Abstract
Actin waves that travel on the planar membrane of a substrate-attached cell underscore the capability of the actin system to assemble into dynamic structures by the recruitment of proteins from the cytoplasm. The waves have no fixed shape, can reverse their direction of propagation and can fuse or divide. Actin waves separate two phases of the plasma membrane that are distinguished by their lipid composition. The area circumscribed by a wave resembles in its phosphoinositide content the interior of a phagocytic cup, leading us to explore the possibility that actin waves are in-plane phagocytic structures generated without the localized stimulus of an attached particle. Consistent with this view, wave-forming cells were found to exhibit a high propensity for taking up particles. Cells fed rod-shaped particles produced elongated phagocytic cups that displayed a zonal pattern that reflected in detail the actin and lipid pattern of free-running actin waves. Neutrophils and macrophages are known to spread on surfaces decorated with immune complexes, a process that has been interpreted as “frustrated” phagocytosis. We suggest that actin waves enable a phagocyte to scan a surface for particles that might be engulfed.
Key words: actin waves; Dictyostelium; membrane tension; pattern formation; phagocytosis; PI3-kinase; PI(3,4,5)P3; self-organization
Introduction
The actin cortex of a cell is a dynamic organelle capable of forming a variety of membrane-anchored structures specialized in cell motility, substrate-interaction, division or intracellular transport. The differentiation of the cell cortex into distinct areas can be induced by external signals, but it can also occur autonomously. For instance, the differentiation of a motile cell into a front and a tail does not require any signal from the environment,1 although the orientation of this differentiation may be determined by external gradients. Filopodia are also spontaneously formed in the absence of a trigger at the site of their protrusion, and actin waves arise spontaneously along the plasma membrane of a cell on a planar surface.2 On the other hand, the formation of a phagocytic cup is locally induced by the attachment of a particle to the cell surface and mediated by trans-membrane signals.3,4
The goal of the present report is to link two activities of the actin system to each other, the self-organization of actin waves and the induction of phagocytic cups. In phagocytosis a particle may be taken up as a nutrient or as a stimulant of immune responses. Engulfment of the particle is initiated by the accumulation of actin in a protrusion of the cell surface, the phagocytic cup, which extends along and finally closes around the particle. Actin filaments are enriched at the extending phagocytic cup, especially at its rim, and disassemble when the particle is completely engulfed.5–9 The differentiation of the cell cortex into a central area delineated by the rim of the cup is directed by the site of particle attachment, and the size of this area is controlled by the volume of the particle. Quite in contrast, actin waves are formed on the planar membrane area of a substrate-attached cell. They are propagated in changing directions over the inner surface of the membrane in contact with a uniform substrate, which means that the waves have no fixed position or shape.2,10 In addressing the role of these dynamic actin structures, we compare cells in a state of extensive wave generation with cells that are in the process of phagocytosing yeast particles.
Our studies were conducted using cells of Dictyostelium as professional phagocytes. In these microbial cells actin assembly and phosphoinositide signaling play similar roles in phagocytosis as in mammalian macrophages. Phosphoinositides are concentrated at the cytosolic face of membranes, where they act as key players in signaling pathways controlling phagocytosis and other processes that involve remodeling of the cytoskeleton and membranes.11–14 By direct interaction with specific binding modules, e.g., pleckstrin homology (PH) domains, each phosphoinositide recruits a different group of signaling proteins to its resident membrane. Two phosphoinositides, phosphatidylinositol 4,5-bisphosphate and phosphatidylinositol 3,4,5-trisphosphate, hereafter called PI(4,5)P2 and PIP3, play important and distinct roles in regulating actin dynamics.12,15 PI(4,5)P2, the predominant phosphoinositide of the plasma membrane, is the substrate of PI3-kinases, which convert PI(4,5)P2 to PIP3. In phagocytic cups, PIP3 is transiently enriched together with PI(3,4)P2, the product of its degradation by inositol 5-phosphatases.16
The domain of CRAC, a cytosolic regulator of adenylyl cyclase in Dictyostelium,17 binds with high specificity to PIP3 and PI(3,4) P2.16,18 Thus, PHcrac-GFP, a fusion of GFP to the PH domain of CRAC, labels the membrane of forming phagosomes and disappears shortly after the phagosome seals.18–20
Using PHcrac-GFP together with mRFP-LimEΔ, a probe for filamentous actin,21 we examined cells in the process of generating actin waves on a planar surface. We found that these waves separate two different areas of the cell surface: an inner area that bound PHcrac-GFP, corresponding to the interior of a phagocytic cup, and an external area that was not labeled by PHcrac-GFP, corresponding to the plasma membrane external to a phagocytic cup. In order to define the patterns of actin and PHcrac along the length of a cup, we fed cells of Dictyostelium discoideum with a mutant of Saccharomyces cerevisiae that forms rod-shaped particles.22 When these long yeast particles were taken up, the probes bound to the surrounding phagosome membrane in an axial direction, along a membrane tube extending from the rim to the base of the cup. Furthermore, very long particles were often not completely engulfed but eventually released, making pattern dynamics recognizable during both the extension and retraction of a cup. We show that the complex 3-dimensional patterns generated in phagocytosis are paralleled by the planar wave patterns, and propose that the waves' function is to search for particles to be taken up.
Results
Analogous PIP3 and actin patterns in waves and phagocytic cups.
