Abstract
BACKGROUND
U19/Eaf2 is an androgen-response gene and its downregulation is frequently observed in advanced human prostate cancer. U19/Eaf2 interacts with ELL, a fusion partner of MLL in the (11;19) (q23;p13.1) translocation in acute myeloid leukemia. U19/Eaf2 overexpression induces apoptosis and suppresses xenograft tumor growth.
METHODS
Transfection and colony formation were used to assay for apoptosis and growth suppression of various U19/Eaf2 mutants. Co-immunoprecipitation was performed to test the interaction between the U19/Eaf2 constructs and ELL.
RESULTS
The region of U19/Eaf2 essential for apoptosis and growth suppression was mapped to amino acids 68–113. This region was necessary and sufficient for binding ELL. Co-expression of U19/Eaf2 and ELL in 293 cells lead to significant increase in cell death and growth suppression.
CONCLUSIONS
These observations argue that the interaction with ELL is essential for the induction of apoptosis and growth suppression by U19/Eaf2.
Keywords: U19/Eaf2, ELL, apoptosis, growth suppression, prostate cancer
INTRODUCTION
Androgens are essential for the development and maintenance of the prostate. The biologically active androgens, testosterone and dihydrotestosterone (DHT), function by binding to the androgen receptor (AR), a ligand-regulated transcription factor [1,2]. AR binds to androgen response elements of the target genes and induces their expression, leading to cell proliferation, differentiation, or apoptosis [3,4].
U19/Eaf2 was initially identified as a gene upregulated by androgens in the rat ventral prostate [3]. It is a putative transcription factor with a transactivation domain in its C-terminal region [5]. Further studies revealed the role of U19/Eaf2 as a potential tumor suppressor in vivo and an inducer of apoptosis upon overexpression [6]. Downregulation, allelic loss, homozygous deletion, and promoter methylation of U19/Eaf2 in human prostate cancer specimens are consistent with its growth inhibitory and tumor suppressive role [6]. The escape from growth inhibition due to U19/Eaf2 inactivation may be an essential step in prostate cancer progression. U19/Eaf2 is suggested to play a role in other organs. During mouse embryogenesis, U19/Eaf2 is developmentally regulated, with spatially regulated patterns in the developing lens, suggesting a role in lens maturation [7]. Loss of U19/Eaf2 function in Xenopus resulted in loss of eyes, further providing evidence for its role in development [8].
U19/Eaf2 was recently reported to interact with Eleven-nineteen Lysine rich Leukemia (ELL) in vivo [5]. ELL is a gene that frequently undergoes chromosomal translocation and fuses to Mixed Lineage Leukemia (MLL) in Acute Myeloid Leukemia (AML) [9]. The MLL-ELL fusion protein is capable of immortalizing myeloid progenitors and causing AML in mice [10-12]. The ELL protein was shown to function as an RNA Polymerase II transcription elongation factor that facilitates the processivity of transcription by suppressing transient pausing by RNA Polymerase II [13]. ELL plays an essential role in animal development because ELL -/- was reported to cause embryonic lethality in the mouse model [14]. In Drosophila, mutations in ELL result in abnormal embryonic segmentation and the protein is important in early development [15,16]. Consistent with its essential role in animal development, ELL was reported to regulate proliferation, survival, and apoptosis in cultured cells [17].
ELL and the U19/Eaf2 homolog EAF1 co-localize in Cajal bodies [18,19], which are nuclear suborganelles involved in various functions including post-mitotic nucleolar formation, pre-mRNA splicing, feedback regulation of snRNA transcription, histone mRNA 3′ end processing, pre-assembly of general transcription machineries, as well as the sorting, (re)assembly, and transport of small ribonucleoproteins [20,21]. Eaf1 and U19/Eaf2 have also been reported to stimulate the ELL-mediated transcription elongation [22]. When cotransfected, U19/Eaf2 and ELL co-localize into distinct nuclear bodies and are tightly associated [5,23]. ELL binding also resulted in stabilization of U19/Eaf2 and enhanced transactivation by U19/Eaf2 [23]. These findings support the importance of the interaction with ELL in the function of U19/Eaf2 as a transcription factor.
In addition to the ELL binding via its N-terminus, U19/Eaf2 has other interesting features. Its C-terminus contains a transactivation domain and a domain that is highly enriched in serine, aspartic acid, and glutamic acid residues that is also present in AF4 and ENL [5]. This suggests that U19/Eaf2 induction of apoptosis could be mediated via different mechanisms. Domain mapping of U19/Eaf2 is likely to provide mechanistic insights into the U19/Eaf2-induced apoptosis.
In this report, we describe the importance of ELL in U19/Eaf2-mediated apoptosis and growth suppression. Amino acids 68-113 of U19/Eaf2, which is required for the interaction with ELL, is sufficient for the induction of apoptosis. Co-expression of U19/Eaf2 and ELL enhances apoptosis and growth suppression. These results suggest the essential role of ELL in the function of U19/Eaf2 as an inducer of apoptosis and suppressor of cell growth.
