Abstract
Background and objectives: Chronic inflammation may play a role in chronic kidney disease (CKD) progression. CRP gene polymorphisms are associated with serum C-reactive protein (CRP) concentrations. It is unknown if CRP polymorphisms are associated with CKD progression or modify the effectiveness of anti-hypertensive therapy in delaying CKD progression.
Design, setting, participants, & measurements: We genotyped 642 participants with CKD from the African American Study of Kidney Disease and Hypertension (AASK), selecting five tag polymorphisms: rs2808630, rs1205, rs3093066, rs1417938, and rs3093058. We compared the minor allele frequencies (MAF) of single nucleotide polymorphisms (SNPs) in AASK to MAFs of African Americans from NHANES III. Among AASK participants, we evaluated the association of SNPs with CRP levels and prospectively with a composite: halving the GFR, ESRD, or death.
Results: The MAF was higher for the rs2808630_G allele (P = 0.03) and lower for the rs1205_A allele (P = 0.03) in the AASK compared with NHANES III. Among AASK participants, the rs3093058_T allele predicted higher CRP concentrations (P < 0.0001) but not CKD progression. The rs2808630_GG genotype was associated with higher risk of the composite endpoint compared with the AA genotype (P = 0.002). Participants with the rs2808630_GG genotype on angiotensin converting enzyme inhibitors (ACEIs) versus β blockers had increased risk of progression (P = 0.03).
Conclusion: CRP SNPs that were associated with higher levels of CRP did not predict CKD progression. The rs2808630_GG genotype was associated with higher risk of CKD progression, and in patients with this genotype, ACEIs did not slow progression.
Familial clustering of chronic kidney disease (CKD) and ESRD has been reported in populations throughout the world for most types of nephropathy (1–6). This genetic predisposition to ESRD seems to be strongly associated with race (7,8). Compared with people with no family history of kidney disease, African Americans with a first-degree relative with ESRD have a nine-fold increase in the risk of ESRD compared with a three- to five-fold increase in whites (8). Recently, the candidate gene MYH9 has been identified as associated with nondiabetic ERSD in African Americans, and this association explains some of the disparity in incidence of ESRD observed between whites and African Americans (7,9). However, it is possible that additional genetic variants, such as those related to inflammatory pathways, may also be associated with ESRD.
Biomarkers of inflammation, including C-reactive protein (CRP), are increased even in early stages of CKD and have been linked to the risk of CKD progression (10–15). These observations have led to studies examining the genetic basis of inflammation and identification of several candidate genes for ESRD susceptibility (16–19). Recently, several large population-based studies showed that plasma CRP levels are under genetic influence (20–25). Some of these polymorphisms have been consistently associated with CRP levels (higher levels associated with rs3093058_T and lower levels associated with rs1205_A and rs2808630_G) and the risk of cardiovascular events (rs3093058_T) in African Americans (23).
CRP gene polymorphisms that affect CRP concentrations may reflect lifetime exposure to CRP more accurately than single time point measurements of serum CRP concentrations. The primary goal of this study was to characterize CRP gene polymorphisms and evaluate their association with CKD progression. We hypothesized that polymorphisms associated with higher levels of CRP would be associated with higher risk of CKD progression. Additionally, we examined whether these polymorphisms modify the renoprotective effects of angiotensin converting enzyme inhibitors (ACEIs), a drug class known to have anti-inflammatory effects (26–28). We hypothesized that patients with polymorphisms associated with higher levels of CRP would benefit most from ACEIs.
Materials and Methods
Study Participants
This study included participants from the African-American Study of Kidney Disease and Hypertension (AASK) trial and The Third National Health and Nutrition Examination Survey (NHANES III). The AASK design and results have been previously described (9,29). Briefly, participants were self-identified African American, 18 to 70 yr, with hypertension defined by diastolic BP >95 mmHg and a GFR between 20 and 65 ml/min per 1.73 m2, measured by I125 iothalamate clearance. Exclusion criteria included (1) identifiable cause of CKD other than hypertension; (2) diabetes mellitus (DM); (3) urine protein/creatinine ratio >2.5; (4) accelerated or malignant hypertension within 6 mo; (5) secondary hypertension, (6) serious illness; and (7) specific contraindication to a study drug. The protocol for genetic testing was approved by the institutional review board at each center, and written informed consent was obtained. Similarly, an Ethical Review Committee from the CDC was obtained for the NHANES III genetic information.
The AASK trial followed a 3 × 2 factorial design. Participants were randomized to β blockers (BBs; metoprolol 50 to 200 mg/d), calcium channel blockers (CCBs; amlodipine, 5 to 10 mg/d), or an ACEI (ramipril, 2.5 to 10 mg/d) and to two levels of BP: low (mean arterial pressure [MAP] goal of <92 mmHg) or usual (MAP 102 to 107 mmHg) (29). Data were collected for the following comorbidities: cancer, liver disease, congestive heart failure, left ventricular hypertrophy, arrhythmias, and cardiovascular disease. An interim analysis at 3 yr halted the amlodipine arm because patients with proteinuria (Upr/Cr > 0.22) taking amlodipine had faster GFR decline. DNA samples were available for 670 AASK participants that entered the trial. Serum CRP concentrations were measured in the AASK participants by nephelometry using a high-sensitivity assay (Dade Behring BN I). All measurements were done by a central laboratory at the Cleveland Clinic laboratory.
The NHANES are surveys conducted by the Centers for Disease Control to provide national estimates of the health of the U.S. civilian noninstitutionalized population. In the second phase of NHANES III conducted between 1991 and 1994, DNA samples were collected from 7159 consenting participants. We used genetic data from NHANES III participants that were self-identified African American, had GFR >60 ml/min, did not have DM, and had a UP/Cr ratio <2.5. For the NHANES III, we defined DM as self-reported physician diagnosis of DM, use of DM medications, fasting glucose >126 mg/dl, or a random glucose ≥200 mg/dl.
