects. This problem clearly requires the application of new technologies, such as reverse genetics systems, for the design and generation of safe and efficacious vaccine candidates and therapeutics based on the use of genetically manipulated viruses.
II. BUNYAVIRUSES
Viruses within the Bunyaviridae family are classified into five genera: Orthobunyavirus, Nairovirus, Phlebovirus, Hantavirus and Tospovirus (Nichol et al., 2005; Schmaljohn & Hooper, 2001; Shimshony, 1999). Bunyaviruses are characterized by a tripartite negative stranded RNA genome comprised of Large (L), Medium (M), and Small (S) segments (Schmaljohn & Hooper, 2001). The L segment encodes for the viral RNA-dependent RNA polymerase - RdRp (L), the M segment for a protein precursor which is post-translationally processed into the mature glycoproteins GN and GC and the S segment for the nucleoprotein (N). Some bunyaviruses express additional nonstructural proteins from the S and M segments, NSs and NSm, respectively (see Fig. 1A). General features of viruses classified in the Bunyaviridae family are similar to other segmented negative-stranded RNA virus families (Elliott, 1996).
Fig. 1. RVFV genome organisation.
A: Schematic representation of the three genomic segments and coding strategy. The arrows indicate the open reading frames in each segment with the cleavage sites generating the mature glycoproteins.
B: RVFV M segment based mRNA with the five in frame AUG start codons at its 5′ terminus. The proteins expressed from the first and the second AUG are displayed.
Following envelope glycoprotein-mediated virus binding to a receptor on a permissive cell, viral and plasma membranes fuse and viral RNA is transferred into the cytoplasm. The immediate early primary transcription step then begins, allowing the viral genomic RNAs (vRNA), in association with N, to be transcribed into mRNAs by the RNP-associated viral RNA polymerase L. (Bellocq & Kolakofsky, 1987; Ikegami et al., 2005b; Kolakofsky et al., 1987; Vialat et al., 2000). Genome replication takes place in the cytoplasm via a complementary full length template RNA (cRNA), also called antigenomic RNA. The vRNAs generated from this replication step provide the template for additional mRNA synthesis (secondary transcription step) and serve as genome segments for progeny virions which are released by budding into Golgi vesicles (Elliott, 1996; Gerrard & Nichol, 2002; 2007). Maturation and budding of particles through the Golgi apparatus is a property of bunyaviruses. mRNA synthesis is initiated using capped oligonucleotides, captured from cellular mRNAs through a cap-snatching mechanism, similar to the one described for orthomyxoviruses except that all steps take place in the cytoplasm. In contrast with most mRNAs, bunyavirus mRNAs are not polyadenylated and represent an incomplete copy of the vRNA template.
III. RIFT VALLEY FEVER VIRUS
RVFV is an arthropod-borne member of the Phlebovirus genus that causes recurrent outbreaks affecting humans and ruminants predominantly in Subsaharan Africa, but spread to Egypt in 1977 and to the Arabian Peninsula, including Saudi Arabia and Yemen in 2000 (Al-Hazmi et al., 2003; Anonymous, 2000; Balkhy & Memish, 2003; Madani et al., 2003; Shoemaker et al., 2002), and re-emerged in four Governorates in Egypt in 2003 (Balkhy & Memish, 2003).
The largest RVFV epidemic-epizootic outbreak affected Egypt along the Nile River (1977–1979), afflicting approximately 200,000 persons with 594 deaths (Meegan, 1979; Meegan et al., 1979), followed by recurrent epidemics in 1993 and 1997 (Abd el-Rahim et al., 1999; Abu-Elyazeed et al., 1996; Arthur et al., 1993). Other severe outbreaks in the past twenty years have affected West African nations in Senegal-Mauritania, (1987) where an outbreak affected an estimated 89,000 victims, resulting in 220 deaths (Digoutte & Peters, 1989). An outbreak in East Africa occured from 1997–1998 in the Garissa District of Northeastern Kenya, affecting an estimated 27,500 individuals with 170 hemorrhagic fever-associated deaths (Woods et al., 2002). RVFV has spread to Madacascar which experienced its own RVFV epidemics in 1990–91 (Morvan et al., 1992; Morvan et al., 1991) and 2008, resulting in only a few deaths in 1990–1991 but 476 cases and 19 deaths in 2008 (Reynes, 2009). Finally, as described above, RVFV spread to the Arabian peninsula in 2000 (Saudi Arabia and Yemen), where an estimated 2,000 people were hospitalized or severely sickened with RVF-related symptoms, including acute hepatic necrosis, hepatitis, delayed-onset encephalitis and retinitis, resulting in at least 245 deaths (CFR 12%) (Al-Hazmi et al., 2003; Alrajhi et al., 2004; Madani et al., 2003). During the past two years (2007–2008) RVFV has circulated in East Africa causing serious epidemics in Kenya, Tanzania, Somalia and Sudan in 2007, and subsequently expanded to Madagascar and South Africa in 2008. RVF activity was also reported in the Comoros Islands (LaBeaud et al., 2008; WHO, 2007).
Simple immunoglobulin (Ig) G testing through ELISA has shown that in endemic areas such as Kenya, RVFV seroprevalence has soared to 32% in high-risk regions (Woods et al., 2002). During interepidemic periods, RVFV seroprevalence rates can vary between 6 and 20% in the urban and rural settings, respectively, with an overall RVFV seropositivity rate of 13% (LaBeaud et al., 2008; LaBeaud et al., 2007).
Disease
In ruminants, RVF is characterized by substantial die-offs of young animals (especially of lambs), fetal deformities and abortion (Flick & Bouloy, 2005; Gerdes, 2004; Swanepoel & Coetzer, 2003). In humans the disease is often associated with benign fever but can lead to more complicated cases such as retinal vasculitis, encephalitis, neurologic deficits, hepatic necrosis, or fatal hemorrhagic fever (Meegan, 1979).
Although the lethality for humans infected with RVFV has been reported to be below 2%, the case fatality rate in recent outbreaks, 23–45%, is substantially higher (LaBeaud et al., 2008; WHO, 2007). Although not addressed by these reports, it is likely that morbidity is also substantially increased. It has yet to be determined if a new, more virulent strain is causing this sudden increase of fatal human cases.
Transmission
RVFV is readily transmitted through a broad range of mosquito genera including Aedes, Anopheles, Culex, Eretmapoites and Mansonia, and by other vectors including sand flies (Fontenille et al., 1998). Thus transmission could rapidly occur in many Western countries, including the United States (US) (Turell et al., 2008) or Europe (Moutailler et al., 2008). Recent studies have illustrated the ability of RVFV to utilize the dominant mosquito species of a given geographical location (Moutailler et al., 2008; Turell & Kay, 1998; Turell & Perkins, 1990), which indicates that there is no natural blockade to protect naive countries from the spread of the virus. By analogy, West Nile virus (WNV) was not considered as a threat to the US prior to its emergence on the US mainland (New York) in 1999 (Hayes, 2001). However, within six years, WNV had become endemic across the US. Interestingly, while the WNV transmission route is limited to two mosquito genera (Culex and Aedes; (Sardelis et al., 2001)) and has a limited effective host range, RVFV can be transmitted by at least five mosquito species and has a large host range. RVFV has been shown to infect most livestock species (with young animals most susceptible) and reach sufficient titers to allow transmission from animal to insect vector (Turell et al., 2008), insect vector to animal, animal-to-animal and animal-to-human (Easterday, 1965).
RVFV is only able to cause epizootics under the appropriate climatological conditions (e.g., wet seasons and resulting flooding required for Aedes mosquito breeding). While RVFV can appear to be absent in an arid region, the virus is capable of lying dormant in the eggs of infected mosquitoes.
It has been suggested that between periods of epizootics, RVFV utilizes wild rats as a reservoir host, replicating to titers that are sufficient for transmission while at subclinical levels (Pretorius et al., 1997). However, this is still a matter of debate and needs confirmation from the field. The spread of RVFV is not limited to vector-borne transmissions, as RVFV is readily passed via bodily fluids (Balkhy & Memish, 2003; Lutwick & Nierengarten, 2002) http://www.cdc.gov/ncidod/dvrd/spb/mnpages/dispages/rvf.htm, and by aerosol transmission (Hoogstraal et al., 1979; Lutwick & Nierengarten, 2002; Meadors et al., 1986). RVFV can be transmitted amongst slaughter-house workers, animal handlers, and has been recently reported to have been inadvertently transmitted amongst veterinarians, veterinary students and laboratory staff at a South African veterinary college after performing necropsies on infected animals.
The recent expansion of RVFV’s geographical range clearly indicates that RVFV is not solely an African problem, but rather is a geographically rapidly expanding disease that is a threat to other countries. It has been proposed that a single infected person or animal (live or dead) that enters a naive country is sufficient for the initiation of a major outbreak before RVFV would ever be detected. This transmission scenario is becoming increasingly more probable due to the expansion of worldwide trade and travel.
Bioterrorism threat
RVFV is a prototype of emerging/re-emerging pathogens and is classified as a BSL-3 agent in Europe and BSL-3Ag agent in the US. RVFV was weaponized by the US offensive biological warfare program prior to its termination in 1969 (Anonymous, 2009), illustrating that the real threat and utility of RVFV as a biological weapon is clearly recognized (Jones et al., 2002). The lack of prophylactic and therapeutic measures, the transmission by many species of mosquitoes and the significant threat to livestock associated with RVFV make this pathogen a serious public health concern not only for endemic, developing countries, but also for many non-endemic developed countries due to the possibility of bioterrorist attacks, based on a report released on Dec. 4th, 2008 by the Commission on the Prevention of Weapons of Mass Destruction and Terrorism (Graham, 2008). Considered a potential bioterrorism agent (Sidwell & Smee, 2003), RVFV is on the Center for Disease Control (CDC) Bioterror Agent list (http://www.bt.cdc.gov/agent/agentlist-category.asp#a) and is classified as a Department of Health and Human Services (HHS), United States Department of Agriculture (USDA) overlap Select Agent (USDA, 2005). Rift Valley Fever Virus is also classified as a Category A High Priority Pathogen by the National Institute for Allergy and Infectious Diseases (NIAID) (http://www3.niaid.nih.gov/topics/BiodefenseRelated/Biodefense/research/CatA.htm). High-priority agents pose a risk to national security because they can be easily disseminated and can cause high mortality and morbidity, potentially resulting in a major impact on public health. Ultimately, this high priority pathogen could lead to widespread public panic and social disruption.
IV. RVFV GENOME ORGANIZATION AND CODING STRATEGY
Over the past two decades, the ability to genetically manipulate viral genomes has facilitated both our understanding of viral pathogenesis and studies of the interactions between viral and cellular components. The establishment of a reverse genetics system opened the door for many studies into the molecular biology of RVFV and mechanisms involved in pathogenicity that were unthinkable some years ago. The important role of non-structural proteins in viral pathogenesis has been gleaned from such investigations. The RVFV genome encodes three nonstructural proteins, NSm1 (78k) and NSm2 (14K) expressed from the M segment-derived polyprotein precursor, and NSs expressed from the S segment (Elliott, 1996) (see Fig. 1A). The alternate utilization of the first two in-frame AUGs of the M segment (five different AUG start codons exist prior to the signal peptidase cleavage site of the mature GN protein) generates NSm1 and NSm2, respectively (see Fig. 1B). Impaired cleavage of the GN signal peptide insures that NSm1 contains the pre-glycoprotein sequence followed by GN. NSs is encoded in positive sense orientation on the ambi-sense RVFV S segment, separated from the N-ORF by an intergenic region. Both NSm and NSs nonstructural genes have been found to be dispensible in the replication cycle, but have been shown to exhibit properties of virulence factors.
V. REVERSE GENETICS SYSTEMS
Reverse genetics technology makes it possible to manipulate viral RNA molecules using complementary DNA (cDNA) copies so one can study the effects of genetic changes on the biology of the virus. This technology was applied to the modification of plus-stranded RNA virus genomes and to the recovery (rescue) of infectious virus from cDNA because these viruses are able to utilize the host cell DNA replication machinery to initiate their life cycles. Thus, plasmid-encoded or in vitro synthesized genomic RNA of these viruses is infectious when introduced into permissive cells. On the other hand, recovery of negative-strand RNA viruses from either cDNA components or synthetic RNA was a substantial challenge because, unlike the plus-stranded RNA viruses, replication initiation requires de novo protein synthesis mediated by viral RNA-dependent RNA polymerase, and the input genomic or antigenomic RNA needs to be encapsidated with the viral nucleoprotein before it can serve as a functional template to initiate transcription/replication. The ultimately successful development and application of reverse genetics technology for the manipulation of negative-strand RNA virus genomes has considerably affected the field of RNA virology. Much has been elucidated about the molecular characteristics and pathogenesis of these viruses, and the insights obtained from such studies has provided new impetus for the development of rationally designed vaccines and antiviral agents.
Basic strategies and technologies
The successful establishment of reverse genetics systems for a variety of different negative strand RNA viruses (segmented and non-segmented) demonstrates that different technologies and strategies can be employed for the generation of recombinant viruses. The following briefly summarizes the common critical tools used for such reverse genetics endeavors.
The first step in the generation of recombinant virus using reverse genetics systems is the introduction of required viral components (expression plasmids: template for viral proteins to drive viral replication and transcription processes; transcription plasmids, PCR fragments or in vitro transcribed RNA: templates for the generation of viral genomes) into select mammalian cells. A variety of liposome-based transfection reagents as well as electroporation methodologies for a variety of different mammalian cells have been successfully demonstrated
The most challenging task for the establishment of an infectious clone system is certainly the generation of viral RNA genomes/genome segments, due to the fact that many viruses require precise termini to provide suitable templates for their viral polymerase to perform replication and transcription steps. Both prokaryotic (e.g., phage T7 polymerase) and eukaryotic promoters (e.g., Pol I and Pol II) are successfully employed to generate such full-length viral genome transcripts. Different eukaryotic promoters are described for the successful expression of structural (glycoproteins, nucleocapsid protein, matrix proteins, phosphoproteins, viral polymerase) and nonstructural components of the infectious recombinant virions. Additionally, optimization of viral protein expression levels can be achieved by using translation initiation enhancers (e.g., Kozak sequence) and codon-optimized gene sequences.
Reporter genes are often used during the establishment of a reverse genetics system (e.g., minigenome rescue systems) and are often included in recombinant viruses to allow for ease of detection (e.g., monitoring intracellular or in vivo localization, plaque detection). The type and location of reporter genes for each respective system depends upon the questions to be assessed. For minigenome rescue systems, the viral ORF is replaced by a reporter gene that retains the flanking non-coding regions containing viral promoter, encapsidation and packaging signals and other regulatory cis-acting elements (Freiberg et al., 2008). The use of eGFP is preferred for system optimization, allowing the monitoring of reporter gene expression in live cells. However, other reporter genes including luciferase and chloramphenicol acetyltransferase have also been successfully employed, especially in systems in which highly sensitive detection techniques were required due to suboptimal expression levels. Reporter genes can be also used to generate viral fusion proteins allowing the intracellular tracking of proteins of interest.
One of the first applications of reverse genetics for a negative-strand virus was to influenza virus (Luytjes et al., 1989), followed by the successful rescue of recombinant rabies virus (Schnell et al., 1994). Rescue of other full-length RNA viruses soon followed, and included vesicular stomatitis Indiana virus (Lawson et al., 1995; Whelan et al., 1995), measles virus (Radecke et al., 1995), and many others (Kawaoka, 2005; Neumann & Kawaoka, 2004; Neumann et al., 2002; Walpita & Flick, 2005). A similar methodology was used to recover the bunyavirus Bunyamwera, which was also the first of the segmented negative strand RNA viruses to be recovered entirely from cloned cDNA (Bridgen & Elliott, 1996). The key challenge in these studies was to generate the RNA genome/genome segments in eukaryotic cells while also providing required trans-acting factors including the viral RNA-dependent RNA polymerase, nucleoprotein, and a viral phosphoprotein specifically for non-segmented RNA viruses. The strong bacteriophage T7 promoter was initially used to generate the required components for subsequent assembly of recombinant infectious viruses. While the methodology based on the bacteriophage T7 polymerase has been used mostly for non-segmented negative strand RNA viruses (e.g., filo-, rhabdo-, paramyxoviruses), an alternative technique employing the cellular DNA-dependent RNA polymerase I (Pol I) to transcribe transfected plasmids into viral RNA genome (segments) was initially developed for influenza viruses (for review, see (Neumann et al., 1999; Walpita & Flick, 2005)). The choice of method was mostly dictated by the intracellular location of viral replication: T7-based method was initially used for those viruses that replicate in the cytoplasm (e.g., bunyaviruses), and the Pol I-based methodology was used for those which have a nuclear replication strategy (e.g. orthomyxoviruses).
