Abstract
Background and Aims
Consensus higher-level molecular phylogenies present a compelling case that an ancient divergence separates eukaryotic green algae into two major monophyletic lineages, Chlorophyta and Streptophyta, and a residuum of green algae, which have been referred to prasinophytes or micromonadophytes. Nuclear DNA content estimates have been published for less than 1% of the described green algal members of Chlorophyta, which includes multicellular green marine algae and freshwater flagellates (e.g. Chlamydomonas and Volvox). The present investigation summarizes the state of our knowledge and adds substantially to our database of C-values, especially for the streptophyte charophycean lineage which is the sister group of the land plants. A recent list of 2C nuclear DNA contents for isolates and species of green algae is expanded by 72 to 157.
Methods
The DNA-localizing fluorochrome DAPI (4′,6-diamidino-2-phenylindole) and red blood cell (chicken erythrocytes) standard were used to estimate 2C values with static microspectrophotometry.
Key Results
In Chlorophyta, including Chlorophyceae, Prasinophyceae, Trebouxiophyceae and Ulvophyceae, 2C DNA estimates range from 0·01 to 5·8 pg. Nuclear DNA content variation trends are noted and discussed for specific problematic taxon pairs, including Ulotrichales–Ulvales, and Cladophorales–Siphonocladales. For Streptophyta, 2C nuclear DNA contents range from 0·2 to 6·4 pg, excluding the highly polyploid Charales and Desmidiales, which have genome sizes of up to 14·8 and 46·8 pg, respectively. Nuclear DNA content data for Streptophyta superimposed on a contemporary molecular phylogeny indicate that early diverging lineages, including some members of Chlorokybales, Coleochaetales and Klebsormidiales, have genomes as small as 0·1–0·5 pg. It is proposed that the streptophyte ancestral nuclear genome common to both the charophyte and the embryophyte lineages can be characterized as 1C = 0·2 pg and 1n = 6.
Conclusions
These data will help pre-screen candidate species for the on-going construction of bacterial artificial chromosome nuclear genome libraries for land plant ancestors. Data for the prasinophyte Mesostigma are of particular interest as this alga reportedly most closely resembles the ‘ancestral green flagellate’. Both mechanistic and ecological processes are discussed that could have produced the observed C-value increase of >100-fold in the charophyte green algae whereas the ancestral genome was conserved in the embryophytes.
Key words: ‘Ancestral green flagellate’ (AGF), C-value enigma, chlorophyta, DNA C-values, nuclear genome size, Streptophyta
INTRODUCTION
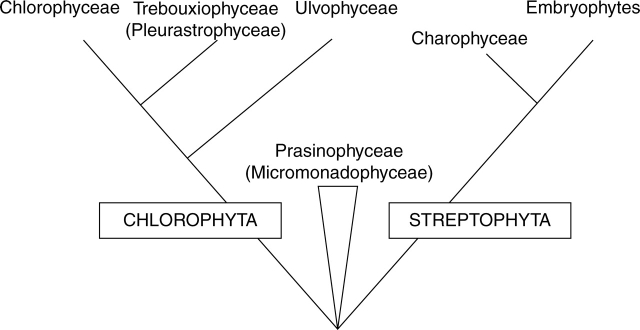
Chlorophyta include the eukaryotic green algae, which possess chlorophyll a and b and starch stored inside plastids with stacks of 2–6 thylakoids per band (Bold and Wynne, 1985). Approximately 425 genera and 6500 species have been described (Alexopoulos and Bold, 1967). Simplicity and antiquity of green algae (Chlorophyta) have long been accepted as evidence of their apparent ancestry to land plants (McCourt, 1995). Phylogenetic analyses of molecular markers including 18S rRNA and rbcL (Bhattacharya et al., 1994; McCourt et al., 1995, 1996; Huss and Kranz, 1997; Katana et al., 2001) and morphological data (Mattox and Stewart, 1984) contradict this view and present a compelling case that an ancient and deep divergence separates green plants into two major monophyletic lineages: Chlorophyta and Streptophyta (Turmel et al., 1999; Karol et al., 2001) (Fig. 1).
Fig. 1.
Summary results of combined analysis using morphological, ultrastructural and large- and small-subunit rRNA gene sequences for chlorophyte and streptophyte algae (McCourt, 1995).
A third polyphyletic green plant lineage, branching at the base of the Chlorophyta–Streptophyta divergence, includes a residuum of related unicellular micromonadophytes (=prasinophytes) (Kantz et al., 1990; Steinkötter et al., 1994; Karol et al., 2001) (Fig. 1). Although the exact relationship of the prasinophytes to land plants remains unclear (Qiu and Palmer, 1999), it seems likely that a prasinophyte-like scaly flagellate was the common ancestor of both green plant divisions (Friedl, 1997). Nuclear genome size and organization remain largely unknown in the prasinophytes. A 2C genome size estimate of about 10 Mbp reported for Ostreococcus tauri (Prasinophyceae) is one of the smallest among free-living eukaryotic organisms (Courties et al., 1998; Derelle et al., 2002). It is assumed that this small genome is evolutionarily derived rather than ancestral (Courties et al., 1994) as other members of Mamiellaceae (Prasinophyceae) are reported to represent secondarily reduced forms (Daugbjerg et al., 1995). Previously, it was suggested that this apparent miniaturization of prasinophyte nuclear genomes may defeat attempts to use them as a model in reconstruction of an ancstral land plant nucleotype (Cunningham et al., 1998; Oakley and Cunningham, 2000). Consequently, the early branching groups in the charophycean lineage (Soltis et al., 1999), including Chlorokybales, Klebsormidiales and Coleochaetales, may provide the best opportunity for gaining these insights, but there are few published estimates of DNA contents in target members of these orders.
The Second Plant Genome Size workshop and Discussion Meeting [hosted by the Royal Botanic Gardens (RBG), Kew, 8–12 September, 2003] identified major gaps (systematic, regional and plant type) in our knowledge of plant DNA amounts (Bennett and Leitch, 2005a). It was noted that no database was available for algae. This major gap was addressed with a compilation of genome size estimates for 247 species of red, green and brown macroscopic algae (Kapraun, 2005). This report included nuclear DNA content estimates for 95 isolates and species of multicellular green marine algae, which are almost exlusively members of Ulvophyceae (Kapraun, 2005). These data are incorporated into a database of plant genome sizes (Kapraun et al., 2004) available online on the RBG Kew website (http://www.rbgkew.org.uk/cval/homepage.html). In addition, nuclear DNA content data for green algae, both from our continuing investigations and from the literature, are regularly updated at http://www.uncw.edu/people/kapraund/DNA (see links there to ‘Table I. Chlorophyta’ and ‘Appendix I. Chlorophyta’). The present paper includes nuclear genome size estimates for 72 additional isolates and species of green algae. Of this total, 33 resulted from our ongoing research. Unicellular freshwater microalgae that were under-represented or excluded previously (Kapraun, 2005) are emphasized here, especially members of Chlorophyceae and Trebouxiophyceae. A significant effort was made to expand our database for streptophyte algae, which are critical in efforts to reconstruct a hypothetical ancestral nuclear genome for the green algal ancestor of the land plants.
Inclusion of published nuclear DNA content data for green algae in the present report was sometimes problematic. The Second Plant Genome Size workshop and Discussion Meeting (Bennett et al., 2000; Bennett and Leitch, 2005b) identified ‘best practice’ methodology for nuclear genome size estimation in plant tissues. Virtually none of the published genome size data for algae resulted from investigations adhering to all of the best practice recommendations, primarily because measurement of the relatively small algal nuclear genomes requires standard species different from those specified as appropriate for vascular plants (Kapraun, 2005). Flow cytometry of oceanic picoeukaryotes and phytoplankton has resulted in nuclear DNA content estimates from whole cells for numerous algae including members of Chlorophyceae and Prasinophyceae (Simon et al., 1994; Veldhuis et al., 1997). In general, only estimates based on isolated nuclei were included in the present study. A comprehensive discussion of standard species and methods is included in the notes related to the Appendix at the end of this paper.
METHODS
Algal material was fixed in Carnoy's solution and stored in 70 % ethanol at 4 °C. Preserved material was rehydrated in water and softened in 5 % w/v EDTA (Goff and Coleman, 1990) for 30 min to 3 h. Specimens were transferred to cover slips treated with subbing solution, air dried and stained with DAPI (0·5 µg mL−1 4′,6-diamidino-2-phenylindole; Sigma Chemical Co.,` St. Louis, MO, USA) as previously described (Goff and Coleman, 1990; Kapraun and Nguyen, 1994). Detailed procedures for microspectrophotometry with DAPI and requirements for reproducible staining have been specified previously (Kapraun, 1994; Kapraun and Nguyen, 1994) using a protocol modified after Goff and Coleman (1990). Microspectrophotometric data for Gallus [chicken erythrocytes or red blood cells (RBC)] with a DNA content of 2·4 pg (Clowes et al., 1983) were used to quantify mean fluorescence intensity (If) values for algal specimens (Kapraun, 1994). DAPI binds by a non-intercalative mechanism to adenine/thymine- (A/T-) rich regions of DNA (Portugal and Waring, 1988). Consequently, RBC are best used as a standard for estimating amounts of DNA when the A/T contents of both standard and experimental DNA are equivalent (Coleman et al., 1981). Gallus has a nuclear DNA base composition of 42–43 mol% G+C (Marmur and Doty, 1962). Limited published data indicate similar mean values of 46 mol% for Chlorophyta (Sueoka, 1961; Olsen et al., 1987; Freshwater et al., 1990; Kooistra et al., 1992; Le Gall et al., 1993; Simon et al., 1994). Algae investigated in this study are assumed to have a similar range of base pair compositions, and linearity is accepted between DAPI–DNA binding in both RBC and algal samples (Le Gall et al., 1993). Nuclear DNA contents were estimated by comparing the If values of the RBC standard and algal sample (Kapraun, 1994). Chlorophyta include taxa with some or all of their cells being multinucleate or endopolyploid (Kapraun and Nguyen, 1994) as well as taxa that exhibit a nuclear ‘incremental size decrease associated with a cascading down of DNA contents’ (Kapraun, 1994). Specific methodologies were developed for specimens to permit assignment of C level and interpretation of If data. Details of materials and methods, as well as information for collection locations, and data for numbers of algal nuclei examined in each sample and estimates of nuclear genome size (pg,±s.d.) are available at http://www.uncw.edu/people/kapraund/DNA.
RESULTS
The present investigation adds nuclear DNA content estimates for 72 species and isolates of green algae to our database of C-values, emphasizing the streptophyte charophycean lineage that is the sister group of land plants. A previous list of 2C nuclear DNA contents in green algae (Kapraun, 2005) is expanded to 157. DNA content estimates, both from the present research and from published information, are presented as picograms (pg) and as megabase pairs (Mbp) (Appendix).
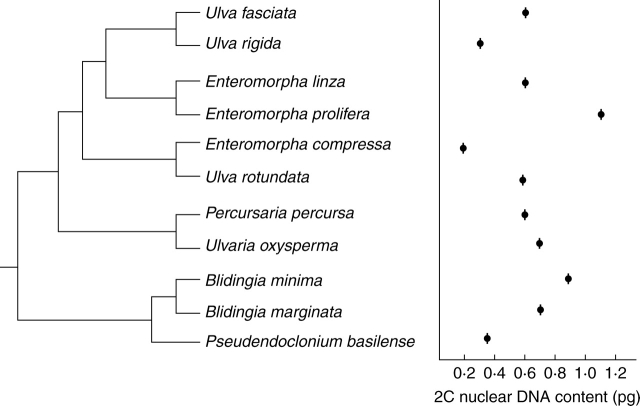
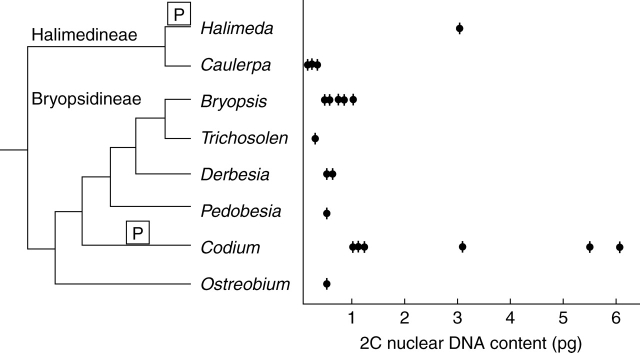
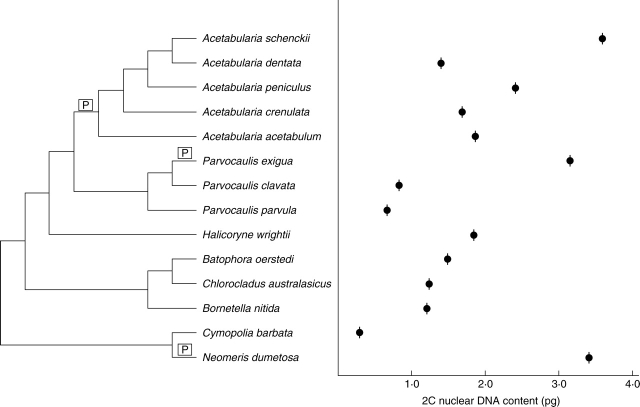
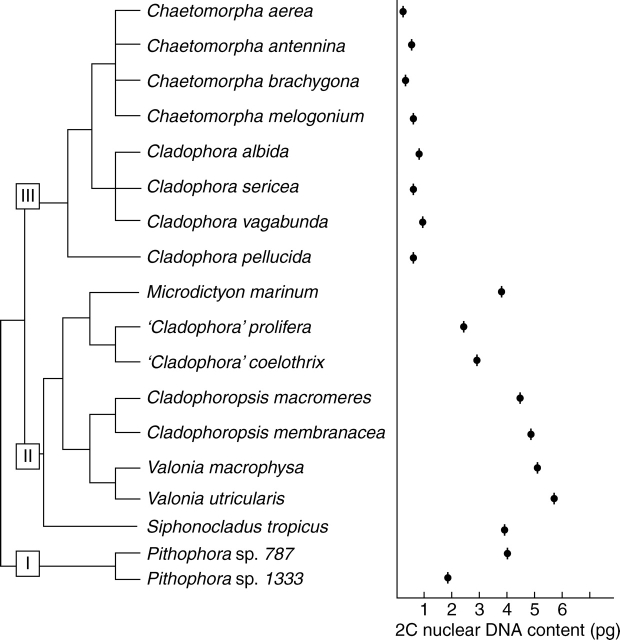
In Chlorophyta, including Chlorophyceae, Trebouxiophyceae, Ulvophyceae and Prasinophyceae (Figs 1–3), 2C DNA estimates range from 0·01 to 5·8 pg. Significant new data are included in this study (Appendix) for all of the major groups of Ulvophyceae (Fig. 4). In Ulotrichales (Fig. 5), nuclear DNA content data are available for seven species of this large and diverse order (Table 1 online, see Introduction) and suggest that it is characterized by small 2C values of 0·2–0·5 pg (Appendix). Estimates of nuclear DNA contents for species of Ulvales (Fig. 6) range from 2C = 0·14 to 1·1 pg (Appendix). In Trentepohliales, nuclear DNA content estimates of 2C = 1·1–4·1 pg appear to be correlated with a molecular phylogenetic tree derived from small-subunit rRNA gene sequence analysis (Fig. 7). Published 2C nuclear DNA content estimates range from 0·1 to 1·0 pg for most members of Caulerpales. Only species of Halimeda and Codium have substantially larger genomes of 1·0–6·1pg (Fig. 8). Previously published (Kapraun, 2005) and present nuclear DNA content estimates for members of Dasycladales indicate a range of 0·7–3·8 pg (Fig. 9). In the Cladophorales/Siphonocladales complex, nuclear DNA content estimates indicate that members of clade III have relatively small genomes (2C = 0·2–0·7 pg) whereas members of clade II, including the bulk of Siphonocladales, have much larger genomes of 2C = 2·0–5·7 pg (Fig. 10). Isolates of Pithophora spp. have genome sizes of 2C = 1·9 pg (UTEX 1333) and 4·1 pg (UTEX 787), respectively.
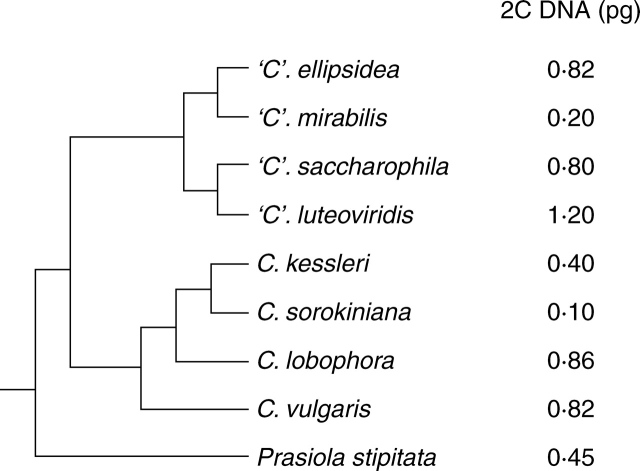
Fig. 3.
Estimated 2C nuclear DNA contents superimposed on a phylogenetic tree for Trebouxiophyceae based on 18S rRNA gene sequence analyses (Friedl, 1995; Krienitz et al., 2004).