CRP Polymorphism Genotyping
Five tag single nucleotide polymorphisms (SNPs) were selected based on variation data for 23 European Americans and 24 African Americans resequenced by Seattle SNPs (pga.gs.washington.edu) (21,30) using LDSelect at default settings (r2 > 0.64): rs1417938, rs1205, rs2808630, rs3093058, and rs3093066. We also selected rs1800947 given previous reports of its association with decreased CRP levels. Genotyping was performed using TaqMan Assays By Design (Applied Biosystems, Foster City, CA) under standard conditions. All markers had a 95% or higher genotyping efficiency.
Outcomes
The main outcome for the AASK and for this study was a composite endpoint of (1) GFR event defined as halving of the baseline GFR or a drop in the GFR ≥ 25 ml/min per 1.73 m2, (2) ESRD, or (3) death. GFR was measured at baseline, at 3 mo, and every 6 mo thereafter. The follow-up of the trial ranged from 3.5 to 6.5 yr. Additionally, we compared at baseline the allele frequencies of each SNP in the AASK participants with those in the African Americans from the NHANES III with normal kidney function as described previously (21).
Statistical Analysis
All analyses were completed using SAS/Genetics 9.1 (SAS Institute, Cary, NC) and STATA 9 (College Station, TX). Each of the CRP SNPs was assessed to determine whether the observed genotype frequencies were in Hardy-Weinberg equilibrium. Marker-marker linkage disequilibrium was assessed using r2, calculated for all pairwise SNP comparisons in the AASK dataset. We considered significance for a two-sided α = 0.05 for all analyses and adjusted α α/n 0.05/11 = 0.0045 after Bonferroni's correction for the number of genotype comparisons. CRP was log-transformed, and the distribution was checked for normality. Cox proportional hazards and Kaplan Meier plots were used to test the association between SNP and the composite outcome. Variables were selected a priori for the multivariate analysis. Haplotypes were inferred from tagSNPs using the expectation-maximization algorithm provided by SAS/Genetics 9.1, which also provides the probabilities for each individual's inferred haplotype pair for subsequent regression analyses. Common haplotypes (>5% frequency) were determined from the most likely pair of haplotypes inferred by the expectation-maximization algorithm. This study was an ancillary study to AASK, and as such, the analyses were not performed by the AASK Data Coordinating Center.
Results
Participant baseline characteristics are shown in Table 1. One hundred fifty-two individuals (24%) reached the composite endpoint: 109 experienced a GFR event, 80 reached ESRD, and 12 patients died.
Table 1.
Baseline characteristics of AASK participants
| Characteristics | (n = 642) |
|---|---|
| Age (yr; mean ± SD) | 54 ± 10.5 |
| Gender (females) | 41% |
| Serum creatinine [mg/dl; median (IQR)] | 1.8 (1.5, 2.3) |
| Mean baseline GFR | 47 ± 13.5 |
| Systolic BP (mmHg; mean ± SD) | 150 ± 24 |
| MAP BP (mmHg; mean ± SD) | 114 ± 16.1 |
| Diastolic BP (mmHg; mean ± SD) | 96 ± 14 |
| Urine protein/creatinine ratio (mean ± SD) | 0.31 ± 0.50 |
| Mean urine 24-h proteinuria (mean ± SD) | 0.50 ± 0.94 |
| Body mass index (mean ± SD) | 31.1 ± 6.7 |
| Current smokers | 28% (n = 180) |
| CRP, [mg/dl; median (IQR)] | 0.5 (0.21, 0.93) |
| Incident ESRD | 12% (n = 80) |
| Number of patients halving the GFR | 17% (n = 109) |
| Death | 1.8% (n = 12) |
| Combined endpoint | 24% (n = 152) |
| Mean rate of decline ml/min per 1.73 m2 | −1.9 (−0.39, −3.5) |
| Blood pressure assignment | Low: 50.15% (n = 322); Usual: 49.9% (n = 320) |
| Drug assignment | |
| ACE inhibitor | 41% (n = 265) |
| Low BP: 52%(n = 138)/Usual BP: 48%(n = 127) | |
| β blocker | 39% (n = 252) |
| Low BP: 50%(n = 125)/Usual BP: 50%(n = 127) | |
| Calcium channel blockers | 19% (n = 125) |
| Low BP: 47%(n = 59)/Usual BP: 53%(n = 66) |
CRP Allele and Genotype Frequencies
The observed genotype distribution for the six CRP SNPs was consistent with Hardy-Weinberg equilibrium. The allele frequencies for CRP SNPs in AASK and NHANES III cohorts are presented in Table 2. Participants in the AASK trial had a higher minor allele frequency (MAF) for rs2808630 (G allele: 17 versus 14% P = 0.03) and a lower MAF for rs1205 (A allele: 18 versus 21%, P = 0.03) compared with the African Americans from the NHANES III with normal kidney function, no DM, and a UP/Cr ratio <2.5; however, this association did not remain significant after adjusting for multiple comparisons (α/n 0.05/6 = 0.008). AASK participants also had a higher MAF for rs3093058 (T allele: 19 versus 16%); however, this did not reach statistical significance (P = 0.08). Table 3 shows the genotype frequencies for each polymorphism for NHANES III and AASK participants.
Table 2.
CRP SNP allele frequencies for African Americans from NHANES III and AASK participants
| CRP SNPs | Location | Alleles | Allele Frequencies |
P Valuea | |
|---|---|---|---|---|---|
| NHANES III (n = 450) | AASK (n = 642) | ||||
| rs2808630 | 3'-flanking region | A | 86% | 83% | 0.03b |
| G | 14% | 17% | |||
| rs1205 | 3'-flanking region | A | 21% | 18% | 0.03b |
| G | 79% | 82% | |||
| rs3093066 | 5'-flanking region | A | 24% | 22% | 0.3 |
| C | 76% | 78% | |||
| rs1417938 | Intron 1 | A | 87% | 88% | 0.3 |
| T | 13% | 12% | |||
| rs3093058 | 5'-flanking region | A | 84% | 81% | 0.08 |
| T | 16% | 19% | |||
| rs1800947c | Exon 2 | C | 1% | 1% | 0.6 |
| G | 99% | 99% | |||
Unadjusted for multiple comparisons.