This paradigm was recently revisited when Flick et al. (2001) demonstrated that a Pol I-driven approach is a viable technique for the successful rescue of Uukuniemi (a RVFV-related phlebovirus) artificial genome segments (minigenomes) expressing reporter genes (Flick & Pettersson, 2001). Over the next several years, Dr. R. Flick’s team demonstrated the ability of the Pol I system to generate functional viral genome segments for reverse genetics studies with other bunyaviruses including Hantaan virus (Flick et al., 2003a) and Crimean-Congo hemorrhagic fever virus (Flick et al., 2003b).
Recently, other studies have shown that both T7- and Pol I-based reverse genetic systems are equally efficient for minigenome rescue of Reston ebolavirus, a filovirus (Groseth et al., 2005) and the recovery of lymphocytic choriomeningitis (LCMV), an arenavirus (Flatz et al., 2006; Sanchez & de la Torre, 2006), both having a cytoplasmic strategy for transcription and replication. Furthermore, the T7 promoter-driven system, in addition to the originally established Pol I-driven rescue system (Fodor et al., 1999; Neumann et al., 1999), can also be successfully applied to the generation of influenza A virus which replicates in the nucleus (de Wit et al., 2007).
Taken together, many different technologies and strategies have been employed to successfully establish reverse genetics systems for the generation of infectious recombinant viruses that can be used for basic research applications and as potential vaccine candidates.
Minigenome rescue systems for bunyaviruses
Rescue of minigenomes, which is a preliminary but recommended step before recovery of infectious virus, has been shown to be efficient with both T7- and Pol I-driven systems for bunyaviruses (Fig. 3). Minigenome transcription driven by T7 or Pol I promoters in combination with T7- or Pol II-driven expression plasmids of the corresponding bunyaviral nucleoprotein N and the polymerase L enable the reconstitution of RNP templates (Blakqori et al., 2003; Bridgen & Elliott, 1996; Dunn et al., 1995; Flick et al., 2003a; Flick & Bouloy, 2005; Flick et al., 2003b; Flick & Pettersson, 2001; Gauliard et al., 2006; Ikegami et al., 2005a; Kohl et al., 2004; Lopez et al., 1995; Walpita & Flick, 2005). Such minigenome rescue systems are used intensively to study viral cis-acting elements (e.g., promoter location and structure, packaging signals, transcription termination signals) and the function of trans-acting factors (polymerase, nucleoprotein, non-structural proteins) (Albarino et al., 2007; Barr et al., 2005; Barr & Wertz, 2005; Flick et al., 2004; Flick & Bouloy, 2005; Gauliard et al., 2006; Haferkamp et al., 2005; Ikegami et al., 2007; Kinsella et al., 2004; Walpita & Flick, 2005). These systems are especially valuable for the effective study of negative strand transcription and replication, as well as the identification of potential antiviral compounds for viruses classified for use in high containment facilities (BSL-3 and BSL-4) since they allow for experiments at a lower biosafety level (Flick & Bouloy, 2005; Flick & Whitehouse, 2005; Freiberg et al., 2008; Haferkamp et al., 2005; Walpita & Flick, 2005).
Fig. 3. Comparison of RVFV minigenome rescue systems.
RVFV M segment-based minigenome plasmids under the control of the cellular RNA polymerase I are transfected into suitable mammalian cells (e.g., HEK293). The viral proteins needed for minigenome transcription and replication are provided by either superinfection with RVFV helpervirus (upper panel) or co-transfection of RVFV L and N expression plasmids (lower panel). Cells are harvested and reporter gene expression levels analyzed with suitable detection tools (e.g., CAT assays).
As a proof of concept, Uukuniemi virus (closely related to RVFV) and Nipah virus minigenome rescue systems were optimized to a 96-well high-throughput format in which potential antivirals are identified by their ability to interfere with viral replication and transcription in a time-efficient and cost-effective manner (Flick, 2009; Freiberg et al., 2008). Such high-throughput screening assays involve transfection of cells with minigenome replicons encoding a recombinant gene marker such as chloramphenical acetyl transferase (CAT), luciferase or GFP (Freiberg et al., 2008). Minigenome rescue systems enable the quantification of viral gene activity such that the level of reporter gene activity directly reflects the extent of viral transcription/replication activities (Freiberg et al., 2008). An ELISA-based format allows for a simple readout of differences in reporter gene activity. After distinguishing confirmatory hits from false positives, antiviral candidate compounds that show inhibitory effects through reduced reporter gene activities are considered as potential antiviral compounds that are then further descriminated for their antiviral effects. Both Pol I- and T7-driven minigenome rescue systems can be used to reduce false positive signals by eliminating antiviral activities against Pol I or T7. Finally, antiviral candidate compounds can be further analyzed in such minigenome rescue systems through dose-dependent titration assays to determine effective dose range. Future improvements will involve the use of stable expressing cell lines that constitutively express viral transcription/replication machinery and a minigenome of choice to eliminate the costly and cumbersome transfection step.
Another example in which minigenome rescue systems were used to identify antiviral compounds against the RVFV-related phlebovirus Uukuniemi virus was recently reported by Flick et al. (unpublished data, see below). Uukuniemi virus-specifc antisense peptide-conjugated phosphorodiamidate morpholino oligomers (PPMOs) were used to identify the mechanism and optimal genome target for efficient antiviral activity. A clear dose-dependent reduction in minigenome expression levels and virus titer was observed using Uukuniemi-specific PPMOs (Dr. R. Flick, unpublished data). PPMOs have been shown to provide resistance in vivo and vitro to several viruses including West Nile virus (Deas et al., 2007; Zhang et al., 2008), influenza A, dengue 2, and Zaire ebolavirus (Gabriel et al., 2008; Lupfer et al., 2008; Stein, 2008; Stein et al., 2008).
Infectious clone systems for bunyaviruses
Among the bunyaviruses, Bunyamwera virus was the first to be recovered from plasmids without the need of a helper virus (Bridgen & Elliott, 1996; Bridgen et al., 2001; Lowen et al., 2004), followed by the recovery of infectious LaCrosse and RVF viruses based on the T7 polymerase methodology (Bird et al., 2008; Bird et al., 2007a; Blakqori & Weber, 2005; Bridgen & Elliott, 1996; Bridgen et al., 2001; Gerrard et al., 2007; Ikegami et al., 2006; Lowen et al., 2004). More recently, Pol I-mediated transcription of viral cRNA was applied to the rescue of Akabane orthobunyavirus (Ogawa et al., 2007) and RVFV (Billecocq et al., 2008; Habjan et al., 2008b), indicating that the Pol I-based rescue system could be applied to bunyaviruses utilizing an ambisense coding strategy (Fig. 4). These two latter studies compared the efficiency of the T7- and the Pol I/Pol II-driven systems and concluded that both were similar. It should be noted that the Pol I system requires co-transfection of the bunyaviral N and L expression plasmids, whereas co-transfections are either not necessary or even inhibitory in the T7 system (Bird et al., 2008; Ikegami et al., 2006). Interestingly, NSs co-expression with the L and N proteins inhibits RVFV minigenome rescue and/or the generation of recombinant RVFV (N. Gauliard, M. Bouloy, unpublished results), which is contradictory to the report by Ikegami (Ikegami et al., 2005a) which claims that NSs promotes minigenome viral RNA replication and transcription. However, Ikegami et al. also showed that NSs expression suppresses infectious RVFV rescue in a T7-driven reverse genetics system (Ikegami et al., 2006).
Fig. 4. RVFV infectious clone system.
Plasmids containing the sequence of the three RVFV genome segments (L, M and S) under the control of the cellular RNA polymerase I are transfected into suitable mammalian cells (e.g., HEK293). Bunyaviral genome segments can be transcribed into genomic vRNA (as shown) or antigenomic cRNA, depending on the orientation within the plasmid. The viral proteins needed for RNA genome transcription and replication are provided by co-transfection of RVFV L and N expression plasmids. Supernatant is harvested and recombinant infectious RVFV can be used for subsequent applications (e.g., vaccine candidate).
A disadvantage of T7- compared to Pol I-driven systems is that the T7 polymerase has to be provided in trans, using either eukaryotic expression plasmids, a recombinant vaccinia virus (MVAGKT7, (Kovacs et al., 2003), or T7 polymerase stable-expressing cell lines (BHK21-T7 or BSR-T7/5) constitutively expressing the T7 RNA polymerase (Conzelmann et al., 1991; Kato et al., 2004) to drive the system, whereas Pol I does not have to be added in trans. Although the 5′ triphosphate termini of T7 transcripts strongly activate the antiviral interferon response via the intracellular receptor RIG-I, T7 polymerase stable-expressing cell lines are suitable for the rescue of viruses of several families because they have a compromised RIG-I pathway (Habjan et al., 2008a).
VI. RVF PATHOGENESIS
The role of the NSs protein as an interferon antagonist
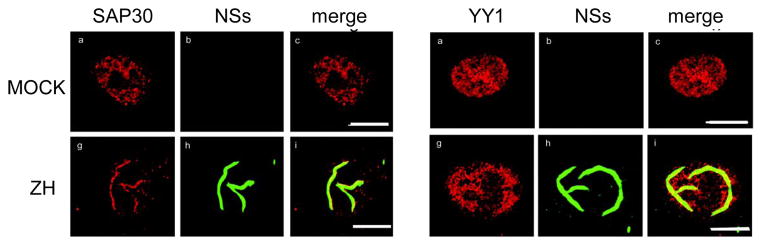
NSs has been identified as a major virulence factor, primarily by acting as an interferon antagonist (Billecocq et al., 2004; Bouloy et al., 2001). Using the established reverse genetic system has been very instrumental to better understand RVFV pathogeneisis. In an attempt to decipher the molecular mechanisms underlying this property, Dr. M. Bouloy’s laboratory identified several cellular proteins that interact with NSs. One of them, SAP30, belongs to the Sin3A/NCoR/HDAC repressor complexes that intervene in the regulation of gene transcription. Moreover, it was shown that SAP30 interacts directly with YY1, a transcription factor involved in regulating the expression of numerous genes, including IFN-β. Through different techniques such as co-immunoprecipitation, confocal microscopy and chromatin immunoprecipitations, Dr. M. Bouloy’s group demonstrated that NSs, SAP30, YY1 and Sin3A-associated corepressor factors are recruited on the IFN-β promoter, inhibiting CBP recruitment, histone acetylation and transcriptional activation (Le May et al., 2008) (see Fig. 2). To ascertain the role of NSs interaction with SAP30 in this mechanism, Le May et al. used reverse genetics to produce a recombinant ZH548 in which the domain of NSs that interacts with SAP30 was deleted from the S segment. In contrast to the virulent ZH548 RVFV, this mutant, ZH548-NSsΔ210–230, is able to induce IFN-β expression but is avirulent in the mouse model.
Fig. 2.
Confocal microscopy of uninfected (MOCK) or infected L929 cells (ZH: with RVFV ZH548). Cells were fixed at 18 h p.i. and stained with antibodies against SAP30, NSs or YY1 and NSs.
During the last few years NSs has been shown to be a multifunctional protein, enabling RVFV to employ several strategies to evade the host antiviral response. One of these relies on its interaction with the p44 subunit of the TFIIH basal transcription factor which is sequestered by the NSs filamentous structure (see Fig. 2), characteristic of RVFV infection (Le May et al., 2004). As a consequence, TFIIH cannot assemble and its concentration drops rapidly, which explains the drastically reduced transcriptional activity of cells infected with NSs-expressing RVFV. Interestingly, the function of NSs as a general inhibitor of transcription (dependent on both RNA polymerase I (Pol I) and II (Pol II) since TFIIH is involved in these two activities) is a relatively late event occurring > 8h p.i., whereas the specific inhibition of IFN-β gene transcription is in place as early as 3 to 4h p.i.
Very recently, a novel function of NSs was described simultaneously by the groups of Drs. S. Makino (Ikegami et al., 2009) and Dr. F. Weber (Habjan et al., 2009b): NSs promotes post transcriptional downregulation of protein kinase PKR and inhibits eIF2–α phosphorylation. This phenomenon occurs through the proteasome-dependent degradation of PKR in cells infected with NSs-expressing RVFV, whereas PKR is clearly detected and activated after infection with the NSs-deleted Clone 13. It was also observed that this activity is specific to RVFV NSs but not shared with other phleboviruses including Sandfly fever Sicilian virus or the orthobunyavirus LaCrosse, since a recombinant RVFV ZH548 in which the NSs sequence was replaced by the heterologous NSs from the phlebovirus or the orthobunyavirus, results in a virus lacking this ability.
Altogether, it appears that NSs has multiple functions to counteract the action of interferon, either at the transcriptional level or at the translational level, by degrading PKR which therefore facilitates translation of the viral products.
The role of the NSm protein as a suppressor of virus-induced apoptosis
To determine the biological function of NSm1 and NSm2, an M segment deletion mutant arMP12-del 21–384 was produced in which the first three initiating AUGs were deleted, resulting in a virus that is unable to synthesize the two nonstructural NSm proteins. Although its growth was similar to that of the parental virus arMP12, arMP12-del 21–384 induced extensive cell death and produced larger plaques than the parent (Won et al., 2007). Further analyses indicated that the deletion mutant triggered apoptosis through the caspase 3, 8 and 9 pathways, thus revealing that NSm has an anti-apoptotic function and contributes to pathogenesis. Interestingly, a study by Pollitt and colleagues with Maguari virus showed that the NSm protein of this orthobunyavirus, which has no sequence homology with the NSm protein of phleboviruses (Nakitare & Elliott, 1993), could be deleted without affecting virus viability (Pollitt et al., 2006). It is not yet known if the NSm proteins from orthobunyaviruses and phleboviruses share the same anti-apoptotic function. However, if so, the same evolutionary pathway could have occurred to conserve the functions of both NSm and NSs among bunyavirus genera.
The function of the RVFV NSs and NSm proteins could not have been elucidated without the indispensable tools and methods of reverse genetics technology that allow the generation of recombinant viruses.
VII. HISTORIC AND CURRENTLY AVAILABLE RVF VACCINES
As discussed above, aside from being endemic to many countries, RVFV is a rapidly expanding disease that may quickly become a worldwide threat. Unfortunately, the development of safe and efficacious RVFV vaccines has proven to be quite difficult (see Table 1). The urgent need for an effective vaccine for RVFV is illustrated by the statistics from recent outbreaks: From 13th Jan to 8th May 2007, a total of 290 cases including 117 deaths [case fatality ratio (CFR) 40%] of RVF were reported in Tanzania; from 30th Nov 2006 to 12th Mar 2007, a total of 684 cases including 155 deaths (CFR 23%) of RVF were reported in Kenya. From 19th Dec 2006 to 20th Feb 2007, a total of 114 cases including 51 deaths (CFR 45%) of RVF were reported in Somalia [World Health Organization (WHO), CSR, Disease Outbreak News, Wed 9 May 2007]. Moreover, the observed increase in RVFV-associated case fatality ratio, between 23–45% in naive populations, further illustrates the continuing and emerging threat RVFV presents. Taken together, the significant increase in RVFV case fatality ratios and a broadening umbrella of epidemics beyond the traditional sub-Saharan African geographical origins underlies the impending urgency for outbreak containment.
Table 1.
