Abstract
Common sequence variants within a gene often generate important differences in expression of corresponding mRNAs. This high level of local (allelic) control—or cis modulation—rivals that produced by gene targeting, but expression is titrated finely over a range of levels. We are interested in exploiting this allelic variation to study gene function and downstream consequences of differences in expression dosage. We have used several bioinformatics and molecular approaches to estimate error rates in the discovery of cis modulation and to analyze some of the biological and technical confounds that contribute to the variation in gene expression profiling. Our analysis of SNPs and alternative transcripts, combined with eQTL maps and selective gene resequencing, revealed that between 17 and 25% of apparent cis modulation is caused by SNPs that overlap probes rather than by genuine quantitative differences in mRNA levels. This estimate climbs to 40–50% when qualitative differences between isoform variants are included. We have developed an analytical approach to filter differences in expression and improve the yield of genuine cis-modulated transcripts to ∼80%. This improvement is important because the resulting variation can be successfully used to study downstream consequences of altered expression on higher-order phenotypes. Using a systems genetics approach we show that two validated cis-modulated genes, Stk25 and Rasd2, are likely to control expression of downstream targets and affect disease susceptibility.
VARIATION in gene expression contributes to phenotypic diversity and has an impact on disease susceptibility. Early biochemical studies revealed heritable variation in levels of β-glucuronidase between strains of mice and their intercross progeny (Morrow et al. 1949; Law et al. 1952). The first linkage study of this type demonstrated that esterase activity in maize was modulated by a locus tightly linked to the esterase gene itself (Schwartz 1962). Three decades later, Damerval et al. (1994) made a breakthrough by applying proteomic methods to the same fundamental problem. They quantified expression differences of 72 proteins in an F2 intercross and simultaneously mapped 40 quantitative trait loci (QTL) that modulated the expression of different isoforms. The advent of high-throughput microarray technology made it practical to apply sophisticated QTL mapping methods to simultaneously map expression quantitative trait loci (eQTL) that potentially control the expression of thousands of transcripts in yeast, plant, and animal populations (Cavalieri et al. 2000; Karp et al. 2000; Brem et al. 2002; Schadt et al. 2003; Monks et al. 2004; Morley et al. 2004; Chesler et al. 2005; Hubner et al. 2005; Dixon et al. 2007; Goring et al. 2007; Stranger et al. 2007).
Genetic variation in expression is produced by mechanisms that act either in cis or in trans. Cis effects refer to local polymorphisms or alleles that influence the synthesis or stability of a gene's own message. When this type of variation is mapped, the regulatory locus coincides with the position of the source gene. Previous studies of gene expression have shown that a great majority of transcripts that have highly significant QTL are potentially cis-modulated (e.g., Chesler et al. 2005; Peirce et al. 2006). Surprisingly, in our own work we found that there was more than a twofold excess of cis-modulated transcripts in which the allele associated with high expression was inherited from strain C57BL/6J—the strain used for almost all genome sequencing, EST discovery, and array design. In contrast, we did not detect any significant allelic imbalance for QTL associated with trans effects. This imbalance highlighted a systematic bias in the detection of a problematic subclass of cis-modulated transcripts associated with sequence differences that affect probe–target hybridization.
In this study we have applied several systematic approaches to estimate error rates in the discovery of cis-modulated transcripts and to explore some of the biological and technical confounds that contribute to expression variation. While this study is by no means the first to point out the potential difficulty of sorting out sources of variation in array hybridization (Schadt et al. 2003; Alberts et al. 2005, 2007, 2008; Doss et al. 2005; Chen et al. 2009), we have extended previous work in three ways. First, we provide a comparatively quantitative assessment of the severity of the problem for two of the preeminent array platforms (Affymetrix and Illumina) by exploiting several large expression data sets and allele-specific expression protocols. Second, we have found that variation in 3′-UTR structure generates serious technical confounds even in the absence of sequence differences that overlap probe sequences. Third, we introduce a relatively simple protocol that can significantly increase the detection of true cis-expression differences. Our analysis is motivated by two factors. First, once cis-modulated transcripts have been validated, they become molecular resources to study downstream consequences of altered gene expression. Second, these variants are particularly amenable to detailed molecular analysis and their dissection will lead to a more complete understanding of the allelic modulation of gene expression.
MATERIALS AND METHODS
Microarray expression profiling:
Measurement of mRNA expression relied on multiple brain tissues collected from strains C57BL/6J (B6) and DBA/2J (D2), their reciprocal F1 hybrids (B6D2F1 and D2B6F1), and BXD recombinant inbred (RI) strains. We used data sets consisting of 39 BXD strains for the whole brain (Peirce et al. 2006), 67 for the hippocampus (Overall et al. 2009), and 54 for the striatum (Rosen et al. 2009). These studies exploited the most widely used mouse arrays: the Affymetrix Mouse Genome 430A–430B array pair to profile the whole brain, the Affymetrix Mouse Genome 430 2.0 array to profile the hippocampus, and the Illumina MouseWG-6 v1.1 array to profile the striatum. Affymetrix microarray data used in this study were normalized using the position-dependent nearest neighbor (PDNN) method (Zhang et al. 2004) but data normalized using robust multiarray average (RMA) and microarray analysis system 5 (MAS 5) procedures give similar results. We have also used an unpublished but open data set from the hippocampus of 72 BXD strains, the 2 parental strains, and F1 hybrids profiled using the Affymetrix Mouse Exon 1.0 ST array (see www.genenetwork.org/dbdoc/UMUTAffyExon_0209_RMA.html). Illumina microarray data were normalized using the vendor's rank invariant method. Additional information about the BXD panel, experimental procedures, and data sets used can be obtained from GeneNetwork information pages (www.genenetwork.org).
Genotyping and QTL mapping:
We used 3785 informative SNPs and microsatellite markers to map expression traits in the BXD strain panel. QTL mapping was performed using QTL Reaper as described (Peirce et al. 2006). The location of the probes was identified using the February 2006 assembly and the University of California, Santa Cruz (UCSC) genome browser (http://genome.ucsc.edu).
SNPs data set:
We used SNPs extracted from the SNPs database and variant browser of GeneNetwork (www.genenetwork.org/webqtl/snpBrowser.py) to analyze the impact of SNPs on expression level. This database incorporates the majority of previously known Celera and Perlegen SNPs available in NCBI Entrez dbSNP. The number of informative SNPs between B6 and D2 used in our screening was ∼1.8 million, although we have recently added an additional set of >1 million SNPs from an ongoing sequencing project.
