Abstract
α-Synuclein (α-syn), a protein implicated in Parkinson’s disease, is structurally diverse. In addition to its random-coil state, α-syn can adopt an α-helical structure upon lipid membrane binding or a β-sheet structure upon aggregation. We used yeast biology and in vitro biochemistry to detect how sequence changes alter the structural propensity of α-syn. The N-terminus of the protein, which adopts an α-helical conformation upon lipid binding, is essential for membrane binding in yeast, and variants that are more prone to forming an α-helical structure in vitro are generally more toxic to yeast. β-Sheet structure and inclusion formation, on the other hand, appear to be protective, possibly by sequestering the protein from the membrane. Surprisingly, sequential deletion of residues 2 through 11 caused a dramatic drop in α-helical propensity, vesicle binding in vitro, and membrane binding and toxicity in yeast, part of which could be mimicked by mutating aspartic acid at position 2 to alanine. Variants with distinct structural preferences, identified here by a reductionist approach, provide valuable tools for elucidating the nature of toxic forms of α-syn in neurons.
Keywords: α-synuclein, Parkinson’s disease, secondary structure, N-terminus, yeast toxicity
Introduction
Parkinson’s disease (PD), the most prevalent neurodegenerative motor disorder, is characterized by progressive loss of dopaminergic neurons and formation of proteinaceous cytoplasmic inclusions (Lewy bodies).1 The major fibrillar constituent of Lewy bodies is α-synuclein (α-syn). Missense mutations in the wild-type α-syn gene (A30P, A53T, and E46K) and allele multiplication of the wild-type gene are linked to early-onset PD. It is unclear whether α-syn aggregation drives disease progression and, if so, how it leads to neuronal cell degeneration.
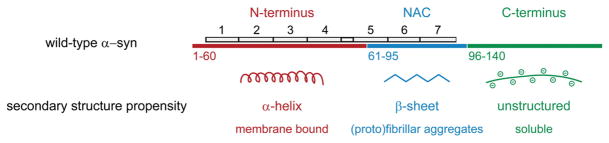
The α-syn monomer is natively unfolded;2 that is, it does not assume a single stable conformation in solution. Apart from its random-coil state, the protein can adopt a β-sheet conformation upon aggregation in vivo1 and in vitro,3 or an α-helical conformation upon binding to membranes.4 α-Syn can be divided into three regions (Fig. 1): (i) the N-terminus (residues 1–60), which consists of amphipathic repeats that fold into an extended or broken α-helix (residues 3–94)5 upon binding to phospholipid vesicles and bicelles, respectively;6 (ii) the middle hydrophobic region [non-Aβ component of amyloid plaques (middle region of α-syn) (NAC); residues 61–95], which is prone to β-sheet formation and fibrillization; and (iii) the C-terminus (residues 96–140), which is rich in acidic residues and prolines.
Fig. 1.
Wild-type α-syn can be divided into three regions. The N-terminus (red) adopts an α-helical structure upon binding to lipids (the seven 11-mer repeats are depicted in gray), the NAC (blue) is hydrophobic and prone to forming β-sheet aggregates, whereas the C-terminus (green) is largely negatively charged and promotes protein solubility.
α-Syn membrane binding is probably required for its normal function, but may also promote toxicity or formation of toxic species. At the membrane, the local protein concentration is high, possibly favoring protein–protein interactions, leading to aggregation. α-Syn forms β-sheet-rich spherical oligomers (protofibrils) in vitro, which subsequently assemble into mature fibrils.7 The observation that acceleration of oligomerization, not fibrillization, is a common characteristic of the pathogenic A30P and A53T α-syn mutants suggests that protofibrils, not fibrils, are toxic.8 Both mutants and wild type form annular protofibrils, which bind and permeabilize lipid vesicles, forming pores resembling those of bacterial toxins.9,10 Recent results in yeast show that non-fibrillar α-syn mediates toxicity.11 Overexpressed monomeric α-helical α-syn may coat the internal membrane, thereby nonspecifically disrupting membrane-based processes and membrane homeostasis. Although the secondary structure and oligomerization state of the toxic species remain elusive, the results above support a key role for membrane binding in α-syn pathogenesis.
In order to more easily study the relationship between α-syn structure and neurotoxicity, generation of variants with distinct structural preferences (α-helix versus β-sheet versus random coil) is required. For this purpose, we constructed a series of α-syn variants, in which the recognized sequence domains (N-terminus, NAC, and C-terminus) were elongated, truncated, or deleted (Fig. 1). The baker’s yeast Saccharomyces cerevisiae is an appealing screening tool, given that (i) it is a simple well-established system,12 (ii) toxicity is a sensitive measure of membrane binding,11 and (iii) protein localization inside the cell can be easily monitored microscopically. The toxicity and localization of our variants were examined in yeast, and selected sequences were characterized in vitro.
α-Syn binds to artificial membranes in vitro4 and to plasma membranes in yeast,12 but is predominately cytosolic in neurons; its localization to the nerve terminal and association with synaptic vesicles13 are mediated by transient, rapidly reversible interactions.14 Wild-type α-syn and the A53T mutant initially localize at the yeast plasma membrane before forming cytoplasmic inclusions in a concentration-dependent manner.12 Both proteins, when overexpressed, strongly inhibit yeast growth. In contrast, the A30P mutant is nontoxic to yeast and is dispersed throughout the yeast cytoplasm, in line with its poor ability to bind to vesicles from neuroblastoma cells15 and rat brain.16 Although S. cerevisiae does not endogenously express the α-syn protein, it reproduces certain aspects of α-syn biology observed in higher eukaryotes12 such as proteasomal dysfunction,17–19 vesicle trafficking impairment,20 and lipid droplet accumulation.21 We report here that α-helical propensity and membrane binding are essential for α-syn toxicity to yeast, whereas β-sheet propensity and cytosolic inclusions are protective. Whether these findings are unique to the yeast system or reflective of neurotoxicity remains to be seen.