RVFV vaccine candidates
| Vaccine Platform | Species | Vaccine Dose | Vaccination Route | Challenge Dose/Virus | Challenge Route | Protection Levels | PRNT80 | Reference |
|---|---|---|---|---|---|---|---|---|
| Recombinant baculovirus/Sf9 cell lysate | Mice | 1×106 disrupted cells (2–10μgRVFV protein) | SC | 1×103 LD50/ZH501 | IP | 100% | PRNT50: 1:40–640 | (Schmaljohn et al., 1989) |
| TSI GSD-200 | Human | Incremental doses (0.1 –1 ml), 3 doses | sc | N/A | N/A | N/A | Low dose (0.1 ml): 1:48 High dose (1 ml): 1:436 | (Krak et al., 1982) |
| Human | Incremental doses (0.1–1 ml) | SC | N/A | N/A | Only localized erythema at the site of injection. | 1:1280(1 month) >1:40 (12 months) |
(Meadors et al., 1986) | |
| Human | 0.5 ml, 3 doses, 6 month booster | sc | N/A | N/A | 90.3% show titer 9.7% show no titer |
Responders: ≥ 1:40 Nonresponders: <1:40 |
(Pittman et al.,1999) | |
| MP-12 | Pregnant ewes Lambs |
1×105 pfu ZH548 1×105 pfu MP12 |
sc | N/A | SC | Abortion rate: ZH548: 100% MP12: 0% |
Ewes: 1:80 to 1:320 Lambs (post parturition): ≥1:80 |
(Morrill et al., 1987) |
| Ewes Lambs |
Ewes: 5×103pfu Lambs:5×103 pfu or 5×105 pfu |
Ewes: SC Lambs: SC |
1.6×105pfu/ZH501 5×105pfu/ZH501 | SC | Abortion rate (ewes): Unvaccinated: 100% Vaccinated: 0% Lambs: 100%survival |
Ewes:1:320 to 1:10,240 Lambs: 1:160 to 1:2,560 |
(Morrill et al., 1991) | |
| Lactating cows Dams Yearling stears |
Lactating cows: 1×105 pfu Dams: 1×105 pfu Yearling stears: 1,2,3 and 4 log 10 pfu. |
SC | 1×1057pfu/ZH501 | sc | Abortion rate (dams): Unvaccinated: 100% Vaccinated: 0% 26% of vaccinated dams developed transient viremia |
Lactating cows: 1:80 to 2,560 Dams’ serum: 1:80 to 1:2,560 colostral: 1:320 to 10,240 Steers: 1:160 to 1:5,120 |
(Morrill et al., 1997a) | |
| Dams Bovine fetus | 1×105 pfu | in utero, IM | 1×1057pfu/ZH501 | sc | 100% survival (vaccine safety) |
In utero vaccinated calves: ≥1:20 to 1:40 Dams’ serum: >1:80 colostrum:>1:320 Calves (post parturition): 1:80 to 1:10,240 |
(Morrill et al., 1997b) | |
| Pregnant ewes | 1 × 106pfu | SC | N/A | N/A | Teratogenic effects | N/A | (Hunter et al, 2002) | |
| Rhesus Macaques | 1×107 pfu | IM, IT, IS | N/A | N/A | N/A | ≥1:640 | (Morrill & Peters, 2003) | |
| Human | 1×105 pfu | IM | N/A | N/A | No viremic side effects | > 1:100 | (Bettingeret al.,2009) | |
| Clone 13 | Mice | 1×10−2 to 1×107 pfu | IP | 1×104LD50/ZH501 | IP | 100% | 1:1,613–2,280 | (Muller et al., 1995) |
| Hamsters | by mosquitoes | 1×104LD50/ZH501 | IP | 100% | N/A | (Muller et al, 1995) | ||
| R566 | Pregnant ewes | 1 × 102 to 1 ×105 pfu | IM | 1×106MICLD50/Buffalo 1998 | IM | Abortion rate: Unvaccinated: 100% Vaccinated: 12.5% |
N/A | (Hunter & Bouloy, 2001) |
| Recombinant lumpy skin disease virus | Mice (six weeks) | 1st vacc.: 1×107 pfu 2nd vacc.: 1×107 pfu |
SC/SC and IM | 100MLD50/AR20368 | IM | 100% | 1:12 | (Wallace et al., 2006) |
| DNA vaccine | Mice (4 to 6 weeks) | 5μg | Gene Gun | 100LD50/ZH501 | IP | Unvaccinated: 10% Vaccinated: 100% |
PRNT50 1:160–1,1280 | (Spik et al.,2006) |
| Mice (6 to 8 weeks) | 50 μg | Gene Gun | 2.4×103pfu or2.4 × 104pfu/ZH548 | IP | Vaccinated (N): 50% asymplomalic Vaccinated (GN/GC): 62.5% asymptomatic | N:<1:25 GN/GC: 1:25–75 | (Lagerqvist et al, 2009) | |
| Recombinant RVFV ΔNSs and/or ΔNSm) | Rats | 1 × 103 pfu 1×104pfu |
SC | 1×103pfu/ZH501 | SC | Unvaccinated: 40% Vaccinated: 100% |
PRNT50: 1×103: 1:640–1,120 1×104: 1:640–7,040 |
(Bird et al., 2008) |
| Sindbis virus replicon | Mice (6 weeks) | 1stvacc.: 1×105 IU 2ndvacc.:1×105 IU |
footpad | 100 MLD50/VRL688/78 | IP or IN | Vaccinated: 100% | 1:8–1:128 | (Heise et at, 2009) |
| Sheep | 1stvacc.:2.5×105 IU 2ndvacc.:2.5×105IU |
SC | N/A | N/A | Neutralizing antibodies | 1:32–1:128 | (Heise et at, 2009) | |
| Virus like particles (VLP) | Mice | 1×105 or 1×106 | IP | 2.4 × 104 pfu/ZH 548 | IP | Unvaccinated: 8% Vaccinated: 1×105: 50%; 1×106: 92% |
1×105:<1:50 1×106: 1:250–1,250 | (Naslund et al., 2009) |
| Mice | 3×6μg (total protein) | SC | 1×103 | IP | 70% | >1:640 | (Mandell et al., 2009) | |
| Rats | 60μg (total protein) | SC | 1×105 | SC | 100% | N/A | (Mandell et al., 2009) |
SC: subcutaneous, IM: intramuscular, IP: intraperitoneal, IN: intranasal, IT: intrathalamic, IS: intraspinal, IV: intravenous, PFU: plaque forming units, PRNT50/80:Plaque reduction neutralization titer by 50% or 80%, ZH501: virulent Zagazig Hospital RVFV strain 501, ZH548: Zagazig strain 548, NA: not applicable, IU: international units, LD50: 50% lethal dose, MICLD50: 50% mouse intracerebral lethal dose, MLD50: 50% mouse lethal dose.
RVFV vaccines for livestock
Smithburn strain
The Smithburn RVFV strain utilized for vaccine development is a derivative of the Entebbe strain isolated in 1944 from a mosquito in Uganda which was mouse neuroadapted by 82 intracerebral passages in suckling mice (Smithburn et al., 1948). This Smithburn modified live virus (SMLV) was further passaged in mice and embryonated eggs before being utilized as a vaccine. Although extensively used for the immunization of livestock, the Smithburn vaccine is only partially attenuated and leads to high abortion rates or teratology in a significant proportion (8.8 to 28%) of pregnant animals (Botros et al., 2006; Coetzer & Barnard, 1977; Swanepoel R, 2004b). Importantly, the concern that SMLV could revert to full virulence precludes its use in countries where RVFV is not known to be endemic (Botros et al., 2006; Swanepoel R, 2004b).
Formalin-inactivated vaccine
Formalin-inactivated derivatives of the Smithburn vaccines, available commercially from Onderstepoort Biological Products LTD (South Africa) as a veterinary vaccine, have been extensively tested in livestock (Barnard, 1979; Barnard & Botha, 1977; Swanepoel R, 2004a). These formalin-inactivated vaccines are more expensive to produce and require multiple inoculations and regular boosters to induce and maintain effective immunity (Barnard, 1979; Swanepoel R, 2004a). Consequently, it is less useful in livestock than live-attenuated viruses, particularly when there is a need to attain rapid immunization in the face of an outbreak.
RVFV vaccines for human use
TSI-GSD-200
The only RVFV vaccine presently available for use in humans is the formalin-inactivated vaccine TSI-GSD-200 (Pittman et al., 1999). This vaccine is derived from RVF virus vaccine (NDBR-103), originally developed by Randall et al. (1962) in African green monkey kidney cultures (Randall et al., 1964; Randall et al., 1962). While NDBR-103 has a stable phenotype and was used to protect laboratory personnel for over two decades, it is plagued with substantial inter-lot variation (Coetzer & Barnard, 1977). NDBR-103 was therefore further developed and standardized by the Salk Institute (1978–79) under contract with the United States Army Medical Research Institute for Infectious Diseases (USAMRIID) (Pittman et al., 1999). This technology was improved and a more modern inactivated RVFV vaccine, TSI-GSD-200, is now produced in diploid fetal rhesus lung cells and is available for veterinarians working in endemic areas, high containment laboratory workers and others at high risk for contracting RVFV (Pittman et al., 1999). Unfortunately, this vaccine is (i) expensive, (ii) difficult to produce, (iii) in short supply, (iv) requires larger dose relative to an attenuated vaccine and three initial inoculations followed by a 6-month booster, (v) and requires continued annual boosters (Frank-Peterside, 2000; Kark et al., 1982; 1985; Niklasson et al., 1985; Pittman et al., 1999).
MP12
Derived by mutagenesis (5-flourouracil) of a RVFV strain (ZH548) that was isolated during the 1977 outbreak in Egypt and grown in MRC5 human diploid fibroblast cells, MP-12 is efficacious in livestock (Caplen et al., 1985; Hunter et al., 2002; Morrill et al., 1991; Morrill et al., 1987; Morrill et al., 1997a; b; Morrill & Peters, 2003). MP-12 neutralizing antibodies are passed to unvaccinated neonates after suckling colostrum, giving rise to detectable titers.
Recently, MP12 was tested in a Phase II clinical trial to determine adverse effects in humans using a single injection dose escalation protocol (Bettinger et al., 2009). MP-12 responders injected with 1×105 pfu were asymptomatic, had a 95% antibody seroconversion rate and developed plaque reduction neutralization test antibody titers (PRNT80) greater than 1:100. Those injected with formalin-inactivated TSI-GSD-200 (as a control) showed 90% seroconversion but PRNT80 titers of only ≤1:28. Genetic analysis of MP-12 isolated from vaccinated individuals showed no reversions of vaccine virus (in attenuated regions) to wild type RVFV.
Clone 13/R566
To counteract the undesireable side effects of live attenuated RVFV vaccine candidates described above (e.g., SMLV strain), two alternative live-attenuated RVFV vaccines are being developed: (i) the natural isolate Clone 13 (Muller et al., 1995) and (ii) the reassortant R566 (Bouloy et al., unpublished data). RVFV Clone 13 is characterized by a large deletion of the NSs gene from the S segment which prevents the virus from hijacking the type 1 IFN pathway (Billecocq et al., 2004; Bouloy et al., 2001). As described above, NSs is an interferon antagonist as well as a general transcription inhibitor (Le May et al., 2004), and its deletion thus allows the vaccinated host to launch an effective immune response (Bouloy et al., 2001). R566 is a reassortant of the L and M segments of MP-12 and the S segment of Clone 13 that combines attenuation markers from both strains.
Clone 13 has been tested in sheep and cattle (Hunter, 2001), and all vaccinated animals elicit immune responses protective against challenge with virulent RVFV. Concerning R566, a vaccination trial was carried out in sheep and pregnant ewes in Senegal using a single dose of R566 ranging from 102 to 106 pfu. None of the vaccinated animals exhibited signs of illness and none of the pregnant ewes aborted. All animals inoculated with 1 × 105 pfu developed antibody responses including the generation of neutralizing antibodies (Y. Thionganne, personal communication). While a challenge experiment was not possible, preliminary experiments in mice show that all animals vaccinated with a dose of 100 pfu are protected against challenge with virulent virus (M. Bouloy et al., unpublished data). Because the deletion of NSs is responsible for attenuation, both Clone 13 and R566 can be considered safe from reversion to a replication-competent, fully infectious virus.
Lumpy skin disease vector
Wallace et al. (2006) expressed RVFV GN and GC in a lumpy skin disease virus (LSDV) and found that mice vaccinated twice with 1×107 PFU recombinant LSDV develop neutralizing antibodies and are fully protected from a 100 LD50 lethal viral challenge (Wallace et al., 2006).
Alphavirus replicon
Alphavirus replicon vectors that express the RVFV glycoproteins have been developed by the US Army Medical Research Institute of Infectious Diseases (USAMRIID) (Heise et al., 2009). These vectors elicit RVFV-specific immune responses and are 100% protective against a 100 LD50 intraperitoneal and intranasal RVFV lethal challenge in a mouse model. Furthermore, this vaccine candidate also elicits a RVFV neutralizing antibody response in vaccinated sheep (Heise et al., 2009). These encouraging data suggest that the use of alphavirus replicon particles (Roy & Noad, 2008)presents a promising vaccine platform for the development of RVFV vaccine candidates.
Baculovirus/Insect Cells
Recombinant baculovirus systems present another promising platform for RVFV vaccine development (Roy & Noad, 2008). Sf9 (Spodoptera frugiperda) insect cells have been used to produce RVFV glycoproteins after infection of recombinant baculoviruses encoding the RVFV M segment ORF (Liu et al., 2008; Noad & Roy, 2003; Schmaljohn et al., 1989). One of the key advantages of this system is the potential for ease of scale-up. Sf9 lysates containing expressed RVFV glycoproteins used for a single subcutaneous inoculation protect mice from lethal (100 LD50) challenge (Schmaljohn et al., 1989). However, 100% protection requires the use of Freund’s Complete Adjuvant in the vaccine formulation and only 50% protection was achieved when Sf9 cell lysates were delivered in PBS alone (Schmaljohn et al., 1989).
Virus-like particles
The use of virus like particles (VLPs) is another promising alternative approach, for the development of a safe and efficient RVFV vaccine. VLPs may be more immunogenic than recombinant proteins as they represent a more natural format that maintains conformational epitopes that can induce neutralizing antibodies (Grgacic, 2006). It has been previously shown that the expression of recombinant GN and GC glycoproteins of a closely related phlebovirus (to RVFV), Uukuniemi, leads to the assembly and budding of VLPs from transfected eukaryotic cells (human embryonic kidney 293 cells) (Overby et al., 2007a; Overby et al., 2006; Overby et al., 2007b). The incorporation of reporter gene-encoding minigenomes into VLPs enables the monitoring of VLP production and also allows the study of packaging signals located on these minigenomes (Flick et al., unpublished data).
Efficient generation of RVF VLPs has been demonstrated by Näslund et al. (2009) and Habjan et al. (2009) in human embryonic kidney 293 cells using transfected DNA encoding the complete RVFV M segment and both the RNA polymerase L and a reporter minireplicon construct (Habjan et al., 2009a; Naslund et al., 2009). The addition of a GFP-expressing minigenome to the VLP producing cells allows minigenome packaging into the budding VLPs and subsequent measurement of VLP titers by quantifying GFP expression in susceptible cell lines expressing the required RVFV transcription machinery (L and N) (Habjan et al., 2009a). Näslund et al. (2009) showed that these RVF VLPs can be used as vaccines. Three i.p. injections of 1×106 RVF VLPs in mice induces antibody titers from 1:300 to 1:900 against GN and GC proteins but does not result in the development of N-specific antibodies. Importantly, these VLPs protect 11 of 12 vaccinated mice from lethal virus challenge (2.4×104 pfu) whereas only 1 of 12 survived in the unvaccinated control group (Naslund et al., 2009).
Recombinant baculovirus system-derived VLPs are an attractive and potentially cost-effective approach for the development of an effective vaccine strategy against RVFV infection because of the potential for high yield production of VLPs in insect cells using the recombinant baculovirus expression system and the ease of scaleup for manufacturing under GMP conditions. Liu et al. (2008) demonstrated the efficient generation of VLPs in insect cells using a single recombinant baculovirus that expresses the RVFV glycoproteins together with the N protein using a dual expression baculovirus vector (Liu et al., 2008). The purified VLPs exhibit enveloped structures that are similar to wild-type RVFV.
Using two separate animal models, Mandell et al. (2009) showed that RVF VLPs generated from 293 cells induce RVFV-specific humoral and cellular immune responses and protect mice from lethal challenge with RVFV (Mandell et al., 2009). Splenocytes from mice injected with RVF VLPs secrete IL-2, IL-4, IL-5 and IFN-γin response to RVFV antigens, consistent with both humoral (TH2) and cellular (TH1) responses. Additionally, vaccination with RVF VLPs elicits RVFV neutralizing antibody titers of >1:640 based on PRNT80 assays even 6 months post vaccination.
Interestingly, Mandell et al. (2009) were able to generate RVF VLPs lacking N, demontrating that the glycoproteins can form VLPs without the need of any other viral proteins (Mandell et al., 2009). This is oposed to findings from others claiming that N is absolutely required for RVF VLP generation (Habjan et al., 2009a).
Differences in survival rates were observed when RVF VLPs containing or lacking the N protein are used in mouse and rat lethal challenge studies. While mice vaccinated with RVF VLPs survived lethal challenge, RVF VLPs containing N provide substantially higher protection rates (~70% vs ~20% without N) (Mandell et al., 2009), consistent with results seen for other vaccine platforms (e.g., DNA, see below) suggesting that RVFV N plays an important role in eliciting a protective immune response. Since rats are often considered more relevant than mice as a model for RVFV disease (Anderson et al., 1991; Bird et al., 2007b) a subsequent rat challenge study was performed. 100% of RVF VLP-vaccinated animals survived a high-dose (105 pfu ZH501) lethal challenge, while all unvaccinated animals died with the first 5 days, post infection (Mandell et al., 2009).
DNA vaccines
DNA-based vaccine platforms have been used to induce immunity against RVFV. Spik et al. (2006) showed that a DNA vaccine encoding the RVFV M segment (with NSm), administered by gene gun technology, displays poor immunogenicity and is only marginally efficacious (<50% survival) in vaccinated mice upon lethal RVFV challenge (Spik et al., 2006). However, DNA vaccines encoding the M segment without the NSm is very immunogenic, and three gene gun-mediated vaccines delivered in three week intervals (5μg total DNA each) leads to 100% survival from a 100 LD50 lethal challenge (Spik et al., 2006).
Three inoculations of sheep with 400μg of a RVFV glycoprotein-expressing DNA vaccine using cationic-liposomes has also shown utility in stimulating RVFV-specific antibodies and boosting proliferating memory cell populations (Lorenzo et al., 2008). The immune response was improved by vaccination with a combination of the above described DNA vaccine with an N-expressing DNA vaccine (Lorenzo et al., 2008).