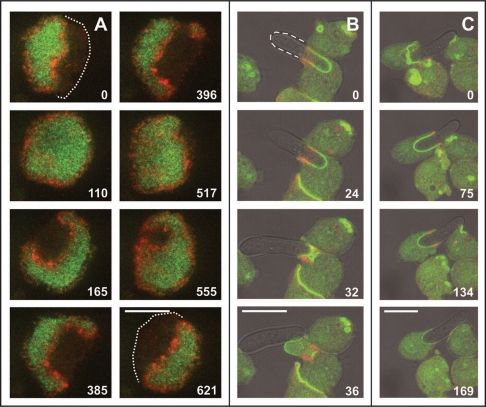
Actin waves are one of several actin-based structures present during normal movement of Dictyostelium cells on a planar glass surface.2 An efficient method to enhance wave formation is the depolymerization of actin by incubating cells with latrunculin A; during the reorganization of actin structure and function after removal of the drug, the cells pass through a period of excessive wave formation before normal cell motility recovers.10,23 Figure 1A shows that under the conditions of wave formation, the cell cortex differentiates into two phases sharply separated from each other by the actin waves. The membrane area circumscribed by a wave is strongly decorated with PHcrac (green). The actin wave (red) forms a boundary between this inner area and the external one, which does not bind PHcrac (see also movie 1).
Figure 1.
Actin wave dynamics (A) compared with the uptake and release of a particle (B and C). The cells express the PHcrac-GFP label for PIP3 (green) and the mRFP-LimEΔ label for filamentous actin (red). Numbers indicate seconds after the beginning of each sequence. (A) actin wave formed at the substrate-attached surface of a cell recovering from actin depolymerization. The actin wave expands over the entire substrate-attached area of the cell surface (frames 0 to 110), retracts (frames 165 and 385), splits into two (frame 396), and changes shape repeatedly (frames 517 to 621). The PHcrac label always coincides in size and shape with the area circumscribed by the wave. The almost unchanged boundary of the cell outside the wave is indicated in the first and last frame by dotted lines. (B) lateral view of a phagocytic cup formed around a long yeast particle, showing an actin-rich ring around the rim of the cup and decoration of the entire membrane area of the cup with PHcrac. The shape of the particle is seen in black-white bright-field illumination and delineated in the 0 s frame. Frames 0 to 36 show release of a particle that has been half-way taken up by a cell. Excess membrane is kept together and wrinkled within the actin ring (frame 32) and eventually turned inside out as a bleb. (C) subsequently the same particle is half-way ingested by a second cell (frames 0 and 75). Other cells in the field show the known enrichment of the PHcrac label at leading edges or at regions of cell-cell contact (frames 24–36 of (B)). The image sequence of (A) was obtained by total internal reflection fluorescence (TIRF) microscopy visualizing selectively the cell cortex up to a depth of about 100 nm above the substrate surface. The sequences of (B and C) were recorded by confocal scanning microscopy. Bars, 10 µm.
Since PHcrac recognizes both PI(3,4,5)P3 and PI(3,4)P2, we also examined cells labeled with GFP-TAPP, which binds specifically to PI(3,4)P2.18 This probe displayed no difference in labeling of the inner and external areas, or the waves separating them, indicating that PI(3,4,5)P3 is the phosphoinositide that distinguishes these two areas in wave-forming cells. When the waves propagate in one or the other direction, the inner and external areas undergo reciprocal changes in size with negligible changes in the total area of the substrate-attached plasma membrane. This interconversion of membrane areas implies up and downregulation of PIP3 at the sites of actin waves.
The zonal arrangement of PIP3 and actin labels in wave patterns can be viewed as a 2-dimensional projection of their arrangement in phagocytic cups. The inner leaflet of the cup membrane is enriched in PIP3, and a ring of filamentous actin at the rim of the cup is equivalent to a wave in separating the nascent phagosome membrane from the outer area of the cell surface (Fig. 1B and C, and movie 2). The two cells shown in Figure 1B and C expelled a long yeast particle after partial uptake. Concomitantly the cup area decorated with PHcrac expanded and shrank before the actin band disappeared.
Actin waves and rings form at the edges of PIP3-enriched areas.
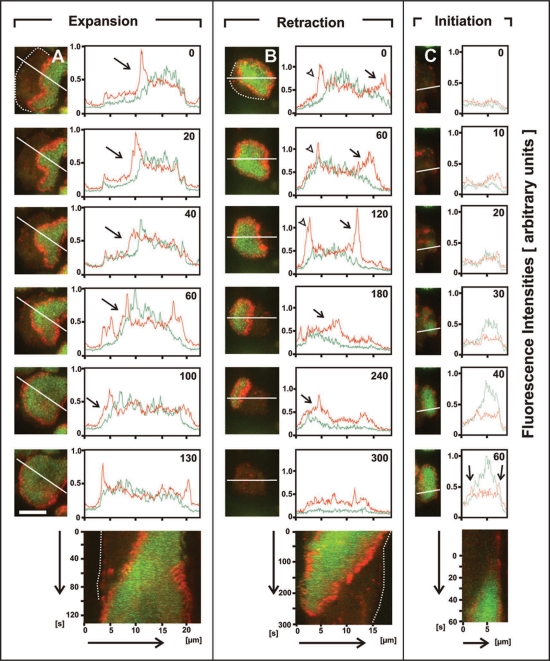
The actin waves always co-localize with the edge of the PHcrac-decorated area, where PIP3 forms a steep gradient between the inner and external areas. This rule holds for waves propagating in any direction and either expanding or diminishing the size of the inner area (Fig. 2A and B). It also holds for the de-novo formation of a circular wave, which is initiated by the focal appearance of PIP3 in association with actin. While the PIP3-enriched area expands, an actin wave is built up at its border (Fig. 2C). The concerted evolution of PIP3 and polymerized actin suggests an autocatalytic circuit of wave initiation, as previously demonstrated for the formation of a leading edge.24 Indeed, PIP3 production and formation of actin waves are interdependent. The plasma membrane became depleted of PIP3 upon the depolymerization of actin with latrunculin A, and regained PIP3 in the form of bursts when actin polymerization was allowed to recover from this treatment, as shown in Figure 2C. Similarly, actin waves and the high PIP3 enrichment of the inner territory disappeared within 2 minutes after addition of the PIP3-kinase inhibitor LY-294002, and both recovered within 3 minutes after removal of the drug (data not shown).