MATERIALS AND METHODS
Plasmid Construction
Full length or deletion mutants of U19 were amplified by PCR and cloned into pEGFP-N3, pM, or pCMV-myc (Clontech, Mountain View, CA). ELL was cloned into pEGFP-N3, pDSRed2-C1, pVP16, pCMV-myc (Clontech), or pcDNA3.1 (Invitrogen, Carlsbad, CA) by PCR. MLL-ELL was cloned into pcDNA3.1 by PCR.
Tissue Culture and Transfection
PC3, LNCaP, DU145, HeLa, and 293 cells were obtained from the American Type Culture Collection (ATCC, Rockville, MD). COS-7 cells were obtained from Dr. Kathleen Rundell (Northwestern University). PC3, LNCaP, and DU145 cells were grown in PRMI1640 medium supplemented with 10% FBS, 1% Penicillin/Streptomycin, and 2 mM L-Glutamine. COS-7, HeLa, and 293 cells were grown in DMEM supplemented with 10% FBS and 1% Penicillin/Streptomycin. Cells were transfected with plasmid DNA using the Fugene6 reagent (Roche, Indianapolis, IN) according to the manufacturer’s instructions. Stable selection of transfected cells was carried out in the presence of G418 at 1 μg/μl.
Mammalian Two-Hybrid Assay
Various regions of U19/Eaf2 were cloned into pM. ELL was cloned into pVP16. PC3 cells grown in 12-well culture plates were co-transfected with the U19/Eaf2 constructs, ELL, and luciferase reporter vectors pFR-Luc (Stratagene, La Jolla, CA) and pRL-TK (Promega, Madison, WI). Twenty-four hours after transfection, cells were washed once with PBS, lysed, and assayed for luciferase activity using the Dual-Luciferase Reporter Assay System (Promega) according to the manufacturer’s instructions.
Co-Immunoprecipitation and Immunoblot
COS-7 cells cultured in 10 cm culture dishes were transfected with 10 μg of each plasmid DNA. Twenty-four hours after transfection, cells were washed with PBS, trypsinized and whole cell lysates were prepared by incubating in lysis buffer (50 mM Tris-HCl, pH7.4; 150 mM NaCl; 1 mM EDTA; 1% Triton-X-100; 1 mM Na3VO4) containing protease inhibitor cocktail (Roche) for 20 min on ice. Cell lysates were incubated with anti-myc agarose (Sigma, St. Louis, MO) overnight at 4°C. Bound proteins were eluted by boiling in SDS loading buffer and separated by SDS–PAGE. The proteins were transferred onto a nitrocellulose membrane (Schleicher and Schuell, Keene, NH), blocked in Tris-buffered saline (TBS) containing 5% nonfat milk and 0.05% Tween-20, and probed with monoclonal antibodies against myc (Clontech) or GFP (Abcam, Cambridge, MA). For immunoblotting, monoclonal antibody against β-actin and polyclonal antibodies against ELL or U19/Eaf2 was used.
Real-Time RT-PCR
Total RNA from various cell lines was prepared using the RNeasy RNA purification system (Qiagen, Valencia, CA). cDNA was synthesized from the total RNA using the SuperScript III First-Strand Synthesis System (Invitrogen). Primer pairs for the amplification of U19/Eaf2, ELL and the human L19 gene are as follows; ELL forward: ACTGCATCCAGCAGTATGTCTCCA, reverse: TCCGAAACTGAACCTTCTTGCCCA, U19/Eaf2 forward: AGGTGAACAGGTGACCA-TAACTCTGC, reverse: GGGAGTCCTGGCTGAATTCCACATTT, L19 forward: CATGAGGTATGCTCAGGCTTCAGA, reverse: AGGTACAGGCTGTGATACATGTGG. The cDNAs and primer pairs were mixed with iQ SYBR Green Supermix (Bio-Rad, Hercules, CA) and the genes were amplified by PCR. The levels of amplified product were normalized to L19.
Colony Formation Assay
Cells (293) were grown in 12-well culture plates. Cells were transfected with 1 μg plasmid DNA. Twenty-four hours after transfection, cells in each well were expanded into 3 × 10 cm dishes. Stable transfectants were selected in medium containing 500 μg/ml G418 for 3 weeks. After 3 weeks, surviving cells were washed once with PBS and stained with 0.5% crystal violet in methanol. Colonies with a diameter larger than 1 mm were counted.
