Abstract
Background
Cardiac electromechanical dyssynchrony causes regional disparities in workload, oxygen consumption, and myocardial perfusion within the left ventricle. We hypothesized that such dyssynchrony also induces region-specific alterations in the myocardial transcriptome that are corrected by cardiac resynchronization (CRT).
Methods and Results
Adult dogs underwent left bundle branch ablation (LBBB) and right atrial pacing at 200 bpm for either 6 weeks (dyssynchronous heart failure, DHF, n=12) or 3 weeks followed by 3 weeks of resynchronization by bi-ventricular pacing at the same pacing rate (CRT, n=10). Control animals without LBBB were not paced (NF, n=13). At 6 weeks, RNA was isolated from the anterior and lateral LV walls and hybridized onto canine-specific 44K microarrays. Echocardiographically, CRT led to a significant decrease in the dyssynchrony index, while DHF and CRT animals had a comparable degree of LV dysfunction. In DHF, changes in gene expression were primarily observed in the anterior LV, resulting in increased regional heterogeneity of gene expression within the left ventricle. Dyssynchrony-induced expression changes in 1050 transcripts were reversed by CRT to levels of NF hearts (false discovery rate <5%). CRT remodeled transcripts with metabolic and cell signaling function and greatly reduced regional heterogeneity of gene expression compared with DHF.
Conclusions
Our results demonstrate a profound effect of electromechanical dyssynchrony on the regional cardiac transcriptome, causing gene expression changes primarily in the anterior LV wall. CRT corrected the alterations in gene expression in the anterior wall, supporting a global effect of biventricular pacing on the ventricular transcriptome that extends beyond the pacing site in the lateral wall.
Keywords: Cardiac Resynchronization Therapy, Heart Failure, Gene Expression, Microarray
Introduction
Nearly 5 million Americans suffer from heart failure (HF) and more than 250,000 die annually.1 Asymmetrical contraction resulting from an intraventricular conduction delay is present in ~30% of patients with HF2 and has been identified as an independent predictor of mortality in HF patients.3-5 A left bundle branch block (LBBB) decreases regional loading, contractile work, myocardial blood flow and oxygen consumption in the early-activated anterior myocardium, while these parameters are increased in the late-activated lateral LV.6, 7 Bi-ventricular stimulation, or cardiac resynchronization therapy (CRT) has been developed to treat this disorder. It improves contractile synchrony, systolic function, re-homogenizes regional work load, and in patients, improves clinical symptoms and survival.8, 9
We first reported that dyssynchronous HF also leads to regional disparities of protein expression, notably in stress-response kinases and cytokines,10, 11 with enhanced levels in the higher stress (late-activated) lateral wall. More recently, we showed that CRT can re-homogenize these changes.10 However, our previous analysis was focused on individual proteins, and most likely missed a much broader impact of dyssynchrony and CRT on regional molecular expression patterns. To test this, we used a global gene expression profiling approach in a recently developed canine model of dyssynchronous HF and CRT,10 examining regional disparities in the cardiac transcriptome in dyssynchronous HF, and determining the capacity of CRT to ameliorate these abnormalities.
Methods
Cardiac Resynchronization in a Canine Tachypacing-Induced Heart Failure Model
Details of the animal model have been described previously.10, 12-14 Briefly, adult male mongrel dogs (n=22) underwent left bundle branch radiofrequency ablation, and later received bipolar epicardial leads (Medtronic, Minneapolis, MN) implanted on the right atrium, right ventricular free wall, and lateral LV. For 3 weeks, all dogs were subjected to rapid atrial pacing (200 min-1) to induce dyssynchronous heart failure (DHF). Dogs were subsequently divided into two groups for an additional pacing period of three weeks: While DHF animals (n=12) continued to receive atrial tachypacing, CRT dogs (n=10) received bi-ventricular tachypacing at the same rate during the latter half of the pacing protocol. Control animals without LBBB were not paced (NF, n=13). All protocols followed USDA and NIH guidelines, and were approved by our institution's Animal Care and Use Committee.
Echocardiography and Hemodynamic Recordings
Chamber function was assessed by two-dimensional echocardiography with tissue Doppler imaging (at 3 and 6 week time points), and by invasive catheterization at the time of sacrifice. The details have been previously reported.10
Microarray Hybridization and Statistical Analysis
Total RNA was isolated with TRIzol reagent from the subendocardium of the anterior and lateral LV walls in the distribution of the LAD and left circumflex artery, respectively. Following a one-color design in 11 NF, 10 DHF and 9 CRT animals, RNA was labeled with Cy3 and hybridized onto Agilent 44K canine-specific microarrays. Agilent's RNA spike-ins were mixed with the sample and co-hybridized to the arrays following the manufacturer's instructions. The quality of the microarray hybridizations was verified by controlling for the dynamic range, saturation, pixel noise, grid misalignment and signal-to-noise ratio.
