Abstract
The zebrafish nuclear progestin receptor (nPR; official symbol PGR) was identified and characterized to better understand its role in regulating reproduction in this well-established teleost model. A full-length cDNA was identified that encoded a 617-amino acid residue protein with high homology to PGRs in other vertebrates, and contained five domains characteristic of nuclear steroid receptors. In contrast to the multiplicity of steroid receptors often found in euteleosts and attributed to probable genome duplication, only a single locus encoding the full-length zebrafish pgr was identified. Cytosolic proteins from pgr-transfected cells showed a high affinity (Kd = 2 nM), saturable, single-binding site specific for a native progestin in euteleosts, 4-pregnen-17,20beta-diol-3-one (17,20beta-DHP). Both 17,20beta-DHP and progesterone were potent inducers of transcriptional activity in cells transiently transfected with pgr in a dual luciferase reporter assay, whereas androgens and estrogens had little potency. The pgr transcript and protein were abundant in the ovaries, testis, and brain and were scarce or undetectable in the intestine, muscle, and gills. Further analyses indicate that Pgr was expressed robustly in the preoptic region of the hypothalamus in the brain; proliferating spermatogonia and early spermatocytes in the testis; and in follicular cells and early-stage oocytes (stages I and II), with very low levels within maturationally competent late-stage oocytes (IV) in the ovary. The localization of Pgr suggests that it mediates progestin regulation of reproductive signaling in the brain, early germ cell proliferation in testis, and ovarian follicular functions, but not final oocyte or sperm maturation.
Keywords: hypothalamus, nuclear progestin receptor, oocyte, ovarian follicle, ovary, pgr, PR, progesterone, progesterone receptor, testis, zebrafish
The localization of nuclear progestin receptor suggests that it mediates progestin regulation of reproductive signaling in the brain, early germ cell proliferation in testis, and ovarian follicular functions, but does not play a role in final oocyte or sperm maturation.
INTRODUCTION
The physiological roles and tissue expression of the nuclear progestin receptor (PGR) have been well studied in mammalian models; however, few studies have characterized and localized the nuclear PGR in early derived vertebrates [1–5]. Previously, Pgr has been reported and characterized in a primitive teleost species, Japanese eel (Anguilla japonica) [6, 7], and in a frog species, Xenopus laevis [8–10]. In contrast to PGRs in other vertebrate species, which generally encode two PGR proteins (PGR-A and PGR-B) from a single locus varying only in length, both Japanese eel and Xenopus transcribe two Pgr proteins from separate loci that differ considerably in their amino acid sequences.
Although the functions of PGRs have been well studied during the past 35–40 yr, many additional roles of the PGRs in reproductive tissues, including the oocytes, testis, and brain, remain unclear and are difficult to study in current models [1–5]. The complexity of mammalian reproductive models complicates the thorough investigation of Pgr functions, and comparative analysis in eel and Xenopus is hindered by the presence of seemingly unique, species-specific Pgr isoforms. Progestins have been recently identified as essential factors for initiating meiosis in the testis of the Japanese eel [11]. However, the specific location and identity of progestin receptors mediating these proliferative activities in the testis have not been determined. In addition, no information is available on the expression and localization of the Pgr in the fish brain, even though the feedback control of neural functions in the fish brain by progestins during reproductive events has been well established [12–15]. Furthermore, it has been suggested that Pgr is involved in rapid nongenomic progestin actions during Xenopus oocyte maturation [8, 9]. Overexpression of Pgr increased nongenomic signaling through activation of MAPK and cell cycle regulators, such as cyclins, in Xenopus [8, 9, 16, 17]. However, the lack of Xenopus Pgr expression in the oocyte membrane is not consistent with a physiological role for Pgr during oocyte maturation [1].
To better understand Pgr's roles in reproduction, we have characterized Pgr and localized its expression in zebrafish. Zebrafish provides a unique model for the study of Pgr because it spawns daily and has oocytes that can undergo growth, maturation, and ovulation in vitro [18]. In this paper, we identified a single locus for pgr in the zebrafish genome and isolated a full-length cDNA for zebrafish pgr. Subsequently, we determined specific 4-pregnen-17,20β-diol-3-one (17,20β-DHP) binding and transcription activities of zebrafish Pgr. We also examined the tissue and cellular expressions of the Pgr in zebrafish oocytes, testis, and brain using specific antibodies and PCR primers. This study provides a foundation for using zebrafish as a model for studying genomic and nongenomic actions of Pgr.
MATERIALS AND METHODS
Cloning of pgr cDNA
Zebrafish, Danio rerio, were obtained from a local pet store and euthanized with MS-222 (200 mg/L in buffered solution). Experiment protocols were approved by the Institutional Animal Care and Use Committee at East Carolina University. Ovaries were removed and placed immediately in 1 ml of TRIzol reagent (Gibco). After homogenization, an aliquot of 200 μl of chloroform was added, and the solution was vortexed vigorously. The mixture was centrifuged, the aqueous layer was transferred to a clean RNase-free microcentrifuge tube, and total RNA was precipitated and redissolved in 100 μl of water. First-strand cDNA was synthesized from 1 μg of total RNA using the GeneRacer kit (Invitrogen), in accordance with the manufacturer's instructions. The PCR was performed using a set of primers (Supplemental Table S1 and Supplemental Fig. S1; all Supplemental Data are available online at www.biolreprod.org) designed by comparing published pgr sequences found in other species to the zebrafish genome (Ensembl Zv 7). The first PCR was performed in 20-μl aliquots using a gradient Eppendorf Mastercycler. After a 2-min denaturation at 94°C, the PCR cycle was repeated 30 times with a denaturation at 94°C for 30 sec, annealing at 45°C–55°C for 30 sec, and elongation at 72°C for 1 min. The amplified partial cDNA products were separated by agarose gel electrophoresis and ligated into a pGEM T-Easy vector (Promega). After the vector was transformed in XL1-Blue-competent cells (Stratagene, La Jolla, CA), positive clones were selected by blue-white screening. Plasmid DNA was purified from bacterial cells using the QIAprep Spin Plasmid kit (Qiagen). All plasmid DNAs were sequenced with forward and reverse universal primers using the Big-Dye Terminator kit and an ABI Prism 377 DNA sequencer (Perkin-Elmer, Wellesley, MA). Thereafter, gene-specific oligonucleotide primers were designed and synthesized from the partial pgr sequences. The 5′ and 3′ rapid amplifications of cDNA ends (RACEs) were performed using the GeneRacer kit per the manufacturer's directions. Nested PCR was performed, and products were separated by agarose gel electrophoresis. The products were ligated into the TOPO TA vector (Invitrogen) and sequenced with forward and reverse vector-specific primers. Sequence data were compiled using Sequence Navigator (ABI, Foster City, CA). To obtain the full-length pgr cDNA, gene-specific primers were designed for pgr. The amplified products were gel purified and ligated into a TA vector (pGEM-T Easy vector). Multiple clones were selected and sequenced.
Phylogenetic Analysis
All previously identified PGR ligand-binding domain sequences were retrieved from GenBank or assembled from Ensembl (for accession/ID numbers, see Supplemental Table S2) and were aligned with the zebrafish pgr sequence by CLUSTALW. The phylogenetic tree was constructed using the maximum-likelihood method implemented in the PhyML program (v3.0 aLRT) with a bootstrap value of 1000 trials for each position and was rooted by the zebrafish androgen receptor (Ar).
Expression of Recombinant PGR in Human Embryonic Kidney Cells
For reporter assay and steroid-binding assay of the pgr, the coding region of the pgr was amplified by PCR with the addition of a Kozak sequence and BamHI cut site on each end (forward: 5′-AACGGATCCCACCATGGACACGGT-3′; reverse: 5′-TCGGATCCCATTTGTGGTG-3′). The PCR products were digested with BamHI, ligated into the pcDNA3.1 C vector (Invitrogen), and verified by DNA sequencing.
Human embryonic kidney cells (HEK293; American Type Culture Collection, Manassas, VA) were plated at 90% confluency with 10% charcoal-stripped fetal bovine serum (CSFBS) media in six-well plates at 24 h prior to transfection. Cells were transiently transfected with 100 ng of pgr-pcDNA3.1 using Lipofectamine 2000 (Invitrogen) according to the manufacturer's directions. Cells were assayed for pgr expression and receptor binding 48 h after transfection.