Figure 2.
Concerted evolution of PIP3 and actin in wave patterns. Cells double-labeled with PHcrac-GFP (green) and mRFP-LimEΔ for filamentous actin (red) were imaged during recovery from actin depolymerization using TIRF microscopy. Temporal evolution of patterns is shown in three cells that represent different types of wave dynamics: (A) expanding actin wave, (B) retracting wave, (C) wave initiation. For each cell is shown a time series of images (left), paralleled by scans along the lines indicated in these images (right), and a kymograph illustrating the temporal evolution of pattern along the scan (bottom). In scans (A) and (C), the propagating wave fronts are indicated by arrows. The retracting front in (B) indicated by arrow is accompanied by an expanding front indicated by arrowhead. Fluorescence intensities in the scans are plotted in arbitrary units, setting the highest intensity of GFP-CRAC in each sequence to 1. Numbers in the scans are seconds after beginning of the kymograph. Cell borders are demarcated by dotted lines where appropriate. Bar in (A), 10 µm; this bar applies also to the images of (B and C).
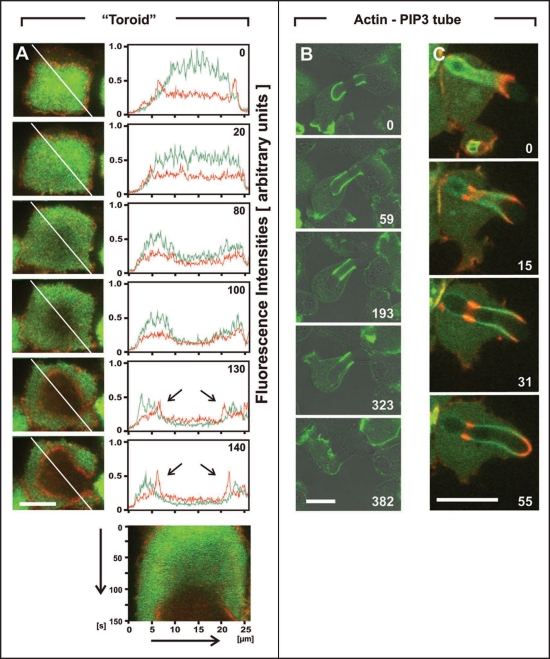
The rule that actin waves are linked to a gradient of PIP3 is best demonstrated by the toroid-like configuration shown in Figure 3A and movie 3. At the beginning of this time series, most of the large area of the cell surface was decorated with PHcrac, indicating it was enriched in PIP3. Gradually the label declined in the middle of the area until a critical steepness was reached and a new actin wave was inserted. This co-evolution of PIP3 and actin patterns shows two distinctive features: (1) the formation of an actin wave is coupled to a steep PIP3 gradient, and (2) there is a size limit for a uniform inner territory beyond which it becomes unstable.
Figure 3.
Toroid-like wave pattern and a length limit in phagocytic cups. (A) a large area circumscribed by an actin wave is subdivided by a second wave into a ring decorated with PHcrac and a central area depleted of this label. The arrangement is the same as in Figure 2: the time series of TIRF images and line scans of GFP-PHcrac and mRFP-LimEΔ fluorescence intensities are presented on top, and the kymograph on bottom. (B) phagocytic cup formed around a rod-shaped particle, showing the actin label (green) to be depleted of the base of the cup. In frames 0 and 59, two cells attempt to engulf the particle. Later on, only one cell is further engaged. As the cup elongates, actin is depleted from its base, forming a tube extending 8 µm from the border to the interior of the cup. This tube disappears when the particle is eventually released. (C) a long phagocytic cup that finally closes to incorporate the particle. This cup is double-labeled with PHcrac (green) and mRFP-LimEΔ (red), as in the wave patterns of (A). Time is indicated in seconds. Bars, 10 µm.
Assuming that the inner territory of the wave pattern corresponds to the membrane area of a phagocytic cup, one would predict that the cup area might also have a size limit, and its expansion beyond this limit would result in the insertion of an actin band similar to the wave pattern in Figure 3A and movie 3. To test this, we have fed Dictyostelium cells with highly elongated yeast particles and recorded the patterns of actin and PIP3 along the large cups shaped by these particles. In Figure 3B and movie 4, the two ends of an elongated particle have been partially ingested by two cells that are expressing only the marker for filamentous actin. Initially, the phagosome surrounding the left-hand end of the particle was completely labeled with actin, but as the particle was drawn deeper into the cell, the actin label disappeared from the bottom of the phagosome (59- and 193-second frames), generating a tube of actin with two strongly labeled ends. This particle was eventually released by both cells.
Figure 3C and movie 5 show the successful uptake of a long particle by a cell expressing markers for both actin and PIP3. This particle, 16 µm in length, was drawn into the cell to a depth of 10 µm until it contacted the cortex on the far side, whereupon the cell completed ingestion by extending its own length to encompass the particle. Throughout the process of phagocytosis, actin filaments were highly enriched at the growing edge of the cup. During the initial stage of uptake, actin filaments diminished along the portion of the phagosome membrane that had already been internalized. However, in the later stage of uptake, a strongly labeled new band of actin filaments formed at the phagosome membrane deep within the cell (Fig. 3C, 15- and 31-second frames); this band persisted until uptake had been completed (Fig. 3C, 55-second frame). A PHcrac-labeled tube of phagosome membrane connected the two bands of actin, while the basal part of the cup below the second actin band was depleted of PHcrac. Thus, PHcrac was enriched between two bands of actin about 8 µm separated from each other, resembling the toroid configuration in a planar wave.
Actin waves correlate with myosin-II independent force production for particle uptake.