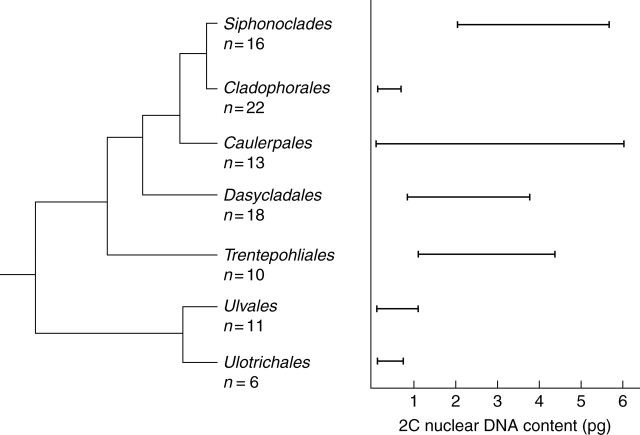
Fig. 4.
Estimated 2C nuclear DNA contents superimposed on a cladogram of Ulvophyceae inferred from small-subunit rRNA gene sequence data (Zechman, 1990).
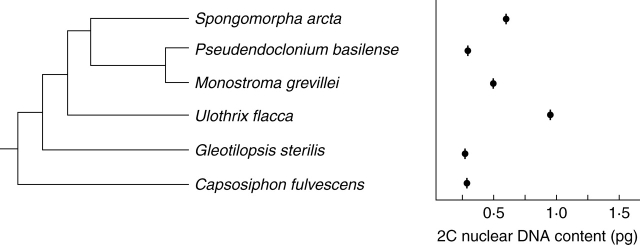
Fig. 5.
Phylogenetic tree of Ulotrichales inferred from 18S rRNA and rbcL gene sequence analysis (Hayden and Waaland, 2002).
Fig. 6.
Phylogenetic tree of the Ulvales inferred from 18S rRNA and rbcL gene sequence analysis (Hayden and Waaland, 2002).
Fig. 7.
Estimated 2C nuclear DNA content estimates superimposed on a cladogram of Trentepohliales inferred from small-subunit rRNA genes (López-Bautista and Chapman, 2003, 2005).
Fig. 8.
Estimated 2C nuclear DNA contents superimposed on a phylogenetic tree for Caulerpales based on cladistical analyses (Vroom et al., 1998; Woolcott et al., 2000). Proposed polyploidy events are indicated by [P].
Fig. 9.
Estimated 2C nuclear DNA contents superimposed on a phylogenetic tree for Dasycladales based on rbcL gene sequence analyses (Zechman, 2003; Berger et al., 2003). Proposed polyploidy events are indicated by [P].
Fig. 10.
Estimated 2C nuclear DNA contents superimposed on a phylogenetic tree for the Cladophorales/Siphonocladales complex based on 18S rRNA gene sequence analysis (Hanyuda et al., 2002).
In Prasinophyceae (Appendix), nuclear genome size estimates of 0·2 and 0·7 pg were obtained for species of Pyramimonas and Tetraselmis, respectively (Appendix), suggesting that at least in these prasinophycean algae, nuclear genome sizes more closely approximate those reported in many other green algae (Kapraun, 2005).
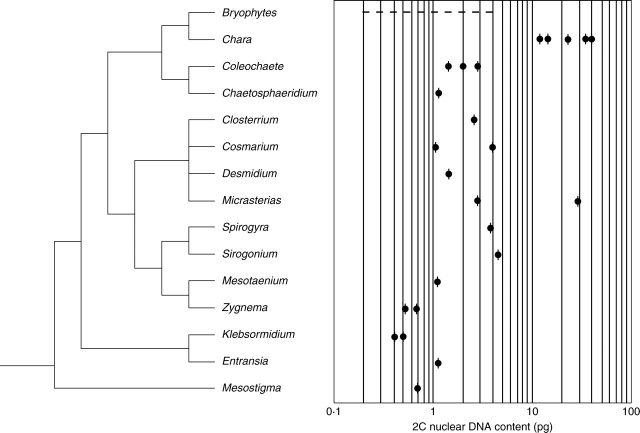
In Streptophyta, 2C nuclear DNA contents range from 0·2 to 6·4 pg, excluding the highly polyploid Charales and Desmidiales, which have genome sizes of up to 14·8 and 46·8 pg, respectively. Nuclear DNA content data for Streptophyta superimposed on a contemporary molecular phylogenetic tree indicate that early diverging lineages, including some members of Chlorokybales, Coleochaetales and Klebsormidiales, have genomes as small as 0·1–0·5 pg. In Mesostigmatales, a genome size of 0·74 pg was estimated for an isolate identified as representing Mesostigama viride (Appendix). In Coleochaete (Coleochaetales) isolates investigated, nuclear DNA content estimates of 1·4, 3·0 and 5·5 pg approximate a doubling sequence, consistent with values that would result from polyploidy. In the present study, a 2C nuclear genome size of 1·2 pg was estimated for Chaetosphaeridium globosum (Appendix). In Klebsormidiales, 2C nuclear genome sizes for Klebsormidium flaccidum and Entransia fimbriata were estimated to be 0·4 and 1·1 pg, respectively (Appendix). An isolate of Klebsormidium nitens from Argentina was found to have an estimated nuclear DNA content of 2C = 0·55 pg (Appendix) or 539 Mbp (using 1 pg = 980 Mbp, Bennett et al., 2000).
In the present study, nuclear DNA content estimates obtained with DAPI microspectrophotometry for five additional species extend the upward range for presumptive 2C nuclei to 46 pg. In a Netrium digitus isolate (UTEX 599), nuclei were too large to be accommodated by the photometer aperture system, but nuclear volume (NV) calculations, as described in the Appendix notes, resulted in a nuclear DNA content estimate of more than 125 pg. This is by far the largest nuclear genome size reported in any green alga (Kapraun, 2005). The present investigation extends the upward range for presumptive 2C nuclei to 7·2 pg in an isolate of Mougeotia transeaui (Fig. 11). The 2C nuclear DNA contents in Charales range from 14·0 to 39·2 pg (Fig. 11).
Fig. 11.
Ranges of 2C nuclear DNA contents superimposed on a consensus molecular phylogenetic tree for the streptophyte green algae and bryophytes based on sequence analyses (Bhattacharya et al., 1994; McCourt et al., 2000; Denboh et al., 2001; Karol et al., 2001; Cimino and Delwiche, 2002; Delwiche et al., 2002; Turmel et al., 2002c; Gontcharov et al., 2003). DNA data for streptophyte algae are from Kapraun (2005) and the Appendix, and for the bryophytes from Renzaglia et al. (1995) and Voglmayr (2000).
DISCUSSION
Chlorophyta
Chlorophyta contain the classical ‘green algae’, primarily Chlorophyceae, Trebouxiophyceae (=Pleurastrophyceae) and Ulvophyceae (Mishler et al., 1994; Watanabe et al., 2001) (Fig. 1). All members of this clade have swimming cells with two or four anterior flagellae arising from basal bodies which are arranged cruciately (O'Kelly and Floyd, 1984). Combined analysis of morphology and molecular data strongly supports the monophyly of these three groups (Mishler et al., 1994) and the phylogenetic topology followed below (Krienitz et al., 2004).
Chlorophycean algae
Chlorophyceae apparently arose during the later stages of green algal evolution and are not an early branching lineage (Watanabe et al., 2001). This group includes a predominance of freshwater taxa such as Sphaeropleales (= Chlorococcales; e.g. Scenedesmus), and many of the familiar flagellates such as Volvox and Chlamydomonas (Volvocales). Recently, molecular techniques were used to re-examine phylogenetic relationships in Volvocales (Buchheim et al., 1996, 1997, 2002) as previously deduced by morphological and gene sequence data (Nozaki et al., 1995; Angeler et al., 1999). Volvox has been considered to be the limit of the colonial development in the volvocine series, in which Gonium and unicellular forms such as Chlamydomonas are early branching lineages (Larson et al., 1992). Although much of the evolution in the colonial Volvocales superficially appears to constitute a gradual progression in colonial complexity and in types of sexual reproduction, as in the traditional volvocine lineage hypothesis, reverse evolution must be considered for the origin of certain species of Pleodorina (Nozaki et al., 2000). Apparently, both colonial and unicellular morphotypes have evolved independently in several clades (Nozaki et al., 1995, 1999; Nozaki and Krienitz, 2001). Thus, the traditional view of a unidirectional, monophyletic progression from unicellular, through colonial, to multicellular forms, represented by the sequence Chlamydomonas, Gonium, Pandorina, Eudorina, Pleodorina and Volvox, is not supported by gene sequence analyses (Larson et al., 1992; Lewis and McCourt, 2004).
In Chlorophyceae, published DNA content estimates for Sphaeropleales (= Chlorococcales), a widely distributed and much investigated group of algae (Buchheim et al., 2005; McManus and Lewis, 2005), are limited to an early study which cited Scenedesmus obliquus as having a nuclear genome size of 0·4 pg (Charles, 1977). Few nuclear DNA content estimates have been published for Volvocales (Fig. 2). The pioneering investigation of Holm-Hansen (1969) using ‘fluorometric measurement’ estimated 0·6 pg for Dunaliella tertiolecta. Flow cytometry of whole cells gave an estimate of 0·015 pg for this alga (Veldhuis et al., 1997). DNA content estimates of 0·01–0·2 pg are given in the Appendix for three additional species of volvocine green algae.
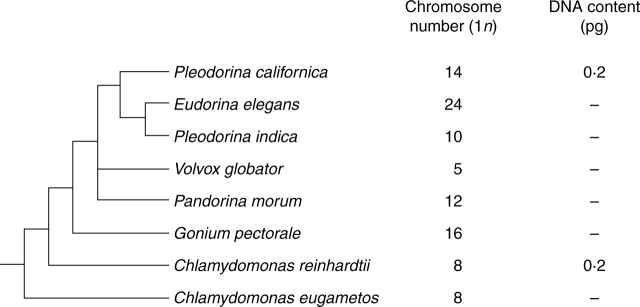
Fig. 2.
Estimated 2C nuclear DNA contents and 2n chromosome complements (Sarma, 1982) superimposed on a phylogenetic tree for Volvocales based on multiple plastid gene sequences (Nozaki et al., 1999, 2000).
Although relatively few nuclear DNA content estimates have been published for Volvocales (Fig. 2), chromosome numbers have been published for many species. These range from 2n = 8 to 76, with the vast majority of reported numbers being between 16 and 24 (Sarma, 1982). In general, higher chromosome numbers are associated with colonial forms, and lower chromosome numbers with unicellular forms. Phylogenetic lines giving rise to Volvox include both low (n = 5) and high (n = 22) chromosome complements (Fig. 2). It would be a matter of great interest to augment the small amount of nucleotype data currently available for Volvocales with a more comprehensive investigation comparing nuclear genome size and chromosome numbers with speciation patterns (Schagerl et al., 1999; Coleman 2001). Specifically, do recognized volvocine morphotypes correlate with nucleotype parameters including genome size and chromosome number?
Trebouxiophycean algae
Trebouxiophyceae (=Pleurastrophyceae) includes those Chlorophyta with non-flagellate vegetative cells which are typically unicellular, sarcinoid or filamentous (Friedl, 1995; Henley et al., 2004; Krienitz et al., 2004; Ueno et al., 2005). Phylogenetic analysis of 18S rRNA gene sequences has demonstrated a monophyletic origin for these algae, and a sister group relationship to Chlorophyceae (Lewis, 1997; Handa et al., 2003; Lokhorst et al., 2004) (Fig. 3). Many of the better known members of this group exist in lichen associations, e.g. Dictyochloropsis and Trebouxia (Lewis and McCourt, 2004).
In Trebouxiophyceae, most nuclear DNA content estimates for Chlorellales (Fig. 3) resulted from an investigation using reassociation kinetics and were published as base pairs (Dörr and Huss, 1990). Pulse field gel electrophoresis (Higashiyama and Yamada, 1991), flow cytometry (Wilhelm et al., 1982; Veldhuis et al., 1997; Yamamoto et al., 2001) and microfluorometric analysis (Cattolico and Gibbs, 1975) have been used as well. The genus Nannochloris, and related genera, includes some of the smallest and ultrastructurally simplest phototrophic eukaryotes, with genomes as small as 12·6 Mbp (Arai et al., 1998; Yamamoto et al., 2001). Using 1 pg = 980 Mbp (after Bennett et al., 2000), 2C genome sizes (reported as Mbp) range from 0·02 pg in Picochlorum atomus (as Nannochloris atomus) (Veldhuis et al., 1997) to 1·06 pg in Chlorella fusca (Dörr and Huss, 1990).
Chlorellales
Recent molecular studies have resulted in the dispersal of species traditionally referred to Chlorella over two classes of chlorophytes, Trebouxiophyceae and Chlorophyceae (Huss et al., 1999; Katana et al., 2001; Henley et al., 2004; Krienitz et al., 2004). In the present investigation, Chlorella species included in the Appendix are considered to be trebouxiophycean algae (Huss et al., 1999). Phylogenetic analysis of some ‘Nannochloris-like’ algae resulted in their transfer to other genera, including Marvania and Picochlorum (Henley et al., 2004).
Prasiolales
Recent molecular data support transfer of Prasiola, formerly in the Ulvophyceae, to Trebouxiophyceae (Sherwood et al., 2000; Friedl and O'Kelly, 2002; Naw and Hara, 2002). This development is particularly notable as Prasiola, and the closely related Rosenvingiella (Rindi et al., 2004), often develop morphotypes which mimic Enteromorpha and Ulva (Ulvales). In the present study, the nuclear genome size estimate of 0·9 pg for Prasiola stipitata is assumed to represent the 4C value as blades in this species are reported to be diploid (Cole and Akintobi, 1963). Nuclear DNA content estimates range from 2C = 0·1 to 1·2 pg and indicate no apparent correlation between genome size and phylogeny (Fig. 3).
Ulvophycean algae
Ulvophyceae are primarily marine species, most with larger and more complex morphologies than typically found in Chlorophyceae. Molecular data support a model for Ulvophyceae sensu Mattox and Stewart (1984) with two separate lineages: a clade including Ulothrichales and Ulvales (Hayden and Waaland, 2002; O'Kelly et al., 2004) and a clade with Caulerpales, Cladophorales/Siphonocladales complex, Dasycladales and Trentepohliales (Zechman et al., 1990; Hanyuda et al., 2002) (Fig. 4). In the present report, presentation of orders reflects a consensus of contemporary phylogenetic treatments (Zechman et al., 1990).
Ulotrichales
Ulotrichales as presently delimited (Floyd and O'Kelly, 1990) have been expanded to include Acrosiphoniaciae (sensu Kornmann and Sahling, 1977; Sussmann et al., 1999). Molecular analyses using 18S rDNA and 18S rRNA gene sequence data have confirmed this placement (Watanabe et al., 2001; Lindstrom and Hanic, 2005). Species of Capsosiphon and Monostroma, included in Ulvales by Bliding (1963, 1968), appear to be more closely related to Ulotrichales (Fig. 5). Nuclear DNA content data, available for seven species of this large and diverse order (table 1 online), suggest that it is characterized by small 2C values of 0·2–0·5 pg (Appendix). No correlation between nuclear genome size and phylogenetic position is apparent (Fig. 5). The relatively small genome sizes reported for species in this order are complemented by relatively small chromosome numbers of 2n = 8–24 (Kapraun, 1993).
Ulvales
The emended order Ulvales (O'Kelly et al. 2004) is monophyletic (Fig. 6), but circumscription of several genera, including the familiar Ulva and Enteromorpha (Hayden and Waaland, 2002, 2004; Hiraoka et al., 2003) and Blidingia (Lindstrom and Golden, 2006), remains problematic. In both Ulva and Enteromorpha, blade and tubular thallus morphotypes apparently arose independently several times throughout the evolutionary diversification of the group (Blomster et al., 1998, 1999; Tan et al., 1999; Shimada et al., 2003). Although results from molecular and culture studies suggest that Ulva, Enteromorpha and Chloropelta should not be recognized as separate genera (Hayden et al., 2003; Matsuo et al., 2003, 2005), these familiar taxonomic epithets are retained here for convenience (Appendix). The absence of a correlation between nuclear genome size and chromosome number in these species (Kapraun and Bailey, 1992) suggests a significant role of aneuploidy in their evolution (Kapraun, 2005).
Trentepohliales
Molecular investigations place members of this order with the second lineage of Ulvophyceae (Mishler et al., 1994; Chapman et al., 1995; Lopez-Bautista and Chapman, 2003), which are otherwise almost exclusively marine. Nuclear DNA content estimates of 2C = 1·1–4·1 pg appear to be correlated with a molecular phylogenetic tree derived from small subunit rRNA gene sequence analysis (Fig. 7). Large-scale discontinuous variation in both reported chromosome complements (n = 4–28) (Appendix) and nuclear DNA contents are indicative of polyploidy in this order (Lopez-Bautista et al., 2000).
Caulerpales
Cladistic analysis of morphological and molecular data supports separation of Caulerpales (Codiales sensu Taylor, 1960) into two clades: (1) the homoplastidic Bryopsidineae, which are generally characterized by diplobiontic life histories and non-holocarpic production of gametes (e.g. Bryopsis and Codium); and (2) the heteroplastidic Halimedineae, which are generally characterized by haplobiontic and diploid life histories and holocarpic production of gametes (e.g. Caulerpa and Halimeda) (Zechman et al., 1990; Vroom et al., 1998; Woolcott et al., 2000). Published 2C nuclear DNA content estimates range from 0·1 to 1·0 pg for most of these algae. Only species of Halimeda and Codium have substantially larger genomes (1·0–6·1pg; Fig. 8). As these two genera are placed in separate clades, it appears that large-scale whole genome increase (polyploidy) arose independently at least twice in the evolution of Caulerpales (Fig. 8).