RESULTS
Amino Acids 68-113 of U19/Eaf2 Are Sufficient for the Induction of Apoptosis in PC3 Cells
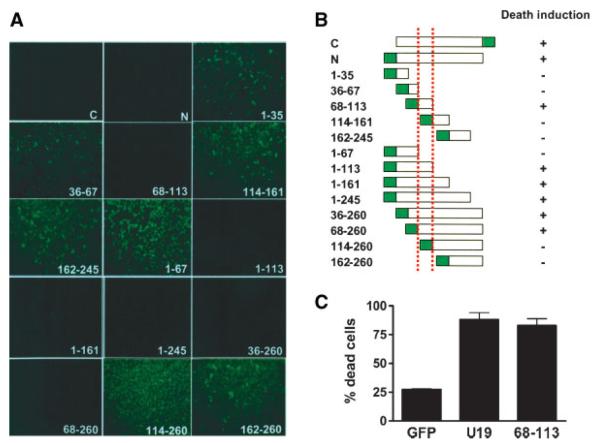
We have recently reported that U19/Eaf2 is able to induce apoptosis in prostate cancer cells upon overexpression [6]. To define the region of U19/Eaf2 responsible for the induction of apoptosis, we generated various deletion mutants of human U19/Eaf2 tagged to enhanced green fluorescent protein (EGFP). Different regions of the U19/Eaf2 cDNA were inserted into the expression vector pEGFP-N3 (Fig. 1B). PC3 cells were transfected with the recombinant plasmids and expression of the EGFP-fusion protein was monitored by fluorescence microscopy. To test the ability of U19/Eaf2 deletion mutants to induce cell death, transfected cells were selected with high doses of G418. After 3 weeks of drug selection, surviving colonies were monitored for the GFP-fusion protein expression. Cells transfected with amino acids 68–113 or constructs containing this region were unable to form colonies (Fig. 1A,B), indicating cell death induction by these constructs. Cells transfected with constructs outside amino acids 68–113 were able to grow and form colonies that express the GFP-fusion proteins (Fig. 1A,B), indicating that transfected cells are able to survive. We next tested whether transient overexpression of amino acids 68–113 was sufficient to induce apoptosis of PC3 cells. Two days after transfection, cells expressing the EGFP-fusion protein were counted and categorized into dead or live cells according to their morphology, with apoptotic cells exhibiting a rounded morphology. Amino acid sequence 68–113 was able to induce death of over 80% of transfected cells, demonstrating the importance of the region in cell death (Fig. 1C). These results show that amino acids 68–113 of U19/Eaf2 are sufficient for the induction of apoptosis and suppression of growth in PC3 cells.
Fig. 1.
Mapping of amino acid sequences of U19/Eaf2 essential for apoptosis induction and growth suppression. A: Mapping the domain of U19/Eaf2 responsible for growth suppression of PC3 cells.Different regions of U19/Eaf2 were cloned into GFP expression vectors and transfected into PC3 cells. Transfected cells were selected with G418 for 3 weeks. Colonies expressing the GFP fusion protein were photographed under a fluorescence microscope. Pictures show representative colonies. B: Schematic representation of the regions of U19/Eaf2 responsible for growth suppression. Dotted red lines indicate amino acids 68–113. C: Amino acids 68–113 induce apoptosis in PC3 cells. PC3 cells were transfected with GFP, U19-GFP, or 68-113-GFP. Percentage of dead cells was counted 3 days after transfection.
Amino Acids 68–113 of U19/Eaf2 Are Responsible for the Interaction With ELL
U19/Eaf2 and ELL have been previously reported to interact in vitro and in vivo [5]. When co-transfected in PC3 cells, U19/Eaf2 and ELL co-localize into nuclear bodies and induce apoptosis. This raised a possibility that U19/Eaf2-induced apoptosis is associated with its ability to bind ELL. Also, amino acids 68–113 of U19/Eaf2 are located at the C-terminal side of the ELL interaction domain, previously mapped to amino acids 17–104 within U19/Eaf2 [5]. To determine whether amino acids 68–113 of U19/Eaf2 are sufficient to interact with ELL, we performed a mammalian two-hybrid assay. Various regions of U19/Eaf2 were tagged to the Gal4 DNA-binding domain in the pM vector. The resulting constructs were co-transfected into PC3 cells with the pVP16 activation domain vector and luciferase reporter vector pFR-luc, along with pRL-TK as an internal control. Twenty-four hours after transfection, cells were harvested and the lysates were assayed for luciferase activity. All constructs harboring amino acids 68–113 were able to interact with ELL, as determined by an increase in luciferase activity (Fig. 2). Minimal or no increase in luciferase activity was detected outside of amino acids 68–113. Interestingly, the regions that interact with ELL were identical to the regions responsible for the induction of apoptosis.
Fig. 2.