Receptor-Binding Assays
The [3H]-labeled 17,20β-DHP was enzymatically converted from [1,2,6,73H]-17α hydroxyprogesterone (94.Ci/mmol; Amersham) with 20β-hydroxysteroid dehydrogenase [19]. The cytoplasmic steroid receptor-binding assay was performed according to the protocol published previously [20], with minor modifications. Briefly, the cells that were transfected with zebrafish pgr cDNA were harvested and washed twice with ice-cold PBS, then homogenized in TEDM buffer (10 mM Tris, 1 mM ethylenediaminetetraacetic acid [EDTA], 5 mM dithiothreitol, and 10 mM Na-molybdate) and centrifuged at 1000 × g for 7 min. The supernatant containing the cytosolic and plasma membrane fractions was centrifuged again at 100 000 × g for 1 h at 4°C, and the supernatant comprising the cytosolic fraction was used for the binding assays. An aliquot (125 μl) of the cytosolic fraction (1 mg/ml protein) in TEDM buffer was added to each assay tube containing 125 μl of 0.75, 1.5, 3, 6, or 12 nM [3H]-17,20β-DHP in the same buffer with or without the addition of 100-fold excess of nonlabeled 17,20β-DHP to measure total and nonspecific binding, respectively. After an overnight incubation at 4°C, 250 μl of a charcoal suspension (1% charcoal, 0.5% Dextran T-70 [Sigma] in TEDM buffer) was added to each tube, and the samples were incubated at 4°C for 5 min. All the samples then were centrifuged at 5000 × g for 5 min, and the supernatant in each tube was transferred to a scintillation tube and counted in a scintillation counter. The specific binding of each sample was calculated by subtracting the nonspecific binding from total binding, and the saturation and Scatchard analyses were performed with GraphPad Prism software.
Dual Luciferase Transactivation/Transcription Assay
The luciferase transactivation assay was conducted as described previously [7], with few modifications. In short, HEK293 cells were plated at 90% confluency with 10% CSFBS media in 96-well plates at 24 h prior to transfection. Cells were transiently transfected with 10 ng of pgr-pcDNA3.1 along with 50 ng of a progestin receptor-activated response element linked to firefly luciferase (PRE2-TATA-Luciferase [21], MMTV-Luciferase [22], or GRE-luciferase; Mercury Pathway Profiling System; BD Biosciences, Palo Alto, CA) and 1 ng of pRL-TK (Promega) control vector expressing Renilla luciferase per well using Lipofectamine 2000 (Invitrogen). Approximately 12 h after transfection, media were replaced with fresh CSFBS media containing various concentrations of steroids (dissolved in 100% EtOH; Steraloids) and 0.1% ethanol. The cells were further incubated for an additional 48 h for the production of the reporter proteins. Cells then were assayed for firefly and Renilla luciferase using Promega's dual luciferase assay system. Luminescent values were standardized using the pRL-TK intensity and reported as fold increase over the nonactivated control.
Reverse Transcription-PCR
To determine the expression of the pgr transcript, total RNA was extracted immediately from various tissue samples (brain, ovary, testis, muscle, intestine, and gill) with TRIzol reagent (Invitrogen), homogenized using a sonicator (Sonic Dismembrator Model 100; Fisher Scientific), and purified following the manufacturer's instructions. Then, total RNA (600 ng) was reverse transcribed into cDNA in a 10-μl reaction using SuperScript II (Invitrogen). The PCR was conducted on the cDNA template for 30 cycles with an annealing temperature of 62°C using pgr-specific primers (forward: 5′-GGATCACCTTTCTGCGCT-3′; reverse: 5′-GACAACCAGAAGCCTC-3′) and β-actin primers (forward: 5′-TTCGAGACCTTCAACACCC-3′; reverse: 5′-TGGTGGTGAAGCTGTAGCC-3′). As a negative control, an equal volume of water was added into the PCR mixture instead of cDNA. The PCR products were run on a 2% agarose gel and imaged using a Fluor Chem 8900 imaging station (Alpha Innotech). To determine whether Pgr was expressed in the follicular cells and/or oocytes, we developed a procedure for the separation of follicular cell layers from the oocytes. For removal and isolation of the follicular cells, oocytes were collected in 50% L-15 media (Sigma), chilled on ice for 30 min, and then manually removed with fine forceps. Removal of the follicular cell layer was verified by staining oocytes for 10 min with propidium iodide (20 μg/ml).
Antibody Production and Western Blot Analysis
Polyclonal antibodies were generated in rabbits against four synthetic peptides corresponding either to the N-terminal (amino acids 74–87, amino acids 100–112) or C-terminal (amino acids 466–479, amino acids 519–532) part of zebrafish Pgr. The peptides were designed to have low/no homology to other zebrafish proteins, and linked to keyhole limpet hemocyanin (KLH) to increase the size of the antigen to ensure strong immunoresponses. After six intradermal injections of KLH-linked peptides (50–100 mg per injection in Freund adjuvant), antibody specificity was verified by detection of Pgr protein in transiently transfected HEK293 and zebrafish tissue samples as described below. Only the N-terminal antibody (amino acids 100–112) was confirmed to be specific and subsequently used for Western blotting, immunohistochemistry (IHC) localization, and immunofluorescent staining. Tissue samples of zebrafish brain, ovary, testis, gill, muscle, and intestine were isolated from three fish and analyzed separately for protein expression on at least three separate days.
Western blotting was performed as described previously [19]. In brief, tissue samples were collected from adult zebrafish by cervical dislocation after an appropriate anesthetic overdose (MS-222; 200 mg/L in buffered solution). Samples were immediately transferred into ice-cold PBS and sonicated with 10 short bursts on a sonicator (Sonic Dismembrator; Fisher Scientific). The homogenate was centrifuged for 10 min at 20 000 × g, and the resulting supernatant was removed, mixed with an equal amounts of 2× SDS sample buffer (0.125 M Tris HCl, pH 6.8; 4% SDS; 20% glycerol; and 10% 2-mercaptoethanol), boiled for 5 min, and then cooled on ice.
Each sample was loaded with 40 μg of total protein (determined by the RC DC protein assay; Bio-Rad, Hercules, CA) onto an SDS/6.5% PAGE gel and transferred to a nitrocellulose membrane. The membrane was blocked with 5% nonfat milk in TBST (50 mM Tris, 100 mM NaCl, and 0.1% Tween 20, pH 7.4) for 1 h. The membrane was further incubated with Pgr antibody (1:2500 dilution) overnight, washed five times for 5 min with TBST, incubated for 1 h with horseradish peroxidase conjugated to goat anti-rabbit antibody (1:5000 dilution; Cell Signaling Technology, Beverly, MA), and finally washed five times for 5 min with TBST. The blots then were developed using Super Signal West Extended Dura Substrate (Pierce, Rockford, IL) and visualized using a Fluor Chem 8900 imaging station (Alpha Innotech). Protein size was determined by comparing blotted protein size to a biotinylated protein ladder (Cell Signaling Technology) following the manufacturer's directions.
Immunohistochemistry
Ovary and testis were removed from adult zebrafish and immediately fixed in 10% buffered formalin phosphate (Fisher Scientific) overnight. Tissues then were dehydrated for 5 min with 75%, 90%, 100% ethanol, 100% xylene, mounted in paraffin wax cassettes, which were cut into 8-μm-thick sections and placed on glass slides (Fisher Scientific). The sections then were deparaffinized by 5-min washes with 100% xylene, 100% ethanol, 95% ethanol, 70% ethanol, and twice with PBS. Sections were incubated with Pgr antibodies (1:500 dilution) using the Vectastain ABC kit (Vector Laboratories) according to the manufacturer's protocol.
In Situ Hybridization
In situ hybridization was performed as described previously [23]. Full-length zebrafish pgr cDNAs cloned into the pSPORT1 vector were linearized with BamHI or SmaI to generate templates for synthesizing sense or antisense probes, respectively. Sense and antisense digoxigenin-labeled probes were generated using T7 or SP6 RNA polymerases by in vitro transcription according to the instructions of the manufacturer (Roche Applied Science, Indianapolis, IN). Zebrafish ovaries were removed and fixed immediately in 4% paraformaldehyde dissolved in PBS overnight. The tissues were dehydrated, embedded in paraffin, cut into 8-μm-thick serial sections, and mounted on Superfrost slides (Fisher Scientific). Sections were processed, rehydrated, postfixed with 4% paraformaldehyde for 20 min, and then treated with 200 mM HCl for 10 min, followed by 15-min digestion with 10 μg/ml protein kinase K. Hybridization was conducted with 10 ng of antisense probe per slide in 60 μl of buffer (50% deionized formamide/2× SSC [where 1× SSC is 0.15 M NaCl/0.015 M sodium citrate]/10% dextran sulfate/0.01% yeast RNA/0.02% SDS) overnight at 55°C. Sections were washed in high-stringency buffer (1× SSC/50% formamide) at 55°C and 1× SSC at room temperature. Sections were equilibrated with TBS (50 mM Tris, pH 7.5; 150 mM NaCl) and blocked with 1× Roche blocking solution containing 10% fetal bovine serum and 1% sheep serum in maleic acid buffer (100 mM maleic acid/150 mM NaCl, pH 7.5) for 1 h at room temperature. Sections then were incubated for 1 h at room temperature with anti-digoxigenin FAB fragment conjugated to alkaline phosphatase (diluted 1:2000 with 1× Roche blocking solution containing 10% fetal calf serum and 1% sheep serum; Roche Applied Science). Sections were washed with TBS, equilibrated in AP buffer, and incubated with Nitro blue tetrazolium/5-bromo-4-chloroindol-3-yl phosphate substrate (Roche Applied Science) overnight at 4°C in a dark, humid chamber. Reactions were stopped by washing with TE buffer (10 mM Tris, pH 7.5; 1 mM EDTA) twice; sections then were fixed with 4% paraformaldehyde, washed twice with TE buffer, and mounted with 90% glycerol in PBS.