To validate results from one-color microarray experiments where RNA isolated from the anterior and lateral walls from the same heart was hybridized onto two separate arrays, experiments were also performed using a two-color design in a subset of animals (6 NF, 5 DHF and 5 CRT dogs), partially overlapping those used for the one-color design. In these experiments, corresponding anterior and lateral samples from the same LV were labeled with Cy3 and Cy5 (including dye swap experiments) and hybridized onto the same array to achieve a direct comparison of the relative gene expression in different regions of the same heart. Additional validation included comparison of the data from this study to two publicly available datasets of dyssynchronous heart failure in dogs with RV pacing-induced LV dysfunction (Gene Expression Omnibus accession numbers GSE5247 and GSE9794).15, 16
Preprocessing and most of the statistical analysis were done using R (www.r-project.org) and Bioconductor (www.bioconductor.org). Microarray data were normalized using quantile normalization implemented in Bioconductor's “affy”-package (for one-color data) or loess normalization implemented in Bioconductor's “limma”-package (for two-color microarray data). Complying with MIAME standards (Minimum Information about a Microarray Experiment),17 microarray data has been submitted to a public repository (Gene Expression Omnibus, GEO; the SuperSeries accession number GSE14661 includes GSE14327 (1-color design) and GSE14338 (2-color design data)).
To determine differentially expressed genes, multiclass and unpaired two-class Significance Analysis of Microarrays (SAM) was used.18 Differences in gene expression were regarded as statistically significant if a false discovery rate (FDR) of q<0.05 was achieved. Functional annotation of differentially expressed genes was based on the Kyoto Encyclopedia of Genes and Genomes (KEGG) pathways database. Overrepresentation of specific KEGG pathways in a gene set was statistically analyzed by “FatiGO+”19 and the Database for Annotation, Visualization and Integrated Discovery (DAVID).20
An on-line data supplement containing one figure and two tables appears at http://circ.ahajournals.org. The authors had full access to and take full responsibility for the integrity of the data. All authors have read and agree to the manuscript as written.
Results
Dyssynchronous Heart Failure and Biventricular Pacing
The hemodynamic features of the canine DHF and CRT model have been reported previously,10 and results obtained more recently from a larger cohort are summarized in Table 1. Compared with normal controls, DHF ventricles display marked dyssynchrony that was restored to normal levels with CRT. Significant differences were observed in ejection fraction (increase of ~10%), stroke volume, and left ventricular contractility assessed by dP/dTmax normalized to instantaneous developed pressure. End-diastolic pressure was similarly elevated, and both DHF and CRT hearts remained dilated and had evidence of failure.
Table 1. Phenotypic characterization. Echocardiographic and invasive hemodynamic measurements of DHF (n=17), CRT (n=15) and non-failing (n=6) dogs.
| Control | DHF | CRT | ANOVA | |
|---|---|---|---|---|
| Dyssynchrony Index (td) | 30±1.2 | 68±4.6 * | 31.3±5.1 | <0.0001 |
| EF (%) | 66.7±3.1 | 24.8±2.6† | 33.1±2.6†‡ | <0.001 |
| Stroke Volume (mL) | 34±2.7 | 21.5±2.4 | 31.9±3.8† | <0.03 |
| End-diastolic Pressure (mmHg) | 6.2±1.4 | 30.9±2.5* | 28.8±2.5* | <0.0001 |
| dP/dTmx/IP (sec-1) | 27.6±1.4 | 13.2±0.6† | 16.9±1.2‡ | <0.0001 |
Regional regulation of gene expression in DHF
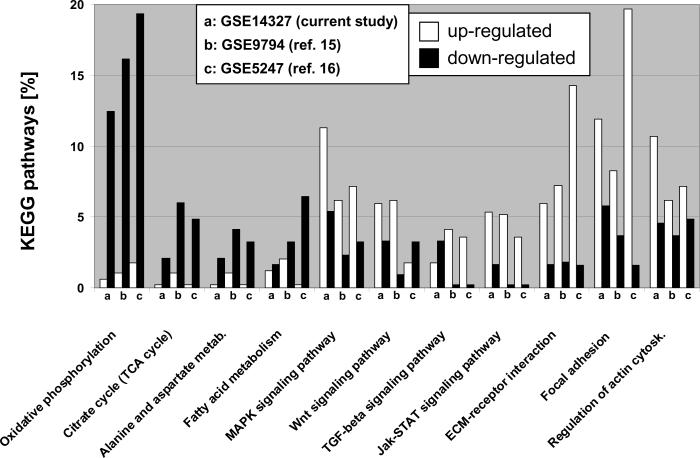
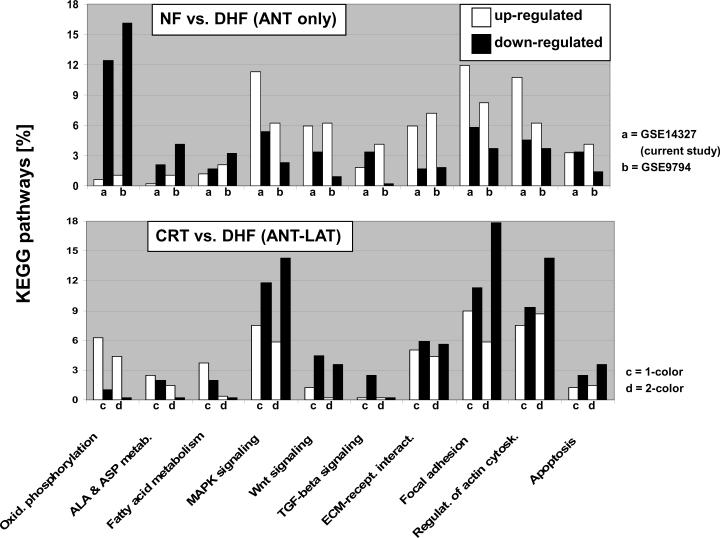
Figure 1 shows the relative distribution of significantly up- and down-regulated transcripts for eleven KEGG pathways in the anterior myocardium comparing non-failing and paced (DHF) animals. Energy-deriving processes including oxidative phosphorylation, tricarboxylic acid cycle (TCA), fatty acid and amino acid metabolism were concomitantly down-regulated in failing myocardium, while various cell signaling pathways and extracellular matrix components were up-regulated. To validate the reproducibility of the results obtained with the Agilent microarray platform, we compared our results to two publicly available datasets of pacing-induced (single site, and also dyssynchronous) HF in dogs, generated with Affymetrix microarrays with tissue from the anterior LV wall (FDR<5% for all studies).15, 16 The high concordance of gene expression patterns between these three studies underscores the reproducibility of our findings and highlights the robust genomic fingerprint of DHF in the anterior myocardium.
