Abstract
Two new species of Amniculicola, A. immersa sp. nov. and A. parva sp. nov. from submerged wood in a freshwater environment in Denmark and France are respectively described and illustrated. In addition, partial 28S rDNA sequence data is analysed to investigate their phylogenetic relationships with other pleosporalean taxa. All presently known Amniculicola species, A. immersa, A. lignicola and A. parva, form a robust clade together with the anamorphic species Anguillospora longissima, Spirosphaera cupreorufescens and Repetophragma ontariense. These six species, which are all from freshwater and mostly from Europe, constitute a well-supported group containing Pleospora rubicunda and Massariosphaeria typhicola. This putative monophyletic assemblage may represent an aquatic group in the Pleosporales. It is also pertinent that all five ascomycete taxa in this group stain their host substrates purple.
Keywords: anamorphs, freshwater fungi, phylogeny, pseudoparaphyses, rDNA
INTRODUCTION
There has been much recent interest in lignicolous freshwater fungi and many new species have been described from France and Belgium (Cai et al. 2008, Zhang et al. 2008a, b, in press). Amniculicola was introduced to accommodate A. lignicola, which was isolated from submerged wood in freshwater in France (Zhang et al. 2008b). Amniculicola is characterised by ascomata with slit-like ostioles, thin, branching and anastomosing hamathecia, cylindrical asci, and hyaline, 1–3-septate ascospores. During our studies of lignicolous freshwater fungi in France (Zhang et al. 2008a, b, in press), we collected several other taxa including two new species of Amniculicola. These two new species as well as Massariosphaeria typhicola and Pleospora rubicunda stain their woody substrates purple, a character that is rarely discussed but may have taxonomic importance.
In this paper we introduce two new species of Amniculicola, i.e., A. immersa and A. parva based on both morphological and phylogenetic characters. The close phylogenetic association of Amniculicola to several freshwater anamorphic fungi is also discussed.
MATERIALS AND METHODS
Sample collection and specimen examination
Fresh fungal specimens were collected on submerged wood in Denmark and France in 2006 and 2007 by J. Fournier, and returned to the laboratory in plastic bags for study. Ascomata were visible on the natural wood without prior incubation. Samples were processed and examined following the method described in Tsui et al. (2000). Observations and photographs were prepared from material mounted in water, Cotton blue or Indian ink. Other cultures used in this study were obtained from the Centraalbureau voor Schimmelcultures in the Netherlands (CBS). Newly obtained nucleotide sequences were deposited in GenBank.
Fungal isolates and DNA extraction
Isolates were grown on potato-dextrose agar (PDA) and malt extract agar (MEA) (Crous et al. 2009) and total genomic DNA was extracted from mycelia following the protocols as outlined by Cai et al. (2006) and Shenoy et al. (2007). The Forensic Kit (UltraClean™ Forensic Kit, Cambio) was used to directly extract DNA from specimens.
DNA amplification and sequencing
DNA amplification was performed by PCR. For partial large subunit (28S) rDNA amplification, LROR and LR5 primers (Vilgalys & Hester 1990) were used. Amplification reactions were performed in a 50 μL reaction volume as outlined by Shenoy et al. (2007). The purified PCR products were sequenced using the above mentioned primers in an Applied Biosystem 3730 DNA analyser at the Genome Research Centre of the University of Hong Kong.
Sequence alignment and phylogenetic analyses
Sequences generated from different primers were analysed with other sequences obtained from the GenBank. A Blast search was performed to find the possible sister groups of the two new species. In addition, fungal members from different families of the Pleosporales were also included in the analyses. Multiple alignment was done in BioEdit (Hall 2005) and Clustal X (Thompson et al. 1997) and analyses were performed in PAUP v4.0b10 (Swofford 2002). Prior to phylogenetic analysis, ambiguous sequences at the start and the end were deleted and gaps manually adjusted to optimise alignment. Maximum Parsimony (MP) was conducted using heuristic searches as implemented in PAUP, with the default options method. Analyses were done under different parameters of maximum parsimony criteria as outlined in Zhang et al. (2008a). Clade stability was assessed in a bootstrap analysis with 1 000 replicates, random sequence additions with maxtrees set to 1 000 and other default parameters as implemented in PAUP. Trees were viewed in Treeview (Page 1996). The terminal taxa of the phylogenetic tree were colour coded following Zhang et al. (2008a).