Results and Discussion
C-terminal alterations in α-syn do not affect yeast toxicity
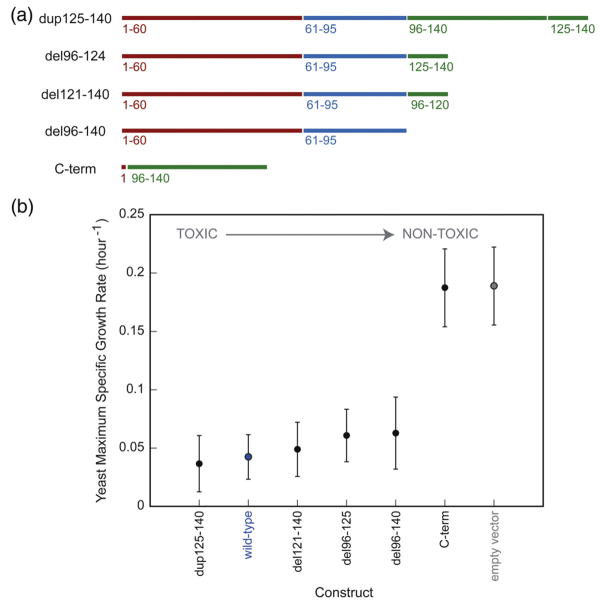
The design of α-syn analogs was based on published studies. The C-terminus of α-syn is composed of two repeats, the second of which (residues 125–140) displays chaperone activity.22 The C-terminally truncated variant del96–124 contains only the second repeat, whereas the elongated dup125–140 variant contains three repeats (duplicated second repeat; Fig. 2a). C-terminally truncated forms of α-syn have been found in brain extracts as a result of proteolytic cleavage23 or alternative splicing.24 Del121–140 α-syn, which aggregates faster than wild type25 and increases the susceptibility of neuroblastoma cells to reactive oxygen species,26 was studied in yeast. In addition to variants with altered C-termini, we also tested a variant lacking this region altogether, as well as the C-terminus in isolation.
Fig. 2.
C-terminal alterations and their effect on yeast toxicity. (a) We created variants in which the C-terminal region was extended, truncated, or deleted. (b) Maximum specific growth rates of yeast cells expressing the α-syn constructs from the high-copy-number p426GAL1 plasmid were measured; they are inversely proportional to toxicity. Wild-type α-syn is labeled in blue, and the empty vector control is labeled in gray. The genes are listed in order of decreasing yeast toxicity.
α-Syn variants were cloned into the 2 μ (high-copy-number) p426GAL1 plasmid and transformed into W303-1a yeast. Cell growth was monitored, and the data were fitted to the Gompertz equation, out of which the maximum specific growth rate (inversely proportional to the doubling time) was extracted.11 Each gene was studied in at least seven independent trials. Like wild-type α-syn, C-terminal alterations (deletion, truncation, or duplication) were highly toxic to yeast (Fig. 2b). Given that α-syn wild-type overexpression causes a dramatic decrease in yeast growth,11,12 small growth differences between these proteins (if any) may not be detectable under these conditions. When expressed independently, the C-terminus is not toxic to yeast (Fig. 2b).
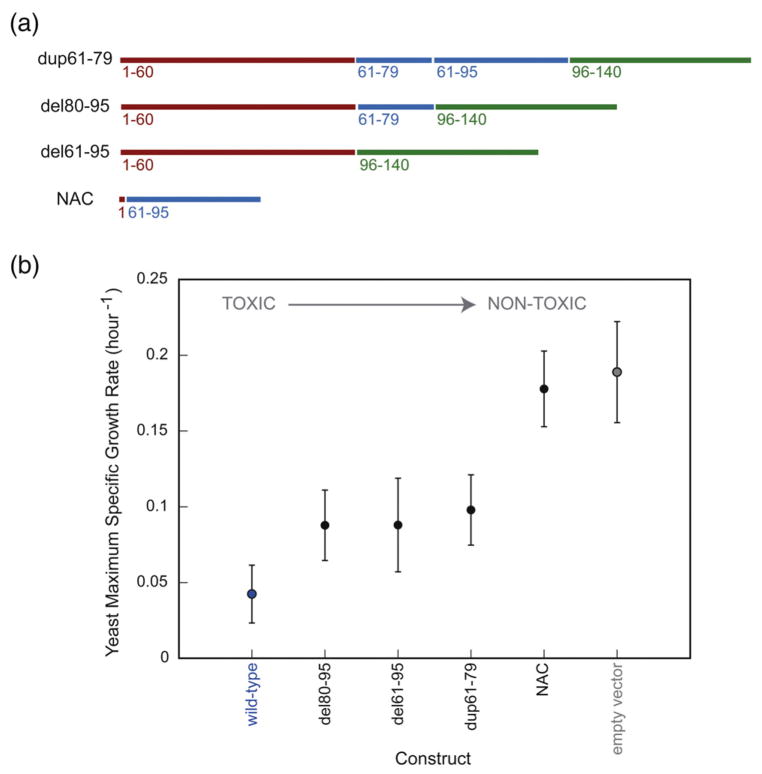
Variants with modified NAC regions display intermediate toxicities
Residues 61–79 comprise the most hydrophobic portion of the NAC region. This region was duplicated in the dup61–79 variant and was retained in the del80–95 variant, which lacks all other NAC residues (Fig. 3a). A variant lacking the entire NAC region (del61–95) was also studied in yeast. Although the NAC is essential for α-syn toxicity in dopaminergic neurons in vitro27 and in a Drosophila model of PD,28 alterations in this domain caused only a moderate reduction in yeast toxicity (Fig. 3b). In accord with our results, β-syn, lacking 11 amino acids (73–83) from the NAC region, has been reported to be somewhat toxic to yeast.11 As a control, NAC was expressed alone in yeast and was found to be nontoxic (Fig. 3b). We also replaced the NAC in α-syn with the more fibrillogenic amyloid β 42 peptide (Aβ42) sequence. Interestingly, NACsubAβ was less toxic than wild type and formed fewer cytoplasmic inclusions (Supplementary Fig. 1), possibly because amyloid β peptide (Aβ) lacks amphipathic repeats and is less prone to adopting an α-helical structure (Supplementary Fig. 2). When the NAC was substituted with the nonfibrillogenic Aβ F19S/L34P double mutant,29 similar results were obtained (Supplementary Figs. 1 and 2).
Fig. 3.
Alterations in the NAC region of α-syn (a) and their effect on yeast toxicity (b).
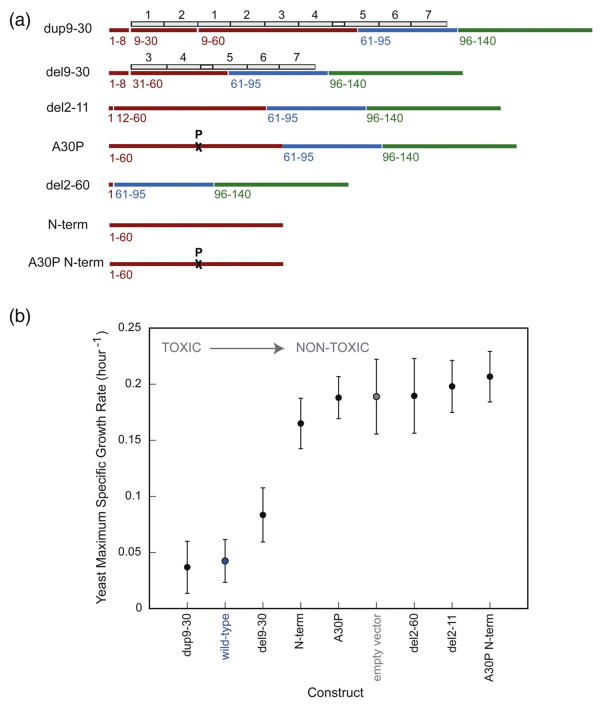
The N-terminus is essential for α-syn toxicity in yeast
The number of N-terminal amphipathic repeats modulates the propensity of α-syn to adopt an α-helical conformation in vitro.30 Selection of N-terminal sequence modifications was based on results showing that variants with two additional (dup9–30) or fewer (del9–30) repeats (Fig. 4a) have very different biophysical properties: the first is prone to forming an α-helical structure, whereas the latter is prone to forming a β-sheet structure, including amyloid fibrils.30 Additionally, we probed the role of the most N-terminal amino acids, which are not part of the amphipathic repeats.