Figure 1. KEGG pathway analysis in tachycardia-pacing induced heart failure.
Transcripts up- and down-regulated by ventricular tachypacing in canine ventricular myocardium are represented by white and black columns, respectively and shown as percentage of eleven major KEGG pathways (studies a-c). Study (a) shows the comparison of anterior samples between NF and DHF hearts in the current Agilent-based microarray study. Studies (b) and (c) show the results of two publicly available Affymetrix microarray datasets (Gene Expression Omnibus accession numbers 9794 and 5247, respectively) comparing myocardial tissue derived from the anterior LV wall from non-paced animals to dogs that were tachypaced for at least 3 weeks. Normalized data was downloaded from Gene Expression Omnibus and analyzed using the same methods employed in this study (a). For all KEGG pathways shown, a p-value of <0.05 (Fisher's exact test implemented in the “FatiGO+” tool)19 was achieved in at least two of the three different canine tachypacing studies presented. It is evident that energy-deriving processes including oxidative phosphorylation and TCA cycle are greatly down-regulated in pacing-induced HF, while various cell signaling pathways and extracellular matrix components are up-regulated. The high concordance of disease-specific gene expression patterns across independent studies and different microarray platforms serves as an independent validation of our results and suggests a common disease-specific genomic fingerprint.
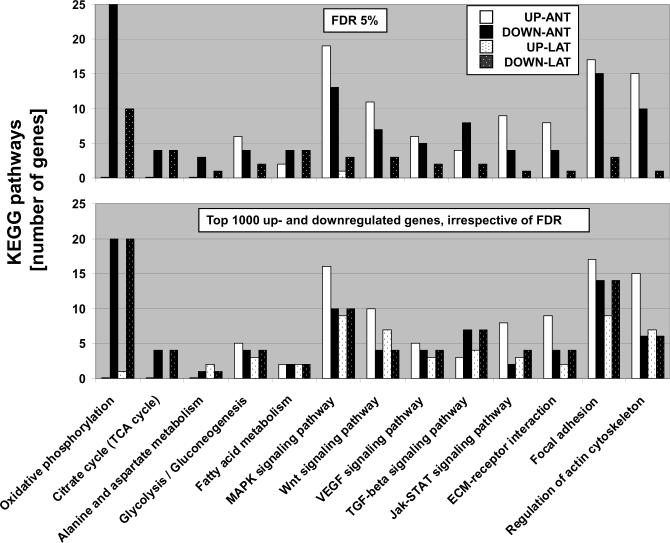
Dyssynchronous contraction of the LV associated with LBBB imposes greater stress on the lateral wall compared to the anterior LV. To test the hypothesis that DHF alters gene expression patterns in a region-specific fashion, samples from anterior and lateral LV myocardium were examined separately in non-failing and DHF animals. Separate analysis of the regional transcriptome from the same hearts (NF n=11, DHF n=10), identified over six-times as many genes that were differentially expressed between NF and DHF hearts in anterior compared to lateral LV myocardium (2173 vs. 346 transcripts, respectively; SAM with FDR<5%; Table 2 and Online Figure 1). Besides these quantitative differences, important qualitative differences were evident for DHF-induced gene expression changes in the anterior and lateral myocardium. While down-regulation of energy-deriving processes including oxidative phosphorylation and TCA cycle was more pronounced in the anterior compared to the lateral wall, regulation of cell signaling pathways and extracellular matrix components displayed a different pattern in the lateral and anterior LV myocardium in dyssynchronous HF. For instance, cell signaling pathways were up-regulated in the anterior LV wall, but predominantly down-regulated in the lateral LV wall (Figure 2, upper panel). The small number of regulated genes found in lateral myocardium of DHF hearts limited the statistical power of the KEGG pathway analysis, thus an additional analysis was performed where KEGG pathways of the first 1000 up- and down-regulated transcripts were compared in both regions, irrespective of the significance level. This approach was employed to determine if the changes in gene expression in the lateral wall differ only quantitatively from those in the anterior LV wall. The metabolic gene classes were still down-regulated; however, the pattern of gene expression in cell signaling pathways and extracellular matrix components differed considerably from the robust genomic fingerprint observed in anterior myocardium in DHF (Figure 1 and lower panel of Figure 2), suggesting a quantitatively and qualitatively different transcriptomic response to electromechanical dyssynchrony in the anterior and lateral LV.