RESULTS
Taxonomy
Amniculicola Yin. Zhang & K.D. Hyde, Mycol. Res. 112: 1189. 2008.
Anamorphs. Possibly Anguillospora longissima (Sacc. & P. Syd.) Ingold, Spirosphaera cupreorufescens Voglmayr and Repetophragma ontariense (Matsush.) W.P. Wu.
Added characters — Besides the characters outlined by Zhang et al. (2008b), the purple staining of the woody substrates is another character regarded as typical for this group of fungi.
KEY TO SPECIES OF AMNICULICOLA
-
1.
Immersed and rarely purplish ascomata with a slightly compressed papilla, and less melanised peridium……………A. immersa
-
1.
Nearly superficial ascomata at maturity with remarkable compressed papilla and elongated ostiole, peridium distinctly melanised……………2
-
2.
Asci longer than 140 μm, larger ascomata (350–450 × 300–500 μm), thicker peridium (40–55 μm) and broader ascospores (6–)8(–9) μm……………A. lignicola
-
2.
Asci shorter than 130 μm, smaller ascomata (260–380 μm high × 260–360 μm diam), thinner peridium, 20–35(–40) μm, and narrower ascospores, 4.5–6.5 μm…………… A. parva
Amniculicola immersa Yin. Zhang, J. Fourn., Crous & K.D. Hyde, sp. nov. — MycoBank MB512915; Fig. 1, 2
Fig. 1.
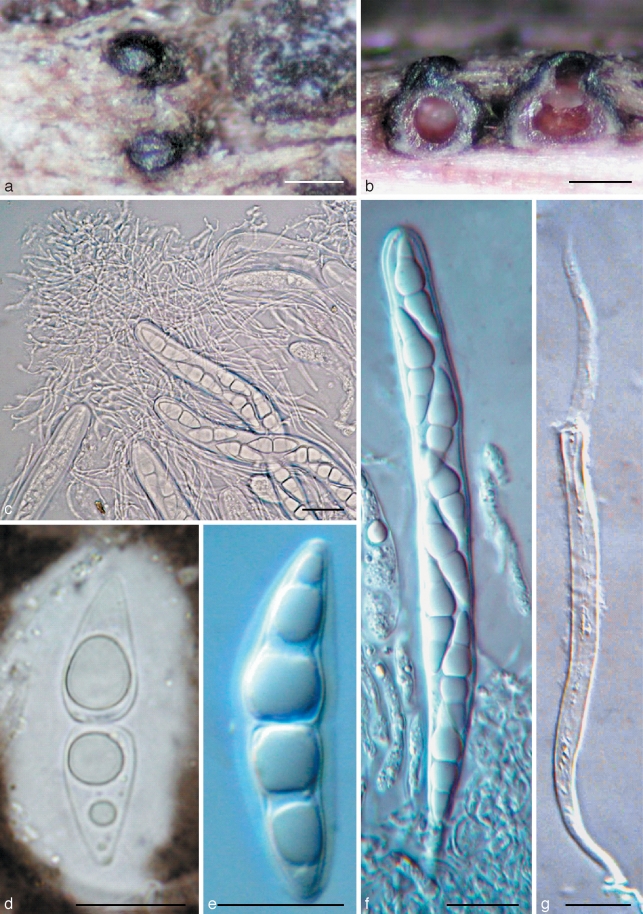
Amniculicola immersa (from holotype). a. Ascomata on host surface; b. habit section of ascomata. Note the purple colour in the ascomata and substrate; c. asci in pseudoparaphyses; d, e. ascospores with sheath; f. ascus; g. dehiscent ascus. c, e–g in water; d in Indian ink. — Scale bars: a = 0.5 mm; b = 200 μm; c–g = 20 μm.
Fig. 2.