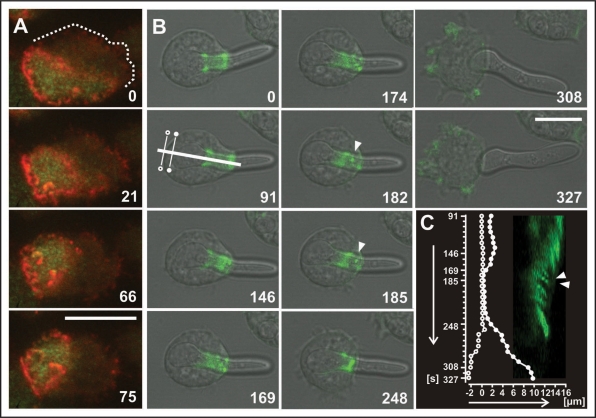
Actin waves can produce force to push the plasma membrane forward,10,23 and phagocytes need to produce force to extend the membrane area around a particle. Figure 4 relates actin waves to the generation of force in a phagocytic cup. On a planar substrate we often observed patterns in which wave fronts escaped from the major wave and propagated independently into the inner area (Fig. 4A). A similar behavior is displayed by the phagocytic cup shown in Figure 4B and movie 6. When the time series began, the Dictyostelium cell had partially ingested an elongated yeast particle. Actin filaments were enriched in two bands, one at the rim of the phagocytic cup and the other deeper in the cell, with a lesser amount of actin along the phagosome membrane between the two bands but none detectable below the second band (Fig. 4B, 0 s frame). Over the next several minutes, waves formed along the actin-rich zone of the phagosome membrane and moved toward the rim of the cup (Fig. 4B, 169 to 185 s frames). Simultaneously, the yeast particle was pushed in the opposite direction against the back of the cell, indicating that during the period of wave generation force was produced that facilitated uptake of the particle (Fig. 4C). Eventually, the cell failed to engulf this particle, which turned out to be too long; wave formation ceased, and the particle was released.
Figure 4.
Sub-structures of waves and force generation in phagocytic cups. (A) wave pattern in a cell labeled with GFP-PHcrac (green) and mRFP-LimEΔ for actin (red). This pattern shows an often observed phenomenon,10 the separation of sub-structures from a wave and their migration into the inner area decorated with PHcrac. (B) sub-structures of actin (green) during the attempted uptake of a long particle. At the beginning (frames 0 and 91) the phagosome shows two actin-rich rings interconnected by an actin tube as in Figure 3B. Subsequently, sub-structures are formed that propagate along this tube toward the border of the cup (arrowheads in frames 182 to 185; see also movie 6). Simultaneously the particle is pushed in opposite direction toward the bottom of the cell. Finally, the particle is expelled while the actin at the cup disassembles. In this case, the excess membrane is converted into actin-rich protrusions of the cell surface (frames 308 and 327). (C) movement of the partially ingested particle (white symbols) and kymograph showing propagation of actin structures (green). Direction and length of the scan in the 91 s frame of (B) is plotted on the x-axis (0 = position of the bottom of the cell in the 91 s frame as indicated). Seconds on the y-axis correspond to those of the images in (B). The plot shows bumping of the proximal end of the particle (closed circles) against the bottom of the cell (open circles) during the 169 to 230 s period, and thereafter release of the particle. The apparent extension of the cell border toward the left is due to actin-rich protrusions that are formed all over the cell surface during and after release of the particle, as shown in the 308 and 327 s frames of (B). The kymograph reveals that during pushing of the particle toward the cell bottom, actin structures propagate from the cell body toward the rim of the phagocytic cup. The two arrowheads indicate movement of the discrete actin patch demarcated in the 182 and 185 s frames in (B). Numbers in (A and B) indicate seconds. In (C) seconds after the 0 s frame in (B) are plotted on the vertical axis from top to bottom. Bars, 10 µm.
Phagocytosis does not require myosin-II,25,26 and the formation of actin waves is independent of this conventional myosin.10 To determine whether the same is true for the actin pattern along phagocytic cups, we fed myosin-II-null cells with long yeast particles. Movie 7 shows that actin initially decorated the entire cup and subsequently condensed into a tube. Finally, the inner ring moved like a wave toward the rim of the cup. In summary, the actin dynamics resembled in all respects that in wild-type cells.
Endocytic activity recovers early after the depolymerization of actin.
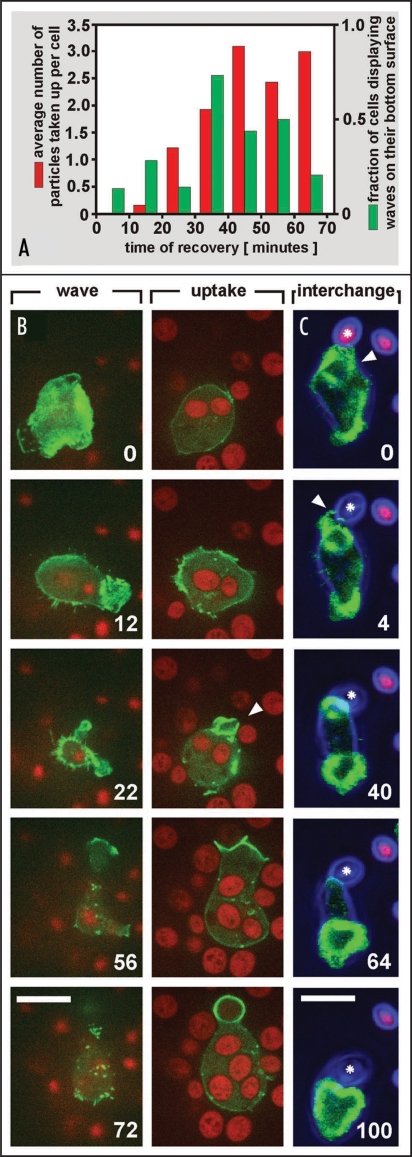
The analogous patterns of 2-dimensional waves and 3-dimensional phagocytic cups suggest that phagocytic structures are spontaneously generated on a planar substrate surface. If this view is correct, the period of extensive wave formation should be a period of high phagocytic activity. This implies that, after the depolymerization of actin, phagocytosis would recover earlier than normal cell motility. To test this hypothesis, we supplied cells at early stages of recovery with yeast particles and recorded uptake of the yeast (Fig. 5 and movies 8 to 11).