Dasycladales
Recent molecular investigations based on analyses of rbcL (Zechman, 2003) and 18S rRNA (Berger et al., 2003) gene sequence data revealed three well-supported clades for which morphological synapomorphies exist, but which are not completely in accordance with previous generic concepts. A Polyphysa clade is distinguished from Acetabularia on the basis of cap morphotype and initiation (Berger and Kaever, 1992; Sawitzky et al., 1998). The range of nuclear DNA content estimates for members of Dasycladales (0·7–3·8 pg) (Fig. 9), which suggests large-scale discontinuous variation in all major clades, almost certainly reflects multiple polyploidy events (Kapraun and Buratti, 1998; Kapraun, 2005). It seems noteworthy that similarly large values (approx. 3·2–3·8 pg) were found in the early diverging Neomeris clade as well as in the highly derived Parvocaulis (=Polyphysa) and Acetabularia clades. No correlation is apparent between cap morphotypes (Sawitzky et al., 1998), cap morphogenesis (Kratz et al., 1998) and ‘polyploid’ nucleotypes in Dasycladales.
The Cladophorales/Siphonocladales complex
Cladophorales and Siphonocladales are a related lineage sharing a gradation of ‘architectural’ morphological types (Olsen-Stojkovich et al., 1986; Van den Hoek et al., 1988; Bakker et al., 1994). Contemporary molecular studies (Zechman et al., 1990; Hanyuda et al., 2002) have identified three well-supported clades in this complex: (I) an early diverging assemblage of mostly freshwater species of cladophoracean genera, including Aegagropila, Arnoldiella, Pithophora and Wittrockiella; (II) species belonging primarily to Siphonocladales sensu Børgesen (1913); and (III) the most derived assemblage including species belonging to the cladophoracean genera Chaetomorpha, Cladophora and Rhizoclonium (Fig. 10). Confusingly, the characteristic morphologies associated with the genera Chaetomorpha, Cladophora and Rhizoclonium appear to have evolved several times, independently, in all three clades. Thus, these genera as presently circumscribed are clearly polyphyletic.
Karyological studies indicate that species in the most highly derived cladophoracean clade III (Rhizoclonium, Chaetomorpha and Cladophora sensu stricto), without exception, share a unique constellation of karyotype features including: (1) six basic chromosomes, three of which have median and the other three have submedian centromeres; and (2) almost universal polyploidy, resulting in chromosome complements in most species of x = 12, 18, 24, 30, 36, etc. (Wik-Sjöstedt, 1970; Kapraun and Gargiuolo, 1987a, b; Miyaji, 1999). Species in the siphonocladacean clade II have (1) various combinations of both metacentric and acrocentric chromosomes (Kapraun and Breden, 1988; Bodenbender and Schnetter, 1990; Kapraun and Nguyen, 1994), and (2) chromosome complements consistent with an aneuploid origin: 1n = 8, 12, 14, 16, 18 and 20 (Kapraun, 1993; Kapraun and Nguyen, 1994). Karyological data for basal clade I taxa appear to be limited to species of Pithophora, which have chromosome complements of 18, 24, 30 and 36 (Noor, 1968; Verma, 1979; Sarma, 1982) and relatively long chromosomes, up to 3 µm (Godward, 1966). Nuclear DNA content estimates indicate that members of clade III have relatively small genomes (2C = 0·2–0·7 pg) whereas members of clade II, including the bulk of Siphonocladales, have much larger genomes of 2C = 2·0–5·7 pg (Fig. 10). Isolates of Pithophora spp., representative of clade I, the earliest diverging among Cladophorales/Siphonocladales investigated, have genome sizes of 2C = 1·9 pg (UTEX 1333) and 4·1 pg (UTEX 787). These values almost certainly represent elevated DNA contents in a polyploid sequence, approximating to 2, 4, 8 and 16 pg (Appendix). As it was beyond the scope of the present investigation to determine chromosome numbers for these isolates, the exact ploidy level remains unknown.
Although the cladophoracean morphotype is reported to have evolved independently in all of the clades, the combination of karyotype pattern (n = 6) and nuclear genome size (2C = 0·2–0·7 pg) characteristic of the core clade III of Cladophorales appears to be unique and diagnostic, and may represent synapomorphies (Kapraun, 2005). If it is assumed that small genome size and symmetric karyotype with 1n = 6 is the ancestral condition in the Cladophorales/Siphonocladales complex, then these features have been most faithfully retained in the core cladophoracean algae in clade III. Clade II Siphonocladales, with a genome size increase of approximately an order of magnitude, and specialization of the karyotype by unequal reciprocal translocations (Kapraun and Breden, 1988), appear to be highly derived. Because nuclear volume is strongly correlated with cell size and cell cycle lengths in higher plants (Shuter et al., 1983) it is not surprising that these algae with their large, multinucleate cells and relatively long cell generation times have relatively large genomes (Kapraun and Nguyen, 1994). Clade I taxa (e.g. Pithophora) appear to have retained the ancestral karyotype, but not the theoretical small genome size. Their substantial increase in nuclear genome size, probably through whole genome duplication or polyploidy, may reflect their reliance on asexual reproduction by multinucleate propagules or akinetes (O'Neal and Lembi, 1983).
Prasinophycean algae
The prasinophytes or micromonads are primarily marine green flagellates. Some of the more commonly recognized taxa include Halosphaera, Pyramimonas and Tetraselmis. They generally have a single plastid, and often possess the accessory pigment prasinoxanthin. The cell membrane of most forms is covered with one or more layers of scales. There are no unique and defining characteristics for the group other than the above constellation of features (Sym and Pienaar, 1993; Graham and Wilcox, 2000). Prasinophytes have been characterized as the form of cell most closely representing the first green alga, or ‘ancestral green flagellate’ (AGF; Lewis and McCourt, 2004) (Fig. 1). Molecular analyses of 18S rRNA gene sequence data have identified at least seven separate lineages that form a grade at the base of the green algal tree of life (Steinkötter et al., 1994; Fawley et al., 2000; Zignone et al., 2002). Mamiellales include some of the smallest eukaryotes known, e.g. Crustomastix (Lewis and McCourt, 2004). There is some evidence that these algae represent secondarily reduced forms (Daugbjerg et al., 1995).
Nuclear genome size and organization remain largely unknown in Prasinophyceae (Appendix). Pulse field gel electrophoresis was used to obtain 2C nuclear genome size estimates for Ostreococcus tauri of 10·2 Mbp (Courties et al., 1998) and 9·7 Mbp (Derelle et al., 2002), or about one-quarter of the value reported for Chlorella (Higashiyama and Yamada, 1991). Using 1 pg = 980 Mbp (Bennett et al., 2000), Ostreococcus has a 2C genome size of about 0·01 pg. Flow cytometry analysis of isolated nuclei resulted in genome size estimates for Micromonas pusilla of 0·027 pg DNA per cell (Veldhuis et al., 1997) and 0·03 pg DNA per cell (Simon et al., 1994) and for Bathycoccus prasinos of 0·02 pg DNA per cell (Simon et al., 1994). These values were published as femtograms (10−15), but are expressed as picograms (10−12) here for consistency. Similar small genome size estimates based on whole cell flow cytometry measurements published for several additional prasinophyte taxa (Simon et al., 1994; Veldhuis et al., 1997) are not included in the Appendix as whole cell staining induces some non-specific background fluorescence, resulting in DNA content overestimates, typically by a factor of 3 or less (Veldhuis et al., 1997).
Streptophyta
Streptophyta (Bremer et al., 1987) include the charophycean lineage along with bryophytes and tracheophytes (Mishler et al., 1994; Turmel et al., 2002a, b). Numerous ultrastructural and molecular synapomorphies support this charophyte clade (Lewis and McCourt, 2004). However, the topology of the charophyte tree remains elusive (Friedl, 1997; Delwiche et al., 2002). Identification of the charophycean lineage as the sister group of land plants (Manhart and Palmer, 1990) suggests that their common ancestor was a branched, filamentous organism (Cook, 2004) with a haplontic life cycle and oogamous reproduction (Karol et al., 2001).
Charophycean algae
The charophycean lineage (Fig. 11) includes Chlorokybales (Qiu and Palmer, 1999), Klebsormidiales (Karol et al., 2001), Conjugophyta (Desmidiales and Zygnematales) (Hoshaw et al., 1990; McCourt et al., 2000; Denboh et al., 2001), Coleochaetales (Bhattacharya et al., 1994; McCourt, 1995; Cimino and Delwiche, 2002) and Charales (Surek et al., 1994; McCourt et al., 1996). The precise relationship of Mesostigmatales to the charophycean lineage remains unclear (Delwiche et al., 2002). The most important characters that define charophycean algae are the unilateral flagellar root in zooids and open mitosis with a persistent telophase spindle (Mattox and Stewart, 1984). In the present report, presentation of orders reflects a consensus of contemporary phylogenetic treatments (Delwiche et al., 2002; Lewis and McCourt, 2004).
Mesostigmatales
For many years the question of the nature of the AGF, the hypothetical ancestor of all land plants, has been controversially discussed (O'Kelly, 1992). Some researchers associate the AGF with a polyphyletic green plant lineage at the base of the split of Chlorophyta and Streptophyta (Fig. 1), which includes the green alga Mesostigma viride (Turmel et al., 2002a). The exact placement of Mesostigma remains controversial as some phylogenetic analyses include this species with Prasinophyceae (Lemieux et al., 2000; Turmel et al., 2002b), whereas others consider it to be ‘basal’ in the charophycean lineage (Palmer et al., 2004). Whatever the exact position of Mesostigma, there is no doubt that this alga belongs to a deeply diverging lineage given that it represents the first branch in trees inferred from sequences of land plants and all five orders of charophytes (Karol et al., 2001). A significant body of research centred around Mesostigma is emerging that provides insights into the timing of events that restructured both mitochondrial (mtDNA) and plastid DNA genomes during the evolution of green algae (Turmel et al., 2002b) and the transition from charophytes to land plants (Turmel et al., 2002a). Mesostigma plastid DNA is highly similar in size (118–360 bp) and gene organization to the plastid DNAs of land plants (Lemieux et al., 2000). Apparently plastid gene loss is an ongoing process in streptophytes, with independent losses occurring in multiple lineages. By contrast, Mesostigma mtDNA differs greatly at the levels of size (42–424 bp), gene organization and intron content from the bryophytes and land plants sequenced to date. During the evolutionary transition from Mesostigma to the liverwort Marchantia, mtDNA underwent a four-fold increase in size, was rearranged extensively and gained many introns while maintaining a similar gene content (Turmel et al., 2002b).
In the present study, a genome size of 0·74 pg was estimated for an isolate identified as Mesostigama viride (Appendix). In previous investigations of nuclear genome sizes in green algae, data from microspectrophotometry were corroborated with genome size estimates derived from NV data (Kapraun and Nguyen, 1994). Assuming a plant NV of 15 µm3 = 1·0 pg (Sparrow and Nauman, 1973; Kapraun et al., 1988), the NV of 12·6 µm3 calculated for Mesostigma is equivalent to a nuclear genome size of 0·8 pg, which closely approximates the present estimates.
Chlorokybales
This order is characterized by thalli of sarcinoid packets of cells that grow subaerially (Rogers et al., 1980). Chlorokybus atmosphyticus, one of the earliest diverging members of Charophyceae (Karol et al., 2001; Delwiche et al., 2002) (Fig. 11), has a plastid DNA sequence of 149–681 bp that closely resembles the Mesostigma plastid DNA in showing a high degree of putatively ancestral features (Turmel et al., 2002b). Unfortunately, no nuclear DNA content estimates have been published for any member of this important order.
Coleochaetales
Published chromosome numbers for three Coleochaete species (Sarma, 1982) include 1n complements of 24, 36 and 42, which almost certainly represent a polyploid sequence derived from a basic complement of x = 12. It was beyond the scope of the present study to determine chromosome numbers for the Coleochaete isolates investigated. However, the nuclear DNA content estimates of 1·4, 3·0 and 5·5 pg, which approximate a doubling sequence, are consistent with values that would be found in a polyploid sequence.
Molecular evidence includes Chaetosphaeridium globosum in Coleochaetales (Karol et al., 2001). In the present study, a 2C nuclear genome size of 1·2 pg was estimated for this species using microspectrophotometry (Appendix) and 1·23 pg using NV calculations. In Chaetosphaeridium, as in Coleochaete, the ploidy level of the isolates used in this study was not confirmed with chromosome counts.
Klebsormidiales
This order includes species of Klebsormidium and Entransia, which have simple, unbranched filaments with parietal laminate or lobed plastids (Lewis and McCourt, 2004). In the present study, 2C nuclear genome sizes for Klebsormidium flaccidum and Entransia fimbriata were estimated to be 0·4 and 1·1 pg, respectively (Appendix). An isolate of Klebsormidium nitens from Argentina, with a reported chromosome complement of n = 6 (Sánchez-Puerta and Leonardi, 2001), was investigated in the present study and found to have a nuclear DNA content of 2C = 0·55 pg (Appendix) or 539 Mbp. Thus, this species may have most nearly retained the proposed small ancestral nucleotype among extant streptophyte algae sampled.
Desmidiales and Zygnematales
Both the true desmids and the filamentous Zygnemataceae have undergone explosive speciation, resulting in thousands of described species (Prescott et al., 1972, 1977, 1981; Hoshaw and McCourt, 1988) from every continent. These algae represent the most species-rich group of charophytes with approximately 4000 species described (Gerrath, 2003). Two morphological synapomorphies unite the group: (1) the complete absence of flagellae in any life history stage, and (2) sexual reproduction by conjugation (McCourt et al., 2000). Molecular sequence data analyses confirm that the placoderm desmids are monophyletic, and constitute a group separate from the ‘false desmids’ and filamentous forms (McCourt et al., 2000; Gontcharov et al., 2003). Apparently, more complex forms evolved from simple filaments, and morphological switching from unicells to filaments occurred several times (Lewis and McCourt, 2004) (Fig. 11). Recent analysis of rbcL gene sequence data supports monophyly of Spirogyra and Sirogonium, two of the largest genera of filamentous Zygnemataceae (Drummond et al., 2005). The relationship of Desmidiales and Zygnematales to other streptophyte algae remains uncertain. Some molecular studies imply a close relationship between land plants (bryophytes) and conjugating green algae (Turmel et al., 2002a, b) whereas others suggest a more distant, sister group relationship (McCourt et al., 2000; Karol et al., 2001; Cimino and Delwiche, 2002; Delwiche et al., 2002).
Previous published nuclear DNA content estimates for the true desmids (Desmidiales) range from 2C = 1·1 to 20·7 pg (Hamada et al., 1985; Kapraun, 2005). Desmidiales are characterized by extensive polyploidy, with both inter- and intraspecific variation in chromosome complements reported (Hoshaw and McCourt, 1988). For example, published chromosome complements for Netrium digitus range from 1n = approx. 30 to approx. 592 (Sarma, 1982). Consequently, assignment of a C-value to specific DNA content estimates is arbitrary as chromosome counts were not made in this study for any of the isolates used in DNA quantification. In a previous investigation of Desmidiales (Kapraun, 2005), chromosome complements and nuclear DNA contents were found to be highly correlated (r2 = 0·7897), providing circumstantial evidence for the pervasive role of polyploidy in the evolution of this group of charophycean algae.
Previously published 2C nuclear DNA contents in Zygnematales ranged from 0·5 to 4·2 pg (Kapraun, 2005). As in Desmidiales, assignment of a C-value to specific DNA content estimates for isolates of Zygnematales is arbitrary. In general, both chromosome numbers (Sarma, 1982) and nuclear DNA contents (Fig. 11) are smaller in Zygnematales than in Desmidiales. Ploidy level in conjugating green algae may be of taxonomic significance as cell dimensions are considered to be diagnostic (Hoshaw and McCourt, 1988), and cell dimensions are highly correlated with genome size (Wang et al., 2005). Although the nucleotype role in gene expression remains poorly understood (Gregory, 2001, 2005a, b), it seems plausible that some of the described taxa of both desmid and filamentous conjugating green algae represent ploidy races, especially in species-rich genera characterized by a large variation in chromosome complements. It is widely assumed that many desmid genera are probably artificial and often display transitional forms to other genera. A recent molecular investigation of the desmid Staurastrum concluded that (1) the taxonomic significance of some morphological characters has been greatly overestimated, and (2) phylogenetic relationships were in conflict with previous formal and informal classification schemes (Gontcharov and Melkonian, 2005).
Charales
The exact relationship of the charophytes to land plants continues to be a subject of investigation. Several recent multigene phylogenies place Charales as the sister taxon to land plants (Karol et al., 2001; Cimino and Delwiche, 2002; Delwiche et al., 2002). Charales, commonly known as stoneworts or brittleworts, flourish in fresh and brackish water habitats throughout the world (Bold and Wynne, 1985). Charophytes are prone to calcification and have left an abundant fossil record to the Cretaceous and perhaps beyond (Grambast, 1974; Feist et al., 2003). The order is well circumscribed and includes six extant genera (McCourt et al., 1996; Sakayama et al., 2005), remnants of a once diverse but now largely extinct group (Feist et al., 2003). Analysis of 18S rRNA gene sequences indicated a poor correlation between molecular phylogeny and traditional hypotheses based on morphological criteria (Meiers et al., 1999). The base chromosome number for Chara is n = 7 and for Nitella is n = 3. However, many species exhibit polyploidy, with chromosome complements to n = 70 reported (Sarma 1982). Published nuclotype data are limited to five species of Chara (Maszewski and Kołodziejczyk, 1991; Kunachowicz et al., 2001) with 2C nuclear DNA contents ranging from 14·0 to 39·2 pg (Fig. 11). Two of these species, with 2n = 28, have 2C DNA contents of about 14 pg. Cytophotometric measurements of DNA content revealed differences in 1C DNA content between female (7·0 pg) and male (7·4 pg) isolates of Chara tomentosa (Kunachowicz et al., 2001).