Isoform mRNA detection:
Our analysis is based on several sources of data: mouse genome assembly and EST sequences were downloaded from UCSC (ftp://hgdownload.cse.ucsc.edu/goldenPath/mm8/), and mouse full-length cDNAs were obtained from NCBI (www.ncbi.nlm.nih.org) and RIKEN databases (http://genome.gsc.riken.go.jp). We used the following steps to identify mRNA isoforms: (1) we aligned the ESTs and cDNA sequences to the mouse genome sequence using BLAT (Kent 2002), (2) we extracted the genome sequences that matched each EST/cDNA sequence and up to 5 kb of the flanking sequence, (3) we aligned the EST/cDNA sequences with the highest BLAT scores to the genome sequence using SIM4 software (Florea et al. 1998) to infer exon/intron boundaries, and (4) we compared the predicted gene structures for each EST/cDNA and identified potential mRNA isoforms.
Allelic specific expression analysis:
We carried out allelic specific expression (ASE) analysis in reciprocal F1 individuals (C57BL/6J × DBA/2J) by combining RT–PCR with SNaPshot (Applied Biosystems, Foster City, CA). PCR primers were designed in the same exon flanking the informative SNPs using Primer 3 (supporting information, Table S1) (Rozen and Skaletsky 2000). We selected the most appropriate side of the SNPs to design SNaPshot extension primers. Hippocampal tissue was collected from F1 mice averaging 60 days old as in the previous microarray experiments (Chesler et al. 2005; Peirce et al. 2006). RNA was isolated from pools of two to four hippocampi using RNA-STAT 60 (Tel-Test) and quantified by a NanoDrop ND-1000 spectrophotometer. We set up four RNA pools, one per each sex and reciprocal cross. The number of individuals in each RNA pool varied from four to eight. We isolated individual DNA from the spleen of four different heterozygote individuals using a standard phenol-chloroform procedure. These DNA samples were used as controls and tested at the same time with the RNA pools. We treated the RNA with Turbo DNase (Ambion) to degrade any traces of genomic DNA. The absence of genomic DNA contamination was confirmed by PCR across small introns.
First-strand cDNA synthesis (GE Healthcare) of the RNA pools was followed by PCR of the cDNA and genomic DNA samples (GoTaq Flexi DNA polymerase; Promega, Madison, WI). The PCR products were purified using ExoSap-IT (United States Biochemical, Cleveland) and used in the SNaPshot reaction. SNaPshot extension products were purified by calf intestinal phosphatase (New England Biolabs, Beverly, MA) and separated by capillary electrophoresis using ABI3130 (Applied Biosystems). We analyzed the quantification results using GeneMapper v4.0 software (Applied Biosystems). We used the peak heights corresponding to each allele as a measure of allelic abundance. We tested for the presence of ASE difference using a t-test to compare the logarithm of the allelic fold change between cDNA and the genomic DNA. We estimated fold difference in expression as the ratio of allelic fold difference in cDNA to the genomic DNA.
qRT-PCR analysis:
Dorsal striatum was dissected from four C57BL/6J and DBA/2J mice and total RNA was extracted using RNA-STAT 60. We treated the RNA pools with Turbo DNase. cDNA synthesis was performed on equal amounts of total RNA, using the First-Strand cDNA Synthesis kit and the NotI-d(T)18 primer (GE Healthcare). We selected the qRT-PCR assays using the Universal Probe Library (www.universalprobelibrary.com, Roche Diagnostics) (Table S2). Two assays were designed for each candidate transcript: one that targets the putative alternative region and one that targets an exon common in both transcripts. Most of the assays designed in the nonpolymorphic exons span an intron while assays targeting alternative regions were designed close to the location of the microarray probe that captured a cis effect. We used cyclophilin D as a reference gene for comparative threshold quantification. This gene is not differentially expressed between strains. qRT-PCR was performed using the LightCycler 480 System (Roche Diagnostics) according to the standard protocol for the LC480 Probes Master. Standard curves for five 10-fold dilutions between 2500 and 0.25 ng of reverse-transcribed RNA samples were run for all assays to determine PCR efficiency under experimental conditions for reference and selected genes. The mean crossing thresholds (CT) for the technical duplicates of different amplicons were used to calculate mean normalized expression (MNE) values that reflect the relative expression of the target gene compared to the reference gene by taking the efficiencies of the PCR into account (Simon 2003). MNE values for C57BL/6J and DBA/2J were log10 transformed and compared by t-tests. Mean fold difference values were calculated using the expression of C57BL/6J as a reference.
RESULTS AND DISCUSSION
SNPs overlapping probes affect the rate of discovery of cis QTL:
Recent linkage studies of gene expression have used commercial short oligomer probes to measure steady-state gene expression levels in a wide variety of tissues and species. The Affymetrix platform is based on sets of 25-mer probes (typically 11 probes per probe set), whereas the Illumina platforms exploit longer 50-mer probes. We used both platforms and systematically mapped cis- and trans-acting loci in multiple tissues of the BXD strains. A cis-acting locus is defined here as a QTL with a likelihood-ratio statistic (LRS) >15 (LOD > 3.26) and located within 3 Mb of the source gene. We detected a total of 3431 Affymetrix probe sets corresponding to 2640 potentially cis-modulated genes and 2944 Illumina probes corresponding to 2587 potentially cis-modulated genes in two large CNS expression data sets.
The size and direction of additive effects for these cis-acting QTL were imbalanced in both tissues and using both array platforms. Approximately 65% of cis QTL were associated with higher expression of the C57BL/6J allele (B) relative to the DBA/2J allele (D). This is close to a 2:1 imbalance in favor of B alleles. If QTL are ranked on the basis of a LRS, the imbalance is even more pronounced among the top 10% of cis QTL: a 3:1 ratio among Affymetrix probe sets and a 6.7:1 ratio among Illumina probes. In contrast, the imbalance is modest among the bottom 10% of cis QTL: 1.3:1 for both Affymetrix and Illumina platforms. In marked contrast, trans-modulated transcripts, which by definition are controlled by distant sequence variants, are almost perfectly balanced (1:1) on both platforms with respect to effect size and polarity.
It would be important to know how many of these cis-acting candidate QTL are genuine and how many are caused by qualitative differences between isoforms. As has been established for several years, SNP position and the number of SNPs overlapping the microarray probes have a significant impact on the discovery rate and the size of apparent expression differences for cis- (Schadt et al. 2003; Alberts et al., 2005, 2007; Doss et al. 2005; Chen et al. 2009) and trans-modulated transcripts (Chen et al. 2009). Our analysis found that the longer Illumina 50-mers have approximately the same sensitivity to sequence differences as the shorter Affymetrix 25-mer probes (Figure S1 and Figure S2). In both cases, SNPs that overlap at the 5′ and 3′ ends of the probes perturb measurements only slightly, whereas those that overlap more central positions have a strong effect (Affymetrix P < 0.00002; Illumina P < 0.02). As expected, the average size of cis effects increases as a function of the number of SNPs (P < 0.0001). In contrast, the effect size of trans QTL is not influenced at all by SNPs that overlap probes.
