Abstract
Human immunodeficiency virus (HIV)-associated dementia (HAD) is a syndrome occurring in HIV-infected patients with advanced disease that likely develops as a result of macrophage and microglial activation as well as other immune events triggered by virus in the central nervous system. The most relevant experimental model of HAD, rhesus macaques exhibiting simian immunodeficiency virus (SIV) encephalitis (SIVE), closely reproduces the human disease and has been successfully used to advance our understanding of mechanisms underlying HAD. In this study we integrate gene expression data from uninfected and SIV-infected hippocampus with a human protein interaction network and discover modules of genes whose expression patterns distinguish these two states, to facilitate identification of neuronal genes that may contribute to SIVE/HIV cognitive deficits. Using this approach we identify several downregulated candidate genes and select one, EGR1, a key molecule in hippocampus-related learning and memory, for further study. We show that EGR1 is downregulated in SIV-infected hippocampus and that it can be downregulated in differentiated human neuroblastoma cells by treatment with CCL8, a product of activated microglia. Integration of expression data with protein interaction data to discover discriminatory modules of interacting proteins can be usefully used to prioritize differentially expressed genes for further study. Investigation of EGR1, selected in this manner, indicates that its downregulation in SIVE may occur as a consequence of the host response to infection, leading to deficits in cognition.
Introduction
Human immunodeficiency virus (HIV)-associated dementia (HAD) is characterized by motor disorders, behavioral changes, and cognitive dysfunction, including deficits in learning and memory (Diesing et al., 2002). Since HIV does not infect neurons, possible pathogenic mechanisms include direct toxicity of HIV proteins and indirect toxicity mediated by HIV-infected or -activated cells. Several HIV proteins have been associated with neurotoxicity (Mattson et al., 2005); however, clinical signs of HAD correlate better with microglial activation than with quantitative measures of brain infection (Kaul et al., 2005). These observations have suggested that HAD likely develops as a secondary neuronal response to macrophage/microglial activation precipitated by HIV infection (Diesing et al., 2002; Kaul et al., 2005).
Simian immunodeficiency virus (SIV) infection of rhesus macaques is a well established model for HIV infection, particularly useful for studying HAD (Rausch et al., 1999; Roberts et al., 2003). Infected monkeys may develop the same clinical disturbances as people with HAD, and they display similar neuropathologic changes, termed SIV encephalitis (SIVE) (Roberts et al., 2003). Furthermore, transient depletion of CD8+ cells soon after infection ensures that SIVE develops rapidly in a large proportion of the infected animals (Roberts et al., 2003), thus facilitating investigation.
Our previous microarray studies of infected human and rhesus frontal lobe using the Affymetrix HG_U95Av2 (human) chip detected both upregulated and downregulated human genes (Masliah et al., 2004), and upregulated rhesus genes, particularly interferon-induced genes (Roberts et al., 2003, 2004). Microarray studies, while useful, are subject to the limitations imposed by incomplete knowledge of pathway membership and insensitivity to pathway members not regulated via transcriptional control. One means to address this limitation is to integrate microarrays with other genomic-level data, such as protein interaction networks, and to extract modules of physically interacting genes with coherent expression patterns (Ideker et al., 2002; Segal et al., 2003; Chen and Yuan, 2006; Chuang et al., 2007) considered likely to reflect known or novel pathways or complexes. In protein interaction networks, nodes represent proteins and are connected to their interacting partners by edges, which represent interactions identified through either small-scale or high-throughput experiments.
We used the Affymetrix Rhesus Genome chip to compare hippocampal gene expression in uninfected rhesus macaques versus those with histopathologic evidence of SIVE. We mapped Affymetrix probesets to human gene identifiers and integrated the expression data with a human protein interaction network constructed from publicly available datasets. We were particularly interested to determine whether downregulated genes might be detectable under these conditions. We anticipated that greater efficiency of rhesus gene detection using the rhesus chip [efficiency ∼70% using the human chip (Roberts and Fox, 2005)], coupled with use of hippocampal tissue (relatively enriched for neurons) and integration of protein interaction data, would facilitate detection of neuronal genes whose downregulation contributes to HAD. We found that EGR1, an early response gene involved in memory and learning (Jones et al., 2001; Bozon et al., 2002; Knapska and Kaczmarek, 2004), was significantly downregulated and present in a differentially expressed network module, and we further investigated how EGR1 may become downregulated in SIVE.
Materials and Methods
Ethics statement.
Experiments were performed under the approval and guidelines of The Scripps Research Institute's Animal Care and Use Committee. Anesthetic and analgesic agents were used to alleviate any pain or discomfort during procedures.
Human protein interaction network.
The human protein interaction network was compiled using the following sources: Human Protein Reference Database (HPRD, 01JAN07) (Peri et al., 2003), Biomolecular Interaction Network Database (BIND, 25MAY06) (Bader et al., 2001), REACTOME (28FEB07) (Joshi-Tope et al., 2005), Database of Interacting Proteins (DIP, 19FEB07) (Xenarios et al., 2000), published human yeast two-hybrid (Rual et al., 2005; Stelzl et al., 2005) and mass spectrometry (Ewing et al., 2007) datasets, and orthology-predicted PPI (Ramani et al., 2005). All interactions for which both proteins could be mapped to Entrez Gene IDs were included. Complexes identified in HPRD and BIND were treated using the matrix paradigm (Bader and Hogue, 2002) to provide consistency with REACTOME's treatment of complexes. After removing redundancies, the final network contains 10,906 proteins (nodes) and 81,211 interactions (edges). Although high-throughput protein-interaction data are known to be noisy (Bader et al., 2004), we elected to avoid prefiltering the network using probabilistic edge weights, opting instead to filter by means of data integration, as described below.
Rhesus macaques.
We used 18 male rhesus macaques free of SIV, type D simian retrovirus and herpes B virus obtained from Covance, LABS of Virginia, and Charles River Laboratories. Experiments were performed under the approval and guidelines of The Scripps Research Institute's Animal Care and Use Committee. Nine monkeys were intravenously inoculated with a cell-free stock of a derivative of SIVmac251 that had been subjected to in vivo passage (Watry et al., 1995; Burdo et al., 2005); four of these monkeys additionally underwent a CD8+ cell depleting regimen in the acute period after viral inoculation (Roberts et al., 2003). Two of the control monkeys were similarly treated with anti-CD8 antibody. The SIV-infected monkeys were killed after development of neurological signs of simian acquired immune deficiency syndrome, after a range of 56–132 (mean, 92; median, 93) days after inoculation. Necropsy was performed after terminal anesthesia with ketamine, xylazine, and pentobarbital, followed by intracardiac perfusion with sterile PBS containing 1 U/ml heparin to clear the brain of blood-borne cells. All SIV-infected monkeys had neuropathological evidence of SIVE on postmortem histopathological examination. A portion of hippocampus was preserved frozen at necropsy for later RNA extraction. A summary of the monkeys used in this study is presented in supplemental Table S1 (available at www.jneurosci.org as supplemental material).
Microarray.
Total RNA was purified from hippocampal samples using TRIzol Reagent (Invitrogen), centrifuged to remove cellular debris, and then purified using the RNeasy mini kit (Qiagen). RNA quantity was assessed by 260 nm UV absorption and quality verified by Agilent BioAnalyzer analysis of ribosomal RNA bands. RNA was reverse transcribed and hybridized to the Affymetrix Rhesus Genome Array. This platform contains 52,024 rhesus probesets representing ∼47,000 transcripts. Each sample was tested in duplicate and results were processed using Affymetrix software (MAS 5.0) to produce CEL files, which were submitted to R/Bioconductor Robust Multi-Chip Analysis (RMA) (Irizarry et al., 2003) to yield normalized log2 expression (intensity) values for each probeset in the 36 arrays. Fold change and minimum expression (intensity) thresholds were not applied to facilitate detection of genes that might be significantly regulated in only a subset of cells in the mixed cell population present in the hippocampus samples. Differential gene expression using mean subject values was determined using Cyber T (Baldi and Long, 2001) software, which calculates significance using a Bayesian approach to regularize t tests and also calculates a posterior probability of differential expression (PPDE), the probability that a gene at a given p value is differentially expressed [http://cybert.microarray.ics.uci.edu/help/index.html (Allison et al., 2002)]. Differential expression p values were calculated by Cyber T using log-transformed data, and a p value of 0.001 corresponded to a PPDE of ∼95%. The microarray data have been deposited in NCBI's Gene Expression Omnibus and are accessible through GEO Series accession number GSE13824 (http://www.ncbi.nlm.nih.gov/geo/query/acc.cgi).
To facilitate integration of the microarray results with the human PPI network, it was necessary to associate Affymetrix probeset identifiers with human Entrez gene identifiers. Affymetrix probesets were mapped to human Entrez Gene IDs by reciprocally blasting probeset target sequences (downloaded from the Affymetrix website) and the human genome (sequences downloaded from Refseq ftp site) keyed to Entrez ID and selecting only the best reciprocal hits that had bit scores ≥50. This approach was taken, rather than using Inparanoid (Remm et al., 2001) software, to avoid multiple rounds of mapping between the Ensembl ID used by Inparanoid, and Entrez Gene ID. Using this method, it was possible to map 18,313 Affymetrix probesets to human Entrez Gene IDs, including 10,209 of the 10,906 proteins present in the human PPI network. Seventeen mappings had bit scores <60 with e values ranging between 3 × 10−5 and 5 × 10−8 and percentage identity 80.43–100%; all other mappings had e values ≤4 × 10−8. Applying a Cyber T p value cutoff of 0.001 identified 826 genes (Affymetrix probesets mapped to Entrez Gene IDs) as significantly differentially expressed, of which 106 were downregulated and 720 were upregulated.