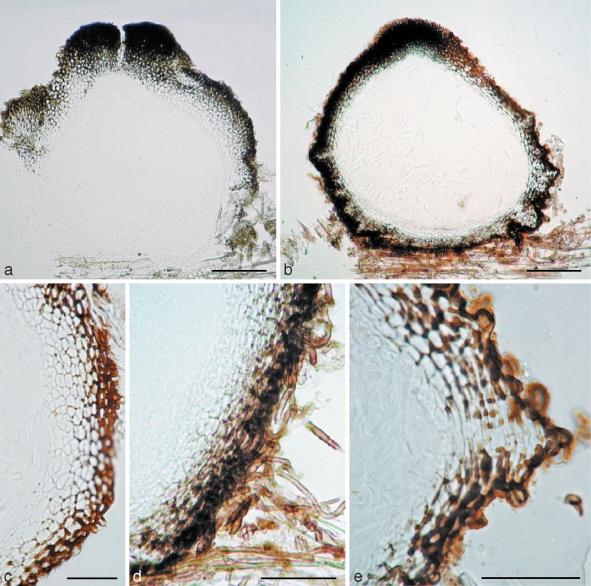
a–c. Amniculicola immersa (from holotype). a. Section of ascoma. Note the thick apical wall; b. section of ascoma. Note the purple colour in the ascomata and substrate; c. section of ascoma. Note the peridium structure. — d, e. Amniculicola parva (from holotype). d. Section of ascoma; e. section of the peridium near base. Note the tubercule. — Scale bars: a, b = 0.1 mm; c–e = 50 μm.
Ascomata 280–400(–450) μm alta × 300–420(–500) μm diam, globosa vel subglobosa, nigra, solitaria vel dispergere, erumpent. Peridium 45–65 μm crassum, bitapetum. Trabeculae 1–1.8 μm latae, hyalinae, gelatine circumdatae. Asci 150–180 × 13–18 μm, 8-spori, cylindrico-fusiformis, fissitunicati, pedicellati. Ascosporae 27–31(–36) × 7.5–8.5(–10) μm, uniseriatae, fusiformes, hyalinae, tunica gelatinosa praeditae.
Etymology. From Latin ‘immersa’, in reference to the mostly immersed ascoma.
Ascomata 280–400(–450) μm high × 300–420(–500) μm diam, solitary, scattered, or in small groups of 2–3, mostly immersed (immersion > 1/2) with a bluntly rounded apex slightly raised above host surface, coriaceous, appearing broadly ellipsoid from above, 120–170 μm long × 80–120 μm broad, globose to subglobose, black, roughened, apex with a inconspicuous slit-like ostiole, oriented in the axis of the wood fibres; underlying wood stained pale purple (Fig. 1a, b). Peridium 45–65 μm thick laterally, 30–35 μm thick at base, 80–100 μm thick at the apex, 2-layered, outer layer composed of small, heavily pigmented, thick-walled cells of textura angularis, cells 4–7 μm diam, cell wall 2–3 μm thick, cells at apex smaller and walls thicker, inner layer composed of hyaline thin-walled cells of textura angularis, 8–12 μm diam (Fig. 2a–c). Hamathecium of dense, very long trabeculate pseudoparaphyses, 1–1.8 μm broad, embedded in mucilage, anastomosing between and above the asci (Fig. 1c). Asci 150–180 × 13–18 μm (av. = 162.5 × 15.5 μm, n = 10), mostly 8-spored, rarely 4- or 6-spored, bitunicate, fissitunicate, cylindrical to narrowly fusiform, with a short, narrowed, twisted, bifurcate pedicel which is 15–20 μm long, with a truncate ocular chamber (Fig. 1d, e). Ascospores 27–31(–36) × 7.5–8.5(–10) μm (av. = 31.3 × 8.2 μm, n = 10), obliquely uniseriate and partially overlapping, broadly fusiform to fusiform with narrowly rounded ends, hyaline, 1–3-septate, deeply constricted at the median septum, the upper cell often longer and broader than the lower one, smooth, surrounded by an hyaline gelatinous sheath, 4–6 μm thick, visible in water and Indian ink, senescent ascospores have a pale brown verrucose wall (Fig. 1f, g).
Cultural characteristics — Colonies reaching 5 cm diam after 20 d growth on PDA at 25 °C, flat, with irregular to rhizoidal margin, white, reverse off-white to straw-yellow to orange-brown, the medium is stained straw-yellow after 2 mo.