We removed from our analysis all of the probes that overlap known SNPs but the bias in favor of B alleles was still present among cis-acting QTL. For example, removing all Illumina probes that overlap known SNPs reduced the bias from 65 to 57%. This represents a drop in the number of cis QTL by 17%. The residual imbalance could be readily explained by unknown polymorphisms that overlap probes or by isoform variation.
Alternative splicing, initiation, and termination of transcription—qualitative changes affecting gene expression measurements:
Differences in alternative splicing, initiation, and termination of transcription have an important contribution to variation in gene expression. Recently Kwan et al. (2008) employed exon arrays and showed that 55% of expression differences in a HapMap population are due to isoform variations caused by alternative splicing, initiation, and termination of transcription. Expression analysis by multiple probes that profile the same transcript provides a comprehensive picture of the complexity of variation. Expression of 37% of genes profiled in striatum by the Illumina array (9200 of ∼25,000 genes) is measured by two or more probes. Expression of 1100 genes is characterized by a moderately strong cis effect (LRS > 15) and measured by multiple probes that are not affected by known SNP variants. This enabled us to assess the consistency with which a cis effect was detected by multiple probes. Twenty-two percent of these 1100 genes with companion probes had discordant cis QTL effects—strong cis QTL detected by one or more probes and insignificant cis effects by other probes. This discordance may be either due to unknown SNPs overlapping probes or due to isoform variation. One example of the latter effect is an apparent difference in expression of ADP-ribosylation factor related protein 1 (Arfrp1) that is detected by a probe that targets an alternatively spliced exon but not by a probe that targets an exon common in all isoforms (Figure 1).
Figure 1.—
Variation in alternative splicing and the expression difference of Arfrp1 in striatum (UTHSC Illumina 6.1: November07 data set). Variation in expression was detected only by a probe that targets the alternatively spliced exon (ILM6290594) but not by a probe that targets the 3′-UTR (ILM1450102). This discrepancy is probably a result of variation in the abundance of an alternatively spliced isoform among BXD strains.
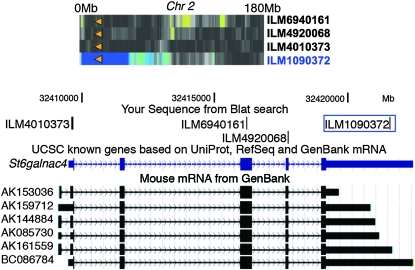
The question at this point is what fraction of observed cis effects is caused by unknown SNPs affecting hybridization or due to isoform variants? To address this we resequenced a random set of 16 genes with discordant probe pairs. We identified new SNPs overlapping probes in only 5 of the genes (31%). In the rest of the genes from this group we identified high levels of cis variation specific to 3′-UTRs. Two independent quantitative real-time RT–PCR (qRT-PCR ) assays confirmed strain-specific differences specific to 3′-UTRs for 5 of the 11 tested genes (Table S3). For the remaining 6 genes the differences were either not specific to the 3′-UTR or undetectable by qRT-PCR. For example, analysis of array data uncovered a cis effect specific to a probe that targets the distal end of the longest 3′-UTR variant of St6galnac4 (Figure 2). In contrast, there is no cis effect for probes that target other exons. There are at least three putative poly(A) sites in St6galnac4 that could generate variation in 3′-UTR length (http://polya.umdnj.edu/polyadb) (Lee et al. 2007).
Figure 2.—
The effect of alternative 3′-UTR processing on the expression of St6galnac4 in striatum (UTHSC Illumina 6.1: November07). A difference in expression was detected only by a probe that targets the distal long 3′-UTR transcript (ILM1090372, LRS = 73.3, 1.2-fold difference) but not by probes that target the 5′-UTR (ILM4010373), exon 3 (ILM6940161), or exon 4 (ILM4920068). This discrepancy is the result of isoform variation among BXD strains due to multiple poly(A) sites. The heat maps are represented in “gray plus blue plus red” in which more intense colors mark chromosomal regions with comparatively high linkage statistics and the spectrum encodes the allelic effect. The blue–green regions are those in which the B allele is associated with higher trait values, whereas the red–yellow regions are those in which the D allele is associated with higher trait values. Gray and black regions have insignificant linkage between traits and DNA markers. The physical location of the gene is represented by an orange triangle.
Exon microarrays provide a more high-throughput alternative to resolve the confound between quantitative and qualitative differences in mRNA expression. We profiled the hippocampus of the BXD panel using the Affymetrix Mouse Exon 1.0 ST array to map loci responsible for quantitative and qualitative variation in expression. In genes that are profiled by at least six probe sets that target multiple exons, we found consistent evidence (>50%) of cis and trans modulation in 282 and 271 genes, respectively (UMUTAAffy Hippocampus Exon Feb09 RMA). In contrast, we detected significant differences in the putative regulation of gene expression that was measured across different exons in many of the genes. For example, a cis effect for solute carrier family 35, member E1 (Slc35e1) was uncovered only by probe sets that target the distal region of the 3′-UTR and not by the rest of the probes. These data provide additional evidence of increased transcriptome diversity in BXD.
As considered above, variation in transcription termination and alternative splicing can give rise to discrete quantitative differences in the steady-state mRNA level of different isoforms between strains. When these mRNA isoform variants (e.g., long or short 3′-UTRs) map as local QTL, we can be sure that the effect is due to a local DNA sequence variant that will not necessarily overlap probes but may nonetheless produce quantitative variation among isoforms. These combined qualitative and quantitative differences are interesting to study and can affect phenotypes in multiple ways—by altering message stability, translation efficiency, protein sequence, and function. For example, a 3′-UTR long-form variant in the μ-opioid receptor in CXB RI strains causes a low response to morphine agonists and higher pain sensitivity (Ikeda et al. 2001; Han et al. 2006).