Not statistically significance at an α = 0.008 level after applying a Bonferroni correction (α = n/6).
Data for rs1800947 are for all NHANES III African-Americans participants with DNA (n = 2108) because the data user agreement does not allow outputs with counts less than 5.
Table 3.
Genotype in NHANES III and AASK participants for each of the CRP polymorphisms
| Genotype Frequency (%) |
||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| rs2808630 |
rs1205 |
rs3093066 |
rs1417938 |
rs3093058 |
rs1800947a |
|||||||||||||
| AA | AG | GG | GG | AG | AA | CC | AC | AA | AA | AT | TT | AA | AT | TT | GG | CG | CC | |
| NHANES | 74 | 24 | 2 | 62 | 33 | 4 | 58 | 36 | 6 | 76 | 23 | 2 | 71 | 27 | 3 | 98 | 2 | 0 |
| AASK | 69 | 28 | 3 | 67 | 30 | 3 | 61 | 17 | 5 | 77 | 21 | 1 | 66 | 31 | 4 | 98 | 2 | 0 |
Data for rs1800947 are for all NHANES III African-Americans participants with DNA (n = 2108) beause the data user agreement does not allow outputs with counts less than 5.
CRP Genotypes and CRP Levels
The median CRP levels in the AASK cohort for the AA, AT, and TT rs3093058 genotypes were 0.40 (interquartile range [IQR] = 0.19, 0.81), 0.69 (IQR: 0.31, 1.13), and 1.14 mg/dl (IQR: 0.45, 1.81), respectively. The rs3093058 genotypes AT and TT were associated with significantly higher levels of CRP in the AASK cohort (P < 0.001) compared with the AA genotypes (Table 4). The P value remained significant after Bonferroni's correction for 11 comparisons. No other tested CRP polymorphism was associated with CRP levels.
Table 4.
Association between serum CRP levels and CRP SNPs in the AASK participants
| CRP SNPs and Genotypes | N | CRP Level Median (IQR) | β Coefficient (95% CI) Log CRP | P Valuea |
|---|---|---|---|---|
| rs2808630 | ||||
| AA | 439 | 0.51 (0.23, 0.95) | Ref | |
| AG | 192 | 0.44 (0.19, 0.92) | −0.17 (−0.37, 0.02) | 0.08 |
| GG | 16 | 0.34 (0.09, .43) | −0.21 (−0.77, 0.35) | 0.46 |
| rs1205 | ||||
| GG | 447 | 0.52 (0.23, 0.96) | Ref | |
| AG | 179 | 0.44 (0.21, 0.83) | −0.11 (−0.30, 0.09) | 0.27 |
| AA | 25 | 0.06 (−0.42, 0.54) | −0.06 (−0.42, 0.54) | 0.81 |
| rs3093066 | ||||
| CC | 399 | 0.52 (0.23, 0.95) | Ref | |
| AC | 213 | 0.40 ((0.20, 0.90) | −0.20 (−0.39, −0.01) | 0.04 |
| AA | 37 | 0.66 (0.24, 1.01) | −0.04 (−0.34, 0.43) | 0.82 |
| rs1417938 | ||||
| AA | 503 | 0.51 (0.22, 0.94) | Ref | |
| AT | 141 | 0.43 (0.23, 0.85) | −0.02 (−0.23, 0.19) | 0.85 |
| TT | 6 | 0.55 (0.38, 1.22) | 0.49 (−0.38, 1.4) | 0.27 |
| rs3093058 | ||||
| AA | 423 | 0.40 (0.19, 0.81) | Ref | |
| AT | 204 | 0.69 (0.31, .13) | 0.40 (0.21, 0.59) | <0.001b |
| TT | 20 | 1.14 (0.45, 1.81) | 0.86 (0.37, 1.37) | <0.001b |
| rs1800947 | ||||
| GG | 644 | 0.5 (0.22, 0.92) | Ref | |
| CG | 7 | 0.21 (0.12, 0.74) | −0.50 (−1.31, 0.30) | 0.22 |
| CC | NA | NA | NA | NA |
NA, not applicable.
Unadjusted for multiple comparisons.
Statistical significance at an α = 0.0045 after Bonferroni correction for 11 genotypes.
CRP Genotypes and Clinical Outcomes
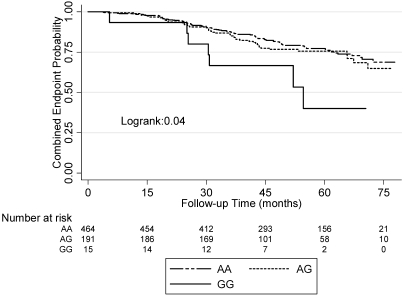
Among AASK participants, the rs2808630_GG genotype was associated with an increased risk of reaching the composite endpoint (adjusted hazard ratio [HR], 3.62; 95% confidence interval [CI], 1.62 to 8.23; P = 0.002) compared with the AA genotype (Table 5; Figure 1). The P value remained significant after Bonferroni's correction for 11 comparisons (adjusted significance level α/n 0.05/11 = 0.0045). Potential confounding variables were determined a priori, including age, gender, comorbidities, BP treatment assignment, baseline GFR, proteinuria, body mass index, smoking, and CRP levels. None of the other tested polymorphisms were associated with the composite endpoint. There were no statistically significant differences in the demographic or clinical characteristics among the different rs2808630 genotypes (Table 6). Other independent predictors for the composite endpoint in the model for the SNP rs2808630 included ACEI use, smoking, baseline proteinuria, and baseline GFR (Table 7).
Table 5.