Immunohistochemistry.
Formalin-fixed, paraffin-embedded tissue blocks containing the hippocampus from four monkeys in each group were sectioned at 5 μm thickness and picked up on glass slides. Following deparaffinization, antigen retrieval was performed by heating to 95°C in 0.01 m citrate buffer, pH 6.39, for 40 min, then left for 20 min to steep. Sections were blocked in 0.5% casein, followed by the addition of a 1/50 dilution of rabbit monoclonal anti-EGR1 antibody (clone 15F7, Cell Signaling Technology) overnight at 4°C. Following washes, signal was detected using the SuperPicture broad spectrum secondary antibody-horseradish peroxidase polymer reagent (Invitrogen) and developed with the NovaRed chromogen (Vector Laboratories), followed by a hematoxylin counterstain (Sigma-Aldrich). Controls included omission of the primary antibody and use of irrelevant primary antibodies. Image capture was performed with a Spot RT Color CCD camera with Spot RT software (Spot Diagnostic Instruments) using a Leica Diaplan microscope (Leica). Adobe Photoshop CS3 (Adobe Systems) was used for quantification. Photomicrographs were converted to black and white using the green filter, inverted, and for each monkey the signal in 20 consecutive neuronal nuclei from the CA1 region was quantified using the mean histogram density measurement. Background was determined for each section, and was subtracted from the average nuclear signal, yielding the average staining signal from each monkey. The two groups were compared using a unpaired two-tailed t test.
Data integration.
To integrate differential gene expression with protein interaction data we used a recently described algorithm (Chuang et al., 2007) that identifies network modules whose expression significantly discriminates between two experimental conditions. The algorithm overlays gene expression values on corresponding network proteins and, starting from every protein (seed) in the protein network, greedily adds interactions to identify the module starting from each seed whose mean expression calculated for each sample best discriminates between the two sample types. This process yields as many candidate modules as there are proteins in the network; nonsignificant subnetworks are then filtered out using three types of permutation testing. This method was shown to improve classification of metastatic versus nonmetastatic breast cancer and to capture well established prognostic markers not regulated at the level of transcription (Chuang et al., 2007). Parameters were chosen as described by Chuang et al. (2007), except that t test rather than mutual information was used to determine significance. We ran the algorithm five times and selected modules that scored as significant in at least three of the runs. The algorithm is available as PinnacleZ, a plugin to Cytoscape (Shannon et al., 2003).
Cell culture.
SK-N-MC human neuroblastoma cells were purchased from ATCC and maintained in culture as recommended. For each experiment, cells were plated a minimum of 9 h or overnight at 2.5 × 105 cells/ml in six-well plates (chemokine experiments; 2 ml/well) or 100 cm dish (supernatant experiment; 14 ml/dish), treated for 4 or 5 d with 10 μm trans-retinoic acid (EMD Biosciences), and then switched to Neurobasal (NB, Invitrogen) medium (without glutamate) supplemented with B-27 (Invitrogen). Cells were kept in supplemented NB medium for 2–3 h or overnight before reagent addition. Macrophage supernatant was prepared as described below; recombinant human β-NGF (Millipore Bioscience Research Reagents), and recombinant human CCL2, CCL8, CXCL8 (IL8), and CXCL10 (Peprotech) were purchased.
Flow cytometry.
SK-N-MC cells were plated overnight at 2 × 105/ml (2 ml/six-well plate well), treated for 4 d with 10 μm trans-retinoic acid (EMD Biosciences) then switched to Neurobasal (NB) medium (without glutamate) supplemented with B-27 (Invitrogen) for overnight culture. Cells were harvested in Versene, centrifuged, and resuspended at 1 × 107/ml in flow buffer (PBS, pH 7.2, containing 2% fetal bovine serum and 0.2% sodium azide). Cells were then incubated for 30 min at 4°C in the presence of fluorescently labeled antibodies (1 μg/106 cells). The antibodies used in the assays were as follows: biotin-mouse anti-human CCR-2 (R&D Systems catalog #FAB151B); PE-mouse anti-human CD193 (CCR3) (BD Biosciences catalog #558165); FITC-mouse anti-human CD181 (CXCR1) (BD Biosciences catalog #555939); APC-mouse anti-human CD182 (CXCR2) (BD Biosciences catalog #551127); PE-mouse anti-human CD183 (CXCXR3) (BD Biosciences catalog #557185); PE-mouse anti-human TrkA (high-affinity NGF receptor) (RD Systems catalog #FAB1751P); biotin-mouse anti-human CD271 (low-affinity NGF receptor) (BD Biosciences catalog #557195). Following a wash, step wells containing biotinylated antibodies were further incubated for 20 min with streptavidin-APC or streptavidin-PE (BD PharMingen). Propidium Iodide was used to differentiate viable cells and acquisition was immediately performed in a FACScalibur (BD Biosciences). Cells were analyzed in FlowJo software (Tree Star).
Macrophage supernatant.
Macrophages/microglia were isolated by Percoll gradient from disrupted, collagenase-digested brains of rhesus macaques with SIVE and maintained in RPMI 1640 supplemented with MCSF at 50 ng/ml (Peprotech) up to 30 d. After day 7, supernatant was harvested daily, and samples with p27 >1 ng/ml were filtered (MW cutoff 30 kDa) and filtrates stored at −80°C.
Western blot.
Following treatment, cells were washed twice in ice-cold PBS, scraped, centrifuged, and then resuspended in ice-cold RIPA lysis buffer [50 mm HEPES buffer, 1 mm sodium orthovanadate, 10 mm sodium pyrophosphate, 10 mm NaF, 1% NP-40, 30 mm p-nitrophenyl phosphate (MP Biomedicals), 1 mm PMSF, 1× complete protease inhibitor cocktail (Roche Diagnostics)]. Samples were incubated on ice for 30 min with periodic vortexing, then centrifuged at high speed for 25 min at 4°C, after which supernatants containing soluble proteins were collected. Total protein concentration was determined with the bicinchoninic acid assay, using bovine serum albumin standards. Proteins were separated on a 4–12% Bis-Tris gradient gel (Invitrogen) and transferred electrophoretically to 0.45 μm pore PVDF membrane (Invitrogen). Nonspecific antibody binding was blocked by 5% nonfat dried milk for 1 h at room temperature. Immunoblotting was performed with antibody against EGR1 (1:1000, Cell Signaling Technology mAb 15F7), followed by secondary antibody (1:10,000 HRP-conjugated anti rabbit IgG; GE Healthcare). The blot was developed with 1:1 solution of Super Signal West Pico Chemiluminescent Substrate and Luminol/Enhancer (Thermo Fisher Scientific). The blot was then stripped using ReStore Western blot stripping buffer (Thermo Fisher Scientific) and reprobed for GAPDH (loading control; Millipore Bioscience Research Reagents, MAB374).
RT-PCR.
Probes (labeled with FAM and TAMRA) and primers were designed to detect human and/or rhesus genes, as appropriate and were obtained from Eurogentec. For the housekeeping genes (18S, GAPDH, and TBP), the sequences used are the same as given in (Wikoff et al., 2008). For the other genes, the sequences (given as forward—for, reverse—rev, and probe—pro) were as follows: EGR1: for ACGCCGAACACTGACATTT, rev GCTGTGGAAACAGGTAGTCG, pro TCCAGTACCCGCCTCCTGCC; PTN: for AATGGCAGTGGAGTGTGTGT, rev CTTCATGGTTTGCTTGCACT, pro CGGCTCCAGTCCGAGTGCCT; NR4A1: for TTCCTGGAGCTCTTCATCCT, rev GAAGATGAGCTTGCCCTCTC, pro CGCCTGGCATACAGGTCTAAGCC; HES5: for AAGCCAGTGGTGGAGAAGAT, rev TAGCTGACAGCCATCTCCAG, pro CGACCGCATCAACAGCAGCA; CNTNAP1: for CTTCAAGACCGAGGAGAAGG, rev CAGCTCGATCGTCACGTAGT, pro ACGGGCTTCTGCTGCACGC. RT-PCR was performed using the Stratagene MXPro 3000 instrument. Figures show gene expression normalized by housekeeping genes in terms of relative units; statistical testing was performed on log-transformed data.
Results
Expression analysis of SIV-infected macaques
Hippocampal RNA from nine uninfected and nine SIV-infected rhesus macaques was hybridized to Affymetrix Rhesus Macaque Genome chips and expression values calculated using the Robust Multichip Average (Irizarry et al., 2003) (see Materials and Methods). A summary of the treatment history of the monkeys studied is provided in supplemental Table S1 (available at www.jneurosci.org as supplemental material). Of 18,313 macaque genes that could be unequivocally mapped to human Entrez Gene IDs by best reciprocal Blast (see Materials and Methods); 720 were considered to be significantly upregulated and 106 significantly downregulated [CyberT (Baldi and Long, 2001) p ≤ 0.001, PPDE ∼95%; see Materials and Methods] (supplemental Table S2, available at www.jneurosci.org as supplemental material). Analysis of the differentially expressed genes for known functions and pathway membership using DAVID (Dennis et al., 2003) showed significant enrichment of upregulated genes for KEGG pathways and/or GO Biological Processes relating to immune, inflammatory, and stress responses, apoptosis, cell proliferation, and signaling cascades (JAK-STAT, NFκB), but little functional enrichment of downregulated genes.