Testicular Culture and Bromodeoxyuridine Labeling
Using methods adapted from previously described eel testicular culture [24], the involvement of Pgr and the effect of progestins on testicular germ cell proliferation were investigated. Bromodeoxyuridine (BrdU) was incorporated into DNA of dividing germ cells in testicular cultures and immunohistochemically detected as an indicator of germ cell proliferation. Testes from each fish were cut into at least four pieces, and the testicular explants were cultured in 1 ml of basal medium with or without 10 nM 17,20β-DHP or 11-ketotestosterone (11-KT). The culture medium was replaced with fresh medium containing same concentration of steroids at the third day of culture. At 6 days of culture, testicular fragments were labeled with BrdU for 12 h according to the manufacturer's instructions (Amersham Pharmacia Bioscience) to detect DNA replication and then immediately fixed in Bouin solution and prepared for IHC using anti-BrdU antibody (1:1000 dilution; Amersham Pharmacia Bioscience). Testicular sections were counterstained with hematoxylin to distinguish testis cell type. Adjacent sections were stained alternately with BrdU or Pgr antibodies for colocalization. The numbers of the spermatogonia and early spermatocytes in a 250 × 250 μm grid in three different sections from same testicular explants of same treatment were counted. In total, six replicates of six different testes per treatment were setup over three different culture periods. The BrdU index was average percentage of the number of immunolabeled germ cells over the total number of spermatogonia and early spermatocytes from six different testes.
Immunofluorescent Staining
Tissue sections (12 μm) prepared with a cryostat from frozen adult female zebrafish brains (n = 5) were used and stained as described previously [25]. In brief, tissue sections were first rinsed in PBS, and nonspecific binding was blocked for 1 h at room temperature in PBS containing 0.2% triton and 0.5% powdered milk. Sections then were incubated with Pgr antibody (1:1000 to 1:5000) overnight at room temperature. After several washes in PBS 0.2% triton, tissue sections were incubated with the appropriate secondary antibodies (goat anti-rabbit Alexa 594; Invitrogen), and slides then were rinsed in PBS-triton. Then, cell nuclei were visualized with a 4′,6′-diamidino-2-phenylindole (DAPI) counterstaining (Vectashield with DAPI; Vector Laboratories). Omission of the primary antibodies on brain sections and Western blots of brain samples confirmed specificity of the Pgr antibody reaction. Observations were carried out on an Olympus Provis photomicroscope equipped with a DP50 digital camera. Images were acquired in TIFF format and merged using the Olympus Cell Software. Plates were prepared using the Adobe Photoshop 7 software with minimum light or contrast adjustment.
Statistical Analyses
All experiments were conducted at least three times. Differences between groups were considered significant when P < 0.05 as analyzed by a Student t-test.
RESULTS
Cloning and Characterization of Zebrafish pgr
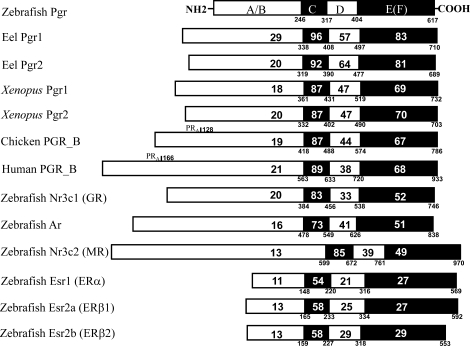
Supplemental Figure S1 illustrates the full-length pgr cDNA (accession number EF155644) cloned from teleost ovarian cDNAs using (5′/3′) RACE and gene-specific primers (Supplemental Table S1). The zebrafish pgr has 3199 nucleotides encoding 617 amino acids. The cDNA has 56 nucleotides in the 5′ untranslated region (UTR) and 1289 nucleotides in the 3′ UTR, multiple poly-A sites, and a poly-A tail (Supplemental Fig. S2). The zebrafish Pgr has five typical steroid receptor domains and shows highest sequence homology in the ligand- and DNA-binding domains to those of eel Pgr (Fig. 1). The ligand-binding domain of the zebrafish Pgr shares high homology (67%–83% amino acid identity) with known PGRs of other vertebrate classes, and low homology (27%–52% amino acid identity) with other steroid receptors of zebrafish, including glucocorticoid receptor (Gr; or official symbol Nr3c1), Ar, mineralocorticoid receptor (Mr; or official symbol Nr3c2), and estrogen receptors (ESRs; Fig. 1). There was no homology with the previously described membrane progestin receptors (mPRs; or official symbol Paqr) in zebrafish [26, 27].
FIG. 1.
Amino acid identities between the various domains of zebrafish nuclear progestin receptor (Pgr) and those of PGRs in other vertebrates are indicated by the numbers in the respective domain. For comparison, the other zebrafish nuclear steroid receptors, including Ar, glucocorticoid (Gr or Nr3c1), mineralocorticoid (Mr or Nr3c2), and estrogen receptors (Esr), are also included. The numbers below the line indicate positions of amino acids from the translation starting site.
Data mining was conducted to find other potential Pgr isoforms in the zebrafish genome. The expected value (E-value) was used for sorting and scoring all of the potential matches during the Blast search because E-value is a parameter that describes the number of hits one can “expect” to see by chance when searching a database of a particular size. The E-value decreases exponentially as the score (S) of the match increases. The lower the E-value, or the closer it is to zero, the more “significant” the match is. Only one locus on chromosome 18 of the zebrafish genome (Ensembl Zv8) with the lowest E-value was found to encode the pgr using peptide sequences of zebrafish Pgr and eel Pgr. The next three hits with low E-values were orphan steroid receptors, including estrogen-related receptor delta (esrrd), nuclear receptor subfamily 2 group F member 2 (nr2f2), and nuclear receptor subfamily 1 group H member 4 (nr1h4), located on the same chromosome. Subsequent hits with low E-values were identified as zebrafish ar, nr3c1(gr), nr3c2(mr), and esr2a (ERβ1), which were located on chromosomes 5, 14, 1, and 20, respectively. These results indicate that the zebrafish genome has only one pgr locus. We also found a single locus of pgr in the genomes of medaka, Takifugu, Tetraodon, and stickleback.
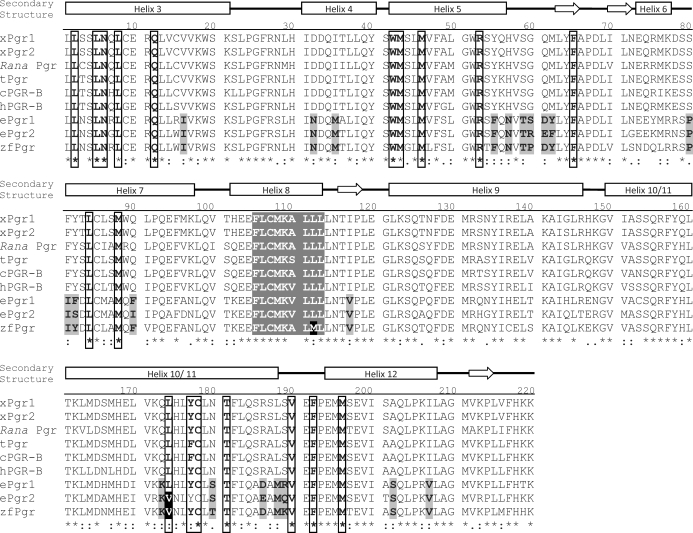
As expected, almost all Pgr residues critical for progestin binding [28] were conserved in the ligand-binding domain of the zebrafish Pgr (18 of 19; Fig. 2). The only exception was a variation from a leucine to a valine residue in zebrafish Pgr and eel Pgr (human L887 to zebrafish V571). Approximately 21 amino acid residues in the ligand-binding domains of the teleost Pgr from zebrafish and eel differed from conserved residues found across all other vertebrate species (Fig. 2). Phylogenetic analysis indicated that zebrafish Pgr clustered together with those of other fish species, including eel, stickleback, Takifugu, and medaka (Fig. 3).