Association of CRP polymorphisms and renal outcomes
| CRP SNPs and Genotype | N | Composite of Halving Your GFR, Reaching ESRD, or Death |
|
|---|---|---|---|
| Hazard Ratio (95% CI) | P Valuesa | ||
| rs2808630 | |||
| AA | 439 | Ref | |
| AG | 191 | 0.87 (0.59–1.29) | 0.49 |
| GG | 15 | 3.65 (1.62–8.23) | 0.002b |
| rs1205 | |||
| GG | 447 | Ref | |
| AG | 179 | 1.14 (0.74–1.52) | 0.48 |
| AA | 25 | 0.90 (0.36–2.27) | 0.84 |
| ss3093066 | |||
| CC | 399 | Ref | |
| AC | 213 | 1.02 (0.6–1.5) | 0.89 |
| AA | 37 | 1.23 (0.6–2.5) | 0.56 |
| rs1417938 | |||
| AA | 503 | Ref | |
| AT | 141 | 0.89 (0.58–1.37) | 0.62 |
| TT | 6 | 1.07 (0.14–7.89) | 0.94 |
| rs3093058 | |||
| AA | 423 | Ref | |
| AT | 204 | 0.81 (0.55–1.19) | 0.28 |
| TT | 20 | 0.92 (0.28–3.02) | 0.89 |
| rs1800947 | |||
| GG | 644 | Ref | |
| CG | 7 | 0.19 (0.03–1.14) | 0.11 |
| CC | NA | NA | |
Composite outcome = decrease in GFR by 50% or a drop in the GFR ≥25 ml/min per 1.73 m2, ESRD, or death.
Hazard ratios are adjusted for age, gender, BP treatment assignment, drug treatment assignment, body mass index, baseline GFR, baseline proteinuria, smoking, and number of comorbidities.
Ref, reference group homozygous for the major allele; NA, not applicable.
Unadjusted for multiple comparisons.
Statistical significance at an α=0.0045 after Bonferroni correction for 11 genotypes.
Figure 1.
Survival without experiencing the combined endpoint of halving the GFR. ESRD or death in CKD patients with different genotypes of the rs2808630.
Table 6.
Clinical Characteristics of AASK participants by rs2808630 genotypes
| Rs2808630 Genotypes |
P Value | |||
|---|---|---|---|---|
| GG | AG | AA | ||
| N | 15 | 191 | 436 | |
| Age (yr) | 48.42 ± 12.55 | 54.7 ± 9.86 | 53.82 ± 10.70 | 0.23 |
| Gender (% male) | 40% | 59% | 60% | 0.3 |
| Serum creatinine (mg/dl) | 1.8 (1.4, 2.1) | 1.8 (1.5, 2.4) | 1.8 (1.5, 2.3) | 0.79 |
| C-reactive protein, mg/dl, median (IQR) (P for log CRP) | 0.34 (0.09, 1.43) | 0.44 (0.19, 0.92) | 0.51 (0.23, 0.95) | 0.54 |
| Mean baseline GFR | 46 11.01 | 45.4 13.25 | 47.49 13.65 | 0.53 |
| Years with hypertension | 13.2 ± 8.73 | 13.9 ± 9.47 | 14.2 ± 9.9 | 0.67 |
| 24-h urine protein at baseline (g/d) | 0.44 ± 0.73 | 0.54 ± 0.53 | 0.48 ± 0.92 | 0.4 |
| Systolic BP at randomization (mmHg) | 149.4 ± 23.36 | 150.5 ± 25.23 | 150.2 ± 23.7 | 0.5 |
| Diastolic BP at randomization (mmHg) | 100 ± 16 | 95.4 ± 14 | 96 ± 14.4 | 0.09 |
| History of heart disease | 40% | 50% | 50% | 0.74 |
| Body mass index (kg/m2) | 33.93 ± 6.4 | 30.5 ± 6.7 | 31.3 ± 6.64 | 0.09 |
| Number of comorbidities | 0 (0,1) | 1 (0,1) | 1 (0,1) | 0.09 |
| Smoking | 6 (40%) | 56 (29%) | 186 (27%) | 0.58 |
| GFR slope (ml/min per m2) | −3.03 IQR −5.2, −0.22 | −2.18 IQR: −3.6, −0.26 | −1.87 IQR: −3.3, −0.26 | 0.07 |
Table 7.
Multivariate Cox proportional hazards analysis of association of the rs2808630 genotypes and composite outcomes and other clinical predictors
| Variable | Hazard Ratio (95% CI) | P Valuea |
|---|---|---|
| Genotype | ||
| rs2808630_AA | Ref | |
| rs2808630_AG | 0.87 (0.59, 1.29) | 0.49 |
| rs2808630_GG | 3.65 (1.62, 8.23) | 0.002b |
| BP assignmentc | 0.78 (0.55, 1.12) | 0.19 |
| Drug assignment | ||
| Calcium channel blockers | Ref | |
| β blockers | 0.72 (0.44, 1.17) | 0.19 |
| ACE inhibitors | 0.56 (0.34, 0.91) | 0.02 |
| Log CRP (mg/dl) | 0.96 (0.81, 1.14) | 0.65 |
| Age | 1.01 (0.99, 1.02) | 0.53 |
| Genderd | 1.04 (0.71, 1.52) | 0.83 |
| Body mass index (kg/m2) | 0.98 (0.95, 1.02) | 0.32 |
| Mean baseline GFR (ml/min) | 0.96 (0.94, 0.97) | <0.0001 |
| Urine protein (g/d) | 1.88 (1.64, 2.17) | <0.0001 |
| Number of comorbidities | 1.04 (0.85, 1.26) | 0.72 |
| Smoking (current versus never or former) | 1.63 (1.11, 2.40) | 0.01 |
Adjustement done for baseline covariates.
BP assignment: usual: MAP 102–107 mmgh; low: MAP <92 mmgh.
Mean baseline GFR: the mean of two GFR measurements by isothalamate.
Unadjusted for multiple comparisons.