Fig. 4.
N-terminal alterations (a) and their effect on yeast toxicity (b).
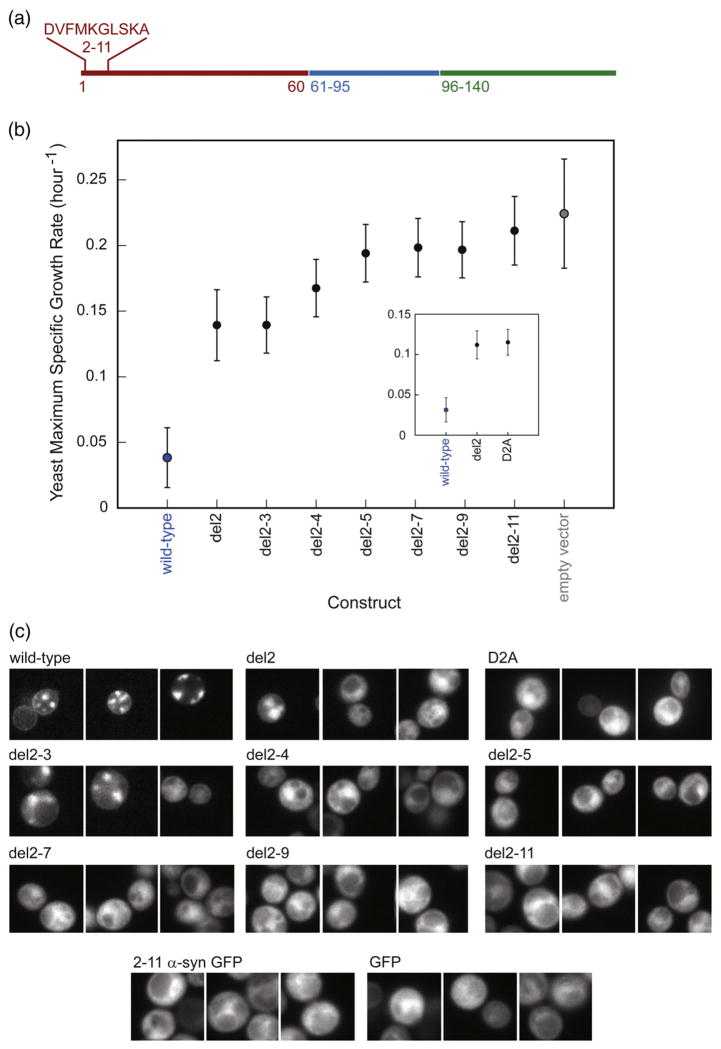
Dup9–30 was more toxic to yeast than del9–30 (Fig. 4b), while deletion of the entire N-terminus rendered the protein nontoxic. The N-terminus is, however, by itself, not toxic to yeast. Surprisingly, deletion of as few as 10 N-terminal residues (amino acids 2–11: DVFMKGLSKA; Fig. 5a) dramatically reduced yeast toxicity. To identify the minimal number of N-terminal amino acid deletions required to alleviate yeast toxicity, we created a series of variants lacking one (del2), two (del2–3), three (del2–4), four (del2–5), six (del2–7), and eight (del2–9) N-terminal amino acids (the starting Met residue was retained). Deletion of as few as one to two amino acids from the α-syn N-terminus significantly decreased yeast toxicity, whereas deletion of more than four amino acids made the proteins nontoxic (Fig. 5b). The mutation D2A decreased yeast toxicity similarly to del2 (Fig. 5b, insert).
Fig. 5.
N-terminal deletions dramatically reduce yeast toxicity. (a) The location and identity of the deleted amino acids are shown in wild-type α-syn. (b) Yeast maximum specific growth rates of the variants, listed in order of increasing deletion size. The D2A mutant behaves like del2 (insert). (c) Microscopy of GFP-tagged N-terminally truncated α-syns. Deletion of Asp2 or D2A substitution reduces membrane localization and inclusion formation. Variants lacking three or more amino acids from the N-terminus localize to the cytoplasm. GFP bearing α-syn amino acids 2–11, unlike GFP-tagged α-syn, does not associate with the plasma membrane, suggesting that the N-terminal amino acids are not a membrane localization sequence.
Fluorescence microscopy was used to localize α-syn variants in yeast
To gain insight into how each protein region (N-terminus, NAC, and C-terminus) affects the sub-cellular localization of α-syn, we visualized yeast cells expressing all constructs fused to the green fluorescent protein (GFP). Cytoplasmic α-syn variants were less toxic than membrane-bound variants (Supplementary Fig. 3), in agreement with previous results.11 Del2 and del2–3 formed significantly fewer inclusions than wild type, whereas all other N-terminally truncated variants were dispersed throughout the cytosol (Fig. 5c). The 10 N-terminal amino acids alone failed to direct GFP to the plasma membrane; similar results were obtained when 2–11 α-syn-GFP was expressed at lower levels (one chromosomally integrated copy, instead of a 2 μ plasmid; data not shown). GFP was nontoxic to yeast and did not alter the yeast toxicity profile of the α-syn variants when attached to their C-termini (Supplementary Fig. 4). The fluorescence intensity of GFP fusions increased with increasing deletion size (Fig. 5c), demonstrating that reduced toxicity is not caused by a lowered expression level. This result is consistent with previous data showing that yeast cells expressing toxic α-syn variants bear fewer plasmid copies (selective advantage) than yeast cells expressing nontoxic variants.11 The difference in growth rate between toxic and nontoxic variants is, therefore, smaller than it would have been if all cells contained a fixed number of gene copies (more than 1).
In a recent genetic screen, inactivation of the N-terminal acetyltransferase NatB decreased α-syn membrane binding and toxicity in yeast.31 In contrast, disruption of NatA and NatC activities did not show such an effect. Wild-type α-syn is a potential substrate for NatB, which (unlike its NatA and NatC counterparts) targets Met-Glu or Met-Asp sequences.32 N-terminal acetylation (removal of the N-terminal amine-positive charge) stabilizes the helix dipole33 and likely promotes α-helix formation coupled to membrane binding, whereas lack of such a posttranslational modification may account for the more cytoplasmic distribution of del2 and D2A compared to wild type (Fig. 5c).