Table 2. Results of Statistical Analysis of Microarrays (SAM), False Discovery Rate (FDR) <5%.
The multiclass analysis of SAM is comparable to an ANOVA test that is corrected for multiple testing by the false discovery rate (FDR). Anterior-lateral stands for the difference in gene expression between the anterior and lateral left ventricle.
| 1-color design | anterior | lateral | anterior-lateral |
|---|---|---|---|
| NF vs. DHF | 2173 | 346 | 1146 |
| DHF vs. CRT | 4 | 3 | 639 |
| multiclass (NF vs. DHF vs. CRT) | 1773 | 578 | 1050 |
Figure 2. Regional KEGG pathway analysis in tachycardia-pacing induced heart failure.
KEGG pathways are plotted as numbers of up- and down-regulated transcripts that were differentially expressed between non-failing and failing myocardium. Transcripts up- and down-regulated by ventricular tachypacing are represented by white and black columns (anterior wall), or textured white and black bars (lateral wall), respectively. The upper panel compares 11 KEGG pathways identified by SAM analysis in anterior and lateral myocardium, respectively. The small number of regulated genes found in lateral myocardium of DHF hearts limited the statistical power of the KEGG pathway analysis, thus an additional analysis was performed where KEGG pathways of the first 1000 up- and down-regulated transcripts were compared in both regions, irrespective of the significance level (lower panel). This approach was employed to determine if the changes in gene expression in the lateral wall differ only quantitatively from those in the anterior LV wall. The metabolic gene classes were still down-regulated; however, the pattern of gene expression for cell signaling pathways and extracellular matrix components differed considerably from the robust genomic fingerprint observed in anterior myocardium in DHF, suggesting also qualitatively different transcriptomic responses to electromechanical dyssynchrony in the anterior and lateral LV.
Biventricular Pacing Reverses Gene Expression Changes
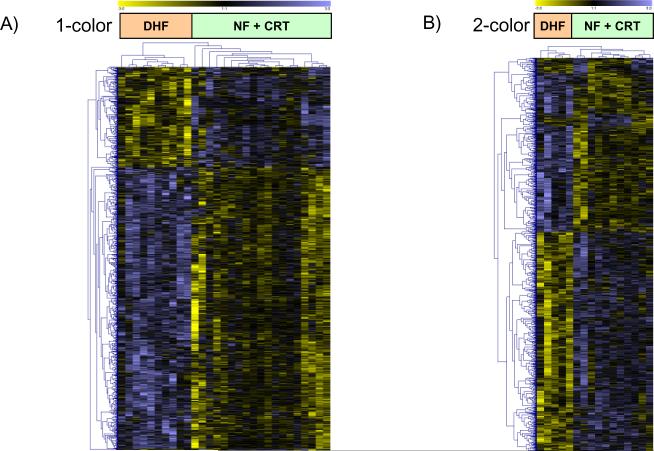
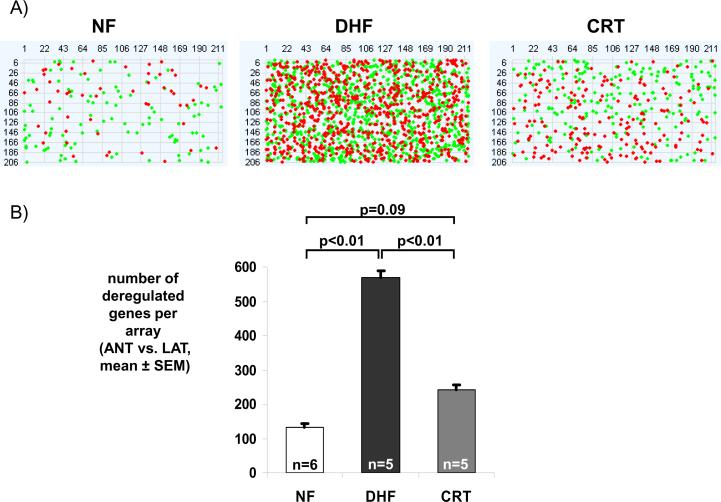
Separate analysis of gene expression in anterior and lateral LV samples identified only 7 differentially expressed transcripts between DHF and CRT hearts (Table 2 and Online Table 1). However, when anterior and lateral samples from the same hearts were paired by examining the difference in gene expression between the two regions (anterior minus lateral LV wall), a large number of transcripts were found to be differentially expressed between DHF and CRT hearts (Table 2). An unsupervised clustering of 1050 transcripts, identified by SAM multiclass analysis of regional differences in gene expression between anterior and lateral wall for NF, DHF and CRT hearts, revealed that CRT hearts clustered with NF, rather than with gene expression patterns from DHF samples (Figure 3A; the gene list is provided in Online Table 2). An identical picture emerged when data from the two-color microarray design were clustered (Figure 3B). The latter experiments also revealed few gene expression changes between the anterior and lateral regions in the NF myocardium (Figure 4). This was substantiated by SAM, with only 2 transcripts being differentially expressed between anterior and lateral regions in non-failing myocardium (LIM domain only protein 3 in anterior myocardium and glutathione S-transferase P in lateral myocardium, FDR5% in two-color array data). In contrast, there was a marked heterogeneity in gene expression between anterior and lateral LV regions in DHF that was reduced by CRT to levels comparable to NF hearts (Figure 4). Moreover, CRT partially reversed DHF-induced gene expression changes (Figure 5), as evidenced by KEGG pathway analysis; transcripts of metabolic activity were up-regulated in the anterior wall while transcripts encoding for cell signaling pathways and extracellular matrix components were down-regulated. Therefore, CRT restored the relative balance of gene expression between the anterior and lateral LV, e.g. expression of MAPK pathway signaling and extracellular matrix components was reduced in the anterior wall but increased in lateral LV (Figure 5).