Statistical significance at an α = 0.0045 after Bonferroni correction for 11 genotypes.
Reference group low BP.
Reference group male.
CRP Levels and Clinical Outcomes
The median CRP in the AASK participants involved in this study was 0.50 mg/dl (IQR, 0.22, 0.93). The median CRP levels were higher in women compared with men (0.60 versus 0.41 mg/dl, P < 0.0001), consistent with observations in the general population (31). In this study, CRP level did not predict CKD progression as reflected by the combined endpoint (HR, 0.94; 95% CI, 0.80 to 1.10; P = 0.4).
CRP Genotypes and ACEI Use
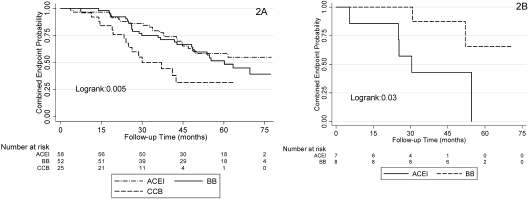
Pharmacogenomic analysis was performed to evaluate the effect of ACE inhibition within each rs2808630 genotype. There was no statistical difference between the three drugs and their effect on the composite outcome. We restricted our analyses to individuals with UP/Cr >0.22 based on the findings from the original trial in which the benefit of ACEI was the strongest in this group (9,29). Patients randomized to ACEI therapy in the setting of a baseline UP/Cr >0.22 experienced fewer composite endpoints than those assigned to CCBs (46 versus 54%; HR, 0.53; 95% CI, 0.31 to 0.91; P = 0.02) and not significantly different than those assigned to BBs (46 versus 51%; HR, 0.89; 95% CI, 0.57 to 1.30; P = 0.6). For the most common genotype, rs2808630 AA with UP/Cr >0.22 (n = 128), ACEI therapy was superior to CCBs (HR, 0.33; 95% CI, 0.17 to 0.67; P = 0.002) and similar to BBs (HR, 1.35; 95% CI, 0.75 to 2.43; P = 0.3; Figure 2A). For all individuals with the rs2808630_GG genotype (n = 15), those allocated to ACEI therapy had a higher rate of progression than those allocated to BBs (HR, 5.34; 95% CI, 1.00 to 28.3; P = 0.04; Figure 2B). We were unable to divide the rs2808630_GG genotype by protein excretion because of the small number of individuals with UP/Cr >0.22 and the genotype of interest (n = 6). There was statistical evidence to support effect modification by an interaction between ACEI and rs2808630 genotype and the composite outcome (P for the interaction = 0.007).
Figure 2.
Effect of different antihypertensive agents in meeting the composite endpoint for participants homozygous for the major allel rs2808630AA and baseline urinary protein-to-creatinine ratio (UP/Cr) >0.22 (above, panel 2A) and for participants homozygous for the minor allele rs2808630CC (below, panel 2B).
Haplotype Analyses, CRP Levels, and Clinical Outcomes
Six common haplotypes were inferred from six CRP tagSNPs in the AASK participants (Table 8). The rs1800947 SNP was dropped from the model because of the low MAF (<1%). We tested for an association between CRP levels and CRP haplotypes, and the results of the haplotype-based analyses mimic the single SNP analyses. The haplotype tagged by the T allele of rs3093058 (H2) was associated with higher levels of CRP (P < 0.0001). No CRP haplotype was associated with the composite endpoint.
Table 8.
Association between CRP haplotypes and plasma CRP concentration
| Haplotype | Allele |
Estimated Frequency (%) | Log CRP |
||||||
|---|---|---|---|---|---|---|---|---|---|
| rs1800947 G/C | rs2808630 A/G | rs1205 G/A | rs3093066 C/A | rs1417938 A/T | rs3093058 A/T | β-Coefficient | P Value | ||
| H1 | A | G | A | A | A | 22.2% | −0.73 | 0.2 | |
| H2 | A | G | C | A | T | 18.7% | 0.44 | <0.0001 | |
| H3 | A | A | C | A | A | 17.8% | −0.07 | 0.4 | |
| H4 | G | G | C | A | A | 17.2% | −0.14 | 0.1 | |
| H5 | A | G | C | A | A | 12.5% | −0.14 | 0.2 | |
| H6 | A | G | C | T | A | 11.7% | −0.01 | 0.8 | |
Haplotypes with estimated frequency of <5% were excluded from analysis.
Discussion
The primary goal of this study was to evaluate the role of CRP polymorphisms in CKD progression. AASK represents an ideal population to study these associations because CKD progression occurs four times faster in African Americans (32). Although our study suggests that some common mechanisms/genetic susceptibilities to ESRD may share a common pathway, it is important to acknowledge that distinct renal diseases may involve other independent factors. Our study is performed in a cohort of African Americans with hypertensive kidney disease. Our results showed that CRP SNP rs3093058 was associated with increased levels of CRP in the CKD population (TT versus AT and AA genotypes) and is consistent with what has been shown in the general population (20–24).
Although the frequency of the rs3093058 T allele, which is associated with increased CRP levels, was marginally higher in the AASK population than community controls, this was not statistically significant. More importantly, we found no association between this allele and CKD progression. On the other hand, we showed a strong association between the rs2808630_GG genotype, which has been associated with lower levels of CRP (21,22), and both baseline CKD (higher in AASK than in controls) and CKD progression, as reflected by the composite endpoint. Additionally, we showed that individuals with this genotype assigned to ACEI progressed faster than those assigned to BBs.
The lack of association observed between the T allele of the rs3093058, which tags a promoter variation rs3093062 (20,21), and CKD progression could be explained by a lack of statistical power to detect the small increase in risk of CKD in African Americans. Alternatively, it is possible that rs3093058 is not a key pathogenic mediator for CKD in this population. Although epidemiologic data support the role of inflammation, including CRP in the pathogenic process (11,13–15), CRP is a surrogate marker of the inflammatory process (33). CKD patients have a high burden of comorbidities, and reverse causation may explain the associations with CRP. However, studies of non-CRP inflammatory cytokine gene polymorphisms could provide a link between inflammatory genes and pathways with CKD.