SUMMARY AND CONCLUSIONS
DNA C-value remains a key character in biology, biodiversity and molecular investigations as genome size has many important practical implications (Bennett et al., 2000). In general, genome size directly influences the cost and difficulty of sequencing projects, and is therefore a primary consideration in choosing future sequencing subjects (Gregory, 2005a, b). Species with large DNA amounts can make use of standard fingerprinting techniques including AFLP problematic (Fay et al., 2005). Low DNA content (genomes approx. 100 Mpb) has been a major criterion in the selection of algae for genomic and genetic analyses (Peters et al., 2004; Waaland et al., 2004), including bacterial artificial chromosome (BAC) cloning technology, used for large-scale physical mapping and genomic sequencing (Wang et al., 2005). To date, candidate macroalgal (multicellular) species have genomes in the range 127–300 Mpb (Waaland et al., 2004). The present study can provide a great service both by expanding the list of target green algal species with appropriately small genome sizes, and by cautioning against use of taxa that may meet many other criteria for genomics investigations (Waaland et al., 2004), but have genome sizes too large for available technologies.
Recently, construction and characterization of large-insert BAC libraries has shown promise in the study of how important features in land plants originated and diversified (Liang et al., 2004). Unfortunately, the draft genome sequence available for the green alga Chlamydomonas reinhardtii (Wang et al., 2005) is of limited value in attempts to identify conserved ancestral genes because of the great phylogenetic distance between this chlorophyte and the streptophytes. The present study has indentified several streptophycean algae, including Klebsormidium and Coleochaete, with genome sizes that make them comparably tractable for genomic investigations.
The availability of a DNA C-values database and a consensus higher-level phylogenetic tree for green algae has opened the way for determining evolutionary trends in DNA amounts. Identification of the chlorophytes, streptophytes and prasinophytes as three of the most important extant green algal lineages provides an opportunity to suggest DNA content transformations which have accompanied their evolution, and to call attention to processes which may be unique or diagnostic for each group. Specific trends observed in this study are summarized below.
Summary for Ulvophyceae
Nuclear DNA content data from previously published investigations (Kapraun, 2005) and present research indicate that clade I of Ulvophyceae (Ulotrichales and Ulvales; O'Kelly et al., 2004) is characterized by relatively small nuclear genome sizes, whereas clade II (Caulerpales, Dasycladales, Cladophorales/Siphonocladales and Trentepohliales; Hanyuda et al., 2002) is characterized by substantially larger nuclear genome sizes (Fig. 4). It has been suggested that these differences in genome size ranges reflect the results of aneuploidy vs. polyploidy in clades I and II, respectively (Kapraun, 2005).
Summary for the charophycean lineage
The charophycean green algae are a sister group to land plants (Mishler et al., 1994; McCourt, 1995; McCourt et al., 2000; Lewis and McCourt, 2004). Ultrastructural data are overwhelmingly supported by the analyses of molecular data from nuclear ribosomal repeat units, mainly the small (18S) subunit, but including 5S and large-subunit (25S) rDNA sequences. Plastid genes including rbcL and small- and large-subunit rRNA yield similar results (Lewis and McCourt, 2004). Bryophytes are thought to comprise a grade of three monophyletic lineages (mosses, liverworts and hornworts) of uncertain relationship to each other (Shaw and Renzaglia, 2004) and to vascular plants (Palmer et al., 2004; Groth-Malonek and Knoop, 2005). Although the topology of the charophyte tree remains elusive (Friedl, 1997; Delwiche et al., 2002), comparison of the nucleotypes of charophycean green algae with those of early branching land plants, including bryophytes (Kenrick and Crane, 1997), could provide useful insights into the reconstruction of a common ancestral (generalized) nuclear genome (Pryer et al., 2002).
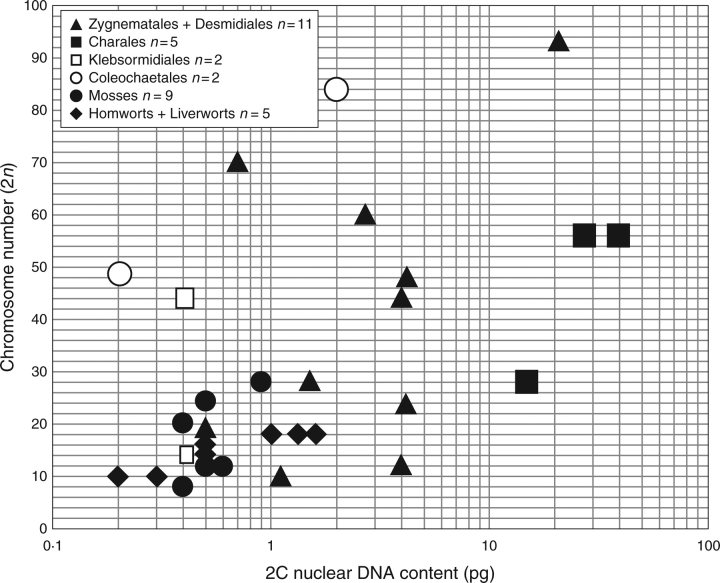
Hornworts, liverworts and mosses, in general, have nucleotypes characterized by small chromosomes (Inoue and Uchino, 1969) and chromosome complements of 2n ≥ 30 and/or 2C nuclear DNA contents >1 pg (Renzaglia et al., 1995; Voglmayr, 2000) (Fig. 12). Although greater values for both parameters are known in bryophytes, they appear to be restricted to polyploid species and do not contradict the generalization. For example, more than 80 % of the nuclear DNA C-values in mosses were reported to occur in a narrow peak between 0·25 and 0·6 pg (Voglmayr, 2000). It has been suggested that the small DNA amounts and low C-value variation are linked to the biflagellate nature of bryophyte sperm cells (Renzaglia et al., 1995). As nuclear genome size and sperm cell size are tightly correlated (Murray, 2005), and sperm cells are thought to lose motility drastically with increasing size, a strong selection pressure against larger sperm, and therefore also against larger DNA amounts, is hypothesized (Voglmayr, 2000).
Fig. 12.
Comparison of chromosome numbers and nuclear genome sizes in the charophycean and embryophyte lineages. Closed circles: DNA data for streptophyte algae from Kapraun (2005) and the Appendix, and for the bryophytes from Renzaglia et al. (1995) and Voglmayr (2000). Open circles: DNA data from Mandoli (2001).
These observations gain additional significance in the context of the suggestion that the common ancestor of all angiosperms may have possessed a small genome (Leitch et al., 1998, 2005). Small genome size in angiosperms appears to be correlated with phenotypic characteristics such as rapid seedling establishment, short minimum generation times, reduced cost of reproduction and an increased reproductive rate (Bennett, 1987; Midgley and Bond, 1991; Bennett and Leitch, 2005a, b). Consequently, small genome size may permit greater evolutionary flexibility (Leitch et al., 1998), whereas larger size and amplification may lead to ‘genomic obesity’ (Bennetzen and Kellogg, 1997; Bennetzen, 2002; Gregory, 2005a; Knight et al., 2005). It is recognized that although nuclear genome size is highly correlated with many cellular and ecological parameters, ‘correlation’ and ‘causation’ are far from interchangeable (Gregory, 2005a, b). The many complex causal factors behind these observations remain obscure.
Published molecular studies have implicated both the conjugating green algae (Turmel et al., 2002a, b) and the charophytes (Karol et al., 2001; Cimino and Delwiche, 2002; Delwiche et al., 2002) as the closest sister group to land plants. Consequently, it is worth noting that published nucleotype data for these charophycean algae suggest that they share a unique constellation of karyotype features that differ substantially from the most closely related bryophytes. Many taxa in Charales, Desmidiales and Zygnematales are highly polyploid (Sarma, 1982; Hoshaw and McCourt, 1988; Kunachowicz et al., 2001) and can be characterized either by chromosome complements of 2n>30 or 2C nuclear DNA contents >1 pg, or both (Fig. 12). In addition, both filamentous and unicellular (desmid) forms of the conjugating green algae have chromosomes with an ‘absence of localized centromeres’ (Godward, 1966) or ‘polycentric chromosomes’ (King, 1960; Sarma, 1982; Hoshaw and McCourt, 1988). Karyotype analyses also indicate an extraordinary range in chromosome lengths, from 1 to 20 µm (King, 1960). Both of these green algal groups typically have asymmetric (specialized) karyotypes and large chromosomes up to 12 µm long (Sarma, 1982; Hoshaw and McCourt, 1988; Kunachowicz et al., 2001), suggesting a structural distinction with land plant karyotypes.
In contrast to the conjugating green algae and charophytes, some extant members of other groups of streptophytes, including Coleochaetales and Klebsormidiales, have an unspecialized nucleotype with both a small genome size and a small chromosome complement. In the present investigation, nuclear DNA content estimates of <1 pg were found in several isolates of Coleochaete, Entransia and Klebsormidium (Appendix). Unfortunately, published karyotype data are unavailable for many of these taxa. Comparison of haploid genome (1n chromosome complement and 2C DNA content) data for these streptophytes suggests that some species may have retained the proposed small ancestral genome size. For example, the chromosome complement of n = 6 reported for Klebsormidium nitens (Sánchez-Puerta and Leonardi, 2001) with an estimated nuclear DNA content of 2C = 0·6 pg (Appendix) is particularly exciting as it closely approximates both the small genome size and chromosome complement (Fig. 12) reported in the bryophytes (Kapraun, 2005).
Did the relatively large nuclear genomes found in many extant charophycean algae reflect independent, sequential polyploidy events that conferred an advantage associated with increased genome size in the absence of specific upward size constraints? Specifically, large genome size and the correlated large cell size could have been advantageous in buoyant aquatic environments. In addition, in an ancient atmosphere with low levels of oxygen and UV-absorbing ozone, highly polyploid, redundant genomes may have conferred significant survival value. However, as atmospheric conditions changed, their relatively large genomes and large cell sizes, specialized for aquatic habitats, rendered them unsuitable contenders for the colonization of land (Graham, 1993).
ACKNOWLEDGEMENTS
I thank Professor M. D. Bennett and Dr I. J. Leitch for their encouragement to compile published information on algal nuclear genomes and to continue these investigations to expand our database, and for providing an opportunity to present this information at the Second Plant Genome Size Workshop, 2003 (Bennett and Leitch, 2005). Many of the data in the Appendix resulted from student research at UNC-W. Consequently, I recognize contributions from the following graduate and undergraduate students who conducted phycological investigations under my direction: Dr J. C. Bailey, Dr P. W. Boone, P. C. Breden, J. R. Buratti, M. F. Capecchi, R. A. Criswell, J. T. Dunwoody, J. A. Dutcher, Dr D. W. Freshwater, Dr G. M. Gargiulo, T. K. Hinson, Dr J. Lopez-Bautista, M. Marlowe, Dr D. J. Martin, M. N. Nguyen, W. Purvis, M. J. Shipley. I thank Drs J. C. Bailey, C. Delwiche, D. W. Freshwater, M. V. Sanchez-Puerta, G. Saunders and J. West for providing algal specimens used in this study. I am particularly indebted to the staff of the University of Texas Culture Collection of algae (UTEX) who provided numerous cultures at no charge. I acknowledge Dr G. Chandler, Robert York and my son, Dustin Kapraun, for technical assistance in producing the computer-generated graphics. Financial support is gratefully acknowledged from UNC-W Cahill Awards during 2003 and 2005.
APPENDIX
Notes on chromosome numbers and nuclear DNA content estimates in species of green algae
Taxa are listed alphabetically.
Most chromosome numbers have been published as haploid (1n) values for the Chlorophyta (Kapraun, 1993), and 2n values given here are extrapolated from 1n numbers (and ranges of probable 1n numbers).
Most DNA amounts in the literature are given in picograms (pg). Unless otherwise indicated, Mbp values here are derived from estimates for 2C or 4C values using the expression 1 pg = 985 Mbp (Cavalier-Smith, 1985; Bennett et al., 2000). These DNA content values should be considered accurate only to 0·1 pg (Kapraun, 2005). Values for some prasinophycean green algae (Simon et al., 1994) were published as femtograms (10−15), but are expressed as picograms (10−12) here for consistency. DNA amounts originally published as base pairs (Mbp = megabase pairs or mbp = million base pairs) are indicated with a dagger (†) whereas value published as pg are indicated with an asterisk (*). Most of these base pair values were derived from reassociation kinetics (Bot et al., 1989a, b, 1990, 1991; Kooistra et al., 1992; Olsen et al., 1987), but LeGall et al. (1993) used ethidium bromide with RBC standard and flow cytometry. Additional base pair values were determined from pulsed field gel electrophoresis (e.g. Higashiyama and Yamada, 1991; Courties et al., 1998) and flow cytometry (Yamamoto et al., 2001).
Algal life histories typically are characterized by an alternation of haploid gametophyte and diploid sporophyte generations (Kapraun, 1993). Thus, DNA content (pg) measurements could be based on either or both 2C replicated haploid (1n) nuclei or 4C replicated diploid (2n) nuclei. In practice, most published DNA content (pg) values are for 2C diploid nuclei and most 1C and 4C values are extrapolated. Here, the original published DNA content (pg) value for each species is indicated with an asterisk (*). In some samples, ploidy level could not be determined with certainty and assignment of DNA content to specific C-level is speculative.
Previously unpublished data are indicated by an asterisk (*).
Standard species. Species used as a calibration standard for algal research are listed in table 1 (online at http://people.uncw.edu/kapraund/DNA/Chlorophyta.htm). The vast majority of nuclear DNA estimates for algae have used chicken red blood cells or erythrocytes (RBC) for a DNA standard with 2·4 pg being a generally accepted value for the 4C DNA content of Gallus gallus (Clowes et al., 1983; Riechmann et al., 2000). Mouse (Mus) sperm was used as a standard by Hamada et al. (1985), the fish Betta splendens was used as a standard by Spring et al. (1978) and Allium cepa was used by Maszewski and Kołodziejczyk (1991). The green alga Chara tomentosa was used as a standard by Kunachowicz et al. (2001). Initial investigations in our laboratory utilized a standard line based on the fluorescence intensity of an alga with a known DNA content and an angiosperm: Antirrhinum majus L. (e.g. Kapraun & Shipley, 1990; Hinson & Kapraun, 1992; Kapraun & Bailey, 1992) or Impatiens balsamina L. (e.g. Kapraun & Shipley, 1990). Saccharomyces cerivisiae (yeast) was used as a standard in investigations of the Chlorellales by Yamamoto et al. (2001) who assumed a DNA content of 13·4 Mbp for Saccharomyces. It should be noted that Kumar and Snyder (2001) calculates the nuclear genome of Saccharomyces to be 12 Mbp.
Methods. Some of the earliest estimates of nuclear genome size in green algae used microfluorometric analysis (MFA) (Cattolico and Gibbs, 1975). Most contemporary research utilizes flow cytometry (FC) (LeGall et al., 1993) and microspectrophotometry (MI) (Kapraun, 1994; Kapraun & Buratti, 1998), which have been shown to be reliable methods for quantification of nuclear DNA contents in green algae. Feulgen microdensitometry (FE) was used by Maszewski and Kołodziejczyk (1991). Reassociation kinetics (RK) has been used successfully as well (Bot et al., 1989a, b, 1990, 1991; Dörr and Huss, 1990; Kooistra et al., 1992; Olsen et al., 1987). Pulse-field gel electrophoresis (PFGE) or electrophoretic karyotyping has been used successfully with Chlorella (Higashiyama and Yamada, 1991).
In the present study, selected nuclear genome size estimates derived from If data were corroborated using nuclear volume (NV) estimates, assuming a plant nuclear volume of 15 µm3 = 1·0 pg (Sparrow and Nauman, 1973). Detailed methodology for estimating nuclear genome size from NV data in green algae using the expression NV = (πd3)/6, where d = cell diameter (Kapraun et al., 1988) has been published previously (Kapraun and Nguyen, 1994).
Several DNA-localizing fluorochromes have been used in published investigations. DAPI (4′,6-diamidino-2-phenylindole) is certainly the most popular, especially in recent studies (Kapraun, 1994; Kapraun & Buratti, 1998). Hydroethidine (H) (Kapraun & Bailey, 1992), ethidium bromide (EB) (Le Gall et al., 1993; Kunachowicz et al., 2001) and propidium iodide (PI) (Spring et al., 1978) were used in selected green algal investigations.
A key to the references cited for chromosome complements and DNA values in Chlorophyta appears below the table.