Wild-type α-syn inclusion formation was induced in yeast by fibril-prone peptides
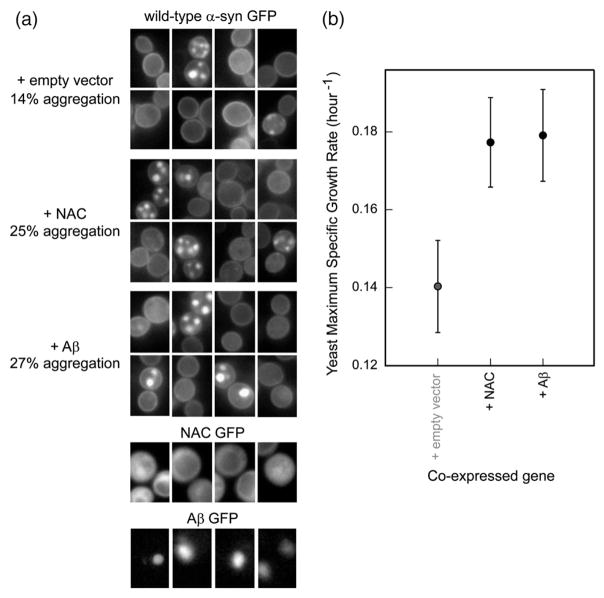
α-Syn inclusion formation in yeast is initiated at the plasma membrane.12,34 To directly study the effect of α-syn inclusion formation (decoupled from membrane binding) on yeast toxicity, an external factor that influences the inclusion formation profile of the wild-type protein without influencing yeast growth must be applied. Given the known affinity between Aβ42 and NAC in vivo35 and in vitro,36 we studied the effects of Aβ42 (high-copy-number expression) on α-syn-GFP (low-copy-number expression) localization. Indeed, Aβ42 promoted α-syn inclusion formation in yeast (Fig. 6a). The NAC peptide also increased the number of punctuate inclusions, compared to control. The nature and/or origin of α-syn inclusions, formed in the presence or in the absence of the peptides, may be different. Cells expressing α-syn together with the Aβ42 or the NAC peptide grew faster than cells expressing α-syn alone (Fig. 6b); neither Aβ42 nor NAC is toxic to yeast when expressed individually (Fig. 3b; Supplementary Fig. 1b). α-Syn inclusion formation in the presence of Aβ42 or NAC therefore reduces yeast toxicity. In analogy to our results, pharmacological promotion of inclusion formation reduces α-syn toxicity in neuroglioma cells.37 Microscopic images of Aβ42 and NAC GFP fusions showed that the former produced cytosolic inclusions, whereas the latter was evenly distributed throughout the yeast cytoplasm; neither one localized to the plasma membrane (Fig. 6a). The Aβ42 and NAC peptides may rescue yeast toxicity by ‘capturing’ α-syn in the yeast cytoplasm, thereby reducing binding to the plasma membrane.
Fig. 6.
NAC and Aβ42 promote the formation of cytosolic inclusions of wild-type α-syn in yeast and reduce toxicity. (a) NAC and Aβ42 were expressed from the high-copy-number p426GAL1 plasmid, whereas α-syn-GFP was expressed from the low-copy-number p414GAL1 plasmid. To calculate the percentages of inclusion formation, photographs of ca 1000 yeast cells coexpressing an empty vector, NAC, or Aβ42 with α-syn-GFP were taken; those containing punctuate bodies were counted and divided by the total number of cells. Microscopic images of GFP-tagged NAC and Aβ42 (p426GAL1 plasmid) are shown for comparison; the first is cytosolic, whereas the latter forms inclusions. Unlike α-syn, neither NAC nor Aβ42 localized at the yeast plasma membrane. (b) Maximum specific growth rates of yeast cells coexpressing wild-type α-syn from centromeric p414GAL1 plasmid with empty vector, NAC, or Aβ42 from a 2 μm p426GAL1 plasmid. NAC and Aβ42 reduce α-syn-induced yeast toxicity possibly by sequestering the protein from the membrane.
α-Helical propensity generally correlates with yeast toxicity
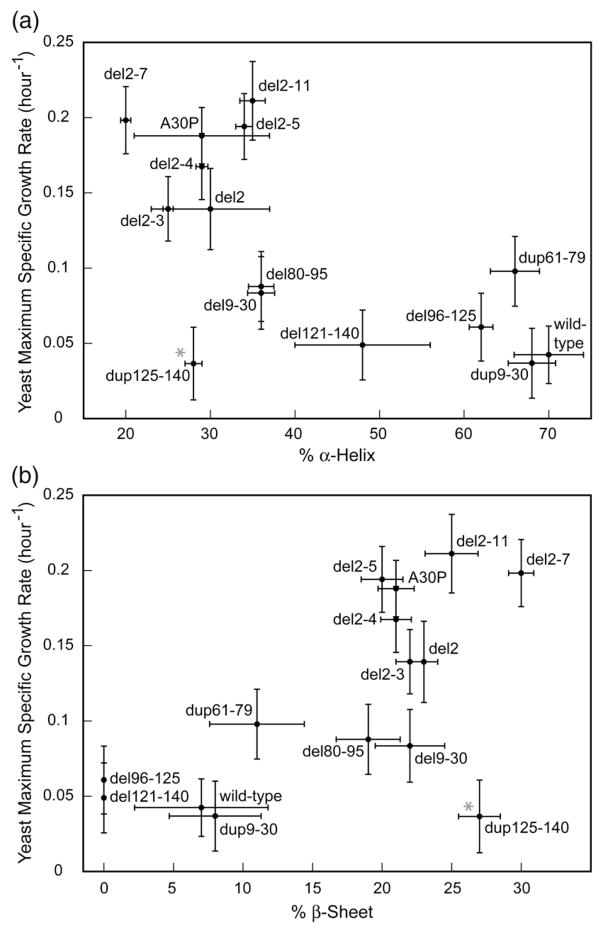
The α-syn variants were overexpressed in Escherichia coli, as previously described.11 The secondary structure propensity of the purified proteins was assessed by circular dichroism (CD) spectroscopy. α-Syn forms an α-helical structure in 4% 1,1,1,3,3,3-hexafluoro-2-propanol (HFIP), an environment that resembles the membrane surface.38 Wild-type and dup9–30 α-syns adopted an α-helical conformation, whereas del9–30 formed a β-sheet structure, in accord with previous results.30 CD spectra of all variants (Supplementary Fig. 5) were analyzed using the software CONTIN.39 Toxic variants generally had a higher propensity to form an α-helical structure (Fig. 7a), in line with data showing a correlation between α-syn toxicity in yeast and membrane binding.11 In contrast, nontoxic variants were more prone to forming a β-sheet structure under the same conditions (4% HFIP; Fig. 7b).