Specimen examined. Denmark, Sjæland, Frederikskilde, Suserup Skove, Tystrup Lake, on submerged wood of Salix sp., 25 May 2007, leg., det. J. Fournier CBS H-20226 holotype, culture ex-type CBS 123083.
Amniculicola parva Yin. Zhang, J. Fourn., Crous & K.D. Hyde, sp. nov. — MycoBank MB512916; Fig. 2, 3
Fig. 3.
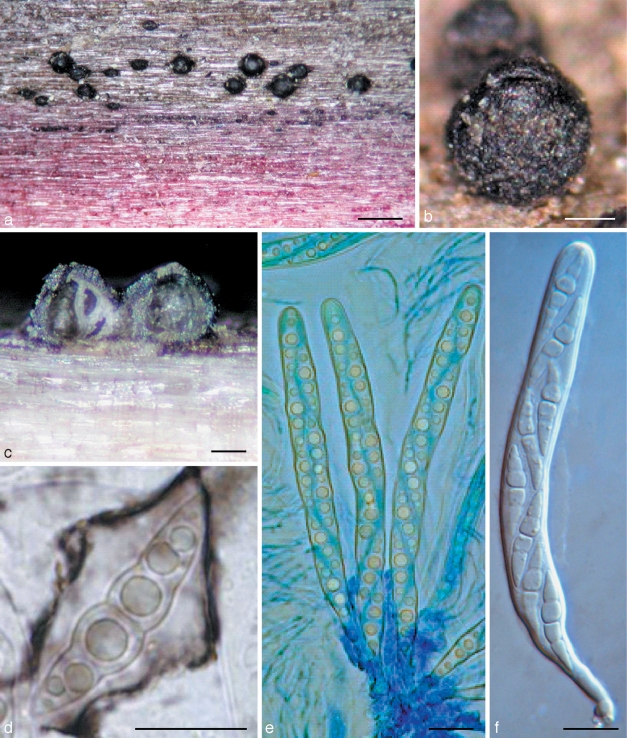
Amniculicola parva (from holotype). a, b. Ascomata on host surface. Note the purple substrate; c. habit section of ascoma; d. ascospores with sheath; e. asci in thin anastomosing pseudoparaphyses; f. ascus. d in Indian ink; e in cotton blue; f in water. — Scale bars: a = 0.5 mm; b, c = 0.1 mm; d–f = 10 μm.
Ascomata 260–380 μm alta × 260–360 μm diam, subglobosa vel conica, nigra, solitaria vel gregariculus, superficialra. Apex fissura. Peridium 20–35 (–40) μm crassum. Trabeculae 0.8–1.5 μm latae, hyalinae, gelatine circumdatae. Asci 100–130 × 8–12 μm, 8-spori, cylindrico-fusiformis, fissitunicati, pedicellati. Ascosporae (23–)27–32 × 4.5–6.5 μm, uniseriatae, fusiformes, uniseptatae, hyalinae, tunica gelatinosa praeditae.
Etymology. From Latin ‘parva’, in reference to the ‘small ascomata’.
Ascomata 260–380 μm high × 260–360 μm diam, solitary to gregarious, superficial, coriaceous, with basal wall remaining immersed in host tissue, subglobose, broadly or narrowly conical, often laterally flattened, with a flattened base not easily removed from the substrate, black, with tubercules, often bearing remnants of wood fibres, the underlying wood occasionally stained purple; broadly round apex with a slit-like ostiole, 110–150 μm long (Fig. 3a–c). Peridium 20–35(–40) μm thick at the sides, up to 55 μm thick at the apex, thinner at the base, peridium at sides textura angularis, 2-layered, outer layer dark-brown, 8–20 μm thick, composed of several rows of small thick-walled cells; cell wall 2–3 μm thick, with dark brown hyphal filaments 5–6 μm broad penetrating the wood at base; inner layer 25–35 μm thick, composed of larger cells 8–12 μm diam, irregularly pigmented, with columns of flattened hyaline cells forming ridges at base angles; apex composed of small, thick-walled, almost opaque cells (Fig. 2d, e). Hamathecium of dense, very long trabeculate pseudoparaphyses, 0.8–1.5 μm broad, embedded in mucilage (Indian Ink), anastomosing and branching between and above the asci. Asci 100–130 × 8–12 μm (av. = 116.5 × 9.3 μm, n = 10), 8-spored, bitunicate, fissitunicate, cylindrical to narrowly fusiform, with a short, narrowed, twisted, bifurcate pedicel which is 15–20 μm long, with a truncate ocular chamber (Fig. 3d, e). Ascospores (23–)27–32 × 4.5–6.5 μm (av. = 28 × 5.4 μm, n = 10), obliquely uniseriate and partially overlapping, narrowly fusiform to fusiform with narrowly rounded to acute ends, hyaline, 1(–3)-septum, deeply constricted at the median septum, often curved, the upper cell often shorter and broader than the lower one, smooth, with a wide gelatinous sheath 2–6 μm thick, visible in both water and India ink, at times drawn out at both ends; senescent ascospores have a pale brown, verrucose wall (Fig. 3f).