The underlying mechanisms regarding the association between the GG genotype of the rs2808630 and the composite endpoint in this study remains unclear. Despite adjustment for multiple comparisons, it is possible that this association represents a false-positive finding. A potential explanation is that the GG genotype of the rs2808630 is in linkage disequilibrium with another functional SNP associated with ESRD not genotyped directly in our study. Using the Genome Variation Server (gvs.gs.washington.edu/GVS/), we surveyed a 50-kb region surrounding rs2808630 for SNPs in high linkage disequilibrium with the rs2808630 in the West African samples from the International HapMap Project (34). No SNP was in high linkage disequilibrium (defined as r2 > 0.80), and 12 intergenic SNPs were in moderate linkage disequilibrium (defined at r2 > 0.50). Fine mapping and functional studies will be needed to identify the causal SNP underlying the association described here.
It is possible that rs2808630 is the causal SNP and affects the response to reno-protective interventions. Our hypothesis was that individuals with chronic inflammation will benefit the most from ACEIs (26–28,35). Albeit small sample size, individuals with the most common genotype AA and UP/Cr ratio >0.22 benefited from ACEIs. Individuals with the less common genotype, GG (low inflammatory profile), were at an increased risk for the primary endpoint even when allocated to ACEIs compared with BBs (9). These findings should be considered preliminary and need to be confirmed in larger studies.
The main strength of our study is that it was performed in an randomized controlled trial primarily designed to address CKD progression, and therefore, outcome ascertainment was optimal. GFR, for example, was performed using the gold standard iothalamate clearance. Also, the longitudinal nature of the study allowed us to evaluate the genotype effect in disease progression among high-risk patients with CKD stages 3 and 4.
Our study is not without limitations. First, our study results are limited to African Americans with hypertensive CKD and cannot be extrapolated to other populations or other diseases. In addition, neither the AASK nor the NHANES III dataset currently has ancestry informative markers to adjust for admixture. Our main focus is CKD progression within the AASK cohort, which was a multicenter study across the nation, and ancestry background should be similar. However, admixture markers will need to be performed in the AASK cohort for future comparisons with other populations.
Finally, a limitation of our study is the lack of replication for the association of CRP polymorphisms and CKD progression in other independent cohorts. However, our study represents an independent replication for the association of CRP polymorphisms with CRP levels (21,22,24). Independent replication studies in racially diverse CKD progression cohorts will be necessary to substantiate the new findings.
In conclusion, CRP SNPs that are associated with higher levels of CRP were more common in African Americans with hypertensive CKD than in African Americans without CKD; however, these SNPs were not associated with CKD progression. Additionally, we showed an association between the rs2808630_GG genotype (low inflammatory profile) with both baseline CKD and the risk of CKD progression and potential loss of renal benefit from ACEIs. Larger studies in African Americans are needed to further explore the role of CRP polymorphisms in response to ACEI and CKD progression.
Disclosures
None.
Acknowledgments
We thank C. L. Sanders (currently with Medco Health) and J. McLean from the NCHS at the CDC for assistance with the NHANES III genetic data. This work was supported in part by Clinical Translational Science Award 1UL-1RR024975 from the National Center for Research Resources. The Vanderbilt University Center for Human Genetics Research, Computational Genomics Core provided computational and/or analytical support for this work. All genotyping was performed by the Vanderbilt DNA Resources Core. A.M.H. was supported by a Research Career Development Award and a Clinical Research Center of Excellence from the Department of Veterans Affairs, National Kidney Foundation (NKF) Young Investigator Award and DK077373. K.C. is supported by K23-DK080952–01 and an NKF Young Investigator Award. M.S.L. was supported by Grants DK048689, DK057867, and RR000071. T.A.I. is supported in part by Grants DK62849, AT003844, HL065193, and DK082192.
Footnotes
Published online ahead of print. Publication date available at www.cjasn.org.
References
- 1.Canani LH, Gerchman F, Gross JL: Increased familial history of arterial hypertension, coronary heart disease, and renal disease in Brazilian type 2 diabetic patients with diabetic nephropathy. Diabetes Care 21: 1545–1550, 1998 [DOI] [PubMed] [Google Scholar]
- 2.Gumprecht J, Zychma MJ, Moczulski DK, Gosek K, Grzeszczak W: Family history of end-stage renal disease among hemodialyzed patients in Poland. J Nephrol 16: 511–515, 2003 [PubMed] [Google Scholar]