Fig. 7.
Secondary structural content of α-syn variants in 4% HFIP as a function of yeast maximum specific growth rate. Wild-type α-syn adopts an α-helical conformation under these conditions, which mimics the membrane surface environment.38 CD spectra were obtained at 25 °C with 16 μM protein in 10 mM Tris buffer (pH 7.4), 150 mM NaCl, and 4% HFIP. Proteins samples were incubated with HFIP for 18 h prior to the CD measurements. The data (Supplementary Fig. 5) were analyzed using the CONTIN software.39 Under these conditions, toxic variants generally had a higher propensity to form an α-helical structure (a), whereas nontoxic variants were more prone to adopting a β-sheet structure (b). We performed linear regression analysis on the points shown, with growth rate as the response variable and percent helix/sheet as the explanatory variable (this analysis does not make use of error bars). The R2 value is about 40%, and the slopes of the regression lines are significantly different from zero (p=0.01 in both cases), indicating that there is a significant linear correlation. Data for dup125–140, the sole outlier in both graphs, are marked with a gray asterisk. Growth rate errors were taken from Figs. 2b–5b, and % α-Helix/β-Sheet structure errors were calculated by the CONTIN software.
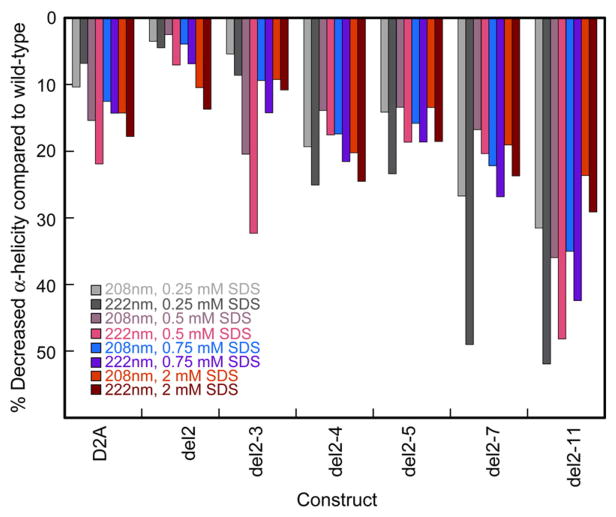
Sequential deletion of α-syn N-terminal amino acids decreased α-helical structure propensity and vesicle binding ability in vitro
To further investigate the effect of N-terminal truncation on α-syn secondary structure, we obtained CD spectra of the variants in the presence of increasing sodium dodecyl sulfate (SDS) concentrations (0.25 mM, 0.5 mM, 0.75 mM, and 2.0 mM); SDS titration induces gradual formation of α-helical structures in wild-type α-syn40 and in all truncated variants. All variants were less α-helical than wild type (based on ellipticity values at 208 nm and 222 nm) at all SDS concentrations (Fig. 8), in line with their weaker membrane binding in yeast (Fig. 5c). Asp2 likely stabilizes the α-helix by diminishing the dipole moment of its N-terminus.33 Consistent with that hypothesis, decreased negative charge at the N-terminus (deletion of Asp2 or mutation to Ala) promoted yeast growth, whereas in a related study,11 increased negative charge near the N-terminus (only amino acids from position 8 and onwards were probed) reduced yeast toxicity by disfavoring interactions with the negatively charged phospholipid headgroups or hydrophobic membrane interior.
Fig. 8.
SDS titration of wild-type, D2A, and N-terminally truncated α-syn monitored by CD spectroscopy. Each bar represents the percent decrease in CD signal of each variant compared to that of wild type [(CD signal of variant–CD signal of wild type)/(CD signal of wild type)×100%] for a given SDS concentration and wavelength (208 nm or 222 nm). We determined the protein concentration of each CD sample by the BCA assay (average of three independent measurements) and corrected the spectra to account for small differences in protein concentrations among the variants.
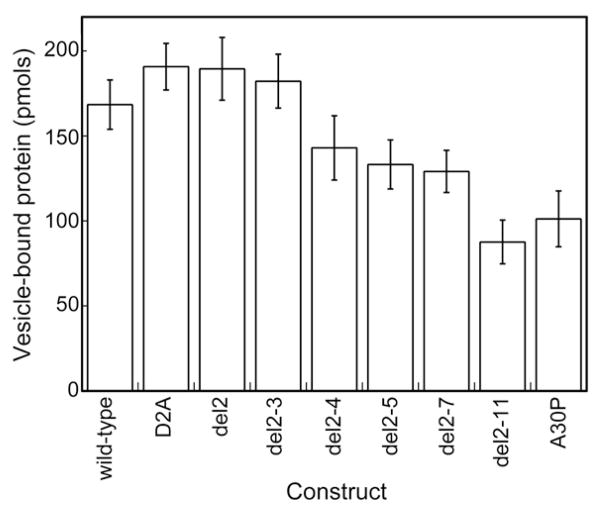
In a complementary experiment, we determined the ability of the truncated variants to bind to negatively charged phospholipid vesicles, using a vesicle sedimentation assay.11 Variants lacking one to two N-terminal amino acids, as well as the D2A mutant, exhibited a vesicle-binding capacity similar to that of wild type, whereas variants lacking more than three amino acids showed a significant decrease in vesicle binding (Fig. 9). The differences in α-helical structure propensity between wild type and D2A, del2, or del2–3 observed by CD may be too subtle to be detected by this method. Del2–11 bound vesicles more weakly than any other variant and was comparable to A30P α-syn, which has impaired membrane-binding ability.41 N-terminal amino acids, therefore, appear to template α-helix formation.
Fig. 9.
Vesicle binding data for wild-type, D2A, and N-terminally truncated α-syn variants. A30P α-syn, which is known to have a membrane binding affinity weaker than that of wild type,41 was included as control.
Concluding Remarks
The membrane-binding ability of α-syn may be required for its normal function, but protein accumulation at the membrane may promote formation of toxic species. Using yeast as a screening tool, we identified intrinsic (extreme N-terminal amino acids) and extrinsic (hydrophobic Aβ42 and NAC peptides) factors that modulate α-syn membrane binding. Future studies will use these variants as tools for elucidating the toxic mechanism in more relevant models of PD.
