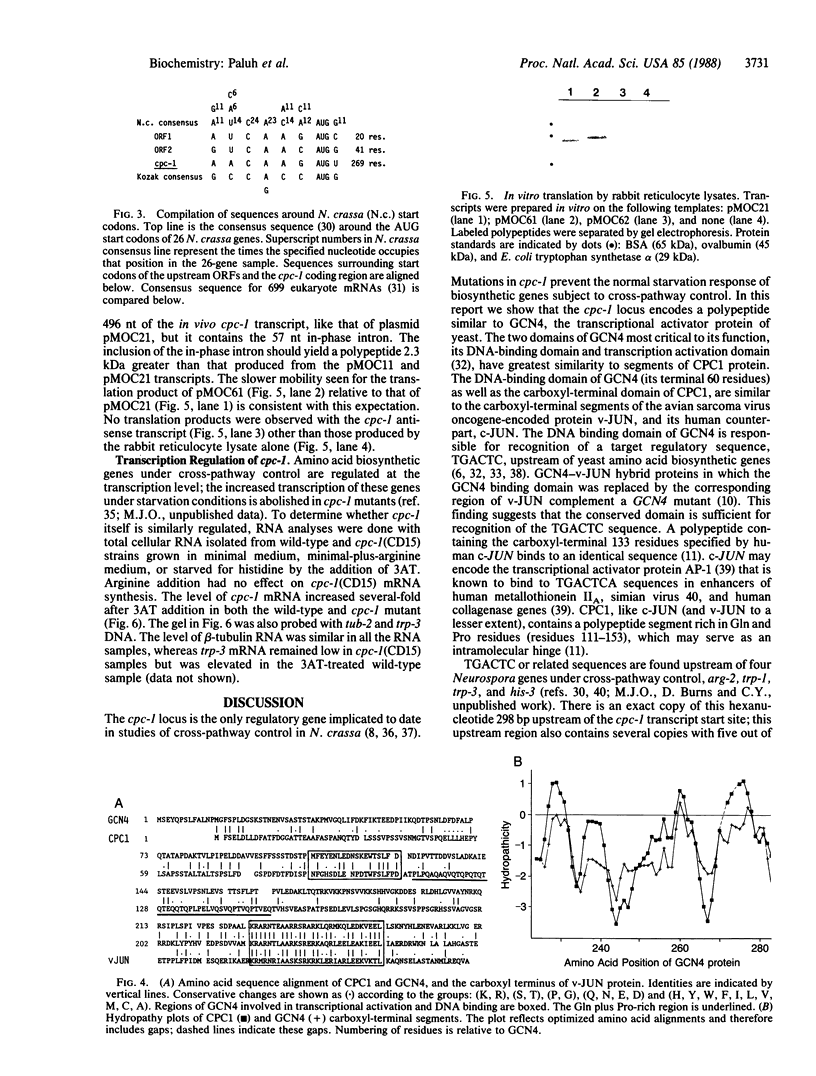
Abstract
Expression of the gene cpc-1 is required for cross-pathway-mediated regulation of amino acid-biosynthetic genes in Neurospora crassa. We have cloned cpc-1 and present an analysis of its structure and regulation. The cpc-1-encoded transcript contains three open reading frames, two of which are located in the 720-nucleotide leader segment preceding the cpc-1 coding region. The two leader open reading frames, if translated, would produce peptides 20 and 41 residues in length. The deduced amino acid sequence of the cpc-1 polypeptide, CPC1, contains segments similar to the DNA-binding and transcriptional activation domains of GCN4, the major cross-pathway regulatory protein of yeast. The structural and functional similarities of CPC1 and GCN4 proteins suggest that cpc-1 encodes the analogous transcriptional activator of N. crassa. Messenger RNA measurements indicate that cpc-1 is transcriptionally regulated in response to amino acid starvation. The segment of CPC1 similar to the DNA-binding domain of GCN4 also is similar to the DNA-binding domains of the avian sarcoma virus oncogene-encoded v-JUN protein and human c-JUN protein.