Figure 3. Clustering of regional differences between anterior and lateral myocardium.
Unsupervised clustering of differentially expressed transcripts identified by SAM (multiclass, false discovery rate <5%) using Euclidean distance for one and two color microarray data (panel A and B, respectively) shows that transcript expression from CRT hearts cluster with NF hearts rather than with DHF samples. Each row represents data for one gene. The gene expression level is color-coded with yellow and blue representing low and high expression, respectively. For one-color data, the difference in gene expression between the anterior and lateral wall from the same heart was compared for NF, DHF and CRT animals.
Figure 4. Dyssynchrony leads to increased regional heterogeneity in gene expression that is partially reduced with CRT.
(A) Pseudoimages of representative microarrays from NF, DHF and CRT hearts with 211 columns and 206 rows (44K array). RNA from the anterior and lateral regions was labeled with Cy3 and Cy5 and hybridized in a two-color design onto one array. Red and green dots represent statistically significant transcripts between anterior and lateral wall, respectively. (B) A bar plot of the number of deregulated genes comparing the anterior and lateral regions in NF, DHF and CRT hearts. In DHF, the number of differentially expressed transcripts between anterior and lateral wall increases 4-fold, while it is greatly reduced by CRT.
Figure 5. Partial correction of pacing-induced gene expression changes by CRT.
A comparison of up- and down-regulated transcripts in KEGG pathways that were differentially expressed between non-failing and failing myocardium (upper panel) and between anterior and lateral LV myocardium (lower panel). Up- and down-regulated transcripts are represented by white and black columns, respectively. CRT partially restores dyssynchrony-induced gene expression changes in failing ventricular myocardium (up regulation of transcripts in oxidative phosphorylation pathways and down regulation of cell signaling pathways in the anterior wall).
Discussion
Using an unbiased and global assessment of transcriptional activity in a large animal model of dyssynchronous heart failure, we found that dyssynchrony-induced changes in gene expression were more pronounced in the anterior compared with the lateral LV. The genes that showed significant heterogeneity in regional expression with dyssynchrony are involved in important processes such as metabolic pathways, extracellular matrix remodeling, and myocardial stress responses. The disparity in the number of regulated transcripts between the early- and late-activated LV regions gave rise to an increased regional heterogeneity of gene expression within the dyssynchronously contracting myocardium. Remarkably, dyssynchrony-induced expression changes were reversed by CRT to levels in NF hearts, as evident by a reduced regional heterogeneity of gene expression and prominent reverse remodeling of transcripts with metabolic and cell signaling function.