Cultural characteristics — Colonies reaching 5 cm diam after 20 d growth on MEA at 25 °C; raised, woolly, grey, with irregular to rhizoidal margin, reverse grey to black.
Specimens examined. France, Ariège, Rimont, Peyrau, 400 m, on submerged wood of Alnus glutinosa, 26 July 2006, leg., det. J. Fournier, CBS H-20227 holotype, culture ex-type CBS 123092; Ariège, Lescure, Bois du Pas du Baup, Volp brook, 500 m, on partly submerged wood of Fraxinus excelsior, 26 Oct. 2006, leg., det. J. Fournier, IFRD2019; Haute Garonne, Martres Tolosane, Balet, artificial lake, 250 m, on submerged wood of Salix sp., 17 May 2008, leg. J. Fournier, X. Besombes & M. Delpont, IFRD 2020.
Phylogenetic analyses
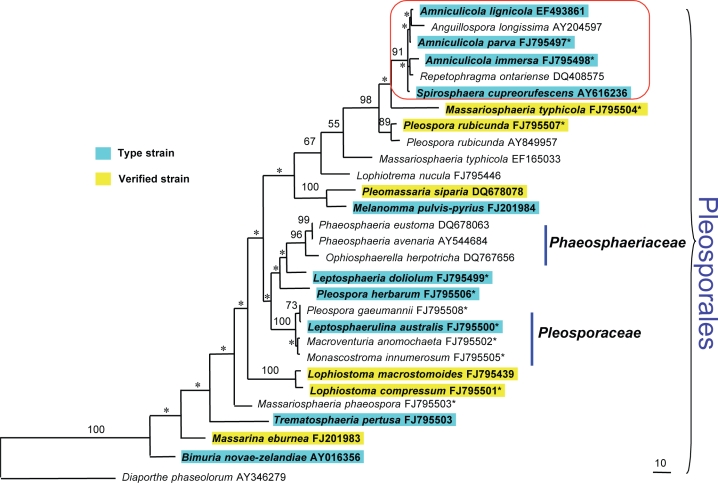
In total, 27 taxa (29 strains) are included in the phylogenetic analysis, with 26 taxa (28 strains) being pleosporalean taxa. In order to indicate the phylogenetic relationship of Amniculicola with other members of Pleosporales, species from 20 genera of Pleosporales are included, as well as morphologically comparable genera such as Lophiostoma, Lophiotrema and Massarina. Of the included taxa, 15 species are generic types. Of the 29 strains used here, 10 are type strains (34 %), and six (21 %) are verified strains (Fig. 4).
Fig. 4.
Maximum parsimony tree generated from partial 28S rDNA and with gaps as missing data. Designated outgroup is Diaporthe phaseolorum (AY346279). Bootstrap support values above 50 % (based on 1 000 replicates) are shown on nodes, and those lower than 50 % are marked as *. Newly generated sequences are marked as * on the right top corner of the GenBank accession numbers.
The 28S rDNA dataset includes 872 characters, out of which 16.7 % are parsimony-informative sites. Thirty-five characters are excluded. Maximum parsimony (MP) analysis with gaps treated as missing data; random addition sequence and TBR branch swapping yielded 1 parsimonious tree of 642 steps with CI, RI, RC and HI of 0.578, 0.707, 0.408 and 0.422, respectively. Diaporthe phaseolorum is the outgroup. Bootstrap values (generated from 1 000 replicates) from maximum parsimony analyses are shown on the upper nodes (Fig. 4). Phylogenies indicate that the three Amniculicola species cluster together with the anamorphic species Anguillospora longissima, Spirosphaera cupreorufescens and Repetophragma ontariense with 91 % MP bootstrap support. They form a well-supported group with Massariosphaeria typhicola and Pleospora rubicunda (Fig. 4).
