Abstract
Small hairpin RNAs (shRNAs) are widely used in RNAi studies and typically consist of a stem of 19–29 base pairs (bp), a loop of at least 4 nucleotides (nt), and a dinucleotide overhang at the 3′ end. Compared with shRNAs with 21–29 bp stems, we have found that shRNAs with 19-bp or shorter stems (sshRNAs) possess some unique structure–activity features that depend on whether the antisense strand is positioned 5′ or 3′ to the loop (L- or R-type sshRNAs, respectively). L sshRNAs can have IC50s in the very low picomolar range, and sshRNAs with nominal loop sizes of 1 or 4 nt were at least as active as those with longer loops. L sshRNAs remained highly potent even when the 3′ end of the antisense strand was directly linked with the 5′ end of the sense strand. In this case, the sense strand can be shorter than the antisense strand, and the loop can be formed entirely by the 3′ end of the antisense strand. Monomer sshRNAs are not processed by recombinant Dicers in vitro. Although they can form dimers that are sometimes Dicer substrates, their RNAi activity is not dependent on the formation of such structures. Our findings have implications for the mechanism of action of sshRNAs, and the ability to design highly potent shRNAs with minimal length is encouraging for the prospects of the therapeutic use of direct-delivered shRNAs.
Keywords: small hairpin RNA, shRNA, sshRNA, Dicer, stem–loop, immunostimulatory
INTRODUCTION
RNA interference (RNAi) is a naturally occurring process whereby a double-stranded RNA (dsRNA) of a specific sequence regulates the expression of genes containing that sequence. RNAi-inducing triggers, such as synthetic or expressed short interfering RNAs (siRNAs), long dsRNAs, and small hairpin RNAs (shRNAs), have been widely used for gene function analysis, pathway mapping, drug target identification and validation, and host–pathogen interactions (Elbashir et al. 2001; Xia et al. 2002; Dorsett and Tuschl 2004; Xia et al. 2004; Harper et al. 2005; Wang et al. 2005; Amarzguioui et al. 2006; Bernards et al. 2006; Chang et al. 2006; Fewell and Schmitt 2006; Ilves et al. 2006; Seyhan et al. 2006, 2007; Vlassov et al. 2007). Among these, shRNA has received considerable attention due to its wide spread use in DNA vector-based applications—such as shRNA libraries for various loss-of-function screens, cell lines, transgenic animals that express silencing triggers against targets of interest, and therapeutic approaches (Xia et al. 2002; Xia et al. 2004; Harper et al. 2005; Li et al. 2005a,b; Grimm et al. 2006). In contrast to expressed shRNA, interest in the direct use of synthetic shRNA has so far been limited.
The basic structure of an ordinary shRNA consists of paired antisense and sense strands connected by a loop of unpaired nucleotides. A duplex stem of 19–29 base pairs (bp), either fully paired or with miRNA-style internal mismatches or loops, is commonly used in vector-expressed shRNAs (Zeng et al. 2002; Cai et al. 2004; Silva et al. 2005; Stegmeier et al. 2005; Chung et al. 2006; Boudreau et al. 2008). Unlike with siRNAs, identification of effective target sequences for shRNAs has not been systematically investigated. It was assumed that the efficacy of siRNAs and shRNAs were governed by the same principles, and consequently, the sequences that were effective with siRNAs were simply placed into the scaffold of shRNA. A recent report (Li et al. 2007), however, suggested that the target sequences that provide the most potent target knockdown with shRNAs may be different from those best for siRNAs. Not surprisingly, the structure of shRNAs has also been found to affect the silencing ability. For instance, shRNAs with fully paired stems and miRNA-based shRNAs have been compared in the context of vector expression, and the former were found to be more potent (Boudreau et al. 2008). Moreover, at least in the case of synthetic shRNAs, the lengths of both stems and loops can affect efficacy (Li et al. 2007; Vlassov et al. 2007). Li and colleagues (Li et al. 2007) reported that in the context of 4-nucleotide (nt) loops, shRNAs with 29-bp stems silenced target gene expression more efficiently than those with 19-bp stems, but 19-bp shRNAs with 9-nt loops outperformed shRNAs with longer stems, including 29-bp shRNAs with 4-nt loops. Our group (Vlassov et al. 2007) found that, in the context of 10-nt loops, 19-bp shRNAs were somewhat more potent than similar 19-bp and 25-bp siRNAs at inhibiting hepatitis C virus (HCV) replicons, and 25-bp shRNAs (in the context of the same 10-nt loop) were less potent than any of the 19-bp shRNAs or siRNAs tested. The presence of short flanking sequences such as a 3′ overhang generally enhanced the efficiency of gene knockdown for shRNAs (Siolas et al. 2005; Vlassov et al. 2007).
The position of the antisense (guide) strand within the hairpin can also affect shRNA activities. Generally, shRNAs are designed to have sense (passenger) strands at the 5′ end of the hairpin (right-hand loop, R-type shRNAs) (Wang et al. 2005; Ilves et al. 2006; Li et al. 2007; Vlassov et al. 2007). Harborth et al. (2003) reported that an shRNA that has the antisense at the 5′ end of the hairpin (left-hand loop, L-type shRNAs) showed comparable silencing efficacy to an R shRNA if the stem length was 21–29 nt. However, when the stem length was shortened to 19 bp (with a 4-nt loop), much less potency was found with R shRNA, whereas L shRNA retained a potency comparable to that of shRNAs with 21–29-bp stems. Similar results were obtained by McManus et al. (2002) with a CD8-specific shRNA. These results suggest that 19-bp shRNAs belong to a special class of hairpin RNAs that may function differently from shRNAs of 21 bp or longer. Indeed, unlike their 29-bp counterparts that can be cleaved by Dicer to generate siRNAs, 19-bp R shRNAs were found not to be Dicer substrates (Siolas et al. 2005). To distinguish these classes, we designate hairpins with a stem length of 19 bp or less as short shRNAs (sshRNAs), and 21-bp or longer hairpins as long shRNAs (lshRNAs).
In view of the unexpected findings that sshRNAs can be highly potent without requiring Dicer processing, and the confusing relationship between potency and R versus L structure, there is clearly a need for a better understanding of the relationship between shRNA structural features, including the size and orientation of the loop as well as the length and base-pairing status of the stem, with silencing activity. To date, comparisons of the activities of L and R sshRNAs have been reported in only two publications (McManus et al. 2002; Harborth et al. 2003), and each report involved only a single target sequence. Thus, it has not been clear whether the reported efficacy differences between L and R sshRNAs hold true in general or if they apply only to certain target sequences. In addition, the relationship between structure (duplex length, loop length, and presence of overhang) and activity has been studied with R but not L sshRNAs. Such a study is important for the prospect of therapeutic applications of shRNAs, since if L sshRNAs are better than R sshRNAs, either inherently or upon structural optimization, lower doses can be used, which in turn reduces the likelihood of RNAi-related side effects, such as cellular inflammatory responses to dsRNA and unintended target silencing. The impact of hairpin structures on the efficiency of target knockdown may also shed light on the mechanism of sshRNA activity.
In this study, we compared the potencies of L and R sshRNAs and investigated in detail the structure–activity relationship of L sshRNAs targeting the HCV internal ribosome entry site (IRES). The requirement of Dicer cleavage in L sshRNA processing, the specificity of the suppression of target mRNA transcription (Ago2-mediated slicing at a specific nucleotide in the target or interferon-mediated nonspecific inhibition), and the impact on shRNA activity of dimerization (a unique feature of synthetic RNA) were also investigated. Consistent with earlier reports (McManus et al. 2002; Harborth et al. 2003), we found that an L sshRNA had better potency than its R counterpart. However, unlike with R sshRNA, the presence of a 3′ overhang was not essential to the activity of L sshRNAs. The connection between the antisense and sense strands could be formed by as few as 1 or 2 nt, or even by just the 3′ part of the antisense strand, without a significant reduction in the potency of L sshRNAs. Neither L nor R shRNA monomers were Dicer substrates. Although sshRNAs can form dimers and multimers that are Dicer substrates, the activity of monomer sshRNAs is not dependent on the formation of such intermolecular structures.