A number of factors have been shown to regulate transcriptional activity in the heart, including contractile activity, stretch, myocardial perfusion and metabolism.21-23 As all these parameters are altered in a region-specific fashion in DHF, they could account for the differential transcriptional response of the anterior and lateral walls. It is well known that cardiac dyssynchrony, whether due to a LBBB or right ventricular free wall pacing, decreases regional loading, contractile work, myocardial blood flow and oxygen consumption in the early-activated anterior myocardium. For instance, the regional pressure-strain loop area which corresponds to the external work performed, is reduced to a greater extent in the anterior compared to the lateral wall in DHF hearts.13 In line with this finding, down-regulation of metabolic transcripts was significantly greater in anterior compared with lateral LV regions. Biventricular pacing improves contractile timing, thereby increasing regional work in the anterior wall, while reducing work in the lateral LV region. Experimentally, this has been shown to couple with rebalancing of glucose metabolism24 and myocardial blood flow25 (rising in the anterior and declining in lateral walls), and such findings are consistent with CRT-associated increases in transcripts levels encoding oxidative phosphorylation and various metabolic pathways in anterior samples observed in this study. In another study performed in these models, we have observed up-regulation of proteins in various metabolic pathways in CRT by examining the myocardial mitochondrial proteome.26 Indeed, nearly 50% of the protein changes involved subunits of the oxidative phosphorylation chain, the majority of which displayed marked up-regulation with CRT. Additionally, key enzymes in anaplerotic pathways such as branched chain amino acid oxidation and pyruvate carboxylation were increased, suggesting that CRT may increase the pool of Krebs cycle intermediates to fuel oxidative phosphorylation.26
While CRT effectively restored dyssynchrony-induced gene expression changes in this model, it did not correct overall HF-induced transcriptomic alterations. The changes in gene expression levels brought about by CRT were only significant when a paired design for anterior and lateral LV region of each heart was employed, i.e. influencing heterogeneity of expression, but far less when anterior and lateral regions in DHF and CRT hearts were compared separately (Table 2). In our view, two main findings suggest that even this small yet significant effect of CRT on the cardiac transcriptome likely has global effects on heart function. First, dyssynchrony-induced gene expression changes tended to aggravate the HF-related transcriptomic signature (Figures 2 and 5). In DHF for example, various cell signaling and extracellular matrix remodeling pathways were up-regulated in the early-activated anterior but down-regulated in the late-activated lateral LV wall. In contrast, biventricular pacing partially reversed cell signaling and extracellular matrix remodeling changes by restoring the relative balance in gene expression levels between anterior and lateral wall (down-regulation in anterior regions with concomitant up-regulation of mRNA levels of these pathways in the lateral wall). Thus, dyssynchrony-induced expression changes could well contribute to the decline in EF in DHF animals between 3 and 6 weeks of pacing, while CRT animals showed a modest increase in EF during the same period (Table 1). Second, dyssynchrony-induced gene expression changes significantly increased the heterogeneity of gene expression within early- and late-activated LV wall regions. It is tempting to speculate that this increased heterogeneity within the LV wall also reflects regionally heterogeneous remodeling of ion channels that exists in this HF model,12 and possibly increased QT dispersion.27, 28 In a study that examined QT interval duration in relation to CRT, Berger et al. found that QT dispersion increased during right ventricular, and decreased during biventricular pacing,29 thus paralleling changes observed for action potential duration with DHF and CRT in isolated cardiomyocytes.12
In summary, we demonstrate a profound effect of electrical activation on the regional cardiac transcriptome and provide unique insights into transcriptome-wide molecular processes underlying transcriptomic remodeling in CRT. The dyssynchronous failing heart is not simply worse heart failure, but a form of disease with profound regional gene expression disparities. Moreover, we show for the first time that by re-coordinating contraction, such expression heterogeneity can be essentially returned to normal, even in a failing heart, on a genome-wide level. This may point to a more biological method to assess the impact of CRT.
Study Limitations
Pacing-induced heart failure is a widely-used animal model of non-ischemic heart failure that shares major electrophysiological (APD prolongation, high incidence of sudden cardiac death, atrial arrhythmias), morphological (biventricular dilatation) and functional (depressed contractility) hallmarks of human heart failure. However, tachypacing-induced HF does not mimic all features of human heart failure, as the changes in myocardial structure occurring with tachypacing are dissimilar to clinical forms of HF due to chronic ischemia or hypertensive disease. Thus, extrapolation of the findings from this heart failure model to clinical forms of HF should be done with caution, as this model will not fully represent the complex clinical spectrum of HF. Additionally, pharmacological interventions known to disrupt the neurohormonal dysfunction in HF (e.g. beta-blockade or ACE-inhibition) were not employed in this model. As this study was designed to examine the effects of CRT with ongoing HF, tachypacing was maintained throughout the study. Thus, if anything, this model might be predicted to delay benefits that might come from re-synchronization itself, though improvement was still clearly documented. Given the higher heart rate necessary to induce and maintain HF in this model, the processes of remodeling and reverse remodeling may differ from the clinical HF syndrome. Additional studies will be needed to differentiate transcriptional changes associated with altered electrical activation of the ventricles and dyssynchronous mechanical contraction independent of heart failure.
Supplementary Material
Acknowledgments
Funding Sources The work was supported by NIH P01 HL 077180, HL 072488 and R33 HL087345 to D.A.K. and G.F.T, NIH T32 HL007227 to A.S.B. and the Abraham and Virginia Weiss Professorship (D.A.K.). G.F.T. is the Michel Mirowski M.D. Professor of Cardiology.
Footnotes
Conflict of Interest Disclosures None.