Full text
PDF



Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Arndt K., Fink G. R. GCN4 protein, a positive transcription factor in yeast, binds general control promoters at all 5' TGACTC 3' sequences. Proc Natl Acad Sci U S A. 1986 Nov;83(22):8516–8520. doi: 10.1073/pnas.83.22.8516. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barthelmess I. B. Mutants affecting amino acid cross-pathway control in Neurospora crassa. Genet Res. 1982 Apr;39(2):169–185. doi: 10.1017/s0016672300020863. [DOI] [PubMed] [Google Scholar]
- Barthelmess I. B. Regulation of amino acid synthetic enzymes in Neurospora crassa in the presence of high concentrations of amino acids. Mol Gen Genet. 1986 Jun;203(3):533–537. doi: 10.1007/BF00422082. [DOI] [PubMed] [Google Scholar]
- Berk A. J., Sharp P. A. Sizing and mapping of early adenovirus mRNAs by gel electrophoresis of S1 endonuclease-digested hybrids. Cell. 1977 Nov;12(3):721–732. doi: 10.1016/0092-8674(77)90272-0. [DOI] [PubMed] [Google Scholar]
- Berlin V., Yanofsky C. Isolation and characterization of genes differentially expressed during conidiation of Neurospora crassa. Mol Cell Biol. 1985 Apr;5(4):849–855. doi: 10.1128/mcb.5.4.849. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Biggin M. D., Gibson T. J., Hong G. F. Buffer gradient gels and 35S label as an aid to rapid DNA sequence determination. Proc Natl Acad Sci U S A. 1983 Jul;80(13):3963–3965. doi: 10.1073/pnas.80.13.3963. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bohmann D., Bos T. J., Admon A., Nishimura T., Vogt P. K., Tjian R. Human proto-oncogene c-jun encodes a DNA binding protein with structural and functional properties of transcription factor AP-1. Science. 1987 Dec 4;238(4832):1386–1392. doi: 10.1126/science.2825349. [DOI] [PubMed] [Google Scholar]
- Carsiotis M., Jones R. F., Lacy A. M., Cleary T. J., Fankhauser D. B. Histidine-mediated control of tryptophan biosynthetic enzymes in Neurospora crassa. J Bacteriol. 1970 Oct;104(1):98–106. doi: 10.1128/jb.104.1.98-106.1970. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carsiotis M., Jones R. F., Wesseling A. C. Cross-pathway regulation: histidine-mediated control of histidine, tryptophan, and arginine biosynthetic enzymes in Neurospora crassa. J Bacteriol. 1974 Sep;119(3):893–898. doi: 10.1128/jb.119.3.893-898.1974. [DOI] [PMC free article] [PubMed] [Google Scholar]
- DAVIS R. H. Consequences of a suppressor gene effective with pyrimidine and proline mutants of Neurospora. Genetics. 1962 Mar;47:351–360. doi: 10.1093/genetics/47.3.351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davis R. H. Genetics of arginine biosynthesis in Neurospora crassa. Genetics. 1979 Nov;93(3):557–575. doi: 10.1093/genetics/93.3.557. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feinberg A. P., Vogelstein B. A technique for radiolabeling DNA restriction endonuclease fragments to high specific activity. Anal Biochem. 1983 Jul 1;132(1):6–13. doi: 10.1016/0003-2697(83)90418-9. [DOI] [PubMed] [Google Scholar]
- Flint H. J. Changes in gene expression elicited by amino acid limitation in Neurospora crassa strains having normal or mutant cross-pathway amino acid control. Mol Gen Genet. 1985;200(2):283–290. doi: 10.1007/BF00425437. [DOI] [PubMed] [Google Scholar]
