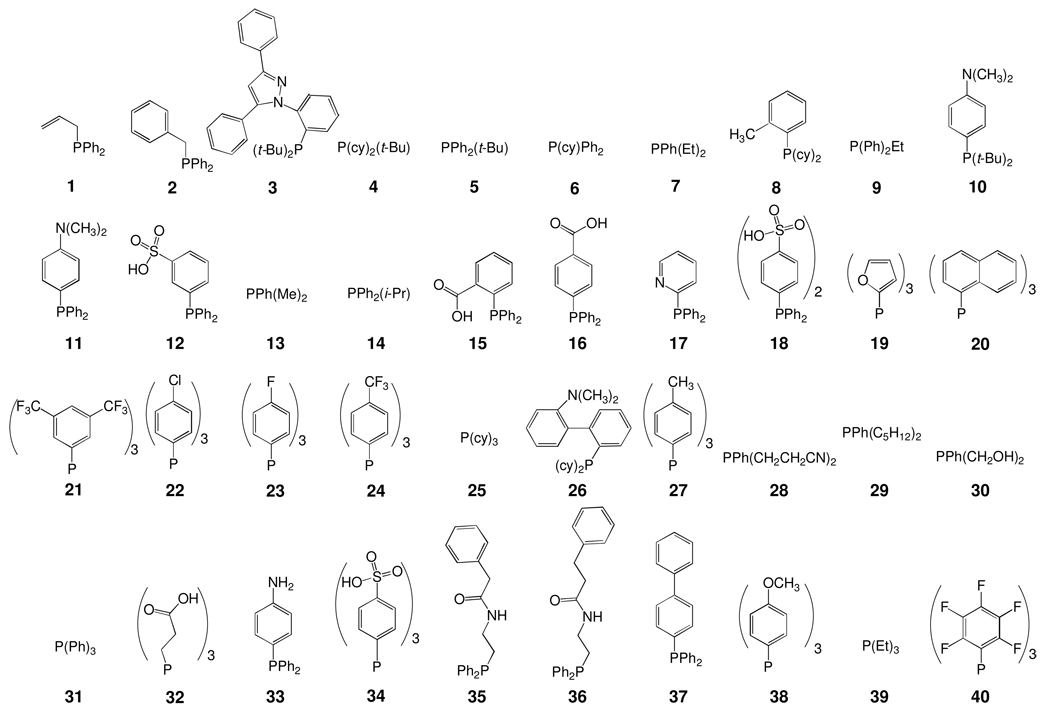
Abstract
Therapeutic inhibition of protein tyrosine phosphatase activity is a compelling yet challenging approach to the treatment of human disease. Towards this end, a library of 40 gold complexes with the general formula R3P–Au–Cl was screened to identify novel inhibitors of PTP activity. The most promising inhibitor obtained for the lymphoid tyrosine phosphatase LYP, (2-pyridine)(Ph2)P–Au–Cl, is one of the most potent and selective LYP inhibitors identified to date with an IC50 of 1.5 ± 0.3 µM, 10-fold selectivity for LYP over PTP-PEST, HePTP and CD45 in vitro and activity in cellular studies as well.
Introduction
Reversible tyrosine phosphorylation results from the balanced action of protein tyrosine kinases (PTKs) and protein tyrosine phosphatases (PTPs) and plays a fundamental role in cellular signaling pathways, controlling many aspects of human physiology from cell growth and differentiation to metabolism and immunology.1, 2 Recent findings have ignited significant interest in PTPs as therapeutic targets. For example, PTP1B inhibitors are highly sought after for the treatment of obesity and type 2 diabetes3, 4 and the lymphoid tyrosine phosphatase (LYP) has recently been identified as a promising new drug target in autoimmunity.5–9 However, PTP inhibitors have yet to reach the clinic due in large part to the difficulty of achieving selectivity for the PTP of interest.10 This difficulty stems from the highly conserved nature of the PTP phosphotyrosine binding pocket.11, 12 Adding to the challenge, many therapeutically relevant PTPs have human homologues whose inhibition would be detrimental; for example, the catalytic domains of PTP1B and LYP exhibit over 70% sequence identity with those of TCPTP and PTP-PEST, respectively.
PTP1B is arguably the most actively sought after PTP as a therapeutic target. Many potent PTP1B inhibitors have been developed over the past 10 years including benzofuran and benzothiophene biphenyls13 as well as trans-β-nitrostyrene derivatives14 among others, which can be found in recent reviews.15, 16 Achieving selective inhibition of PTP1B over the highly similar TCPTP has been met with some success, 17,18 however, this success has not yet reached the clinic. The PTP1B inhibitor ertiprotafib was a promising drug candidate,19 but the compound did not make it past phase II clinical trials due to poor efficacy and undesired side effects.20 On the other hand, the therapeutic inhibition of LYP has only begun to be explored, and only a few LYP inhibitors have been developed.21–23 An inhibitor with modest selectivity for LYP over PTP1B has been reported, but selectivity for LYP over the highly homologous PTP-PEST was not discussed.23 Another recent paper reported a potent submicromolar inhibitor of LYP, but no selectivity information was published for this compound.22 Research conducted in our lab found that Au(I)-N-heterocyclic carbene complexes were relatively potent inhibitors of both LYP and PTP-PEST.21
Gold(I) complexes have long been used to treat the autoimmune disorder rheumatoid arthritis24–26 and have been shown to inhibit the activity PTPs including both LYP and PTP1B.21, 27 PTPs contain a catalytically essential cysteine residue in the active site with a lowered pKa value.28 This cysteine is a likely target of the gold drugs due to the thiophilicity of Au(I).25, 29 Au(I) has been shown to be a competitive, reversible inhibitor of PTP activity, presumably due to reversible binding of Au(I) to the active site cysteine residue.21, 27 Although no PTP-Au(I) structures are available, there is structural precedent for the coordination of Au(I) to nucleophilic cysteine residues in enzyme active sites.30, 31 Because of the ability of Au(I) compounds to inhibit PTP activity, the relevance of Au(I) in the treatment of autoimmunity and the importance of PTP activity in autoimmunity,32 we reasoned that a library of Au(I)-based compounds would be an appropriate starting place for the development of PTP-selective inhibitors. To this end, we designed a library of 40 potential inhibitors, all with the general molecular formula PR3-Au-Cl (Figure 1). This library of gold compounds was derived from the antiarthritic drug auranofin. Here we describe a comparative screen of this Au(I) compound library for novel PTP inhibitors using both a traditional PTP substrate (difluoromethylumbelliferyl phosphate, DiFMUP)33 and fluorogenic, peptide-based PTP substrates designed in our laboratory. Interestingly, while some of the hits exhibited similar potencies and selectivities with both substrates, others showed notable differences. This was true of inhibition of all of the PTPs tested. From these screens, we were able to identify Au(I)-based inhibitors with some selectivity for either LYP, PTP1B and TCPTP. In addition, the most potent and selective LYP inhibitors in vitro were also validated as potent and selective LYP inhibitors in T cells.