References
- 1.Rosamond W, Flegal K, Furie K, Go A, Greenlund K, Haase N, Hailpern SM, Ho M, Howard V, Kissela B, Kittner S, Lloyd-Jones D, McDermott M, Meigs J, Moy C, Nichol G, O'Donnell C, Roger V, Sorlie P, Steinberger J, Thom T, Wilson M, Hong Y. Heart disease and stroke statistics--2008 update: a report from the American Heart Association Statistics Committee and Stroke Statistics Subcommittee. Circulation. 2008;117:e25–146. doi: 10.1161/CIRCULATIONAHA.107.187998. [DOI] [PubMed] [Google Scholar]
- 2.Baldasseroni S, Opasich C, Gorini M, Lucci D, Marchionni N, Marini M, Campana C, Perini G, Deorsola A, Masotti G, Tavazzi L, Maggioni AP. Left bundle-branch block is associated with increased 1-year sudden and total mortality rate in 5517 outpatients with congestive heart failure: a report from the Italian network on congestive heart failure. Am Heart J. 2002;143:398–405. doi: 10.1067/mhj.2002.121264. [DOI] [PubMed] [Google Scholar]
- 3.Bader H, Garrigue S, Lafitte S, Reuter S, Jais P, Haissaguerre M, Bonnet J, Clementy J, Roudaut R. Intra-left ventricular electromechanical asynchrony. A new independent predictor of severe cardiac events in heart failure patients. J Am Coll Cardiol. 2004;43:248–256. doi: 10.1016/j.jacc.2003.08.038. [DOI] [PubMed] [Google Scholar]
- 4.Kass DA, Chen CH, Curry C, Talbot M, Berger R, Fetics B, Nevo E. Improved left ventricular mechanics from acute VDD pacing in patients with dilated cardiomyopathy and ventricular conduction delay. Circulation. 1999;99:1567–1573. doi: 10.1161/01.cir.99.12.1567. [DOI] [PubMed] [Google Scholar]
- 5.Spragg DD, Kass DA. Pathobiology of left ventricular dyssynchrony and resynchronization. Prog Cardiovasc Dis. 2006;49:26–41. doi: 10.1016/j.pcad.2006.05.001. [DOI] [PubMed] [Google Scholar]
- 6.van Oosterhout MF, Prinzen FW, Arts T, Schreuder JJ, Vanagt WY, Cleutjens JP, Reneman RS. Asynchronous electrical activation induces asymmetrical hypertrophy of the left ventricular wall. Circulation. 1998;98:588–595. doi: 10.1161/01.cir.98.6.588. [DOI] [PubMed] [Google Scholar]
- 7.Vernooy K, Verbeek XA, Peschar M, Crijns HJ, Arts T, Cornelussen RN, Prinzen FW. Left bundle branch block induces ventricular remodelling and functional septal hypoperfusion. Eur Heart J. 2005;26:91–98. doi: 10.1093/eurheartj/ehi008. [DOI] [PubMed] [Google Scholar]
- 8.Bristow MR, Saxon LA, Boehmer J, Krueger S, Kass DA, De Marco T, Carson P, DiCarlo L, DeMets D, White BG, DeVries DW, Feldman AM. Cardiac-resynchronization therapy with or without an implantable defibrillator in advanced chronic heart failure. N Engl J Med. 2004;350:2140–2150. doi: 10.1056/NEJMoa032423. [DOI] [PubMed] [Google Scholar]
- 9.Cleland JG, Daubert JC, Erdmann E, Freemantle N, Gras D, Kappenberger L, Tavazzi L. The effect of cardiac resynchronization on morbidity and mortality in heart failure. N Engl J Med. 2005;352:1539–1549. doi: 10.1056/NEJMoa050496. [DOI] [PubMed] [Google Scholar]
- 10.Chakir K, Daya SK, Tunin RS, Helm RH, Byrne MJ, Dimaano VL, Lardo AC, Abraham TP, Tomaselli GF, Kass DA. Reversal of global apoptosis and regional stress kinase activation by cardiac resynchronization. Circulation. 2008;117:1369–1377. doi: 10.1161/CIRCULATIONAHA.107.706291. [DOI] [PubMed] [Google Scholar]
- 11.Spragg DD, Akar FG, Helm RH, Tunin RS, Tomaselli GF, Kass DA. Abnormal conduction and repolarization in late-activated myocardium of dyssynchronously contracting hearts. Cardiovasc Res. 2005;67:77–86. doi: 10.1016/j.cardiores.2005.03.008. [DOI] [PubMed] [Google Scholar]
- 12.Aiba T, Hesketh GG, Barth AS, Liu T, Daya S, Chakir K, Dimaano VL, Abraham TP, O'Rourke B, Akar FG, Kass DA, Tomaselli GF. Electrophysiological consequences of dyssynchronous heart failure and its restoration by resynchronization therapy. Circulation. 2009;119:1220–1230. doi: 10.1161/CIRCULATIONAHA.108.794834. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Chakir K, Daya SK, Aiba T, Tunin RS, Dimaano VL, Abraham TP, Jacques K, Lai EW, Pacak K, Zhu WZ, Xiao RP, Tomaselli GF, Kass DA. Mechanisms of enhanced {beta}-adrenergic reserve from cardiac resynchronization therapy. Circulation. 2009;119:1231–1240. doi: 10.1161/CIRCULATIONAHA.108.774752. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Leclercq C, Faris O, Tunin R, Johnson J, Kato R, Evans F, Spinelli J, Halperin H, McVeigh E, Kass DA. Systolic improvement and mechanical resynchronization does not require electrical synchrony in the dilated failing heart with left bundle-branch block. Circulation. 2002;106:1760–1763. doi: 10.1161/01.cir.0000035037.11968.5c. [DOI] [PubMed] [Google Scholar]