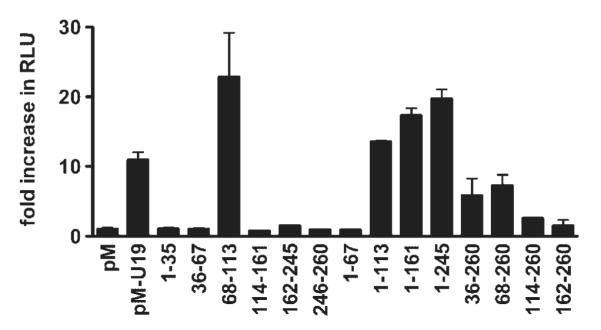
Mapping the regions of U19/Eaf2 that interact with ELL. Mammalian two-hybrid assay was performed to map the regions of U19/Eaf2 that interact with ELL. Indicated regions of U19/Eaf2 were cloned into pM. The entire ELL open reading frame was cloned into pVP16. PC3 cells were co-transfected with the vectors and luciferase reporter vectors. Twenty-four hours after transfection, the cells were harvested, lysed, and luciferase activity was measured using a luminometer.
To further test the physical interaction between ELL and amino acids 68–113 of U19/Eaf2, we performed a co-immunoprecipitation assay. COS7 cells were co-transfected with 68-113-GFP and myc-ELL. Twenty-four hours after transfection, cells were harvested and whole cell lysates were incubated with anti-myc agarose conjugate. Proteins bound to the myc-agarose were separated by SDS–PAGE and probed using a monoclonal antibody against GFP. The 68-113-GFP protein coimmunoprecipitated with myc-ELL, indicating that amino acids 68–113 of U19/Eaf2 are sufficient for the interaction with ELL (Fig. 3).
Fig. 3.
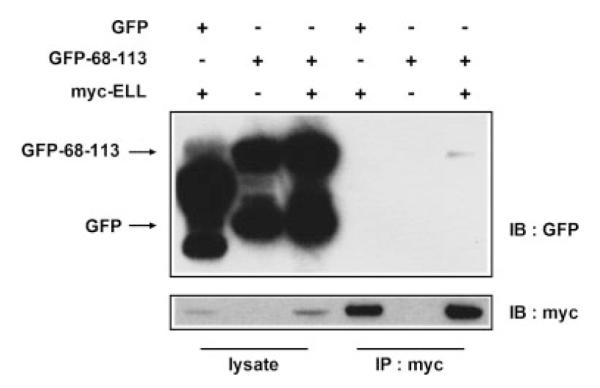
Co-immunoprecipitation of amino acids 68–113 of U19/Eaf2 and ELL. COS-7 cells were co-transfected with GFP-68-113 and myc-ELL. Twenty-four hours after transfection, cells were harvested, lysed, and the lysates were incubated and precipitated with anti-myc agarose. Proteins were separated by PAGE, transferred onto a nitrocellulose membrane and probed with antibodies against myc or GFP.
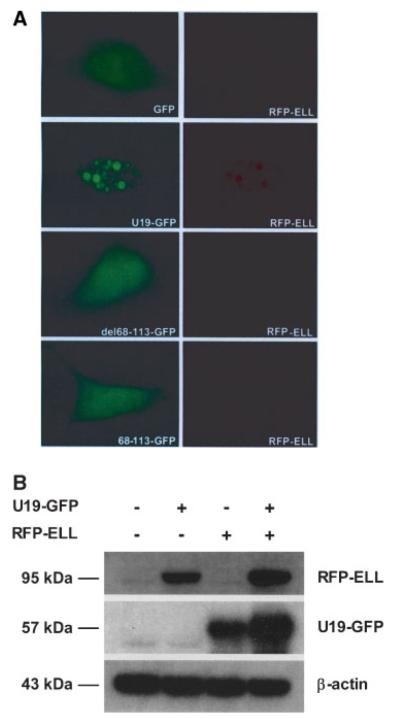
We next tested whether amino acids 68–113 of U19/Eaf2 were necessary for the co-localization with ELL in nuclear bodies. Full length U19/Eaf2 tagged to GFP (U19-GFP) or mutant U19-GFP lacking amino acids 68–113 (U19del68-113-GFP) was co-transfected with RFP-ELL. Full length U19-GFP was co-localized in nuclear bodies with ELL. U19del68-113-GFP did not form nuclear bodies with RFP-ELL. U19del68-113-GFP was localized to the nucleus and cytoplasm while RFP-ELL was localized diffusely throughout the nucleus. The red fluorescence signal of RFP-ELL was much lower when co-transfected with U19del68-113-GFP compared to when co-transfected with U19-GFP. When amino acids 68–113 of U19/Eaf2 tagged to GFP (68-113-GFP) was co-transfected with RFP-ELL, no nuclear bodies were detected, indicating that amino acids 68–113 are necessary but not sufficient for the co-localization with ELL (Fig. 4A). To assess whether increased red fluorescence signal of RFP-ELL upon co-transfection with U19-GFP was due to increased RFP-ELL protein level, Western blot was performed. Cells (293) were co-transfected with RFP-ELL, U19-GFP, or both. The RFP-ELL expression level was higher in the presence of U19-GFP, which can in part explain the increased red fluorescence signal of RFP-ELL in the presence of U19-GFP. U19-GFP levels were significantly increased in the presence of RFP-ELL, which is consistent with our previous finding that ELL stabilizes U19/Eaf2 [23]. Thus, U19/Eaf2 and ELL can increase each other’s expression levels (Fig. 4B).