An alternative DNA vaccine approach was reported by Lagerqvist et al. (2009), who assessed the possible contribution of the RVFV N protein to vaccine immunogenicity. Using a non-lethal challenge of 2.4×103 and 2.4×104 pfu of RVFV ZH548, four of eight and five of eight mice vaccinated with DNA constructs encoding RVFV N or GN/GC, respectively, were aviremic (Lagerqvist et al., 2009). In contrast, all unvaccinated control mice showed clinical manifestations of RVFV. Analysis of neutralizing antibody titers revealed that mice vaccinated with constructs expressing both GN/GC show higher neutralizing levels (PRNT50: 25–75) than those encoding the N protein (PRNT50: <25). These findings suggest that antibody-mediated in vitro neutralization of RVFV might not specifically correlate with vaccine efficacy, and RVFV N might indeed be an important vaccine antigen.
Immune modulation
Since a balanced immune response (humoral and cellular) is required for life long protection (Andrew et al., 1989; Guiso et al., 2007; Petrovsky, 2006; Petrovsky & Aguilar, 2004; Silva et al., 2004; Tangy & Naim, 2005), the use of immune modulators such as the HyperAcute® αGal Technology (Flick et al., 2009; Hellrung et al., 2007; Hellrung et al., 2006; Turell & Bailey, 1987; Unfer et al., 2003) presents an interesting concept for the improvement of RVFV vaccines based on VLPs or other inactivated and/or replication-incompetent platforms. The HyperAcute® αGal Technology exploits the Gal-mediated barrier against pathogens from lower mammals that have incorporated Gal epitopes (Gal α(1–3) Gal-β(1,4) GlcNAc-R) on their cell surfaces (Rossi et al., 2005a; Rossi et al., 2005b; Squinto, 1996). A continual priming of this endogenous antigen is provided by αGal epitopes on bacteria that reside in the intestine. With up to 2% of the entire circulating IgG antibodies targeting the αGal epitope (Galili, 1999) and more than 1% of B lymphocytes producing anti-αGal antibodies (Galili, 1993), vaccine antigens modified with Gal epitopes are naturally primed to elicit a more robust and protective immune response than vaccines displaying the antigen alone (Abdel-Motal et al., 2005; Abdel-Motal et al., 2007; Abdel-Motal et al., 2009; Flick et al., 2009; Galili et al., 1985a; Galili et al., 1985b; Galili et al., 1985c; Galili et al., 2007; Slavin et al., 1985; Thall & Galili, 1990). Such technology could be a major step forward in the improvement of existing vaccines or for the development of efficacious new vaccine candidates (Mandell et al., 2009).
VII. HISTORIC AND CURRENTLY AVAILABLE THERAPIES FOR RVF
While vaccines are the primary defense against viral diseases (Plotkin, 2005), the development of antiviral compounds with therapeutic efficacy against highly pathogenic RNA viruses is also important (Beigel & Bray, 2008; Bray, 2008). Licensed antiviral influenza compounds such as amantadine (Symmetrel, generic) and the neuraminidase inhibitors oseltamivir (Tamiflu, Roche Laboratories, Inc.) and zanamivir (Relenza, GlaxoSmithKline) are available. However, with the exception of ribavirin (Rebetol, Schering Corporation), few compounds are licensed for treatment of hemorrhagic fever viruses (Gowen & Holbrook, 2008). Furthermore, ribavirin has limited utility because of adverse side effects and lack of specificity (Kilgore et al., 1997; McCormick et al., 1986; Monath, 2008). Pyrazinecarboxamides (Toyama Chemical Co., Ltd.) are new antiviral compounds that have been shown to be especially useful for post exposure antiviral therapy as broad spectrum antiviral inhibitors (Furuta et al., 2009) (see Table 2).
Table 2.
Bunyaviral therapeutic candidates
| Therapeutic | Species | Vaccine Dose | Vaccination Route | Challenge Dose/Virus | Challenge Route | Protection Levels | PRNT80 | Reference |
|---|---|---|---|---|---|---|---|---|
| Ribavirin | Rhesus macaques (Adult) | Rhesus: 50 mg/kg, followed by 10 mg/kg at 8 h intervals for 9 days | Rhesus: | Rhesus NHP:1×104.2 pfu/ZH 501 | Rhesus: IV | Rhesus: Unvaccinated: viremia: 100% Vaccinated: 25% |
Rhesus: 1:640 to 1:1280 | (Peters et al., 1986) |
| Hamsters (Syrian golden) | Hamsters: 60 mg/kg twice daily, followed by 20 mg/kg twice daily for 9 days | Hamsters: SC | Hamsters: 10 pfu/ZH 501 | Hamsters: SC | Hamsters: 80% | |||
| C57BL/6N Mice (male, 6 weeks old) | Mice: 75 mg/kg at 8 h post infection and 25 mg/kg twice daily for 10 days | Mice: SC | Mice: 200 pfu/ZH 501 | Mice: SC | Mice: 75% 30% after rechallenge with 5×103 pfu |
|||
| Interferon α | Rhesus macaques | 5 X at 5×105U/kg at 24 h before or 6 h post challenge | IM | 1 × 105 pfu/ZH501 | IV | Neutralizing antibodies | 1:320 –1:5,120 | (Morrill et al., 1989) |
| T-705 pyrazine- carboxamide | Mice | 2 X for 5 days beginning 4 h prior to challenge: 3–30 mg/kg/day | Oral | 500 pfu/PTV | SC |
4 h prior to infection: 30 mg/kg/2X day: 100% 10 mg/kg/2X day: 100% 3 mg/kg/2X day: 10% |
N/A | |
| 2 X for 5 days beginning 24 h after challenge: 10–30 mg/kg/day | 5,000 pfu/PTV |
24 h after infection: 30 mg/kg/2X day: 90% 10 mg/kg/2X day: 30% |
||||||
| 500 pfu/PTV | 30 mg/kg/2X day: 90% 10 mg/kg/2X day: 40% |
(Gowen et al., 2007) | ||||||
| 50 pfu/PTV | 30 mg/kg/2X day: 100% 10 mg/kg/2X day: 0% |
|||||||
| Hamsters | 2 X for 6 days beginning 4 h prior to challenge: 15–60 mg/kg/day | 5 pfu/PTV | 60 mg/kg/2X day: 90% 30 mg/kg/2X day: 90% 15 mg/kg/2X day: 50% |
|||||
SC: subcutaneous, IM: intramuscular, IV: intravenous, PFU: plaque forming units, PRNT50/80:Plaque reduction neutralization titer by 50% or 80%, ZH501: virulent Zagazig Hospital RVFV strain 501, PTV: Punta Toro virus, NA: not applicable.
MxA
Although RVFV is a strong antagonist of IFN production, it is very sensitive to the action of IFN or to IFN inducers (Bouloy et al., 2001). The infection cycle of bunyaviruses is significantly retarded by human inferferon-induced protein MxA by blocking the immediate early primary viral transcription step (Kochs et al., 2002; Reichelt et al., 2004). MxA, a large dynamin-like GTPase, exhibits antiviral capabilities against a wide array of RNA viruses that are beyond the scope of this review (Kochs et al., 2002; Pavlovic et al., 1992; Reichelt et al., 2004).
Ribavirin and polyriboinosinic acid
As a preliminary assessment of antivirals, C.J. Peters et al. (1986) showed that ribavirin and polyriboinosinic acid complexed with poly-L-lysine and carboxymethylcellulose (poly(ICLC)) prevents clinical symptoms of RVFV in mice and hamsters (Peters et al., 1986). Furthermore, RVFV-infected rhesus macaques injected 50 mg/kg of ribavirin followed by daily 10mg/kg injections at 8h intervals over a nine day period showed no RVFV-induced viremia (Peters et al., 1986).
In a more recent study, real time RT-PCR was used to quantify RVFV titers in cell culture in the presence of the antiviral compounds ribavirin, IFN-α, 6-azauridine and glycyrrhizin (Garcia et al., 2001), all of which are commercially available. The maximal tolerated dose of each drug inhibited viral replication by > 4 log RNA copies/ml. The minimal doses that showed antiviral activity were tested with titration of RVFV by TCID50 and RT-PCR. At the lowest tested concentrations, 62.5μg/ml ribavirin, 1IU/ml interferon alpha (IFN-α) and 0.3μg/ml 6-azauridine were still inhibitory, while glycyrrhizin showed no antiviral activity with viral replication at the lowest tested concentration (156μg/μl), with inhibition only at high concentrations (1,250 and 2,500μg/ml). IFN-α and ribavirin showed dose-dependent reduction in viral replication.
Although ribavirin was shown to have prophylactic RVFV activity in vitro, (Peters et al., 1986), it might have unexpected/undesired effects. Ribavirin was used in Saudi Arabia to treat patients suffering from the hemorrhagic icterus form of RVF. Although ribavirin seemed active in treating these hemorrhagic forms, it did not prevent the development of severe meningo-encephalic complications, including hallucinations, lethargy and coma (B Swanepeoel, personnal communication). The ribavirin molecule is too large to pass through the meningeal barrier. New molecules that mimic Ribavirin activity but are small enough to pass the meningeal barrier are in devlopment for RVF treatment (P. Formenty, personal communication).
T-705
Compound T-705, a pyrazinecarboxamide, inhibits RNA-dependent RNA polymerase activity in a dose-dependent manner. It has been suggested that host cell enzymes (cellular kinases) convert T-705 into T-705 ribofuranosyl phosphate (T-705RTP), a form that inhibits virus polymerase without affecting host cellular RNA or DNA synthesis. T-705 protects mice and hamsters against a lethal challenge with Punta Toro virus (PTV) when administered orally 4 hours pre- to 24 hours post-challenge. 10mg/kg administered twice daily over a 5 day post-infection period was sufficient to protect mice, while initiation of T-705 treatment 24 hours post infection requires 30 mg/kg twice daily (Gowen et al., 2007). Hamsters given 30 mg/kg of T-705 orally twice daily beginning 4 hours prior to PTV infection followed by treatment over 6 days showed significant protection (90%). In fact, T-705 was shown to be efficacious in vitro for several viruses in the bunyavirus family including LaCrosse, Punta Toro, Rift Valley fever, sandfly fever Sicilian virus, and in the arenavirus family including Junín, Pichinde, and Tacaribe viruses (Gowen et al., 2007). T-705 was also shown to be less toxic (less of a reduction animals’ body weight and shorter clinical recovery times) compared to ribavirin (Gowen et al., 2008). Antiviral therapeutics such as T-705 may therefore present a more viable choice than ribavirin for combating post exposure treatment of RNA viral infections (Furuta et al., 2009; Julander et al., 2009).
PPMO
Reverse genetics-driven minigenome rescue systems were used recently in Dr. R. Flick’s group to identify antiviral compounds against the RVFV-related phlebovirus Uukuniemi virus (unpublished data). Uukuniemi virus-specifc antisense peptide-conjugated phosphorodiamidate morpholino oligomers (PPMOs) were used to identify the mechanism and optimal genome target for efficient antiviral activity. A clear dose-dependent reduction in minigenome expression levels and virus titer was observed using Uukuniemi-specific PPMOs (unpublished data). PPMOs have been shown to provide resistance in vivo and vitro to several viruses including West Nile virus (Deas et al., 2007; Zhang et al., 2008), Influenza A, Dengue 2, and Zaire ebolavirus (Bergthaler et al., 2006; Buchholz et al., 2000; Flanagan et al., 2001; Gabriel et al., 2008; Lupfer et al., 2008; Stein, 2008; Stein et al., 2008).
VIII. REVERSE GENETICS-DERIVED RVF VACCINE CANDIDATES
Until recently, vaccine trials and basic science investigations have been hampered by the biosafety restrictions surrounding RVFV, but new technology utilizing a minigenome rescue system has facilitated studies of the molecular biology of RVFV (see above)(Flick & Bouloy, 2005; Walpita & Flick, 2005). In addition, established reverse genetics systems (infectious clone systems) have allowed for the generation of attenuated recombinant RVFV vaccine candidates (Billecocq et al., 2008; Gerrard et al., 2007; Habjan et al., 2008b; Ikegami et al., 2006).
RVFV has only one serotype which makes vaccine development relatively straightforward in design. Unlike previous live-attenuated vaccines developed empirically by adapting the virus to a new species (e.g., the attenuated RVFV Smithburn strain; see above), new vaccines are expected to be devoid of toxicity or pathogenic effects. Studies carried out in many laboratories on the immune-evading (e.g., interferon antagonist) role of some viral proteins (mostly nonstructural) have paved the way for the creation of viruses deficient for these proteins using reverse genetics methodologies. In the case of RVFV, it became possible to produce a virus avirulent like Clone 13 but in which NSs is entirely deleted (see Fig. 5) (Billecocq et al., 2004; Bird et al., 2008; Habjan et al., 2009b; Ikegami et al., 2005b).
Fig. 5.
Plaques generated by RVFV rescued from plasmids using reverse genetics. rZH: rescued from wild type ZH548-based plasmids; rZHΔNSs: rescued with a mutated S segment, lacking the NSs coding sequence.
To further investigate the role of RVFV nonstructural protein NSm, recombinant viruses lacking the gene were created by reverse genetics, either in the context of the virulent ZH501 strain or the attenuated MP12 strain (Won et al., 2006). The recombinant ΔNSm ZH501 (rZH501) virus lacks the sequence from the first AUG start codon to the sequence immediately upstream of the fourth AUG, whereas the recombinant MP12 viruses (rMP12) contain mutations in the first and/or the second AUG to generate RVFV lacking the 14k or 78 k NSm protein (or both). These recombinant viruses grow in various cell types, indicating that the NSm proteins are dispensable for the viral replication cycle. Furthermore, studies with highly susceptible Wistar-furth rats which, like mice, are suitable animal models for RVFV vaccine testing in challenge studies, show that ΔNSm-rZH501 virus retains lethality in these animals but is attenuated compared to ZH501 (Anderson et al., 1991; Anderson, 1988; Bird et al., 2007a).
More recently, using an established infectious clone system, Bird et al. (2008) (Bird et al., 2008) employed reverse genetics to generate live attenuated recombinant ZH501 RVFV vaccine candidates by replacing NSs with the reporter gene GFP and by eliminating the non-structural protein NSm (rZH501-ΔNSs:GFP, rZH501-ΔNSs:GFP-ΔNSm). These highly attenuated viruses proved to be asymptomatic and protective, after a single 1×103 or 1×104 pfu vaccination, against viral challenge (1×103 pfu ZH501) with robust seroconversion in a lethal rat model. The use of reverse genetics to develop vaccine prototypes, such as these with precisely engineered disruptions to the NSs and NSm genes, clearly enables the study of fundamental questions regarding the molecular biology and pathogenesis of bunyaviruses.
Interestingly, these viruses, similar to Clone 13, lack some molecular markers as indicated by the absence of anti-NSs antibodies in vaccinated animals. These gene deletions therefore allow for serological differentiation of naturally infected animals from vaccinated animals based upon NSs antibody titers. By assessing the presence or absence of anti-NSs specific antibodies, Bird et al. (2008) demonstrate the applicability of reverse genetics for the development of vaccine candidates providing the ability for differentiating infected from vaccinated animals (DIVA) (see below) (Bird et al., 2008).
IX. FUTURE APPLICATIONS OF REVERSE GENETICS SYSTEMS
Reverse genetics has been successfully used to study the molecular biology of many negative strand RNA viruses. Important discoveries in regard to pathogenicity mechanisms, the analysis of genome packaging and studies of the entry mechanism of viruses into their host cells are a few examples of the power of reverse genetics. Recent successes in the generation of improved (safety and efficacy) vaccine candidates for RVFV by means of reverse genetics further illustrate the utility and importance of such systems.
The use of minigenome rescue systems allows for the determination of the role of individual proteins in the viral replication cycle (Billecocq et al., 2008; Gerrard & Nichol, 2007; Habjan et al., 2008b; Habjan et al., 2009a; Ikegami et al., 2006). These systems are also ideal for the identification of cis-acting elements responsible for the regulation of viral genome encapsidation, transcription, replication and packaging (Albarino et al., 2007; Billecocq et al., 2008; Habjan et al., 2009a; Ikegami et al., 2007). As identified through reverse genetics, the nonstructural proteins of RVFV (NSs and NSm), while accessory to replication, can be fully characterized for their virulence properties and compared to nonstructural proteins of other bunyaviruses. Using reverse genetics systems or classical genetic reassortment, NSs type I IFN antagonistic activity has also been described for another phlebovirus, Punta Toro virus (Perrone et al., 2007), as well as for the orthobunyaviruses Bunyamwera virus (Bridgen et al., 2001) and LaCrosse virus (Blakqori et al., 2007).