Identification of transcriptionally active network modules
To facilitate identification of genes whose downregulation may contribute to neuronal dysfunction in HAD, macaque expression data were integrated with a human protein interaction network (10,906 proteins, 81,211 edges) assembled from several publicly available datasets (see Materials and Methods). The integrated network was then searched to identify “discriminatory network modules” using PinnacleZ (Chuang et al., 2007). “Discriminatory network modules” are connected subnetworks in the protein interaction map whose average mRNA expression level significantly discriminates between two experimental classes; significance is assessed using permutation testing (see Materials and Methods). Figure 1 provides an overview of the experimental and bioinformatic methodology.
Figure 1.
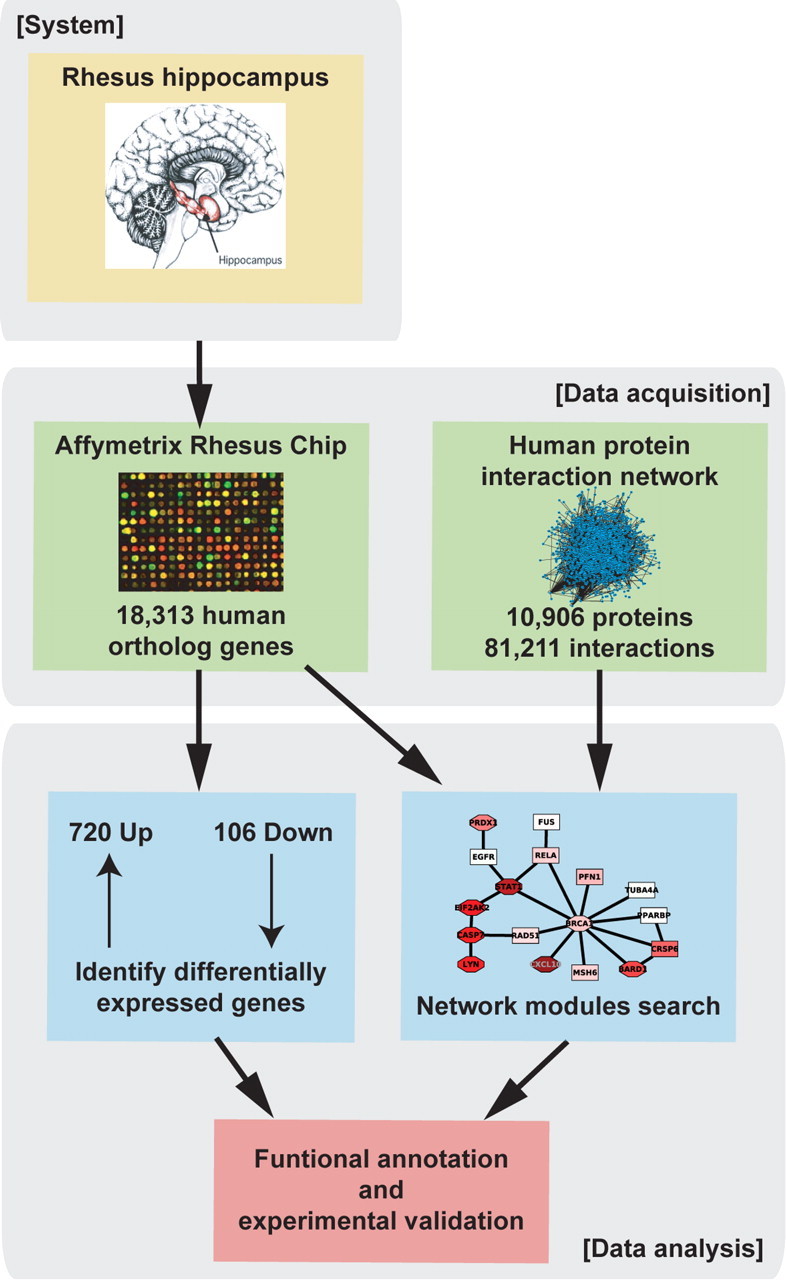
Overview of analysis process. Expression data from uninfected and SIV-infected rhesus hippocampus were integrated with a human protein–protein interaction network assembled from public sources to facilitate identification of novel SIVE-associated gene modules using a greedy search algorithm.
We recovered 447 modules containing a range of 2–20 proteins with discriminatory scores significantly better than those of randomly generated modules. Module expression was largely coherent, as modules contained predominantly upregulated (N = 429) or downregulated (N = 18) proteins in addition to protein products of nondifferentially expressed genes. As shown in supplemental Figure S1 (available at www.jneurosci.org as supplemental material), the effect of averaging expression values of genes in significant modules is to reduce intensity and increase homogeneity within the experimental group (SIVE or non-SIVE) relative to individual gene expression values. We noted that 31 proteins appeared in 5% or more of the significant modules and 30 of these proteins form a connected module within the protein interaction network (Fig. 2A). This group of high-frequency proteins is significantly enriched [Benjamini (Benjamini and Hochberg, 1995)-corrected p < 0.05; DAVID (Dennis et al., 2003)] for intracellular signaling cascade (N = 14), response to stress (N = 13), cell cycle (N = 10), and apoptosis (N = 9), key processes in SIVE. Most of these proteins are either highly connected in the network (>50 interactions; N = 22) or correspond to highly differentially expressed genes (p < 10−5; N = 13), or both (N = 9), providing a rationale for their frequent selection in significant modules. To reduce redundancy among significant modules containing these high-frequency proteins while maximizing recovery of modules containing significantly downregulated genes, we separated the modules into those containing ≥1 downregulated genes and those containing only ≥1 upregulated genes, then in each case removed modules showing substantial overlap with higher-scoring modules (Jaccard index > 0.15). Our final set of 110 modules represents the union of these two filtering procedures and is available in the CellCircuits Database (Mak et al., 2007) at http://www.cellcircuits.org/Gersten2009/.
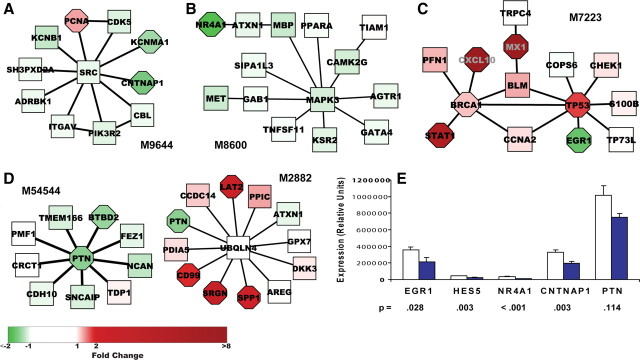
Figure 2.
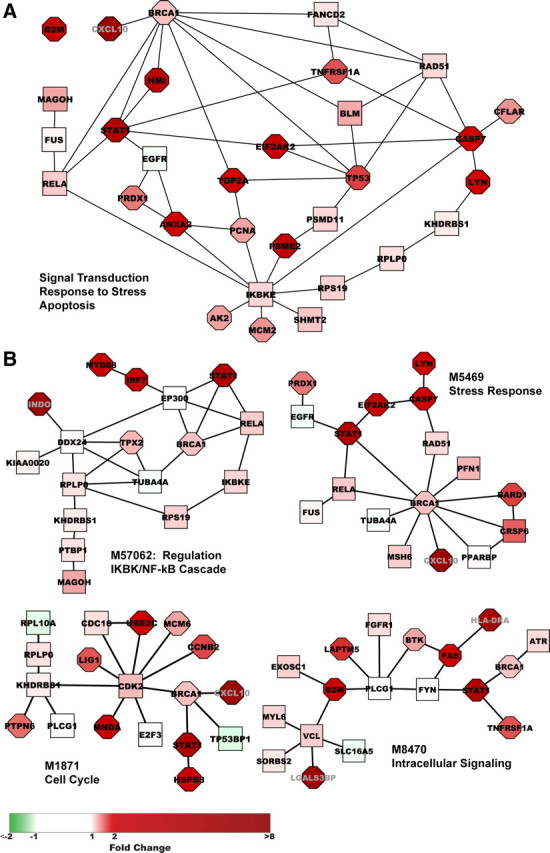
Highest scoring significant modules. A, Thirty of thirty-one proteins present in ≥22 (5%) of the significant modules form a connected module within the PPIN. B, The four highest scoring filtered modules, M57062, M5469, M1871, and M8470, are upregulated modules with top GO annotations (BiNGO plugin to Cytoscape) relating to regulation of I-κB kinase/NF-κB cascade (MYD88, RELA, IKBKE), stress response (CXCL10, PRDX1, EGFR, EIF2AK2, RELA, BRCA1, BARD1, RAD51, MSH6), cell cycle (UBE2C, E2F3, KHDRBS1, CDK2, CDC16, MCM6, CCNB2, LIG1), and intracellular signaling (FAS, ATR, FGFR1, BRCA1, STAT1, FYN, BTK, PLCG1), respectively. Fold change in gene expression relative to uninfected monkeys is depicted by intensity of node color: red (upregulated) or green (downregulated). Significance of differential expression as determined by Cyber T p value is represented by node shape: rectangular (p > 10−3), octagonal (≤10−3).