RESULTS AND DISCUSSION
Low-picomolar IC50s can be achieved with L sshRNAs
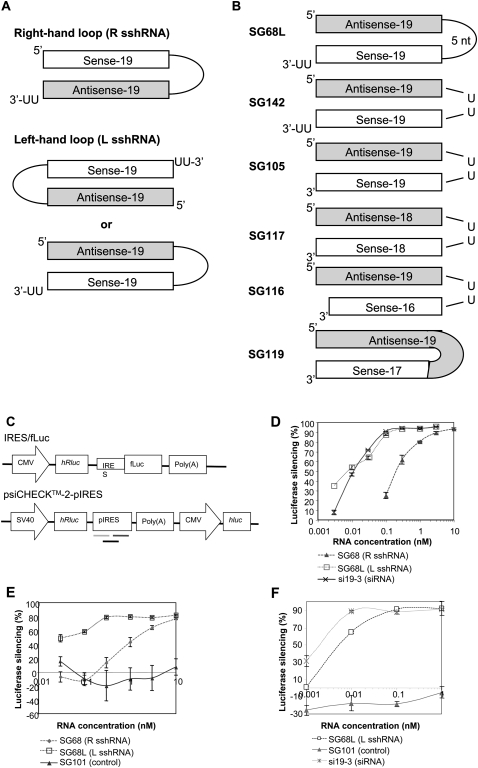
shRNAs are generally employed using the right-hand loop design. In our initial attempt to screen for shRNAs effective against HCV, we also used the R shRNA design with stem lengths of 19 bp and 25 bp. Since better efficacies were reported with two L sshRNAs (specific for Lamin A and CD8) compared with their R counterparts (McManus et al. 2002; Harborth et al. 2003), we compared R and L versions of a sshRNA that we had previously found to strongly inhibit gene expression mediated by the HCV internal ribosome entry site (IRES), in both a luciferase reporter system and an HCV subgenomic replicon system (Vlassov et al. 2007). The structures of these R and L sshRNAs are shown in Figure 1A. Synthetic R and L sshRNAs were transfected at various concentrations into human cells—the kidney line 293FT (Fig. 1D) and the hepatocyte line Huh7 (Fig. 1E)—together with a firefly luciferase (fLuc) reporter plasmid whose expression was driven by the HCV IRES (Fig. 1C, IRES/fLuc plasmid). With appropriate correction using controls without sshRNA or with an unrelated sshRNA (SG101), the expression levels of fLuc with the IRES-specific sshRNAs reflect the target gene knockdown capabilities of the test sshRNAs. No difference in fLuc expression was found between the two controls (data not shown). Consistent with previous reports, in both cell lines, SG68L, the L sshRNA counterpart to SG68, showed a nearly 30-fold higher potency (IC50) than SG68 (R sshRNA) (Fig. 1D,E). SG68L was also more effective than a 25-bp lshRNA whose target sequence encompasses that of SG68L (Supplemental Table S1; data not shown). The dose response curve of si19-3, a siRNA that targets the same sequence as the SG68 and SG68L, was very close to that of SG68L. To eliminate the possibility that this observation is due to the use of this particular IRES-driven reporter system, the targeting sequence was inserted into the 3′ end of the Renilla luciferase in a dual luciferase reporter plasmid that is commonly used to study RNAi (Fig. 1C, psiCHECK-2-pIRES). Si19-3 and SG68 showed similar potencies using this target (Fig. 1F). The high potency with the L form might relate to the fact that its 5′ end, which is the 5′ end of the antisense sequence, has an exposed 5′-phosphate that facilitates binding to Ago2 (Nykanen et al. 2001) in the RNA-induced silencing complex (RISC), whereas the 5′ end of the antisense sequence of R sshRNAs is connected to the loop.
FIGURE 1.
Structure and activity of R and L sshRNAs. (A) General structures of R and L sshRNAs. (B) Representative structures of L sshRNAs, with sense and antisense strands of equal or unequal length. (C) Schematic representation of luciferase reporter plasmids expressing the target for si/shRNAs. The IRES/fLuc plasmid has the near full-length HCV IRES upstream of the firefly luciferase open reading frame. The expression of the firefly luciferase is driven by the HCV IRES. The psiCHECK-2-pIRES plasmid was constructed based on psiCHECK-2 from Promega. The targeting regions (part of the HCV IRES [pIRES]) of si19-3 (black line), si72 (light gray line), and si74 (dark gray line) were individually inserted at the 3′ end of hRluc by site-directed mutagenesis. (D) Comparison of the activities of L and R sshRNAs using IRES/fLuc plasmid in 293FT cells. sshRNAs against the same target were chemically synthesized with right- and left-hand loops (SG68 and SG68L). These shRNAs were cotransfected in triplicate into 293FT cells with IRES-fLuc plasmid. The luciferase expression was measured 48 h post-transfection. At least two independent experiments were performed and similar results were obtained. Curves were plotted using Microsoft Excel software, while IC50s were calculated based on curve fitting using Erithacus GraFit software. (E) Comparison of the activities of L and R sshRNAs in Huh7 cells. Transfection conditions were as in D. SG101 was a nonspecific control for SG68 and SG68L. (F) Comparison of the activities of L sshRNA and siRNA using the dual luciferase reporter plasmid (psiCheck2-pIRES) in 293FT cells. Transfection conditions were as in D.
The influence of loop structure on sshRNA potency
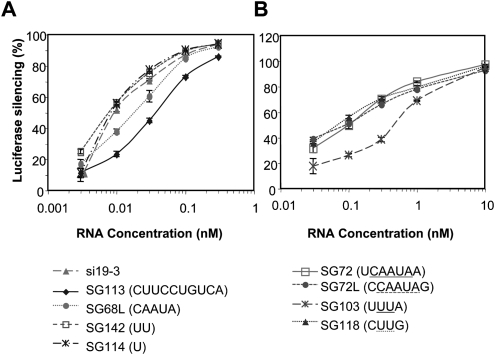
Unlike the previous results with R sshRNAs, for which a larger loop (5 nt or more) sometimes provides greater potency (Li et al. 2007; Vlassov et al. 2007), we find that L sshRNAs have higher potencies when the loop is very small (1 or 2 nt) than when it is larger (5 or 10 nt) (see Fig. 1B for structures; see Fig. 2A for activities). The IC50 of SG142 (L sshRNA with a 5′-UU-3′ loop) was 4.6 pM, slightly more potent than si19-3, which targets the same region. Of the sshRNAs with 4-nt loops, SG118 (L, CUUG loop) had a lower IC50 than SG103 (R, AUUU loop), whereas R and L sshRNAs with a 5-nt loop (CAAUA in SG72 and SG72L) showed similar potencies (Fig. 2B).
FIGURE 2.
Comparison of L sshRNA activity with various loop structures and base pairs adjacent to the loop. (A) Loop length comparison, using L sshRNAs against the same target region as sh68 and si19-3, but with various lengths and sequences of loops. sshRNAs were chemically synthesized and their abilities to inhibit HCV IRES-dependent luciferase expression were compared in 293FT cells. (B) Loop-adjacent base-pair comparison. sshRNAs against the same target region as si72, but with loops of 5 nt (SG72 and SG72L) and 2 nt (SG118 and SG103) and left-loop (SG72L and SG118) and right-loop (SG72 and SG103) orientations were compared for their inhibitor activity in 293FT cells. Loop sequences are underlined.