- 15.Gao Z, Barth AS, DiSilvestre D, Akar FG, Tian Y, Tanskanen A, Kass DA, Winslow RL, Tomaselli GF. Key pathways associated with heart failure development revealed by gene networks correlated with cardiac remodeling. Physiol Genomics. 2008;35:222–230. doi: 10.1152/physiolgenomics.00100.2007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Ojaimi C, Qanud K, Hintze TH, Recchia FA. Altered expression of a limited number of genes contributes to cardiac decompensation during chronic ventricular tachypacing in dogs. Physiol Genomics. 2007;29:76–83. doi: 10.1152/physiolgenomics.00159.2006. [DOI] [PubMed] [Google Scholar]
- 17.Brazma A, Hingamp P, Quackenbush J, Sherlock G, Spellman P, Stoeckert C, Aach J, Ansorge W, Ball CA, Causton HC, Gaasterland T, Glenisson P, Holstege FC, Kim IF, Markowitz V, Matese JC, Parkinson H, Robinson A, Sarkans U, Schulze-Kremer S, Stewart J, Taylor R, Vilo J, Vingron M. Minimum information about a microarray experiment (MIAME)-toward standards for microarray data. Nat Genet. 2001;29:365–371. doi: 10.1038/ng1201-365. [DOI] [PubMed] [Google Scholar]
- 18.Tusher VG, Tibshirani R, Chu G. Significance analysis of microarrays applied to the ionizing radiation response. Proc.Natl.Acad Sci U.S.A. 2001;98:5116–5121. doi: 10.1073/pnas.091062498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Al-Shahrour F, Minguez P, Tarraga J, Medina I, Alloza E, Montaner D, Dopazo J. FatiGO +: a functional profiling tool for genomic data. Integration of functional annotation, regulatory motifs and interaction data with microarray experiments. Nucleic Acids Res. 2007;35:W91–96. doi: 10.1093/nar/gkm260. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Dennis G, Jr., Sherman BT, Hosack DA, Yang J, Gao W, Lane HC, Lempicki RA. DAVID: Database for Annotation, Visualization, and Integrated Discovery. Genome Biol. 2003;4:P3. [PubMed] [Google Scholar]
- 21.Das DK, Maulik N, Moraru Gene expression in acute myocardial stress. Induction by hypoxia, ischemia, reperfusion, hyperthermia and oxidative stress. J Mol Cell Cardiol. 1995;27:181–193. doi: 10.1016/s0022-2828(08)80017-x. [DOI] [PubMed] [Google Scholar]
- 22.Kubisch C, Wollnik B, Maass A, Meyer R, Vetter H, Neyses L. Immediate-early gene induction by repetitive mechanical but not electrical activity in adult rat cardiomyocytes. FEBS Lett. 1993;335:37–40. doi: 10.1016/0014-5793(93)80434-v. [DOI] [PubMed] [Google Scholar]
- 23.Schoonjans K, Peinado-Onsurbe J, Lefebvre AM, Heyman RA, Briggs M, Deeb S, Staels B, Auwerx J. PPARalpha and PPARgamma activators direct a distinct tissue-specific transcriptional response via a PPRE in the lipoprotein lipase gene. Embo J. 1996;15:5336–5348. [PMC free article] [PubMed] [Google Scholar]
- 24.Nowak B, Sinha AM, Schaefer WM, Koch KC, Kaiser HJ, Hanrath P, Buell U, Stellbrink C. Cardiac resynchronization therapy homogenizes myocardial glucose metabolism and perfusion in dilated cardiomyopathy and left bundle branch block. J Am Coll Cardiol. 2003;41:1523–1528. doi: 10.1016/s0735-1097(03)00257-2. [DOI] [PubMed] [Google Scholar]
- 25.Valzania C, Gadler F, Winter R, Braunschweig F, Brodin LA, Gudmundsson P, Boriani G, Eriksson MJ. Effects of cardiac resynchronization therapy on coronary blood flow: evaluation by transthoracic Doppler echocardiography. Eur J Heart Fail. 2008;10:514–520. doi: 10.1016/j.ejheart.2008.03.011. [DOI] [PubMed] [Google Scholar]
- 26.Agnetti GE, Agnetti ST, Kane LA, Yung C, Chakir K, Samantapudi D, Guamieri C, Caldarera CM, Kass DA, Van Eyk JE. Effects of cardiac resynchronization therapy on the mitochondrial proteome in a canine model of heart failure. Circulation. 2007;116:664–664. [Google Scholar]
- 27.Bugra Z, Koylan N, Vural A, Erzengin F, Umman B, Yilmaz E, Meric M, Buyukozturk K. Left ventricular geometric patterns and QT dispersion in untreated essential hypertension. Am J Hypertens. 1998;11:1164–1170. doi: 10.1016/s0895-7061(98)00144-7. [DOI] [PubMed] [Google Scholar]
- 28.Harjai KJ, Samal A, Shah M, Edupuganti R, Nunez E, Pandian NG. The relationship between left ventricular shape and QT interval dispersion. Echocardiography. 2002;19:641–644. doi: 10.1046/j.1540-8175.2002.00641.x. [DOI] [PubMed] [Google Scholar]
- 29.Berger T, Hanser F, Hintringer F, Poelzl G, Fischer G, Modre R, Tilg B, Pachinger O, Roithinger FX. Effects of cardiac resynchronization therapy on ventricular repolarization in patients with congestive heart failure. J Cardiovasc Electrophysiol. 2005;16:611–617. doi: 10.1046/j.1540-8167.2005.40496.x. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.