APPENDIX
Chromosome number and nuclear DNA content in chlorophycean and charophycean algae
| Entry number | Species(a) | 2n(b) | Original ref. for 2n | DNA amount | Original ref. for C-value(e) | Standard species(f) | Method(g) | |||
|---|---|---|---|---|---|---|---|---|---|---|
| 1C (Mbp)(c) | 1C (pg)(d) | 2C (pg)(d) | 4C (pg)(d) | |||||||
| Charophycean Green Algae | ||||||||||
| CHARALES | ||||||||||
| Characeae | ||||||||||
| 1a | Chara tomentosa Linnaeus (male) | 28 | 19 | 7252 | 7·4* | 14·8 | 29·6 | 19 | Chara | MI:EB |
| 1b | Chara tomentsoa Linnaeus(female) | 28 | 19 | 6860 | 7·0* | 14·0 | 28·0 | 19 | Chara | MI:EB |
| COLEOCHAETALES | ||||||||||
| Coleochaetaceae | ||||||||||
| 2 | Chaetosphaeridium globosum (Nordstedt) Klebahn | 588 | 0·6 | 1·2* | 2·4 | * | Gallus | MI:DAPI | ||
| 3 | Coleochaete nitellarum Jost | 84 | 22 | 343 | 0·35 | 0·7 | 1·4* | * | Gallus | MI:DAPI |
| 4 | Coleochaete orbicularis Pringsheim | 48 | 22 | 686 | 0·7 | 1·5 | 3·0* | * | Gallus | MI:DAPI |
| 5 | Coleochaete scutata Brébisson | 1287 | 1·3 | 2·7 | 5·5* | * | Gallus | MI:DAPI | ||
| DESMIDIALES1 | ||||||||||
| Desmidiaceae | ||||||||||
| 6 | Cosmocladium perissum Roy et Bisset | 15288 | 15·6 | 31·2* | 62·4 | * | Gallus | MI:DAPI | ||
| 7 | Euastrum pectinatum (Brébisson) ex Brébisson | 22932 | 23·4 | 46·8* | 93·6 | * | Gallus | MI:DAPI | ||
| Peniaceae | ||||||||||
| 8 | Gonatozygon monotaenium de Bary | c. 34 | 18 | 8624 | 8·8 | 17·6* | 35·2 | * | Gallus | MI:DAPI |
| KLEBSORMIDIALES | ||||||||||
| 9 | Entransia fimbriata Hughes | 539 | 0·55 | 1·1* | 2·2 | * | Gallus | MI:DAPI | ||
| 10 | Klebsormidium flaccidum (Kützing) P.C. Silva, K. Mattox et W. Blackwell | 44 | 18 | 198 | 0·2 | 0·4* | 0·8 | * | Gallus | MI:DAPI |
| 11 | Klebsormidium nitens (Meneghini) Lokhorst | 12 | 21 | 2695 | 0·28 | 0·55* | 1·1 | * | Gallus | MI:DAPI |
| MESOSTIGMATALES | ||||||||||
| Mesostigmataceae | ||||||||||
| 12 | Mesostigma viride Lauterborn | 343 | 0·35 | 0·7* | 1·4 | * | Gallus | MI:DAPI | ||
| ZYGNEMATALES | ||||||||||
| Mesotaeniaceae | ||||||||||
| 13 | Mesotaenia kramstae Lemmermann | 539 | 0·55 | 1·1* | 2·2 | * | Gallus | MI:DAPI | ||
| 14 | Roya anglica G. S. West | 784 | 0·8 | 1·6* | 3·2 | * | Gallus | MI:DAPI | ||
| Zygnemataceae | ||||||||||
| 15 | Mougeotia transeaui Collins | 3136 | 3·2 | 6·4* | 12·8 | * | Gallus | MI:DAPI | ||
| Chlorophycean Green Algae | ||||||||||
| SPHAEROPLEALES | ||||||||||
| Scenedesmaceae | ||||||||||
| 17 | Scenedesmus obliquus (Turpin) Kuetzing | 12 | 22 | 196 | 0·2 | 0·40* | 0·8 | 6 | MFA | |
| VOLVOCALES2 | ||||||||||
| Chlamydomonaceae | ||||||||||
| 18 | Brachiomonas sp. | 4·9 | 0·005 | 0·01* | 0·02 | 26 | FC | |||
| 19 | Chlamydomonas reinhardtii P. A. Dangeard | 16 | 3 | 88 | 0·09 | 0·19* | 0·38 | 4 | ||
| 20 | Dunaliella tertiolecta Butcher | 294 | 0·3 | 0·6* | 1·2 | 13 | ||||
| Volvocaceae | ||||||||||
| 21 | Pleodorina californica Shaw (as Eudorina californica) | 28 | 5 | 78 | 0·08 | 0·17* | 0·34 | 25 | ||
| Prasinophycean Green Algae | ||||||||||
| CHLORODENDRALES | ||||||||||
| Chlorodendraceae | ||||||||||
| 22 | Tetraselmis suecica (Kylin) Butcher | 343 | 0·35 | 0·7* | 1·4 | * | Gallus | MI:DAPI | ||
| Halosphaeraceae | ||||||||||
| 23a | Micromonas pusilla (Butcher) Manton et Parke | 15 | 0·015 | 0·03* | 0·06 | 23 | FC | |||
| 23b | Micromonas pusilla | 13 | 0·0135 | 0·027* | 0·05 | 26 | FC | |||
| MAMIELLALES | ||||||||||
| Mamiellaceae | ||||||||||
| 24 | Bathycoccus prasinos Eikrem et Throndsen | 10 | 0·01 | 0·02 | 0·04 | 23 | FC | |||
| 25a | Ostreococcus tauri Courties et Chretiennot-Dinet | 28 | 10 | 10·2† | 0·005 | 0·01 | 0·02 | 9 | PFGE | |
| 25b | Ostreococcus tauri | 9·7† | 0·005 | 0·01 | 0·02 | 10 | PFGE | |||
| PYRAMIMONADALES | ||||||||||
| Pyramimonadaceae | ||||||||||
| 26 | Pyramimonas parkeae Norris et Pearson | 67 | 0·07 | 0·15* | 0·3 | * | Gallus | MI:DAPI | ||
| Trebouxiophycean Green Algae | ||||||||||
| CHLORELLALES3 | ||||||||||
| Chlorellaceae | ||||||||||
| 27 | *Chlorella ellipsoidea Gerneck | 18 | 12 | 400† | 0·41 | 0·82 | 1·64 | 12 | PFGE | |
| 28 | Chlorella fusca var. vacuolata Shihira et Krauss | 521† | 0·53 | 1·06 | 1·12 | 11 | RK | |||
| 29 | Chlorella homosphaera Skuja | 418† | 0·2 | 0·4 | 0·8 | 11 | RK | |||
| 30a | Chlorella kessleri Fott et Nováková | 196† | 0·2 | 0·4 | 0·8 | 11 | RK | |||
| 30b | Chlorella kessleri | 48† | 0·05 | 0·1 | 0·2 | 28 | FC | |||
| 31 | Chlorella lobophora Andreeva | 426† | 0·43 | 0·86 | 1·72 | 11 | RK | |||
| 32 | *Chlorella luteoviridis Chodat | 593† | 0·6 | 1·2 | 2·4 | 11 | RK | |||
| 33 | *Chlorella minutissima Fott et Novakova | 126† | 0·13 | 0·26 | 0·52 | 11 | RK | |||
| 34 | *Chlorella mirabilis Andreeva | 98† | 0·1 | 0·2* | 0·4 | 11 | RK | |||
| 35 | Chlorella protothecoides Krüger | 195† | 0·2 | 0·4 | 0·8 | 11 | RK | |||
| 36 | *Chlorella saccharophila var. ellipsoidea (Gerneck) Fott et Nováková | 808† | 0·8 | 1·6 | 3·2 | 11 | RK | |||
| 37 | Chlorella saccharophila var. saccharophila (Krüger) Migula, Fott et Nováková | 394† | 0·4 | 0·8 | 1·6 | 11 | RK | |||
| 38a | Chlorella sorokiana Shihira et Krauss | 49 | 0·05 | 0·11* | 0·22 | 4 | MFA | |||
| 38b | Chlorella sorokiana | 597† | 0·61 | 1·2 | 2·4 | 11 | RK | |||
| 39a | Chlorella vulgaris M. Beijerinck | 16 | 12 | 400† | 0·41 | 0·82 | 1·64 | 12 | ||
| 39b | Chlorella vulgaris | 140† | 0·14 | 0·28 | 0·56 | 11 | RK | |||
| 40 | *Chlorella zofingiensis Dönz | 413† | 0·4 | 0·8 | 1·6 | 11 | RK | |||
| 41 | Marvania coccoides (Naumann) Henley et al. ( = Nannochloris coccoides Naumann) | 18† | 0·02 | 0·04 | 0·08 | 28 | yeast | FC | ||
| 42a | Nannochloris bacillaris Naumann | 14 | 2 | 98 | 0·1 | 0·2* | 0·4 | 12 | PFGE | |
| 42b | Nannochloris bacillaris | 20† | 0·02 | 0·04 | 0·08 | 28 | yeast | FC | ||
| 43 | Picochlorum atomus (Butcher) Henley et al. | |||||||||
| (as Nannochloris atomus Butcher) | 9·8 | 0·01 | 0·02* | 0·04 | 26 | FC | ||||
| (as Nannochloris atomus) | 47† | 0·05 | 0·1 | 0·2 | 28 | yeast | FC | |||
| 44 | Picochlorum eukaryotum (Wilhelm, Eisenbeis, Wild et Zahn) Henley et al. | |||||||||
| (as Nanochlorum eucaryotum (Wilhem, Eisenbeis, Wild et Zahn) Henley et al.) | 59 | 0·06* | 0·12 | 0·24 | 27 | FC | ||||
| (as Nanochlorum eucaryotum) | 23† | 0·02 | 0·04 | 0·08 | 28 | yeast | FC | |||
| 45 | Picochlorum maculatum (Butcher) Henley et al. (as Nannochloris maculatus Butcher) | 14† | 0·01 | 0·02 | 0·04 | 28 | yeast | FC | ||
| PRASIOLALES | ||||||||||
| Prasiolaceae | ||||||||||
| 46 | Prasiola stipitata Suhr in Jessen | 16 | 8 | 221 | 0·23 | 0·45 | 0·9* | * | Gallus | MI:DAPI |
| Ulvophycean Green Algae | ||||||||||
| CAULERPALES | ||||||||||
| Codiaceae | ||||||||||
| 47 | Codium fragile subsp. tomentosoides (van Goor) P.C. Silva | 20 | 17 | 284 | 2·9 | 5·8 | 11·6* | * | Gallus | MI:DAPI |
| 48 | Codium lucasii Setchell | 882 | 0·9 | 1·8* | 3·6 | * | Gallus | MI:DAPI | ||
| 49 | Codium prostratum Levring | 833 | 0·8 | 1·7* | 3·4 | * | Gallus | MI:DAPI | ||
| CLADOPHORALES/SIPHONOCLADALES COMPLEX4 | ||||||||||
| 50 | Cladophora coelothrix Kützing | 1421 | 1·45 | 2·9 | 5·8 | * | Gallus | MI:DAPI | ||
| 51 | Pithora sp. (UTEX 787) | 2009 | 2·05 | 4·1 | 8·2* | * | Gallus | MI:DAPI | ||
| 52 | Pithophora sp. (UTEX 1333) | 882 | 0·9 | 1·9 | 3·8* | * | Gallus | MI:DAPI | ||
| DASYCLADALES5 | ||||||||||
| Dasycladaceae | ||||||||||
| 53 | Neomeris dumetosa Lamouroux | 1862 | 1·9 | 3·8* | 7·6 | * | Gallus | MI:DAPI | ||
| 54 | Parvocaulis exigua (Solms-Laubach) S. Berger et al. (=Polyphysa exigua (Solms-Laubach) M. J. Wynne) | 1568 | 1·6 | 3·2* | 6·4 | * | Gallus | MI:DAPI | ||
| TRENTEPOHLIALES | ||||||||||
| Trentepohliaceae | ||||||||||
| 55 | Cephaleuros parasiticus Karsten | 1911 | 1·95 | 3·9* | 7·2 | 20 | Gallus | MI:DAPI | ||
| 56 | Cephaleuros virescens Kunze in Fries | 36 | 14 | 980 | 1·0 | 2·0* | 4·0 | 20 | Gallus | MI:DAPI |
| 57 | Physolinum monile (De Wildeman) Printz | 22 | 7 | 2009 | 2·05 | 4·1* | 8·2 | 20 | Gallus | MI:DAPI |
| 58 | Trentepohlia arborum (Agardh) Hariot | 1470 | 1·5 | 3·0* | 6·0 | 20 | Gallus | MI:DAPI | ||
| 59a | Trentepohlia aurea (Linnaeus) Martius | 32,34 | 1, 24 | 588 | 0·6 | 1·2* | 2·4 | 20 | Gallus | MI:DAPI |
| 59b | Trentepohlia aurea | 701 | 0·71 | 1·43* | 2·86 | * | Gallus | MI:DAPI | ||
| 60 | Trentepohlia iolithus (Linnaeus) Wallroth | 980 | 1·0 | 2·0* | 4·0 | * | Gallus | MI:DAPI | ||
| 61 | Trentepohlia odorata (Wiggers) Wittrock | 539 | 0·55 | 1·1* | 2·2 | 20 | Gallus | MI:DAPI | ||
| 62 | Trentepohlia umbrina (Kützing) Bornet | 24 | 22 | 657 | 0·67 | 1·34 | 2·68 | * | Gallus | MI:DAPI |
| ULOTRICHALES6 | ||||||||||
| Incertae sedis | ||||||||||
| 63 | Gleotilopsis sterilis Deason | 108 | 0·11 | 0·23* | 0·46 | * | Gallus | MI:DAPI | ||
| 64 | Pseudendoclonium basilense Vischer | 167 | 0·17 | 0·34* | 0·68 | * | Gallus | MI:DAPI | ||
| Monostromaceae | ||||||||||
| 65 | Capsosiphon fulvescens (C.Agardh) Setchell et N.L.Gardner | 157 | 0·16 | 0·33* | 0·6 | * | Gallus | MI:DAPI | ||
| ULVALES7 | ||||||||||
| Incertae sedis | ||||||||||
| 66 | Pseudendoclonium basilense Vischer | 196 | 0·17 | 0·34* | 0·6 | * | Gallus | MI:DAPI | ||
| Ulvaceae | ||||||||||
| 67 | Percursaria percursa (C.Agardh) Rosenvinge | 294 | 0·3 | 0·6* | 1·2 | * | Gallus | MI:DAPI | ||
| 68 | Ulva compressa Linnaeus | |||||||||
| (as Enteromorpha compressa (Linnaeus) Greville) | 20 | 16 | 26·8* | 0·07 | 0·14* | 0·28 | 15 | Arabidopsis | FC | |
| 69 | Ulva rotundata Bliding | 294 | 0·3 | 0·6* | 1·2 | * | Gallus | MI:DAPI | ||
1Traditional taxonomic lists often grouped all conjugating green algae within one order, Zygnematales (Conjugales) (Bold and Wynne, 1985). Results of recent molecular studies support recognition of two orders, Desmidiales and Zygnematales (McCourt et al., 2000; Denboh et al., 2001).
2Molecular data demonstrate that Chlamydomonas is not monophyletic (Nozaki et al., 2000; Nozaki and Krienitz, 2001) and that revision of the circumscription of these genera will be required (Larson et al., 11992). Dunalliella tertiolecta, included here in Chlamydomonaceae, is part of a polyphyletic complex that may warrant recognition as a separate order (Nakayama et al., 1966).
3Recent molecular studies have demonstrated that Chlorella taxa are dispersed over two classes: Trebouxiophyceae and Chlorophyceae (Krienitz et al., 2004). Chlorellales included here are considered to be trebouxiophycean algae (Huss et al., 1999).
4Molecular data clearly demonstrate that classifications of the genus Cladophora should be revised (Hanyuda et al., 2002). Circumscription of families in this complex will require sequence data for additional cladophoralean algae.
5Recent molecular investigations indicate that genera of Dasycladaceae are well delineated, but this does not hold true for genera of Polyphysaceae (=Acetabulariaceae). 18S rRNA gene sequence data support transfer of Acicularia schenckii and Polyphysa peniculus to the genus Acetabularia (Berger et al., 2003).The familiar binomials are retained here for convenience until a complete taxonomic revision of Dasycladales is available.
6Recent phylogenetic investigations have redefined the boundary between Ulotrichales and Ulvales (O'Kelly et al., 2004). Species of Monostroma appear to be more closely related to Ulotrichales than to the Ulvales (Hayden and Waaland, 2002). No contemporary characterization of families is available for this newly circumscribed order.
7Characters used to separate the genera Ulva and Enteromorpha lack taxonomic significance (Tan et al., 1999; Shimada et al., 2003). The familiar binomials have been retained here in the absence of formal reassignment of species (Hayden and Waaland, 2003, 2004). Exact placement of Blidingia in Ulvales remains uncertain (Incertae sedis) as no contemporary characterization of the emended family Monostromaceae is available.