Materials and Methods
DNA synthesis
Genes encoding the α-syn variants were constructed using an approach reminiscent of DNA shuffling.42 The wild-type α-syn gene was used as template to produce three ‘modules’: the N-terminus, the NAC, and the C-terminus. Synthetic oligonucleotides encoding amino acids 61–79 and 125–140 were used. Oligonucleotides with complementary sequences to the 3′ and 5′ ends of two sequential modules were designed to link them together. Modules and oligonucleotides were mixed, and the corresponding genes were assembled. The construction of dup9–30 and del9–30 variants has been previously described.30 The Aβ42 gene was assembled from two overlapping synthetic oligonucleotides. All genes were PCR-amplified with forward primer 5′-GACTTCTAACTAGTAAAAAATGGATGTATTCATGAAAGGAC-3′ (contains a SpeI site and a yeast consensus translation sequence before the start codon) and reverse primers 5′-TCCGGACTCGAGTTAGGCTTCAGGTTCGTAG-TCTTGATACC-3′ (contains a XhoI site and a stop codon) and 5′-TCCGGAAAGCTTGGCTTCAGGTTCGTAGTCTTGATACCCTTC-3′ (for preparing GFP fusion proteins; contains a HindIII site and lacks a stop codon). The proteins are fused to GFP with a short linker.11 N-terminally (del2 to del2–11) and C-terminally (del121–140) truncated variants were constructed using appropriately modified forward and reverse primers, respectively. The PCR products were digested, gel-purified, and ligated into the correspondingly digested vector p426GAL1 lacking and bearing GFP.11 The ligation products were transformed into chemically competent DH5a E. coli cells, and the transformations were grown on LB-agar plates supplemented with ampicillin. All sequences (see Supplementary Material) were verified by DNA sequencing.
Yeast transformation and growth studies
S. cerevisiae haploid strain W303-1a was transformed with high-copy-number p426GAL1 plasmids bearing the α-syn genes, as previously described.11 The transformation was plated onto a synthetic noninducing (glucose) medium lacking uracil (yeast-nitrogen-based media were obtained from Difco, and CSM dropout mixtures were obtained from Qbiogene) and incubated at 30 °C for 3–4 days. Single colonies were inoculated into 1 mL of synthetic inducing (galactose) media lacking uracil, in each well of sterile 12-well polystyrene plates. Yeast cells coexpressing α-syn (low-copy-number p414GAL1 plasmid) and Aβ42 or NAC (high-copy-number p426GAL1 plasmid) were grown in media lacking uracil and tryptophan. Yeast growth was monitored as reported, and the data were fitted to the Gompertz growth equation, out of which the maximum specific growth rate was extracted.11 The data were analyzed using an additive two-way analysis of variance model (synuclein genes and independent experiments). Error bars represent simultaneous Tukey–Kramer comparison intervals (p< 0.05). MATLAB software was used for statistical analysis; each gene was studied in at least seven independent measurements.
Yeast microscopy
Protein localization was monitored by GFP fluorescence. Yeast cells grown for 48 h in galactose media lacking uracil were examined with a Zeiss Axioskop 2 plus microscope, using a 100× objective. Representative photographs are shown in Fig. 5c, Supplementary Fig. 3 (exposure time, 100 ms), and Fig. 6a (exposure time, 250 ms). Given that the sample exposure time was identical within each experimental set, the data are directly comparable.
Protein production and purification
Gene variants were subcloned into a pT7-7 vector, and their sequences were verified by DNA sequencing. E. coli BL21-Gold(DE3) cells (Stratagene) were transformed with the corresponding pT7-7 plasmids and plated on LB media supplemented with ampicillin. One hundred milliliters of LB ampicillin (150 mg/L) media was inoculated with single colonies, and the cultures were grown at 37 °C and 250 rpm to an OD600 nm of ca 0.75–1.5. The cultures were then induced with 1 mM IPTG and supplemented with additional ampicillin (100 mg/L). After 3 h at 37 °C and 250 rpm, the cells were harvested, and the pellet was resuspended in 0.75 mL of buffer [50 mM Tris (pH 8.0), 10 mM ethylenediaminetetraacetic acid, and 150 mM NaCl] and frozen at −80 °C. The proteins were purified as previously reported,11 lyophilized, and stored in a desiccator (over P2O5) in the dark.
CD experiments
A few milligrams of lyophilized powder were resuspended in 80 mL of Tris buffer [10 mM Tris (pH 7.4) and 150 mM NaCl]. The pH was adjusted to ca 7.4 (using pH paper) by addition of 1 M NaOH (several microliters). α-Syn is acidic and poorly soluble at low pH. The proteins were dialyzed (10 kDa; Pierce Slide-A-Lyzer mini dialysis units) against 2 L of Tris buffer at 4 °C overnight, and then against another 2 L of Tris buffer for 2 h. After dialysis, the samples were spun through ultrafilters (100 kDa; Microcon) for 7 min to sterilize and remove oligomers and higher-order aggregates. The proteins were analyzed by SDS-PAGE gel electrophoresis (16.5% N-[2-hydroxy-1,1-bis(hydroxymethyl)ethyl]glycine gel, Coomassie staining; Bio-Rad): their purity was generally >95%, and each band corresponded to the expected molecular weight (Supplementary Fig. 6). Protein concentrations were determined by the BCA assay (Pierce) and standardized against the absorbance of wild-type α-syn (ε280 nm =6500 M −1 cm−1). CD spectra were recorded at 25 °C on an Aviv Biomedical spectrometer (model 410) equipped with a Peltier temperature controller. Three scans from 190 nm to 260 nm for the HFIP experiments, and from 195 nm to 245 nm for the SDS titration experiments, in 1-nm increments with 0.33 s of averaging time, were collectively averaged to obtain each spectrum, using a 1-mm pathlength cell and 16 μM protein.
Vesicle binding experiments
Proteins were extensively dialyzed (10 kDa; Pierce Slide-A-Lyzer mini dialysis units) against 2 L of HBS buffer [10 mM Hepes (pH 7.4) and 145 mM KCl] at 4 °C overnight, and then against another 2 L of HBS buffer for 2 h. Vesicles were prepared using egg phosphatidylglycerol (Avanti Polar Lipids), as previously described.11 Purified synuclein (25 μL) in HBS and 10 mM CaCl2 were incubated with 25 μL of lipid solution for 1 h at room temperature. The final concentration of protein in the lipid mixture was either ca 75 μM or 38 μM. Using an established protocol,11 we measured the total and vesicle-bound proteins. Each measurement was repeated three times at high and low concentrations, and the data were averaged and interpolated to 50 μM protein. The experiment was performed four times, yielding the statistical significance shown in Fig. 9.
Supplementary Material
Acknowledgments
We are grateful to Dr. Michael S. Wolfe and his laboratory members for helpful discussions. This work was supported by a Morris K. Udall Parkinson’s Disease Research Center of Excellence grant (NS038375; P.T.L.) and a postdoctoral fellowship from the Swiss National Science Foundation (K.V.).