Systematic identification of genuine cis-modulated transcripts:
We developed an efficient analytical approach that combines bioinformatics and molecular methods to filter out artifactual expression differences. In general, a robust expression-profiling platform will uncover a large number of cis-regulated candidates in a segregating population. Initially, a large fraction of apparent cis QTL are excluded on the basis of significance of the linkage, position, and fold difference in expression. All these steps are common to most expression genetics analyses. In one of the CNS data sets (Hippocampus Consortium M430v2 June06), we identified 3810 cis-acting QTL candidates (LRS > 20) closely mapped to the source gene (±3 Mb) (Table 1, step A). The size and direction of additive effects were imbalanced with ∼66% of cis QTL associated with higher expression of the B alleles relative to the D alleles. A more relaxed stringency of effect size (LRS > 15) or distance between QTL and the source gene (±5 Mb) did not correct the imbalance in favor of B alleles (61%:39% to 67%:33%). If we apply the highest stringencies (LRS > 20, ±3 Mb from source), 750 of 3810 cis QTL have robust differences in gene expression (fold difference >1.68). These 750 candidate cis QTL modulate the expression of 630 genes. The imbalance in this reduced set of candidates favored the B over the D alleles (60%:40%) (Table 1, step B).
TABLE 1.
Filtering process of the cis QTL candidates in the hippocampus of BXD
| Steps of the filtering process | No. of probe sets (genes) | Additive effect (B vs. D ratio, %) |
|---|---|---|
| Affymetrix M430 v2.0 array data set | 45,101 (23,080) | 47:53 |
| A. cis QTL (LRS > 20, QTL mapped ± 3 Mb from source) | 3,810 (2,856) | 66:34 |
| cis QTL (LRS > 20, QTL mapped ± 5 Mb from source) | 4,117 (3,064) | 67:33 |
| cis QTL (LRS > 15, QTL mapped ± 3 Mb from source) | 4,416 (3,279) | 61:39 |
| cis QTL (LRS > 15, QTL mapped ± 5 Mb from source) | 4,844 (3,548) | 65:35 |
| B. cis QTL (LRS > 20, QTL mapped ± 3 Mb from source) fold difference between allelic expression >1.7 | 750 (630) | 60:40 |
| C. Probes associated with potentially true cis QTL—filtering out the remaining 440 cis QTL was based on: | 190 (166) | 49:51 |
| Potential SNP overlapping probes (70%) | 308 (302) | 74:23 |
| Alternative 3′-UTR processing (14%) | 62 (62) | 76:24 |
| Alternative splicing (10%) | 44 (44) | 72:28 |
| Other (6%) |
26 (25) |
64:36 |
We analyzed each of these transcripts at the level of probes and probe sets to evaluate likely sources of expression differences (Table 1, step C). We were interested in identifying expression variation that is solely quantitative and not a result of sequence differences overlapping probes or due to alternative splicing, initiation, and termination of transcription. First, we identified probes overlapping known SNPs by comparing flanking positions of probes with positions of all informative SNPs. We included this information and flagged problematic probes, using the GeneNetwork “probe tool” page created for each probe set. This procedure is frequently used by most expression genetics studies (Stranger et al. 2007). Other studies simply assess the impact of SNPs on the rate of discovery of cis-modulated transcripts by sequencing the complementary area of probes associated with the highest effects (Druka et al. 2008).
We are aware that a large number of sequence variants have not been yet identified (especially indels) and some of these can overlap probes. We used QTL Reaper and a more refined linkage analysis of individual probe data—the single 25-mers as opposed to the full probe set—and examined the consistency of linkage across individual probes and those generated by the Affymetrix full probe set. Alberts et al. (2007) developed a different approach of identifying problematic probes by decomposing the signal provided by the probe set and analyzing the deviation of the individual probes from the entire set. A computational protocol that flags probes that may overlap SNPs is freely available in R (Alberts et al. 2008). The end result of both approaches is similar and neither of the methods requires a priori information on SNP location. Our approach and resequencing data revealed many examples of artifactual expression variation due to overlaps between probes and sequence variants.
We assumed that a large fraction of expression differences are due to isoform variation as a result of events such as alternative splicing, initiation, or termination of the transcription. We used EST/cDNA sequences from various sources, identified their position in the mouse genome, and compared the predicted gene structures at each locus for detection of possible isoform variants. We determined that a considerable fraction of the probe sets target areas that are polymorphic between isoforms, which vary between strains, especially at the distal end of the 3′-UTR. In CNS tissues this can become an important confound, because genes tend to express longer transcripts and 3′-UTRs (Zhang et al. 2005) and exhibit higher levels of alternative splicing than other tissues (Yeo et al. 2004).
Following all these bioinformatics analyses we predicted that the expression differences of 190 of the 750 candidates (25%) are solely quantitative and a result of cis modulation. Probes that target these transcripts will map cis irrespective of their location relative to different regions of the transcript. In this group of 190 candidates we were able to regain close to a 1:1 balance between B and D alleles (B vs. D, 49.2–50.8%, Table 1). In contrast, we predicted that the remaining 440 potentially cis-modulated transcripts are mainly a result of SNPs or other sequence variation (known and unknown) that overlap probes (70%), 3′-UTR processing, (14%) and alternative splicing (10%). The imbalance in the direction of the additive effect is significantly higher in favor of B alleles in all of these categories that affect expression measurements. We excluded all cis QTL candidates on the basis of probes that overlap known SNPs or probes with inconsistent linkage across the set due to potential unknown sequence variants. We filtered out the probes that detect combined qualitative and quantitative differences between isoforms in a distinct group and used them separately in validation studies and downstream analyses.
A large number of genuine cis-regulated transcripts were probably excluded by our filtration process due to overly stringent criteria. For example, the number of candidate cis-modulated transcripts could be increased to 5000 by simply selecting all transcripts with a local cis LRS > 10 or by extending the allowed distance between the source gene and the QTL. While this procedure will boost the final number of candidates, it will also increase the false discovery rate (FDR). The FDR at other stages of our filtration process could be determined empirically by measuring allelic expression or by similar procedures using sets of randomly selected transcripts from each step.
Validation of cis variation by ASE:
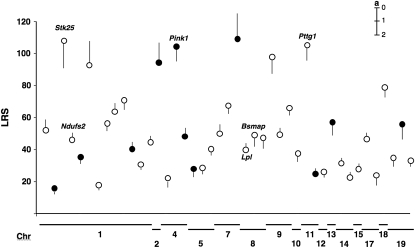
The ASE approach provides a relatively unambiguous measure of expression variation generated by cis-acting factors (Doss et al. 2005). From the short list of 190 putative cis-acting QTL described above we selected 43 candidates for ASE analysis (Figure 3 and Table S4). These candidates vary in their effect and significance and cover most chromosomes (Figure 3). They also provide sufficient statistical power to predict the true distribution of the 190 cis candidates. ASE of at least 40 candidate QTL is required to predict with sufficient accuracy the true distribution of the entire set of 190 cis QTL (95% confidence interval ≥|0.15|).