LITERATURE CITED
- Alexopoulos CJ, Bold HC. Algae and Fungi. New York: Macmillan Company; 1967. [Google Scholar]
- Angeler DG, Schagerl M, Coleman AW. Phylogenetic relationships among isolates of Eudorina species (Volvocales, Chlorophyta) inferred from molecular and biochemical data. Journal of Phycology. 1999;35:815–823. [Google Scholar]
- Arai S, Takahashi H, Takano H, Sakai A, Kawano S. Isolation, characterization and chromosome mapping of an actin gene from the primitive green alga, Nannochloris bacillaris (Chlorophyceae) Journal of Phycology. 1998;34:477–485. [Google Scholar]
- Bakker FT, Olsen JL, Stam WT, Hoek van den C. The Cladophora complex (Chlorophyta): new views based on 18S rRNA gene sequences. Molecular Phylogenetics and Evolution. 1994;3:365–382. doi: 10.1006/mpev.1994.1043. [DOI] [PubMed] [Google Scholar]
- Bennett MD. Variation in genomic form in plants and its ecological implications. New Phytologist. 1987;106(Supplement):177–200. [Google Scholar]
- Bennett MD, Leitch IJ. Nuclear DNA amounts in angiosperms: progress, problems and prospects. Annals of Botany. 2005a;95:45–90. doi: 10.1093/aob/mci003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bennett MD, Leitch IJ. Plant genome size research: a field in focus. Annals of Botany. 2005b;95:1–6. doi: 10.1093/aob/mci001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bennett MD, Bhandol P, Leitch IJ. Nuclear DNA amounts in angiosperms and their modern uses-807 new estimates. Annals of Botany. 2000;86:859–909. [Google Scholar]
- Bennetzen JL. Mechanisms and rates of genome expansion and contraction in flowering plants. Genetica. 2002;115:29–36. doi: 10.1023/a:1016015913350. [DOI] [PubMed] [Google Scholar]
- Bennetzen JL, Kellogg EA. Do plants have a one-way ticket to genomic obesity? The Plant Cell. 1997;9:1509–1514. doi: 10.1105/tpc.9.9.1509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berger SL, Kaever MJ. Dasycladales: an illustrated monograph of a fascinating algal order. Stuttgart: Georg Thieme Verlag; 1992. New York: Oxford University Press. [Google Scholar]
- Berger S, Fettweiss U, Gleissberg S, Liddle LB, Richter U, Sawitzky, Zuccarello C. 18S rDNA phylogeny and evolution of cap development in Polyphysaceae (formerly Acetabulariaceae: Dasycladales, Chlorophyta) Phycologia. 2003;42:506–561. [Google Scholar]
- Bhattacharya D, Surek B, Rüsing M, Damberger S, Melkonian M. Group I introns are inherited through common ancestry in the nuclear-encoded rRNA of Zygnematales (Charophyceae) Proceedings of the National Academy of Sciences of the USA. 1994;91:9916–9920. doi: 10.1073/pnas.91.21.9916. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bliding C. A critical survey of European taxa in the Ulvales. Part I. Capsosiphon, Percursaria, Blidingia, Enteromorpha. Opera Botanica. 1963;8:1–160. [Google Scholar]
- Bliding C. A critical survey of the European taxa in the Ulvales II. Botaniska Notiser. 1968;121:535–629. [Google Scholar]
- Blomster J, Maggs CA, Stanhope MJ. Molecular and morphological analysis of Enteromorpha intestinalis and E. compressa (Chlorophyta) in the British Isles. Journal of Phycology. 1998;34:319–340. [Google Scholar]
- Blomster J, Maggs CA, Stanhope MJ. Extensive intraspecific morphological variation in Enteromorpha muscoides (Chlorophyta) revealed by molecular analysis. Journal of Phycology. 1999;35:575–586. [Google Scholar]
- Bodenbender S, Schnetter R. Nuclear behavior during the life cycles of Chaetomorpha, Ernodesmis and Struvea (Ulvophyceae, Chlorophyta) under culture conditions. Cryptogamic Botany. 1990;1:340–354. [Google Scholar]
- Bold HC, Wynne MJ. Introduction to the algae. 2nd edn. New Jersey: Prentice Hall; 1985. [Google Scholar]
- Børgesen F. The Marine Algae of the Danish West Indies. Part I. Chlorophyceae. Det Kongelige Danske Videnskabernes Selskab Biologiske Meddeleser. 1913;20:1–158. [Google Scholar]
- Bot PVM, Holton RW, Stam WT, van den Hoek C. Molecular divergence between North Atlantic and indo-West Pacific Cladophora albida (Cladophorales, Chlorophyta) isolates as indicated by DNA–DNA hybridization. Marine Biology, Berlin. 1989a;102:307–313. [Google Scholar]
- Bot PVM, Stam WT, Boele-Bos SA, van den Hoek C, van Selden W. Biogeographic and phylogenetic studies in three North Atlantic species of Cladophora (Cladophorales, Chlorophyta) using DNA–DNA hybridization. Phycologia. 1989b;28:159–168. [Google Scholar]
- Bot PVM, Stam WT, van den Hoek C. Genotypic relations between geographic isolates of Cladophora laetevirens and C. vagabunda. Botanica Marina. 1990;33:441–446. [Google Scholar]
- Bot PVM, Brussaard CPD, Stam WT, van den Hoek C. Evolutionary relationships between four species of Cladophora (Cladophorales, Chlorophyta) based on DNA–DNA hybridization. Journal of Phycology. 1991;27:617–623. [Google Scholar]
- Bremer K, Humphries CJ, Mishler BD, Churchill SP. On cladistic relationships in green plants. Taxon. 1987;36:339–349. [Google Scholar]
- Buchheim MA, Lemieux C, Otis C, Gutell RR, Chapman RL, Turmel M. Phylogeny of the Chlamydomonadales (Chlorophyceae): a comparison of ribosomal RNA gene sequences from the nucleus and the chloroplast. Molecular Phylogenetics and Evolution. 1996;5:391–402. doi: 10.1006/mpev.1996.0034. [DOI] [PubMed] [Google Scholar]
- Buchheim MA, Buchheim JA, Chapman RL. Phylogeny of the VLE-14 Chlamydomonas (Chlorophyceae) group: a study of 18S rRNA gene sequences. Journal of Phycology. 1997;33:1024–1030. [Google Scholar]
- Buchheim MA, Buchheim JA, Carloson T, Kugrens P. Phylogeny of Lobocharacium (Chlorophyceae) and allies: a study of 18S and 26S rDNA data. Journal of Phycology. 2002;38:376–383. [Google Scholar]
- Buchheim M, Buchheim J, Carlson T, Braband A, Hepperle D, Krienitz L, et al. Phylogeny of the Hydrodictyaceae (Chlorophyceae): inferences from rDNA data. Journal of Phycology. 2005;41:1039–1054. [Google Scholar]
- Cattolico RA, Gibbs SP. Rapid filter method for the microfluorometric analysis of DNA. Analytical Biochemistry. 1975;69:572–582. doi: 10.1016/0003-2697(75)90162-1. [DOI] [PubMed] [Google Scholar]
- Cavalier-Smith T. Cell volume and evolution of eukaryotic genome size. In: Cavalier-Smith T, editor. The evolution of genome size. New York: John Wiley; 1985. pp. 105–184. [Google Scholar]
- Chapman RL, Waters D, Lopez-Bautista J. Phylogenetic affinities of the Trentepohliales (Chlorophyta) inferred from small subunit rRNA sequences. Journal of Phycology. 1995;31(suppl):7. [Google Scholar]
- Charles D. Isolation and characterization of DNA from unicellular algae. Plant Science Letters. 1977;8:35–44. [Google Scholar]
- Cimino MT, Delwiche CF. Molecular and morphological data identify a cryptic species complex in endophytic members of the genus Coleochaete Bréb. (Charophyta: Coleochaetaceae) Journal of Phycology. 2002;38:1213–1221. [Google Scholar]
- Clowes AW, Reidy MA, Clowes MM. Kinetics of cellular proliferation after arterial injury. I. Smooth muscle growth in absence of endothelium. Laboratory Investigations. 1983;49:327–333. [PubMed] [Google Scholar]
- Cole K, Akintobi S. The life cycle of Prasiola meridionalis Setchell and Gardner. Canadian Journal of Botany. 1963;4:661–668. [Google Scholar]
- Coleman AW. Biogeography and speciation in the Pandorina/Volvulina (Chlorophyta) superclade. Journal of Phycology. 2001;37:836–851. [Google Scholar]
- Coleman AW, Maguire M.J, Coleman JR. Mithramycin- and 4′–6 diamidino-2-phenolindole (DAPI)-staining for fluorescence microspectrophotometric measurement of DNA in nuclei, plastids, and virus particles. Journal of Histochemistry and Cytochemistry. 1981;29:959–968. doi: 10.1177/29.8.6168681. [DOI] [PubMed] [Google Scholar]
- Cook ME. Cytokinesis in Coleochaete orbicularis (Charophyceae): an ancestral mechanism inherited by plants. American Journal of Botany. 2004;91:313–320. doi: 10.3732/ajb.91.3.313. [DOI] [PubMed] [Google Scholar]
- Courties C, Vaquer A, Troussellier M, Lautier J, Chrétiennot-Dinet M-J, Neveux J, et al. Smallest eukaryotic organism. Nature. 1994;370:255. [Google Scholar]
- Courties C, Perasso R, Chrétiennot-Dinet M-J, Gouy M, Guillou L, Troussellir M. Phylogenetic analysis and genome size of Ostreococcus tauri (Chlorophyta, Prasinophyceae) Journal of Phycology. 1998;34:844–849. [Google Scholar]
- Cunningham CW, Omland KE, Oakley TH. Reconstructing ancestral character states: a critical reappraisal. Trends in Ecology and Evolution. 1998;13:361–366. doi: 10.1016/s0169-5347(98)01382-2. [DOI] [PubMed] [Google Scholar]
- Daugbjerg N, Moestrup Ø, Arctander P. Phylogeny of genera of Prasinophyceae and Pedinophyceae (Chlorophyta) deduced from molecular analysis of the rbcL gene. Phycological Research. 1995;43:203–213. [Google Scholar]
- Delwiche CF, Karol KG, Cimino MT, Systma KJ. Phylogeny of the genus Coleochaete (Coleochaetales, Charophyta) and related taxa inferred by analysis of the chloroplast gene rbcL. Journal of Phycology. 2002;38:394–403. [Google Scholar]
- Denboh T, Hendrayanti D, Ichimura T. Monophyly of the genus Closterium and the Order Desmidiales (Charophyceae, Chlorophyta) inferred from nuclear small subunit rDNA data. Journal of Phycology. 2001;37:1063–1072. [Google Scholar]
- Derelle E, Ferraz C, Lagoda P, Eychenie S, Cooke R, Regad F, et al. DNA libraries for sequencing the genome of Ostreococcus tauri (Chlorophyta, Prasinophyceae): the smallest free-living eukaryotic cell. Journal of Phycology. 2002;38:1150–1156. [Google Scholar]
- Dörr R, Huss VAR. Characterization of nuclear DNA in 12 species of Chlorella (Chlorococcales, Chlorophyta) by DNA reassociation. BioSystems. 1990;24:145–155. doi: 10.1016/0303-2647(90)90007-n. [DOI] [PubMed] [Google Scholar]
- Drummond CS, Hall J, Karol KG, Delwiche CF. Phylogeny of Spirogyra and Sirogonium (Zygnematophyceae) based on rbcL sequence data. Journal of Phycology. 2005;41:1055–1064. [Google Scholar]
- Fawley MW, Yun Y, Qin M. Phylogenetic analyses of 18s rDNA sequences reveal a new coccoid lineage of the Prasinophyceae (Chlorophyta) Journal of Phycology. 2000;36:387–393. [Google Scholar]
- Fay MF, Cowan RS, Leitch IJ. The effects of nuclear DNA content (C-value) on the quality and utility of AFLP fingerprints. Annals of Botany. 2005;95:237–246. doi: 10.1093/aob/mci017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feist M, Génot P, Grambast-Fessard N. Ancient Dasycladales and Charophyta: convergences and differences, with special attention to Munieria baconica. Phycologia. 2003;42:123–132. [Google Scholar]
- Floyd GL, O'Kelly JC. Ulvophyceae. In: Margulis L, Corliss JO, Melkonian M, Chapman DJ, editors. Handbook of Protoctista. Boston: Jones and Bartlett Publishers; 1990. pp. 617–635. [Google Scholar]
- Freshwater DW, Dutcher JA, Kapraun DF, Sizemore RK. Variation in nuclear DNA base composition (mol% G+C) in three orders of marine green algae. Hydrobiologia. 1990;204/205:167–172. [Google Scholar]
- Friedl T. Inferring taxonomic positions and testing genus level assignments in coccoid green lichen algae: a phylogenetic analysis of 18S ribosomal RNA sequences from Dictyochloropsis reticulata and from members of the genus Myrmecia (Chlorophyta, Trebouxiophyceae cl. nov.) Journal of Phycology. 1995;31:632–639. [Google Scholar]
- Friedl T. The evolution of the Green Algae. Plant Systematics and Evolution. 1997;11(Supplement):87–101. [Google Scholar]
- Friedl T, O'Kelly CJ. Phylogenetic relationship of green algae assigned to the genus Planophila (Chlorophyta): evidence from 18S rDNA sequence data and ultrastructure. European Journal of Phycology. 2002;37:373–384. [Google Scholar]
- Gerrath JF. Conjugating green algae and desmids. In: Wehr JD, Sheath RG, editors. Freshwater algae of North America. Amsterdam: Academic Press; 2003. pp. 353–381. [Google Scholar]
- Godward MBE. The chromosomes of the Algae. London: Edward Arnold; 1966. [Google Scholar]
- Goff LJ, Coleman AW. DNA: microspectrofluorometric studies. In: Cole KM, Sheath RG, editors. Biology of the red algae. New York: Cambridge University Press; 1990. pp. 43–72. [Google Scholar]
- Gontcharov AA, Melkonian M. Molecular phylogeny of Staurastrum meyen ex Ralfs and related genera (Zygnematophyceae, Streptophyta) based on coding and noncoding rDNA sequence comparisons. Journal of Phycology. 2005;41:887–899. [Google Scholar]
- Gontcharov AA, Marin B, Melkonian M. Molecular phylogeny of conjugating green algae (Zygnemophyceae, Streptophyta) inferred from SSU rDNA sequence comparisons. Journal of Molecular Evolution. 2003;56:89–103. doi: 10.1007/s00239-002-2383-4. [DOI] [PubMed] [Google Scholar]
- Graham LE. Origin of land plants. New York: John Wiley and Sons; 1993. [Google Scholar]
- Graham LE, Wilcox LW. Algae. Upper Saddle River, NJ: Prentice Hall; 2000. [Google Scholar]
- Grambast LJ. Phylogeny of the Charophyta. Taxon. 1974;23:463–481. [Google Scholar]
- Gregory TR. Coincidence, coevolution, or causation? DNA content, cell size, and the C-value enigma. Biological Revue. 2001;76:65–101. doi: 10.1017/s1464793100005595. [DOI] [PubMed] [Google Scholar]
- Gregory TR. The C-value enigma in plants and animals: a review of parallels and an appeal for partnership. Annals of Botany. 2005a;95:133–146. doi: 10.1093/aob/mci009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gregory TR. Synergy between sequence and size in large-scale genomics. Nature Reviews. 2005b;6:699–708. doi: 10.1038/nrg1674. [DOI] [PubMed] [Google Scholar]
- Groth-Malonek M, Knoop V. Bryophytes and other basal land plants: the mitochondrial perspective. Taxon. 2005;54:293–297. [Google Scholar]
- Hamada J, Saito M, Ishida MR. Nuclear phase in vegetative and gamete cells of Closterium ehrenbergii: fluorescence microspectrophotometry of DNA content. Annual reports of the Research Reactor Institute. 1985;18:56–61. [Google Scholar]
- Handa S, Nakahara M, Tsubota H, Deguchi H, Nakano T. A new aerial alga, Stichococcus ampulliformis sp. nov. (Trebouxiophyceae, Chlorophyta) from Japan. Phycological Research. 2003;51:203–210. [Google Scholar]
- Hanyuda T, Wakana I, Arai S, Miyaji K, Watano Y, Ueda K. Phylogenetic relationships within Cladophorales (Ulvophyceae, Chlorophyta) inferred from 18S rRNA gene sequences, with special reference to Aegagropila linnaei. Journal of Phycology. 2002;38:564–571. [Google Scholar]
- Hayden HS, Waaland JR. Phylogenetic systematics of the Ulvaceae (Ulvales, Ulvophyceae) using chloroplast and nuclear DNA sequences. Journal of Phycology. 2002;38:1200–1212. [Google Scholar]
- Hayden HS, Waaland JR. A molecular systematic study of Ulva (Ulvaceae, Ulvales) from the northeast Pacific. Phycologia. 2004;43:364–382. [Google Scholar]
- Hayden HS, Blomster J, Maggs CA, Silva PC, Stanhope MJ, Waaland JR. Linnaeus was right all along: Ulva and Enteromorpha are not distinct genera. European Journal of Phycology. 2003;38:277–294. [Google Scholar]
- Henley WJ, Hironaka JL, Guillou L, Buchheim MA, Buchheim JA, Fawlley MW, Fawley KP. Phylogenetic analysis of the “Nannochloris-like” algae and diagnoses of Picochlorum oklahomensis gen. et sp. nov. (Trebouxiophyceae, Chlorophyta) Phycologia. 2004;43:641–652. [Google Scholar]
- Higashiyama T, Yamada T. Electrophoretic karyotyping and chromosomal gene mapping of Chlorella. Nucleic Acids Research. 1991;19:6191–6195. doi: 10.1093/nar/19.22.6191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hinson TK, Kapraun DF. Karyology and nuclear DNA quantification of four species of Chaetomorpha (Cladophorales, Chlorophyta) from the western Atlantic. Helgoländer Meersuntersuchungen. 1992;45:273–285. [Google Scholar]
- Hiraoka M, Shimada S, Uenosono M, Masuda M. A new green-tide forming alga, Ulva ohnoi Hiraoka et Shimada sp. nov. (Ulvales, Ulvophyceae) from Japan. Phycological Research. 2003;51:17–29. [Google Scholar]
- Holm-Hansen O. Algae: amounts of DNA and organic carbon in single cells. Science. 1969;163:87–88. doi: 10.1126/science.163.3862.87. [DOI] [PubMed] [Google Scholar]
- Hoshaw RW, McCourt RM. The Zygnemataceae (Chlorophyta): a twenty-year update of research. Phycologia. 1988;27:511–548. [Google Scholar]
- Hoshaw RW, McCourt RM, Wang J-C. Phylum Conjugophyta. In: Margulis L, Corliss JO, Melkonian M, Chapman DJ., editors. Handbook of Protoctista. Boston: Jones and Bartlett Publishers; 1990. pp. 119–131. [Google Scholar]
- Huss VAR, Kranz HD. Charophyte evolution and the origin of land plants. In: Bhattacharya D, editor. The origins of the algae and their plastids. Vienna: Springer-Verlag; 1997. pp. 103–114. [Google Scholar]
- Huss VAR, Frank C, Hartmann EC, Hirmer M, Kloboucek A, Seidel BM, et al. Biochemical taxonomy and molecular phylogeny of the genus Chlorella sensu lato (Chlorophyta) Journal of Phycology. 1999;35:587–598. [Google Scholar]
- Inoue S, Uchino A. Karyological studies on mosses. VI. Karyotypes of fourteen species including some species with the intraspecific euploid and aneuploid. Botanical Magazine (Tokyo) 1969;82:359–367. [Google Scholar]
- Kantz TS, Theriot EC, Zimmer EA, Chapman RL. The Pleurastrophyceae and Micromonadophyceae: a cladistic analysis of nuclear rRNA sequence data. Journal of Phycology. 1990;26:711–721. [Google Scholar]
- Kapraun DF. Karyology of marine green algae. Phycologia. 1993;32:1–21. [Google Scholar]
- Kapraun DF. Cytophotometric estimation of nuclear DNA contents in thirteen species of the Caulerpales (Chlorophyta) Cryptogamic Botany. 1994;4:410–418. [Google Scholar]
- Kapraun DF. Nuclear DNA content estimates in multicellular eukaryotic green, red and brown algae: phylogenetic considerations. Annals of Botany. 2005;95:7–44. doi: 10.1093/aob/mci002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kapraun DF, Bailey JC. Karyology and cytophotometric estimation of nuclear DNA variation in seven species of Ulvales (Chlorophyta) Japanese Journal of Phycology. 1992;40:15–26. [Google Scholar]
- Kapraun DF, Breden PC. Karyological studies of Cladophoropsis (Siphonocladales, Chlorophyta) from Bermuda. Botanica Marina. 1988;31:515–520. [Google Scholar]
- Kapraun DF, Buratti JR. Evolution of genome size in the Dasycladales (Chlorophyta) as determined by DAPI cytophotometry. Phycologia. 1998;37:176–183. [Google Scholar]
- Kapraun DF, Gargiulo GM. Karyological studies of three species of Cladophora (Cladophorales, Chlorophyta) species from Bermuda. Giornale Botanico Italiano. 1987a;121:165–176. [Google Scholar]
- Kapraun DF, Gargiulo GM. Karyological studies of four Cladophora (Cladophorales, Chlorophyta) species from coastal North Carolina. Giornale Botanico Italiano. 1987b;121:1–26. [Google Scholar]
- Kapraun DF, Nguyen MN. Karyology, nuclear DNA quantification and nucleus-cytoplasmic domain variations in some multinucleate green algae. Phycologia. 1994;33:42–52. [Google Scholar]
- Kapraun DF, Shipley MJ. Karyology and nuclear DNA quantification in Bryopsis (Chlorophyta) from North Carolina. USA. Phycologia. 1990;29:443–453. [Google Scholar]
- Kapraun DF, Gargiulo GM, Tripodi G. Nuclear DNA and karyotype variation in species of Codium (Codiales, Chlorophyta) from the North Atlantic. Phycologia. 1988;27:273–282. [Google Scholar]
- Kapraun DF, Leitch IJ, Bennett MD. Algal DNA C-values database (release 1.0, December 2004) 2004. http://www.rgbkew.org.uk/cval/homepage.html .