Fig. 4.
A: Co-localization analysis of ELL with wild-type and mutant U19/Eaf2. PC3 cells were co-transfected with pEGFP-N3 and pRFP-ELL (top row), pU19-GFP and pRFP-ELL (second row), pU19-del68-113-GFP and pRFP-ELL (third row), or p68-113-GFP and pRFP-ELL (fourth row). Pictures were taken 24hr after transfection. U19/Eaf2-GFP and RFP-ELL co-localize in nuclear bodies while U19-del68-113-GFP or 68-113-GFP do not co-localize with RFP-ELL. B: Western blot analysis of U19/Eaf2 effect on ELL protein level. PC3 cells were cotransfected with U19-GFP and RFP-ELL. Cells were harvested and lysed. Western blot was performed using antibodies against U19/Eaf2 or ELL.
Co-expression of U19/Eaf2 and ELL Leads to Enhanced Death of 293 Cells
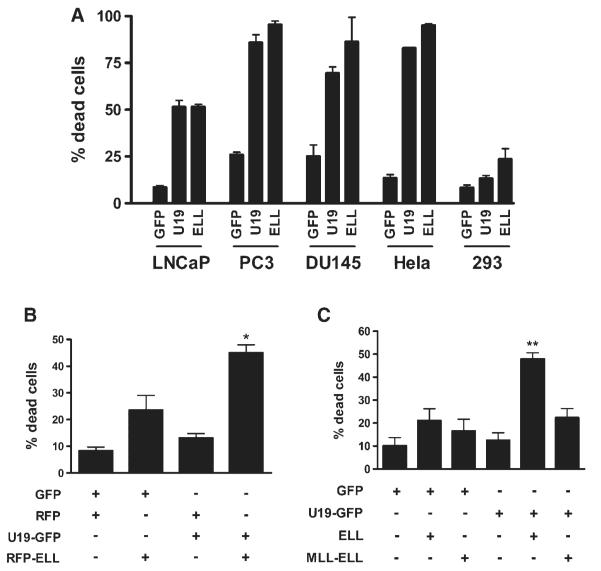
U19/Eaf2 and ELL have been shown to be potent inducers of apoptosis, inducing death in all of the cell lines that we have transfected, including PC3, LNCaP, DU145, and HeLa, except the human embryonic kidney cell line 293. In 293 cells, transfection of ELL resulted in death of 30% of cells while the effect of U19/Eaf2 transfection was virtually the same as the GFP control (Fig. 5A). Therefore, 293 cells were much less sensitive to ELL or U19/Eaf2 individually and were used as a model to study the potential cooperation between the two proteins in the induction of cell death and growth suppression.
Fig. 5.
Induction of apoptosis by over expression of U19/Eaf2, ELL, or both. A:U19/Eaf2 and ELL induce death in various cell lines. Indicated cell lines were transfected with U19-GFP or ELL-GFP. Cells expressing the fusion proteins were counted 3days after transfection. B: Co-expression of U19/Eaf2 and ELL result in enhanced cell death. Cells (293) were transfected with U19-GFP, RFP-ELL, or both.Percentage of dead cells expressing both proteins was counted. *, P < 0.05. C: Effect of MLL-ELL expression on U19/Eaf2-mediated cell death. Cells (293) were co-transfected with U19-GFP and MLL-ELL. Cells were counted 3 days after transfection. **, P < 0.01.
To test whether the overexpression of both proteins results in increased cell death, 293 cells were co-transfected with U19-GFP and RFP-ELL. Compared to cells transfected with U19-GFP or RFP-ELL alone, significantly greater percentage of cell death occurred when both proteins were expressed (Fig. 5B). To test the importance of U19/Eaf2-ELL interaction in the enhanced apoptosis, we co-transfected U19/Eaf2 with the MLL-ELL fusion protein, which lacks the first 45 amino acids of ELL, and is unable to interact with U19/Eaf2 [5,9]. Cells co-transfected with U19/Eaf2 and MLL-ELL exhibited no significant difference in cell death relative to cells transfected with U19/Eaf2 or MLL-ELL alone (Fig. 5C).
Colony formation assay was performed to determine whether co-transfection of U19/Eaf2 and ELL would lead to enhanced growth suppression. Cells (293) were co-transfected with pELL-EGFP and pMyc-U19. After 3 weeks of G418 selection, surviving colonies of cells were stained with crystal violet and counted. ELL transfection caused inhibition of colony formation, which agrees with ELL’s ability to induce some apoptosis in 293 cells. U19/Eaf2 transfection did not inhibit colony formation, which is consistent with its inability to induce cell death. Compared to U19/Eaf2 or ELL alone, significantly fewer colonies were formed when transfected with both genes (Fig. 6). This provides evidence that there is cooperative action between U19/Eaf2 and ELL in suppression of growth in 293 cells.