Figure 3.—
Chromosomal distribution and the significance of self-linkages for 43 transcripts selected for ASE quantification. The size and the direction of the additive genetic effect (a) were estimated using “INIA Brain mRNA M430: January06” data and are represented by lines next to the LRS circles. The additive effect is measured on a log2 scale in which one unit corresponds approximately to a twofold difference. A line above the LRS circles indicates that D alleles increase trait values. In contrast, a line below the circles indicates that B alleles increase the trait. The fold difference in expression between parental alleles ranges from 0.29 to 3.23. The significance of the linkage varies from 16 to 110. The selected genes are located in all but three of the somatic chromosomes (Table S4). Solid circles represent genes that failed to show by ASE a significant difference in expression in F1 individuals.
We used reciprocal C57BL/6J × DBA/2J F1 hybrids and SNPs in which the expression of both alleles can be quantified simultaneously in single samples. If the variation in expression of a particular transcript is a result of genuine cis modulation, the expression of B and D alleles will be significantly different in F1 hybrids. If the variation in expression across BXD strains is a result of trans modulation or is simply an artifact, no difference in allelic expression will be observed in hybrids. While the trans effects remain in F1 individuals, the difference in allelic expression will not be detected since both allelic trans factors (B and D) are part of the same cell and have equal access to their downstream targets. We intentionally selected SNPs that are shared by most or all mRNA isoforms, to capture quantitative differences in expression rather than structural differences among isoforms.
Fold difference in expression and the significance of cis modulation vary considerably among transcripts. We confirmed that 33 (77%) of 43 genes are genuinely cis modulated at P < 0.05 and that 81% are cis modulated at P < 0.1. If we apply the properties of the binomial distribution, the 95% confidence interval of the validation rate in the entire set of 190 genes will be between 66 and 88%. We performed a statistical evaluation of the LRS score effect on true positive prediction of cis QTL. While at first glance a LRS threshold of at least 60 seems to improve the number of genuine cis QTL (Figure 3), we did not identify any significant statistical relationship between LRS and the validation results. Directions of allelic effects agreed in all cases with our array data. Furthermore, the percentage of validation was independent of the polarity of the effect (77% for B and 76% for D alleles).
We estimated the fraction of genuine cis-modulated transcripts by applying our ASE validation rate with the assumption that the bias in favor of B-allele effects is generated entirely by sequence variants that affect probe signal. In the original set of 750 cis-acting QTL the imbalance favored B over D QTL by a ratio of ∼3/2. This imbalance indicates that one-third of B-positive cis effects are false, assuming that all D-positive cis effects are true. After we apply the ASE validation rate, the estimated fraction of true cis-modulated transcripts is ∼51% for B-positive and ∼77% for D-positive alleles.
Sequence variation and the universal biases in genetic expression studies:
It is important to note that even in the absence of an explicit imbalance in allelic effects seen in the BXD panel, the same problems and challenges apply with equal force in other population resources. For example, in the LXS panel there is a perfect balance between cis QTL with high levels of L vs. high levels of S alleles since both L and S haplotypes share roughly the same fraction of the B haplotype used to design microarrays. However, in this cross, there is a similar fraction of artifactual expression variation, but the source is shared equally by both parental strains. After taking into account the sample size, parental strain genetic differences, and the choice of microarray platform, we estimate that in the hippocampus data set of 3810 candidate cis QTL there are at least 1000 genuine QTL responsible for a fold difference in expression >50% (Hippocampus Consortium M430v2 June06).
Downstream analysis of cis-modulated variation:
The possibility of quantifying gene expression in segregating populations provides an opportunity to infer relationships between genes and their transcripts. Another objective of this study is to show the potential of system genetics to identify downstream targets of cis modulation and the impact on higher-order phenotypes.
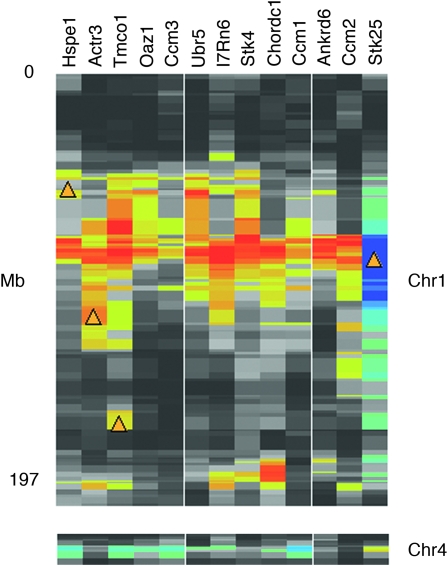
Some of the cis-modulated transcripts validated by ASE have roles in CNS development and neurotransmission and are potential candidates for behavioral traits and neurological diseases. It would be interesting to know if any of these expression differences are large enough to lead to quantitative differences at the protein level that could ultimately trigger variation in behavior and disease susceptibility. For example, serine threonine kinase 25 (Stk25) is known to be involved in a pathway that leads to cerebral cavernous malformation (CCM) in humans (Voss et al. 2007). Human STK25 interacts directly with the products of two genes associated with CCM: CCM2 and CCM3 (Voss et al. 2007). In agreement with these results we detected moderate to high correlations between expression of Stk25 and expression of three mouse orthologs of the CCM genes (Table 2). We uncovered a locus responsible for the variation in the expression of all three transcripts—Krit1 (Ccm1), Ccm2, and Pdcd10 (Ccm3)—that maps to chromosome (Chr) 1 between 90 and 100 Mb, the precise location of Stk25, strongly implicating Stk25 as the source of expression difference (Figure 4). We then used a global method to generate a list of other downstream targets of Stk25 (Table S5). All of the targets presented in Table 2 have a correlation with Stk25 of at least 0.45 in one of the CNS tissues and at least 0.25 in three or more tissues. These potential targets have moderate to strong QTL that overlap Stk25. Finally, all targets have strong correlations with Stk25 purely on the basis of an analysis of their shared literature (Homayouni et al. 2005). For example, Stk25 is related by expression, by QTL, and by literature with Stk4.
TABLE 2.