FIG. 2.
Comparison of the ligand-binding domains of the nuclear progestin receptor (Pgr) in representative vertebrate species. Residues previously determined to be important for P4 binding are boxed. The sole variable amino acid change from leucine to valine in the binding boxes of teleosts is highlighted in a white letter with a black background. Residues that uniquely vary in teleosts from other species in the ligand-binding domain of the receptor are in black letters with a gray background. Putative membrane localization motif (FXCXXXXLL) is shown in white letters with a gray background, whereas the variation in zebrafish is shown with a black background. Common secondary-structural elements of the Pgr ligand-binding domain, including alpha-helices and beta-sheets (arrows), are marked above the sequence information. xPgr1, X. laevis Pgr1; xPgr2, X. laevis Pgr2; Rana Pgr, Rana dybowskii Pgr; tPgr, turtle Pgr; cPGR-B, chicken PGR; hPGR-B, human PGR-B; zfPgr, zebrafish Pgr.
FIG. 3.
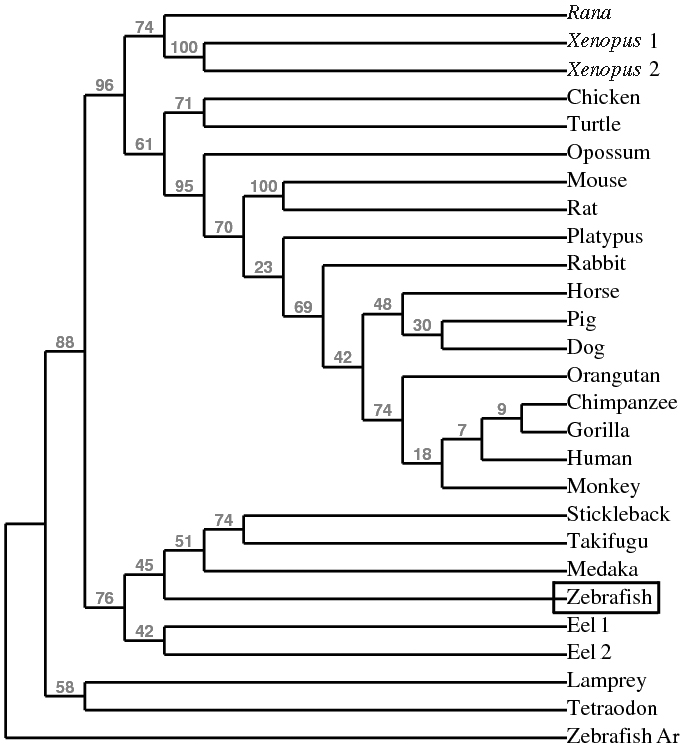
Phylogenetic analysis of vertebrate nuclear progestin receptor (Pgr) using conserved ligand-binding domains. Node labels above branches show percent bootstrap values.
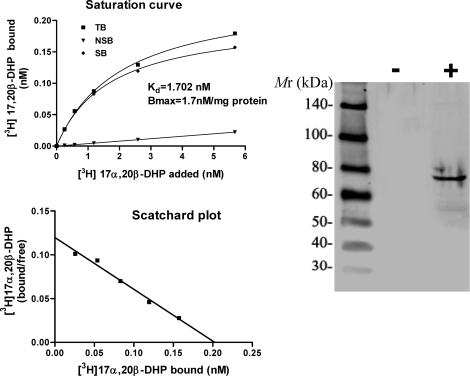
Using zebrafish Pgr antibodies developed in our lab, a specific band at ∼69 kDa corresponding to the calculated molecular mass of zebrafish Pgr was observed in HEK293 cells transfected with zebrafish Pgr, whereas no immunoreaction was observed in HEK293 cells transfected with a carrier vector (Fig. 4). Cytosolic proteins from pgr-transfected cells showed a high-affinity (Kd = 2 nM), saturable, single-binding site specific for the native progestin in zebrafish, 17,20β-DHP, whereas negligible specific 17,20β-DHP binding was observed in nontransfected cells (Fig. 4).
FIG. 4.
Expression and steroid binding specificity of recombinant zebrafish nuclear progestin receptor (Pgr) expressed in HEK293 cells transiently transfected with zebrafish Pgr (+) or a carrier vector without Pgr insert (−).
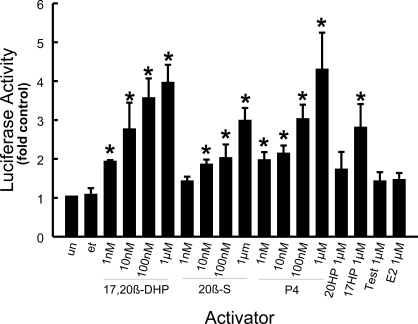
Specific progestins (1 nM to 1 μM) activated zebrafish Pgr-mediated transcription of luciferase, whereas estrogen (E2) and testosterone (Test) did not (Fig. 5). Of the progestins tested, the maturation-inducing steroid (MIS), 17,20β-DHP, and progesterone (P4) were the most potent activators of this of Pgr-mediated transcription. Minimal steroid-activated transcription of luciferase was detected in HEK293 cells that were not transfected with Pgr. HEK293 cells transfected with zebrafish Pgr along with MMTV-luciferase or GRE-luciferase reporters exhibited similar P4 responsiveness (data not shown).
FIG. 5.
Transactivation activity of zebrafish nuclear progestin receptor (Pgr) in transfected HEK293 cells exposed to various concentrations of steroids (17,20β-DHP, 20β-S, P4, 4-pregnen-20β-ol-3-one [20HP], 4-pregnen-17-ol-3,20-dione [17-hydroxyprogesterone; 17HP], E2, and Test). Luciferase activity was assayed and expressed as ratios of the activity of untransfected control cells, which was set at 1. Data show mean ± SEM from triplicate samples and represent the results of three separate experiments. Asterisks indicate statistical difference (P < 0.05) from controls (un, untransfected cell; et, transfected cells treated with 0.1% ethanol).
RT-PCR and Western Blot Analysis of Pgr Expression in Zebrafish Tissues
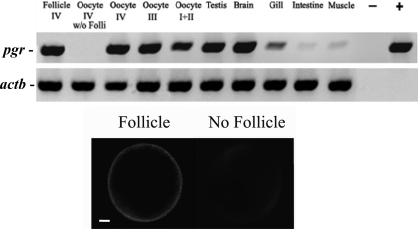
The pgr transcript was expressed strongly in oocyte-follicular cell complexes at all stages of oocyte development, as well as in the testis and brain, and weakly in the gill, intestine, and muscle (Fig. 6). Further analyses indicated that the source of the pgr transcript in the late-stage oocyte-follicular cell complex was restricted to the follicular cell component, because the pgr transcript was undetectable in denuded stage IV oocytes (Fig. 6).
FIG. 6.
The RT-PCR analysis of zebrafish nuclear progestin receptor (pgr) and β-actin (actb) in follicular cell layers without oocytes (Follicle IV), denuded late stage IV oocytes (w/o folli, without follicular cells), follicular cell-enclosed oocytes (stages I–IV), testis, brain, gill, intestine, and muscle. Minus sign indicates negative control without DNA template for PCR; plus sign, positive control with Pgr plasmid as a template for PCR. Staining of the intact oocyte with follicular cells (bottom left) and oocytes without follicular cells (bottom right) with propidium iodide. Bar = 100 μm.
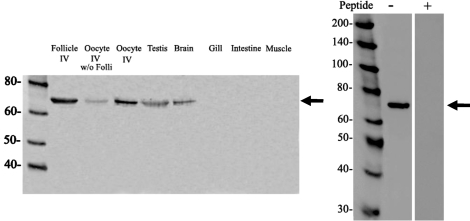
One specific band at ∼69 kDa corresponding to the calculated molecular mass of Pgr was observed in the extracts of oocytes by Western blotting using the zebrafish Pgr antibody (Fig. 7). The ∼69-kDa band was not observed using the Pgr antibody preabsorbed with the antigens (Fig. 7), thus confirming the specificity of the antibody. The majority of the Pgr immunoreactive protein was observed in the follicular layer surrounding the oocyte, although weak immunoreactivity was also detected in denuded oocytes by Western blotting (Fig. 7). The highest levels of Pgr protein were observed in the ovary, followed by testis and brain, with little or no expression in the gill, intestine, and muscle (Fig. 7).
FIG. 7.
Western blot analysis of the expression of zebrafish nuclear progestin receptor protein in follicular cell layers without oocytes (Follicle IV), denuded stage IV oocytes (w/o folli, without follicular cells), stage IV follicular cell-enclosed oocytes, testis, brain, gill, intestine, and muscle (left). Analyses of antibody specificity with (+) or without (−) preabsorption of antibodies with the synthetic peptides used for antibody generation (right). Arrows indicate expected size of Pgr. Values are in kDa.