Fig. 6.
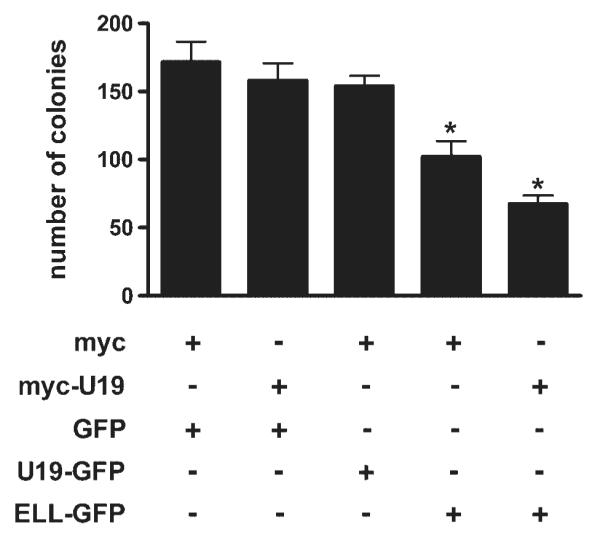
Colony formation (293) by transfection of U19/Eaf2, ELL, or both. Cells (293) were co-transfected with indicated vectors. Stable transfectants were selected with G418 for 3 weeks. Surviving colonies were stained with crystal violet. Colonies with diameters over 1 mm were counted. *, P < 0.05.
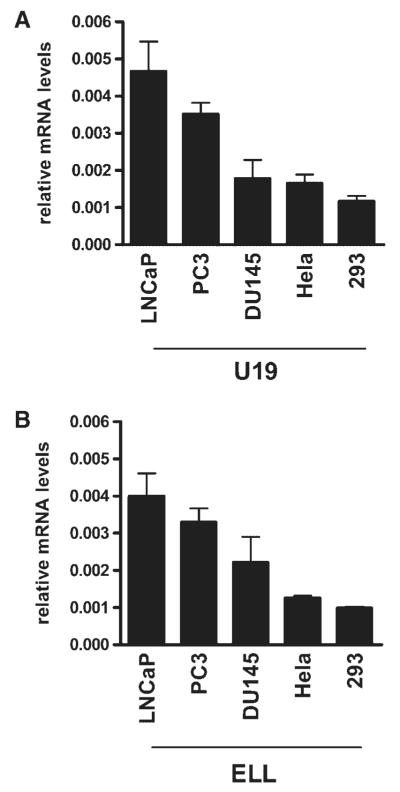
Because the interaction with ELL was an important event in U19/Eaf2-induced cell death, the expression level of ELL in a cell may be one of the factors that regulate its sensitivity to U19/Eaf2-induced cell death or growth suppression. Real-time RT-PCR was performed to quantitate the mRNA levels of ELL and U19/Eaf2 in the various cell lines used in this study. Both U19/Eaf2 and ELL were expressed at the lowest levels in 293 cells (Fig. 7A,B). The low ELL level in 293 cells may contribute to the resistance of the cell against U19/Eaf2-mediated apoptosis.
Fig. 7.
Expression levels of U19/Eaf2 and ELL in cultured cells. Real-time RT-PCR was performed to quantify them RNA levels of (A) U19/Eaf2 or (B) ELL in LNCaP, PC3, DU145, HeLa, and 293 cells.
DISCUSSION
In this study, we provide evidence that amino acids 68–113 of U19/Eaf2 are sufficient to induce apoptosis in prostate cancer cells and that this region is important in ELL binding. Co-expression of U19/Eaf2 and ELL in 293 cells led to a decrease in cell survival compared to that of either gene alone, suggesting a significant role of their interaction in growth suppression.
U19/Eaf2 consists of at least three structural/functional domains, the ELL-binding domain, transactivation domain and the serine, aspartic acid, and glutamic acid-rich domain, suggesting that U19 could have multiple targets. To elucidate the mechanism of U19/Eaf2 induction of apoptosis, we have mapped the domain within U19/Eaf2 responsible for the induction of apoptosis. Amino acids 68–113 of U19/Eaf2, located at the reported ELL-binding domain (amino acids 17–104), were both sufficient and necessary for the induction of cell death. Colony formation assays further showed that the overexpression of amino acids 68–113 suppresses the growth of cells.