Importantly, these systems also facilitate the identification of pathogenic factors and viral mechanisms used to evade the innate immune system that are crucial for targeted vaccine development (Bird et al., 2007a; Ikegami et al., 2006). Similar approaches to eliminate viral proteins involved in immune evasion via reverse genetics could result in additional promising vaccine candidates. Furthermore, such minigenome rescue systems can also be used for the identification of potential antiviral agents in an high-throughput screening assay (Flick, 2009; Freiberg et al., 2008), especially considering the recent progress in the development of sensitive minigenome rescue systems and the establishment of a reverse genetics-driven infectious clone system for RVFV
Minigenome rescue systems were also used to identify other potential antivirals (Flick, 2009; Freiberg et al., 2008). Based on successful reduction of viral titer using virus-specific PPMO’s for several virus families, a similar approach can be applied to the identification of PPMOs with RVFV-specific inhibitory activities. Especially for viruses classified as high-containment agents, minigenome rescue systems eliminate the cumbersome work required under biosafety level 3 and 4 conditions.
The establishment of an infectious clone system for RVFV via reverse genetics allowed the generation of recombinant RVFV which were successfully used as promising RVFV vaccines. Reverse genetics allows the change of a single nucleotide in the RVFV genome to generate new live attenuated vaccine candidates. But it also enables the exchange or deletion of a whole viral protein, e.g., NSs or NSm, resulting in safe vaccine candidates.
A major drawback for commercially available RVFV vaccines for livestock, which are based on live attenuated vaccine, is the inability to differentiate infected from vaccinated animals. Vaccines that allow for the differentiating infected from vaccinated animals (DIVA) can be applied to livestock in enzootic regions of countries listed by the World Organization for Animal Health (WOAH) (IOE, 2007). The use of live attenuated vaccine strains, such as genetically engineered RVFV recombinants without NSm or NSs genes, or naturally derived Clone 13 lacking most of its NSs ORF, shown to protect vaccinated animals, allows for the serologic differentiation of naturally or experimentially infected subjects by the absence of anti-NSs antibodies (Bird et al., 2008; Muller et al., 1995). Another RVFV vaccine candidate following the DIVA concept is based on VLPs (Mandell et al., 2009; Naslund et al., 2009). Since RVF VLPs do not induce a detectable anti-N response in contrast to natural infection, vaccinated animals can be easily differentiated from naturally infected individuals by the presence or absence of anti-N antibodies.
Reverse genetics may also enable a broad expansion of tissue specific detection of non segmented and segmented minus strand RNA viruses including those that infect specific target organs. Generating chimeric proteins comprised of the viral protein of interest fused to a reporter gene, e.g., GFP, replacing a viral gene with a GFP gene, or inserting such reporter gene into a viral genome using reverse genetics technology can generate useful tools to monitor virus replication and protein trafficking.
For example, the intracellular localization of the CCHFV glycoproteins GN and GC have been clearly defined by expression of GFP fusion proteins. Haferkamp et al. (2005) used recombinant glycoprotein-GFP chimeras to identify Golgi and endoplasmic reticulum retention signals in the cytoplasmic and ectodomains of the GN and GC glycoproteins, respectively (Haferkamp et al., 2005). Similar intracellular trafficking signals have been identified for other bunyaviruses including Uukuniemi (Andersson & Pettersson, 1998), Punta Toro (Chen & Compans, 1991; Chen et al., 1991; Matsuoka et al., 1996), RVFV (Gerrard & Nichol, 2002), Bunyamwera (Lappin et al., 1994) and Sin Nombre (Spiropoulou et al., 2003).
Replication-incompetent viral vectors including retroviral, adeno- and adeno-associated vectors that encode reporter genes, including GFP, have been extensively used to show tissue-specific expression of heterologous transgenes. GFP is the reporter gene of choice for tracking tissue specific expression including the pancreas, cornea, liver, kidney, small intestine, and peripheral blood in mouse models (Cheng et al., 2004; Fu et al., 2003; Thalmeier & Huss, 2001; Thalmeier et al., 2001). Flow cytometry, immunofluorescence, direct tissue fluorescence and immunohistochemical staining are all options for GFP detection if the proper controls are implemented (Guo et al., 2007; Swenson et al., 2007). These in vivo models are of particular relevence for studies on tissue specific tropism for RVFV induced hepatic necrosis in the liver, hemorrhagic damage in the peripheral blood system, and neurogenic disorders in the brain, (e.g. delayed onset encephalitis) and eye (severe retinitis).
Additionally, reverse genetic technologies can be applied to the development of sensitive diagnostic tools. For example, a recombinant rabies virus encoding the GFP gene is the key component of a new high-throughput, rapid fluorescent focus inhibition test (RFFIT) that eliminates the need for direct immunofluorescence with anti-rabies antibodies to measure infection/neutralization (Khawplod et al., 2005; Khawplod et al., 2006).
An example of a GFP-tagged replication-competent RVFV is described by Bird et al., (2008) who replaced NSs with a GFP gene. This construct allows for tracking of the tissue distribution of RVFV load, in vivo, without the need for invasive procedures. This live attenuated vaccine candidate can also serve as an immunological indicator of vaccination because it is easily distinguished from infection by wild type virus since immunized animals do not generate antibodies against NSs. In contrast, animals infected with wt RVFV generate anti-NSs antibodies. Therefore, this recombinant vaccine provides a platform that differentiates naturally infected and vaccinated animals (DIVA).
Thus, recombinant viruses developed by reverse genetics encoding the GFP gene or other reporters can be used for intra- and extracellular, tissue-specific tracking of viruses and/or specific viral proteins, as an efficient diagnostic for antiviral compounds and as tools for the development of highly sensitive, cost-effective assays for diagnostic screening.
The importance of reverse genetics systems can not be overstated. The established reverse genetics systems for RVFV opened the doors to previously unreachable research areas, including (i) RVFV-specific pathogenicity, (ii) innate immune system evasion mechanisms, (iii) a detailed dissection of the molecular biology of the virus and (iv) rational design of safe and efficient vaccine candidates. Importantly, the tools developed by reverse genetics for RVFV are readily applicable to other human pathogenic viruses.
Acknowledgments
The authors would like to thank the members of their laboratories for helpful discussions, especially Robert B. Mandell and William R. Staplin for contributions to and critical review of the manuscript and P. Formenty for valuable comments.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Abd el-Rahim IH, Abd el-Hakim U, Hussein M. An epizootic of Rift Valley fever in Egypt in 1997. Rev Sci Tech. 1999;18:741–8. doi: 10.20506/rst.18.3.1195. [DOI] [PubMed] [Google Scholar]
- Abdel-Motal UM, Gillis J, Manson K, Wyand M, Montefiori D, Stefano-Cole K, Montelaro RC, Altman JD, Johnson RP. Kinetics of expansion of SIV Gag-specific CD8+ T lymphocytes following challenge of vaccinated macaques. Virology. 2005;333:226–38. doi: 10.1016/j.virol.2004.12.030. [DOI] [PubMed] [Google Scholar]
- Abdel-Motal UM, Guay HM, Wigglesworth K, Welsh RM, Galili U. Immunogenicity of influenza virus vaccine is increased by anti-gal-mediated targeting to antigen-presenting cells. J Virol. 2007;81:9131–41. doi: 10.1128/JVI.00647-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Abdel-Motal UM, Wigglesworth K, Galili U. Mechanism for increased immunogenicity of vaccines that form in vivo immune complexes with the natural anti-Gal antibody. Vaccine. 2009;27:3072–82. doi: 10.1016/j.vaccine.2009.03.019. [DOI] [PubMed] [Google Scholar]
- Abu-Elyazeed R, el-Sharkawy S, Olson J, Botros B, Soliman A, Salib A, Cummings C, Arthur R. Prevalence of anti-Rift-Valley-fever IgM antibody in abattoir workers in the Nile delta during the 1993 outbreak in Egypt. Bull World Health Organ. 1996;74:155–8. [PMC free article] [PubMed] [Google Scholar]
- Al-Hazmi M, Ayoola EA, Abdurahman M, Banzal S, Ashraf J, El-Bushra A, Hazmi A, Abdullah M, Abbo H, Elamin A, Al-Sammani el T, Gadour M, Menon C, Hamza M, Rahim I, Hafez M, Jambavalikar M, Arishi H, Aqeel A. Epidemic Rift Valley fever in Saudi Arabia: a clinical study of severe illness in humans. Clin Infect Dis. 2003;36:245–52. doi: 10.1086/345671. [DOI] [PubMed] [Google Scholar]
- Albarino CG, Bird BH, Nichol ST. A shared transcription termination signal on negative and ambisense RNA genome segments of Rift Valley fever, sandfly fever Sicilian, and Toscana viruses. J Virol. 2007;81:5246–56. doi: 10.1128/JVI.02778-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alrajhi AA, Al-Semari A, Al-Watban J. Rift Valley fever encephalitis. Emerg Infect Dis. 2004;10:554–5. doi: 10.3201/eid1003.020817. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Anderson GW, Jr, Rosebrock JA, Johnson AJ, Jennings GB, Peters CJ. Infection of inbred rat strains with Rift Valley fever virus: development of a congenic resistant strain and observations on age-dependence of resistance. Am J Trop Med Hyg. 1991;44:475–80. doi: 10.4269/ajtmh.1991.44.475. [DOI] [PubMed] [Google Scholar]
- Anderson GW, Jr, Peters CJ. Viral determinants of virulence for Rift Valley fever (RVF) in rats. Microb Pathog. 1988;5:241–250. doi: 10.1016/0882-4010(88)90096-4. [DOI] [PubMed] [Google Scholar]
- Andersson AM, Pettersson RF. Targeting of a short peptide derived from the cytoplasmic tail of the G1 membrane glycoprotein of Uukuniemi virus (Bunyaviridae) to the Golgi complex. J Virol. 1998;72:9585–96. doi: 10.1128/jvi.72.12.9585-9596.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Andrew ME, Coupar BE, Boyle DB. Humoral and cell-mediated immune responses to recombinant vaccinia viruses in mice. Immunol Cell Biol. 1989;67 (Pt 5):331–7. doi: 10.1038/icb.1989.48. [DOI] [PubMed] [Google Scholar]
- Anonymous Outbreak of Rift Valley fever, Yemen, August-October 2000. Wkly Epidemiol Rec. 2000;75:392–5. [PubMed] [Google Scholar]
- Anonymous . Chemical and Biological Weapons: Possession and Programs Past and Present. CNS, James Martin Center for Nonproliferation Studies; 2009. [Google Scholar]
- Arthur RR, el-Sharkawy MS, Cope SE, Botros BA, Oun S, Morrill JC, Shope RE, Hibbs RG, Darwish MA, Imam IZ. Recurrence of Rift Valley fever in Egypt. Lancet. 1993;342:1149–50. doi: 10.1016/0140-6736(93)92128-g. [DOI] [PubMed] [Google Scholar]
- Balkhy HH, Memish ZA. Rift Valley fever: an uninvited zoonosis in the Arabian peninsula. Int J Antimicrob Agents. 2003;21:153–7. doi: 10.1016/s0924-8579(02)00295-9. [DOI] [PubMed] [Google Scholar]
- Barnard BJ. Rift Valley fever vaccine--antibody and immune response in cattle to a live and an inactivated vaccine. J S Afr Vet Assoc. 1979;50:155–7. [PubMed] [Google Scholar]
- Barnard BJ, Botha MJ. An inactivated rift valley fever vaccine. J S Afr Vet Assoc. 1977;48:45–8. [PubMed] [Google Scholar]
- Barr JN, Rodgers JW, Wertz GW. The Bunyamwera virus mRNA transcription signal resides within both the 3′ and the 5′ terminal regions and allows ambisense transcription from a model RNA segment. J Virol. 2005;79:12602–7. doi: 10.1128/JVI.79.19.12602-12607.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barr JN, Wertz GW. Role of the conserved nucleotide mismatch within 3′- and 5′-terminal regions of Bunyamwera virus in signaling transcription. J Virol. 2005;79:3586–94. doi: 10.1128/JVI.79.6.3586-3594.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beigel J, Bray M. Current and future antiviral therapy of severe seasonal and avian influenza. Antiviral Res. 2008;78:91–102. doi: 10.1016/j.antiviral.2008.01.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bellocq C, Kolakofsky D. Translational requirement for La Crosse virus S-mRNA synthesis: a possible mechanism. J Virol. 1987;61:3960–7. doi: 10.1128/jvi.61.12.3960-3967.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bergthaler A, Gerber NU, Merkler D, Horvath E, de la Torre JC, Pinschewer DD. Envelope exchange for the generation of live-attenuated arenavirus vaccines. PLoS Pathog. 2006;2:e51. doi: 10.1371/journal.ppat.0020051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bettinger GE, Peters CJ, Pittman P, Morrill JC, Ranadive M, Kormann RN, Lokukamage N. Rift Valley Fever MP-12 Vaccine: A University, Government, & Industry Collaborative Development. Rift Valley Fever Workshop.2009. [Google Scholar]
- Billecocq A, Gauliard N, Le May N, Elliott RM, Flick R, Bouloy M. RNA polymerase I-mediated expression of viral RNA for the rescue of infectious virulent and avirulent Rift Valley fever viruses. Virology. 2008;378:377–84. doi: 10.1016/j.virol.2008.05.033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Billecocq A, Spiegel M, Vialat P, Kohl A, Weber F, Bouloy M, Haller O. NSs protein of Rift Valley fever virus blocks interferon production by inhibiting host gene transcription. J Virol. 2004;78:9798–806. doi: 10.1128/JVI.78.18.9798-9806.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bird BH, Albarino CG, Hartman AL, Erickson BR, Ksiazek TG, Nichol ST. Rift valley fever virus lacking the NSs and NSm genes is highly attenuated, confers protective immunity from virulent virus challenge, and allows for differential identification of infected and vaccinated animals. J Virol. 2008;82:2681–91. doi: 10.1128/JVI.02501-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bird BH, Albarino CG, Nichol ST. Rift Valley fever virus lacking NSm proteins retains high virulence in vivo and may provide a model of human delayed onset neurologic disease. Virology. 2007a;362:10–5. doi: 10.1016/j.virol.2007.01.046. [DOI] [PubMed] [Google Scholar]
- Bird BH, Khristova ML, Rollin PE, Ksiazek TG, Nichol ST. Complete genome analysis of 33 ecologically and biologically diverse Rift Valley fever virus strains reveals widespread virus movement and low genetic diversity due to recent common ancestry. J Virol. 2007b;81:2805–16. doi: 10.1128/JVI.02095-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blakqori G, Delhaye S, Habjan M, Blair CD, Sanchez-Vargas I, Olson KE, Attarzadeh-Yazdi G, Fragkoudis R, Kohl A, Kalinke U, Weiss S, Michiels T, Staeheli P, Weber F. La Crosse bunyavirus nonstructural protein NSs serves to suppress the type I interferon system of mammalian hosts. J Virol. 2007;81:4991–9. doi: 10.1128/JVI.01933-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blakqori G, Kochs G, Haller O, Weber F. Functional L polymerase of La Crosse virus allows in vivo reconstitution of recombinant nucleocapsids. J Gen Virol. 2003;84:1207–14. doi: 10.1099/vir.0.18876-0. [DOI] [PubMed] [Google Scholar]