- Flint H. J., Wilkening J. Cloning of the arg-12 gene of Neurospora crassa and regulation of its transcript via cross-pathway amino acid control. Mol Gen Genet. 1986 Apr;203(1):110–116. doi: 10.1007/BF00330391. [DOI] [PubMed] [Google Scholar]
- Henikoff S. Unidirectional digestion with exonuclease III creates targeted breakpoints for DNA sequencing. Gene. 1984 Jun;28(3):351–359. doi: 10.1016/0378-1119(84)90153-7. [DOI] [PubMed] [Google Scholar]
- Hill D. E., Hope I. A., Macke J. P., Struhl K. Saturation mutagenesis of the yeast his3 regulatory site: requirements for transcriptional induction and for binding by GCN4 activator protein. Science. 1986 Oct 24;234(4775):451–457. doi: 10.1126/science.3532321. [DOI] [PubMed] [Google Scholar]
- Hinnebusch A. G. Evidence for translational regulation of the activator of general amino acid control in yeast. Proc Natl Acad Sci U S A. 1984 Oct;81(20):6442–6446. doi: 10.1073/pnas.81.20.6442. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hinnebusch A. G., Fink G. R. Positive regulation in the general amino acid control of Saccharomyces cerevisiae. Proc Natl Acad Sci U S A. 1983 Sep;80(17):5374–5378. doi: 10.1073/pnas.80.17.5374. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hinnebusch A. G. The general control of amino acid biosynthetic genes in the yeast Saccharomyces cerevisiae. CRC Crit Rev Biochem. 1986;21(3):277–317. doi: 10.3109/10409238609113614. [DOI] [PubMed] [Google Scholar]
- Hope I. A., Struhl K. Functional dissection of a eukaryotic transcriptional activator protein, GCN4 of yeast. Cell. 1986 Sep 12;46(6):885–894. doi: 10.1016/0092-8674(86)90070-x. [DOI] [PubMed] [Google Scholar]
- Hope I. A., Struhl K. GCN4 protein, synthesized in vitro, binds HIS3 regulatory sequences: implications for general control of amino acid biosynthetic genes in yeast. Cell. 1985 Nov;43(1):177–188. doi: 10.1016/0092-8674(85)90022-4. [DOI] [PubMed] [Google Scholar]
- Hope I. A., Struhl K. GCN4, a eukaryotic transcriptional activator protein, binds as a dimer to target DNA. EMBO J. 1987 Sep;6(9):2781–2784. doi: 10.1002/j.1460-2075.1987.tb02573.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hudson G. S., Davidson B. E. Nucleotide sequence and transcription of the phenylalanine and tyrosine operons of Escherichia coli K12. J Mol Biol. 1984 Dec 25;180(4):1023–1051. doi: 10.1016/0022-2836(84)90269-9. [DOI] [PubMed] [Google Scholar]
- Kozak M. An analysis of 5'-noncoding sequences from 699 vertebrate messenger RNAs. Nucleic Acids Res. 1987 Oct 26;15(20):8125–8148. doi: 10.1093/nar/15.20.8125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kozak M. Bifunctional messenger RNAs in eukaryotes. Cell. 1986 Nov 21;47(4):481–483. doi: 10.1016/0092-8674(86)90609-4. [DOI] [PubMed] [Google Scholar]
- Kyte J., Doolittle R. F. A simple method for displaying the hydropathic character of a protein. J Mol Biol. 1982 May 5;157(1):105–132. doi: 10.1016/0022-2836(82)90515-0. [DOI] [PubMed] [Google Scholar]
- Laemmli U. K. Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature. 1970 Aug 15;227(5259):680–685. doi: 10.1038/227680a0. [DOI] [PubMed] [Google Scholar]
- Lee W., Mitchell P., Tjian R. Purified transcription factor AP-1 interacts with TPA-inducible enhancer elements. Cell. 1987 Jun 19;49(6):741–752. doi: 10.1016/0092-8674(87)90612-x. [DOI] [PubMed] [Google Scholar]