Figure 1.
Chemical structures of the phosphine ligands included in the library of Au(I) complexes with the general formula R3P–Au–Cl.
Results and Discussion
Commercially available phosphine ligands were chosen such that we could achieve a diverse set of compounds with varying charge, electronic and steric properties (Figure 1). The PR3-Au(I)-Cl compounds were initially synthesized in parallel in a similar fashion to a previously published procedure for individual compounds34 and tested as isolated without further purification. For the screening, both the traditional fluorogenic PTP substrate DiFMUP and the peptide substrates Ac-ARLIEDNE-(pCAP)-TAREG-NH2 (peptide 1, used for LYP and PTP-PEST) and Ac-DIDE-(pCAP)-LAA-NH2 (peptide 2, used for PTP1B and TCPTP) were used. It has been proposed that substrates such as peptides displaying extended interactions with the enzyme outside the active site would be optimal substrates for PTP inhibitor screening.12 The sequence of peptide 1 is based on the sequence surrounding the reversibly phosphorylated tyrosine residue 394 in Lck, a known substrate of LYP that may also be turned over by PTP-PEST in vitro.32, 35 The sequence of peptide 2 is derived from the sequence surrounding tyrosine 992 in the EGF receptor, a known substrate of PTP1B and TCPTP, and has previously been validated as a substrate of TCPTP in our laboratory.36 In both cases, the central phosphotyrosine residue is replaced with the fluorogenic phosphotyrosine mimic, phosphocoumaryl propionic acid (pCAP), which we have previously shown to be an excellent substrate of PTPs with kinetic parameters very similar to those of phosphotyrosine.37
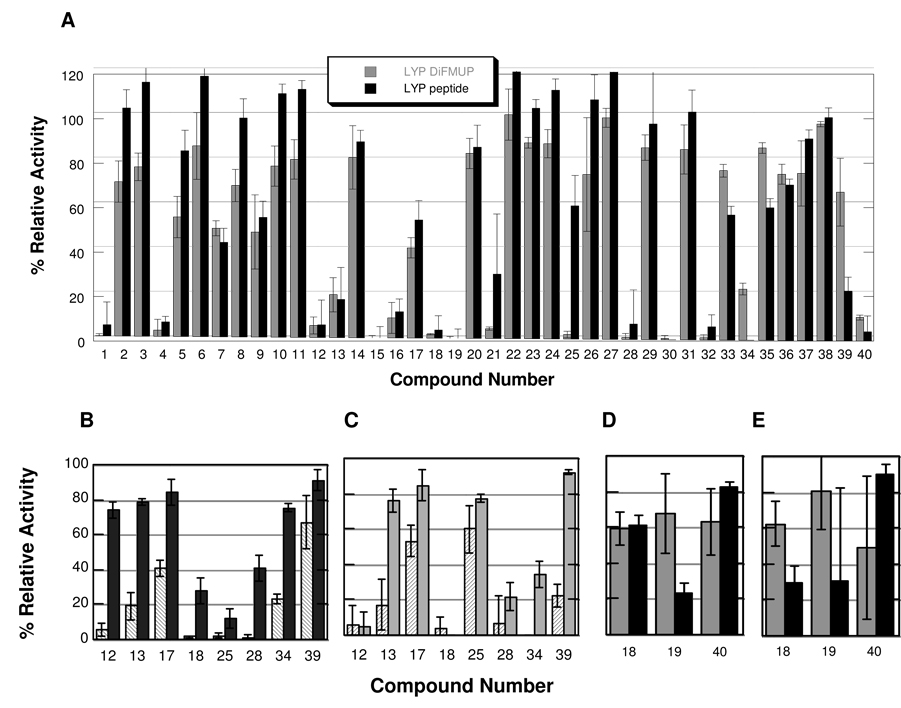
The objectives of this study were two-fold: first, to identify gold-based, PTP-selective inhibitors and second, to investigate potential differences between the fluorogenic peptide-based assay and the small-molecule fluorogenic assay in inhibitor screening. A screening for LYP inhibition by the library of gold compounds using DiFMUP and peptide 1 can be seen in Figure 2A where approximately 40% of all compounds tested inhibited LYP activity by more than 60%. Because the library of inhibitors was relatively small, a second comparative screen for PTP-PEST inhibition was carried out and graphed next to the LYP screening results (selected inhibitors are shown in Figures 2B and 2C, full results can be found in Figure S1 in the supporting information). While many compounds inhibit both LYP and PTP-PEST equally (Figure S1), several of the hits shown in Figure 2B and C demonstrate significant LYP selective inhibition. Complexes 13, 17 and 34 were found to be more potent inhibitors of LYP than PTP-PEST regardless of the substrate used, while other complexes such as 12 and 25 demonstrated substrate-dependent inhibition differences and appear LYP-selective when DiFMUP is used in the assay, but not when the peptide is used. Finally, compound 39 appears LYP selective with peptide 1, but not with DiFMUP (Figure 2B and C) as it seems to inhibit LYP more potently with peptide substrate (Figure 2A). One clear pattern observed is that gold inhibitors with negatively charged sulfates on their phosphine ligands such as 12, 18, and 34 were good inhibitors of PTP activity, which is not surprising considering many previously reported anionic PTP inhibitors.15 Notably, in the absence of Au(I), none of the phosphine ligands inhibited LYP activity up to 100 µM.
Figure 2.
Results of the comparative library screens with all potential inhibitors at a nominal concentration of 5 µM. A) Screening for LYP inhibition using the substrate DiFMUP (grey) and peptide 1 (black). B) Selected hits from a DiFMUP-based screen for LYP (striped) and PTP-PEST (shaded) inhibitors. C) Selected hits from a peptide 1-based screen for LYP (striped) and PTP-PEST (shaded) inhibitors. D) Selected hits from a PTP1B inhibitor screen using the substrates DiFMUP (grey) and peptide 2 (black). E) Selected hits from a TCPTP inhibitor screen using the substrates DiFMUP (grey) and peptide 2 (black).
Comparative screens for PTP1B and TCPTP inhibitors yielded considerably fewer hits than the LYP/PTP-PEST screens (see Figure S2 in the supplementary information for full details). Compound 18 appears to be slightly TCPTP selective with peptide 2 (Figures 2D and 2E), selectivity that would not have been observed in a DiFMUP-only screen. Also comparing substrate screens with PTP1B as seen in Figure 1D, compound 19 appears to inhibit PTP1B better with peptide 2 than with DiFMUP while the reverse seems to be true for compound 40.