Functional enrichment of active network modules
Figure 2B presents the four top-scoring modules, all upregulated, which share STAT1 and BRCA1 but include other proteins yielding top GO annotations relating to regulation of I-κB kinase/NF-κB cascade (M57062), stress response (M5469), cell cycle (M1871), and intracellular signaling (M8470), respectively [Benjamini (Benjamini and Hochberg, 1995)-corrected p < 0.05; BiNGO plugin (Maere et al., 2005) to Cytoscape (Shannon et al., 2003)]. Figure 3A presents the top-scoring downregulated module, M9644, which is significantly enriched for genes participating in signal transduction. Notably, this module includes three neuronal signaling proteins [CNTNAP1 (Peles et al., 1997), CDK5 (Chen et al., 2007), and ITGAV (Taga et al., 2002)] and two ion channels (KCNB1 and KCNMA1). Proteins in this module are connected via the tyrosine kinase SRC; several of these proteins, or related proteins, have been predicted (CBL, CDK5, PIK3R2, KCNB2, and ITGB1,3,4; http://networkin.info/search.php) (Linding et al., 2007) or demonstrated (SH3PXD2A) (Lock et al., 1998) to be SRC substrates. Most of the genes are only minimally, if at all, downregulated; nevertheless, expression of the module protein members, taken as a unit, significantly distinguishes between uninfected animals and those with SIVE. The full set of 110 modules contains 749 proteins, including 189 differentially expressed genes (175 upregulated and 14 downregulated), which represents 33% of the 570 differentially expressed genes present in the protein interaction network. The large number of nondifferentially expressed genes in the modules provide connectivity among differentially expressed genes and in some cases may reflect masking of neuronal differential expression as a result of expression in non-neuronal hippocampal cells and/or functionality not regulated via transcription. For example, the activity of SRC (Fig. 3A) is modulated by phosphorylation (Lieser et al., 2005).
Figure 3.
Significant modules containing selected downregulated genes and RT-PCR confirmation. A–D, Significant modules in which downregulated genes appear. Module M9644 (Fig. 3A) was the highest scoring downregulated module and is significantly enriched for genes participating in signal transduction (PCNA, ITGAV, ADRBK1, CDK5, CBL, CNTNAP1, PIK3R2, SRC). Fold change in gene expression relative to uninfected monkeys is depicted by intensity of node color: red (upregulated) or green (downregulated). Significance of differential expression as determined by Cyber T p value is represented by node shape: rectangular (p > 10−3), octagonal (≤10−3). E, Hippocampal tissue from each of the nine uninfected and nine SIVE monkeys was tested by RT-PCR for four downregulated genes present in significant modules and HES5. Normalization was done using housekeeping genes 18S, TBP, and GAPDH. Figure shows mean expression ± SEM in terms of relative units; t test was performed on log-transformed data. Unfilled bars show control data; solid blue bars show SIVE data.
Network module analysis implicates EGR1 in SIVE-induced dementia
Past SIV/HIV microarray studies have focused on upregulated genes which are more striking in their differential expression and hence easier to confirm using small-scale experiments. However, these upregulated genes have predominantly reflected inflammatory responses, whereas many neuron-related functional annotations in a prior human study were associated with downregulated genes (Masliah et al., 2004). To identify novel HAD-related candidate genes we focused on downregulated genes present in significant modules. Of the 14 genes with significant p values ≤0.001, six had a fold-reduction ≥1.5 (∼33% reduction). We selected four of these genes, which have been associated with brain physiology, as well as HES5, a significantly downregulated gene with more than twofold reduction not included in a significant module, and performed RT-PCR to confirm the downregulation seen in the microarray data. Figure 3A–D presents the modules in which these gene products appear. CNTNAP1 (M9644) is a transmembrane protein that may act as a signaling subunit of contactin (Peles et al., 1997) and is required, together with contactin, for normal organization of paranodal junctions between glia and axons in myelinated fibers (Bhat et al., 2001). NR4A1 (M8600) is a nuclear receptor and an early response gene that is specifically upregulated during memory consolidation (von Hertzen and Giese, 2005). It is connected to the module via ATXN1, whose absence in Sca1-null mice is associated with deficits in learning (Matilla et al., 1998). EGR1 (see below) is an early response transcription factor critical for certain types of memory and learning (Jones et al., 2001; Bozon et al., 2002; Knapska and Kaczmarek, 2004). PTN (M54544, M2882) is an extracellular matrix-associated protein that promotes neurite outgrowth (Marchionini et al., 2007) and that has been implicated in both reduced LTP and faster learning in mice (Pavlov et al., 2002). Among its neighbors in M54544, PTN is connected to SNCAIP, which associates with α-synuclein and has been identified as a component of Lewy bodies and other neurodegenerative-associated protein inclusions (Krüger, 2004). As shown in Figure 3E, RT-PCR confirmed significant downregulation in SIVE hippocampus for four of the five genes tested (EGR1, NR4A1, CNTNAP1, and HES5, but not PTN).
We were particularly intrigued by the inclusion of EGR1 (also known as Zif268, KROX-24, and NGFI-A), a zinc finger transcription factor, among the group of downregulated genes in the discriminatory modules. In addition to being downregulated in the current SIVE hippocampus dataset, it has been found to be downregulated in gene expression studies of experimental models of other neurodegenerative diseases, including Huntington's (Crocker et al., 2006), age-related cognitive impairment (Blalock et al., 2003), and scrapie (Booth et al., 2004). As shown in Figure 3C, EGR1 appears in a module (M7223) which contains mostly upregulated proteins involved in regulation of transcription, response to stress, programmed death, and cell cycle. EGR1 is linked to the module via TP53, a transcription factor which contributes to HAD via induction of apoptotic pathways and perhaps other more subtle effects (Garden and Morrison, 2005). The seed protein for this module, TRPC4, is a member of a family of nonselective cation channels; it was found to be highly expressed in several brain regions of the adult rodent, including frontal cortex, hippocampus, and dentate gyrus, and may be involved in learning and memory (Fowler et al., 2007). The association of downregulated EGR1 with other proteins that may impact memory is of interest, given the well described role of EGR1 in synapse plasticity, memory, and learning (Jones et al., 2001; Bozon et al., 2002; Knapska and Kaczmarek, 2004), and led us to select EGR1 for further study.
EGR1 expression in neurons
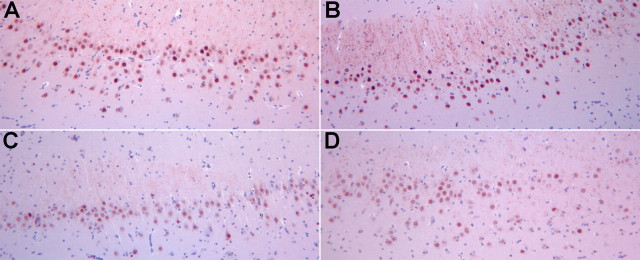
After confirming by RT-PCR downregulation of EGR1 in SIVE hippocampus (Fig. 3E), we asked whether the observed downregulation of EGR1 was occurring in neurons. Tissue from the hippocampus from four of the animals from each was available for examination of EGR1 protein expression by immunohistochemistry. Examination of the hippocampus revealed a distinctly reduced expression of EGR1 within hippocampal neurons (Fig. 4). Quantification revealed a significant reduction of EGR1 expression by 42% in SIVE relative to control (average ± SD expression in the CA1 region in control brains 78.24 ± 17.33, in SIVE brains 45.69 ± 7.62, p = 0.014). Therefore the reduction in EGR1 mRNA in the hippocampus is indeed reflective of reduced protein expression in neurons.
Figure 4.
EGR1 expression in hippocampal neurons. Deparaffinized hippocampal sections from control monkeys (A, B) and monkeys with SIVE (C, D) were immunohistochemically stained for EGR1 content by immunohistochemistry. Compared with control sections, staining of sections from SIVE monkeys is less intense and includes fewer heavily stained cells. Quantification (see Materials and Methods) revealed a 42% reduction of EGR1 in SIVE relative to control (p = 0.014).
CCL8 downregulates EGR1 in differentiated SK-N-MC cells
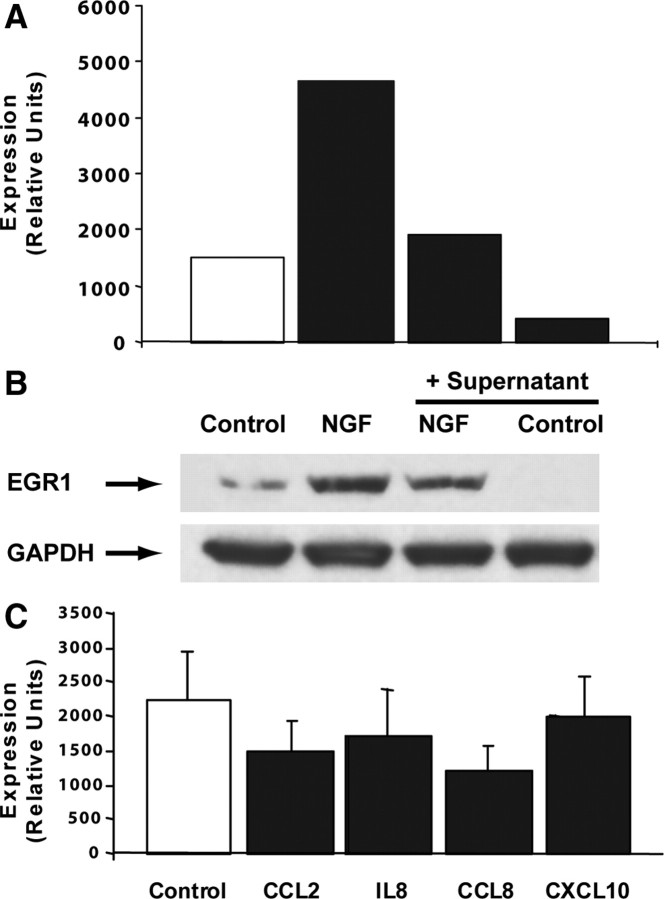
We next questioned whether neuronal downregulation of EGR1 might be a result of microglial activation, thereby contributing to impaired cognition. To facilitate experimental manipulation we used SK-N-MC, a human neuroblastoma cell line which can be differentiated toward a neuronal phenotype by exposure to retinoic acid (Caffrey et al., 2006). We first evaluated the effect of supernatant prepared from cultured SIV-infected macrophages (see Materials and Methods). As shown in Figure 5A, retinoic acid-differentiated SK-N-MC cells showed a decrease in both constitutive and NGF-stimulated EGR1 expression after 2-day pretreatment with a 1:50 dilution of supernatant; this decrease in expression was associated with a decrease in EGR1 protein, as shown in Figure 5B.