DISCUSSION
Amniculicola was introduced as a monotypic genus represented by A. lignicola, which is characterised by ascomata with slit-like ostioles, thin, branching and anastomosing pseudoparaphyses, and 1-septate, strongly constricted, fusiform ascospores, with a wide gelatinous sheath (Zhang et al. 2008b). The two newly described species, Amniculicola immersa and A. parva, have all of the above characters. The immersed ascomata, less pigmented peridium, thicker apical peridium and broader asci morphologically distinguish A. immersa from A. lignicola (Table 1), while the smaller ascomata with thinner peridium and narrower ascospores typically swollen above the median septum can easily distinguish A. parva from the two other Amniculicola species (Table 1).
Table 1.
Morphological comparison of Amniculicola immersa, A. lignicola and A. parva (dimensions in μm).
| Taxa | Ascomata |
Asci | Ascospores | Peridium |
||
|---|---|---|---|---|---|---|
| Apex | Side | |||||
| A. immersa | Immersed | 280–400(–450) × 300–420(–500) | 150–180 × 13–18 | 27–31(–36) × 7.5–8.5(–10) | 80–100 | 45–65 |
| A. lignicola | Erumpent to nearly superficial | 350–450 × 300–500 | 140–184 × 9–10 | (20.5–)28–32 × (6–)8(–9) | 120 | 40–55 |
| A. parva | Superficial | 260–380 × 260–360 | 100–130 × 8–12 | (23–)27–32 × 4.5–6.5 | 55 | 20–35(–40) |
Phylogenetic analysis of 28S rDNA (Fig. 4) reveals that Amniculicola is related to Anguillospora longissima, Spirosphaera cupreorufescens and Repetophragma ontariense (anamorphic fungi), and they are unequivocally positioned within the Pleosporales (Fig. 4). According to Willoughby & Archer (1973), the teleomorph of Anguillospora longissima is an undescribed species of ‘Massarina’, which is characterised by the superficial ascomata (c. 500 μm diam), a dark peridium c. 20 μm thick; numerous, cylindrical, 8-spored asci (130–142 × 9–11 μm), and fusiform 1-septate ascospores (22–28 × 7–9 μm). The ostiole structure is unclear as it was described by Willoughby & Archer (1973) as ‘wide’, but all other morphological characters agree well with Amniculicola, but do not fit any of the presently described species. Thus the teleomorph of Anguillospora longissima may well be another new species of Amniculicola.
All of the three Amniculicola species, i.e., A. immersa, A. lignicola and A. parva, were collected from submerged wood in freshwater (Zhang et al. 2008b). Anguillospora longissima is also a common aquatic hyphomycete (Baschien et al. 2008). The strain of A. longissima (CCM F-00980) used here is from freshwater in Germany (Baschien et al. 2008), and Spirosphaera cupreorufescens is an aeroaquatic taxon from Austria (Voglmayr 2004). Although the location of the Repetophragma ontariense strain used here is unknown, the aquatic fungal status of R. ontariense is verified (Wu & Zhuang 2005). In addition, Massariosphaeria typhicola (CBS 123126) and Pleospora rubicunda (voucher-IFRD 2017) used here are also from submerged wood collected in freshwater in France. This might indicate that the freshwater environment is an important ecological character for this group of fungi.
The conspecificity of the two strains of Massariosphaeria typhicola used here is not well indicated (Fig. 4). The other strain of M. typhicola (CBS 609.86) used here was collected from Switzerland by A. Leuchtmann in 1981. Leuchtmann (1984) monographed Phaeosphaeria, and defined ascospores of M. typhicola as having 6–10 septa, while only 7 septa were observed in the ascospores of our collection (Fig. 5). The voucher specimen of CBS 609.86 could not be located; while the phylogenetic separation of these two strains might be because of M. typhicola s.l. was used by Leuchtmann (1984). Almost all of the strains in the Amniculicola clade are from Europe, and our results are in agreement with the hypothesis that ‘the distribution of fungi’ appears to be ‘parallel with their phylogenetic structure’ (Lumbsch et al. 2008).