Several of the most interesting hits from our initial screens were purified, characterized and carried forward into secondary dose-response screens and counterscreened against other PTPs to obtain data on selectivity. As seen in Table 1, several compounds including 13, 17, 18, 28, 34 and 39 were selective for LYP over the ~ 70% identical PTP-PEST in vitro, confirming the results from the library screen. These compounds, with the exception of 39, were low µM inhibitors of LYP with both substrates and were also selective for LYP over HePTP and CD45, two other PTPs involved in T-cell receptor signaling.38 Compound 25 exhibited a similar trend to that seen in the original screening, inhibiting LYP activity slightly better with DiFMUP compared to peptide 1 substrate as well as exhibiting modest selectivity over PTP-PEST, but was not selective when counterscreened against HePTP. The IC50 data for compound 12 also mirror the trend seen in the initial screen, displaying 35-fold selectivity for LYP over PTP-PEST with DiFMUP but only 2-fold selectivity with peptide 1 as the substrate. Compound 39 was found to be a moderately potent inhibitor that is nearly 4-fold selective for LYP over PTP-PEST with peptide 1 as the substrate, but also showed significant inhibition of CD45. The secondary screening of PTP1B and TCPTP hits is shown in Table 2. Compounds 18 and 40 showed no significant selectivity with DiFMUP as the substrate, but are approximately 2-fold selective for TCPTP and 10-fold selective for PTP1B, respectively, with peptide 2 as the substrate. In addition, compound 19 is selective for PTP1B with both substrates. Although interesting differences in inhibitory potency were observed with PTP1B and TCPTP and the two substrates, other more potent PTP1B inhibitors have been reported in the literature15,16 and the inhibitors obtained in this study were not of sufficient potency to warrant further investigation.
Table 1.
IC50 data for hits from the LYP/PTP-PEST screen in µM.
| Cmpd # | Phosphine | LYP (DiFMUP) |
LYP (peptide 1) |
PEST (DiFMUP) |
PEST (peptide 1) |
HePTP (DiFMUP) |
CD45 (DiFMUP) |
|---|---|---|---|---|---|---|---|
| 12 | P(Ph)2(m-SO3H-Ph) | 2.0±0.1 | 1.9±0.2 | 70±2 | 4.0±0.2 | 40±1 | > 70 |
| 13 | P(Me)2(Ph) | 3.5±0.3 | 2.0±0.3 | 70±2 | 30±2 | > 50 | > 100 |
| 17 | P(Ph)2(2-pyridine) | 5.0±0.2 | 1.5±0.3 | 76±3 | 43±3 | 35±2 | > 150 |
| 18 | P(p-SO3H-Ph)2(Ph) | 2.0±0.1 | 0.85±0.05 | 39±1 | 10±2 | 16±2 | 26±1 |
| 25 | P(cy)3 | 25±1 | 61±1 | 45±1 | > 80 | 20±2 | > 70 |
| 28 | P(EtCN)2(Ph) | 3.5±0.1 | 2.5±0.6 | > 80 | 33±1 | > 50 | > 70 |
| 34 | P(p-SO3H-Ph)3 | 1.5±0.1 | 0.75±0.15 | 15±1 | 3.2±0.2 | 13±3 | 23±1 |
| 39 | P(Et)3 | 33±3 | 26±2 | >150 | 93±9 | 120±5 | 33±3 |
Table 2.
IC50 data for hits from the PTP1B/TCPTP screen in µM.
| Cmpd # | Phosphine | PTP1B (DiFMUP) |
PTP1B (peptide 2) |
TCPTP (DiFMUP) |
TCPTP (peptide 2) |
|---|---|---|---|---|---|
| 18 | P(p-SO3H-Ph)2(Ph) | 16±1 | 15±1 | 16±1 | 7.5±0.3 |
| 19 | P(2-furyl)3 | 0.33±0.02 | 0.20±0.02 | 2.4±0.2 | 2.2±0.3 |
| 40 | P(F5Ph)3 | 7.5±0.2 | 5.4±0.3 | 6.7±0.2 | 57±1 |
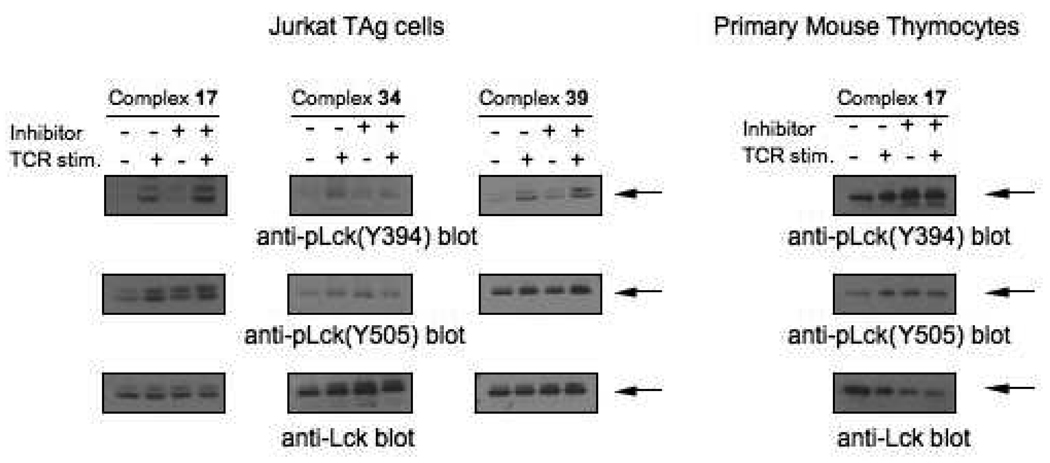
To date, there have been few LYP inhibitors reported21–23 and although some exhibit moderate selectivity for LYP over other PTPs, no inhibitor with selectivity for LYP over the highly homologous PTP-PEST has been reported. Gold(I) compounds are competitive, reversible inhibitors of PTP activity21, 27 and are likely to interact directly with cysteine residue(s) in the active site. In order to more fully characterize the biological relevance of Au(I)-mediated PTP inhibition, three representative compounds from the library of Au(I) phosphine compounds were chosen for further investigation in cellular inhibition studies. Compound 17 showed considerable potency and was one of the most selective inhibitors identified with approximately 10-fold selectivity for LYP over PTP-PEST, HePTP and CD45 in vitro. Compound 34 was the most potent LYP inhibitor identified and compound 39 would not have been identified as a hit in a conventional, DiFMUP-only screen but is a moderately potent LYP inhibitor when the more biologically relevant peptide 1 is used as the substrate. These compounds were introduced into cell-based LYP inhibition screens in the Jurkat T Antigen (JTAg) human T cell line. Early stage T cell receptor (TCR) signaling involves the negative regulation of the kinase Lck by dephosphorylation at position Y394 by LYP as well as its activation by dephosphorylation at Y505 by CD45.35, 39 Therefore, the inhibition of cellular LYP and CD45 activity can be monitored by following the phosphorylation at these two sites on Lck as previously described.21 Figure 3A shows the western blot results of cells incubated in the presence and absence of inhibitor, with and without TCR stimulation. It is clear from these blots that in the TCR stimulated cells (lanes 2 and 4), an increase in phosphorylation at Y394 is seen in the presence of compounds 17 and 39 (lane 4), demonstrating their ability to inhibit LYP activity in cells. There is no change in phosphorylation levels at position Y505 of Lck in the presence of 17, but there is an increase in this band when 39 is used, supporting the in vitro data which indicates that 17 does not significantly inhibit CD45 activity but 39 does. Compound 34 has little effect on the phosphorylation levels of Lck in cells, likely because it does not appreciably penetrate the cells due to its negative charge. As seen in Figure 3B, compound 17 appears to inhibit LYP activity and not CD45 activity in primary mouse thymocytes as well. The data in Figure 3 clearly demonstrate significant, selective inhibition of LYP activity in both cultured and primary T cells, confirming the selective inhibition of LYP activity by compound 17.