- Blakqori G, Weber F. Efficient cDNA-based rescue of La Crosse bunyaviruses expressing or lacking the nonstructural protein NSs. J Virol. 2005;79:10420–8. doi: 10.1128/JVI.79.16.10420-10428.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Botros B, Omar A, Elian K, Mohamed G, Soliman A, Salib A, Salman D, Saad M, Earhart K. Adverse response of non-indigenous cattle of European breeds to live attenuated Smithburn Rift Valley fever vaccine. J Med Virol. 2006;78:787–91. doi: 10.1002/jmv.20624. [DOI] [PubMed] [Google Scholar]
- Bouloy M, Janzen C, Vialat P, Khun H, Pavlovic J, Huerre M, Haller O. Genetic evidence for an interferon-antagonistic function of rift valley fever virus nonstructural protein NSs. J Virol. 2001;75:1371–7. doi: 10.1128/JVI.75.3.1371-1377.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bray M. Highly pathogenic RNA viral infections: challenges for antiviral research. Antiviral Res. 2008;78:1–8. doi: 10.1016/j.antiviral.2007.12.007. [DOI] [PubMed] [Google Scholar]
- Bridgen A, Elliott RM. Rescue of a segmented negative-strand RNA virus entirely from cloned complementary DNAs. Proc Natl Acad Sci U S A. 1996;93:15400–4. doi: 10.1073/pnas.93.26.15400. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bridgen A, Weber F, Fazakerley JK, Elliott RM. Bunyamwera bunyavirus nonstructural protein NSs is a nonessential gene product that contributes to viral pathogenesis. Proc Natl Acad Sci U S A. 2001;98:664–9. doi: 10.1073/pnas.98.2.664. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Buchholz UJ, Granzow H, Schuldt K, Whitehead SS, Murphy BR, Collins PL. Chimeric bovine respiratory syncytial virus with glycoprotein gene substitutions from human respiratory syncytial virus (HRSV): effects on host range and evaluation as a live-attenuated HRSV vaccine. J Virol. 2000;74:1187–99. doi: 10.1128/jvi.74.3.1187-1199.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Caplen H, Peters CJ, Bishop DH. Mutagen-directed attenuation of Rift Valley fever virus as a method for vaccine development. J Gen Virol. 1985;66 (Pt 10):2271–7. doi: 10.1099/0022-1317-66-10-2271. [DOI] [PubMed] [Google Scholar]
- Chen SY, Compans RW. Oligomerization, transport, and Golgi retention of Punta Toro virus glycoproteins. J Virol. 1991;65:5902–9. doi: 10.1128/jvi.65.11.5902-5909.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen SY, Matsuoka Y, Compans RW. Golgi complex localization of the Punta Toro virus G2 protein requires its association with the G1 protein. Virology. 1991;183:351–65. doi: 10.1016/0042-6822(91)90148-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheng K, Fraga D, Zhang C, Kotb M, Gaber AO, Guntaka RV, Mahato RI. Adenovirus-based vascular endothelial growth factor gene delivery to human pancreatic islets. Gene Ther. 2004;11:1105–16. doi: 10.1038/sj.gt.3302267. [DOI] [PubMed] [Google Scholar]
- Coetzer JA, Barnard BJ. Hydrops amnii in sheep associated with hydranencephaly and arthrogryposis with wesselsbron disease and rift valley fever viruses as aetiological agents. Onderstepoort J Vet Res. 1977;44:119–26. [PubMed] [Google Scholar]
- Conzelmann KK, Cox JH, Thiel HJ. An L (polymerase)-deficient rabies virus defective interfering particle RNA is replicated and transcribed by heterologous helper virus L proteins. Virology. 1991;184:655–63. doi: 10.1016/0042-6822(91)90435-e. [DOI] [PubMed] [Google Scholar]
- de Wit E, Spronken MI, Vervaet G, Rimmelzwaan GF, Osterhaus AD, Fouchier RA. A reverse-genetics system for Influenza A virus using T7 RNA polymerase. J Gen Virol. 2007;88:1281–7. doi: 10.1099/vir.0.82452-0. [DOI] [PubMed] [Google Scholar]
- Deas TS, Bennett CJ, Jones SA, Tilgner M, Ren P, Behr MJ, Stein DA, Iversen PL, Kramer LD, Bernard KA, Shi PY. In vitro resistance selection and in vivo efficacy of morpholino oligomers against West Nile virus. Antimicrob Agents Chemother. 2007;51:2470–82. doi: 10.1128/AAC.00069-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Digoutte JP, Peters CJ. General aspects of the 1987 Rift Valley fever epidemic in Mauritania. Res Virol. 1989;140:27–30. doi: 10.1016/s0923-2516(89)80081-0. [DOI] [PubMed] [Google Scholar]
- Dunn EF, Pritlove DC, Jin H, Elliott RM. Transcription of a recombinant bunyavirus RNA template by transiently expressed bunyavirus proteins. Virology. 1995;211:133–43. doi: 10.1006/viro.1995.1386. [DOI] [PubMed] [Google Scholar]
- Easterday BC. Rift valley fever. Adv Vet Sci. 1965;10:65–127. [PubMed] [Google Scholar]
- Elliott RM. The Bunyaviridae. Plenum Press; New York and London: 1996. [Google Scholar]
- Flanagan EB, Zamparo JM, Ball LA, Rodriguez LL, Wertz GW. Rearrangement of the genes of vesicular stomatitis virus eliminates clinical disease in the natural host: new strategy for vaccine development. J Virol. 2001;75:6107–14. doi: 10.1128/JVI.75.13.6107-6114.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Flatz L, Bergthaler A, de la Torre JC, Pinschewer DD. Recovery of an arenavirus entirely from RNA polymerase I/II-driven cDNA. Proc Natl Acad Sci U S A. 2006;103:4663–8. doi: 10.1073/pnas.0600652103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Flick K, Hooper JW, Schmaljohn CS, Pettersson RF, Feldmann H, Flick R. Rescue of Hantaan virus minigenomes. Virology. 2003a;306:219–24. doi: 10.1016/s0042-6822(02)00070-3. [DOI] [PubMed] [Google Scholar]
- Flick K, Katz A, Overby A, Feldmann H, Pettersson RF, Flick R. Functional analysis of the noncoding regions of the Uukuniemi virus (Bunyaviridae) RNA segments. J Virol. 2004;78:11726–38. doi: 10.1128/JVI.78.21.11726-11738.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Flick R. 2009 unpublished data. [Google Scholar]
- Flick R, Bouloy M. Rift Valley fever virus. Curr Mol Med. 2005;5:827–34. doi: 10.2174/156652405774962263. [DOI] [PubMed] [Google Scholar]
- Flick R, Flick K, Feldmann H, Elgh F. Reverse genetics for crimean-congo hemorrhagic fever virus. J Virol. 2003b;77:5997–6006. doi: 10.1128/JVI.77.10.5997-6006.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Flick R, Mandell RB, Staplin WR, Kaniewski LD, Carzoli AK, Manuszak RP, Wang J, Rossi GR, Vahanian N, Link CJ. The alphaGal HyperAcute® Technology: Enhancing Immunogenicity of Antiviral Vaccines by Exploiting the Natural alphaGal-mediated Zoonotic Blockade. J Zoonoses and Public Health. 2009 doi: 10.1111/j.1863-2378.2008.01191.x. in press. [DOI] [PubMed] [Google Scholar]
- Flick R, Pettersson RF. Reverse genetics system for Uukuniemi virus (Bunyaviridae): RNA polymerase I-catalyzed expression of chimeric viral RNAs. J Virol. 2001;75:1643–55. doi: 10.1128/JVI.75.4.1643-1655.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Flick R, Whitehouse CA. Crimean-Congo hemorrhagic fever virus. Curr Mol Med. 2005;5:753–60. doi: 10.2174/156652405774962335. [DOI] [PubMed] [Google Scholar]
- Fodor E, Devenish L, Engelhardt OG, Palese P, Brownlee GG, Garcia-Sastre A. Rescue of influenza A virus from recombinant DNA. J Virol. 1999;73:9679–82. doi: 10.1128/jvi.73.11.9679-9682.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fontenille D, Traore-Lamizana M, Diallo M, Thonnon J, Digoutte JP, Zeller HG. New vectors of Rift Valley fever in West Africa. Emerg Infect Dis. 1998;4:289–93. doi: 10.3201/eid0402.980218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frank-Peterside N. Response of laboratory staff to vaccination with an inactivated Rift Valley fever vaccine--TSI-GSD 200. Afr J Med Med Sci. 2000;29:89–92. [PubMed] [Google Scholar]
- Freiberg A, Dolores LK, Enterlein S, Flick R. Establishment and characterization of plasmid-driven minigenome rescue systems for Nipah virus: RNA polymerase I- and T7-catalyzed generation of functional paramyxoviral RNA. Virology. 2008;370:33–44. doi: 10.1016/j.virol.2007.08.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fu H, Muenzer J, Samulski RJ, Breese G, Sifford J, Zeng X, McCarty DM. Self-complementary adeno-associated virus serotype 2 vector: global distribution and broad dispersion of AAV-mediated transgene expression in mouse brain. Mol Ther. 2003;8:911–7. doi: 10.1016/j.ymthe.2003.08.021. [DOI] [PubMed] [Google Scholar]
- Furuta Y, Takahashi K, Shiraki K, Sakamoto K, Smee DF, Barnard DL, Gowen BB, Julander JG, Morrey JD. T-705 (favipiravir) and related compounds: Novel broad-spectrum inhibitors of RNA viral infections. Antiviral Res. 2009;82:95–102. doi: 10.1016/j.antiviral.2009.02.198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gabriel G, Nordmann A, Stein DA, Iversen PL, Klenk HD. Morpholino oligomers targeting the PB1 and NP genes enhance the survival of mice infected with highly pathogenic influenza A H7N7 virus. J Gen Virol. 2008;89:939–48. doi: 10.1099/vir.0.83449-0. [DOI] [PubMed] [Google Scholar]
- Galili U. Evolution and pathophysiology of the human natural anti-alpha-galactosyl IgG (anti-Gal) antibody. Springer Semin Immunopathol. 1993;15:155–71. doi: 10.1007/BF00201098. [DOI] [PubMed] [Google Scholar]
- Galili U. Evolution of alpha 1,3galactosyltransferase and of the alpha-Gal epitope. Subcell Biochem. 1999;32:1–23. doi: 10.1007/978-1-4615-4771-6_1. [DOI] [PubMed] [Google Scholar]
- Galili U, Clark MR, Shohet SB. Excessive binding of the natural anti-alpha-galactosyl IgG to sickle red cells: enhancement of red cell destruction by a physiological process. Trans Assoc Am Physicians. 1985a;98:158–65. [PubMed] [Google Scholar]
- Galili U, Flechner I, Rachmilewitz EA. A naturally occurring anti-alpha-galactosyl IgG recognizing senescent human red cells. Prog Clin Biol Res. 1985b;195:263–78. [PubMed] [Google Scholar]
- Galili U, Macher BA, Buehler J, Shohet SB. Human natural anti-alpha-galactosyl IgG. II. The specific recognition of alpha (1----3)-linked galactose residues. J Exp Med. 1985c;162:573–82. doi: 10.1084/jem.162.2.573. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Galili U, Wigglesworth K, Abdel-Motal UM. Intratumoral injection of alpha-gal glycolipids induces xenograft-like destruction and conversion of lesions into endogenous vaccines. J Immunol. 2007;178:4676–87. doi: 10.4049/jimmunol.178.7.4676. [DOI] [PubMed] [Google Scholar]
- Garcia S, Crance JM, Billecocq A, Peinnequin A, Jouan A, Bouloy M, Garin D. Quantitative real-time PCR detection of Rift Valley fever virus and its application to evaluation of antiviral compounds. J Clin Microbiol. 2001;39:4456–61. doi: 10.1128/JCM.39.12.4456-4461.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gauliard N, Billecocq A, Flick R, Bouloy M. Rift Valley fever virus noncoding regions of L, M and S segments regulate RNA synthesis. Virology. 2006;351:170–9. doi: 10.1016/j.virol.2006.03.018. [DOI] [PubMed] [Google Scholar]
- Gerdes GH. Rift Valley fever. Rev Sci Tech. 2004;23:613–23. doi: 10.20506/rst.23.2.1500. [DOI] [PubMed] [Google Scholar]
- Gerrard SR, Bird BH, Albarino CG, Nichol ST. The NSm proteins of Rift Valley fever virus are dispensable for maturation, replication and infection. Virology. 2007;359:459–65. doi: 10.1016/j.virol.2006.09.035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gerrard SR, Nichol ST. Characterization of the Golgi retention motif of Rift Valley fever virus G(N) glycoprotein. J Virol. 2002;76:12200–10. doi: 10.1128/JVI.76.23.12200-12210.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gerrard SR, Nichol ST. Synthesis, proteolytic processing and complex formation of N-terminally nested precursor proteins of the Rift Valley fever virus glycoproteins. Virology. 2007;357:124–33. doi: 10.1016/j.virol.2006.08.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gowen BB, Holbrook MR. Animal models of highly pathogenic RNA viral infections: hemorrhagic fever viruses. Antiviral Res. 2008;78:79–90. doi: 10.1016/j.antiviral.2007.10.002. [DOI] [PubMed] [Google Scholar]
- Gowen BB, Smee DF, Wong MH, Hall JO, Jung KH, Bailey KW, Stevens JR, Furuta Y, Morrey JD. Treatment of late stage disease in a model of arenaviral hemorrhagic fever: T-705 efficacy and reduced toxicity suggests an alternative to ribavirin. PLoS ONE. 2008;3:e3725. doi: 10.1371/journal.pone.0003725. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gowen BB, Wong MH, Jung KH, Sanders AB, Mendenhall M, Bailey KW, Furuta Y, Sidwell RW. In vitro and in vivo activities of T-705 against arenavirus and bunyavirus infections. Antimicrob Agents Chemother. 2007;51:3168–76. doi: 10.1128/AAC.00356-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Graham R, Talent Jim, Allison Graham, Cleveland Robin, Rademaker Steve, Roemer Tim, Sherman Wendy, Sokolski Henry, Verma Rich. vintage books. A Division of Random House, Inc; 2008. WORLD AT RISK: The Report of the Commission on the Prevention of WMD Proliferation and Terrorism. [Google Scholar]
- Grgacic E, Anderson D. Virus-like particles; passport to immune recognition. Methods. 2006:40. doi: 10.1016/j.ymeth.2006.07.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Groseth A, Feldmann H, Theriault S, Mehmetoglu G, Flick R. RNA polymerase I-driven minigenome system for Ebola viruses. J Virol. 2005;79:4425–33. doi: 10.1128/JVI.79.7.4425-4433.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guiso N, Njamkepo E, Vie le Sage F, Zepp F, Meyer CU, Abitbol V, Clyti N, Chevallier S. Long-term humoral and cell-mediated immunity after acellular pertussis vaccination compares favourably with whole-cell vaccines 6 years after booster vaccination in the second year of life. Vaccine. 2007;25:1390–7. doi: 10.1016/j.vaccine.2006.10.048. [DOI] [PubMed] [Google Scholar]
- Guo JK, Cheng EC, Wang L, Swenson ES, Ardito TA, Kashgarian M, Cantley LG, Krause DS. The commonly used beta-actin-GFP transgenic mouse strain develops a distinct type of glomerulosclerosis. Transgenic Res. 2007;16:829–34. doi: 10.1007/s11248-007-9107-x. [DOI] [PubMed] [Google Scholar]
- Habjan M, Andersson I, Klingstrom J, Schumann M, Martin A, Zimmermann P, Wagner V, Pichlmair A, Schneider U, Muhlberger E, Mirazimi A, Weber F. Processing of genome 5′ termini as a strategy of negative-strand RNA viruses to avoid RIG-I-dependent interferon induction. PLoS ONE. 2008a;3:e2032. doi: 10.1371/journal.pone.0002032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Habjan M, Penski N, Spiegel M, Weber F. T7 RNA polymerase-dependent and -independent systems for cDNA-based rescue of Rift Valley fever virus. J Gen Virol. 2008b;89:2157–66. doi: 10.1099/vir.0.2008/002097-0. [DOI] [PubMed] [Google Scholar]
- Habjan M, Penski N, Wagner V, Spiegel M, Overby AK, Kochs G, Huiskonen JT, Weber F. Efficient production of Rift Valley fever virus-like particles: The antiviral protein MxA can inhibit primary transcription of bunyaviruses. Virology. 2009a;385:400–8. doi: 10.1016/j.virol.2008.12.011. [DOI] [PubMed] [Google Scholar]