To assess the potential involvement of ELL in the apoptotic and growth suppressive activity of U19/Eaf2, we tested whether the apoptosis-induction domain of U19/Eaf2 is required for ELL binding. Our results show that amino acids 68–113, which is the region crucial for the induction of apoptosis, is also the region critical for the interaction with ELL. U19/Eaf2 constructs containing amino acids 68–113 were sufficient both for the induction of cell death and the interaction with ELL. Wild-type U19/Eaf2 was able to co-immunoprecipitate to ELL while deletion of amino acids 68–113 disrupted the interaction. The deletion also resulted in the loss of ability for the two proteins to co-localize in nuclear bodies. The formation of nuclear bodies is not necessary for apoptosis induction by U19/Eaf2, because amino acids 68–113, which is apoptotic, were not sufficient for the co-localization with ELL and the formation of nuclear bodies. It is possible that other regions of U19/Eaf2 influence its localization into nuclear bodies.
To provide evidence that the interaction between U19/Eaf2 and ELL leads to growth suppression of cells, colony formation assay was performed. Because both ELL and U19/Eaf2 possess apoptotic activity in virtually all the cell lines, it was difficult to assay their potential cooperativeness in the induction of apoptosis. However, the human embryonic kidney cell line 293 provided a useful system, as it was resistant to U19/Eaf2-induced death and partially resistant to ELL-induced death. Real-time RT-PCR results indicate that both ELL and U19/Eaf2 are expressed at very low levels in this cell line, compared to all other cell lines used in our study. This may account in part for the lack of sensitivity to U19/Eaf2-mediated cell death. Expression of both genes resulted in significant reduction in surviving cells compared to either gene alone, suggesting their cooperative action in the induction of apoptosis and inhibition of cell growth. This cooperativeness was absent between U19/Eaf2 and MLL-ELL. The MLL-ELL fusion protein, which is frequently found in Acute Myeloid Leukemia, lacks the U19/Eaf2-interaction domain of ELL [9]. It is possible that the inability of MLL-ELL to bind U19/Eaf2 and mediate U19/Eaf2 action contributes to the progression of Acute Myeloid Leukemia.
Our results suggest a mechanism of cell growth suppression involving two proteins that are associated with two major human cancers, prostate cancer and leukemia. In the normal prostate, androgen upregulation of U19/Eaf2 may lead to increased interaction with ELL, leading to growth suppression and the maintenance of the prostate. The downregulation of U19/Eaf2 in prostate cancer may result in reduced interaction with ELL, favoring the promotion of growth. In leukemia, MLL-ELL translocation disrupts normal interaction between ELL and U19/Eaf2, which may contribute to uncontrolled cell growth. Although links between prostate cancer and AML have not been reported, our data suggest that U19/Eaf2 and ELL may play key roles in the suppression of these two types of cancers.
ACKNOWLEDGMENTS
We thank Dr. Junkui Ai and the members of the Wang laboratory for critical review of the manuscript. This study was supported by NIH R01 DK51193 and NIH P50 CA90386 Prostate Cancer SPORE. Zhou Wang is the O’Connor Family Research Professor of Urology.
Grant sponsor: NIH; Grant numbers: R01 DK51193, P50 CA90386.
REFERENCES
- 1.Chang CS, Kokontis J, Liao ST. Molecular cloning of human and rat complementary DNA encoding androgen receptors. Science. 1988;240:324–326. doi: 10.1126/science.3353726. [DOI] [PubMed] [Google Scholar]
- 2.Zhou ZX, Wong CI, Sar M, Wilson EM. The androgen receptor: An overview. Recent Prog Horm Res. 1994;49:249–274. doi: 10.1016/b978-0-12-571149-4.50017-9. [DOI] [PubMed] [Google Scholar]
- 3.Wang Z, Tufts R, Haleem R, Cai X. Genes regulated by androgen in the rat ventral prostate. Proc Natl Acad Sci USA. 1997;94:12999–13004. doi: 10.1073/pnas.94.24.12999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Jiang F, Wang Z. Identification of androgen-responsive genes in the rat ventral prostate by complementary deoxyribonucleic acid subtraction and microarray. Endocrinology. 2003;144:1257–1265. doi: 10.1210/en.2002-220718. [DOI] [PubMed] [Google Scholar]
- 5.Simone F, Luo RT, Polak PE, Kaberlein JJ, Thirman MJ. ELL-associated factor 2 (EAF2), a functional homolog of EAF1 with alternative ELL binding properties. Blood. 2003;101:2355–2362. doi: 10.1182/blood-2002-06-1664. [DOI] [PubMed] [Google Scholar]