Figure 3.
Intracellular inhibition of LYP by gold complexes. (A) Top panels: anti-pLck(Tyr394) immunoblots of lysates of Jurkat TAg cells treated with 50 µM of complexes 17 and 34 and 25 µM of complex 39 (lanes 3 and 4 in each panel) or untreated (lanes 1 and 2 in each panel) and either left unstimulated (lanes 1 and 3 in each panel) or stimulated (lanes 2 and 4 in each panel) with C305 antibodies for 2 min. Middle panels: anti-pLck(Tyr505) blots of the same samples. Bottom panels: anti-Lck blot of the same samples. Arrows indicate the position of Lck (56 KDa) in each panel. (B) Top panel: anti-pLck(Tyr394) immunoblot of lysates of mouse thymocytes treated with 50 µM of compound 17 (lanes 3 and 4 in each panel) or untreated (lanes 1 and 2 in each panel) and either left unstimulated (lanes 1 and 3 in each panel) or stimulated (lanes 2 and 4 in each panel) with biotinylated anti-CD3 and anti-CD4, followed by cross-linking with streptavidin for 1.5 min. Middle panel: anti-pLck(Tyr505) blot of the same sample. Bottom panel: anti-Lck blot of the same sample.
Conclusions
The promise of PTP-targeted therapeutics has yet to be fulfilled due in part to significant challenges in obtaining selective PTP inhibitors as the catalytic domains of this family of enzymes contain high degree of similarity. The data presented here indicate that there may be an advantage to using peptide-based substrates in PTP inhibitor screens. In addition, several LYP-selective inhibitors were identified from a library of Au(I)-phosphine complexes. Interestingly, several of these inhibitors showed significant selectivity for LYP over the highly homologous PTP-PEST, something that had not been reported to date. Structural characterization of the Au(I)-LYP adduct is underway to provide more insight into this selectivity. One of the best LYP inhibitors, compound 17, also demonstrated potency and selectivity in both JTAg cells as well as primary mouse thymocytes. The leads and information gained through this study should aid in the further development of LYP-selective inhibitors for therapeutic application.
Experimental Section
General Considerations
All reagents were obtained from commercial sources and used without further purification. The library of Au(I)-phosphine complexes was synthesized using a FlexChem Synthesis System with a 48-well polypropylene block (SciGene). The pCAP residue and pCAP-containing peptides were synthesized as described previously.37 All 1H and 31P NMR spectra were recorded on a Mercury 400 MHz spectrometer. 1H NMR spectra were referenced to residual solvent peaks or TMS (0.00 ppm) and 31P NMR spectra were referenced to an 85% phosphoric acid external standard (0.00 ppm). Column chromatography was performed on EMD silica gel (60–200 mesh). Thin layer chromatography was performed on EMD silica gel 60 aluminum precoated plates (0.2mm thickness). The compounds tested in cells (17, 34 and 39) were all greater than 95% pure as evidenced by elemental analysis (Atlantic Microlab, Norcross, GA, compounds 17 and 34) or certified by the supplier (Sigma Aldrich, compound 39). The catalytic domains of human recombinant CD45, TCPTP and PTP1B were obtained from Biomol. The modified pBAD plasmid encoding the catalytic domain of HePTP (aa 44–339) in frame with a non-cleavable 6×His tag was a kind gift of Lutz Tautz.37 cDNA fragments encoding the catalytic domains of LYP (aa 2–309) and PTP-PEST (aa. 2–323) were cloned between the BamH1 and the Xho1 sites of the pET28a plasmid (Novagen) in frame with a cleavable N-terminal 6×His-tag. Recombinant proteins were purified from lysates of IPTG-induced E. coli BL21 cells by affinity chromatography on Ni-nitrilotriacetic acid columns. 6×HisHePTP was eluted using 250 mM imidazole. Untagged LYP and PTP-PEST were eluted by incubating columns with thrombin, followed by removal of thrombin from the protein preparation by a second chromatography step on benzamidine columns. Fluorescence data were collected on a Molecular Devices Spectramax M5 multimode plate reader with excitation and emission at 360 nm and 455 nm, respectively.
Au(I)-Phosphine Library Synthesis
The following general procedure was used in the synthesis of complexes 1–32. An approximately 80 mg/mL stock solution of aqueous NaAuCl4 was prepared. Approximately 15 mL of a 0.325 M aqueous solution of 2,2′-thiodiethanol was added dropwise over 20 min and the yellow solution was allowed to stir at room temperature until it turned colorless, indicating that all of the Au(III) was reduced to Au(I). The final concentration of gold(I) was 20 mg/mL. Then, a 0.497 mL (0.05 mmol Au) aliquot of the Au(I)/ 2,2′-thiodiethanol solution was added to each well of a 48-well polypropylene FlexChem Synthesis block. Phosphine ligands (0.05 mmol) dissolved in acetone (1–11, 13–17, 19, and 21–31), distilled water (12, 18, and 32), or chloroform (20) were then added dropwise to the individual wells while the block was gently rocking. The synthesis block was then sealed and the reactions were allowed to mix at room temperature on a rocking platform for 18 h with additional random inverting and manual shaking. Samples were filtered through the medium porosity frit of the polypropylene block, the wells were washed twice with acetone, water, or chloroform and filtered again. The combined filtrates were diluted to approximately 30 mL with water and lyophilized to dryness. The complexes were characterized by 31P NMR. A reaction was considered successful if the 31P NMR revealed complete consumption of the starting material with a new peak corresponding to the Au(I)-phosphine complexes appearing downfield relative to the reduced phosphine starting material, distinct from that of the oxidized phosphine. Approximately 20 mM stock solutions of the complexes were made in DMSO. Other complexes were added to our library as well. Complexes 33–38 and 40 were previously synthesized and purified by our lab40 and complex 39 was purchased from Aldrich.