The effects of 3′ overhang and stem lengths on the efficacy of sshRNA
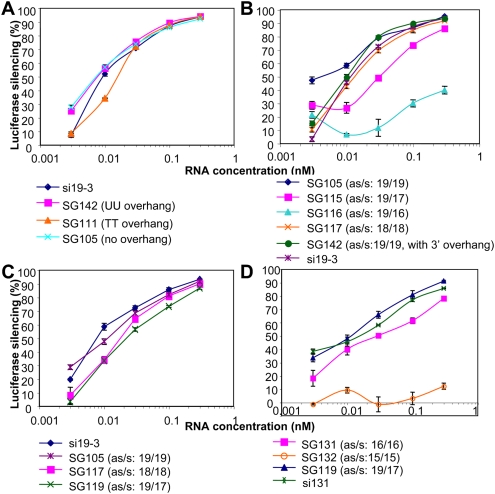
L sshRNAs with (SG142) and without (SG105) 3′-UU overhangs were compared and no significant difference in gene-specific silencing was found (Fig. 1B, structures; Fig. 3A, activities). We also found that the ribonucleotide UU at the 3′ overhang could be replaced with the deoxyribonucleotide TT without significantly affecting the activity (Fig. 3A).
FIGURE 3.
Comparison of the activities of L sshRNAs differing in 3′ overhang and stem length. (A) Overhang comparison. L sshRNAs against the same target region as si19-3 and SG142, but with or without a 3′ overhang were chemically synthesized and their ability to inhibit HCV IRES-dependent luciferase expression was compared in 293FT cells. (B) Stem length comparison. L sshRNAs (UU-loop) against the same target region as SG142 and si19-3, but with different stem lengths were compared for their inhibitory activity in 293FT cells. SG105 differs from SG142 in lacking a 3′-UU overhang. (C) SG119 and SG117 comparison in 293FT cells. SG119 has a 19-nt antisense strand directly linked with a 17-nt sense strand, probably forming a 16-bp duplex and 4-nt loop (UGCA). (D) Stem length comparison with SG119 derivatives. SG131 and SG132 have UU to connect the 3′ end of the antisense and the 5′ end of the sense strands. They were compared with SG119 for target knockdown in 293FT cells. si131 has two complementary strands 16 nt in length and UU overhangs at the 3′ ends. Values labeled “as/s” represent the nucleotide lengths of antisense and sense strands.
The effect of stem length on sshRNA activity was also investigated. Shortening the sense strand from its 3′ end to 17 or 16 nt, while maintaining the length of the antisense strand at 19 nt significantly reduced gene silencing activity while maintaining the length of the antisense strand at 19 nt (SG116) (see Fig. 1B), significantly reduced gene silencing activity (Fig. 3B), suggesting the importance of a duplex structure at the 5′ end of the antisense sequence. However, when nucleotides adjacent to the loop were deleted, for example, in SG117 (one base pair deleted at the 3′ end of the antisense/the 5′ end of the sense strand), no significant loss in potency was observed (Fig. 1B, structure; Fig. 3B, activity).
To better understand how short the stem of a shRNA can be before activity is lost, we tested SG119, a sshRNA with a stem of 16 bp consisting of a 19-nt antisense strand connected directly to a 17-nt sense strand (Fig. 1B). SG119 showed similar potency to SG117, a sshRNA with 18 nt in each strand joined by a UU linker, which probably forms a 17-bp stem with a GUUC loop (Fig. 3C). However, SG131, with 16 nt in each strand connected by UU (which probably forms a 15-bp stem with a GUUC loop), showed somewhat lower activity, and SG132, with an effective stem of probably 14 bp, had very little activity (Fig. 3D). These results indicate that potent silencing activity requires a hairpin with a duplex length of at least 16 bp, or at least 17 bp if the G and C residues of the GUUC loops are paired. Interestingly, an siRNA with a 16-bp duplex and UU overhangs at the 3′-end of both strands (si131) showed similar but slightly less potency than its near-cognate sshRNA (SG119) and an siRNA with a 19-bp duplex (si19-3) (Fig. 3D; data not shown). This finding is in contrast to the report of Chu and Rana (2008) that a 16-bp siRNA was a more potent RNAi trigger than its 19-bp counterpart, suggesting that the effect of stem length can be modulated by sequence, at least in the case of siRNAs.
To summarize the effects of stem length and 3′ overhang, whereas 19-bp R sshRNAs require the presence of a 3′-UU overhang for maximal efficacy (Siolas et al. 2005; Li et al. 2007; Vlassov et al. 2007), for L sshRNAs, a 3′ overhang is not essential for their activity. L sshRNAs can tolerate a linker as short as 1 or 2 nt connecting the antisense strand and sense strand, or even a direct connection between the two strands, without a significant effect on their inhibitory efficiency; and both antisense and sense strands can be shortened to 18 nt (SG117). Finally, the sense strand can be shortened to 17 nt when directly connected to the 3′ end of the 19-nt antisense strand (SG119) without significant loss of activity. These results are consistent with recent crystal structure studies (Wang et al. 2008) showing that archaeal Ago2 can form stable ternary complexes with a 21-nt guide strand and a short complementary (target) RNA. In this published structure, the 3′ end of the guide strand is released from binding to the PAZ domain of the protein, suggesting flexibility in the length of the guide strand required for efficient binding.
The sshRNA structure–activity relationship is somewhat sequence dependent
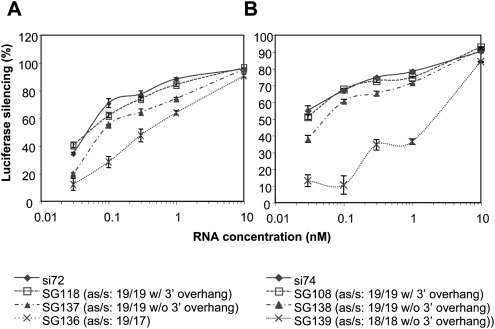
To further examine whether the requirements regarding the 3′ overhang and stem length are sequence specific or can be generalized to shRNAs of different target sequences, two targets were selected with nonoverlapping sequences (Fig. 4A,B). siRNAs against these two targets (si72 and si74) (see Fig. 1C for target locations) were used as positive controls for dose response experiments. As shown in Figure 4, A and B, L shRNAs having antisense and sense strands 19 nt in length, with 3′-UU overhangs and dinucleotides connecting the two strands, showed potencies similar to those of their respective siRNA controls for each target sequence (SG118 versus si72; SG108 versus si74). However, removing the 3′ overhang from these two sshRNAs resulted in a slight but reproducible loss in activity. By comparison, for the other target sequence, SG142 (with 3′-overhang) and SG105 (without overhang) had similar potencies (Fig. 3A). In addition, a significant reduction of efficacy was seen when the stem length was reduced (Fig. 4A,B). For example, SG136, with a 19-nt antisense strand directly connected with a 17-nt sense strand, had an IC50 of 393.1 pM, whereas SG118, with 19 nt in both strands and a UU connector, had an IC50 of 51.9 pM. This discrepancy in the structure–activity relationship among sshRNAs against three different targets clearly indicates the existence of sequence-related effects.
FIGURE 4.
Comparison of the activities of L sshRNAs against two different targets. (A) Comparison of L sshRNAs specific for the same target as siRNA si72. SG118 and SG137 have a dinucleotide UU to link the antisense and sense strands (both are 19 nt in length). SG136 has a 19-nt antisense strand directly connected with the 5′ end of a 17-nt sense strand. (B) Comparison of L sshRNAs specific for the same target as siRNA si74. All three sshRNAs have dinucleotide UU as a connection between antisense and sense strands. Values labeled “as/s” represent the nucleotide lengths of antisense and sense strands.
It is noteworthy that many published studies examining structure–activity relationships have focused on siRNAs or shRNAs specific for only one or two targets. Some of the structure–activity correlations derived from these studies may thus be sequence specific and not generalizable to other target sequences. Because different target sequences are often associated with large differences in potency, structure–activity correlations may be influenced by the inherent potency associated with a target site. Thus, we find that sshRNAs having IC50 less than 10 pM, such as SG68L and SG142, are less sensitive to structural perturbations than less potent sshRNAs, such as SG118 and SG108 (IC50 ∼20–60 pM). Where potency may be related to the affinity of Ago2 or other processing components for the hairpin or its antisense strand, the sensitivity to structure can be understood on thermodynamic grounds. For example, if Ago2 binds to the antisense strand with high affinity, the binding energy may dominate over the influence of structural elements, such as the 3′-end overhang and the identity of the 3′-most nucleotides of the antisense strand. However, if the binding of Ago2 is weaker, the influence of those structural elements on the interaction may be relatively greater.