- Legerton T. L., Yanofsky C. Cloning and characterization of the multifunctional his-3 gene of Neurospora crassa. Gene. 1985;39(2-3):129–140. doi: 10.1016/0378-1119(85)90306-3. [DOI] [PubMed] [Google Scholar]
- Maki Y., Bos T. J., Davis C., Starbuck M., Vogt P. K. Avian sarcoma virus 17 carries the jun oncogene. Proc Natl Acad Sci U S A. 1987 May;84(9):2848–2852. doi: 10.1073/pnas.84.9.2848. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Metzenberg R. L., Stevens J. N., Selker E. U., Morzycka-Wroblewska E. Identification and chromosomal distribution of 5S rRNA genes in Neurospora crassa. Proc Natl Acad Sci U S A. 1985 Apr;82(7):2067–2071. doi: 10.1073/pnas.82.7.2067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mueller P. P., Hinnebusch A. G. Multiple upstream AUG codons mediate translational control of GCN4. Cell. 1986 Apr 25;45(2):201–207. doi: 10.1016/0092-8674(86)90384-3. [DOI] [PubMed] [Google Scholar]
- Norrander J., Kempe T., Messing J. Construction of improved M13 vectors using oligodeoxynucleotide-directed mutagenesis. Gene. 1983 Dec;26(1):101–106. doi: 10.1016/0378-1119(83)90040-9. [DOI] [PubMed] [Google Scholar]
- Orbach M. J., Porro E. B., Yanofsky C. Cloning and characterization of the gene for beta-tubulin from a benomyl-resistant mutant of Neurospora crassa and its use as a dominant selectable marker. Mol Cell Biol. 1986 Jul;6(7):2452–2461. doi: 10.1128/mcb.6.7.2452. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perkins D. D., Radford A., Newmeyer D., Björkman M. Chromosomal loci of Neurospora crassa. Microbiol Rev. 1982 Dec;46(4):426–570. doi: 10.1128/mr.46.4.426-570.1982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sanger F., Nicklen S., Coulson A. R. DNA sequencing with chain-terminating inhibitors. Proc Natl Acad Sci U S A. 1977 Dec;74(12):5463–5467. doi: 10.1073/pnas.74.12.5463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schechtman M. G., Yanofsky C. Structure of the trifunctional trp-1 gene from Neurospora crassa and its aberrant expression in Escherichia coli. J Mol Appl Genet. 1983;2(1):83–99. [PubMed] [Google Scholar]
- Struhl K. The DNA-binding domains of the jun oncoprotein and the yeast GCN4 transcriptional activator protein are functionally homologous. Cell. 1987 Sep 11;50(6):841–846. doi: 10.1016/0092-8674(87)90511-3. [DOI] [PubMed] [Google Scholar]
- Thireos G., Penn M. D., Greer H. 5' untranslated sequences are required for the translational control of a yeast regulatory gene. Proc Natl Acad Sci U S A. 1984 Aug;81(16):5096–5100. doi: 10.1073/pnas.81.16.5096. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vogt P. K., Bos T. J., Doolittle R. F. Homology between the DNA-binding domain of the GCN4 regulatory protein of yeast and the carboxyl-terminal region of a protein coded for by the oncogene jun. Proc Natl Acad Sci U S A. 1987 May;84(10):3316–3319. doi: 10.1073/pnas.84.10.3316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vollmer S. J., Yanofsky C. Efficient cloning of genes of Neurospora crassa. Proc Natl Acad Sci U S A. 1986 Jul;83(13):4869–4873. doi: 10.1073/pnas.83.13.4869. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Werner M., Feller A., Messenguy F., Piérard A. The leader peptide of yeast gene CPA1 is essential for the translational repression of its expression. Cell. 1987 Jun 19;49(6):805–813. doi: 10.1016/0092-8674(87)90618-0. [DOI] [PubMed] [Google Scholar]