Figure 5.
CCL8 downregulated EGR1 in differentiated SK-N-MC cells. A, Culture supernatant from SIV-infected macrophages inhibited both constitutive and NGF-induced (30 min treatment) EGR1 expression in 4 d differentiated SK-N-MC cells after 2 d pretreatment at 1:50 supernatant dilution. Normalization was done using housekeeping genes 18S and GAPDH; the figure shows expression in terms of relative units. B, In cells cultured at the same time and under the same conditions as for A, constitutive and NGF-induced (2 h treatment) EGR1 protein was also reduced by 2 d pretreatment with 1:50 dilution of supernatant from infected macrophages. C, Mean EGR1 expression (normalized to 18S and GAPDH; relative units) ± SEM of five experiments in which 5 d differentiated SK-N-MC cells were treated for 2.5–3 h with one of the indicated chemokines at 100 ng/ml is presented. Inhibition of EGR1 by CCL8 was significant at p < 0.01 as assessed by paired sample one-way ANOVA (Prism: GraphPad) performed on log-transformed data followed by Dunnett's posttest.
Since chemotactic cytokines have been linked to both macrophage infiltration into the brain and effects on neurons in HAD (Bajetto et al., 2002; Ambrosini and Aloisi, 2004), we examined chemokine expression in our dataset to identify microglia products that might be present in the supernatant and responsible for EGR1 downregulation. CCL2, CCL8, IL8 (CXCL8), and CXCL10 were the most significantly upregulated chemokines in monkey SIVE-hippocampus (23.6-, 16.8-, 4.8-, and 36.3-fold, respectively) and are known to bind to receptors CCR2, CCR2/CCR3, CXCR1/CXCR2, and CXCR3, respectively. We confirmed that retinoic acid-treated SK-N-MC express these chemokine receptors (supplemental Fig. S2, available at www.jneurosci.org as supplemental material). To eliminate potential secondary effects, we evaluated the effect of these chemokines on EGR1 expression after a brief (2.5–3 h) exposure, using a standard chemokine concentration (100 ng/ml, ∼12 nm). In these experiments, cells were exposed to chemokines shortly (2–3 h) after removal from serum-containing medium. As shown in Figure 5C, CCL2, CCL8, and IL8 each appear to reduce EGR1 expression by ∼30%, similar in magnitude to the 40% reduction seen in SIV-infected hippocampus, whereas no reduction is seen after treatment with CXCL10. Paired sample one-way ANOVA using Prism (GraphPad) was performed on log-transformed data and confirmed significant differences among treatment groups (p = 0.0065); however, Dunnett's posttest revealed only CCL8 as significantly different from control (p < 0.01).
Discussion
Integration of hippocampal gene expression data from nine SIV-infected and nine uninfected monkeys with human protein interaction data recovered significant upregulated modules enriched for well described SIVE/HIVE processes, as well as several downregulated modules which include candidate neuronal genes whose downregulation may contribute to the neurologic dysfunction observed in monkeys and humans with SIV/HIV infection. Notably, many of the genes included in these modules are themselves not significantly differentially expressed, but taken together, each module significantly distinguishes between control and infected animals. This is considered to reflect perturbation of the same molecular pathways in different SIVE monkeys, even as individual animals may differ in perturbed expression of specific genes within those pathways. Dysregulation of molecular networks are responsible for functional pathology, leading to the change in state from normality to disease.
We selected several of the downregulated genes for quantitative RT-PCR confirmation and were able to confirm significant downregulation of EGR1, NR4A1, and CNTNAP1 in infected monkey hippocampus, despite the modest (less than twofold) downregulation of these of the genes. In addition to verifying that these genes are indeed downregulated in vivo, this confirmation provides support for our method of mapping Affymetrix rhesus probesets to the human protein network.
One of these genes, EGR1, has been well described as a participant in hippocampal functions of learning and memory (Jones et al., 2001; Bozon et al., 2002; Knapska and Kaczmarek, 2004) and it seems plausible, given the impairment of these functions in SIVE/HAD, that the observed downregulation occurs in neurons, rather than in other hippocampus cell types. Indeed, examination of hippocampal tissue sections revealed reduced expression in neurons in animals with SIVE.
Supporting this finding, others have reported that the HIV-Tat protein reduces NGF-stimulated EGR1 expression in SK-N-MC cells, the neuroblastoma cell line used in our studies, acting via inhibition of the MAPK/Erk1/2 pathway (Darbinian-Sarkissian et al., 2006). Since EGR1 is reported as downregulated in models of several other neurodegenerative diseases (Blalock et al., 2003; Booth et al., 2004; Crocker et al., 2006), we asked whether EGR1 downregulation might be a consequence, at least in part, of macrophage/microglia activation present in many of these diseases, rather than the result of SIV infection per se. We evaluated several chemokines that were highly upregulated in SIV-infected hippocampus and demonstrated that acute treatment with CCL8 reduces EGR1 expression in differentiated SK-N-MC cells by ∼30%. Although this appears to be a modest reduction, it is similar to the ∼50% reduction seen in heterozygous mouse EGR1 knock-out mutants which display the same signs of cognitive impairment (defects of late LTP and spatial learning) as those seen in homozygous mutants (Jones et al., 2001; Bozon et al., 2002).
Chemokines bind to G-protein-coupled receptors and activate different signaling pathways, including MAPK, PKB/AKT, and JAK/STAT pathways, depending on receptor(s) activated and cell type (Cho and Miller, 2002; Ragozzino, 2002). Although EGR1 is activated via the MAPK1/2 pathway (Darbinian-Sarkissian et al., 2006) and would therefore be expected to be induced by chemokine stimulation, inhibition of this pathway could occur via “RAF1-AKT crosstalk,” in which RAF1 kinase activity (and downstream MAPK/ERK activation) is suppressed by PKB/AKT phosphorylation of Ser259 in a ligand-, concentration-, and time course-dependent manner (Zimmermann and Moelling, 1999; Moelling et al., 2002) (Fig. 6B). This inhibitory mechanism could be particularly effective after CCL8 stimulation, as it has been reported that binding of CCL8 to CCR2, in contrast to CCL2 binding, fails to induce nuclear translocation of activated ERK1 and downstream gene induction (O'Boyle et al., 2007). AKT activity has been associated with neuroprotection in models of corticosterone- (Moosavi et al., 2008) and ischemia-induced (Fukunaga and Kawano, 2003) cell death. AKT activity is also required for CXCR2-mediated survival of cerebellar granule cells (Limatola et al., 2002), and is induced after CCL2 stimulation of spinal cord neurons (Gosselin et al., 2005). The latter two observations underscore the ability of chemokines to mediate neuroprotective, as well as neuroinflammatory functions. Downregulation of EGR1 might then occur as part of a neuroprotective response, with the associated untoward effect in hippocampus of reduced neuroplasticity.
Figure 6.
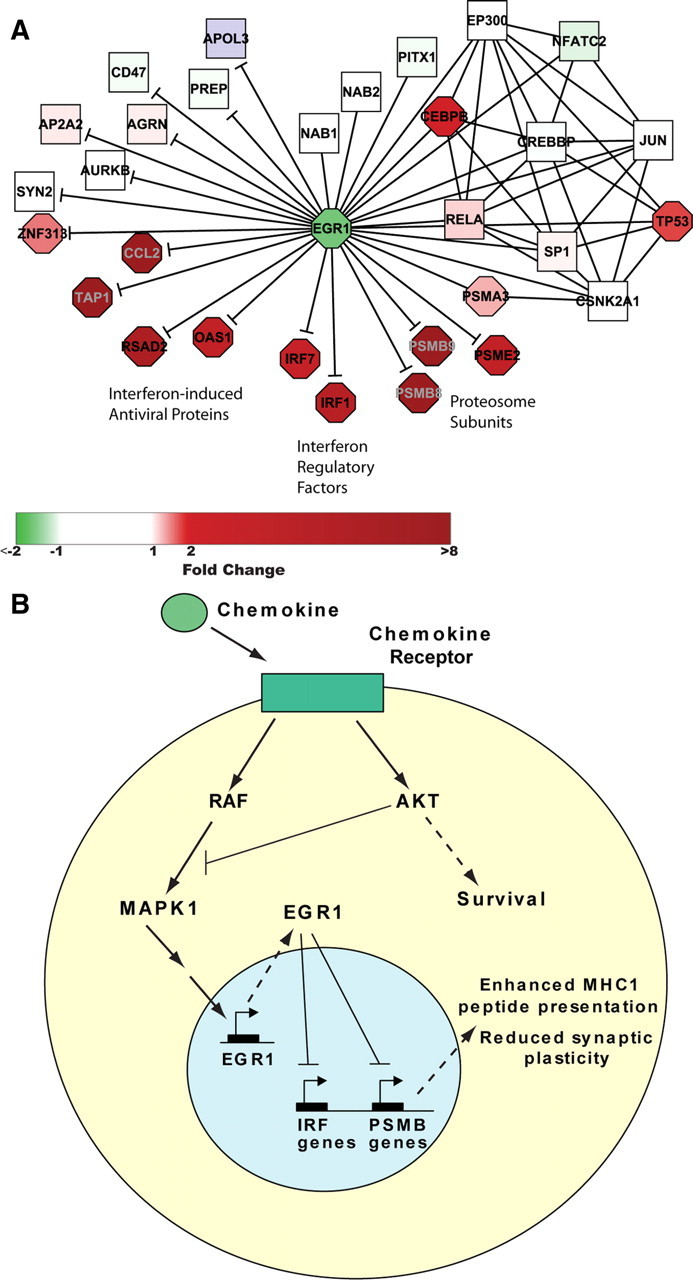
Proposed model of EGR1 downregulation in SIVE. A, EGR1 protein binding partners in the protein interaction network (—) and high-confidence gene targets (—|) taken from James et al. (2005) (their Table 1), in which Irf7 appears twice, once with p = 0.0025 and once with p = 0.05. Rat gene names were associated with human Entrez Gene IDs using Entrez Gene Symbols and aliases. B, Schematic depiction of proposed mechanism of EGR1 downregulation by chemokines in SIV-infected hippocampus and consequent reduced neuroplasticity.