- Karol KG, McCourt RM, Cimino MT, Delwiche CF. The closest living relatives of land plants. Science. 2001;294:2351–2353. doi: 10.1126/science.1065156. [DOI] [PubMed] [Google Scholar]
- Katana A, Kwiatowski J, Spalik K, Zakryś B. Phylogenetic position of Koliella (Chlorophyta) as inferred from nuclear and chloroplast small subunit rDNA. Journal of Phycology. 2001;37:443–451. [Google Scholar]
- Kenrick P, Crane PR. The origin and early diversification of land plants—a cladistic study. Washington: Smithsonian Institution Press; 1997. [Google Scholar]
- King GC. The cytology of desmids: the chromosomes. New Phytologist. 1960;59:65–72. [Google Scholar]
- Knight CA, Molinari NA, Petrov DA. The large genome constraint hypothesis: evolution, ecology and phenotype. Annals of Botany. 2005;95:177–190. doi: 10.1093/aob/mci011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kooistra WHCF, Boele-Bos SA, Stam WT, Van den Hoek C. Biogeography of Cladophoropsis membranacea (Siphonocladales, Chlorophyta) as revealed by single copy DNA distances. Botanica Marina. 1992;35:329–336. [Google Scholar]
- Kornmann P, Sahling P.-H. Meeresalgen von Helgoland, Benthische Grün-, Graun- und Rotalgen. Helgoländer Meeresuntersuchungen. 1977;29:1–289. [Google Scholar]
- Kratz RF, Young PA, Mandoli DF. Timing and light regulation of apical morphogenesis during reproductive development in wild-type populations of Acetabularia acetabulum (Chlorophyceae) Journal of Phycology. 1998;34:138–146. [Google Scholar]
- Krienitz L, Hegewald EH, Hepperle D, Huss VAR, Rohr T, Wolf M. Phylogenetic relationship of Chlorella and Parachlorella gen. nov. (Chlorophyta, Trebouxiophyceae) Phycologia. 2004;43:529–542. [Google Scholar]
- Kunachowicz A, Luchniak P, Olszewska MJ, Sakowicz T. Comparative karyology, DNA methylation and restriction pattern analysis of male and female plants of the dioecious alga Chara tomentosa (Charophyceae) European Journal of Phycology. 2001;36:29–34. [Google Scholar]
- Larson A., Kirk MM, Kirk DL. Molecular phylogeny of the volvocine flagellates. Molecular Biology and Evolution. 1992;9:85–105. doi: 10.1093/oxfordjournals.molbev.a040710. [DOI] [PubMed] [Google Scholar]
- Le Gall Y, Brown S, Marie D, Mejjad M, Kloareg B. Quantification of nuclear DNA and G-C content in marine macroalgae by flow cytometry of isolated nuclei. Protoplasma. 1993;173:123–132. [Google Scholar]
- Leitch IJ, Chase MW, Bennett MD. Phylogenetic analysis of DNA C-values provides evidence for a small ancestral genome size in flowering plants. Annals of Botany. 1998;82(Supplement A):85–94. [Google Scholar]
- Leitch IJ, Soltis DE, Soltis PS, Bennett MD. Evolution of DNA amounts across land plants (Embryophyta) Annals of Botany. 2005;95:207–217. doi: 10.1093/aob/mci014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lemieux C, Otis C, Turmel M. Ancestral chloroplast genome in Mesostigma viride reveals an early branch of green plant evolution. Nature. 2000;403:649–652. doi: 10.1038/35001059. [DOI] [PubMed] [Google Scholar]
- Lewis AL. Diversity and phylogenetic placement of Bracteacoccus Tereg (Chlorophyceae, Chlorophyta) based on 18S ribosomal RNA gene sequence data. Journal of Phycology. 1997;33:279–285. [Google Scholar]
- Lewis LA, McCourt RM. Green algae and the origin of land plants. American Journal of Botany. 2004;91:1535–1556. doi: 10.3732/ajb.91.10.1535. [DOI] [PubMed] [Google Scholar]
- Liang C, Xi Y, Shu J, Li J, Yang J, Che K, et al. Construction of a BAC library of Physcomitrella pattens and isolation of a LEA gene. Plant Science. 2004;167:491–498. [Google Scholar]
- Lindstrom SC, Hanic LA. The phylogeny of North American Urospora (Ulotrichales, Chlorophyta) based on sequence analysis of nuclear ribosomal genes, introns and spacers. Phycologia. 2005;44:194–201. [Google Scholar]
- Lindstrom SC, Golden L. Studies of the green alga Percursaria dawsonii (=Blidingia dawsonii comb. nov., Kornmanniaceae, Ulvales) in British Columbia. Phycological Research. 2006;54:40–56. [Google Scholar]
- Lokhorst GM, Star W, Zuccarello GC. New genus Koliellopsis (Trebouxiophyceae, Chlorophyta): its phylogenetic position inferred from ultrastructure and nuclear ribosomal DNA sequences. Phycological Research. 2004;52:235–243. [Google Scholar]
- Lopez-Bautista JM, Chapman RL. Phylogenetic affinities of the Trentepohliales inferred from small-subunit rDNA. International Journal of Systematic and Evolutionary Microbiology. 2003;53:2099–2106. doi: 10.1099/ijs.0.02256-0. [DOI] [PubMed] [Google Scholar]
- Lopez-Bautista JM, Kapraun DF, Waters DA, Chapman RL. Nuclear DNA quantification and the life cycle in Cephaleuros parasiticus (Trentepohliales, Chlorophyta) American Journal of Botany. 2000;87(6, supplement):248. [Google Scholar]
- Manhart JR, Palmer JD. The gain of two chloroplast tRNA introns marks the green algal ancestors of land plants. Nature. 1990;345:268–270. doi: 10.1038/345268a0. [DOI] [PubMed] [Google Scholar]
- Marmur J, Doty P. Determination of the base composition of deoxyribonucleic acid from its thermal denaturation temperature. Journal of Molecular Biology. 1962;5:109–118. doi: 10.1016/s0022-2836(62)80066-7. [DOI] [PubMed] [Google Scholar]
- Maszewski J, Kołodziejczyk P. Cell cycle duration in antheridial filaments of Chara spp. (Characeae) with different genome size and heterochromatin content. Plant Systematics and Evolution. 1991;175:23–38. [Google Scholar]
- Matsuo Y, Suzuki M, Kasai H, Shizuri Y, Harayama S. Isolation and phylogenetic characterization of bacteria capable of inducing differentiation in the green alga Monostroma oxyspermum. Environmental Microbiology. 2003;5:25–35. doi: 10.1046/j.1462-2920.2003.00382.x. [DOI] [PubMed] [Google Scholar]
- Matsuo Y, Imagawa H, Nishizawa M, Shizuri Y. Isolation of an algal morphogenesis inducer from a marine bacterium. Science. 2005;307:1598. doi: 10.1126/science.1105486. [DOI] [PubMed] [Google Scholar]
- Mattox KR, Stewart KD. Classification of the green algae: a concept based on comparative cytology. In: Irvine DEG, John DM, editors. Systematics of the Green Algae. London: Academic Press; 1984. pp. 29–72. [Google Scholar]
- McCourt RM. Green algal phylogeny. Trends in Ecology and Evolution. 1995;10:159–163. doi: 10.1016/s0169-5347(00)89027-8. [DOI] [PubMed] [Google Scholar]
- McCourt RM, Karol KG, Kaplan S, Hoshaw RW. Using rbcL sequences to test hypotheses of chloroplast and thallus evolution in conjgating green algae (Zygnematales, Charophyceae) Journal of Phycology. 1995;31:989–995. [Google Scholar]
- McCourt RM, Karol KG, Guerlesquin M, Feist M. Phylogeny of extant genera in the family Characeae (Charales, Charophyceae) based on rbcL sequences and morphology. American Journal of Botany. 1996;83:125–131. [Google Scholar]
- McCourt RM, Karol KG, Bell J, Helm-Bychowski M, Grajewska A, Wojciechowski MF, Hoshaw RW. Phylogeny of the conjugating green algae (Zygnemophyceae) based on rbcL sequences. Journal of Phycology. 2000;36:747–758. doi: 10.1046/j.1529-8817.2000.99106.x. [DOI] [PubMed] [Google Scholar]
- McManus HA, Lewis LA. Molecular phylogenetics, morphological variation and colony-form evolution in the family Hydrodictyaceae (Sphaeropleales, Chlorophyta) Phycologia. 2005;44:582–595. [Google Scholar]
- Meiers ST, Proctor VW, Chapman RL. Phylogeny and biogeography of Chara (Charophyta) inferred from 18S rDNA sequences. Australian Journal of Botany. 1999;47:347–360. [Google Scholar]
- Midgley JJ, Bond WJ. Ecological aspects of the rise of angiosperms: a challenge to the reproductive superiority hypotheses. Biological Journal of the Linnean Society. 1991;44:81–92. [Google Scholar]
- Mishler BD, Lewis LA, Buchheim MA, Renzaglia KS, Garbary DJ, Delwiche CF, et al. Phylogenetic relationships of the “Green Algae” and “Bryophytes”. Annals of the Missouri Botanic Garden. 1994;81:451–483. [Google Scholar]
- Miyaji K. A new type of pyrenoid in the genus Rhizoclonium (Cladophorales, Chlorophyta) Phycologia. 1999;38:267–276. [Google Scholar]
- Murray BG. When does intraspecific C-value variation become taxonomically significant? Annals of Botany. 2005;95:119–125. doi: 10.1093/aob/mci007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Naw MWD, Hara Y. Morphology and molecular phylogeny of Prasiola sp. (Prasiolales, Chlorophyta) from Myanmar. Phycological Research. 2002;50:175–182. [Google Scholar]
- Noor MN. A new record on the cytology of Pithophora oedogonia Wittrock from India. Phykos. 1968;7:58–61. [Google Scholar]
- Nozaki H, Krienitz L. Morphology and phylogeny of Eudorina minodii (Chodat) Nozaki et Krienitz, comb. nov. (Volvocales, Chlorophyta) from Germany. European Journal of Phycology. 2001;36:23–28. [Google Scholar]
- Nozaki H, Itoh M, Sano R, Uchida H, Watanabe MM, Kuroiwa T. Phylogenetic relationships within the colonial Volvocales (Chlorophyta) inferred from rbcL gene sequence data. Journal of Phycology. 1995;31:970–979. [Google Scholar]
- Nozaki H, Ohta N, Takano H, Watanabe MM. Reexamination of phylogenetic relationships within the colonial Volvocales (Chlorophyta): an analysis of atpB and rbcL gene sequences. Journal of Phycology. 1999;35:104–112. [Google Scholar]
- Nozaki H, Misawa K, Kajita T, Kono M, Nohara S, Watanabe MM. Origin and evolution of the colonial Volvocales (Chlorophyceae) as inferred from multiple, chloroplast gene sequences. Molecular Phylogenetics and Evolution. 2000;17:256–268. doi: 10.1006/mpev.2000.0831. [DOI] [PubMed] [Google Scholar]
- Oakley TH, Cunningham CW. Independent contrasts succeed where ancestor reconstruction fails in a known bacteriophage phylogeny. Evolution. 2000;54:397–405. doi: 10.1111/j.0014-3820.2000.tb00042.x. [DOI] [PubMed] [Google Scholar]
- O'Kelly JC. Flagellar apparatus architecture and the phylogeny of ‘green’ algae: chlorophytes, euglenoids, glaucophytes. In: Menzel D, editor. The cytoskeleton of the algae. Boca Raton: CRC Press; 1992. pp. 315–345. [Google Scholar]
- O'Kelly JC, Floyd GL. Correlations among patterns of sporangial structure and development, life histories and ultrastructural features in the Ulvophyceae. In: Irvine DEG, John DM, editors. Systematics of the Green Algae. London: Academic Press; 1984. pp. 121–156. [Google Scholar]
- O'Kelly JC, Bellows WK, Wysor B. Phylogenetic position of Bolbocoleon piliferum (Ulvophyceae, Chlorophyta): evidence from reproduction, zoospore and gamete ultrastructure, and small subunit rRNA gene sequences. Journal of Phycology. 2004;40:209–222. [Google Scholar]
- O'Neal SW, Lembi CA. Physiological changes during germination of Pithophora oedogonia (Chlorophyceae) akinetes. Journal of Phycology. 1983;19:193–199. [Google Scholar]
- Olsen JL, Stam WT, Bot PVM, van den Hoek C. ScDNA–DNA hybridization studies in Pacific and Caribbean isolates of Dictyosphaeria cavernosa (Chlorophyta) indicate a long divergence. Helgoländer Meeresuntersuchungen. 1987;41:377–383. [Google Scholar]
- Olsen-Stojkovich JL, West JA, Lowenstein JM. Phylogenetics and biogeography in the Cladophorales complex (Chlorophyta): some insights from immunological distance data. Botanica Marina. 1986;29:239–249. [Google Scholar]
- Palmer JD, Soltis DE, Chase MW. The plant tree of life: an overview and some points of view. American Journal of Botany. 2004;91:1437–1445. doi: 10.3732/ajb.91.10.1437. [DOI] [PubMed] [Google Scholar]
- Peters AF, Marie D, Scornet D, Kloareg B, Cock JM. Proposal of Ectocarpus siliculosus (Ectocarpalaes, Phaeophyceae) as a model organism for brown algal genetics and genomics. Journal of Phycology. 2004;40:1079–1088. [Google Scholar]
- Prescott GW, Croasdale HT, Vinyard WC. Desmidiales. I. Saccodermae, Mesotaeniaceae. North American flora series. New York: New York Botanical Garden; 1972. II (6)Bronx: 84. [Google Scholar]
- Prescott GW, Croasdale HT, Vinyard WC. A synopsis of North American Desmids. II. Desmidiaceae: Placodermae, Sec. I. Lincoln, NE: University of Nebraska Press; 1977. p. 275. [Google Scholar]
- Prescott GW, Croasdale HT, Vinyard WC, Bicudo CE de M. A synopsis of North American Desmids. II. Desmidiaceae: Placodermae, Sec. 5. Lincoln, NE: University of Nebraska; 1981. p. 117. [Google Scholar]
- Portugal J, Waring M. Assignment of DNA binding sites for DAPI and bisbenzimide (Hoeschst 33258). Comparative footprinting study. Biochimica et Biophysica Acta. 1988;949:158–168. doi: 10.1016/0167-4781(88)90079-6. [DOI] [PubMed] [Google Scholar]
- Pryer KM, Schneider H, Zimmer EA, Banks JA. Deciding among green plants for whole genome studies. Trends in Plant Science. 2002;7:550–554. doi: 10.1016/s1360-1385(02)02375-0. [DOI] [PubMed] [Google Scholar]
- Qiu Y-L, Palmer JD. Phylogeny of early land plants: insights from genes and genomes. Trends in Plant Science. 1999;4:26–30. doi: 10.1016/s1360-1385(98)01361-2. [DOI] [PubMed] [Google Scholar]
- Renzaglia KS, Rasch EM, Pike LM. Estimates of nuclear DNA content in bryophyte sperm cells: phylogenetic considerations. American Journal of Botany. 1995;82:18–25. [Google Scholar]
- Riechmann JL, Heard J, Martin G, Reuber L, Jiang C.-Z, Keddle J, et al. Arabidopsis transcription factors: genome-wide comparative analysis among eukaryotes. Science. 2000;290:2105–2110. doi: 10.1126/science.290.5499.2105. [DOI] [PubMed] [Google Scholar]
- Rindi F, McIvor L, Guiry MD. The Prasiolales (Chlorophyta) of Atlantic Europe: an assessment based on morphological, molecular, and ecological data, including the characterization of Rosenvingiella radicans (Kützing) comb. nov. Journal of Phycology. 2004;40:977–997. [Google Scholar]
- Rogers CE, Mattox KR, Stewart KD. The zoospore of Chlorokybus atmophyticus, a charophyte with sarcinoid growth habit. American Journal of Botany. 1980;67:774–783. [Google Scholar]
- Sakayama H, Miyaji K, Nagumo T, Kato M, Hara Y, Nozaki H. Taxonomic reexamination of the 17 species of Nitella subgenus Tieffallenia (Charales, Charophyceae) based on internal morphology of the oospore wall and multiple DNA marker sequences. Journal of Phycology. 2005;41:195–211. [Google Scholar]