Figure 5.
Phagocytic activity of wave-forming cells recovering from actin depolymerization. Waves and phagocytic cups were visualized by LimEΔ-GFP (green), particles of heat-killed Saccharomyces cerevisiae were labeled with TRITC (red). (A) Uptake of particles and wave formation monitored concomitantly by z-scanning. The particles were added immediately after the removal of latrunculin A. At different times thereafter, the number of wave-forming cells and the average number of particles that have been taken up were determined. For each time interval of 10 minutes, 11 to 34 cells were counted. Within the first 10 minute period, only a single wave after 7 minutes was observed. (B) a cell forming a large wave had already taken up four particles, and engulfed a fifth one while the wave regressed. Transiently a rosette-shaped wave was formed in an area that contained no particles (12 s frame). A macropinocytic cup is indicated by arrowhead. Wave formation and particle uptake were monitored in parallel by switching between the substrate-attached cell surface (left) and a plane 2.8 µm above through the cell body (right), using a spinning-disc confocal microscope. (C) a wave that was converted into a phagocytic cup (arrowheads) while a second wave continued to propagate. The phagocytosed particle is demarcated by asterisks. Confocal microscopy visualizing wave and phagocytic cup formation in one plane was combined with bright-field imaging (blue) to trace the particle out of plane. Time is indicated in seconds. Bars, 10 µm.
During the stage of wave-formation, cells proved to be highly active phagocytes. This is indicated by the increase in the number of particles ingested during the period of intense wave formation at 20 to 50 minutes of recovery from actin depolymerization (Fig. 5A). Often, but not always, a wave regressed when a particle was taken up. The wave-forming cell of Figure 5B had already taken up four yeast particles, and took up a fifth one as the wave regressed (Movie 8). A direct link between wave formation and particle uptake is evident in Figure 5C, which shows the conversion of a wave into a cup upon contact with a particle (see also movie 10).
Another actin-dependent activity involving membrane internalization is fluid uptake. For cells in the wave-forming state we observed both extensive formation of macropinocytic cups (Fig. 5B) and clathrin-associated micropinocytosis (unpublished data). Together these data indicate that the capacity for endocytosis is one of the earliest activities to recover during the repolymerization of actin. Since in the time window of extensive wave formation the cells have a high potential of forming phagocytic structures even on a planar surface, they can be used to study this activity by viewing their substrate-attached surface essentially independent of cell motility.
Discussion
The results presented here link a phenomenon of self-organization in the actin system to the established biological function of this system in phagocytosis. We show that the structure of a phagocytic cup is mimicked by the relationship of planar actin waves to PIP3-enriched areas of the plasma membrane, as outlined in Figure 6A. An interrelation of actin polymerization and PIP3 localization in the plasma membrane has been noted for several actin-based cellular functions: chemotaxis,27,28 phagocytosis,18–20 cytokinesis29,30 and actin wave propagation. In the keratocyte-like cells of a Dictyostelium mutant, but also in wild-type cells, actin waves were found to be correlated with PIP3 waves.31 In these cases, the actin waves were usually chased by PIP3 waves, which suggested that effectors recruited by PIP3 may induce the depolymerization of actin filaments. Under our conditions the actin waves separated two areas of the plasma membrane, an inner area enriched in PIP3 from an external area depleted of this lipid. When a wave expanded, PIP3 followed its back; when the wave retracted, PIP3 disappeared at its front (Figs. 2A and B, and movie 1).
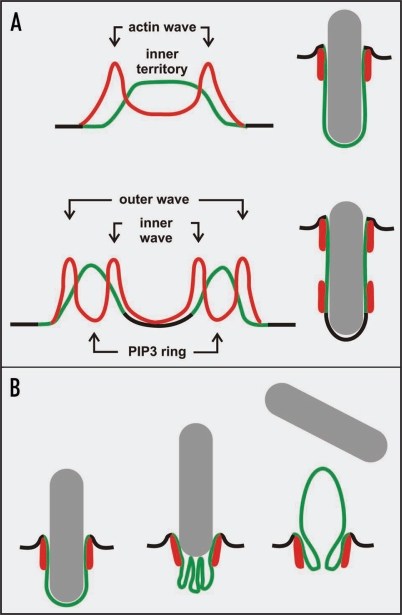
Figure 6.
Diagram of patterns in planar waves and phagocytic cups. Green: PIP3-enriched membrane areas. Red: Waves or rings of filamentous actin. (A) comparison of planar wave patterns (left) with the 3-dimensional patterns in phagocytic cups shaped by long particles (right). The wave patterns are based on the scans in Figures 2 and 3A, the cup patterns on Figures 1B and C and 3B and C. (B) conversion of a phagocytic cup into a bleb. When a particle is rapidly expelled, the excess cup membrane within the actin ring may be folded and eventually everted to cover the bleb (Fig. 1B).
The peak of actin polymerization along a spatial gradient of PIP3, illustrated in Figures 2 and 3A and movie 3, may be relevant not only to phagocytosis. It may also be instrumental in translating a PIP3 gradient formed in response to chemoattractant into a directional movement of the cell. In the membrane of a cell exposed to a gradient of chemoattractant, PIP3 synthesis is induced in the form of a crescent with the highest concentration pointing to the source of attractant and gradual decline toward both sides.27 The actin-driven response of cells in a shallow gradient of attractant is characterized by split pseudopod formation.32 Similarly, in the absence of external cues, cells move persistently by split pseudopodia that are extended alternately right/left/right from the previous one.33 Thus, it appears that the actin-based formation of pseudopodia is focused on the lateral slopes of a PIP3-enriched membrane area, similar to the actin waves on a planar cell surface and the actin rings in a phagocytic cup (Fig. 6A).