Although only CCL8 produced statistically significant EGR1 downregulation in our study, CCL2 and IL8 also appeared to inhibit EGR1 expression, and failure to observe significance for CCL2 and IL8 could reflect the dose dependency of chemokine effects. The possibility of EGR1 downregulation by IL8 is particularly interesting, as IL8 has been reported both to mediate neuroprotection (Limatola et al., 2002) and to reduce rat hippocampal long-term potentiation (LTP) (Xiong et al., 2003); suppression of EGR1 might then work in tandem with the mechanism proposed by the authors, i.e., reduction of Ca2+ currents by IL8 (Xiong et al., 2003).
We selected EGR1 for further study based on its inclusion in significant protein-interaction modules; however, as a transcription factor, the effects of EGR1 downregulation may be better understood in terms of its target genes, amplifying the effect of its altered expression through its control of expression of many other genes. Interestingly, a recent microarray study of rat PC12 neuronal cells transfected with an EGR1 expression vector reported putative targets of EGR1 regulation (James et al., 2005), some of which were later confirmed experimentally (James et al., 2006). All but three of the genes were downregulated by EGR1 overexpression. We analyzed the putative targets reported to have at least one EGR response element in the promoter region (excluding MHC genes, which could not be reliably mapped to a human Entrez ID) for their expression in our SIVE dataset. Of the 100 genes mapped to a human Entrez ID (see supplemental Table S3, available at www.jneurosci.org as supplemental material) 94 had expression data, and 25 were differentially expressed. Of the 24 upregulated genes, all were reported as downregulated in the EGR1-overexpressing in PC12 cells; the single downregulated gene was EGR1, reported to upregulate itself. In this SIVE setting of downregulated EGR1, the set of putative EGR1-downregulated target genes is highly enriched for upregulated genes (hypergeometric p = 2.5 × 10−12) with respect to hippocampal SIVE gene expression in the complete set of mapped genes, providing support for their identification by James et al. (2005) as targets of EGR1 repression in neurons.
We then focused on a high-confidence subset of putative EGR1 targets whose differential expression was reported as significant at p < 0.05, all 17 of which were downregulated in the EGR1-overexpressing in PC12 cells, and added them to the human network subset of EGR1-protein interactions as directed interactions. As shown in Figure 6A, expression of these genes in the SIVE dataset, in the context of downregulated EGR1, was either significantly upregulated (N = 10), unchanged (N = 6), or not determined; in no case was expression significantly downregulated (significant enrichment for upregulated genes: hypergeometric p = 7.3 × 10−11). This high-confidence set of upregulated putative targets of EGR1 regulation includes three clusters, interferon-induced antiviral proteins, OAS1 and RSAD2, interferon regulatory factors, IRF1 and IRF7, and three interferon gamma-induced proteasome subunits, PSMB8, PSMB9, and PSME2, as well as TAP1, essential for MHC class I antigen processing. IRF comprise a family of transcription factors that not only induce type I interferon transcription but also regulate transcription of interferon-induced genes, as well as play diverse roles in a variety of cell types (Ozato et al., 2007; Paun and Pitha, 2007). In particular, IRF1 regulates interferon gamma-stimulated induction of proteosome immunosubunits PSMB8, PSMB9, and PSMB10 whose replacement in the proteosome facilitates MHC class I antigen processing and antiviral immunity (Namiki et al., 2005). Proteasome activity has also been implicated in neuronal plasticity, associated with both a facilitating (Lopez-Salon et al., 2001) and suppressive (Speese et al., 2003) effect, with the overall outcome likely related to the precise balance of active subunits influencing specific protein degradation (James et al., 2006).
These results, together, suggest a model in which downregulation of EGR1 in SIVE is a consequence of chemokine expression, brought about in the host for immune stimulation and protection of the CNS. In turn, downregulation of EGR1 promotes changes, both directly and via interferon regulatory factors, in proteasomal subunit selection that are detrimental to neuronal plasticity and cognition (Fig. 6B). In any case, by linking activated/infected macrophages to cognitive deficit in SIVE via downregulation of EGR1, an essential transcription factor in hippocampal neuronal function, our studies have identified a major factor in the indirect toxicity of microglia/macrophages on neurons.
Footnotes
We acknowledge National Institutes of Health Grants MH062261 and MH073490 for support of this work. We also thank Han-Yu Chuang for assistance with the PinnacleZ software, Samad Lotia for implementing PinnacleZ as a plug-in to Cytoscape, and James Buescher for assistance with immunohistochemistry. This is manuscript #06 from the Center for Integrative and Translational Neuroscience at the University of Nebraska Medical Center.
References
- Allison DB, Gadbury GL, Heo M, Fernandez JR, Lee C-K, Prolla TA, Weindruch R. A mixture model approach for the analysis of microarray gene expression data. Comput Stat Data Anal. 2002;39:1–20. [Google Scholar]
- Ambrosini E, Aloisi F. Chemokines and glial cells: a complex network in the central nervous system. Neurochem Res. 2004;29:1017–1038. doi: 10.1023/b:nere.0000021246.96864.89. [DOI] [PubMed] [Google Scholar]
- Bader GD, Hogue CW. Analyzing yeast protein-protein interaction data obtained from different sources. Nat Biotechnol. 2002;20:991–997. doi: 10.1038/nbt1002-991. [DOI] [PubMed] [Google Scholar]
- Bader GD, Donaldson I, Wolting C, Ouellette BF, Pawson T, Hogue CW. BIND—the Biomolecular Interaction Network Database. Nucleic Acids Res. 2001;29:242–245. doi: 10.1093/nar/29.1.242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bader JS, Chaudhuri A, Rothberg JM, Chant J. Gaining confidence in high-throughput protein interaction networks. Nat Biotechnol. 2004;22:78–85. doi: 10.1038/nbt924. [DOI] [PubMed] [Google Scholar]
- Bajetto A, Bonavia R, Barbero S, Schettini G. Characterization of chemokines and their receptors in the central nervous system: physiopathological implications. J Neurochem. 2002;82:1311–1329. doi: 10.1046/j.1471-4159.2002.01091.x. [DOI] [PubMed] [Google Scholar]
- Baldi P, Long AD. A Bayesian framework for the analysis of microarray expression data: regularized t-test and statistical inferences of gene changes. Bioinformatics. 2001;17:509–519. doi: 10.1093/bioinformatics/17.6.509. [DOI] [PubMed] [Google Scholar]
- Benjamini Y, Hochberg Y. Controlling the false discovery rate: a practical and powerful approach to multiple testing. J R Stat Soc Series B Stat Methodol. 1995;57:289–300. [Google Scholar]
- Bhat MA, Rios JC, Lu Y, Garcia-Fresco GP, Ching W, St Martin M, Li J, Einheber S, Chesler M, Rosenbluth J, Salzer JL, Bellen HJ. Axon-glia interactions and the domain organization of myelinated axons requires neurexin IV/Caspr/Paranodin. Neuron. 2001;30:369–383. doi: 10.1016/s0896-6273(01)00294-x. [DOI] [PubMed] [Google Scholar]
- Blalock EM, Chen KC, Sharrow K, Herman JP, Porter NM, Foster TC, Landfield PW. Gene microarrays in hippocampal aging: statistical profiling identifies novel processes correlated with cognitive impairment. J Neurosci. 2003;23:3807–3819. doi: 10.1523/JNEUROSCI.23-09-03807.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Booth S, Bowman C, Baumgartner R, Sorensen G, Robertson C, Coulthart M, Phillipson C, Somorjai RL. Identification of central nervous system genes involved in the host response to the scrapie agent during preclinical and clinical infection. J Gen Virol. 2004;85:3459–3471. doi: 10.1099/vir.0.80110-0. [DOI] [PubMed] [Google Scholar]
- Bozon B, Davis S, Laroche S. Regulated transcription of the immediate-early gene Zif268: mechanisms and gene dosage-dependent function in synaptic plasticity and memory formation. Hippocampus. 2002;12:570–577. doi: 10.1002/hipo.10100. [DOI] [PubMed] [Google Scholar]