Abbreviations used
- α-syn
α-synuclein
- PD
Parkinson’s disease
- NAC
non-Aβ component of amyloid plaques (middle region of α-syn)
- Aβ42
amyloid β 42 peptide
- Aβ
amyloid β peptide
- GFP
green fluorescent protein
- HFIP
1,1,1,3,3,3-hexafluoro-2-propanol
Footnotes
Supplementary Data: Supplementary data associated with this article can be found, in the online version, at doi:10.1016/j.jmb.2009.03.021
References
- 1.Spillantini MG, Schmidt ML, Lee VM, Trojanowski JQ, Jakes R, Goedert M. Alpha-synuclein in Lewy bodies. Nature. 1997;388:839–840. doi: 10.1038/42166. [DOI] [PubMed] [Google Scholar]
- 2.Weinreb PH, Zhen W, Poon AW, Conway KA, Lansbury PT., Jr NACP, a protein implicated in Alzheimer’s disease and learning, is natively unfolded. Biochemistry. 1996;35:13709–13715. doi: 10.1021/bi961799n. [DOI] [PubMed] [Google Scholar]
- 3.Conway KA, Harper JD, Lansbury PT., Jr Fibrils formed in vitro from alpha-synuclein and two mutant forms linked to Parkinson’s disease are typical amyloid. Biochemistry. 2000;39:2552–2563. doi: 10.1021/bi991447r. [DOI] [PubMed] [Google Scholar]
- 4.Davidson WS, Jonas A, Clayton DF, George JM. Stabilization of alpha-synuclein secondary structure upon binding to synthetic membranes. J Biol Chem. 1998;273:9443–9449. doi: 10.1074/jbc.273.16.9443. [DOI] [PubMed] [Google Scholar]
- 5.Ulmer TS, Bax A, Cole NB, Nussbaum RL. Structure and dynamics of micelle-bound human α-synuclein. J Biol Chem. 2005;280:9595–9603. doi: 10.1074/jbc.M411805200. [DOI] [PubMed] [Google Scholar]
- 6.Georgieva ER, Ramlall TF, Borbat PP, Freed JH, Eliezer D. Membrane-bound α-synuclein forms an extended helix: long-distance pulsed ESR measurements using vesicles, bicelles, and rodlike micelles. J Am Chem Soc. 2008;130:12856–12857. doi: 10.1021/ja804517m. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Volles MJ, Lansbury PT., Jr Zeroing in on the pathogenic form of alpha-synuclein and its mechanism of neurotoxicity in Parkinson’s disease. Biochemistry. 2003;42:7871–7878. doi: 10.1021/bi030086j. [DOI] [PubMed] [Google Scholar]
- 8.Conway KA, Lee SJ, Rochet J-C, Ding TT, Williamson RE, Lansbury PT., Jr Acceleration of oligomerization, not fibrillization, is a shared property of both alpha-synuclein mutations linked to early-onset Parkinson’s disease: implications for pathogenesis and therapy. Proc Natl Acad Sci USA. 2000;97:571–576. doi: 10.1073/pnas.97.2.571. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Lashuel HA, Hartley D, Petre BM, Walz T, Lansbury PT., Jr Neurodegenerative disease: amyloid pores from pathogenic mutations. Nature. 2002;418:291. doi: 10.1038/418291a. [DOI] [PubMed] [Google Scholar]
- 10.Volles MJ, Lansbury PT., Jr Vesicle permeabilization by protofibrillar alpha-synuclein is sensitive to Parkinson’s disease-linked mutations and occurs by a pore-like mechanism. Biochemistry. 2002;41:4595–4602. doi: 10.1021/bi0121353. [DOI] [PubMed] [Google Scholar]
- 11.Volles MJ, Lansbury PT., Jr Relationships between the sequence of alpha-synuclein and its membrane affinity, fibrillization propensity, and yeast toxicity. J Mol Biol. 2007;366:1510–1522. doi: 10.1016/j.jmb.2006.12.044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Outeiro TF, Lindquist S. Yeast cells provide insight into alpha-synuclein biology and pathobiology. Science. 2003;302:1772–1775. doi: 10.1126/science.1090439. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Iwai A, Masliah E, Yoshimoto M, Ge N, Flanagan L, de Silva HA, et al. The precursor protein of non-A beta component of Alzheimer’s disease amyloid is a presynaptic protein of the central nervous system. Neuron. 1995;14:467–475. doi: 10.1016/0896-6273(95)90302-x. [DOI] [PubMed] [Google Scholar]
- 14.Fortin D, Nemani V, Voglmaier S, Anthony M, Ryan T, Edwards R. Neural activity controls the synaptic accumulation of α-synuclein. J Neurosci. 2005;25:10913–10921. doi: 10.1523/JNEUROSCI.2922-05.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Kim YS, Laurine E, Woods W, Lee SJ. A novel mechanism of interaction between alpha-synuclein and biological membranes. J Mol Biol. 2006;360:386–397. doi: 10.1016/j.jmb.2006.05.004. [DOI] [PubMed] [Google Scholar]
- 16.Jensen PH, Nielsen MS, Jakes R, Dotti CG, Goedert M. Binding of alpha-synuclein to brain vesicles is abolished by familial Parkinson’s disease mutation. J Biol Chem. 1998;273:26292–26294. doi: 10.1074/jbc.273.41.26292. [DOI] [PubMed] [Google Scholar]
- 17.Stefanis L, Larsen KE, Rideout HJ, Sulzer D, Greene LA. Expression of A53T mutant but not wild-type alpha-synuclein in PC12 cells induces alterations of the ubiquitin-dependent degradation system, loss of dopamine release, and autophagic cell death. J Neurosci. 2001;21:9549–9560. doi: 10.1523/JNEUROSCI.21-24-09549.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Tanaka Y, Engelender S, Igarashi S, Rao RK, Wanner T, Tanzi RE, et al. Inducible expression of mutant alpha-synuclein decreases proteasome activity and increases sensitivity to mitochondria-dependent apoptosis. Hum Mol Genet. 2001;10:919–926. doi: 10.1093/hmg/10.9.919. [DOI] [PubMed] [Google Scholar]
- 19.Snyder H, Mensah K, Theisler C, Lee J, Matouschek A, Wolozin B. Aggregated and monomeric alpha-synuclein bind to the S6′ proteasomal protein and inhibit proteasomal function. J Biol Chem. 2003;278:11753–11759. doi: 10.1074/jbc.M208641200. [DOI] [PubMed] [Google Scholar]
- 20.Cooper AA, Gitler AD, Cashikar A, Haynes CM, Hill KJ, Bhullar B, et al. Alpha-synuclein blocks ER-Golgi traffic and Rab1 rescues neuron loss in Parkinson’s models. Science. 2006;313:324–328. doi: 10.1126/science.1129462. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Cole NB, Murphy DD, Grider T, Rueter S, Brasaemle D, Nussbaum RL. Lipid droplet binding and oligomerization properties of the Parkinson’s disease protein alpha-synuclein. J Biol Chem. 2002;277:6344–6352. doi: 10.1074/jbc.M108414200. [DOI] [PubMed] [Google Scholar]