In many of the published studies of the effects of structure on activity, the siRNAs or shRNAs were tested only at one or two concentrations. As can be seen from the dose response curves presented above, if only one concentration of sshRNA is tested and it is not near the IC50 of the inhibitors being compared, little difference may be seen. For example, the level of inhibition at 0.3 nM is generally much more revealing of potency differences than is inhibition at 10 nM (e.g., Figs. 2B, 4). Therefore, a dose response curve of at least three concentrations covering from close to 100% to less than 50% suppression is essential in understanding structure–activity relationships. Taken together, our results indicate that, for drug development, an extensive optimization for each target site of interest should be performed, whereas for research purposes, an L sshRNA with a 19-bp stem, 5-nt loop, and UU overhang at the 3′ end is likely to have good activity at many sites.
The effect of single-nucleotide mismatches on shRNA activity
Several considerations motivated us to examine the impact of single-nucleotide mismatches on the activity of shRNAs. First, the hairpin structures of all known pre-microRNAs have mismatches in their duplex regions. It has been shown that a single-nucleotide mismatch is generally tolerable to the function of siRNAs and lshRNAs as long as the mismatch is not close to the slicer site (Jackson et al. 2003; Du et al. 2005). However, it is not clear a priori whether the same is true for sshRNAs, given that shorter duplex regions are more likely to be destabilized by single mismatches than longer duplexes. Second, since both the antisense and sense strands of a siRNA can be incorporated into RISC, the selection of the sense strand by Ago2 could induce undesired effects if part of the sense strand (particularly the seed region, nucleotides 2–7) is complementary to the coding region or 3′-UTR of nontarget mRNAs (Doench et al. 2003; Jackson et al. 2003; Khvorova et al. 2003; Schwarz et al. 2003; Scacheri et al. 2004; Lin et al. 2005). It might be desirable to alter a nucleotide in positions 2–7 of the sense strand to abrogate a specific off-target effect without affecting the on-target activity, and this change would introduce a mismatch into the shRNA. It is not clear whether such a strategy could be tolerated by sshRNAs.
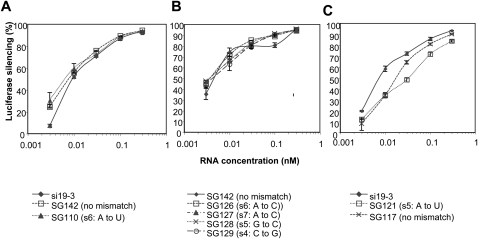
We thus investigated the effect of single-nucleotide mismatches at various positions within the potential seed region of the sense strand of SG142. This construct is an L sshRNA with 19-nt antisense and sense strands, a 3′-UU overhang, and a UU loop. As shown in Figure 5, A and B, these changes resulted in no significant loss of activity, regardless of the type of mismatch. For example, SG110, with a U–U mismatch, and SG126, with a U–C mismatch at position 6 from the 5′ end of the sense strand have similar efficacy, suggesting that this trait is sequence independent.
FIGURE 5.
Effect of single mismatches in the seed region of the sense strand of L sshRNAs on inhibitory activity. (A,B) Effect of position of mismatch. SG142-based L sshRNAs with single mismatches at positions 4–7 from the 5′ end of the sense strand were chemically synthesized and their abilities to inhibit HCV IRES-dependent luciferase expression were compared in 293FT cells. (C) Effect of a single mismatch at position 5 from the 5′ end of the sense strand on inhibition of SG117-based sshRNAs (s/as = 18/18), compared with the fully matched SG117 and si19-3.
Interestingly, when the single-nucleotide mismatch was introduced into the sense strand of SG117, an L sshRNA with 18 nt in both sense and antisense strands, a UU linker, and no overhang, the activity is slightly reduced (Fig. 5C). The difference in behavior of SG117 from SG142 may reflect the ability of mismatches to destabilize shorter duplexes more than longer ones.
sshRNAs are not Dicer substrates in vitro
As with the processing of miRNAs in the cytoplasm, 21–29-bp lshRNAs are recognized and cleaved by Dicer to generate siRNAs with strand lengths of 21–23 nt (Siolas et al. 2005). For 19-bp sshRNAs with 4-nt loops, no cleavage by Dicer is seen in either cell-free systems or cell cultures (Siolas et al. 2005). To investigate whether our synthetic sshRNAs with a duplex length of 19 bp or less can be cleaved by Dicer, several of the sshRNAs tested above were incubated in vitro with recombinant Dicer. A 25-bp lshRNA (Fig. 6, labeled as sh1), used as a positive control, generated a product of a size between 20 and 25 bp after Dicer treatment. All other sshRNAs tested, including one R sshRNA with a stem of 19 bp (SG68), two L sshRNAs with a stem of 19 bp (SG142, SG105), and one L sshRNA with a stem of 18 bp (SG117), showed no cleavage products in both nondenaturing (Fig. 6A) and denaturing PAGE (Fig. 6B). Consistent with the previous report (Siolas et al. 2005), our results indicate that sshRNAs with a duplex length of 19 bp or less are not substrates for Dicer alone.
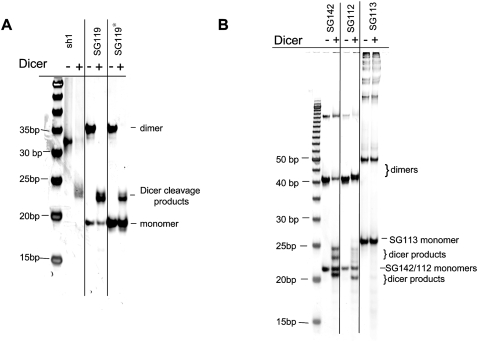
FIGURE 6.
sshRNAs monomers are not Dicer substrates. Eight picomoles of each synthetic shRNA (after heating and snap-cooling) were incubated in a 10 μL reaction in the presence of 1 U of recombinant Dicer enzyme (Stratagene) and buffer containing 150 mM NaCl, 20 mM Tris-HCl (pH 8), and 2.5 mM MgCl2 for 18 h at 37°C. Control reactions that contained each shRNA but lacked Dicer were incubated in parallel. Samples were analyzed by (A) nondenaturing 10% PAGE and (B) denaturing 12% PAGE (8 M urea/20% formamide) and were stained with SYBR Gold. Note that SYBR Gold did not stain single-strand RNA as strongly as double-strand RNA.
It has been shown that some longer dsRNAs (e.g., with 27-bp stems) have better target knockdown activities than corresponding 21–23 nt siRNAs (Rose et al. 2005). This was attributed to Dicer processing of the 27-bp dsRNAs that generated more potent molecules. However, sshRNAs as monomers were found not to be Dicer substrates in vitro, despite the fact that most of the sshRNAs tested in this study showed comparable potency to siRNAs having the same antisense sequence and better potency than a lshRNA (sh1, 25-bp stem length) that contains the same antisense sequence (data not shown). These results suggest that Dicer processing is not generally required to achieve high potency with at least some sshRNAs.
Siolas et al. (2005) suggested that some single-strand-specific ribonuclease may be involved in the processing of 19-bp shRNAs in the loop because of the detection of 21-nt and 23-nt cleavage products inside RISC. However, it is not clear whether such processing is a prerequisite for RISC loading. Our data appear to argue against a requirement for a nonspecific ribonuclease cleavage on several accounts. First, changes in the size of the loop from 10 down to 1 nt and in its sequence did not significantly affect target knockdown efficiency for L sshRNAs. Second, SG119 has a 17-nt sense strand directly linked to the 3′ end of a 19-nt antisense strand. Based on known minimal sizes for hairpin loops in RNA, SG119 may adopt hairpin structures in which the loop size is between 2 and 4 nt. It would presumably recruit 2–3 nt from the 3′ end of the antisense strand and 0–1 nt from the 5′ end of the sense sequence. If this loop were randomly cleaved by a single-strand-specific ribonuclease, the silencing activity of the sshRNA would likely to be affected due to the incorporation of a shortened antisense strand into the RISC. However, this was not the case (Fig. 3C,D). Third, a nonspecific cleavage at the loop would generate multiple products, with weak inhibitors diluting the effect of the potent ones. However, we found that the best sshRNAs are at least as potent as the cognate siRNA, where no such dilution would occur.