A relationship of cell spreading on a planar surface to phagocytosis of a curved particle has repeatedly been proposed (reviewed in ref. 34). Neutrophils adhere to immune complexes along a surface that they cannot phagocytose. In response to interaction with the functionalized surface, lysosomal constituents, which are normally delivered into a closed phagosome, are released into the environment. Under these conditions, the attempted phagocytosis has been considered to be “frustrated” by the large size of the surface.35 Frustrated phagocytosis has also been reported for macrophages that interact through their Fc receptors with ligands on a surface.36 Here we consider the propagation of actin waves as a mechanism that enables a phagocyte to scan a surface for particles to be taken up. As scanning machines the actin waves propagate in different directions along the entire substrate-attached membrane of a cell by the incorporation of actin monomers and the depolymerization of actin filaments in a defined 3-dimensional pattern.10
Actin waves have been shown to apply force on the cell border to push the membrane forward.23 Likewise, the actin structures in a phagocytic cup appear to apply force for internalization of the particle against membrane tension. The long particles employed in this study required a substantial area of the membrane for their uptake; in fact the particles were often too long to be completely engulfed. A rough estimate assuming a spherical cell with a smooth surface suggests for a phagocytosing cell, like the one in Figure 4B, that at least 20 percent of the total membrane area is reversibly pulled into the phagocytic cup.
Upon cessation of actin mediated force production at the cup surface, the pressure for internalization is relieved and the particle is expelled. We observed two modes of redistribution of the excess membrane after the often forceful release of a particle: global formation of surface ruffles (Fig. 4, 308 and 327 s frames) and less often local bleb formation within the boundary of an actin ring (Fig. 1B and movie 2) as diagrammed in Figure 6B. The sudden conversion of an invaginated membrane area confined by an actin ring into a protrusion illustrates most clearly that the uptake machinery has acted against tension. In all cases that we recorded the particle was either completely engulfed or eventually released. Thus, phagocytes must have mechanisms to recognize that a particle is not suitable for uptake. The decision for release might be based on a clock counting the time spent in the state of an open cup, as suggested by Figure 4B or on a safety-valve opened when internal pressure exceeds a certain limit, as suggested by Figure 1B.
In conclusion, the patterns generated by wave forming cells of Dictyostelium correspond to two-dimensional projections of phagocytic cups, indicating that the cup structure can be spontaneously generated on a planar surface by differentiation of the cell cortex into two areas separated by actin waves. The inner area is distinguished by its high PIP3 content from the external area. These areas expand or shrink reciprocally when the waves propagate into one direction or the other, paralleling the increase and decrease of cup area during the uptake and expulsion of a particle. The temporal evolution of actin wave and phagocytic cup patterns indicates that spatial gradients of PIP3 are translated into sharp peaks of polymerized actin.
Materials and Methods
Cell strains and experimental conditions.
PHcrac-GFP27 and mRFP-LimEΔ,21 LimEΔ-GFP37only, or GFP-TAPP18 and mRFP-LimEΔ were expressed in Dictyostelium discoideum strain AX2-214. The strain used for Figures 3B and 4B carried a VacuolinB-null mutation that is not relevant to the work presented. The AX2-derived mutant HS2205 deficient in myosin-II heavy chains38 was transfected with LimEΔ-GFP. Cells were cultivated in Petri dishes with nutrient medium and washed for imaging in 17 mM K/Na-phosphate buffer, pH 6.0 (PB).10 Experiments were performed at 23 ± 2°C. For recovery of actin polymerization, cells were incubated for 15–20 minutes with 5 µM latrunculin A (Invitrogen), and imaged after replacement of the drug with PB. For treatment of wave forming cells with Ly 294002 (Sigma L9908), a 30 mM stock solution in DMSO was diluted with PB to 50 µM. After incubation for 5 to 10 minutes, the drug was replaced with PB.
For phagocytosis assays, cells were fed with long particles of the conditional cdc42 S. cerevisiae mutant DDY 1306,22 grown in YPD at 27°C with 3 percent formamide or, for Figure 5, with spherical S. cerevisiae particles (Sigma YSC2) boiled for 30 minutes in a waterbath and labeled with TRITC. For Figure 5A, 1 × 109 yeast particles per ml of PB were vortexed and filtered through gauze of 8 µm pore size to remove aggregates. 10 µl of this suspension and 1 ml of PB were added to a coverslip within a ring of 20 mm diameter on which the Dictyostelium cells were attached. To visualize waves on the bottom of the cells together with all particles taken up, confocal images were acquired in 4 planes separated from each other by 2 µm.
Optical techniques.
Wave patterns in Figures 1A, 2 and 3A and 4A were imaged using through-objective TIRF microscopy essentially as described.10 GFP and mRFP were simultaneously excited at 491 nm such that emissions of both fluorophores were recorded from the same evanescent field. All other images were obtained by confocal scanning fluorescence microscopy. For Figure 3C, cells were imaged using an Ultraview ERS spinning disc confocal set (Perkin Elmer LAS) on a Nikon TE-2000 inverted microscope equipped with a Plan Apo VC 100x/1.4 NA objective. In this system, channels were recorded sequentially onto an EM-CCD camera using 488 nm excitation, 527/55 nm emission and 568 nm excitation, 615/70 nm emission for the GFP and mRFP labels, respectively. For Figure 5A and B, an Eclipse TE300 microscope with a 100x/1.4 oil Plan Apo objective (Nikon), an Ultraview spinning disc scanner (Perkin Elmer), an ORCA ER model C4742-95-ERG camera (Hamamatsu Photonics) was used. Figures 1B and C, 3B and 4B were recorded on a Zeiss LSM 510 as described,20 and Figure 5C was acquired on a Zeiss LSM 410 confocal microscope.