- 22.Kim TD, Paik SR, Yang CH. Structural and functional implications of C-terminal regions of alpha-synuclein. Biochemistry. 2002;41:13782–13790. doi: 10.1021/bi026284c. [DOI] [PubMed] [Google Scholar]
- 23.Li W, West N, Colla E, Pletnikova O, Troncoso JC, Marsh L, et al. Aggregation promoting C-terminal truncation of alpha-synuclein is a normal cellular process and is enhanced by the familial Parkinson’s disease-linked mutations. Proc Natl Acad Sci USA. 2005;102:2162–2167. doi: 10.1073/pnas.0406976102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Ueda K, Saitoh T, Mori H. Tissue-dependent alternative splicing of mRNA for NACP, the precursor of non-A beta component of Alzheimer’s disease amyloid. Biochem Biophys Res Commun. 1994;205:1366–1372. doi: 10.1006/bbrc.1994.2816. [DOI] [PubMed] [Google Scholar]
- 25.Murray IV, Giasson BI, Quinn SM, Koppaka V, Axelsen PH, Ischiropoulos H, et al. Role of alpha-synuclein carboxy-terminus on fibril formation in vitro. Biochemistry. 2003;42:8530–8540. doi: 10.1021/bi027363r. [DOI] [PubMed] [Google Scholar]
- 26.Kanda S, Bishop JF, Eglitis MA, Yang Y, Mouradian MM. Enhanced vulnerability to oxidative stress by alpha-synuclein mutations and C-terminal truncation. Neuroscience. 2000;97:279–284. doi: 10.1016/s0306-4522(00)00077-4. [DOI] [PubMed] [Google Scholar]
- 27.Bodles AM, Guthrie DJ, Greer B, Irvine GB. Identification of the region of non-Abeta component (NAC) of Alzheimer’s disease amyloid responsible for its aggregation and toxicity. J Neurochem. 2001;78:384–395. doi: 10.1046/j.1471-4159.2001.00408.x. [DOI] [PubMed] [Google Scholar]
- 28.Periquet M, Fulga T, Myllykangas L, Schlossmacher MG, Feany MB. Aggregated alpha-synuclein mediates dopaminergic neurotoxicity in vivo. J Neurosci. 2007;27:3338–3346. doi: 10.1523/JNEUROSCI.0285-07.2007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Wurth C, Guimard NK, Hecht MH. Mutations that reduce aggregation of the Alzheimer’s Abeta42 peptide: an unbiased search for the sequence determinants of Abeta amyloidogenesis. J Mol Biol. 2002;319:1279–1290. doi: 10.1016/S0022-2836(02)00399-6. [DOI] [PubMed] [Google Scholar]
- 30.Kessler JC, Rochet JC, Lansbury PT., Jr The N-terminal repeat domain of alpha-synuclein inhibits beta-sheet and amyloid fibril formation. Biochemistry. 2003;42:672–678. doi: 10.1021/bi020429y. [DOI] [PubMed] [Google Scholar]
- 31.Zabrocki P, Bastiaens I, Delay C, Bammens T, Ghillebert R, Pellens K, et al. Phosphorylation, lipid raft interaction and traffic of alpha-synuclein in a yeast model for Parkinson. Biochim Biophys Acta. 2008;1783:1767–1780. doi: 10.1016/j.bbamcr.2008.06.010. [DOI] [PubMed] [Google Scholar]
- 32.Polevoda B, Sherman F. N-terminal acetyl-transferases and sequence requirements for N-terminal acetylation of eukaryotic proteins. J Mol Biol. 2003;325:595–622. doi: 10.1016/s0022-2836(02)01269-x. [DOI] [PubMed] [Google Scholar]
- 33.Hol W. The role of the α-helix dipole in protein function and structure. Prog Biophys Mol Biol. 1985;45:149–195. doi: 10.1016/0079-6107(85)90001-x. [DOI] [PubMed] [Google Scholar]
- 34.Soper JH, Roy S, Stieber A, Lee E, Wilson RB, Trojanowski JQ, et al. alpha-Synuclein-induced aggregation of cytoplasmic vesicles in Saccharomyces cerevisiae. Mol Biol Cell. 2008;19:1093–1103. doi: 10.1091/mbc.E07-08-0827. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Brookes AJ, St Clair D. Synuclein proteins and Alzheimer’s disease. Trends Neurosci. 1994;17:404–405. doi: 10.1016/0166-2236(94)90013-2. [DOI] [PubMed] [Google Scholar]
- 36.Han H, Weinreb PH, Lansbury PT., Jr The core Alzheimer’s peptide NAC forms amyloid fibrils which seed and are seeded by beta-amyloid: is NAC a common trigger or target in neurodegenerative disease? Chem Biol. 1995;2:163–169. doi: 10.1016/1074-5521(95)90071-3. [DOI] [PubMed] [Google Scholar]
- 37.Bodner RA, Outeiro TF, Altmann S, Maxwell MM, Cho SH, Hyman BT, et al. Pharmacological promotion of inclusion formation: a therapeutic approach for Huntington’s and Parkinson’s diseases. Proc Natl Acad Sci USA. 2006;103:4246–4251. doi: 10.1073/pnas.0511256103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Munishkina LA, Phelan C, Uversky VN, Fink AL. Conformational behavior and aggregation of alpha-synuclein in organic solvents: modeling the effects of membranes. Biochemistry. 2003;42:2720–2730. doi: 10.1021/bi027166s. [DOI] [PubMed] [Google Scholar]
- 39.Sreerama N, Woody R. Estimation of protein secondary structure from circular dichroism spectra: comparison of CONTIN, SELCON, and CDSSTR methods with an expanded reference set. Anal Biochem. 2000;287:252–260. doi: 10.1006/abio.2000.4880. [DOI] [PubMed] [Google Scholar]
- 40.Bisaglia M, Tessari I, Pinato L, Bellanda M, Giraudo S, Fasano M, et al. A topological model of the interaction between alpha-synuclein and sodium dodecyl sulfate micelles. Biochemistry. 2005;44:329–339. doi: 10.1021/bi048448q. [DOI] [PubMed] [Google Scholar]
- 41.Jo E, Fuller N, Rand RP, St George-Hyslop P, Fraser PE. Defective membrane interactions of familial Parkinson’s disease mutant A30P alpha-synuclein. J Mol Biol. 2002;315:799–807. doi: 10.1006/jmbi.2001.5269. [DOI] [PubMed] [Google Scholar]
- 42.Stemmer WP. DNA shuffling by random fragmentation and reassembly: in vitro recombination for molecular evolution. Proc Natl Acad Sci USA. 1994;91:10747–10751. doi: 10.1073/pnas.91.22.10747. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.