- 3.Harjutsalo V, Katoh S, Sarti C, Tajima N, Tuomilehto J: Population-based assessment of familial clustering of diabetic nephropathy in type 1 diabetes. Diabetes 53: 2449–2454, 2004 [DOI] [PubMed] [Google Scholar]
- 4.Ramirez SP, McClellan W, Port FK, Hsu SI: Risk factors for proteinuria in a large, multiracial, southeast Asian population. J Am Soc Nephrol 13: 1907–1917, 2002 [DOI] [PubMed] [Google Scholar]
- 5.Scolari F, Amoroso A, Savoldi S, Mazzola G, Prati E, Valzorio B, Viola BF, Nicola B, Movilli E, Sandrini M, Campanini M, Maiorca R: Familial clustering of IgA nephropathy: Further evidence in an Italian population. Am J Kidney Dis 33: 857–865, 1999 [DOI] [PubMed] [Google Scholar]
- 6.Vijay V, Snehalatha C, Shina K, Lalitha S, Ramachandran A: Familial aggregation of diabetic kidney disease in type 2 diabetes in south India. Diabetes Res Clin Pract 43: 167–171, 1999 [DOI] [PubMed] [Google Scholar]
- 7.Kao WH, Klag MJ, Meoni LA, Reich D, Berthier-Schaad Y, Li M, Coresh J, Patterson N, Tandon A, Powe NR, Fink NE, Sadler JH, Weir MR, Abboud HE, Adler SG, Divers J, Iyengar SK, Freedman BI, Kimmel PL, Knowler WC, Kohn OF, Kramp K, Leehey DJ, Nicholas SB, Pahl MV, Schelling JR, Sedor JR, Thornley-Brown D, Winkler CA, Smith MW, Parekh RS: MYH9 is associated with nondiabetic end-stage renal disease in African Americans. Nat Genet 40: 1185–1192, 2008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Freedman BI, Soucie JM, Stone SM, Pegram S: Familial clustering of end-stage renal disease in blacks with HIV-associated nephropathy. Am J Kidney Dis 34: 254–258, 1999 [DOI] [PubMed] [Google Scholar]
- 9.Appel LJ, Wright JT, Jr, Greene T, Kusek JW, Lewis JB, Wang X, Lipkowitz MS, Norris KC, Bakris GL, Rahman M, Contreras G, Rostand SG, Kopple JD, Gabbai FB, Schulman GI, Gassman JJ, Charleston J, Agodoa LY: Long-term effects of renin-angiotensin system-blocking therapy and a low blood pressure goal on progression of hypertensive chronic kidney disease in African Americans. Arch Intern Med 168: 832–839, 2008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Descamps-Latscha B, Herbelin A, Nguyen AT, Roux-Lombard P, Zingraff J, Moynot A, Verger C, Dahmane D, de Groote D, Jungers P, Dayer JM: Balance between IL-1 beta, TNF-alpha, and their specific inhibitors in chronic renal failure and maintenance dialysis. Relationships with activation markers of T cells, B cells, and monocytes. J Immunol, 154: 882–892, 1995 [PubMed] [Google Scholar]
- 11.Fried L, Solomon C, Shlipak M, Seliger S, Stehman-Breen C, Bleyer AJ, Chaves P, Furberg C, Kuller L, Newman A: Inflammatory and prothrombotic markers and the progression of renal disease in elderly individuals. J Am Soc Nephrol, 15: 3184–3191, 2004 [DOI] [PubMed] [Google Scholar]
- 12.Ramos LF, Shintani A, Ikizler TA, Himmelfarb J: Oxidative stress and inflammation are associated with adiposity in moderate to severe CKD. J Am Soc Nephrol 19: 593–599, 2008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Tonelli M, Sacks F, Pfeffer M, Jhangri GS, Curhan G: Biomarkers of inflammation and progression of chronic kidney disease. Kidney Int 68: 237–245, 2005 [DOI] [PubMed] [Google Scholar]
- 14.Tong M, Carrero JJ, Qureshi AR, Anderstam B, Heimburger O, Barany P, Axelsson J, Alvestrand A, Stenvinkel P, Lindholm B, Suliman ME: Plasma pentraxin 3 in patients with chronic kidney disease: associations with renal function, protein-energy wasting, cardiovascular disease, and mortality. Clin J Am Soc Nephrol 2: 889–897, 2007 [DOI] [PubMed] [Google Scholar]
- 15.Wolkow PP, Niewczas MA, Perkins B, Ficociello LH, Lipinski B, Warram JH, Krolewski AS: Association of urinary inflammatory markers and renal decline in microalbuminuric type 1 diabetics. J Am Soc Nephrol 19: 789–797, 2008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Balakrishnan VS, Guo D, Rao M, Jaber BL, Tighiouart H, Freeman RL, Huang C, King AJ, Pereira BJ: Cytokine gene polymorphisms in hemodialysis patients: Association with comorbidity, functionality, and serum albumin. Kidney Int 65: 1449–1460, 2004 [DOI] [PubMed] [Google Scholar]
- 17.Nordfors L, Lindholm B, Stenvinkel P: End-stage renal disease—not an equal opportunity disease: The role of genetic polymorphisms. J Intern Med 258: 1–12, 2005 [DOI] [PubMed] [Google Scholar]
- 18.Rao M, Wong C, Kanetsky P, Girndt M, Stenvinkel P, Reilly M, Raj DS: Cytokine gene polymorphism and progression of renal and cardiovascular diseases. Kidney Int 72: 549–556, 2007 [DOI] [PubMed] [Google Scholar]
- 19.Stenvinkel P: New insights on inflammation in chronic kidney disease-genetic and non-genetic factors. Nephrol Ther 2: 111–119, 2006 [DOI] [PubMed] [Google Scholar]
- 20.Carlson CS, Aldred SF, Lee PK, Tracy RP, Schwartz SM, Rieder M, Liu K, Williams OD, Iribarren C, Lewis EC, Fornage M, Boerwinkle E, Gross M, Jaquish C, Nickerson DA, Myers RM, Siscovick DS, Reiner AP: Polymorphisms within the C-reactive protein (CRP) promoter region are associated with plasma CRP levels. Am J Hum Genet 77: 64–77, 2005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Crawford DC, Sanders CL, Qin X, Smith JD, Shephard C, Wong M, Witrak L, Rieder MJ, Nickerson DA: Genetic variation is associated with C-reactive protein levels in the Third National Health and Nutrition Examination Survey. Circulation 114: 2458–2465, 2006 [DOI] [PubMed] [Google Scholar]