Investigation of target mRNA cleavage site
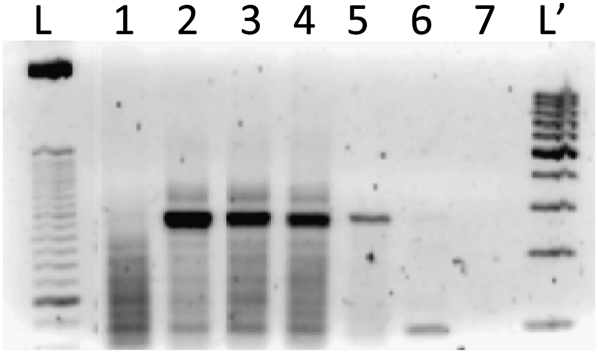
To verify whether the observed gene suppression by sshRNAs is due to RNAi-directed cleavage, a modified 5′-rapid amplification of cDNA ends (5′-RACE) procedure (Soutschek et al. 2004) was performed to identify the specific cleavage site of the target mRNA. This site is expected to be 10 nt downstream from the 5′ end of the guide strand (Rana 2007). As shown in Figure 7, a PCR amplicon of the expected size (∼250 bp) was generated when cells were transfected with either siRNAs or sshRNAs targeting the same mRNA region, but not when cells were left untreated or were transfected with the target alone. Sequencing of the amplified fragments showed that the cleavage site on the target mRNA was indeed 10 nt from the 5′ end of the antisense strand of both sshRNA and siRNA. These results strongly support the involvement of the RNAi machinery in sshRNA-mediated gene knockdown and suggest that the processing of sshRNAs, whether R or L (SG68 versus SG105), long or short (SG105 with an 18-bp stem length; SG119 with a 16-bp stem length), generates guide strands similar to those of standard siRNAs directed at the same target. The questions of whether and how the sshRNAs are processed in the silencing pathway are being addressed in a separate study (Q Ge, H Ilves, A Dallas, P Kumar, J Shorenstein, SA Kazakov, MA Behlke, and BH Johnston, in prep.).
FIGURE 7.
Target mRNA cleavage in vivo. 5′-RACE–PCR amplification of samples with and without target-specific siRNA or sshRNA treatments was analyzed on 2% agarose gels. L, 25-bp ladder; L′, 100-bp ladder; target pSG154m alone (lane 1); SG105 (lane 2); SG119 (lane 3); SG68 (lane 4); si19-3 (lane 5); no transfection control (lane 6); and water control (lane 7).
Interferon response to sshRNAs
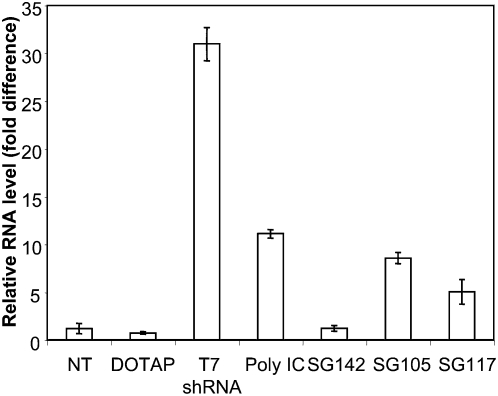
shRNAs and siRNAs have the potential to induce the undesired expression of proinflammatory cytokines, such as type I interferon (IFN). Several factors, including length, sequence, and structure influence this effect, and it has been shown that protein kinase R (PKR), toll-like receptors, and the cytosolic RNA helicase retinoic acid-inducible gene (RIG-I) are involved in the recognition and activation of the IFN pathway (Judge and MacLachlan 2008). To evaluate IFN induction by our molecules, freshly purified human peripheral blood mononuclear cells (PBMCs) were transfected by HCV-specific sshRNAs and the level of mRNA encoding 2′-5′-oligoadenylate synthetase (OAS), an interferon-induced enzyme, was measured. sshRNAs with a 3′ overhang (SG142) did not induce OAS expression, whereas those with blunt ends (SG105 and SG117) induced OAS (Fig. 8) as well as IFN-β expression (data not shown). The effect of blunt ends suggests that IFN induction may be mediated by RIG–I (Marques et al. 2006). When 2′-O-methyl (2′-OMe) modifications were introduced into the sense strand of SG105, induction of inflammatory cytokines, including OAS-1 and IFN-β, was lost (Ge et al. 2010). Importantly, the efficiency of target knockdown in 293FT cells was the same whether shRNAs were modified or not, suggesting that 293FT cells may not have a strong innate immune response to blunt-ended dsRNA. Thus, the silencing of target gene expression by unmodified blunt-ended sshRNAs in 293FT cells shown above is likely to be a specific on-target effect rather than an IFN-mediated off-target effect.
FIGURE 8.
IFN-responsive gene OAS1-induction by sshRNAs in human PBMCs. Synthetic sshRNAs (20 nM) were complexed with DOTAP (Roche) and transfected into freshly isolated human PBMC in triplicate. DOTAP alone, or DOTAP complexed with equivalent amounts of either T7 transcribed shRNA or polyI:C were used as controls. RNA was extracted from cells 24-h post-transfection and quantitative RT-PCR was performed to measure the level of GAPDH and OAS1. The RNA level of OAS1 was normalized to GAPDH and is shown relative to DOTAP alone.
Effect of dimerization on shRNA activity
Dimerization of hairpin RNAs is an intermolecular process that has been documented in retroviral RNAs, tRNAs, and some artificial RNA hairpins (Sun et al. 2007). The propensity of hairpin RNAs to dimerize depends on their loop size, sequence, and concentration as well as how they are handled (Bernacchi et al. 2005; Liu et al. 2005). However, despite the potential of dimerization to alter the RNAi potency of shRNAs and to promote an interferon response, to our knowledge this issue has not been discussed in the context of miRNA- or shRNA-mediated RNAi.
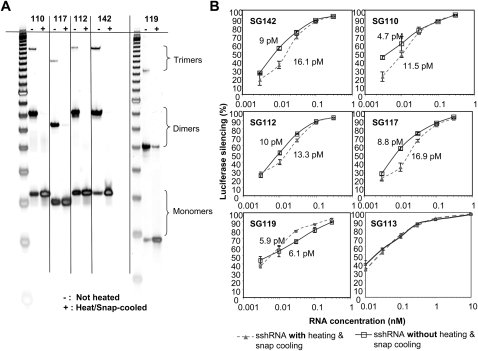
The shRNAs and sshRNAs synthesized and HPLC-purified by IDT, when stored at concentrations of 1–5 μM, were found to comprise at least three major species that resolve in native polyacrylamide gels (Fig. 9A; data not shown). In contrast, under highly denaturing conditions (12% polyacrylamide gel containing 8 M urea and 20% formamide), only a single band was seen (data not shown). When solutions of the shRNAs were heated to 95°C for 4 min and quickly cooled in an ice bath (snap-cooling), the lower-mobility bands in the native PAGE gels largely disappeared (Fig. 9A). Thus, the three major bands seen prior to heating and snap-cooling probably correspond to monomer, dimer, and trimer forms of shRNAs. This phenomenon was found with all the shRNAs synthesized and purified by IDT, irrespective of stem length, loop size, or L versus R loop orientation. However, routine handling of monomer shRNA solutions does not result in formation of dimers or multimers.
FIGURE 9.
L sshRNA is active in monomer form. L sshRNAs were treated with or without heating (95°C for 4 min) and snap-cooling (ice/water bath for 20 min). (A) The presence of monomers and multimers was examined using native 10% polyacrylamide gel electrophoresis and SYBR Gold staining. (B) Comparison of inhibition of IRES-dependent luciferase expression with and without the heat–cool step.