Expression and literature correlations between transcripts with trans QTL located on Chr 1 (90 and 100 Mb) and Stk25
| Symbol | Probe set ID | Gene ID | Correlation with Stk25, probe 1416770_at (P-value) | Literature correlation with Stk25 | QTL Chr 1 LRS |
|---|---|---|---|---|---|
| Stk25 | 1416770_at | 59041 | 1.00 (0) | 1.00 | 96.2 |
| Actr3 | 1452051_at | 74117 | −0.54 (0.00014) | 0.42 | 11.9 |
| Ankrd6 | 1437217_at | 140577 | −0.57 (3.64e-05) | 0.44 | 15.1 |
| Ccm1 | 1448701_a_at | 79264 | −0.48 (1.01e-0.3)S | 0.55 | 9.2 |
| Ccm2 | 1456290_x_at | 216527 | −0.54 (0.00013) | 0.63 | 8.7 |
| Ccm3 | 1448527_at | 56426 | −0.37 (1.39e-02) | N.A. | 8.7 |
| Chordc1 | 1460645_at | 66917 | −0.49 (0.00075) | 0.58 | 14.7 |
| Hspe1 | 1450668_s_at | 15528 | −0.51 (0.00043) | 0.52 | 13.7 |
| l7Rn6 | 1419351_a_at | 67669 | −0.72 (2.67e-09) | 0.51 | 22.5 |
| Oaz1 | 1436292_a_at | 18245 | −0.47 (0.00139) | 0.49 | 13.0 |
| Stk4 | 1436015_s_at | 58231 | −0.47 (0.00034) | 0.78 | 11.7 |
| Tmco1 | 1423759_a_at | 68944 | −0.52 (0.00031) | 0.53 | 14.4 |
|
Ubr5 |
1452718_at |
70790 |
−0.47 (0.00113) |
0.57 |
12.6 |
Figure 4.—
Downstream potential targets of serine threonine kinase 25 (Stk25). Stk25 is located on Chr 1 at 95.4 Mb and is cis-modulated in brain (LRS = 96.1, INIA Brain mRNA M430: January06, probe set 141770_at). Several potential targets of Stk25 have relatively strong trans QTL that overlap Stk25 (heat map conventions are as in Figure 2). All these targets have a correlation with Stk25 of at least 0.45 in whole brain and a shared literature correlation of at least 0.42 (Table 2). Most of these transcripts share a trans QTL located on Chr 4.
Over the years the BXD panel has been extensively phenotyped for many pharmacological, anatomical, and behavioral traits, and these data are available in GeneNetwork. We explored if variation in expression of Stk25 correlates with any of these traits. Since Stk25 is linked to brain vascularization, it is noteworthy that the top correlates of Stk25 are all neural and behavioral traits. The expression of Stk25 is highly correlated with the initial sensitivity to ethanol-induced ataxia (r = 0.70, P < 0.001; GeneNetwork trait ID, 10144) and cerebellum volume (r = −0.48, P < 0.05; trait ID, 10004) and moderately correlated with many traits associated with motor activity in an open field such as vertical activity (r = 0.48, P < 0.0001; trait ID, 11863) or distance traveled in a novel environment (r = 0.36, P < 0.01; trait ID, 11602).
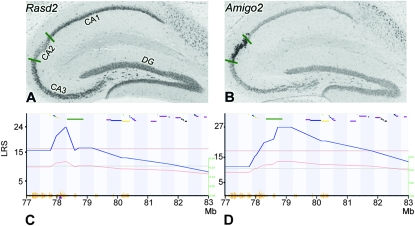
RASD family, member 2 (Rasd2) is another cis-modulated gene that exhibits a significant difference in expression in the BXD panel. Rasd2 is a member of the Ras GTPase family of proteins that controls pathways involved in synaptic plasticity, learning, and memory (Spano et al. 2004). Rasd2 is predominantly expressed in striatum but also in other areas of the brain such as hippocampus. We detected a moderate correlation between the expression of Rasd2 and the adhesion molecule with the Ig-like domain 2 gene (Amigo2, r = 0.41) in hippocampus. Amigo2 is located on Chr 15 at 97.1 Mb and is trans-modulated by a locus on Chr 8 at 78.7 Mb (LRS = 27), very close to Rasd2 (Chr 8, 78.1 Mb). This trans QTL is responsible for ∼15% of the variation in expression of Amigo2 in hippocampus. The confidence limits of the QTL extend from 78 to 79.2 Mb and include only three genes: Rasd2, Mcm5, and 1700007B14Rik. We examined expression patterns of these genes and Amigo2 using in situ data available from Allen Brain Atlas (www.brain-map.org). Mcm5 and 1700007B14Rik have low expression in the brain and are less interesting candidates for the Amigo2 trans QTL. In contrast, variants in Rasd2 could clearly influence Amigo2 expression. Rasd2 exhibits regional expression in the hippocampus, high in both CA1 and CA3, but little to none in CA2. Amigo2 shows complementary regional differences with high expression in CA2, but almost none in CA1 or CA3, supporting a repressive influence of Rasd2 on Amigo2 expression (Figure 5).
Figure 5.—
Expression complementarity between Rasd2 and Amigo2. Expression pattern of Rasd2 and Amigo2 was analyzed using in situ data available from Allen Brain Atlas (www.brain-map.org). (A) The expression of Rasd2 in hippocampus is intense in CA1 and CA3 and weak or absent in CA2 or dentate gyrus (DG). (B) In contrast, the expression of Amigo2 in hippocampus is intense only in the CA2 region. (C) Rasd2 (Chr 8, 78.1 Mb) is cis-modulated in hippocampus of the BXD panel (LRS = 23.7, Hippocampus Consortium M430v2: June06, probe set 1427343_at). (D) Amigo2 is located on Chr 15 at 97.1 Mb and is modulated by a trans QTL on Chr 8 at 78.7 Mb (LRS = 27, probe set 1434601_at) overlapping Rasd2. The left scale represents the LRS. The right scale represents the additive effect: a red line indicates that B6 alleles increase the trait.
To test the hypothesis that Rasd2 expression consistently represses expression of Amigo2 we analyzed 10 other CNS regions in which Rasd2 is expressed. A complementary pattern of high expression of Rasd2 and low to absent expression of Amigo2 was found in 8 of the 10 areas: the pyramidal cell layer of fields CA1 and CA3 of the hippocampus, the dorsal striatum, the mitral layer and periglomerular cells of the main olfactory bulb, the olfactory tubercle, the substantia nigra pars compacta, the entorhinal cortex, and the reticular nucleus of the thalamus. We did not detect the same pattern for the hilus of the hippocampus or for the paraventricular nucleus of the thalamus. The presence of the Amigo2 trans QTL on Chr 8 and the relatively consistent complementary expression pattern and correlation provide cumulatively strong support for the hypothesis that Rasd2 is an inhibitor of Amigo2.