Fig. 5.
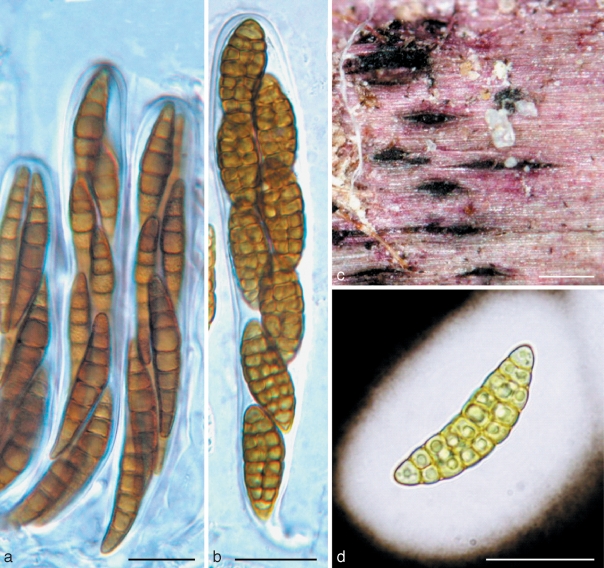
a. Massariosphaeria typhicola (from IFRD 2018). a. Asci in pseudoparaphyses. — b, c, d. Pleospora rubicunda (from IFRD 2017). b. Ascus; c. ascomata on the host surface; d. ascospore with wide sheath. a, b in water; d in Indian ink. — Scale bars: a, b, d = 20 μm; c = 0.5 mm.
The generic types of Massariosphaeria (M. phaeospora) and Pleospora (P. herbarum) are included in the analysis, and both Pleospora rubicunda and Massariosphaeria typhicola are phylogenetically distinct from their generic types and cluster basal to the Amniculicola group. Massariosphaeria is characterised by the heavily pigmented and multiseptate ascospores, and Pleospora is characterised by muriform ascospores, which are very different from the hyaline, 1-septate ascospores of Amniculicola. What is remarkable is that the fruiting bodies of Amniculicola immersa, A. lignicola, A. parva, Massariosphaeria typhicola and Pleospora rubicunda studied here were observed to consistently stain the wood purple, which might indicate the whole purple staining assemblage of taxa to represent a familial rank. The phylogenetic significance of the purple-stained substrate should be further explored, as it might be characteristic of metabolites produced by these fungal taxa. In xylariaceous taxa metabolite production and staining reactions have been shown to be related to their phylogeny (Stadler et al. 2004, Bitzer et al. 2008, Tang et al. 2009), suggesting that more attention should be paid to secondary metabolites in the Dothideomycetes.
Acknowledgments
The University of Hong Kong is thanked for providing Ying Zhang with a Postgraduate Studentship. Shaun Pennycook is thanked for his valuable suggestions on nomenclature. Jacques Fournier is grateful to Dr Thomas Læssøe (Department of Biology, University of Copenhagen) and the Danish Mycological Society for facilitating and funding his trip to Denmark in May 2007.