Acknowledgements
Our work was supported by the Max Planck Society and grants in the SPP 1128 program of the Deutsche Forschungsgemeinschaft to G.G. and Stefan Diez, and by National Science Foundation grant MCB-0344541 to M.C. We acknowledge the equipment and technical support of the Imaging Core Facility at the Oklahoma Medical Research Foundation. We appreciate the advice of Stefan Diez and thank Hans Meinhardt, Tübingen, for stimulating discussions, Jana Prassler for expert assistance, Carol Parent, NIH, for the PHcrac-GFP vector and Cornelis Weijer, Dundee, for the TAPP-GFP vector.
Footnotes
Previously published online as a Cell Adhesion & Migration E-publication: http://www.landesbioscience.com/journals/celladhesion/article/9708
Supplementary Material
References
- 1.Maeda YT, Inose J, Matsuo MY, Iwaya S, Sano M. Ordered patterns of cell shape and orientational correlation during spontaneous cell migration. PloS one. 2008;3734:1–14. doi: 10.1371/journal.pone.0003734. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Bretschneider T, Diez S, Anderson K, Heuser J, Clarke M, Müller-Taubenberger A, et al. Dynamic actin patterns and Arp2/3 assembly at the substrate-attached surface of motile cells. Curr Biol. 2004;14:1–10. doi: 10.1016/j.cub.2003.12.005. [DOI] [PubMed] [Google Scholar]
- 3.Peracino B, Borleis J, Jin T, Westphal M, Schwartz J-M, Wu L, et al. G protein β subunit-null mutants are impaired in phagocytosis and chemotaxis due to inappropriate regulation of the actin cytoskeleton. J Cell Biol. 1998;141:1529–1537. doi: 10.1083/jcb.141.7.1529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Cox D, Dale BM, Kashiwada M, Helgason CD, Greenberg S. A regulatory role for Src homology 2 domain-containing inositol 5′-phosphatase (SHIP) in phagocytosis mediated by Fcγ receptors and complement receptor 3 (αMβ2; CD11b/CD18) J Exp Med. 2001;193:61–71. doi: 10.1084/jem.193.1.61. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Maniak M, Rauchenberger R, Albrecht R, Murphy J, Gerisch G. Coronin involved in phagocytosis: dynamics of particle-induced relocalization visualized by a green fluorescent protein tag. Cell. 1995;83:915–924. doi: 10.1016/0092-8674(95)90207-4. [DOI] [PubMed] [Google Scholar]
- 6.Konzok A, Weber I, Simmeth E, Hacker U, Maniak M, Müller-Taubenberger A. DAip1, a Dictyostelium homologue of the yeast actin-interacting protein 1, is involved in endocytosis, cytokinesis and motility. J Cell Biol. 1999;146:453–464. doi: 10.1083/jcb.146.2.453. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Swanson JA, Johnson MT, Beningo K, Post P, Mooseker M, Araki N. A contractile activity that closes phagosomes in macrophages. J Cell Sci. 1999;112:307–316. doi: 10.1242/jcs.112.3.307. [DOI] [PubMed] [Google Scholar]
- 8.Hoppe AD, Swanson JA. Cdc42, Rac1 and Rac2 display distinct patterns of activation during phagocytosis. Mol Biol Cell. 2004;15:3509–3519. doi: 10.1091/mbc.E03-11-0847. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Clarke M, Maddera L. Phagocyte meets prey: Uptake, internalization and killing of bacteria by Dictyostelium amoebae. Eur J Cell Biol. 2006;85:1001–1010. doi: 10.1016/j.ejcb.2006.05.004. [DOI] [PubMed] [Google Scholar]
- 10.Bretschneider T, Anderson K, Ecke M, Müller-Taubenberger A, Schroth-Diez B, Ishikawa-Ankerhold HC, et al. The three-dimensional dynamics of actin waves, a model of cytoskeletal self-organization. Biophys J. 2009;96:2888–2900. doi: 10.1016/j.bpj.2008.12.3942. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Yeung T, Ozdamar B, Paroutis P, Grinstein S. Lipid metabolism and dynamics during phagocytosis. Curr Opin Cell Biol. 2006;18:429–437. doi: 10.1016/j.ceb.2006.06.006. [DOI] [PubMed] [Google Scholar]
- 12.Di Paolo G, De Camilli P. Phosphoinositides in cell regulation and membrane dynamics. Nature. 2006;443:651–657. doi: 10.1038/nature05185. [DOI] [PubMed] [Google Scholar]
- 13.Lindmo K, Stenmark H. Regulation of membrane traffic by phosphoinositide 3-kinases. J Cell Sci. 2006;119:605–614. doi: 10.1242/jcs.02855. [DOI] [PubMed] [Google Scholar]
- 14.Hawkins PT, Anderson KE, Davidson K, Stephens LR. Signalling through class I PI3Ks in mammalian cells. Biochem Soc Trans. 2006;34:647–662. doi: 10.1042/BST0340647. [DOI] [PubMed] [Google Scholar]
- 15.Janmey PA, Lindberg U. Cytoskeletal regulation: rich in lipids. Nat Rev Mol Cell Biol. 2004;5:658–666. doi: 10.1038/nrm1434. [DOI] [PubMed] [Google Scholar]
- 16.Loovers HM, Kortholt A, de Groote H, Whitty L, Nussbaum RL, van Haastert PJM. Regulation of phagocytosis in Dictyostelium by the inositol 5-phosphatase OCRL homolog Dd5P4. Traffic. 2007;8:618–628. doi: 10.1111/j.1600-0854.2007.00546.x. [DOI] [PubMed] [Google Scholar]
- 17.Insall R, Kuspa A, Lilly PJ, Shaulsky G, Levin LR, Loomis WF, et al. CRAC, a cytosolic protein containing a pleckstrin homology domain, is required for receptor and G protein-mediated activation of adenylyl cyclase in Dictyostelium. J Cell Biol. 1994;126:1537–1545. doi: 10.1083/jcb.126.6.1537. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Dormann D, Weijer G, Dowler S, Weijer CJ. In vivo analysis of 3-phophoinositide dynamics during Dictyostelium phagocytosis and chemotaxis. J Cell Sci. 2004;117:6497–6509. doi: 10.1242/jcs.01579. [DOI] [PubMed] [Google Scholar]