In activity assays, sshRNAs efficiently inhibited IRES-dependent luciferase expression both before and after the heating/snap-cooling procedure (Fig. 9B), even for some sshRNAs that were predominantly dimers before the treatment (Fig. 9A, SG142). These results suggest that the dimers of certain sshRNAs are functional molecules that can be processed and utilized by the RNAi machinery with similar efficiency as the monomers. Any possible effect of the presence of multimers on efficacy is too small to affect any of the structure–activity relationships described above. However, for consistency we adopted a procedure of heating shRNAs to 95°C for 4 min and snap-cooling before each cell transfection experiment (Figs. 1–7).
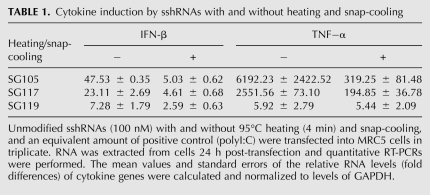
Although Dicer does not appear to be involved in processing monomer sshRNAs (Fig. 6), the dimer conformations of SG119 (a perfect duplex without mismatches), as well as SG112 and SG142 (with two central T–T and U–U mismatches, respectively) can be cleaved by recombinant Dicer in vitro (Fig. 10A,B). However, dimers of SG113, which presumably have a large internal loop, were found not to be Dicer substrates in vitro (Fig. 10B). Considering that the dose response curves of SG113 before and after heat and snap-cool treatments were almost identical (Fig. 9B), these results further indicate that Dicer processing may not be needed for the activity of sshRNAs.
FIGURE 10.
Dicer cleavage of sshRNAs in their dimeric forms. sshRNAs with (*) or without heating and snap-cooling were subjected to recombinant Dicer treatment as indicated. Samples were analyzed by 10% nondenaturing PAGE with SYBR Gold staining. (A) sh1 (25-bp stem; positive control) and SG119. (B) SG 142, SG112, and SG113.
Since the initial results of OAS induction (Fig. 8) were obtained from the mixed sshRNA populations before the treatment, those experiments were repeated using heated and snap-cooled sshRNAs. As shown in Table 1, induction of cytokines such as IFN-β and TNF-α was reduced when sshRNAs were in homogeneous monomer form, suggesting that the length of the duplex (a dimer has a longer duplex length than a monomer) may play a role in the activation of the IFN response. The relative duplex lengths of three sshRNAs targeting the same region as si19-3 are SG105 > SG117 > SG119, whether they are predominantly in dimer or monomer form. Interestingly, the induction of cytokines by these three sshRNAs followed a similar trend regardless of whether they underwent heat/snap-cool cycles, with SG105 the highest and SG119 the lowest (Table 1). This further confirms the influence of duplex length on the immunostimulation capability of hairpins. However, length appears not to be the sole determinant, since SG142—an sshRNA that has the same sequence as SG105 with an additional 3′ overhang—showed no cytokine induction, even as a mixture of dimer and monomer forms (data not shown).
TABLE 1.
Cytokine induction by sshRNAs with and without heating and snap-cooling
Summary: Structure–activity relationship of L sshRNAs
A series of active L sshRNAs have been identified with a stem length of 19 bp or less and a minimal loop, in which the antisense and sense strands are either directly connected or are linked through a UU dinucleotide unrelated to the target. The 5′ end of the antisense strand must be base-paired with the sense strand for maximal activity. A 3′ overhang is not essential for L sshRNAs. Single mismatches in the seed region of the sense strand do not significantly affect the activity of sshRNAs. Unlike the well-studied lshRNAs with a stem of 21 bp or longer, which, if not chemically modified, are recognized and cleaved by Dicer to produce siRNAs and subsequently shuttled into RISC, these sshRNAs are not Dicer substrates in monomer form, whether with left-hand or right-hand loops. Synthetic shRNAs may contain multiple species, including dimers and higher multimers in addition to monomers. At least for some sshRNAs, these mixed populations are almost as active as homogeneous populations of monomer sshRNAs. sshRNAs with blunt ends induced pro-inflammatory cytokine expression in human PBMCs and MRC-5 cells. Higher cytokine production was found when sshRNAs were in dimer/monomer mixture than in homogeneous monomer form, probably owing to the longer duplex length in the dimer form of the hairpin.
The finding that highly potent shRNAs can be designed with short stems and very small or no added loops suggests considerable flexibility on the part of the RNAi machinery, and is encouraging for the development of shRNA therapeutics where high potency is critical and small size is important to keep the cost of manufacture manageable.
MATERIALS AND METHODS
Preparation of shRNAs
shRNAs were chemically synthesized and HPLC purified by Integrated DNA Technologies (IDT) and resuspended in RNase- and pyrogen-free buffer containing 20 mM KCl, 6 mM HEPES-KOH (pH 7.5), and 0.2 mM MgCl2 (Thermo Fisher Scientific, Dharmacon Products). The sequences of all the shRNAs used in this paper are presented in Supplemental Table S1. DNA oligonucleotides were purchased from IDT.
Transfection and reporter gene assays
The human kidney cell line 293FT (Invitrogen) and the human hepatocarcinoma cell line Huh7 (kindly provided by Andrew Simmons, Cell Genesys) were maintained in DMEM (Cambrex) with 10% fetal bovine serum (Hyclone), supplemented with 2 mM of L-glutamine and 1 mM of sodium pyruvate. One day prior to transfection, cells were seeded at 23,000 cells/well in a 96-well plate, resulting in ∼80% cell confluency at the time of transfection. Transfections were performed using Lipofectamine 2000 (Invitrogen) following the manufacturer's instructions. Thirteen nanograms of IRES/fLuc reporter plasmid, 20 ng pSEAP2 control plasmid (BD Biosciences Clontech) as a transfection control, and the indicated amounts of shRNAs were cotransfected into 293FT or Huh7 cells. IRES/fLuc is a dual luciferase expression plasmid in which the HCV IRES is placed between the coding sequences for Renilla and firefly luciferase (fLuc), so that fLuc expression is dependent on the IRES. Forty-eight hours later, the cells were lysed and luciferase activity was measured in a MicroLumat LB 96P luminometer (Berthold Technologies). Unless otherwise indicated, all the siRNA and shRNA samples were tested in triplicate and two or more independent experiments were performed. The IC50 for each dose response curve was calculated using GraFit data analysis software. A similar transfection condition was used when a second luciferase reporter construct, psiCHECK-2-pIRES was tested. psiCHECKTM-2 was purchased from Promega. The targeting sequence for all the si/shRNAs involved in this study (part of the IRES, pIRES) was inserted at the 3′ end of Renilla luciferase by site-directed mutagenesis. The expression of Renilla luciferase of each siRNA or shRNA transfection was normalized to its corresponding firefly expression.
In vitro Dicer cleavage and electrophoresis
shRNA (8 pmol) was incubated in a 10 μL reaction in the presence of 1 U of recombinant Dicer enzyme (Agilent Technology, Stratagene, Catalog no. 240100) and buffer containing 150 mM NaCl, 20 mM Tris-HCl (pH 8), and 2.5 mM MgCl2 for 18 h at 37°C. Control reactions that contained each shRNA, but lacked Dicer, were incubated in parallel. In some experiments, shRNAs were heated to 95°C for 4 min and then were transferred immediately to an ice-water bath to cool for ∼10–20 min before further use, to ensure that all molecules formed hairpin monomers. Samples were analyzed by nondenaturing 10% PAGE and/or denaturing 12% PAGE (8 M urea/20% formamide) and were stained with SYBR Gold (Invitrogen).