- 22.Kathiresan S, Larson MG, Vasan RS, Guo CY, Gona P, Keaney JF, Jr, Wilson PW, Newton-Cheh C, Musone SL, Camargo AL, Drake JA, Levy D, O'Donnell CJ, Hirschhorn JN, Benjamin EJ: Contribution of clinical correlates and 13 C-reactive protein gene polymorphisms to interindividual variability in serum C-reactive protein level. Circulation 113: 1415–1423, 2006 [DOI] [PubMed] [Google Scholar]
- 23.Lange LA, Carlson CS, Hindorff LA, Lange EM, Walston J, Durda JP, Cushman M, Bis JC, Zeng D, Lin D, Kuller LH, Nickerson DA, Psaty BM, Tracy RP, Reiner AP: Association of polymorphisms in the CRP gene with circulating C-reactive protein levels and cardiovascular events. JAMA 296: 2703–2711, 2006 [DOI] [PubMed] [Google Scholar]
- 24.Miller DT, Zee RY, Suk Danik J, Kozlowski P, Chasman DI, Lazarus R, Cook NR, Ridker PM, Kwiatkowski DJ: Association of common CRP gene variants with CRP levels and cardiovascular events. Ann Hum Genet 69: 623–638, 2005 [DOI] [PubMed] [Google Scholar]
- 25.Zacho J, Tybjaerg-Hansen A, Jensen JS, Grande P, Sillesen H, Nordestgaard BG: Genetically elevated C-reactive protein and ischemic vascular disease. N Engl J Med 359: 1897–1908, 2008 [DOI] [PubMed] [Google Scholar]
- 26.Koulouris S, Symeonides P, Triantafyllou K, Ioannidis G, Karabinos I, Katostaras T, El-Ali M, Theodoridis T, Vratsista E, Thalassinos N, Kokkinou V, Nanas I, Stamatelopoulos S, Toutouzas P: Comparison of the effects of ramipril versus telmisartan in reducing serum levels of high-sensitivity C-reactive protein and oxidized low-density lipoprotein cholesterol in patients with type 2 diabetes mellitus. Am J Cardiol 95: 1386–1388, 2005 [DOI] [PubMed] [Google Scholar]
- 27.Lopez Santi RG, Valeff EC, Duymovich CR, Mazziotta D, Mijailovsky NE, Filippa GC, Maltez R, Hernandez VA, Monroy AG, Borzi JG, Acheme RA, Etchegoyen MC: Effects of an angiotensin-converting enzyme inhibitor (ramipril) on inflammatory markers in secondary prevention patients: RAICES Study. Coron Artery Dis 16: 423–429, 2005 [DOI] [PubMed] [Google Scholar]
- 28.Mitrovic V, Klein HH, Krekel N, Kreuzer J, Fichtlscherer S, Schirmer A, Paar WD, Hamm CW: Influence of the angiotensin converting enzyme inhibitor ramipril on high-sensitivity C-reactive protein (hs-CRP) in patients with documented atherosclerosis. Z Kardiol 94: 336–342, 2005 [DOI] [PubMed] [Google Scholar]
- 29.Wright JT, Jr, Bakris G, Greene T, Agodoa LY, Appel LJ, Charleston J, Cheek D, Douglas-Baltimore JG, Gassman J, Glassock R, Hebert L, Jamerson K, Lewis J, Phillips RA, Toto RD, Middleton JP, Rostand SG: Effect of blood pressure lowering and antihypertensive drug class on progression of hypertensive kidney disease: Results from the AASK trial. JAMA 288: 2421–2431, 2002 [DOI] [PubMed] [Google Scholar]
- 30.Crawford DC, Akey DT, Nickerson DA: The patterns of natural variation in human genes. Annu Rev Genomics Hum Genet 6: 287–312, 2005 [DOI] [PubMed] [Google Scholar]
- 31.Lakoski SG, Cushman M, Palmas W, Blumenthal R, D'Agostino RB, Jr, Herrington DM: The relationship between blood pressure and C-reactive protein in the Multi-Ethnic Study of Atherosclerosis (MESA). J Am Coll Cardiol 46: 1869–1874, 2005 [DOI] [PubMed] [Google Scholar]
- 32.US Renal Data System: USRDS 2006 Annual Data Report: Atlas of End-Stage Renal Disease in the United States, Bethesda, National Institutes of Health, National Institute of Diabetes and Digestive and Kidney Diseases, 2006 [Google Scholar]
- 33.Gabay C, Kushner I: Acute-phase proteins and other systemic responses to inflammation. N Engl J Med 340: 448–454, 1999 [DOI] [PubMed] [Google Scholar]
- 34.Frazer KA, Ballinger DG, Cox DR, Hinds DA, Stuve LL, Gibbs RA, Belmont JW, Boudreau A, Hardenbol P, Leal SM, Pasternak S, Wheeler DA, Willis TD, Yu F, Yang H, Zeng C, Gao Y, Hu H, Hu W, Li C, Lin W, Liu S, Pan H, Tang X, Wang J, Wang W, Yu J, Zhang B, Zhang Q, Zhao H, Zhou J, Gabriel SB, Barry R, Blumenstiel B, Camargo A, Defelice M, Faggart M, Goyette M, Gupta S, Moore J, Nguyen H, Onofrio RC, Parkin M, Roy J, Stahl E, Winchester E, Ziaugra L, Altshuler D, Shen Y, Yao Z, Huang W, Chu X, He Y, Jin L, Liu Y, Sun W, Wang H, Wang Y, Xiong X, Xu L, Waye MM, Tsui SK, Xue H, Wong JT, Galver LM, Fan JB, Gunderson K, Murray SS, Oliphant AR, Chee MS, Montpetit A, Chagnon F, Ferretti V, Leboeuf M, Olivier JF, Phillips MS, Roumy S, Sallee C, Verner A, Hudson TJ, Kwok PY, Cai D, Koboldt DC, Miller RD, Pawlikowska L, Taillon-Miller P, Xiao M, Tsui LC, Mak W, Song YQ, Tam PK, Nakamura Y, Kawaguchi T, Kitamoto T, Morizono T, Nagashima A, Ohnishi Y, Sekine A, Tanaka T, Tsunoda T, et al. : A second generation human haplotype map of over 3.1 million SNPs. Nature 449: 851–861, 2007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Wang CH, Li SH, Weisel RD, Fedak PW, Dumont AS, Szmitko P, Li RK, Mickle DA, Verma S: C-reactive protein upregulates angiotensin type 1 receptors in vascular smooth muscle. Circulation 107: 1783–1790, 2003 [DOI] [PubMed] [Google Scholar]