Library Screening with LYP and PTP-PEST
All assays were performed in a buffer containing 50 mM Tris, 100 mM NaCl, 1 mM EDTA, 0.01% Brij 35, 0.1 mM dithiothreitol (DTT) pH 6.5 buffer. Final enzyme concentrations of 5 nM were used for both LYP and PTP-PEST with 1.5 µM 6,8-difluoro-4-methylumbelliferyl phosphate (DiFMUP) and 30 µM of the peptide Ac-ARLIEDNE(pCAP)TAREG-NH2 (peptide 1) used as fluorogenic substrates separately. DMSO stock solutions of inhibitors and substrates were added such that the final amount of DMSO was 5% of the total reaction volume. Prior to the reaction, enzymes were incubated with 0.1 mM DTT for 30 min. Inhibitors were screened at 5 µM in black 96 well plates for 30 min recording fluorescence at 455 nm every 60 sec. Each assay was repeated thrice.
Library Screening with PTP1B and TCPTP
All assays were performed in a buffer containing 50 mM HEPES, 100 mM NaCl, 1 mM EDTA, 0.01% Brij 35, 5 µM DTT, pH 7.1 buffer. Final enzyme concentrations were 0.067 nM for PTP1B and 0.79 nM for TCPTP with 1.5 µM DiFMUP and 60 µM of the peptide Ac-DIDE(pCAP)LAA-NH2 (peptide 2) used as fluorogenic substrates separately. Prior to the reaction, enzymes were incubated with 5 µM DTT for 30 min. All other conditions were the same as described above.
General Individual Complex Synthesis
To a solution of 0.05 mmol NaAuCl4 in 1 mL of distilled water, 0.12 mmol 2,2′-thiodiethanol was added dropwise in about 1 mL distilled water. The reaction was stirred until the solution was colorless. Next, 0.05 mmol of the phosphine was added dropwise in acetone or water. The reaction was allowed to stir for 2 h at room temperature or on ice. The complexes were isolated by vacuum filtration or purified using column chromatography.
Synthesis of 3-(Diphenylphosphino)benzenesulfonic acid-Au(I)-Cl (12)
The aqueous reaction mixture containing complex 12 was lyophilized to dryness. Complex 12 was then purified by column chromatography eluting with 10:1 CHCl3:MeOH, (31% yield). 31P NMR (162 MHz, D2O) δ 33.58 ppm. 1H NMR (400 MHz D2O) δ 6.85–7.50 (12, m), 7.64–7.78 (2H, m). The spectral data obtained are in good agreement with the published data.41
Synthesis of Dimethylphenylphosphine-Au(I)-Cl (13)
The acetone from the reaction mixture was evaporated and the aqueous layer extracted 3× with CHCl3. The organic layer was dried with Na2SO4, filtered and evaporated. The title product was then purified using column chromatography, eluting with hexanes: EtOAc at 2:1/1:1 as a whitish oil (86% yield). 31P NMR (162 MHz, CDCl3) δ 4.58 ppm. 1H NMR (400 MHz CDCl3) δ 1.84–1.86 (3H, s), 1.86–1.88 (3H, s), 7.45–7.59 (3H, m), 7.70–7.79 (2H, m). The spectral data obtained are in good agreement with the published data.42
Synthesis of Diphenyl-2-pyridylphosphine-Au(I)-Cl (17)
Complex 17 was isolated by allowing the acetone to slowly evaporate at room temperature yielding white/colorless crystals (75% yield). 31P NMR (162 MHz, CDCl3) δ 32.92 ppm. 1H NMR (400 MHz CDCl3) δ 7.34–7.42 (1H, m), 7.43–7.57 (6H, m), 7.64–7.74 (4H, m), 7.76–7.85 (1H, m), 7.95–8.04 (1H, t), 8.76–8.81 (1H, d). Anal. Calcd for C17H14AuClNP: C, 41.19; H, 2.85. Found: C, 41.15; H, 2.82.
Synthesis of 4,4′-(Phenylphosphinidene)bis(benzenesulfonic acid)-Au(I)-Cl (18)
Complex 18 was synthesized by stirring 0.05 mmol Au(I)-dimethylsulfide with an equivalent amount of the phosphine ligand in 20:1 MeOH:DCM at room temperature for 24 h. Pure product was then obtained after evaporation of the solvent as a white solid (59% yield). 31P NMR (162 MHz, methanol-d4) δ 33.51 ppm. 1H NMR (400 MHz methanol-d4) δ 7.56–7.68 (9H, m), 7.97–8.02 (4H, dd). The spectral data obtained are in good agreement with the published data.41
Synthesis of Tri-2-furylphosphine-Au(I)-Cl (19)
Complex 19 was isolated by rotary evaporation of the acetone from the reaction mixture and filtering the resulting white solid from the aqueous solution (41% yield). 31P NMR (162 MHz, CDCl3) δ −29.10 ppm. 1H NMR (400 MHz CDCl3) δ 6.52–6.57 (3H, m), 7.15–7.20 (3H, t), 7.77–7.81 (3H, m). Anal. Calcd for C12H9AuClO3P: C, 31.02; H, 1.95. Found: C, 31.28; H, 1.92.
Synthesis of Tricyclohexylphosphine-Au(I)-Cl (25)
Complex 25 was purified in a similar manner to 13 eluting using column chromatography with hexanes: EtOAc at 10:1.5 as a whitish oil (7% yield). 31P NMR (162 MHz, CDCl3) δ 55.06 ppm. 1H NMR (400 MHz CDCl3) δ 1.15–1.36 (10H, m), 1.37–1.57 (7H, m), 1.67–1.78 (3H, m), 1.79–1.91 (6H, m), 1.92–2.06 (7H, m). The spectral data obtained are in good agreement with the published data.43
Synthesis of Bis(2-cyanoethyl)phenylphosphine-Au(I)-Cl (28)
Complex 28 was purified using column chromatography with hexanes: EtOAc at 1:2 and was isolated as an opaque oil (86% yield). 31P NMR (162 MHz, CDCl3) δ 30.24 ppm. 1H NMR (400 MHz CDCl3) δ 2.45–2.81 (8H, m), 7.58–7.72 (3H, m), 7.79–7.87 (2H m). Anal. Calcd for C12H13AuClN2P: C, 32.13; H, 2.92. Found: C, 32.54; H, 2.98.