IHC and In Situ Localization of Pgr in Oocytes
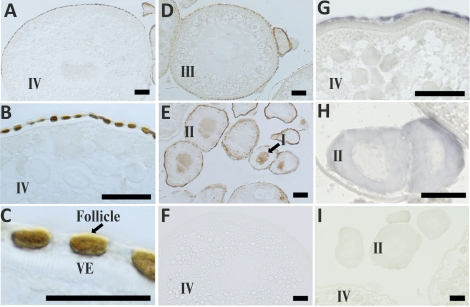
Consistent with the results obtained by RT-PCR and Western blot analyses, strong immunostaining with the Pgr antibody was found once again in the ovary (Fig. 8), testis (Fig. 9), and brain (Fig. 10). Strong immunoreactivity with the Pgr antibody was observed in the nuclei of follicular layer cells surrounding all stages of oocytes (Fig. 8). Immunostaining of the Pgr in the nuclei along with a weak cytoplasmic immunoreaction were also observed in early-stage oocytes (stages I and II; Fig. 8E). However, no immunoreaction of the Pgr antibodies within the cytoplasm, nucleus, yolk, membrane, or vitelline envelope of the oocyte was observed in late-stage oocytes (stages III and IV; Fig. 8, A–D). Consistent with the results obtained by RT-PCR and immunohistochemistry, the pgr transcript also was restricted to the follicular layers, with no detectable signaling within the late stage IV oocytes (Fig. 8G). In marked contrast, the pgr transcript was observed in the cytoplasm of early stage I and II oocytes (Fig. 8H).
FIG. 8.
Immunohistochemical and in situ localization of zebrafish nuclear progestin receptor (Pgr) proteins or transcript in stage I–IV oocytes. A–E) Immunostaining images of a representative stage IV oocyte at different magnifications (A–C), a stage III oocyte (D), or stage I or II oocytes (E). F) Stage IV oocytes stained with preimmune serum. G–I) Representative images of in situ hybridization in stage IV oocytes (G) or stage II oocytes (H) using pgr antisense probe or a sense probe (I). VE, vitelline envelope. Bars = 50 μm (A, B, and D–I) and 25 μm (C).
FIG. 9.
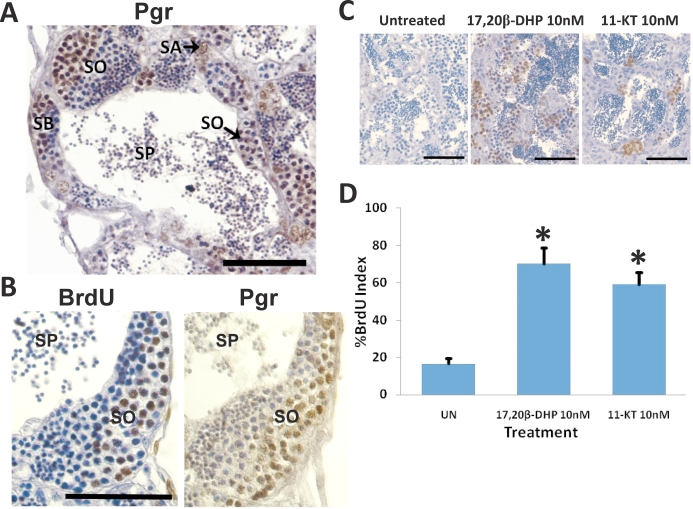
Localization of nuclear progestin receptor (Pgr) and proliferating germ cells in zebrafish testis using Pgr- and BrdU-specific antibodies, respectively. A) Pgr immunostaining. B and C) Adjacent sections were stained with Pgr and BrdU antibodies, respectively. D) Quantification of percent BrdU-positive germ cells. Data show mean ± SEM from six testicular samples and represent the results of three separate experimemts. *P < 0.01. SA, type A spermatogonia; SB, type B spermatogonia; SO, spermatocyte; SP, sperm. Bar = 50 μm.
FIG. 10.
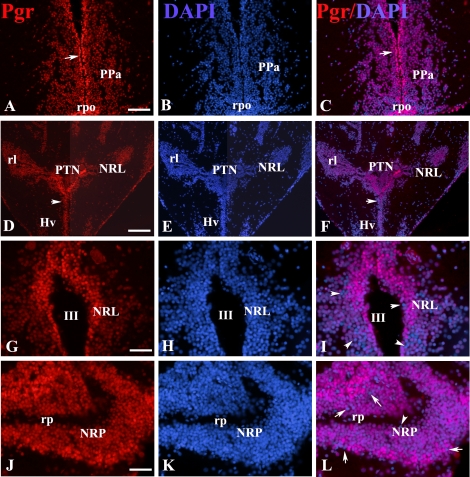
Representative immunoreactivity in the forebrain of an adult female zebrafish at the level of the anterior preoptic area (A–C), hypothalamus (D–I), and caudal line (J–L) for Pgr (red; A, D, G, and J) and DAPI (blue; B, E, H, and K). Superimposed images (C, F, I, and L) are shown. Arrows in A, C, D, and F indicate that the majority of the nuclei located in the periventricular layers have stronger Pgr immunostaining than those in the parenchyma. Pgr-negative nuclei were indicated by arrows in I and L. NPT, nucleus posterioris tuberis; NRL, nucleus recessus lateralis; NRP, nucleus recessus posterioris; PPa, anterior preoptic region; rpo, preoptic recess; III, third ventricle; rl, lateral recess; rp, posterior recess. Bars = 60 μm (A–C), 120 μm (D–F), or 30 μm (G–L).
Localization of Pgr in the Testis and Its Role in Germ Cell Proliferation
Strong Pgr expression was observed in spermatogonia and early spermatocytes, whereas no Pgr expression was detected in spermatids or sperm (Fig. 9). Immunopositive Pgr germ cells were also BrdU positive, suggesting a role for Pgr in proliferation of germ cells in the testis (Fig. 9). Furthermore, the BrdU index increased significantly for testicular germ cells treated with 10 nM 17,20β-DHP in vitro; this was comparable to the increase in the BrdU index observed for similar testicular germ cells exposed to the teleost androgen, 11-KT (Fig. 9).
Localization of Pgr in the Hypothalamus and Preoptic Region
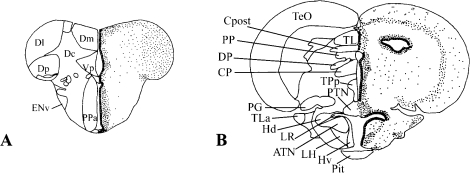
The Pgr staining was localized specifically to the nuclei of cells in all forebrain regions examined. As shown in Figure 10, Pgr-positive cells are extremely numerous and exhibit more intense staining along the midline in the ventricular and periventricular regions (Fig. 10, A–F). In all animals, less intense staining was observed toward more lateral regions. This intriguing observation was observed in most, if not all, brain regions. The only exception was in the nucleus of the lateral recess of the hypothalamus (Fig. 10, I–L). In all of these regions, a majority of cells were positive, but counterstaining with DAPI clearly showed that not all cells express PR (Fig. 10, G–L). Figure 11 [29] provides a clear illustration of the distribution of Pgr in the two main neuroendocrine regions of the brain: the preoptic area (Fig. 11A) and the mediobasal hypothalamus (Fig. 11B).
FIG. 11.
Schematic drawings show a wide distribution of Pgr in the preoptic area (A) and mediobasal hypothalamus (B). The transverse sections of the two brain regions were adopted from the atlas of the brain of zebrafish [29] with permission. Larger dots along the brain ventricles are meant to represent the stronger immunoreactivity of the cell nuclei in the ventricular layer. ATN, anterior tuberal nucleus; CP, central posterior thalamic nucleus; Cpost, commissura posterior; Dc, central zone of D; DI, lateral zone of D; Dm, medial zone of D; Dp, posterior zone of D; DP, dorsal posterior thalamic nucleus; Env, entopeduncular nucleus, ventral part; Hd, dorsal zone of periventricular hypothalamus; LR, lateral recess of diencephalic ventricle; LH, lateral hypothalamic nucleus; Hv, ventral zone of periventricular hypothalamus; PG, preglomerular nucleus; Pit, pituitary; PP, periventricular pretecal nucleus; PPa, parvocellular preoptic nucleus, anterior part; PTN, posterior tuberal nucleus; TeO, tectum opticum; TL, torus longitudinalis; TLa, torus lateralis; TPp, periventricular nucleus of posterior tuberculum; Vp, postcommissural nucleus of V.