- 6.Xiao W, Zhang Q, Jiang F, Pins M, Kozlowski JM, Wang Z. Suppression of prostate tumor growth by U19, a novel testosterone-regulated apoptosis inducer. Cancer Res. 2003;63:4698–4704. [PubMed] [Google Scholar]
- 7.Li M, Wu X, Zhuang F, Jiang S, Jiang M, Liu YH. Expression of murine ELL-associated factor 2 (Eaf2) is developmentally regulated. Dev Dyn. 2003;228:273–280. doi: 10.1002/dvdy.10367. [DOI] [PubMed] [Google Scholar]
- 8.Maurus D, Heligon C, Burger-Schwarzler A, Brandli AW, Kuhl M. Noncanonical Wnt-4 signaling and EAF2 are required for eye development in Xenopus laevis. EMBO J. 2005;24:1181–1191. doi: 10.1038/sj.emboj.7600603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Thirman MJ, Levitan DA, Kobayashi H, Simon MC, Rowley JD. Cloning of ELL, a gene that fuses to MLL in a t(11;19)(q23;p13.1) in acute myeloid leukemia. Proc Natl Acad Sci USA. 1994;91:12110–12114. doi: 10.1073/pnas.91.25.12110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.DiMartino JF, Miller T, Ayton PM, Landewe T, Hess JL, Cleary ML, Shilatifard A. A carboxy-terminal domain of ELL is required and sufficient for immortalization of myeloid progenitors by MLL-ELL. Blood. 2000;12:3887–3893. [PubMed] [Google Scholar]
- 11.Lavau C, Luo RT, Du C, Thirman MJ. Retrovirus-mediated gene transfer of MLL-ELL transforms primary myeloid progenitors and causes acute myeloid leukemias in mice. Proc Natl Acad Sci USA. 2000;97:10984–10989. doi: 10.1073/pnas.190167297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Luo RT, Lavau C, Du C, Simone F, Polak PE, Kawamata S, Thirman MJ. The elongation domain of ELL is dispensable but its ELL-associated factor 1 interaction domain is essential for MLL-ELL-induced leukemogenesis. Mol Cell Biol. 2001;21:5678–5687. doi: 10.1128/MCB.21.16.5678-5687.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Shilatifard A, Lane WS, Jackson KW, Conaway RC, Conaway JW. An RNA polymerase II elongation factor encoded by the human ELL gene. Science. 1996;271:1873–1876. doi: 10.1126/science.271.5257.1873. [DOI] [PubMed] [Google Scholar]
- 14.Mitani K, Yamagata T, Iida C, Oda H, Maki K, Ichikawa M, Asai T, Honda H, Kurokawa M, Hirai H. Nonredundant roles of the elongation factor MEN in postimplantation development. Biochem Biophys Res Commun. 2000;279:563–567. doi: 10.1006/bbrc.2000.3970. [DOI] [PubMed] [Google Scholar]
- 15.Eissenberg JC, Ma J, Gerber MA, Christensen A, Kennison JA, Shilatifard A. dELL is an essential RNA polymerase II elongation factor with a general role in development. Proc Natl Acad Sci USA. 2002;15:9894–9899. doi: 10.1073/pnas.152193699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Khattak S, Im H, Park T, Ahnn J, Spoerel NA. dELL, a drosophila homologue of transcription elongation factor ELL (Eleven-nineteen Lysine rich Leukemia), is required for early development. Cell Biochem Funct. 2002;20:119–127. doi: 10.1002/cbf.960. [DOI] [PubMed] [Google Scholar]
- 17.Johnstone RW, Gerber M, Landewe T, Tollefson A, Wold WS, Shilatifard A. Functional analysis of the leukemia protein ELL: Evidence for a role in the regulation of cell growth and survival. Mol Cell Biol. 2001;21:1672–1681. doi: 10.1128/MCB.21.5.1672-1681.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Polak PE, Simone F, Kaberlein JJ, Luo RT, Thirman MJ. ELL and EAF1 are Cajal body components that are disrupted in MLL-ELL leukemia. Mol Biol Cell. 2003;14:1517–1528. doi: 10.1091/mbc.E02-07-0394. Erratum in: Mol Biol Cell 2003; May 14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Simone F, Polak PE, Kaberlein JJ, Luo RT, Levitan DA, Thirman MJ. EAF1, a novel ELL-associated factor that is delocalized by expression of the MLL-ELL fusion protein. Blood. 2001;98:201–209. doi: 10.1182/blood.v98.1.201. [DOI] [PubMed] [Google Scholar]
- 20.Gall JG. The centennial of the Cajal body. Nat Rev Mol Cell Biol. 2003;4:975–980. doi: 10.1038/nrm1262. [DOI] [PubMed] [Google Scholar]
- 21.Cioce M, Lamond AI. Cajal bodies: A long history of discovery. Annu Rev Cell Dev Biol. 2005;21:105–131. doi: 10.1146/annurev.cellbio.20.010403.103738. [DOI] [PubMed] [Google Scholar]
- 22.Kong SE, Banks CA, Shilatifard A, Conaway JW, Conaway RC. ELL-associated factors 1 and 2 are positive regulators of RNA polymerase II elongation factor ELL. Proc Natl Acad Sci USA. 2005;102:10094–10098. doi: 10.1073/pnas.0503017102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Xiao W, Jiang F, Wang Z. ELL binding regulates U19/Eaf2 intracellular localization, stability, and transactivation. Prostate. 2006;66:1–12. doi: 10.1002/pros.20309. [DOI] [PubMed] [Google Scholar]