- Habjan M, Pichlmair A, Elliott RM, Overby AK, Glatter T, Gstaiger M, Superti-Furga G, Unger H, Weber F. NSs protein of Rift Valley Fever Virus induces the specific degradation of the double-stranded RNA-dependent protein kinase (PKR) J Virol. 2009b doi: 10.1128/JVI.02148-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haferkamp S, Fernando L, Schwarz TF, Feldmann H, Flick R. Intracellular localization of Crimean-Congo Hemorrhagic Fever (CCHF) virus glycoproteins. Virol J. 2005;2:42. doi: 10.1186/1743-422X-2-42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hayes CG. West Nile virus: Uganda, 1937, to New York City, 1999. Ann N Y Acad Sci. 2001;951:25–37. doi: 10.1111/j.1749-6632.2001.tb02682.x. [DOI] [PubMed] [Google Scholar]
- Heise MT, Whitmore A, Thompson J, Parsons M, Grobbelaar AA, Kemp A, Paweska JT, Madric K, White LJ, Swanepoel R, Burt FJ. An alphavirus replicon-derived candidate vaccine against Rift Valley fever virus. Epidemiol Infect. 2009:1–10. doi: 10.1017/S0950268808001696. [DOI] [PubMed] [Google Scholar]
- Hellrung DJ, Kisselev S, Link CJ. Co-expression of alpha(1,3)galactosyltransferase and Bacillus thuringiensis PIPLC enhances hyperacute rejection of tumor cells. Cancer Immunol Immunother. 2007;56:25–34. doi: 10.1007/s00262-006-0163-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hellrung DJ, Rossi G, Link CJ. High-throughput fluorescent screening of transgenic animals: phenotyping and haplotyping. Cytometry A. 2006;69:1092–5. doi: 10.1002/cyto.a.20328. [DOI] [PubMed] [Google Scholar]
- Hoogstraal H, Meegan JM, Khalil GM, Adham FK. The Rift Valley fever epizootic in Egypt 1977–78. 2. Ecological and entomological studies. Trans R Soc Trop Med Hyg. 1979;73:624–9. doi: 10.1016/0035-9203(79)90005-1. [DOI] [PubMed] [Google Scholar]
- Hunter B. Investigation of C13 RVF mutant as a vaccine strain. Proceedings of 5th International sheep veterinary congress; 2001. pp. 21–25. [Google Scholar]
- Ikegami T, Narayanan K, Won S, Kamitani W, Peters CJ, Makino S. Rift Valley fever virus NSs protein promotes post-transcriptional downregulation of protein kinase PKR and inhibits eIF2alpha phosphorylation. PLoS Pathog. 2009;5:e1000287. doi: 10.1371/journal.ppat.1000287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ikegami T, Peters CJ, Makino S. Rift valley fever virus nonstructural protein NSs promotes viral RNA replication and transcription in a minigenome system. J Virol. 2005a;79:5606–15. doi: 10.1128/JVI.79.9.5606-5615.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ikegami T, Won S, Peters CJ, Makino S. Rift Valley fever virus NSs mRNA is transcribed from an incoming anti-viral-sense S RNA segment. J Virol. 2005b;79:12106–11. doi: 10.1128/JVI.79.18.12106-12111.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ikegami T, Won S, Peters CJ, Makino S. Rescue of infectious rift valley fever virus entirely from cDNA, analysis of virus lacking the NSs gene, and expression of a foreign gene. J Virol. 2006;80:2933–40. doi: 10.1128/JVI.80.6.2933-2940.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ikegami T, Won S, Peters CJ, Makino S. Characterization of Rift Valley fever virus transcriptional terminations. J Virol. 2007;81:8421–38. doi: 10.1128/JVI.02641-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Epizootics IOo., editor. IOE. Terrestrial animal health code. XI. 2007. p. 14.1. [Google Scholar]
- Jones S, Flick R, Jensen V, Feldmann H. Lethal Viruses - Viral Hemorrhagic Fevers. Research Advances in Microbiology 2002 [Google Scholar]
- Julander JG, Shafer K, Smee DF, Morrey JD, Furuta Y. Activity of T-705 in a hamster model of yellow fever virus infection in comparison with that of a chemically related compound, T-1106. Antimicrob Agents Chemother. 2009;53:202–9. doi: 10.1128/AAC.01074-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kark JD, Aynor Y, Peters CJ. A rift Valley fever vaccine trial. I. Side effects and serologic response over a six-month follow-up. Am J Epidemiol. 1982;116:808–20. doi: 10.1093/oxfordjournals.aje.a113471. [DOI] [PubMed] [Google Scholar]
- Kark JD, Aynor Y, Peters CJ. A Rift Valley fever vaccine trial: 2. Serological response to booster doses with a comparison of intradermal versus subcutaneous injection. Vaccine. 1985;3:117–22. doi: 10.1016/0264-410x(85)90060-x. [DOI] [PubMed] [Google Scholar]
- Kato M, Ito T, Wagner G, Ellenberger T. A molecular handoff between bacteriophage T7 DNA primase and T7 DNA polymerase initiates DNA synthesis. J Biol Chem. 2004;279:30554–62. doi: 10.1074/jbc.M403485200. [DOI] [PubMed] [Google Scholar]
- Kawaoka Y. Biology of negative stranded RNA viruses: The power of reverse genetics. In: Kawaoka Y, editor. Current Topics in Micro and Immunol. Vol. 283. Springer Verlag; Berlin, Heidelberg, New York: 2005. [Google Scholar]
- Khawplod P, Inoue K, Shoji Y, Wilde H, Ubol S, Nishizono A, Kurane I, Morimoto K. A novel rapid fluorescent focus inhibition test for rabies virus using a recombinant rabies virus visualizing a green fluorescent protein. J Virol Methods. 2005;125:35–40. doi: 10.1016/j.jviromet.2004.12.003. [DOI] [PubMed] [Google Scholar]
- Khawplod P, Shoji Y, Ubol S, Mitmoonpitak C, Wilde H, Nishizono A, Kurane I, Morimoto K. Genetic analysis of dog rabies viruses circulating in Bangkok. Infect Genet Evol. 2006;6:235–40. doi: 10.1016/j.meegid.2005.06.002. [DOI] [PubMed] [Google Scholar]
- Kilgore PE, Ksiazek TG, Rollin PE, Mills JN, Villagra MR, Montenegro MJ, Costales MA, Paredes LC, Peters CJ. Treatment of Bolivian hemorrhagic fever with intravenous ribavirin. Clin Infect Dis. 1997;24:718–22. doi: 10.1093/clind/24.4.718. [DOI] [PubMed] [Google Scholar]
- Kinsella E, Martin SG, Grolla A, Czub M, Feldmann H, Flick R. Sequence determination of the Crimean-Congo hemorrhagic fever virus L segment. Virology. 2004;321:23–8. doi: 10.1016/j.virol.2003.09.046. [DOI] [PubMed] [Google Scholar]
- Kochs G, Janzen C, Hohenberg H, Haller O. Antivirally active MxA protein sequesters La Crosse virus nucleocapsid protein into perinuclear complexes. Proc Natl Acad Sci U S A. 2002;99:3153–8. doi: 10.1073/pnas.052430399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kohl A, Hart TJ, Noonan C, Royall E, Roberts LO, Elliott RM. A bunyamwera virus minireplicon system in mosquito cells. J Virol. 2004;78:5679–85. doi: 10.1128/JVI.78.11.5679-5685.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kolakofsky D, Bellocq C, Raju R. The translational requirement for La Crosse virus S-mRNA synthesis. Cold Spring Harb Symp Quant Biol. 1987;52:373–9. doi: 10.1101/sqb.1987.052.01.043. [DOI] [PubMed] [Google Scholar]
- Kovacs GR, Parks CL, Vasilakis N, Udem SA. Enhanced genetic rescue of negative-strand RNA viruses: use of an MVA-T7 RNA polymerase vector and DNA replication inhibitors. J Virol Methods. 2003;111:29–36. doi: 10.1016/s0166-0934(03)00132-0. [DOI] [PubMed] [Google Scholar]
- LaBeaud AD, Muchiri EM, Ndzovu M, Mwanje MT, Muiruri S, Peters CJ, King CH. Interepidemic Rift Valley fever virus seropositivity, northeastern Kenya. Emerg Infect Dis. 2008;14:1240–6. doi: 10.3201/eid1408.080082. [DOI] [PMC free article] [PubMed] [Google Scholar]
- LaBeaud AD, Ochiai Y, Peters CJ, Muchiri EM, King CH. Spectrum of Rift Valley fever virus transmission in Kenya: insights from three distinct regions. Am J Trop Med Hyg. 2007;76:795–800. [PMC free article] [PubMed] [Google Scholar]
- Lagerqvist N, Naslund J, Lundkvist A, Bouloy M, Ahlm C, Bucht G. Characterisation of immune responses and protective efficacy in mice after immunisation with Rift Valley Fever virus cDNA constructs. Virol J. 2009;6:6. doi: 10.1186/1743-422X-6-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lappin DF, Nakitare GW, Palfreyman JW, Elliott RM. Localization of Bunyamwera bunyavirus G1 glycoprotein to the Golgi requires association with G2 but not with NSm. J Gen Virol. 1994;75 (Pt 12):3441–51. doi: 10.1099/0022-1317-75-12-3441. [DOI] [PubMed] [Google Scholar]
- Lawson ND, Stillman EA, Whitt MA, Rose JK. Recombinant vesicular stomatitis viruses from DNA. Proc Natl Acad Sci U S A. 1995;92:4477–81. doi: 10.1073/pnas.92.10.4477. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Le May N, Dubaele S, Proietti De Santis L, Billecocq A, Bouloy M, Egly JM. TFIIH transcription factor, a target for the Rift Valley hemorrhagic fever virus. Cell. 2004;116:541–50. doi: 10.1016/s0092-8674(04)00132-1. [DOI] [PubMed] [Google Scholar]
- Le May N, Mansuroglu Z, Leger P, Josse T, Blot G, Billecocq A, Flick R, Jacob Y, Bonnefoy E, Bouloy M. A SAP30 complex inhibits IFN-beta expression in Rift Valley fever virus infected cells. PLoS Pathog. 2008;4:e13. doi: 10.1371/journal.ppat.0040013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu L, Celma CC, Roy P. Rift Valley fever virus structural proteins: expression, characterization and assembly of recombinant proteins. Virol J. 2008;5:82. doi: 10.1186/1743-422X-5-82. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lopez N, Muller R, Prehaud C, Bouloy M. The L protein of Rift Valley fever virus can rescue viral ribonucleoproteins and transcribe synthetic genome-like RNA molecules. J Virol. 1995;69:3972–9. doi: 10.1128/jvi.69.7.3972-3979.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lorenzo G, Martin-Folgar R, Rodriguez F, Brun A. Priming with DNA plasmids encoding the nucleocapsid protein and glycoprotein precursors from Rift Valley fever virus accelerates the immune responses induced by an attenuated vaccine in sheep. Vaccine. 2008;26:5255–62. doi: 10.1016/j.vaccine.2008.07.042. [DOI] [PubMed] [Google Scholar]
- Lowen AC, Noonan C, McLees A, Elliott RM. Efficient bunyavirus rescue from cloned cDNA. Virology. 2004;330:493–500. doi: 10.1016/j.virol.2004.10.009. [DOI] [PubMed] [Google Scholar]
- Lupfer C, Stein DA, Mourich DV, Tepper SE, Iversen PL, Pastey M. Inhibition of influenza A H3N8 virus infections in mice by morpholino oligomers. Arch Virol. 2008;153:929–37. doi: 10.1007/s00705-008-0067-0. [DOI] [PubMed] [Google Scholar]
- Lutwick LI, Nierengarten MB. Vaccines for Category A bioterrorism diseases. Expert Opin Biol Ther. 2002;2:883–93. doi: 10.1517/14712598.2.8.883. [DOI] [PubMed] [Google Scholar]
- Luytjes W, Krystal M, Enami M, Parvin JD, Palese P. Amplification, expression, and packaging of foreign gene by influenza virus. Cell. 1989;59:1107–13. doi: 10.1016/0092-8674(89)90766-6. [DOI] [PubMed] [Google Scholar]
- Madani TA, Al-Mazrou YY, Al-Jeffri MH, Mishkhas AA, Al-Rabeah AM, Turkistani AM, Al-Sayed MO, Abodahish AA, Khan AS, Ksiazek TG, Shobokshi O. Rift Valley fever epidemic in Saudi Arabia: epidemiological, clinical, and laboratory characteristics. Clin Infect Dis. 2003;37:1084–92. doi: 10.1086/378747. [DOI] [PubMed] [Google Scholar]
- Mandell RB, Koukuntla R, Staplin WR, Kaniewski LD, Carzoli AK, Freiberg A, Holbrook M, Link CJ, Flick R. A safe replication-incompetent Rift Valley fever vaccine: Chimeric virus-like particles fully protect rodents in a challenge study. PLoS Pathogens submitted 2009 [Google Scholar]
- Matsuoka Y, Chen SY, Holland CE, Compans RW. Molecular determinants of Golgi retention in the Punta Toro virus G1 protein. Arch Biochem Biophys. 1996;336:184–9. doi: 10.1006/abbi.1996.0547. [DOI] [PubMed] [Google Scholar]
- McCormick JB, King IJ, Webb PA, Scribner CL, Craven RB, Johnson KM, Elliott LH, Belmont-Williams R. Lassa fever. Effective therapy with ribavirin. N Engl J Med. 1986;314:20–6. doi: 10.1056/NEJM198601023140104. [DOI] [PubMed] [Google Scholar]
- Meadors GF, 3rd, Gibbs PH, Peters CJ. Evaluation of a new Rift Valley fever vaccine: safety and immunogenicity trials. Vaccine. 1986;4:179–84. doi: 10.1016/0264-410x(86)90007-1. [DOI] [PubMed] [Google Scholar]
- Meegan JM. The Rift Valley fever epizootic in Egypt 1977–78. 1. Description of the epizzotic and virological studies. Trans R Soc Trop Med Hyg. 1979;73:618–23. doi: 10.1016/0035-9203(79)90004-x. [DOI] [PubMed] [Google Scholar]
- Meegan JM, Hoogstraal H, Moussa MI. An epizootic of Rift Valley fever in Egypt in 1977. Vet Rec. 1979;105:124–5. doi: 10.1136/vr.105.6.124. [DOI] [PubMed] [Google Scholar]
- Monath TP. Treatment of yellow fever. Antiviral Res. 2008;78:116–24. doi: 10.1016/j.antiviral.2007.10.009. [DOI] [PubMed] [Google Scholar]
- Morrill JC, Carpenter L, Taylor D, Ramsburg HH, Quance J, Peters CJ. Further evaluation of a mutagen-attenuated Rift Valley fever vaccine in sheep. Vaccine. 1991;9:35–41. doi: 10.1016/0264-410x(91)90314-v. [DOI] [PubMed] [Google Scholar]
- Morrill JC, Jennings GB, Caplen H, Turell MJ, Johnson AJ, Peters CJ. Pathogenicity and immunogenicity of a mutagen-attenuated Rift Valley fever virus immunogen in pregnant ewes. Am J Vet Res. 1987;48:1042–7. [PubMed] [Google Scholar]
- Morrill JC, Mebus CA, Peters CJ. Safety and efficacy of a mutagen-attenuated Rift Valley fever virus vaccine in cattle. Am J Vet Res. 1997a;58:1104–9. [PubMed] [Google Scholar]
- Morrill JC, Mebus CA, Peters CJ. Safety of a mutagen-attenuated Rift Valley fever virus vaccine in fetal and neonatal bovids. Am J Vet Res. 1997b;58:1110–4. [PubMed] [Google Scholar]
- Morrill JC, Peters CJ. Pathogenicity and neurovirulence of a mutagen-attenuated Rift Valley fever vaccine in rhesus monkeys. Vaccine. 2003;21:2994–3002. doi: 10.1016/s0264-410x(03)00131-2. [DOI] [PubMed] [Google Scholar]
- Morvan J, Rollin PE, Laventure S, Rakotoarivony I, Roux J. Rift Valley fever epizootic in the central highlands of Madagascar. Res Virol. 1992;143:407–15. doi: 10.1016/s0923-2516(06)80134-2. [DOI] [PubMed] [Google Scholar]
- Morvan J, Saluzzo JF, Fontenille D, Rollin PE, Coulanges P. Rift Valley fever on the east coast of Madagascar. Res Virol. 1991;142:475–82. doi: 10.1016/0923-2516(91)90070-j. [DOI] [PubMed] [Google Scholar]
- Moutailler S, Krida G, Schaffner F, Vazeille M, Failloux AB. Potential vectors of Rift Valley fever virus in the Mediterranean region. Vector Borne Zoonotic Dis. 2008;8:749–53. doi: 10.1089/vbz.2008.0009. [DOI] [PubMed] [Google Scholar]
- Muller R, Saluzzo JF, Lopez N, Dreier T, Turell M, Smith J, Bouloy M. Characterization of clone 13, a naturally attenuated avirulent isolate of Rift Valley fever virus, which is altered in the small segment. Am J Trop Med Hyg. 1995;53:405–11. doi: 10.4269/ajtmh.1995.53.405. [DOI] [PubMed] [Google Scholar]
- Nakitare GW, Elliott RM. Expression of the Bunyamwera virus M genome segment and intracellular localization of NSm. Virology. 1993;195:511–20. doi: 10.1006/viro.1993.1402. [DOI] [PubMed] [Google Scholar]
- Naslund J, Lagerqvist N, Habjan M, Lundkvist A, Evander M, Ahlm C, Weber F, Bucht G. Vaccination with virus-like particles protects mice from lethal infection of Rift Valley Fever Virus. Virology. 2009;385:409–15. doi: 10.1016/j.virol.2008.12.012. [DOI] [PubMed] [Google Scholar]
- Neumann G, Kawaoka Y. Reverse genetics systems for the generation of segmented negative-sense RNA viruses entirely from cloned cDNA. Curr Top Microbiol Immunol. 2004;283:43–60. doi: 10.1007/978-3-662-06099-5_2. [DOI] [PubMed] [Google Scholar]