Rasd2 is known to be regulated by thyroid hormones (Errico et al. 2008) and interestingly we found the expression of its potential downstream target—Amigo2—to be highly correlated with total serum thyroxine (r = 0.87, P < 0.005; trait ID, 10602). The expression of Amigo2 was shown to promote the survival of cerebellar granule neurons and was considered a candidate for familial Alzheimer's disease type 5 (Ono et al. 2003). We found the expression of Amigo2 to be highly correlated with the granule cell number in hippocampus (r = −0.76, P < 0.001; trait ID, 10337) and with the volume of the dorsal (r = −0.48, P < 0.01; ID, 10755), and ventral hippocampus (r = −0.47, P < 0.01; trait ID, 10757). Rasd2 was found recently to affect another neurogenerative disorder—Huntington's disease—by interacting with the mutant allele of huntingtin (Subramaniam et al. 2009).
Next-generation eQTL studies:
While the methods we have described here provide an effective way to correct for problems associated with array-based measurement of expression, there are more satisfying solutions that are now on the horizon. Massively parallel sequencing of RNA samples (e.g., Mortazavi et al. 2008) represents a more direct way to measure steady-state abundance of mRNAs. RNA sequencing (RNA-seq) has three potential benefits in eQTL studies. First, this method is comparatively insensitive to sequence differences among individuals in segregating populations. SNPs and short indels should not have a significant impact on the alignment and identification of 50-nt mRNA tags. Second, this method can resolve the relative expression of mRNA isoforms and will enable the genetic dissection of splice variants and alternative UTRs. Finally, this method can be multiplexed more easily than array-based methods. It will be practical to sequence pools of RNA from many samples in single runs. This should reduce technical confounds in large experimental designs that are so common in this field. We are now beginning to exploit this approach by profiling expression in the parental strains of the BXD RIs—a step that should allow us to recalibrate first-generation eQTL studies.
In this report we have developed and tested a protocol to systematically extract, study, and validate DNA sequence variants that are responsible for differences in mRNA abundance in a RI population. We have done this by combining gene expression profiling with a collection of bioinformatics routines and molecular techniques, including qRT-PCR, ASE, and resequencing. Our study demonstrates that both quantitative and qualitative variation contributes to the mRNA diversity in segregating populations. In particular, we have focused our work on cis-acting QTL. Validated cis variations are in essence more subtle and natural forms of standard genetically engineered mutants. Rather than completely inactivating gene expression of a gene on a single strain background, these alleles modulate expression over a 2- to 20-fold range across a panel of readily available strains. As we have exemplified above, once validated, these allelic expression differences can be used in reverse genetic studies to generate well-defined hypotheses regarding downstream effects on molecular, cellular, and functional networks.
Acknowledgments
We thank M. Nielsen, T. Cunningham, F. Jiao, S. Li, H. Li, and Z. Sun for technical assistance. This work was supported by the National Institute on Alcohol Abuse and Alcoholism (NIAAA) (U01AA013499, U01AA014425), by the National Institute of Drug Abuse (NIDA) and NIAAA (P20-DA 21131), by the National Cancer Institute (U01CA105417), and by the National Center for Research Resources (U01NR 105417).
Supporting information is available online at http://www.genetics.org/cgi/content/full/genetics.109.107474/DC1.
References
- Alberts, R., P. Terpstra, L. V. Bystrykh, G. de Haan and R. C. Jansen, 2005. A statistical multiprobe model for analyzing cis and trans genes in genetical genomics experiments with short-oligonucleotide arrays. Genetics 171 1437–1439. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alberts, R., P. Terpstra, Y. Li, R. Breitling, J. P. Nap et al., 2007. Sequence polymorphisms cause many false cis eQTLs. PLoS ONE 2 e622. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alberts, R., G. Vera and R.C. Jansen, 2008. affyGG: computational protocols for genetical genomics with Affymetrix arrays. Bioinformatics 24 433–434. [DOI] [PubMed] [Google Scholar]
- Brem, R. B., G. Yvert, R. Clinton and L. Kruglyak, 2002. Genetic dissection of transcriptional regulation in budding yeast. Science 296 752–755. [DOI] [PubMed] [Google Scholar]
- Cavalieri, D., J. P. Townsend and D. L. Hartl, 2000. Manifold anomalies in gene expression in a vineyard isolate of Saccharomyces cerevisiae revealed by DNA microarray analysis. Proc. Natl. Acad. Sci. USA 97 12369–12374. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen, L., G. P. Page, T. Mehta, R. Feng and X. Cui, 2009. Single nucleotide polymorphisms affect both cis- and trans- eQTL. Genomics 93 501–508. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chesler, E. J., L. Lu, S. Shou, Y. Qu, J. Gu et al., 2005. Complex trait analysis of gene expression uncovers polygenic and pleiotropic networks that modulate nervous system function. Nat. Genet. 37 233–242. [DOI] [PubMed] [Google Scholar]
- Damerval, C., A. Maurice, J. M. Josse and D. de Vienne, 1994. Quantitative trait loci underlying gene product variation: a novel perspective for analyzing regulation of genome expression. Genetics 137 289–301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dixon, A. L., L. Liang, M. F. Moffatt, W. Chen, S. Heath et al., 2007. A genome-wide association study of global gene expression. Nat. Genet. 39 1202–1207. [DOI] [PubMed] [Google Scholar]
- Doss, S., E. E. Schadt, T. A. Drake and A. J. Lusis, 2005. Cis-acting expression quantitative trait loci in mice. Genome Res. 15 681–691. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Druka, A., E. Potokina, Z. Luo, N. Bonar, I. Druka et al., 2008. Exploiting regulatory variation to identify genes underlying quantitative resistance to the wheat stem rust pathogen Puccinia graminis f. sp. tritici in barley. Theor. Appl. Genet. 117 261–272. [DOI] [PubMed] [Google Scholar]