- Burdo TH, Marcondes MC, Lanigan CM, Penedo MC, Fox HS. Susceptibility of Chinese rhesus monkeys to SIV infection. AIDS. 2005;19:1704–1706. doi: 10.1097/01.aids.0000186823.76230.33. [DOI] [PubMed] [Google Scholar]
- Caffrey TM, Joachim C, Paracchini S, Esiri MM, Wade-Martins R. Haplotype-specific expression of exon 10 at the human MAPT locus. Hum Mol Genet. 2006;15:3529–3537. doi: 10.1093/hmg/ddl429. [DOI] [PubMed] [Google Scholar]
- Chen J, Yuan B. Detecting functional modules in the yeast protein-protein interaction network. Bioinformatics. 2006;22:2283–2290. doi: 10.1093/bioinformatics/btl370. [DOI] [PubMed] [Google Scholar]
- Chen TC, Lai YK, Yu CK, Juang JL. Enterovirus 71 triggering of neuronal apoptosis through activation of Abl-Cdk5 signalling. Cell Microbiol. 2007;9:2676–2688. doi: 10.1111/j.1462-5822.2007.00988.x. [DOI] [PubMed] [Google Scholar]
- Cho C, Miller RJ. Chemokine receptors and neural function. J Neurovirol. 2002;8:573–584. doi: 10.1080/13550280290101003. [DOI] [PubMed] [Google Scholar]
- Chuang HY, Lee E, Liu YT, Lee D, Ideker T. Network-based classification of breast cancer metastasis. Mol Syst Biol. 2007;3:140. doi: 10.1038/msb4100180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Crocker SF, Costain WJ, Robertson HA. DNA microarray analysis of striatal gene expression in symptomatic transgenic Huntington's mice (R6/2) reveals neuroinflammation and insulin associations. Brain Res. 2006;1088:176–186. doi: 10.1016/j.brainres.2006.02.102. [DOI] [PubMed] [Google Scholar]
- Darbinian-Sarkissian N, Czernik M, Peruzzi F, Gordon J, Rappaport J, Reiss K, Khalili K, Amini S. Dysregulation of NGF-signaling and Egr-1 expression by Tat in neuronal cell culture. J Cell Physiol. 2006;208:506–515. doi: 10.1002/jcp.20675. [DOI] [PubMed] [Google Scholar]
- Dennis G, Jr, Sherman BT, Hosack DA, Yang J, Gao W, Lane HC, Lempicki RA. DAVID: Database for Annotation, Visualization, and Integrated Discovery. Genome Biol. 2003;4:P3. [PubMed] [Google Scholar]
- Diesing TS, Swindells S, Gelbard H, Gendelman HE. HIV-1-associated dementia: a basic science and clinical perspective. AIDS Read. 2002;12:358–368. [PubMed] [Google Scholar]
- Ewing RM, Chu P, Elisma F, Li H, Taylor P, Climie S, McBroom-Cerajewski L, Robinson MD, O'Connor L, Li M, Taylor R, Dharsee M, Ho Y, Heilbut A, Moore L, Zhang S, Ornatsky O, Bukhman YV, Ethier M, Sheng Y, et al. Large-scale mapping of human protein-protein interactions by mass spectrometry. Mol Syst Biol. 2007;3:89. doi: 10.1038/msb4100134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fowler MA, Sidiropoulou K, Ozkan ED, Phillips CW, Cooper DC. Corticolimbic expression of TRPC4 and TRPC5 channels in the rodent brain. PLoS One. 2007;2:e573. doi: 10.1371/journal.pone.0000573. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fukunaga K, Kawano T. Akt is a molecular target for signal transduction therapy in brain ischemic insult. J Pharmacol Sci. 2003;92:317–327. doi: 10.1254/jphs.92.317. [DOI] [PubMed] [Google Scholar]
- Garden GA, Morrison RS. The multiple roles of p53 in the pathogenesis of HIV associated dementia. Biochem Biophys Res Commun. 2005;331:799–809. doi: 10.1016/j.bbrc.2005.03.185. [DOI] [PubMed] [Google Scholar]
- Gosselin RD, Varela C, Banisadr G, Mechighel P, Rostene W, Kitabgi P, Melik-Parsadaniantz S. Constitutive expression of CCR2 chemokine receptor and inhibition by MCP-1/CCL2 of GABA-induced currents in spinal cord neurones. J Neurochem. 2005;95:1023–1034. doi: 10.1111/j.1471-4159.2005.03431.x. [DOI] [PubMed] [Google Scholar]
- Ideker T, Ozier O, Schwikowski B, Siegel AF. Discovering regulatory and signalling circuits in molecular interaction networks. Bioinformatics. 2002;18(Suppl 1):S233–S240. doi: 10.1093/bioinformatics/18.suppl_1.s233. [DOI] [PubMed] [Google Scholar]
- Irizarry RA, Bolstad BM, Collin F, Cope LM, Hobbs B, Speed TP. Summaries of Affymetrix GeneChip probe level data. Nucleic Acids Res. 2003;31:e15. doi: 10.1093/nar/gng015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- James AB, Conway AM, Morris BJ. Genomic profiling of the neuronal target genes of the plasticity-related transcription factor—Zif268. J Neurochem. 2005;95:796–810. doi: 10.1111/j.1471-4159.2005.03400.x. [DOI] [PubMed] [Google Scholar]
- James AB, Conway AM, Morris BJ. Regulation of the neuronal proteasome by Zif268 (Egr1) J Neurosci. 2006;26:1624–1634. doi: 10.1523/JNEUROSCI.4199-05.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jones MW, Errington ML, French PJ, Fine A, Bliss TV, Garel S, Charnay P, Bozon B, Laroche S, Davis S. A requirement for the immediate early gene Zif268 in the expression of late LTP and long-term memories. Nat Neurosci. 2001;4:289–296. doi: 10.1038/85138. [DOI] [PubMed] [Google Scholar]
- Joshi-Tope G, Gillespie M, Vastrik I, D'Eustachio P, Schmidt E, de Bono B, Jassal B, Gopinath GR, Wu GR, Matthews L, Lewis S, Birney E, Stein L. Reactome: a knowledgebase of biological pathways. Nucleic Acids Res. 2005;33:D428–D432. doi: 10.1093/nar/gki072. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaul M, Zheng J, Okamoto S, Gendelman HE, Lipton SA. HIV-1 infection and AIDS: consequences for the central nervous system. Cell Death Differ. 2005;12(Suppl 1):878–892. doi: 10.1038/sj.cdd.4401623. [DOI] [PubMed] [Google Scholar]
- Knapska E, Kaczmarek L. A gene for neuronal plasticity in the mammalian brain: Zif268/Egr-1/NGFI-A/Krox-24/TIS8/ZENK? Prog Neurobiol. 2004;74:183–211. doi: 10.1016/j.pneurobio.2004.05.007. [DOI] [PubMed] [Google Scholar]
- Krüger R. The role of synphilin-1 in synaptic function and protein degradation. Cell Tissue Res. 2004;318:195–199. doi: 10.1007/s00441-004-0953-z. [DOI] [PubMed] [Google Scholar]
- Lieser SA, Shindler C, Aubol BE, Lee S, Sun G, Adams JA. Phosphoryl transfer step in the C-terminal Src kinase controls Src recognition. J Biol Chem. 2005;280:7769–7776. doi: 10.1074/jbc.M411736200. [DOI] [PubMed] [Google Scholar]
- Limatola C, Ciotti MT, Mercanti D, Santoni A, Eusebi F. Signaling pathways activated by chemokine receptor CXCR2 and AMPA-type glutamate receptors and involvement in granule cells survival. J Neuroimmunol. 2002;123:9–17. doi: 10.1016/s0165-5728(01)00472-6. [DOI] [PubMed] [Google Scholar]
- Linding R, Jensen LJ, Ostheimer GJ, van Vugt MA, Jørgensen C, Miron IM, Diella F, Colwill K, Taylor L, Elder K, Metalnikov P, Nguyen V, Pasculescu A, Jin J, Park JG, Samson LD, Woodgett JR, Russell RB, Bork P, Yaffe MB, et al. Systematic discovery of in vivo phosphorylation networks. Cell. 2007;129:1415–1426. doi: 10.1016/j.cell.2007.05.052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lock P, Abram CL, Gibson T, Courtneidge SA. A new method for isolating tyrosine kinase substrates used to identify fish, an SH3 and PX domain-containing protein, and Src substrate. EMBO J. 1998;17:4346–4357. doi: 10.1093/emboj/17.15.4346. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lopez-Salon M, Alonso M, Vianna MR, Viola H, Mello e Souza T, Izquierdo I, Pasquini JM, Medina JH. The ubiquitin-proteasome cascade is required for mammalian long-term memory formation. Eur J Neurosci. 2001;14:1820–1826. doi: 10.1046/j.0953-816x.2001.01806.x. [DOI] [PubMed] [Google Scholar]
- Maere S, Heymans K, Kuiper M. BiNGO: a Cytoscape plugin to assess overrepresentation of gene ontology categories in biological networks. Bioinformatics. 2005;21:3448–3449. doi: 10.1093/bioinformatics/bti551. [DOI] [PubMed] [Google Scholar]
- Mak HC, Daly M, Gruebel B, Ideker T. CellCircuits: a database of protein network models. Nucleic Acids Res. 2007;35:D538–D545. doi: 10.1093/nar/gkl937. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marchionini DM, Lehrmann E, Chu Y, He B, Sortwell CE, Becker KG, Freed WJ, Kordower JH, Collier TJ. Role of heparin binding growth factors in nigrostriatal dopamine system development and Parkinson's disease. Brain Res. 2007;1147:77–88. doi: 10.1016/j.brainres.2007.02.028. [DOI] [PubMed] [Google Scholar]
- Masliah E, Roberts ES, Langford D, Everall I, Crews L, Adame A, Rockenstein E, Fox HS. Patterns of gene dysregulation in the frontal cortex of patients with HIV encephalitis. J Neuroimmunol. 2004;157:163–175. doi: 10.1016/j.jneuroim.2004.08.026. [DOI] [PubMed] [Google Scholar]