DISCUSSION
To our knowledge, this is the first study to identify and characterize the pgr of zebrafish, which is a unique full-length pgr transcript with a typical steroid receptor structure encoded from a single locus. Further, we have shown that the zebrafish Pgr has specific progestin binding and transcriptional activities that validate it as a functional PGR. These results and the observation of tissue-specific expression of Pgr in the ovary, testis, and preoptic region of the hypothalamus strongly support the concept that Pgr plays a role in a variety of reproductive processes in this species, including a role in testicular germ cell proliferation as well as, potentially, oocyte development and ovulation, as in rats [30, 31]. Interestingly, the majority of the zebrafish Pgr protein, and all detectable mRNA, were restricted to the follicular cell layer of stage IV oocytes, diminishing Pgr's potential role in oocyte maturation.
Only one pgr loci was found in the genomes of zebrafish and in other fish species, including medaka, Takeifugu, Tetradon, and stickleback. Our experimental evidence obtained by extensive RT-PCR with multiple primer sets (Supplemental Table S1 and Supplemental Fig. S1) and Western blotting further support the existence of a single full-length pgr transcript/protein produced from a single locus in the zebrafish. Of the 24 species from which a pgr gene could be identified, a single pgr gene has been described from 22 species, whereas only two species (the Japanese eel and the frog species, Xenopus) have two described pgr genes (see Supplemental Table S2). The ancestral genome of ray-finned fish likely contained two pgr loci that were maintained as two active genes in some species, and one pgr locus was lost in others during their evolution. The second locus for pgr in zebrafish was lost, whereas in the Japanese eel both pgr genes were conserved. Likewise, the separate pgr loci found in the tetraploid frog species, Xenopus, were likely conserved from a separate genome duplication event. Separate duplication events are supported by the distinct location of Xenopus and eel on distant nodes of the phylogenetic analysis. Interestingly, zebrafish only have one Ar allele and one estrogen receptor α allele, yet they have two estrogen receptor β alleles [32, 33].
The separation of tetrapods, including Xenopus, and fish species on distant nodes of the phylogenetic tree may also confer a greater likelihood of differences in ligand specificity between the clades: 17,20β-DHP and 4-pregen-17,20β,21-triol-3-one (20β-S) in fish, and P4 in frog and late-derived species. Interestingly, the known biologically active progestin in zebrafish, 17,20β-DHP, and P4 at the physiological relevant range of 1–10 nM were the most potent activators of Pgr-mediated transactivation activities. Similar results have also been observed for eel Pgr [6, 7]. In contrast, P4 is the endogenous ligand for PGRs in mammals and Xenopus. In fact, a number of binding studies with mammalian PGRs report that progestins containing hydroxyl groups at the 17 or 20 position exhibit significantly lower binding than standard P4 [34–36]; this has been discussed in detail in Pinter and Thomas [37].
Alignment of the ligand-binding domains of different vertebrate models confirmed that nearly all the residues important for P4 binding were conserved (18 of 19). The only exception was a variation of a leucine residue to a valine residue in zebrafish Pgr and eel Pgr2 (human L887 to zebrafish V571). The significance of this valine substitution in changing the affinity for 17 and 20 hydroxyl groups of progestins is uncertain, because it is distant from the proposed location/interaction with P4 near carbons 8 and 14 [28]. A number of uniquely varied residues in teleosts (∼21) were identified that differed from conserved residues found across all other species, which could be used as a starting point for analyzing 17,20β-DHP binding in future studies. However, no unique or obvious binding pocket residues that may prefer the 17,20β-DHP ligand could be identified.
This study is the first to report a wide distribution of Pgr in the fish brain, which is similar to the broad expression patterns of PGR reported in diverse brain regions of mammals. The presence of Pgr proteins in the neuroendocrine regions agrees well with progestins' key functions in reproduction. However, the finding that Pgr is also present in other nonreproductive regions of the fish brain suggests that the actions of P4 extend far beyond reproduction in fish [38–40]. In mammals, neuronal survival, neurite outgrowth, and neurogenesis are just some of the recently identified neural functions of progestins. Some of these effects are mediated by neuroprogestins produced de novo in the brain. Indeed, paracrine effects of neurons and glias in the central nervous system have been suggested based on the presence of active steroid enzymes and high concentrations of progestins [39]. Astrocytes are considered to be main production sites of neuroprogestins in the mammalian brains [41]. Whether the same mechanisms occur in the fish brain is currently unknown, because the cellular localizations of the steroidogenic enzymes, with the exception of aromatase in the fish brain, are still unclear [41–45]. Aromatase is only expressed in radial glial cells, progenitors in the adult fish brain [25, 43–47]. It is not known whether PGR is expressed in the radial glial cells, but it is interesting to note that estradiol can increase PGR expression in some mammalian tissues [41, 48, 49]. The abundant expression of Pgr along the brain ventricles, where aromatase expression is maximal in fish brain [44], suggests that estradiol may regulate Pgr expression in fish brain. Further studies are required to validate this hypothesis and identify precise sites for synthesizing progestins in the brain of fish.
In the testis, strong expression of Pgr in zebrafish spermatogonia and spermatocytes suggests a potential role for the receptor in early germ cell development. As in the Japanese eel, 17,20β-DHP was found to be a potent inducer of testicular germ cell proliferation in zebrafish and as potent as the endogenous androgen 11-KT [6, 11, 24]. In eel it has been demonstrated that 17,20β-DHP is an essential factor for meiotic initiation in spermatogenetic cells [11]. Colocalization of zebrafish Pgr in proliferating spermatogonia and early spermatocytes for the first time implies a direct role for Pgr in progestin-mediated germ cell proliferation. Interestingly, deficiencies of PGR expression in human testis have recently been linked to infertility [50]. Further studies should make zebrafish testicular culture a unique system for examining the influence of progestins on testicular germ cell development.
In the ovary, primary expression of the Pgr protein and transcript in the follicular layer surrounding oocytes, similar to the PGR expression found in other vertebrates, implies an important and comparable role in progestin regulation of oocyte development and ovulation through the follicular cell layers [51–55]. Immunostaining of the zebrafish Pgr antibody was observed in nuclei of early stage I and II oocytes but not in the nucleus of late stage III and stage IV immature oocytes, suggesting a role for Pgr in growth and development of early-stage oocytes, and in follicular development in later stages. In early stage I and II oocytes, in situ localization in the cytoplasm suggests that pgr mRNA may be processed into protein in the cytoplasm and transported back into the nucleus.
In addition to its well-known function as a transcription factor, Pgr may also be involved in nongenomic progestin signaling during final oocyte maturation. Interestingly, microinjection of pgr transcripts into immature oocytes accelerated oocyte maturation in zebrafish oocytes (Hanna et al., unpublished results) and in Xenopus oocytes [8, 9]. However, the majority of the zebrafish Pgr protein, and all detectable mRNA, were restricted to the follicular cells associated with stage IV oocytes, diminishing Pgr's potential role in oocyte maturation. Furthermore, membrane localization of progestin receptors is important for their ability to mediate nongenomic progestin signaling and oocyte maturation [1, 56–59]. Interestingly, a recently identified palmitoylation motif, proposed to be important for membrane localization of a number of nuclear steroid receptors (FXCXXXXLL [60]) varied by one amino acid in the zebrafish Pgr ligand-binding domain (FXCXXXXML; Fig. 2), potentially inhibiting Pgr membrane localization and subsequent nongenomic signaling in this species.
As a final point, membrane progestin receptors (mPRs; or official symbol Paqrs) have recently been identified and demonstrated to be mediators of nongenomic progestin signaling in oocyte maturation in spotted seatrout [26], zebrafish [1, 19, 26, 27], goldfish [61], and Xenopus [62]. The zebrafish mPR proteins are localized mainly in the same progestin-responsive reproductive tissues as Pgr proteins, and they are activated by the same ligand, 17,20β-DHP [63], leading to speculation of interactions and cross-talk between mPRs and Pgr. Colocalization, interaction, and cross-talk in signaling between mPR proteins and classical Pgr proteins have been demonstrated in human myometrial cells in a recent study [64]. The interactions between membrane and nuclear receptor signaling have been suggested based on mPR-dependent Pgr transactivation and downregulation of steroid receptor coactivator (SRC2) by mPRα in human myometrial cells [64].
Zebrafish, in which both pgr and mPR genes have now been identified, are a good model for studying nongenomic and genomic actions of progestins. Many unresolved controversies remain, including the extent of signaling involvement of each progestin receptor in reproductive development, the primary progestin receptor involved in nongenomic signaling, and the potential interaction of mPR proteins and Pgr proteins. It is our hope that future studies using the Pgr identified in this paper will help answer some of the critical questions that remain regarding genomic and nongenomic progestin actions.
Supplementary Material
Acknowledgments
We would like to graciously thank Takeshi Miura and Chiemi Miura from Ehime University, Ehime, Japan, for their congenial assistance with culture and staining methods of zebrafish gonads, and Kathryn Horwitz from the University of Colorado, Denver, Colorado, for providing us with the Pre2-TATA-luciferase and MMTV-luciferase vectors.