REFERENCES
- Baschien C, Manz W, Neu TR, Marvanová L, Szewzyk U. 2008. In situ detection of freshwater fungi in an alpine stream by new taxon-specific fluorescence in situ hybridization probes. Applied and Environmental Microbiology 74: 6427 – 6436 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bitzer J, Læssøe T, Fournier J, Kummer V, Decock C, Tichy H-V, Piepenbring M, Peršoh D, Stadler M. 2008. Affinities of Phylacia and the daldinoid Xylariaceae, inferred from chemotypes of cultures and ribosomal DNA sequences. Mycological Research 112: 251 – 270 [DOI] [PubMed] [Google Scholar]
- Cai L, Guo XY, Hyde KD. 2008. Morphological and molecular characterisation of a new anamorphic genus Cheirosporium, from freshwater in China. Persoonia 20: 53 – 58 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cai L, Jeewon R, Hyde KD. 2006. Phylogenetic investigations of Sordariaceae based on multiple gene sequences and morphology. Mycological Research 110: 137 – 150 [DOI] [PubMed] [Google Scholar]
- Crous PW, Verkleij GJM, Groenewald JZ, Samson RA. (eds). 2009. Fungal biodiversity. CBS Laboratory Manual Series 1 Centraalbureau voor Schimmelcultures, Utrecht, Netherlands: [Google Scholar]
- Hall T. Bioedit version 7.0.4. Department of Microbiology, North Carolina State University; 2005. [Google Scholar]
- Leuchtmann A. 1984. Über Phaeosphaeria Miyake und andere bitunicate Ascomyceten mit mehrfach querseptierten Ascosporen. Sydowia 37: 75 – 194 [Google Scholar]
- Lumbsch HT, Buchanan PK, May WM, Mueller GM. 2008. Phylogeography and biogeography of Fungi. Mycological Research 112: 423 – 424 [DOI] [PubMed] [Google Scholar]
- Page RDM. 1996. TreeView: an application to display phylogenetic trees on personal computers. Computer Applications in the Bosciences 12: 357 – 358 [DOI] [PubMed] [Google Scholar]
- Shenoy BD, Jeewon R, Lam WH, Bhat DJ, Than PP, Taylor PWJ, Hyde KD. 2007. Morpho-molecular characterisation and epitypification of Colletotrichum capsici (Glomerellaceae, Sordariomycetes), the causative agent of anthracnose in chilli. Fungal Diversity 27: 197 – 211 [Google Scholar]
- Stadler M, Ju Y-M, Rogers JD. 2004. Chemotaxonomy of Entonaema, Rhopalostroma and other Xylariaceae. Mycological Research 108: 239 – 256 [DOI] [PubMed] [Google Scholar]
- Swofford DL. 2002. PAUP*: Phylogenetic Analysis Using Parsimony (and other methods). Version 4b10 Sinauer Associates, Sunderland, USA: [Google Scholar]
- Tang AMC, Jeewon R, Hyde KD. 2009. A re-evaluation of the evolutionary relationships within the Xylariaceae based on ribosomal and protein-coding gene sequences. Fungal Diversity 34: 127 – 155 [Google Scholar]
- Thompson JD, Gibson TJ, Plewniak F, Jeanmougin F, Higgins DG. 1997. The Clustal X windows interface: flexible strategies for multiple sequence alignment aided by quality analysis tools. Nucleic Acids Research 24: 4876 – 4882 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsui CKM, Hyde KD, Hodgkiss IJ. 2000. Biodiversity of fungi on submerged wood in Hong Kong streams. Aquatic Microbial Ecology 21: 289 – 298 [Google Scholar]
- Vilgalys R, Hester M. 1990. Rapid genetic identification and mapping enzymatically amplified ribosomal DNA from several Cryptococcus species. Journal of Bacteriology 172: 4238 – 4246 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Voglmayr H. 2004. Spirosphaera cupreorufescens sp. nov., a rare aeroaquatic fungus. Studies in Mycology 50: 221 – 228 [Google Scholar]
- Willoughby LG, Archer JF. 1973. The fungal spora of a freshwater stream and its colonization pattern on wood. Freshwater Biology 3: 219 – 239 [Google Scholar]
- Wu WP, Zhuang WY. 2005. Sporidesmium, Endophragmiella and related genera from China. Fungal Diversity Research Series 15: 1 – 351 [Google Scholar]
- Zhang Y, Fournier J, Pointing SB, Hyde KD. 2008a. Are Melanomma pulvis-pyrius and Trematosphaeria pertusa congeneric? Fungal Diversity 33: 47 – 60 [Google Scholar]
- Zhang Y, Jeewon R, Fournier J, Hyde KD. 2008b. Multi-gene phylogeny and morphotaxonomy of Amniculicola lignicola: a novel freshwater fungus from France and its relationships to the Pleosporales. Mycological Research 112: 1186 – 1194 [DOI] [PubMed] [Google Scholar]
- Zhang Y, Wang HK, Fournier J, Crous PW, Pointing SB, Hyde KD. Towards a phylogenetic clarification of Lophiostoma / Massarina and morphologically similar genera in the Pleosporales. Fungal Diversity In press [Google Scholar]