- Matilla A, Roberson ED, Banfi S, Morales J, Armstrong DL, Burright EN, Orr HT, Sweatt JD, Zoghbi HY, Matzuk MM. Mice lacking ataxin-1 display learning deficits and decreased hippocampal paired-pulse facilitation. J Neurosci. 1998;18:5508–5516. doi: 10.1523/JNEUROSCI.18-14-05508.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mattson MP, Haughey NJ, Nath A. Cell death in HIV dementia. Cell Death Differ. 2005;12(Suppl 1):893–904. doi: 10.1038/sj.cdd.4401577. [DOI] [PubMed] [Google Scholar]
- Moelling K, Schad K, Bosse M, Zimmermann S, Schweneker M. Regulation of Raf-Akt cross-talk. J Biol Chem. 2002;277:31099–31106. doi: 10.1074/jbc.M111974200. [DOI] [PubMed] [Google Scholar]
- Moosavi M, Maghsoudi N, Zahedi-Asl S, Naghdi N, Yousefpour M, Trounce IA. The role of PI3/Akt pathway in the protective effect of insulin against corticosterone cell death induction in hippocampal cell culture. Neuroendocrinology. 2008;88:293–298. doi: 10.1159/000150441. [DOI] [PubMed] [Google Scholar]
- Namiki S, Nakamura T, Oshima S, Yamazaki M, Sekine Y, Tsuchiya K, Okamoto R, Kanai T, Watanabe M. IRF-1 mediates upregulation of LMP7 by IFN-gamma and concerted expression of immunosubunits of the proteasome. FEBS Lett. 2005;579:2781–2787. doi: 10.1016/j.febslet.2005.04.012. [DOI] [PubMed] [Google Scholar]
- O'Boyle G, Brain JG, Kirby JA, Ali S. Chemokine-mediated inflammation: identification of a possible regulatory role for CCR2. Mol Immunol. 2007;44:1944–1953. doi: 10.1016/j.molimm.2006.09.033. [DOI] [PubMed] [Google Scholar]
- Ozato K, Tailor P, Kubota T. The interferon regulatory factor family in host defense: mechanism of action. J Biol Chem. 2007;282:20065–20069. doi: 10.1074/jbc.R700003200. [DOI] [PubMed] [Google Scholar]
- Paun A, Pitha PM. The IRF family, revisited. Biochimie. 2007;89:744–753. doi: 10.1016/j.biochi.2007.01.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pavlov I, Võikar V, Kaksonen M, Lauri SE, Hienola A, Taira T, Rauvala H. Role of heparin-binding growth-associated molecule (HB-GAM) in hippocampal LTP and spatial learning revealed by studies on overexpressing and knockout mice. Mol Cell Neurosci. 2002;20:330–342. doi: 10.1006/mcne.2002.1104. [DOI] [PubMed] [Google Scholar]
- Peles E, Nativ M, Lustig M, Grumet M, Schilling J, Martinez R, Plowman GD, Schlessinger J. Identification of a novel contactin-associated transmembrane receptor with multiple domains implicated in protein-protein interactions. EMBO J. 1997;16:978–988. doi: 10.1093/emboj/16.5.978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peri S, Navarro JD, Amanchy R, Kristiansen TZ, Jonnalagadda CK, Surendranath V, Niranjan V, Muthusamy B, Gandhi TK, Gronborg M, Ibarrola N, Deshpande N, Shanker K, Shivashankar HN, Rashmi BP, Ramya MA, Zhao Z, Chandrika KN, Padma N, Harsha HC, et al. Development of human protein reference database as an initial platform for approaching systems biology in humans. Genome Res. 2003;13:2363–2371. doi: 10.1101/gr.1680803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ragozzino D. CXC chemokine receptors in the central nervous system: role in cerebellar neuromodulation and development. J Neurovirol. 2002;8:559–572. doi: 10.1080/13550280290100932. [DOI] [PubMed] [Google Scholar]
- Ramani AK, Bunescu RC, Mooney RJ, Marcotte EM. Consolidating the set of known human protein-protein interactions in preparation for large-scale mapping of the human interactome. Genome Biol. 2005;6:R40. doi: 10.1186/gb-2005-6-5-r40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rausch DM, Murray EA, Eiden LE. The SIV-infected rhesus monkey model for HIV-associated dementia and implications for neurological diseases. J Leukoc Biol. 1999;65:466–474. doi: 10.1002/jlb.65.4.466. [DOI] [PubMed] [Google Scholar]
- Remm M, Storm CE, Sonnhammer EL. Automatic clustering of orthologs and in-paralogs from pairwise species comparisons. J Mol Biol. 2001;314:1041–1052. doi: 10.1006/jmbi.2000.5197. [DOI] [PubMed] [Google Scholar]
- Roberts ES, Fox HS. Transcriptional analysis of inflammatory and glial responses in the central nervous system: development of RNA fingerprints for neuroAIDS and other central nervous system disorders. In: Gendelman HE, Grant I, Everall IP, Lipton SA, Swindells S, editors. The neurology of AIDS. Ed 2. Oxford: Oxford UP; 2005. pp. 225–237. [Google Scholar]
- Roberts ES, Zandonatti MA, Watry DD, Madden LJ, Henriksen SJ, Taffe MA, Fox HS. Induction of pathogenic sets of genes in macrophages and neurons in neuroAIDS. Am J Pathol. 2003;162:2041–2057. doi: 10.1016/S0002-9440(10)64336-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roberts ES, Burudi EM, Flynn C, Madden LJ, Roinick KL, Watry DD, Zandonatti MA, Taffe MA, Fox HS. Acute SIV infection of the brain leads to upregulation of IL6 and interferon-regulated genes: expression patterns throughout disease progression and impact on neuroAIDS. J Neuroimmunol. 2004;157:81–92. doi: 10.1016/j.jneuroim.2004.08.030. [DOI] [PubMed] [Google Scholar]
- Rual JF, Venkatesan K, Hao T, Hirozane-Kishikawa T, Dricot A, Li N, Berriz GF, Gibbons FD, Dreze M, Ayivi-Guedehoussou N, Klitgord N, Simon C, Boxem M, Milstein S, Rosenberg J, Goldberg DS, Zhang LV, Wong SL, Franklin G, Li S, et al. Towards a proteome-scale map of the human protein-protein interaction network. Nature. 2005;437:1173–1178. doi: 10.1038/nature04209. [DOI] [PubMed] [Google Scholar]
- Segal E, Wang H, Koller D. Discovering molecular pathways from protein interaction and gene expression data. Bioinformatics. 2003;19(Suppl 1):i264–i272. doi: 10.1093/bioinformatics/btg1037. [DOI] [PubMed] [Google Scholar]
- Shannon P, Markiel A, Ozier O, Baliga NS, Wang JT, Ramage D, Amin N, Schwikowski B, Ideker T. Cytoscape: a software environment for integrated models of biomolecular interaction networks. Genome Res. 2003;13:2498–2504. doi: 10.1101/gr.1239303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Speese SD, Trotta N, Rodesch CK, Aravamudan B, Broadie K. The ubiquitin proteasome system acutely regulates presynaptic protein turnover and synaptic efficacy. Curr Biol. 2003;13:899–910. doi: 10.1016/s0960-9822(03)00338-5. [DOI] [PubMed] [Google Scholar]
- Stelzl U, Worm U, Lalowski M, Haenig C, Brembeck FH, Goehler H, Stroedicke M, Zenkner M, Schoenherr A, Koeppen S, Timm J, Mintzlaff S, Abraham C, Bock N, Kietzmann S, Goedde A, Toksöz E, Droege A, Krobitsch S, Korn B, et al. A human protein-protein interaction network: a resource for annotating the proteome. Cell. 2005;122:957–968. doi: 10.1016/j.cell.2005.08.029. [DOI] [PubMed] [Google Scholar]
- Taga T, Suzuki A, Gonzalez-Gomez I, Gilles FH, Stins M, Shimada H, Barsky L, Weinberg KI, Laug WE. alpha v-Integrin antagonist EMD 121974 induces apoptosis in brain tumor cells growing on vitronectin and tenascin. Int J Cancer. 2002;98:690–697. doi: 10.1002/ijc.10265. [DOI] [PubMed] [Google Scholar]
- von Hertzen LS, Giese KP. Memory reconsolidation engages only a subset of immediate-early genes induced during consolidation. J Neurosci. 2005;25:1935–1942. doi: 10.1523/JNEUROSCI.4707-04.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Watry D, Lane TE, Streb M, Fox HS. Transfer of neuropathogenic simian immunodeficiency virus with naturally infected microglia. Am J Pathol. 1995;146:914–923. [PMC free article] [PubMed] [Google Scholar]
- Wikoff WR, Pendyala G, Siuzdak G, Fox HS. Metabolomic analysis of the cerebrospinal fluid reveals changes in phospholipase expression in the CNS of SIV-infected macaques. J Clin Invest. 2008;118:2661–2669. doi: 10.1172/JCI34138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xenarios I, Rice DW, Salwinski L, Baron MK, Marcotte EM, Eisenberg D. DIP: the database of interacting proteins. Nucleic Acids Res. 2000;28:289–291. doi: 10.1093/nar/28.1.289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xiong H, Boyle J, Winkelbauer M, Gorantla S, Zheng J, Ghorpade A, Persidsky Y, Carlson KA, Gendelman HE. Inhibition of long-term potentiation by interleukin-8: implications for human immunodeficiency virus-1-associated dementia. J Neurosci Res. 2003;71:600–607. doi: 10.1002/jnr.10503. [DOI] [PubMed] [Google Scholar]
- Zimmermann S, Moelling K. Phosphorylation and regulation of Raf by Akt (protein kinase B) Science. 1999;286:1741–1744. doi: 10.1126/science.286.5445.1741. [DOI] [PubMed] [Google Scholar]