Individual Inhibition Assays
LYP/PTP-PEST and PTP1B/TCPTP leads were investigated under the same conditions used in the initial library screens with the exception that the final concentration of peptide 1 in the reaction mix was 10 µM. Final concentrations were 1 nM for CD45 and HePTP. The concentrations of Au(I) complexes ranged from 0.2–150 µM and the final amount of DMSO was again 5% of the total reaction volume. Assays were conducted in triplicate following the fluorescence at 455 nm every 60 s for 30 min.
Control Studies
Thiodiethanol did not inhibit LYP activity at a concentration of 150 µM. The phosphine ligands for 12, 13, 17, 18, 25 and 28 were tested at 100 µM and none were shown to inhibit LYP or PTP-PEST activity significantly. The ligands for 18, 19, and 40 were tested at 100 µM and did not significantly inhibit PTP1B or TCPTP activity.
Antibodies
The polyclonal anti-pTyr505-Lck and anti-pTyr416-Src (cross-reactive with LckpTyr394) antibodies were obtained from Cell Signaling Technology, Inc., while the monoclonal anti-Lck antibody was from Santa Cruz Biotechnology. The ECL-Plus Chemiluminescence kit was obtained from GE-Amersham Biosciences.
Cell Based Assays
Cells and Cell treatments
Jurkat T leukemia cells expressing SV-40 large T Antigen (JTAg)44 were kept at logarithmic growth in RPMI 1640 medium supplemented with 10% fetal bovine serum, 2 mM L-glutamine, 1 mM sodium pyruvate, 10 mM HEPES pH 7.3, 2.5 mg/mL D-glucose, and 100 units/mL of penicillin and 100 µg/mL streptomycin.
Studies with JTAg cells
Fixed concentrations of inhibitors (25 or 50 µM) or DMSO (control) were added to 20 × 106 cells suspended in 800 µL RPMI 1640, and incubated for 1 h at room temperature. The volume of DMSO added was held constant at less than 2% of the total volume. JTAg cells pre-incubated with DMSO or inhibitor were divided into 400 µL aliquots containing 10 × 106 cells and stimulated with supernatants of C305 hybridomas45 for 2 min or left untreated. Cells were lysed in 20 mM Tris-HCl pH 7.5, 150 mM NaCl, 5 mM EDTA pH 8.0 containing 1% NP-40, 10 µg/mL aprotinin and leupeptin, 10 µg/mL soybean trypsin inhibitor, 1 mM Na3VO4 and 1 mM phenylmethylsulfonyl fluoride after which lysates were clarified by centrifugation at 13,200 rpm for 20 min. The total protein concentration in each cell lysate was determined by the Bradford protein assay (Bio-Rad) in order to normalize the amount of protein used in SDS-PAGE.
Studies with mouse thymocytes
Thymocytes were isolated from the homogenized thymi of 6-week old C57BL/6 mice (Taconic Farms, Inc.) after depletion of red blood cells by incubating with red blood cell lysis buffer (Sigma) for 1 min at room temperature. A fixed concentration of complex 17 or DMSO (control) was added to 20 × 106 cells suspended in 800 µL RPMI 1640 and incubated for 1 h at room temperature. The volume of DMSO added was less than 2% of the total volume. Thymocytes preincubated with DMSO or inhibitor were divided into 400 µL aliquots containing 10 × 106 cells, treated with biotinylated CD3 and CD4 antibodies for 30 min, and stimulated with the cross-linker, streptavidin for 1.5 min, or left untreated. Cells were then lysed under identical conditions as described for JTAg cells and the total protein concentration in the lysates determined with the Bradford protein assay.
SDS-PAGE and immunoblots
Aliquots of lysates were suspended in SDS sample buffer, heated at 95 °C for 5 min and the boiled samples run on 10% SDS-polyacrylamide gels. Proteins resolved by gel electrophoresis were transferred onto nitrocellulose membranes (Hybond ECL, GE Healthcare), using appropriate dilutions per manufacturers instructions of unconjugated primary antibodies followed by horseradish peroxidase-conjugated secondary antibodies (purchased from GE Healthcare). Blots were developed with the enhanced chemiluminescence detection system, ECL-Plus, following manufacturers directions.
Supplementary Material
Acknowledgements
The authors gratefully acknowledge Dr. Shamila Gunatilleke for her assistance in preparing some of the Au(I) complexes. This work was supported in part by funding from NIH grant number DK080165 awarded to NB and AMB.
Abbreviations used
- PTP
protein tyrosine phosphatase
- PTK
protein tyrosine kinase
- LYP
Lymphoid tyrosine phosphatase
- DiFMUP
difluoromethylumbelliferyl phosphate
- pCAP
phosphocoumaryl propionic acid
- JTAg
Jurkat T Antigen cells
- TCR
T cell receptor
- DTT
dithiothreitol
Footnotes
Supporting Information Available: Figures showing full results from the comparative screens are available as supporting information. This material is available free of charge via the Internet at http://pubs.acs.org.