- Errico, F., E. Santini, S. Migliarini, A. Borgkvist, D. Centonze et al., 2008. The GTP-binding protein Rhes modulates dopamine signalling in striatal medium spiny neurons. Mol. Cell. Neurosci. 37 335–345. [DOI] [PubMed] [Google Scholar]
- Florea, L., G. Hartzell, Z. Zhang, G..M. Rubin and W. Miller, 1998. A computer program for aligning a cDNA sequence with a genomic DNA sequence. Genome Res. 8 967–974. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goring, H. H., J. E. Curran, M. P. Johnson, T. D. Dyer, J. Charlesworth et al., 2007. Discovery of expression QTLs using large-scale transcriptional profiling in human lymphocytes. Nat. Genet. 39 1208–1216. [DOI] [PubMed] [Google Scholar]
- Han, W., S. Kasai, H. Hata, T. Takahashi, Y. Takamatsu et al., 2006. Intracisternal A-particle element in the 3′ noncoding region of the mu-opioid receptor gene in CXBK mice: a new genetic mechanism underlying differences in opioid sensitivity. Pharmacogenet. Genomics 16 451–460. [DOI] [PubMed] [Google Scholar]
- Homayouni, R., K. Heinrich, L. Wei and M. W. Berry, 2005. Gene clustering by latent semantic indexing of MEDLINE abstracts. Bioinformatics 21 104–115. [DOI] [PubMed] [Google Scholar]
- Hubner, N., C. A. Wallace, H. Zimdahl, E. Petretto, H. Schulz et al., 2005. Integrated transcriptional profiling and linkage analysis for identification of genes underlying disease. Nat. Genet. 37 243–253. [DOI] [PubMed] [Google Scholar]
- Ikeda, K., T. Kobayashi, T. Ichikawa, T. Kumanishi, H. Niki et al., 2001. The untranslated region of (mu)-opioid receptor mRNA contributes to reduced opioid sensitivity in CXBK mice. J. Neurosci. 21 1334–1339. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karp, C. L., A. Grupe, E. Schadt, S. L. Ewart, M. Keane-Moore et al., 2000. Identification of complement factor 5 as a susceptibility locus for experimental allergic asthma. Nat. Immunol. 1 221–226. [DOI] [PubMed] [Google Scholar]
- Kent, W. J., 2002. BLAT—the BLAST-like alignment tool. Genome Res. 12 656–664. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kwan, T., D. Benovoy, C. Dias, S. Gurd, C. Provencher et al., 2008. Genome-wide analysis of transcript isoform variation in humans. Nat. Genet. 40 225–231. [DOI] [PubMed] [Google Scholar]
- Law, L. W., A. G. Morrow and E. M. Greenspan, 1952. Inheritance of low liver glucuronidase activity in the mouse. J. Natl. Cancer Inst. 12 909–916. [PubMed] [Google Scholar]
- Lee, J. Y., I. Yeh, J. Y. Park and B. Tian, 2007. PolyA_DB 2: mRNA polyadenylation sites in vertebrate genes. Nucleic Acids Res. 35 D165–D168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Monks, S. A., A. Leonardson, H. Zhu, P. Cundiff, P. Pietrusiak et al., 2004. Genetic inheritance of gene expression in human cell lines. Am. J. Hum. Genet. 75 1094–1105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morley, M., C. M. Molony, T. M. Weber, J. L. Devlin, K. G. Ewens et al., 2004. Genetic analysis of genome-wide variation in human gene expression. Nature 430 743–747. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morrow, A. G., E. M. Greenspan and D. M. Carroll, 1949. Liver-glucuronidase activity of inbred mouse strains. J. Natl. Cancer Inst. 10 657–661. [PubMed] [Google Scholar]
- Mortazavi, A., B. A. Williams, K. McCue, L. Schaeffer and B. Wold, 2008. Mapping and quantifying mammalian transcriptomes by RNA-Seq. Nat. Methods 5 621–628. [DOI] [PubMed] [Google Scholar]
- Ono, T., N. Sekino-Suzuki, Y. Kikkawa, H. Yonekawa and S. Kawashima, 2003. Alivin 1, a novel neuronal activity-dependent gene, inhibits apoptosis and promotes survival of cerebellar granule neurons. J. Neurosci. 23 5887–5896. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Overall, R. W., G. Kempermann, J. Peirce, L. Lu, D. Goldowitz et al., 2009. Genetics of the hippocampal transcriptome in mouse: a systematic survey and online neurogenomics resource. Front. Neurogen. doi: 10.3389/neuro.15.003.2009. [DOI] [PMC free article] [PubMed]
- Peirce, J. L., H. Li, J. Wang, K. F. Manly, R. J. Hitzemann et al., 2006. How replicable are mRNA expression QTL? Mamm. Genome 17 643–656. [DOI] [PubMed] [Google Scholar]
- Rosen, G. D., C. J. Pung, C. B. Owens, J. Caplow, H. Kim et al., 2009. Genetic modulation of striatal volume by loci on Chrs 6 and 17 in BXD recombinant inbred mice. Genes Brain Behav. 8 296–308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rozen, S., and H. Skaletsky, 2000. Primer3 on the WWW for general users and for biologist programmers. Methods Mol. Biol. 132 365–386. [DOI] [PubMed] [Google Scholar]
- Schadt, E. E., S. A. Monks, T. A. Drake, A. J. Lusis, N. Che et al., 2003. Genetics of gene expression surveyed in maize, mouse and man. Nature 422 297–302. [DOI] [PubMed] [Google Scholar]
- Schwartz, D., 1962. Genetic studies on mutant enzymes in maize. II. On the mode of synthesis of the hybrid enzymes. Proc. Natl. Acad. Sci. USA 48 750–756. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Simon, P., 2003. Q-Gene: processing quantitative real-time RT-PCR data. Bioinformatics 19 1439–1440. [DOI] [PubMed] [Google Scholar]
- Spano, D., I. Branchi, A. Rosica, M. T. Pirro, A. Riccio et al., 2004. Rhes is involved in striatal function. Mol. Cell. Biol. 24 5788–5796. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stranger, B. E., A. C. Nica, M. S. Forrest, A. Dimas, C. P. Bird et al., 2007. Population genomics of human gene expression. Nat. Genet. 39 1217–1224. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Subramaniam, S., K. M. Sixt, R. Barrow and S. H. Snyder, 2009. Rhes, a striatal specific protein, mediates mutant-huntingtin cytotoxicity. Science 324 1327–1330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Voss, K., S. Stahl, E. Schleider, S. Ullrich, J. Nickel et al., 2007. CCM3 interacts with CCM2 indicating common pathogenesis for cerebral cavernous malformations. Neurogenetics 8 249–256. [DOI] [PubMed] [Google Scholar]
- Yeo, G., D. Holste, G. Kreiman and C. B. Burge, 2004. Variation in alternative splicing across human tissues. Genome Biol. 5 R74. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang, B., D. Schmoyer, S. Kirov and J. Snoddy, 2004. GOTree Machine (GOTM): a web-based platform for interpreting sets of interesting genes using Gene Ontology hierarchies. BMC Bioinformatics 5 16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang, H., J. Y. Lee and B. Tian, 2005. Biased alternative polyadenylation in human tissues. Genome Biol. 6 R100. [DOI] [PMC free article] [PubMed] [Google Scholar]