Footnotes
Supported in part by the National Science Foundation grant nos. IBN-0315349 to Y.Z. and OISE-0813146 to R.N.H.; East Carolina University Research and Creative Activity grant no. 2003-2010 to Y.Z.; East Carolina University Thomas Harriot College of Arts and Sciences Research Award to Y.Z.; East Carolina University Division of Research & Graduate Studies Research 2007 Development Award to Y.Z.; National Institutes of Health grant no. ESO12961 to P.T.; and funding from the Agence Nationale de la Recherche (Contaminants, Ecosystems, Health 2008-11: NEED) and Centre National de la Recherche Scientifique to O.K.
REFERENCES
- Zhu Y, Hanna R, Schaaf M, Spaink H, Thomas P.Candidates for membrane progestin receptors—past approaches and future challenges. Comp Biochem Physiol C Toxicol Pharmacol 2008; 148: 381–389. [DOI] [PubMed] [Google Scholar]
- Rondell P.Role of steroid synthesis in the process of ovulation. Biol Reprod 1974; 10: 199–215. [DOI] [PubMed] [Google Scholar]
- Graham J, Clarke C.Physiological action of progesterone in target tissues. Endocr Rev 1997; 18: 502–519. [DOI] [PubMed] [Google Scholar]
- Mulac-Jericevic B, Mullinax R, DeMayo F, Lydon J, Conneely O.Subgroup of reproductive functions of progesterone mediated by progesterone receptor-B isoform. Science 2000; 289: 1751–1754. [DOI] [PubMed] [Google Scholar]
- Hasegawa J, Yanaihara A, Iwasaki S, Otsuka Y, Negishi M, Akahane T, Okai T.Reduction of progesterone receptor expression in human cumulus cells at the time of oocyte collection during IVF is associated with good embryo quality. Hum Reprod 2005; 20: 2194–2200. [DOI] [PubMed] [Google Scholar]
- Todo T, Ikeuchi T, Kobayashi T, Kajiura-Kobayashi H, Suzuki K, Yoshikuni M, Yamauchi K, Nagahama Y.Characterization of a testicular 17alpha,20beta-dihydroxy-4-pregnen-3-one (a spermiation-inducing steroid in fish) receptor from a teleost, Japanese eel (Anguilla japonica). FEBS Lett 2000; 465: 12–17. [DOI] [PubMed] [Google Scholar]
- Ikeuchi T, Todo T, Kobayashi T, Nagahama Y.A novel progestogen receptor subtype in the Japanese eel, Anguilla japonica. FEBS Lett 2002; 510: 77–82. [DOI] [PubMed] [Google Scholar]
- Bayaa M, Booth R, Sheng Y, Liu X.The classical progesterone receptor mediates Xenopus oocyte maturation through a nongenomic mechanism. Proc Natl Acad Sci U S A 2000; 97: 12607–12612. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tian J, Kim S, Heilig E, Ruderman J.Identification of XPR-1, a progesterone receptor required for Xenopus oocyte activation. Proc Natl Acad Sci U S A 2000; 97: 14358–14363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu X, Ma C, Hamam A, Liu X.Transcription-dependent and transcription-independent functions of the classical progesterone receptor in Xenopus ovaries. Dev Biol 2005; 283: 180–190. [DOI] [PubMed] [Google Scholar]
- Miura T, Higuchi M, Ozaki Y, Ohta T, Miura C.Progestin is an essential factor for the initiation of the meiosis in spermatogenetic cells of the eel. Proc Natl Acad Sci U S A 2006; 103: 7333–7338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oshima K, Gorbman A.Influence of thyroxine and steroid hormones on spontaneous and evoked unitary activity in the olfactory bulb of goldfish. Gen Comp Endocrinol 1966; 7: 482–491. [DOI] [PubMed] [Google Scholar]
- Sundararaj B, Goswami S.Effects of estrogen, progesterone, and testosterone on the pituitary and ovary of catfish, Heteropneustes fossilis (Bloch). J Exp Zool 1968; 169: 211–219. [DOI] [PubMed] [Google Scholar]
- van den Hurk R.Effects of steroids on gonadotropic (GTH) cells in the pituitary of rainbow trout, Salmo gairdneri, shortly after hatching. Cell Tissue Res 1982; 224: 361–368. [DOI] [PubMed] [Google Scholar]
- Mathews S, Khan I, Thomas P.Effects of the maturation-inducing steroid on LH secretion and the GnRH system at different stages of the gonadal cycle in Atlantic croaker. Gen Comp Endocrinol 2002; 126: 287–297. [DOI] [PubMed] [Google Scholar]
- Faivre E, Skildum A, Pierson-Mullany L, Lange C.Integration of progesterone receptor mediated rapid signaling and nuclear actions in breast cancer cell models: role of mitogen-activated protein kinases and cell cycle regulators. Steroids 2005; 70: 418–426. [DOI] [PubMed] [Google Scholar]
- Boonyaratanakornkit V, McGowan E, Sherman L, Mancini M, Cheskis B, Edwards D.The role of extranuclear signaling actions of progesterone receptor in mediating progesterone regulation of gene expression and the cell cycle. Mol Endocrinol 2007; 21: 359–375. [DOI] [PubMed] [Google Scholar]
- Clelland E, Peng C.Endocrine/paracrine control of zebrafish ovarian development. Mol Cell Endocrinol 2009; 312: 42–52. [DOI] [PubMed] [Google Scholar]
- Hanna R, Pang Y, Thomas P, Zhu Y.Cell-surface expression, progestin binding, and rapid nongenomic signaling of zebrafish membrane progestin receptors alpha and beta in transfected cells. J Endocrinol 2006; 190: 247–260. [DOI] [PubMed] [Google Scholar]
- Pinter J, Thomas P.Characterization of a progestogen receptor in the ovary of the spotted seatrout, Cynoscion nebulosus. Biol Reprod 1995; 52: 667–675. [DOI] [PubMed] [Google Scholar]
- Takimoto G, Hovland A, Tasset D, Melville M, Tung L, Horwitz K.Role of phosphorylation on DNA binding and transcriptional functions of human progesterone receptors. J Biol Chem 1996; 271: 13308–13316. [DOI] [PubMed] [Google Scholar]
- Nordeen S.Luciferase reporter gene vectors for analysis of promoters and enhancers. Biotechniques 1988; 6: 454–458. [PubMed] [Google Scholar]
- Zhu Y, Stiller J, Shaner M, Baldini A, Scemama J, Capehart A.Cloning of somatolactin alpha and beta cDNAs in zebrafish and phylogenetic analysis of two distinct somatolactin subtypes in fish. J Endocrinol 2004; 182: 509–518. [DOI] [PubMed] [Google Scholar]
- Miura T, Yamauchi K, Takahashi H, Nagahama Y.Hormonal induction of all stages of spermatogenesis in vitro in the male Japanese eel (Anguilla japonica). Proc Natl Acad Sci U S A 1991; 88: 5774–5778. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pellegrini E, Mouriec K, Anglade I, Menuet A, Le Page Y, Gueguen MM, Marmignon MH, Brion F, Pakdel F, Kah O.Identification of aromatase-positive radial glial cells as progenitor cells in the ventricular layer of the forebrain in zebrafish. J Comp Neurol 2007; 501: 150–167. [DOI] [PubMed] [Google Scholar]
- Zhu Y, Rice C, Pang Y, Pace M, Thomas P.Cloning, expression, and characterization of a membrane progestin receptor and evidence it is an intermediary in meiotic maturation of fish oocytes. Proc Natl Acad Sci U S A 2003; 100: 2231–2236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu Y, Bond J, Thomas P.Identification, classification, and partial characterization of genes in humans and other vertebrates homologous to a fish membrane progestin receptor. Proc Natl Acad Sci U S A 2003; 100: 2237–2242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Williams S, Sigler P.Atomic structure of progesterone complexed with its receptor. Nature 1998; 393: 392–396. [DOI] [PubMed] [Google Scholar]
- Wullimann MF, Rupp B, Reichert H. Neuroanatomy of the Zebrafish Brain: A Topological Atlas. Basel, Switzerland:: Birkhèauser Verlag;; 1996. [Google Scholar]
- Peluso J.Progesterone as a regulator of granulosa cell viability. J Steroid Biochem Mol Biol 2003; 85: 167–173. [DOI] [PubMed] [Google Scholar]
- Richards J, Russell D, Ochsner S, Hsieh M, Doyle K, Falender A, Lo Y, Sharma S.Novel signaling pathways that control ovarian follicular development, ovulation, and luteinization. Recent Prog Horm Res 2002; 57: 195–220. [DOI] [PubMed] [Google Scholar]