Detection of inflammatory cytokine responses
Human PBMCs were prepared from buffy coats (Stanford Blood Center) by density gradient centrifugation, washed, and then seeded in 24-well plates at 5 × 105 cells per well in RPMI 1640 containing 10% heat-inactivated fetal calf serum. Transfections were performed using DOTAP (Roche) following the manufacturer's instructions. Similarly, MRC-5 cells (human fetal lung fibroblast line) were seeded in 24-well plates at 6 × 104 cells per well with DMEM containing 10% heat-inactivated fetal calf serum. Transfections were done using Lipofectamine 2000 following the manufacturer's instruction. shRNAs (20 or 100 nM) were transfected in each well in triplicate. Twenty-four hours later, the cells were lysed in Trizol (Invitrogen) and total RNA was extracted according to the manufacturer's instructions. Quantitative RT-PCR was done using High-Capacity cDNA Reverse Transcription Kits with the TaqMan Universal PCR Master Mix, OAS1 (Hs00242943_m1), IFN-β (Hs01077958_s1), TNFα (Hs99999043_m1), and GAPDH (Hs99999905_m1) TaqMan probes and a Fast 7500 real-time PCR machine (Applied Biosystems) following the manufacturer's instructions.
5′-RACE analysis
293FT cells were transfected with siRNA or sshRNA together with pSG154m. Twelve hours later, total RNA was extracted and mRNA was purified by use of the Oligotex mRNA kit (Qiagen). The mRNA was then subjected to a modified 5′-RACE analysis using the GeneRacer Kit (Invitrogen) following the manufacturer's instruction (Soutschek et al. 2004). Briefly, GeneRacer RNA adaptor (Invitrogen) was ligated to mRNA at its 5′ end. Ligated RNAs were reverse transcribed using a primer 5′-CGCGCCCAACACCGGCATAAAGAATT-3′ and amplified by PCR using primers 5′-GCTTCTGCCAACCGAACGGACATTT-3′ and (adaptor specific) 5′-CGACTGGAGCACGAGGACACTGA-3′. The PCR was started with five cycles of 95°C for 45 sec and 72°C for 30 sec, followed by five cycles of 95°C for 45 sec and 69°C for 30 sec, then by 25 cycles of 95°C for 45 sec, 65°C for 30 sec, and 72°C for 30 sec. The PCR products were revealed by 2% agarose gel electrophoresis, the band with the predicted length of the cleavage product was excised, and the DNA was then purified and cloned for sequencing.
SUPPLEMENTAL MATERIAL
Supplemental material can be found at http://www.rnajournal.org.
ACKNOWLEDGMENTS
We thank Drs. Mark Behlke and John Rossi for critical reading of the manuscript and helpful comments. This project was supported by NIH grants R44AI056611 and R43AI074214 (B.H.J.).
Footnotes
Article published online ahead of print. Article and publication date are at http://www.rnajournal.org/cgi/doi/10.1261/rna.1894510.
REFERENCES
- Amarzguioui M, Lundberg P, Cantin E, Hagstrom J, Behlke MA, Rossi JJ. Rational design and in vitro and in vivo delivery of Dicer substrate siRNA. Nat Protoc. 2006;1:508–517. doi: 10.1038/nprot.2006.72. [DOI] [PubMed] [Google Scholar]
- Bernacchi S, Ennifar E, Toth K, Walter P, Langowski J, Dumas P. Mechanism of hairpin-duplex conversion for the HIV-1 dimerization initiation site. J Biol Chem. 2005;280:40112–40121. doi: 10.1074/jbc.M503230200. [DOI] [PubMed] [Google Scholar]
- Bernards R, Brummelkamp TR, Beijersbergen RL. shRNA libraries and their use in cancer genetics. Nat Methods. 2006;3:701–706. doi: 10.1038/nmeth921. [DOI] [PubMed] [Google Scholar]
- Boudreau RL, Monteys AM, Davidson BL. Minimizing variables among hairpin-based RNAi vectors reveals the potency of shRNAs. RNA. 2008;14:1834–1844. doi: 10.1261/rna.1062908. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cai X, Hagedorn CH, Cullen BR. Human microRNAs are processed from capped, polyadenylated transcripts that can also function as mRNAs. RNA. 2004;10:1957–1966. doi: 10.1261/rna.7135204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chang K, Elledge SJ, Hannon GJ. Lessons from nature: MicroRNA-based shRNA libraries. Nat Methods. 2006;3:707–714. doi: 10.1038/nmeth923. [DOI] [PubMed] [Google Scholar]
- Chu CY, Rana TM. Potent RNAi by short RNA triggers. RNA. 2008;14:1714–1719. doi: 10.1261/rna.1161908. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chung KH, Hart CC, Al-Bassam S, Avery A, Taylor J, Patel PD, Vojtek AB, Turner DL. Polycistronic RNA polymerase II expression vectors for RNA interference based on BIC/miR-155. Nucleic Acids Res. 2006;34:e53. doi: 10.1093/nar/gkl143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Doench JG, Petersen CP, Sharp PA. siRNAs can function as miRNAs. Genes & Dev. 2003;17:438–442. doi: 10.1101/gad.1064703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dorsett Y, Tuschl T. siRNAs: Applications in functional genomics and potential as therapeutics. Nat Rev Drug Discov. 2004;3:318–329. doi: 10.1038/nrd1345. [DOI] [PubMed] [Google Scholar]
- Du Q, Thonberg H, Wang J, Wahlestedt C, Liang Z. A systematic analysis of the silencing effects of an active siRNA at all single-nucleotide mismatched target sites. Nucleic Acids Res. 2005;33:1671–1677. doi: 10.1093/nar/gki312. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Elbashir SM, Harborth J, Lendeckel W, Yalcin A, Weber K, Tuschl T. Duplexes of 21-nucleotide RNAs mediate RNA interference in cultured mammalian cells. Nature. 2001;411:494–498. doi: 10.1038/35078107. [DOI] [PubMed] [Google Scholar]
- Fewell GD, Schmitt K. Vector-based RNAi approaches for stable, inducible, and genome-wide screens. Drug Discov Today. 2006;11:975–982. doi: 10.1016/j.drudis.2006.09.008. [DOI] [PubMed] [Google Scholar]
- Ge Q, Dallas A, Ilves H, Shorenstein J, Behlke MA, Johnston BH. Effects of chemical modification on the potency, serum stability, and immunostimulatory properties of short shRNAs. RNA. 2010 doi: 10.1261/rna.1901810. (this issue). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grimm D, Streetz KL, Jopling CL, Storm TA, Pandey K, Davis CR, Marion P, Salazar F, Kay MA. Fatality in mice due to oversaturation of cellular microRNA/short hairpin RNA pathways. Nature. 2006;441:537–541. doi: 10.1038/nature04791. [DOI] [PubMed] [Google Scholar]
- Harborth J, Elbashir SM, Vandenburgh K, Manninga H, Scaringe SA, Weber K, Tuschl T. Sequence, chemical, and structural variation of small interfering RNAs and short hairpin RNAs and the effect on mammalian gene silencing. Antisense Nucleic Acid Drug Dev. 2003;13:83–105. doi: 10.1089/108729003321629638. [DOI] [PubMed] [Google Scholar]
- Harper SQ, Staber PD, He X, Eliason SL, Martins IH, Mao Q, Yang L, Kotin RM, Paulson HL, Davidson BL. RNA interference improves motor and neuropathological abnormalities in a Huntington's disease mouse model. Proc Natl Acad Sci. 2005;102:5820–5825. doi: 10.1073/pnas.0501507102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ilves H, Kaspar RL, Wang Q, Seyhan AA, Vlassov AV, Contag CH, Leake D, Johnston BH. Inhibition of hepatitis C IRES-mediated gene expression by small hairpin RNAs in human hepatocytes and mice. Ann N Y Acad Sci. 2006;1082:52–55. doi: 10.1196/annals.1348.060. [DOI] [PubMed] [Google Scholar]