- Sánchez-Puerta MV, Leonardi PI. Ciclo de vida, desarrollo y cariología de Klebsormidium nitens (Klebsormidiales, Charophyta) Darwiniana. 2001;39:223–230. [Google Scholar]
- Sarma YSRK. Chromosome numbers in algae. Nucleus. 1982;25:66–108. [Google Scholar]
- Sawitzky H, Gleissberg S, Berger S. Phylogenetic implications of patterns of cap development in selected species of Acetabularia/Polyphysa (Dasycladales, Chlorophyta) Phycologia. 1998;37:478–485. [Google Scholar]
- Schagerl MD, Angeler G, Coleman AW. Infraspecific phylogeny of Pandorina morum (Volvocales, Chlorophyta) inferred from molecular, biochemical and traditional data. European Journal of Phycology. 1999;34:87–93. [Google Scholar]
- Shaw J, Renzaglia K. Phylogeny and diversification of Bryophytes. American Journal of Botany. 2004;91:1557–1581. doi: 10.3732/ajb.91.10.1557. [DOI] [PubMed] [Google Scholar]
- Sherwood AR, Garbary DJ, Sheath RG. Assessing the phylogenetic position of the Prasiolales (Chlorophyta using rbcL and 18S rRNA gene sequence data. Phycologia. 2000;39:139–146. [Google Scholar]
- Shimada S, Hiraoka M, Nabata S, Iima M, Masuda M. Molecular phylogenetic analyses of the Japanese Ulva and Enteromorpha (Ulvales, Ulvophyceae), with special reference to the free-floating Ulva. Phycological Research. 2003;51:99–108. [Google Scholar]
- Shuter BJ, Thomas JE, Taylor WD, Zimmerman AM. Phenotypic correlates of genomic DNA content in unicellular eukaryotes and other cells. American Naturalist. 1983;122:26–44. [Google Scholar]
- Simon N, Barlow RG, Marie D, Partensky F, Vaulot D. Characterization of oceanic photosynthetic picoeukaryotes by flow cytometry. Journal of Phycology. 1994;30:922–935. [Google Scholar]
- Soltis PS, Soltis DE, Wolf PG, Nickrent DL, Chaw S-M, Chapman RL. The phylogeny of land plants inferred from 18S rDNA sequences: pushing the limits of rDNA signal? Molecular Biology and Evolution. 1999;16:1774–1784. doi: 10.1093/oxfordjournals.molbev.a026089. [DOI] [PubMed] [Google Scholar]
- Sparrow AH, Nauman AF. Evolutionary changes in genome and chromosome sizes and in DNA content in the grasses. Brookhaven Symposium in Biology. 1973;25:367–389. [Google Scholar]
- Spring H, Grierson D, Hemleben V, Stohr M, Krohne G, Stadler J, Franke W. DNA contents and numbers of nuclei and pre rRNA-genes in nuclei of gametes and vegetative cells of Acetabularia mediterranea. Experimental Cell Research. 1978;114:203–215. doi: 10.1016/0014-4827(78)90054-x. [DOI] [PubMed] [Google Scholar]
- Steinkötter J, Bhattacharya D, Semmelroth I, Bibeau C, Melkonian M. Prasinophytes form independent lineages within the Chlorophyta: evidence from ribosomal RNA sequence comparisons. Journal of Phycology. 1994;30:340–345. [Google Scholar]
- Sueoka N. Variation and heterogeneity of base composition of deoxyribonucleic acids: a compilation of old and new data. Journal of Molecular Biology. 1961;3:31–40. [Google Scholar]
- Sussmann AV, Mable BK, DeWreede RE, Berbee ML. Identification of green algal endophytes as the alternate phase of Acrosiphonia (Codiales, Chlorophyta) using ITS1 and ITS2 ribosomal DNA sequence data. Journal of Phycology. 1999;35:607–614. [Google Scholar]
- Surek B, Beemelmanns U, Melkonian M, Bhattacharya D. Ribosomal RNA sequence comparisons demonstrate an evolutionary relationship between Zygnematales and charophytes. Plant Systematics and Evolution. 1994;191:171–181. [Google Scholar]
- Sym SD, Pienaar RN. The Class Prasinophyceae. In: Round FE, Chapman DJ, editors. Progress in phycological research. vol. 9. Bristol: Biopress Ltd; 1993. pp. 281–376. [Google Scholar]
- Tan IH, Blomster J, Hansen G, Leskinen E, Maggs CA, Mann DG, et al. Molecular phylogenetic evidence for a reversible morphogenetic switch controlling the gross morphology of two common genera of green seaweeds, Ulva and Enteromorpha. Molecular Biology and Evolution. 1999;16:1011–1018. doi: 10.1093/oxfordjournals.molbev.a026190. [DOI] [PubMed] [Google Scholar]
- Taylor WR. Marine algae of the eastern tropical and subtropical coasts of the Americas. Ann Arbor: University of Michigan Press; 1960. [Google Scholar]
- Turmel M, Lemieux C, Burger G, Lang BF, Otis C, Plante I, Gray MW. The complete mitochondrial DNA sequences of Nephoroselmis olivacea and Pedinomonas minor: two radically different evolutionary patterns within green algae. Plant Cell. 1999;11:1717–1730. doi: 10.1105/tpc.11.9.1717. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Turmel M, Otis C, Lemieux C. The complete mitochondrial DNA sequence of Mesostigma viride identifies this green alga as the earliest green plant divergence and predicts a highly compact mitochondrial genome in the ancestor of all green plants. Molecular Biology and Evolution. 2002a;19:24–38. doi: 10.1093/oxfordjournals.molbev.a003979. [DOI] [PubMed] [Google Scholar]
- Turmel M, Otis C, Lemieux C. The chlorplast and mitochondrial genome sequences of the charophyte Chaetosphaeridium globosum: insights into the timing of the events that restructured organelle DNAs within the green algal lineage that led to land plants. Proceedings of the National Academy of Sciences of the USA. 2002b;99:11275–11280. doi: 10.1073/pnas.162203299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Turmel M, Ehara M, Otis C, Lemieux C. Phylogenetic relationships among streptophytes as inferred from chloroplast small and large subunit rRNA gene sequences. Journal of Phycology. 2002c;38:364–375. [Google Scholar]
- Ueno R, Hanagata N, Urano N, Suzuki M. Molecular phylogeny and phenotypic variation in the heterotrophic green algal genus Prototheca (Trebouxiophyceae, Chlorophyta) Journal of Phycology. 2005;41:1268–1280. [Google Scholar]
- Van den Hoek C, Stam WT, Olsen JL. The emergence of a new chlorophytan system, and dr. Kornmann's contribution thereto. Helgoländer Meeresuntersuchungen. 1988;42:339–383. [Google Scholar]
- Veldhuis MJW, Cucci TL, Sieracki ME. Cellular DNA content of marine phytoplankton using two new fluorochromes: taxonomic and ecological implications. Journal of Phycology. 1997;33:527–541. [Google Scholar]
- Verma BN. Cytological studies in four species of Pithophora Wittr. Cytologia. 1979;44:29–38. [Google Scholar]
- Voglmayr H. Nuclear DNA amounts in mosses (Musci) Annals of Botany. 2000;85:531–546. [Google Scholar]
- Vroom PS, Smith CM, Keeley SC. Cladistics of the Bryopsidales: a preliminary analysis. Journal of Phycology. 1998;34:351–360. [Google Scholar]
- Waaland JR, Stiller JW, Cheney DP. Macroalgal candidates for genomics. Journal of Phycology. 2004;40:26–33. [Google Scholar]
- Wang W, Tanurdzic M, Luo M, Sisneros N, Kim HR, Weng J-K, et al. Construction of a bacterial artificial chromosome library from the spikemoss Selaginella moellendorffii: a new resource for plant comparative genomics. BioMed Central Plant Biology. 2005;5:10. doi: 10.1186/1471-2229-5-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Watanabe S, Kuroda N, Maiwa F. Phylogenetic status of Helicodictyon planctonicum and Desmochloris halophila gen. et comb. nov. and the definition of the class Ulvophyceae (Chlorophyta) Phycologia. 2001;40:421–434. [Google Scholar]
- Wik-Sjöstedt A. Cytogenetic investigations in Cladophora. Hereditas. 1970;66:233–262. [Google Scholar]
- Wilhelm C, Elsenbeis G, Wild A, Zahn R. Nanochlorum eucaryotum: a very reduced coccoid species of marine Chlorophyceae. Zeitschrift für Naturforschung. 1982;37:107–114. [Google Scholar]
- Woolcott GW, Knöller K, King RJ. Phylogeny of the Bryopsidaceae (Bryopsidales, Chlorophyta): cladistic analyses of morphological and molecular data. Phycologia. 2000;39:471–481. [Google Scholar]
- Yamamoto M, Nozaki H, Kawano S. Evolutionary relationships among multiple modes of cell division in the genus Nannochloris (Chlorophyta) revealed by genome size, actin gene multiplicity, and phylogeny. Journal of Phycology. 2001;37:106–120. [Google Scholar]
- Zechman FW. Phylogeny of the Dasycladales (Chlorophyta, Ulvophyceae) based on analyses of RuBisCO large subunit (rbcL) gene sequences. Journal of Phycology. 2003;39:819–827. [Google Scholar]
- Zechman FW, Theriot FC, Zimmer EA, Chapman RL. Phylogeny of the Ulvophyceae (Chlorophyta): cladistic analysis of nuclear-encoded rRNA sequence data. Journal of Phycology. 1990;26:700–710. [Google Scholar]
- Zignone A, Borra M, Brunet C, Forlani C, Kooistra WHCF, Procaccini G. Phylogenetic position of Crustomastix stigmatica sp. nov. and Dolichomastix tenuilepis in relation to the Mamiellales (Prasinophyceae, Chlorophyta) Journal of Phycology. 2002;38:1024–1039. [Google Scholar]
KEY TO REFERENCES
- 1.Abbas A, Godward M. Cytology in relation to taxonomy in Chaetophorales. Journal of the Linnean Society (Botany) 1964;58:499–507. [Google Scholar]
- 2.Arai S, Takahashi H, Takano H, Sakai A, Kawano S. Isolation, characterization, and chromosome mapping of an actin gene from the primitive green alga, Nannochloris bacillaris (Chlorophyceae) Journal of Phycology. 1998;34:477–485. [Google Scholar]
- 3.Buffaloe NP. A comparative cytological study of four species of Chlamydomonas. Bulletin of the Torrey Botanical Club. 1958;85:157–178. [Google Scholar]
- 4.Cattolico RA, Gibbs SP. Rapid filter method for the microfluorometric analysis of DNA. Analytical Biochemistry. 1975;69:572–582. doi: 10.1016/0003-2697(75)90162-1. [DOI] [PubMed] [Google Scholar]
- 5.Cave MS, Pocock MA. Karyological studies in Volvocaceae. American Journal of Botany. 1951;38:800–811. [Google Scholar]
- 6.Charles D. Isolation and characterization of DNA from unicellular algae. Plant Science Letters. 1977;8:35–44. [Google Scholar]
- 7.Chowdary Y. On the cytology and systematic position of Physolinum monilia Printz. Nucleus. 1963;6:44–48. [Google Scholar]
- 8.Cole K, Akintobi S. The life cycle of Prasiola meridionalis Setchell and Gardner. Canadian Journal of Botany. 1963;4:661–668. [Google Scholar]
- 9.Courties C, Perasso R, Chrétiennot-Dinet J-J, Gouy M, Guillou L, Troussellier M. Phylogenetic analysis and genome size of Ostreococcus tauri (Chlorophyta, Presinophyceae) Journal of Phycology. 1998;34:844–849. [Google Scholar]
- 10.Derelle E, Ferraz C, Lagoda P, Eychenie S, Cooke R, Regad F, et al. DNA libraries for sequencing the genome of Ostreococcus tauri (Chlorophyta, Prasinophyceae): the smallest free-living eukaryotic cell. Journal of Phycology. 2002;38:1150–1156. [Google Scholar]
- 11.Dörr R, Huss VAR. Characterization of nuclear DNA in 12 species of Chlorella (Chlorococcales, Chlorophyta) by DNA reassociation. BioSystems. 1990;24:145–155. doi: 10.1016/0303-2647(90)90007-n. [DOI] [PubMed] [Google Scholar]
- 12.Higashiyama T, Yamada T. Electrophoretic karyotyping and chromosomal gene mapping of Chlorella. Nucleic Acids Research. 1991;19:6191–6195. doi: 10.1093/nar/19.22.6191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Holm-Hansen O. Algae: amounts of DNA and organic carbon in single cells. Science. 1969;163:87–88. doi: 10.1126/science.163.3862.87. [DOI] [PubMed] [Google Scholar]
- 14.Jose G, Chowdary Y. Karyological studies on Cephaleuros Kunze. Acta Botanica Indica. 1977;5:114–122. [Google Scholar]
- 15.Kagami Y, Fujishita M, Matsuyama-Serisawa K, Yamamoto M, Kuwano K, Saga N, Kawano S. DNA content of Ulva compressa (Ulvales, Chlorophyta) nuclei determined with laser scanning cytometry. Phycological Research. 2005;53:77–83. [Google Scholar]
- 16.Kapraun DF. Karyology of marine green algae. Phycologia. 1993;32:1–21. [Google Scholar]
- 17.Kapraun DF, Gargiulo GM, Tripodi G. Nuclear DNA and karyotype variatrion in species of Codium (Codiales, Chlorophyta) from the North Atlantic. Phycologia. 1988;27:273–282. [Google Scholar]
- 18.King GC. The cytology of the desmids: the chromosomes. New Phytologist. 1960;59:65–72. [Google Scholar]
- 19.Kunachowicz A, Luchniak P, Olszewska MJ, Sakowicz T. Comparative karyology, DNA methylation and restriction pattern analysis of male and female plants of the dioecious alga Chara tomentosa (Charophyceae) European Journal of Phycology. 2001;36:29–34. [Google Scholar]
- 20.Lopez-Bautista JM, Kapraun DF, Waters DA, Chapman RL. Nuclear DNA quantification and the life cycle in Cephaleuros parasiticus (Trentepohliales, Chlorophyta) American Journal of Botany. 2000;87(6, supplement):248. [Google Scholar]
- 21.Sánchez Puerta MV, Leonardi PI. Ciclo de vida, desarrollo y cariología de Klebsormidium nitens (Klebsormidiales, Charophyta) Darwiniana. 2001;39:223–230. [Google Scholar]
- 22.Sarma YSRK. Chromosome number in algae. Nucleus. 1982;25:66–108. [Google Scholar]
- 23.Simon N, Barlow RG, Marie D, Partensky F, Vaulot D. Characterization of oceanic photosynthetic picoeukaryotes by flow cytometry. Journal of Phycology. 1994;30:922–935. [Google Scholar]
- 24.Suematu S. The somatic nuclear division in Trentepohlia aurea, the aerial alga. Bulletin Liberal Art College Wakayama University of Natural Sciences. 1960;10:111. [Google Scholar]
- 25.Tautvydas KJ. Evidence for chromosome endoreduplication in Eudorina californica, a colonial alga. Differentiation. 1976;5:35–42. doi: 10.1111/j.1432-0436.1976.tb00889.x. [DOI] [PubMed] [Google Scholar]
- 26.Veldhuis MJW, Cucci TL, Sieracki ME. Cellular DNA content of marine phytoplankton using two new fluorochromes: taxonomic and ecological implications. Journal of Phycology. 1997;33:527–541. [Google Scholar]
- 27.Wilhelm C, Elsenbeis G, Wild A, Zahn R. Nanochlorum eucaryotum: a very reduced coccoid species of marine Chlorophyceae. Zeitschrift für Naturforschung. 1982;37:107–114. [Google Scholar]
- 28.Yamamoto M, Nozaki H, Kawano S. Evolutionary relationships among multiple modes of cell division in the genus Nannochloris (Chlorophyta) revealed by genome size, actin gene multiplicity, and phylogeny. Journal of Phycology. 2001;37:106–120. [Google Scholar]