- Neumann G, Watanabe T, Ito H, Watanabe S, Goto H, Gao P, Hughes M, Perez DR, Donis R, Hoffmann E, Hobom G, Kawaoka Y. Generation of influenza A viruses entirely from cloned cDNAs. Proc Natl Acad Sci U S A. 1999;96:9345–50. doi: 10.1073/pnas.96.16.9345. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Neumann G, Whitt MA, Kawaoka Y. A decade after the generation of a negative-sense RNA virus from cloned cDNA - what have we learned? J Gen Virol. 2002;83:2635–62. doi: 10.1099/0022-1317-83-11-2635. [DOI] [PubMed] [Google Scholar]
- Niklasson B, Peters CJ, Bengtsson E, Norrby E. Rift Valley fever virus vaccine trial: study of neutralizing antibody response in humans. Vaccine. 1985;3:123–7. doi: 10.1016/0264-410x(85)90061-1. [DOI] [PubMed] [Google Scholar]
- Noad R, Roy P. Virus-like particles as immunogens. Trends Microbiol. 2003;11:438–44. doi: 10.1016/s0966-842x(03)00208-7. [DOI] [PubMed] [Google Scholar]
- Ogawa Y, Sugiura K, Kato K, Tohya Y, Akashi H. Rescue of Akabane virus (family Bunyaviridae) entirely from cloned cDNAs by using RNA polymerase I. J Gen Virol. 2007;88:3385–90. doi: 10.1099/vir.0.83173-0. [DOI] [PubMed] [Google Scholar]
- Overby AK, Pettersson RF, Neve EP. The glycoprotein cytoplasmic tail of Uukuniemi virus (Bunyaviridae) interacts with ribonucleoproteins and is critical for genome packaging. J Virol. 2007a;81:3198–205. doi: 10.1128/JVI.02655-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Overby AK, Popov V, Neve EP, Pettersson RF. Generation and analysis of infectious virus-like particles of uukuniemi virus (bunyaviridae): a useful system for studying bunyaviral packaging and budding. J Virol. 2006;80:10428–35. doi: 10.1128/JVI.01362-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Overby AK, Popov VL, Pettersson RF, Neve EP. The cytoplasmic tails of Uukuniemi Virus (Bunyaviridae) G(N) and G(C) glycoproteins are important for intracellular targeting and the budding of virus-like particles. J Virol. 2007b;81:11381–91. doi: 10.1128/JVI.00767-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pavlovic J, Haller O, Staeheli P. Human and mouse Mx proteins inhibit different steps of the influenza virus multiplication cycle. J Virol. 1992;66:2564–9. doi: 10.1128/jvi.66.4.2564-2569.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perrone LA, Narayanan K, Worthy M, Peters CJ. The S segment of Punta Toro virus (Bunyaviridae, Phlebovirus) is a major determinant of lethality in the Syrian hamster and codes for a type I interferon antagonist. J Virol. 2007;81:884–92. doi: 10.1128/JVI.01074-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peters CJ, Reynolds JA, Slone TW, Jones DE, Stephen EL. Prophylaxis of Rift Valley fever with antiviral drugs, immune serum, an interferon inducer, and a macrophage activator. Antiviral Res. 1986;6:285–97. doi: 10.1016/0166-3542(86)90024-0. [DOI] [PubMed] [Google Scholar]
- Petrovsky N. Novel human polysaccharide adjuvants with dual Th1 and Th2 potentiating activity. Vaccine. 2006;24(Suppl 2):S2-26-9. doi: 10.1016/j.vaccine.2005.01.107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Petrovsky N, Aguilar JC. Vaccine adjuvants: current state and future trends. Immunol Cell Biol. 2004;82:488–96. doi: 10.1111/j.0818-9641.2004.01272.x. [DOI] [PubMed] [Google Scholar]
- Pittman PR, Liu CT, Cannon TL, Makuch RS, Mangiafico JA, Gibbs PH, Peters CJ. Immunogenicity of an inactivated Rift Valley fever vaccine in humans: a 12-year experience. Vaccine. 1999;18:181–9. doi: 10.1016/s0264-410x(99)00218-2. [DOI] [PubMed] [Google Scholar]
- Plotkin SA. Vaccines: past, present and future. Nat Med. 2005;11:S5–11. doi: 10.1038/nm1209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pollitt E, Zhao J, Muscat P, Elliott RM. Characterization of Maguari orthobunyavirus mutants suggests the nonstructural protein NSm is not essential for growth in tissue culture. Virology. 2006;348:224–32. doi: 10.1016/j.virol.2005.12.026. [DOI] [PubMed] [Google Scholar]
- Pretorius A, Oelofsen MJ, Smith MS, van der Ryst E. Rift Valley fever virus: a seroepidemiologic study of small terrestrial vertebrates in South Africa. Am J Trop Med Hyg. 1997;57:693–8. doi: 10.4269/ajtmh.1997.57.693. [DOI] [PubMed] [Google Scholar]
- Radecke F, Spielhofer P, Schneider H, Kaelin K, Huber M, Dotsch C, Christiansen G, Billeter MA. Rescue of measles viruses from cloned DNA. EMBO J. 1995;14:5773–84. doi: 10.1002/j.1460-2075.1995.tb00266.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Randall R, Binn LN, Harrison VR. Immunization against Rift Valley Fever Virus. Studies on the Immunogenicity of Lyophilized Formalin-Inactivated Vaccine. J Immunol. 1964;93:293–9. [PubMed] [Google Scholar]
- Randall R, Gibbs CJ, Jr, Aulisio CG, Binn LN, Harrison VR. The development of a formalin-killed Rift Valley fever virus vaccine for use in man. J Immunol. 1962;89:660–71. [PubMed] [Google Scholar]
- Reichelt M, Stertz S, Krijnse-Locker J, Haller O, Kochs G. Missorting of LaCrosse virus nucleocapsid protein by the interferon-induced MxA GTPase involves smooth ER membranes. Traffic. 2004;5:772–84. doi: 10.1111/j.1600-0854.2004.00219.x. [DOI] [PubMed] [Google Scholar]
- Reynes JM. RVF OIE Bloemfontein. Institut Pasteur de Madagascar; Feb 16–18, 2009. Rift Valley Fever in Madagascar 2008 Laboratory Surveillance. [Google Scholar]
- Rossi GR, Mautino MR, Unfer RC, Seregina TM, Vahanian N, Link CJ. Effective treatment of preexisting melanoma with whole cell vaccines expressing alpha(1,3)-galactosyl epitopes. Cancer Res. 2005a;65:10555–61. doi: 10.1158/0008-5472.CAN-05-0627. [DOI] [PubMed] [Google Scholar]
- Rossi GR, Unfer RC, Seregina T, Link CJ. Complete protection against melanoma in absence of autoimmune depigmentation after rejection of melanoma cells expressing alpha(1,3)galactosyl epitopes. Cancer Immunol Immunother. 2005b;54:999–1009. doi: 10.1007/s00262-005-0667-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roy P, Noad R. Virus-Like Particles as a Vaccine Delivery System: Myths and Facts. Landes Bioscience/Springer Science; Austin/New York: 2008. [Google Scholar]
- Sanchez AB, de la Torre JC. Rescue of the prototypic Arenavirus LCMV entirely from plasmid. Virology. 2006;350:370–80. doi: 10.1016/j.virol.2006.01.012. [DOI] [PubMed] [Google Scholar]
- Sardelis MR, Turell MJ, Dohm DJ, O’Guinn ML. Vector competence of selected North American Culex and Coquillettidia mosquitoes for West Nile virus. Emerg Infect Dis. 2001;7:1018–22. doi: 10.3201/eid0706.010617. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schmaljohn C, Hooper JW. Bunyaviridae: the viruses and their replication. Vol. 2. Lippincott Williams & Wilkins; Philadelphia: 2001. [Google Scholar]
- Schmaljohn CS, Parker MD, Ennis WH, Dalrymple JM, Collett MS, Suzich JA, Schmaljohn AL. Baculovirus expression of the M genome segment of Rift Valley fever virus and examination of antigenic and immunogenic properties of the expressed proteins. Virology. 1989;170:184–92. doi: 10.1016/0042-6822(89)90365-6. [DOI] [PubMed] [Google Scholar]
- Schnell MJ, Mebatsion T, Conzelmann KK. Infectious rabies viruses from cloned cDNA. Embo J. 1994;13:4195–203. doi: 10.1002/j.1460-2075.1994.tb06739.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shoemaker T, Boulianne C, Vincent MJ, Pezzanite L, Al-Qahtani MM, Al-Mazrou Y, Khan AS, Rollin PE, Swanepoel R, Ksiazek TG, Nichol ST. Genetic analysis of viruses associated with emergence of Rift Valley fever in Saudi Arabia and Yemen, 2000–01. Emerg Infect Dis. 2002;8:1415–20. doi: 10.3201/eid0812.020195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sidwell RW, Smee DF. Viruses of the Bunya- and Togaviridae families: potential as bioterrorism agents and means of control. Antiviral Res. 2003;57:101–11. doi: 10.1016/s0166-3542(02)00203-6. [DOI] [PubMed] [Google Scholar]
- Silva DG, Cooper PD, Petrovsky N. Inulin-derived adjuvants efficiently promote both Th1 and Th2 immune responses. Immunol Cell Biol. 2004;82:611–6. doi: 10.1111/j.1440-1711.2004.01290.x. [DOI] [PubMed] [Google Scholar]
- Slavin S, Or R, Weiss L, Steiner-Zalz D, Cividalli G, Brautbar C, Weshler Z, Galun E, Samuels S, Galili D, et al. Elimination of graft versus host disease in matched allogeneic leukemic transplant recipients using CAMPATH-1. Adv Exp Med Biol. 1985;186:813–8. doi: 10.1007/978-1-4613-2463-8_98. [DOI] [PubMed] [Google Scholar]
- Smithburn KC, Haddow AJ, Gillett JD. Rift Valley fever; isolation of the virus from wild mosquitoes. Br J Exp Pathol. 1948;29:107–21. [PMC free article] [PubMed] [Google Scholar]
- Spik K, Shurtleff A, McElroy AK, Guttieri MC, Hooper JW, SchmalJohn C. Immunogenicity of combination DNA vaccines for Rift Valley fever virus, tick-borne encephalitis virus, Hantaan virus, and Crimean Congo hemorrhagic fever virus. Vaccine. 2006;24:4657–66. doi: 10.1016/j.vaccine.2005.08.034. [DOI] [PubMed] [Google Scholar]
- Spiropoulou CF, Goldsmith CS, Shoemaker TR, Peters CJ, Compans RW. Sin Nombre virus glycoprotein trafficking. Virology. 2003;308:48–63. doi: 10.1016/s0042-6822(02)00092-2. [DOI] [PubMed] [Google Scholar]
- Squinto SP. Xenogeneic organ transplantation. Curr Opin Biotechnol. 1996;7:641–5. doi: 10.1016/s0958-1669(96)80076-0. [DOI] [PubMed] [Google Scholar]
- Stein DA. Inhibition of RNA virus infections with peptide-conjugated morpholino oligomers. Curr Pharm Des. 2008;14:2619–34. doi: 10.2174/138161208786071290. [DOI] [PubMed] [Google Scholar]
- Stein DA, Huang CY, Silengo S, Amantana A, Crumley S, Blouch RE, Iversen PL, Kinney RM. Treatment of AG129 mice with antisense morpholino oligomers increases survival time following challenge with dengue 2 virus. J Antimicrob Chemother. 2008;62:555–65. doi: 10.1093/jac/dkn221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Swanepoel RCJ. Infectious Diseases of Livestock. Oxford University Press; Cape Town: 2004a. [Google Scholar]
- Swanepoel RCJ. Rift Valley Fever. Oxford University Press; Cape Town: 2004b. [Google Scholar]
- Swanepoel R, Coetzer JAW. Rift Valley fever. In: Coetzer JAW, Tustin RC, editors. Infectious diseases of livestock with special reference to South Africa. Oxford University Press; Southern Africa, Cape Town: 2003. [Google Scholar]
- Swenson ES, Price JG, Brazelton T, Krause DS. Limitations of green fluorescent protein as a cell lineage marker. Stem Cells. 2007;25:2593–600. doi: 10.1634/stemcells.2007-0241. [DOI] [PubMed] [Google Scholar]
- Tangy F, Naim HY. Live attenuated measles vaccine as a potential multivalent pediatric vaccination vector. Viral Immunol. 2005;18:317–26. doi: 10.1089/vim.2005.18.317. [DOI] [PubMed] [Google Scholar]
- Thall A, Galili U. Distribution of Gal alpha 1----3Gal beta 1----4GlcNAc residues on secreted mammalian glycoproteins (thyroglobulin, fibrinogen, and immunoglobulin G) as measured by a sensitive solid-phase radioimmunoassay. Biochemistry. 1990;29:3959–65. doi: 10.1021/bi00468a024. [DOI] [PubMed] [Google Scholar]
- Thalmeier K, Huss R. Highly efficient retroviral gene transfer into immortalized CD34(−) cells and organ distribution after transplantation into NOD/SCID mice. Cytotherapy. 2001;3:245–51. doi: 10.1080/146532401317070871. [DOI] [PubMed] [Google Scholar]
- Thalmeier K, Meissner P, Moosmann S, Sagebiel S, Wiest I, Huss R. Mesenchymal differentiation and organ distribution of established human stromal cell lines in NOD/SCID mice. Acta Haematol. 2001;105:159–65. doi: 10.1159/000046559. [DOI] [PubMed] [Google Scholar]
- Turell MJ, Bailey CL. Transmission studies in mosquitoes (Diptera: Culicidae) with disseminated Rift Valley fever virus infections. J Med Entomol. 1987;24:11–8. doi: 10.1093/jmedent/24.1.11. [DOI] [PubMed] [Google Scholar]
- Turell MJ, Dohm DJ, Mores CN, Terracina L, Wallette DL, Jr, Hribar LJ, Pecor JE, Blow JA. Potential for North American mosquitoes to transmit Rift Valley fever virus. J Am Mosq Control Assoc. 2008;24:502–7. doi: 10.2987/08-5791.1. [DOI] [PubMed] [Google Scholar]
- Turell MJ, Kay BH. Susceptibility of selected strains of Australian mosquitoes (Diptera: Culicidae) to Rift Valley fever virus. J Med Entomol. 1998;35:132–5. doi: 10.1093/jmedent/35.2.132. [DOI] [PubMed] [Google Scholar]
- Turell MJ, Perkins PV. Transmission of Rift Valley fever virus by the sand fly, Phlebotomus duboscqi (Diptera: Psychodidae) Am J Trop Med Hyg. 1990;42:185–8. doi: 10.4269/ajtmh.1990.42.185. [DOI] [PubMed] [Google Scholar]
- Unfer RC, Hellrung D, Link CJ., Jr Immunity to the alpha(1,3)galactosyl epitope provides protection in mice challenged with colon cancer cells expressing alpha(1,3)galactosyl-transferase: a novel suicide gene for cancer gene therapy. Cancer Res. 2003;63:987–93. [PubMed] [Google Scholar]
- USDA. Agricultural Bioterrorism Protection Act of 2002; possession, use, and transfer of biological agents and toxins. In: USDA A, editor. Fed Register. 2005. pp. 13241–13292. Part II. 7 CFR Part 331 and 9 CFR Part 121. [Google Scholar]
- Vialat P, Billecocq A, Kohl A, Bouloy M. The S segment of Rift valley fever phlebovirus (Bunyaviridae) carries determinants for attenuation and virulence in mice. J Virol. 2000;74:1538–43. doi: 10.1128/jvi.74.3.1538-1543.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wallace DB, Ellis CE, Espach A, Smith SJ, Greyling RR, Viljoen GJ. Protective immune responses induced by different recombinant vaccine regimes to Rift Valley fever. Vaccine. 2006;24:7181–9. doi: 10.1016/j.vaccine.2006.06.041. [DOI] [PubMed] [Google Scholar]
- Walpita P, Flick R. Reverse genetics of negative-stranded RNA viruses: a global perspective. FEMS Microbiol Lett. 2005;244:9–18. doi: 10.1016/j.femsle.2005.01.046. [DOI] [PubMed] [Google Scholar]
- Whelan SP, Ball LA, Barr JN, Wertz GT. Efficient recovery of infectious vesicular stomatitis virus entirely from cDNA clones. Proc Natl Acad Sci U S A. 1995;92:8388–92. doi: 10.1073/pnas.92.18.8388. [DOI] [PMC free article] [PubMed] [Google Scholar]
- WHO. Outbreaks of Rift Valley fever in Kenya, Somalia and United Republic of Tanzania, December 2006–April 2007. Wkly Epidemiol Rec. 2007;82:169–78. [PubMed] [Google Scholar]
- Won S, Ikegami T, Peters CJ, Makino S. NSm and 78-kilodalton proteins of Rift Valley fever virus are nonessential for viral replication in cell culture. J Virol. 2006;80:8274–8. doi: 10.1128/JVI.00476-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Won S, Ikegami T, Peters CJ, Makino S. NSm protein of Rift Valley fever virus suppresses virus-induced apoptosis. J Virol. 2007;81:13335–45. doi: 10.1128/JVI.01238-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Woods CW, Karpati AM, Grein T, McCarthy N, Gaturuku P, Muchiri E, Dunster L, Henderson A, Khan AS, Swanepoel R, Bonmarin I, Martin L, Mann P, Smoak BL, Ryan M, Ksiazek TG, Arthur RR, Ndikuyeze A, Agata NN, Peters CJ. An outbreak of Rift Valley fever in Northeastern Kenya, 1997–98. Emerg Infect Dis. 2002;8:138–44. doi: 10.3201/eid0802.010023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang B, Dong H, Stein DA, Shi PY. Co-selection of West Nile virus nucleotides that confer resistance to an antisense oligomer while maintaining long-distance RNA/RNA base pairings. Virology. 2008;382:98–106. doi: 10.1016/j.virol.2008.08.044. [DOI] [PMC free article] [PubMed] [Google Scholar]