- Bardet P, Horard B, Robinson-Rechavi M, Laudet V, Vanacker J.Characterization of oestrogen receptors in zebrafish (Danio rerio). J Mol Endocrinol 2002; 28: 153–163. [DOI] [PubMed] [Google Scholar]
- Hossain M, Larsson A, Scherbak N, Olsson P, Orban L.Zebrafish androgen receptor: isolation, molecular, and biochemical characterization. Biol Reprod 2008; 78: 361–369. [DOI] [PubMed] [Google Scholar]
- Kontula K, Jänne O, Vihko R, de Jager E, de Visser J, Zeelen F.Progesterone-binding proteins: in vitro binding and biological activity of different steroidal ligands. Acta Endocrinol (Copenh) 1975; 78: 574–592. [DOI] [PubMed] [Google Scholar]
- McGuire J, Bariso C, Shroff A.Interaction between steroids and a uterine progestogen specific binding macromolecule. Biochemistry 1974; 13: 319–322. [DOI] [PubMed] [Google Scholar]
- Neelima M, Bhaduri A.Progesterone receptor binding of steroidal and nonsteroidal compounds. Prog Drug Res 1986; 30: 151–188. [DOI] [PubMed] [Google Scholar]
- Pinter J, Thomas P.The ovarian progestogen receptor in the spotted seatrout, Cynoscion nebulosus, demonstrates steroid specificity different from progesterone receptors in other vertebrates. J Steroid Biochem Mol Biol 1997; 60: 113–119. [DOI] [PubMed] [Google Scholar]
- Brinton RD, Thompson RF, Foy MR, Baudry M, Wang J, Finch CE, Morgan TE, Pike CJ, Mack WJ, Stanczyk FZ, Nilsen J.Progesterone receptors: form and function in brain. Front Neuroendocrinol 2008; 29: 313–339. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Charalampopoulos I, Remboutsika E, Margioris AN, Gravanis A.Neurosteroids as modulators of neurogenesis and neuronal survival. Trends Endocrinol Metab 2008; 19: 300–307. [DOI] [PubMed] [Google Scholar]
- Wang JM, Irwin RW, Liu L, Chen S, Brinton RD.Regeneration in a degenerating brain: potential of allopregnanolone as a neuroregenerative agent. Curr Alzheimer Res 2007; 4: 510–517. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Micevych P, Sinchak K.Estradiol regulation of progesterone synthesis in the brain. Mol Cell Endocrinol 2008; 290: 44–50. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Forlano PM, Deitcher DL, Myers DA, Bass AH.Anatomical distribution and cellular basis for high levels of aromatase activity in the brain of teleost fish: aromatase enzyme and mRNA expression identify glia as source. J Neurosci 2001; 21: 8943–8955. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Menuet A, Anglade I, Le Guevel R, Pellegrini E, Pakdel F, Kah O.Distribution of aromatase mRNA and protein in the brain and pituitary of female rainbow trout: comparison with estrogen receptor alpha. J Comp Neurol 2003; 462: 180–193. [DOI] [PubMed] [Google Scholar]
- Menuet A, Pellegrini E, Brion F, Gueguen MM, Anglade I, Pakdel F, Kah O.Expression and estrogen-dependent regulation of the zebrafish brain aromatase gene. J Comp Neurol 2005; 485: 304–320. [DOI] [PubMed] [Google Scholar]
- Strobl-Mazzulla PH, Moncaut NP, Lopez GC, Miranda LA, Canario AV, Somoza GM.Brain aromatase from pejerrey fish (Odontesthes bonariensis): cDNA cloning, tissue expression, and immunohistochemical localization. Gen Comp Endocrinol 2005; 143: 21–32. [DOI] [PubMed] [Google Scholar]
- Adolf B, Chapouton P, Lam CS, Topp S, Tannhauser B, Strahle U, Gotz M, Bally-Cuif L.Conserved and acquired features of adult neurogenesis in the zebrafish telencephalon. Dev Biol 2006; 295: 278–293. [DOI] [PubMed] [Google Scholar]
- Lam CS, Marz M, Strahle U.gfap and nestin reporter lines reveal characteristics of neural progenitors in the adult zebrafish brain. Dev Dyn 2009; 238: 475–486. [DOI] [PubMed] [Google Scholar]
- Micevych P, Sinchak K.Synthesis and function of hypothalamic neuroprogesterone in reproduction. Endocrinology 2008; 149: 2739–2742. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Micevych P, Soma KK, Sinchak K.Neuroprogesterone: key to estrogen positive feedback? Brain Res Rev 2008; 57: 470–480. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Abid S, Gokral J, Maitra A, Meherji P, Kadam S, Pires E, Modi D.Altered expression of progesterone receptors in testis of infertile men. Reprod Biomed Online 2008; 17: 175–184. [DOI] [PubMed] [Google Scholar]
- Gava N, Clarke C, Byth K, Arnett-Mansfield R, deFazio A.Expression of progesterone receptors A and B in the mouse ovary during the estrous cycle. Endocrinology 2004; 145: 3487–3494. [DOI] [PubMed] [Google Scholar]
- Natraj U, Richards J.Hormonal regulation, localization, and functional activity of the progesterone receptor in granulosa cells of rat preovulatory follicles. Endocrinology 1993; 133: 761–769. [DOI] [PubMed] [Google Scholar]
- Teilmann S, Clement C, Thorup J, Byskov A, Christensen S.Expression and localization of the progesterone receptor in mouse and human reproductive organs. J Endocrinol 2006; 191: 525–535. [DOI] [PubMed] [Google Scholar]
- Shao R, Markström E, Friberg P, Johansson M, Billig H.Expression of progesterone receptor (PR) A and B isoforms in mouse granulosa cells: stage-dependent PR-mediated regulation of apoptosis and cell proliferation. Biol Reprod 2003; 68: 914–921. [DOI] [PubMed] [Google Scholar]
- Pinter J, Thomas P.Induction of ovulation of mature oocytes by the maturation-inducing steroid 17,20beta,21-trihydroxy-4-pregnen-3-one in the spotted seatrout. Gen Comp Endocrinol 1999; 115: 200–209. [DOI] [PubMed] [Google Scholar]
- Younglai E, Wu Y, Foster W, Lobb D, Price T.Binding of progesterone to cell surfaces of human granulosa-lutein cells. J Steroid Biochem Mol Biol 2006; 101: 61–67. [DOI] [PubMed] [Google Scholar]
- Trant J, Thomas P, Shackleton C.Identification of 17 alpha,20 beta,21-trihydroxy-4-pregnen-3-one as the major ovarian steroid produced by the teleost Micropogonias undulatus during final oocyte maturation. Steroids 1986; 47: 89–99. [DOI] [PubMed] [Google Scholar]
- Thomas P, Trant J.Evidence that 17,20β,21-trihydroxy-4-pregnen-3-one is a maturation-inducing steroid in spotted seatrout. Fish Physiol Biochem 1989; 7: 185–191. [DOI] [PubMed] [Google Scholar]
- Patiño R, Thomas P.Characterization of membrane receptor activity for 17 alpha, 20 beta, 21-trihydroxy-4-pregnen-3-one in ovaries of spotted seatrout (Cynoscion nebulosus). Gen Comp Endocrinol 1990; 78: 204–217. [DOI] [PubMed] [Google Scholar]
- Pedram A, Razandi M, Sainson R, Kim J, Hughes C, Levin E.A conserved mechanism for steroid receptor translocation to the plasma membrane. J Biol Chem 2007; 282: 22278–22288. [DOI] [PubMed] [Google Scholar]
- Tokumoto M, Nagahama Y, Thomas P, Tokumoto T.Cloning and identification of a membrane progestin receptor in goldfish ovaries and evidence it is an intermediary in oocyte meiotic maturation. Gen Comp Endocrinol 2006; 145: 101–108. [DOI] [PubMed] [Google Scholar]
- Josefsberg Ben-Yehoshua L, Lewellyn A, Thomas P, Maller J.The role of Xenopus membrane progesterone receptor beta in mediating the effect of progesterone on oocyte maturation. Mol Endocrinol 2007; 21: 664–673. [DOI] [PubMed] [Google Scholar]
- Hanna R, Zhu Y.Expression of membrane progestin receptors in zebrafish (Danio rerio) oocytes, testis and pituitary. Gen Comp Endocrinol 2009; 161: 153–157. [DOI] [PubMed] [Google Scholar]
- Karteris E, Zervou S, Pang Y, Dong J, Hillhouse E, Randeva H, Thomas P.Progesterone signaling in human myometrium through two novel membrane G protein-coupled receptors: potential role in functional progesterone withdrawal at term. Mol Endocrinol 2006; 20: 1519–1534. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.