References
- 1.Hunter T. Protein Kinases and Phosphatases: The yin and yang of protein phosphorylation and signaling. Cell. 1995;80:225–236. doi: 10.1016/0092-8674(95)90405-0. [DOI] [PubMed] [Google Scholar]
- 2.Hunter T. The Role of Tyrosine Phosphorylation in Cell Growth and Disease. Harvey Lect. 1998;94:81–119. [PubMed] [Google Scholar]
- 3.Elchebly M, Payette P, Michaliszyn E, Cromlish W, Collins S, Loy AL, Normandin D, Cheng A, Himms-Hagen J, Chan C-C, Ramachandran C, Gresser MJ, Tremblay ML, Kennedy BP. Increased Insulin Sensitivity and Obesity Resistance in Mice Lacking the Protein Tyrosine Phosphatase-1B Gene. Science. 1999;283:1544–1548. doi: 10.1126/science.283.5407.1544. [DOI] [PubMed] [Google Scholar]
- 4.Zhang Z-Y. Protein Tyrosine Phosphatases: Prospects for Therapeutics. Curr. Opin. Chem. Biol. 2001;5:416–423. doi: 10.1016/s1367-5931(00)00223-4. [DOI] [PubMed] [Google Scholar]
- 5.Bottini N, Musumeci L, Alonso A, Rahmouni S, Nika K, Rostamkhani M, MacMurray J, Meloni GF, Lucarelli P, Pellecchia M, Eisenbarth GS, Comings D, Mustelin T. A Functional Variant of Lymphoid Tyrosine Phosphatase is Associated with Type I Diabetes. Nat. Genet. 2004;36:337–338. doi: 10.1038/ng1323. [DOI] [PubMed] [Google Scholar]
- 6.Gregersen PK. Gaining Insight into PTPN22 and Autoimmunity. Nat. Genet. 2005;37:1300–1302. doi: 10.1038/ng1205-1300. [DOI] [PubMed] [Google Scholar]
- 7.Hinks A, Barton A, John S, Bruce I, Hawkins C, Griffiths CE, Donn R, Thomson W, Silman A, Worthington J. Association Between the PTPN22 Gene and Rheumatoid Arthritis and Juvenile Idiopathic Arthritis in a UK Population: Further Support that PTPN22 is an Autoimmunity Gene. Arthritis Rheum. 2005;52:1694–1699. doi: 10.1002/art.21049. [DOI] [PubMed] [Google Scholar]
- 8.Mustelin T, Vang T, Bottini N. Protein Tyrosine Phosphatases and the Immune Response. Nat. Immunol. 2005;5:43–57. doi: 10.1038/nri1530. [DOI] [PubMed] [Google Scholar]
- 9.Orru V, Tsai SJ, Rueda B, Fiorillo E, Stanford SM, Dasgupta J, Hartiala J, Zhao L, Ortego-Centeno N, D'Alfanso S, Arnett FC, Wu H, Gonzalez-Gay MA, Tsao BP, Pons-Estel B, Alarcon-Riquelme ME, He Y, Zhang Z-Y, Allayee H, Chen XS, Martin J, Bottini N. A Loss-of-Function Variant of PTPN22 is Associated with Reduced Risk of Systemic Lupus Erythematosus. Hum. Mol. Genet. 2009;18:569–579. doi: 10.1093/hmg/ddn363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Tautz L, Mustelin T. Strategies for Developing Protein Tyrosine Phosphatase Inhibitors. Methods. 2007;42:250–260. doi: 10.1016/j.ymeth.2007.02.014. [DOI] [PubMed] [Google Scholar]
- 11.Alonso A, Sasin J, Bottini N, Friedberg I, Friedberg I, Osterman A, Godzik A, Hunter T, Dixon J, Mustelin T. Protein Tyrosine Phosphatases in the Human Genome. Cell. 2004;117:699–711. doi: 10.1016/j.cell.2004.05.018. [DOI] [PubMed] [Google Scholar]
- 12.Holmes CP, Macher N, Grove JR, Jang L, Irvine JD. Designing Better Coumarin-based Fluorogenic Substrates for PTP1B. Bioorg. Med. Chem. Lett. 2008;18:3382–3385. doi: 10.1016/j.bmcl.2008.04.021. [DOI] [PubMed] [Google Scholar]
- 13.Malamas MS, Sredy J, Moxham C, Katz A, Xu W, McDevitt R, Adebayo FO, Sawicki DR, Seestaller L, Sullivan D, Taylor JR. Novel Benzofuran and Benzothiophene Biphenyls as Inhibitors of Protein Tyrosine Phosphatase 1B with Antihyperglycemic Properties. J. Med. Chem. 2000;43:1293–1310. doi: 10.1021/jm990560c. [DOI] [PubMed] [Google Scholar]
- 14.Park J, Pei D. trans-B-Nitrostyrene Derivatives as Slow-Binding Inhibitors of Tyrosine Phosphatases. Biochemistry. 2004;43:15014–15021. doi: 10.1021/bi0486233. [DOI] [PubMed] [Google Scholar]
- 15.Heneberg P. Use of Protein Tyrosine Phosphatase Inhibitors as Promising Targeted Therapeutic Drugs. Curr. Med. Chem. 2009;16:706–733. doi: 10.2174/092986709787458407. [DOI] [PubMed] [Google Scholar]
- 16.Zhang S, Zhang Z-Y. PTP1B as a Drug Target: Recent Developments in PTP1B Inhibitor Discovery. Drug Discovery Today. 2007;12:373–381. doi: 10.1016/j.drudis.2007.03.011. [DOI] [PubMed] [Google Scholar]
- 17.Asante-Appiah E, Patel S, Desponts C, Taylor JM, Lau C, Dufresne C, Therien M, Friesen R, Becker JW, Leblanc Y, Kennedy BP, Scapin G. Conformation-assisted Inhibition of Protein-tyrosine phosphatase-1B Elicits Inhibitor Selectivity over T-cell Protein-tyrosine Phosphatase. J. Biol. Chem. 2006;281:8010–8015. doi: 10.1074/jbc.M511827200. [DOI] [PubMed] [Google Scholar]
- 18.Shen K, Keng Y-F, Wu L, Guo X-L, Lawrence DS, Zhang Z-Y. Acquisition of a Specific and Potent PTP1B Inhibitor from a Novel Combinatorial Library and Screening Procedure. J. Biol. Chem. 2001;276:47311–47319. doi: 10.1074/jbc.M106568200. [DOI] [PubMed] [Google Scholar]
- 19.Erbe DV, Wang S, Zhang Y-L, Harding K, Kung L, Tam M, Stolz L, Xing Y, Furey S, Qadri A, Klaman LD, Tobin JF. Ertiprotafib Improves Glycemic Control and Lowers Lipids via Multiple Mechanisms. Mol. Pharmacol. 2005;67:69–77. doi: 10.1124/mol.104.005553. [DOI] [PubMed] [Google Scholar]
- 20.Vintonyak VV, Antonchick AP, Rauh D, Waldmann H. The Therapeutic Potential of Phosphatase Inhibitors. Curr. Opin. Chem. Biol. 2009;13:1–12. doi: 10.1016/j.cbpa.2009.03.021. [DOI] [PubMed] [Google Scholar]
- 21.Krishnamurthy D, Karver MR, Fiorillo E, Orru V, Stanford SM, Bottini N, Barrios AM. Gold(I)-Mediated Inhibition of Protein Tyrosine Phosphatases: A Detailed in vitro and Cellular Study. J. Med. Chem. 2008;51:4790–4795. doi: 10.1021/jm800101w. [DOI] [PubMed] [Google Scholar]