- Jackson AL, Bartz SR, Schelter J, Kobayashi SV, Burchard J, Mao M, Li B, Cavet G, Linsley PS. Expression profiling reveals off-target gene regulation by RNAi. Nat Biotechnol. 2003;21:635–637. doi: 10.1038/nbt831. [DOI] [PubMed] [Google Scholar]
- Judge A, MacLachlan I. Overcoming the innate immune response to small interfering RNA. Hum Gene Ther. 2008;19:111–124. doi: 10.1089/hum.2007.179. [DOI] [PubMed] [Google Scholar]
- Khvorova A, Reynolds A, Jayasena SD. Functional siRNAs and miRNAs exhibit strand bias. Cell. 2003;115:209–216. doi: 10.1016/s0092-8674(03)00801-8. [DOI] [PubMed] [Google Scholar]
- Li L, Lin X, Staver M, Shoemaker A, Semizarov D, Fesik SW, Shen Y. Evaluating hypoxia-inducible factor-1α as a cancer therapeutic target via inducible RNA interference in vivo. Cancer Res. 2005a;65:7249–7258. doi: 10.1158/0008-5472.CAN-04-4426. [DOI] [PubMed] [Google Scholar]
- Li MJ, Kim J, Li S, Zaia J, Yee JK, Anderson J, Akkina R, Rossi JJ. Long-term inhibition of HIV-1 infection in primary hematopoietic cells by lentiviral vector delivery of a triple combination of anti-HIV shRNA, anti-CCR5 ribozyme, and a nucleolar-localizing TAR decoy. Mol Ther. 2005b;12:900–909. doi: 10.1016/j.ymthe.2005.07.524. [DOI] [PubMed] [Google Scholar]
- Li L, Lin X, Khvorova A, Fesik SW, Shen Y. Defining the optimal parameters for hairpin-based knockdown constructs. RNA. 2007;13:1765–1774. doi: 10.1261/rna.599107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin X, Ruan X, Anderson MG, McDowell JA, Kroeger PE, Fesik SW, Shen Y. siRNA-mediated off-target gene silencing triggered by a 7 nt complementation. Nucleic Acids Res. 2005;33:4527–4535. doi: 10.1093/nar/gki762. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu HW, Cosa G, Landes CF, Zeng Y, Kovaleski BJ, Mullen DG, Barany G, Musier-Forsyth K, Barbara PF. Single-molecule FRET studies of important intermediates in the nucleocapsid-protein-chaperoned minus-strand transfer step in HIV-1 reverse transcription. Biophys J. 2005;89:3470–3479. doi: 10.1529/biophysj.105.065326. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marques JT, Devosse T, Wang D, Zamanian-Daryoush M, Serbinowski P, Hartmann R, Fujita T, Behlke MA, Williams BR. A structural basis for discriminating between self and nonself double-stranded RNAs in mammalian cells. Nat Biotechnol. 2006;24:559–565. doi: 10.1038/nbt1205. [DOI] [PubMed] [Google Scholar]
- McManus MT, Petersen CP, Haines BB, Chen J, Sharp PA. Gene silencing using micro-RNA designed hairpins. RNA. 2002;8:842–850. doi: 10.1017/s1355838202024032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nykanen A, Haley B, Zamore PD. ATP requirements and small interfering RNA structure in the RNA interference pathway. Cell. 2001;107:309–321. doi: 10.1016/s0092-8674(01)00547-5. [DOI] [PubMed] [Google Scholar]
- Rana TM. Illuminating the silence: Understanding the structure and function of small RNAs. Nat Rev Mol Cell Biol. 2007;8:23–36. doi: 10.1038/nrm2085. [DOI] [PubMed] [Google Scholar]
- Rose SD, Kim DH, Amarzguioui M, Heidel JD, Collingwood MA, Davis ME, Rossi JJ, Behlke MA. Functional polarity is introduced by Dicer processing of short substrate RNAs. Nucleic Acids Res. 2005;33:4140–4156. doi: 10.1093/nar/gki732. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scacheri PC, Rozenblatt-Rosen O, Caplen NJ, Wolfsberg TG, Umayam L, Lee JC, Hughes CM, Shanmugam KS, Bhattacharjee A, Meyerson M, et al. Short interfering RNAs can induce unexpected and divergent changes in the levels of untargeted proteins in mammalian cells. Proc Natl Acad Sci. 2004;101:1892–1897. doi: 10.1073/pnas.0308698100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schwarz DS, Hutvagner G, Du T, Xu Z, Aronin N, Zamore PD. Asymmetry in the assembly of the RNAi enzyme complex. Cell. 2003;115:199–208. doi: 10.1016/s0092-8674(03)00759-1. [DOI] [PubMed] [Google Scholar]
- Seyhan AA, Vlassov AV, Johnston BH. RNA interference from multimeric shRNAs generated by rolling circle transcription. Oligonucleotides. 2006;16:353–363. doi: 10.1089/oli.2006.16.353. [DOI] [PubMed] [Google Scholar]
- Seyhan AA, Alizadeh BN, Lundstrom K, Johnston BH. RNA interference-mediated inhibition of Semliki Forest virus replication in mammalian cells. Oligonucleotides. 2007;17:473–484. doi: 10.1089/oli.2007.0079. [DOI] [PubMed] [Google Scholar]
- Silva JM, Li MZ, Chang K, Ge W, Golding MC, Rickles RJ, Siolas D, Hu G, Paddison PJ, Schlabach MR, et al. Second-generation shRNA libraries covering the mouse and human genomes. Nat Genet. 2005;37:1281–1288. doi: 10.1038/ng1650. [DOI] [PubMed] [Google Scholar]
- Siolas D, Lerner C, Burchard J, Ge W, Linsley PS, Paddison PJ, Hannon GJ, Cleary MA. Synthetic shRNAs as potent RNAi triggers. Nat Biotechnol. 2005;23:227–231. doi: 10.1038/nbt1052. [DOI] [PubMed] [Google Scholar]
- Soutschek J, Akinc A, Bramlage B, Charisse K, Constien R, Donoghue M, Elbashir S, Geick A, Hadwiger P, Harborth J, et al. Therapeutic silencing of an endogenous gene by systemic administration of modified siRNAs. Nature. 2004;432:173–178. doi: 10.1038/nature03121. [DOI] [PubMed] [Google Scholar]
- Stegmeier F, Hu G, Rickles RJ, Hannon GJ, Elledge SJ. A lentiviral microRNA-based system for single-copy polymerase II-regulated RNA interference in mammalian cells. Proc Natl Acad Sci. 2005;102:13212–13217. doi: 10.1073/pnas.0506306102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun X, Li JM, Wartell RM. Conversion of stable RNA hairpin to a metastable dimer in frozen solution. RNA. 2007;13:2277–2286. doi: 10.1261/rna.433307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vlassov AV, Korba B, Farrar K, Mukerjee S, Seyhan AA, Ilves H, Kaspar RL, Leake D, Kazakov SA, Johnston BH. shRNAs targeting hepatitis C: Effects of sequence and structural features, and comparision with siRNA. Oligonucleotides. 2007;17:223–236. doi: 10.1089/oli.2006.0069. [DOI] [PubMed] [Google Scholar]
- Wang Q, Contag CH, Ilves H, Johnston BH, Kaspar RL. Small hairpin RNAs efficiently inhibit hepatitis C IRES-mediated gene expression in human tissue culture cells and a mouse model. Mol Ther. 2005;12:562–568. doi: 10.1016/j.ymthe.2005.04.014. [DOI] [PubMed] [Google Scholar]
- Wang Y, Juranek S, Li H, Sheng G, Tuschl T, Patel DJ. Structure of an argonaute silencing complex with a seed-containing guide DNA and target RNA duplex. Nature. 2008;456:921–926. doi: 10.1038/nature07666. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xia H, Mao Q, Paulson HL, Davidson BL. siRNA-mediated gene silencing in vitro and in vivo. Nat Biotechnol. 2002;20:1006–1010. doi: 10.1038/nbt739. [DOI] [PubMed] [Google Scholar]
- Xia H, Mao Q, Eliason SL, Harper SQ, Martins IH, Orr HT, Paulson HL, Yang L, Kotin RM, Davidson BL. RNAi suppresses polyglutamine-induced neurodegeneration in a model of spinocerebellar ataxia. Nat Med. 2004;10:816–820. doi: 10.1038/nm1076. [DOI] [PubMed] [Google Scholar]
- Zeng Y, Wagner EJ, Cullen BR. Both natural and designed micro-RNAs can inhibit the expression of cognate mRNAs when expressed in human cells. Mol Cell. 2002;9:1327–1333. doi: 10.1016/s1097-2765(02)00541-5. [DOI] [PubMed] [Google Scholar]