- 19.Devreotes P, Janetopoulos C. Eukaryotic chemotaxis: Distinctions between directional sensing and polarization. J Biol Chem. 2003;278:20445–20448. doi: 10.1074/jbc.R300010200. [DOI] [PubMed] [Google Scholar]
- 20.Giorgione J, Clarke M. Heterogeneous modes of uptake for latex beads revealed through live cell imaging of phagocytes expressing a probe for phosphatidylinositol-(3,4,5)-trisphosphate and phosphatidylinositol-(3,4)-bisphosphate. Cell Motil Cytoskeleton. 2008;65:721–733. doi: 10.1002/cm.20293. [DOI] [PubMed] [Google Scholar]
- 21.Fischer M, Haase I, Simmeth E, Gerisch G, Müller-Taubenberger A. A brilliant monomeric red fluorescent protein to visualize cytoskeleton dynamics in Dictyostelium. FEBS Letters. 2004;577:227–232. doi: 10.1016/j.febslet.2004.09.084. [DOI] [PubMed] [Google Scholar]
- 22.Kozminski KG, Chen AJ, Rodal AA, Drubin DG. Functions and functional domains of the GTPase Cdc42p. Mol Biol Cell. 2000;11:339–354. doi: 10.1091/mbc.11.1.339. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Gerisch G, Bretschneider T, Müller-Taubenberger A, Simmeth E, Ecke M, Diez S, et al. Mobile actin clusters and traveling waves in cells recovering from actin depolymerization. Biophys J. 2004;87:3493–3503. doi: 10.1529/biophysj.104.047589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Sasaki AT, Janetopoulos C, Lee S, Charest PG, Takeda K, Sundheimer LW, et al. G protein-independent Ras/PI3K/F-actin circuit regulates basic cell motility. J Cell Biol. 2007;178:185–191. doi: 10.1083/jcb.200611138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Fukui Y, De Lozanne A, Spudich JA. Structure and function of the cytoskeleton of a Dictyostelium myosin-defective mutant. J Cell Biol. 1990;110:367–378. doi: 10.1083/jcb.110.2.367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Shu S, Liu X, Korn ED. Blebbistatin and blebbistatin-inactivated myosin II inhibit myosin II-independent processes in Dictyostelium. Proc Natl Acad Sci USA. 2005;102:1472–1477. doi: 10.1073/pnas.0409528102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Parent CA, Devreotes PN. A cell's sense of direction. Science. 1999;284:765–770. doi: 10.1126/science.284.5415.765. [DOI] [PubMed] [Google Scholar]
- 28.Funamoto S, Meili R, Lee S, Parry L, Firtel RA. Spatial and temporal regulation of 3-phospoinositides by PI3-Kinase and PTEN mediates chemotaxis. Cell. 2002;109:611–623. doi: 10.1016/s0092-8674(02)00755-9. [DOI] [PubMed] [Google Scholar]
- 29.Janetopoulos C, Borleis J, Vazquez F, Iijima M, Devreotes P. Temporal and spatial regulation of phosphoinositide signaling mediates cytokinesis. Dev Cell. 2005;8:467–477. doi: 10.1016/j.devcel.2005.02.010. [DOI] [PubMed] [Google Scholar]
- 30.Janetopoulos C, Devreotes P. Phosphoinositide signaling plays a key role in cytokinesis. J Cell Biol. 2006;174:485–490. doi: 10.1083/jcb.200603156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Asano Y, Nagasaki A, Uyeda TQP. Correlated waves of actin filaments and PIP3 in Dictyostelium cells. Cell Motil Cytoskeleton. 2008;65:923–934. doi: 10.1002/cm.20314. [DOI] [PubMed] [Google Scholar]
- 32.Andrew N, Insall RH. Chemotaxis in shallow gradients is mediated independently of Ptdlns 3-kinase by biased choises between random protrusions. Nat Cell Biol. 2007;9:193–200. doi: 10.1038/ncb1536. [DOI] [PubMed] [Google Scholar]
- 33.Bosgraaf L, Van Haastert PJM. The ordered extension of pseudopodia by amoeboid cells in the absence of external cues. PLoS One. 2009;4:5253. doi: 10.1371/journal.pone.0005253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Grinnell F. Fibroblast spreading and phagocytosis: similar cell responses to different-sized substrata. J Cell Physiol. 1984;119:58–64. doi: 10.1002/jcp.1041190110. [DOI] [PubMed] [Google Scholar]
- 35.Henson PM. Interaction of cells with immune complexes: adherence, release of constituents, and tissue injury. J Exp Med. 1971;134:114–135. [PMC free article] [PubMed] [Google Scholar]
- 36.Takemura R, Stenberg PE, Bainton DF, Werb Z. Rapid redistribution of clathrin onto macrophage plasma membranes in response to Fc receptor-ligand interaction during frustrated phagocytosis. J Cell Biol. 1986;102:55–69. doi: 10.1083/jcb.102.1.55. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Schneider N, Weber I, Faix J, Prassler J, Müller-Taubenberger A, Köhler J, et al. A Lim protein involved in the progression of cytokinesis and regulation of the mitotic spindle. Cell Motil Cytoskeleton. 2003;56:130–139. doi: 10.1002/cm.10139. [DOI] [PubMed] [Google Scholar]
- 38.Manstein DJ, Titus MA, De Lozanne A, Spudich JA. Gene replacement in Dictyostelium: generation of myosin null mutants. EMBO J. 1989;8:923–932. doi: 10.1002/j.1460-2075.1989.tb03453.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.