- 22.Xie Y, Liu Y, Gong G, Rinderspacher A, Deng S-X, Smith DH, Toebben U, Tzilianos E, Branden L, Vidovic D, Chung C, Schurer S, Tautz L, Landry DW. Discovery of a Novel Submicromolar Inhibitor of the Lymphoid Specific Tyrosine Phosphatase. Bioorg. Med. Chem. Lett. 2008;18:2840–2844. doi: 10.1016/j.bmcl.2008.03.079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Yu X, Sun J-P, He Y, Guo X, Liu S, Zhou B, Hudmon A, Zhang Z-Y. Structure, Inhibitor, and Regulatory Mechanism of Lyp, a Lymphoid-Specific Tyrosine Phosphatase Implicated in Autoimmune Diseases. Proc. Natl. Acad. Sci. U.S.A. 2007;104:19767–19772. doi: 10.1073/pnas.0706233104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Forestier J. Rheumatoid Arthritis and its Treatment by Gold Salts. J. Lab. Clin. Med. 1935;20:827–840. [Google Scholar]
- 25.Shaw CF., III Gold-Based Therapeutic Agents. Chem. Rev. 1999;99:2589–2600. doi: 10.1021/cr980431o. [DOI] [PubMed] [Google Scholar]
- 26.Best SL, Sadler PJ. Gold Drugs: Mechanism of Action and Toxicity. Gold Bull. 1996;29:87–93. [Google Scholar]
- 27.Wang Q, Janzen N, Ramachandran C, Jirik F. Mechanism of Inhibition of Protein-Tyrosine Phosphatases by Disodium Aurothiomalate. Biochem. Pharmacol. 1997;54:703–711. doi: 10.1016/s0006-2952(97)00217-7. [DOI] [PubMed] [Google Scholar]
- 28.Zhang Z-Y, Dixon JE. Active Site Labeling of the Yersinia Protein Tyrosine Phosphatase: The Determination of the pKa of the Active Site Cysteine and the Function of the Conserved Histidine 402. Biochemistry. 1993;32:9340–9345. doi: 10.1021/bi00087a012. [DOI] [PubMed] [Google Scholar]
- 29.Fricker SP. Medicinal Uses of Gold Compounds: Past, Present and Future. Gold Bull. 1996;29:53–60. [Google Scholar]
- 30.Urig S, Fritz-Wolf K, Reau R, Herold-Mende C, Toth K, Davioud-Charvet E, Becker K. Undressing of Phosphine Gold(I) Complexes as Irreversible Inhibitors of Human Disulfide Reductases. Angew. Chem. Int. Ed., Engl. 2006:1881–1886. doi: 10.1002/anie.200502756. [DOI] [PubMed] [Google Scholar]
- 31.Weidauer E, Yasuda Y, Biswal BK, Cherny M, James MNG, Brömme D. Effects of Disease-Modifying Anti-Rheumatic-Drugs (DMARDs) on the Activities of Rheumatiod Arthritis-Associated Cathepsins K and S. Biol. Chem. 2007;388:331–336. doi: 10.1515/BC.2007.037. [DOI] [PubMed] [Google Scholar]
- 32.Vang T, Miletic AV, Arimura Y, Tautz L, Rickert RC, Mustelin T. Protein Tyrosine Phosphatases in Autoimmunity. Annu. Rev. Immunol. 2008;26:29–55. doi: 10.1146/annurev.immunol.26.021607.090418. [DOI] [PubMed] [Google Scholar]
- 33.Montalibet J, Skorey KI, Kennedy BP. Protein Tyrosine Phosphatases: Enzymatic Assays. Methods. 2005;35:2–8. doi: 10.1016/j.ymeth.2004.07.002. [DOI] [PubMed] [Google Scholar]
- 34.Gunatilleke SS, Barrios AM. Inhibition of Lysosomal Cysteine Proteases by a Series of Au(I) Complexes: A Detailed Mechanistic Investigation. J. Med. Chem. 2006;49:3933–3937. doi: 10.1021/jm060158f. [DOI] [PubMed] [Google Scholar]
- 35.Wu J, Katrekar A, Honigberg LA, Smith AM, Conn MT, Tang J, Jeffery D, Mortara K, Sampang J, Williams SR, Buggy J, Clark JM. Identification of Substrates of Human Protein-Tyrosine Phosphatase PTPN22. J. Biol. Chem. 2006;281:11002–11010. doi: 10.1074/jbc.M600498200. [DOI] [PubMed] [Google Scholar]
- 36.Mitra S, Barrios AM. Identifying Selective Protein Tyrosine Phosphatase Substrates and Inhibitors from a Fluorogenic, Combinatorial Peptide Library. ChemBioChem. 2008;9:1216–1219. doi: 10.1002/cbic.200800046. [DOI] [PubMed] [Google Scholar]
- 37.Mitra S, Barrios AM. Highly Sensitive Peptide-Based Probes for Protein Tyrosine Phosphatase Activity Utlizing a Fluorogenic Mimic of Phosphotyrosine. Bioorg. Med. Chem. Lett. 2005;15:5142–5145. doi: 10.1016/j.bmcl.2005.08.054. [DOI] [PubMed] [Google Scholar]
- 38.Mustelin T, Rahmouni S, Bottini N, Alonso A. Role of Protein Tyrosine Phosphatases in T Cell Activation. Immunol. Rev. 2003;191:139–147. doi: 10.1034/j.1600-065x.2003.00014.x. [DOI] [PubMed] [Google Scholar]
- 39.Mustelin T, Alonso A, Bottini N, Huynh H, Rahmouni S, Nika K, Louis-dit-Sully C, Tautz L, Togo SH, Bruckner S, Mena-Duran AV, al-Khouri AM. Protein Tyrosine Phosphatases in T Cell Physiology. Mol. Immunol. 2004;41:687–700. doi: 10.1016/j.molimm.2004.04.015. [DOI] [PubMed] [Google Scholar]
- 40.Gunatilleke SS, Barrios AM. Tuning the Au(I)-Mediated Inhibition of Cathepsin B Through Ligand Substitutions. J. Inorg. Biochem. 2008;102:555–563. doi: 10.1016/j.jinorgbio.2007.10.019. [DOI] [PubMed] [Google Scholar]
- 41.Sanz S, Jones LA, Mohr F, Laguna M. Homogenous Catalysis with Gold: Efficient Hydration of Phenylacetylene in Aqueous Media. Organometallics. 2007;26:952–957. [Google Scholar]
- 42.Toronto DV, Weissbart B, Tinti DS, Balch AL. Solid State Structures and Gold-Gold Bonding in Luminescent Halo(dimethylphenylphosphine)gold(I) Complexes. Inorg. Chem. 1996;35:2484–2489. doi: 10.1021/ic951099r. [DOI] [PubMed] [Google Scholar]
- 43.Isab AA, Fettouhi M, Ahmad S, Ouahab L. Mixed Ligand Gold(I) Complexes of Phosphines and Thiourea and X-ray Structure of (Thiourea-kS)( Tricyclohexylphosphine)Gold(I)Chloride. Polyhedron. 2003;22:1349–1354. [Google Scholar]
- 44.Clipstone NA, Crabtree GR. Identification of calcineurin as a key signalling enzyme in T-lymphocyte activation. Nature. 1992;357:695–697. doi: 10.1038/357695a0. [DOI] [PubMed] [Google Scholar]
- 45.Weiss A, Stobo JD. Requirement for the coexpression of T3 and the T cell antigen receptor on a malignant human T cell line. J. Exp. Med. 1984;160:1284–1299. doi: 10.1084/jem.160.5.1284. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.