Abstract
Aggregate results from Genome-Wide Association Studies (GWAS)1-3, such as genotype frequencies for cases and controls were available until recently on public websites4-5 because they were thought to reveal negligible information concerning an individual’s participation in a study. Homer et al.6 suggested a method for forensic detection of an individual’s contribution to an admixed DNA sample could be applied to aggregate GWAS data. Using a likelihood-based statistical framework, we develop an improved statistic that uses genotype frequencies and an individual’s genotypes to infer whether the individual or a close relative participated in the GWAS and, if so, the participant’s phenotype status. Our statistic compares the logarithm of genotype frequencies, in contrast to that of Homer et al.6, which is based on differences in SNP probe intensity or allele frequencies. We derive the theoretical power of the test statistics and explore the empirical performance in scenarios with a varying numbers of randomly chosen or top-associated SNPs.
A recent report by Homer et al.6 demonstrated a method to determine whether a given person contributed a trace amount (<0.1%) of DNA to a pool of DNA from 200 individuals based on allelic probe intensities from the same microarray genotyping technology commonly employed in GWAS. Three sets of probe data are required for Homer’s test statistic, denoted here as Tallele: 1) from a pool to which the individual may or may not have contributed DNA; 2) from a reference pool of DNA sampled from members of the same genetic population; and 3) from DNA obtained from a single individual to be tested for membership in the first pool. Their primary applications were directed toward specific forensic challenges, such as determining whether a DNA sample contributed to a biospecimen of mixed origin. They speculated that their method could be applied to GWAS allele frequency data.
We explored that suggestion and have extended the approach of Homer et al.6 to propose an improved test statistic Tgeno to detect whether an individual contributed genotypes to a GWAS, and, if so, determine their case/control phenotype. Conceptually, Tgeno substitutes frequencies from two groups (e.g., cases and controls) and genotype states of the individual to be tested for pools of admixed DNA and measures of allelic probe intensities in the method of Homer et al.6. Tgeno is a novel likelihood ratio statistic that can detect whether an individual contributed DNA to neither, one, or both groups given only genotypes for the individual to be tested and genotype frequencies from each group. Our statistic is similar to Tallele except that Tgeno contrasts the logarithm of frequencies of the each observed genotype of the tested individual (as opposed to allelic probe intensity or allele frequency in Tallele) between two populations.
The predictive power of Tgeno was derived approximately (see Methods), and evaluated by simulation under a range of scenarios consistent with the size and scope of current GWAS designs. In each scenario, we assumed two independent groups of individuals, denoted “test” and “reference”, drawn from a homogeneous population with genotypes from independent biallelic single-nucleotide polymorphism (SNP) markers in Hardy-Weinberg equilibrium with fixed minor allele frequency (MAF). We varied the size of each group, the number and MAF of independent loci, and genotype error rate. Power was computed by simulation, randomly sampling test and reference groups as well as individuals to be tested and comparing Tgeno to its theoretical null distribution using a two-sided test that assumes Z could be a member of either group.
Table 1 shows simulation results of the sensitivity and specificity of Tgeno for significance levels ranging from 0.05 to 10−6 for scenarios labeled (a)-(e). We observed that the power to detect membership in either group increased as the test group size decreased due to the larger contribution of the individual relative to the size of the test group; power also increased as the reference group size increased due to the improved knowledge of underlying population frequencies. Scenario (c) demonstrated that substantial genotyping error was required to attenuate the power of the method. Power was essentially unaffected with 1% genotyping error and only began to appreciably diminish with rates approaching 10%. Scenario (d) demonstrated that the power to detect membership does not depend on MAF. Scenario (e) emphasized the role of the size of the population that did not contain the individual being tested. Power was dramatically reduced when only 60 individuals were available in the reference group, reaching only 20% at the 0.05 significance level and no power at the 10−6 significance level; power increases to 100% at the 0.05 significance level and to 96% at the 10−6 significance level with 5,000 reference individuals. In each scenario, the predicted theoretical and empirical power matched very closely.
Table 1.
Theoretical power of Tgeno to detect an individual in the Test group.
| a. 1,000 samples/group, number of loci varied |
Power of Tgeno at Significance Level | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Test group | Ref group | Ind. SNPs | Error rate | MAF | 0.05 | 0.01 | 0.001 | 10−4 | 10−5 | 10−6 |
| 1,000 | 1,000 | 5,000 | 0.00 | 0.25 | 0.62 | 0.38 | 0.15 | 0.05 | 0.02 | 0.00 |
| 1,000 | 1,000 | 10,000 | 0.00 | 0.25 | 0.89 | 0.73 | 0.45 | 0.24 | 0.11 | 0.04 |
| 1,000 | 1,000 | 25,000 | 0.00 | 0.25 | 1.00 | 0.99 | 0.96 | 0.86 | 0.72 | 0.54 |
| 1,000 | 1,000 | 50,000 | 0.00 | 0.25 | 1.00 | 1.00 | 1.00 | 1.00 | 1.00 | 0.99 |
| 1,000 | 1,000 | 75,000 | 0.00 | 0.25 | 1.00 | 1.00 | 1.00 | 1.00 | 1.00 | 1.00 |
| b. 5,000 samples/group, number of loci varied |
Power of Tgeno at Significance Level | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Test group | Ref group | Ind. SNPs | Error rate | MAF | 0.05 | 0.01 | 0.001 | 10−4 | 10−5 | 10−6 |
| 5,000 | 5,000 | 10,000 | 0.00 | 0.25 | 0.23 | 0.09 | 0.02 | 0.00 | 0.00 | 0.00 |
| 5,000 | 5,000 | 25,000 | 0.00 | 0.25 | 0.55 | 0.31 | 0.12 | 0.04 | 0.01 | 0.00 |
| 5,000 | 5,000 | 50,000 | 0.00 | 0.25 | 0.92 | 0.78 | 0.53 | 0.30 | 0.15 | 0.07 |
| 5,000 | 5,000 | 75,000 | 0.00 | 0.25 | 0.98 | 0.91 | 0.74 | 0.52 | 0.32 | 0.17 |
| 5,000 | 5,000 | 100,000 | 0.00 | 0.25 | 1.00 | 0.98 | 0.90 | 0.75 | 0.56 | 0.38 |
| 5,000 | 5,000 | 150,000 | 0.00 | 0.25 | 1.00 | 1.00 | 0.98 | 0.93 | 0.83 | 0.69 |
| 5,000 | 5,000 | 200,000 | 0.00 | 0.25 | 1.00 | 1.00 | 1.00 | 0.99 | 0.97 | 0.92 |
| c. 1,000 samples/group, genotype error rate varied |
Power of Tgeno at Significance Level | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Test group | Ref group | Ind. SNPs | Error rate | MAF | 0.05 | 0.01 | 0.001 | 10−4 | 10−5 | 10−6 |
| 1,000 | 1,000 | 50,000 | 0.00 | 0.25 | 1.00 | 1.00 | 1.00 | 1.00 | 1.00 | 0.99 |
| 1,000 | 1,000 | 50,000 | 0.01 | 0.25 | 1.00 | 1.00 | 1.00 | 1.00 | 0.99 | 0.98 |
| 1,000 | 1,000 | 50,000 | 0.05 | 0.25 | 1.00 | 1.00 | 1.00 | 1.00 | 0.98 | 0.94 |
| 1,000 | 1,000 | 50,000 | 0.10 | 0.25 | 1.00 | 1.00 | 1.00 | 0.98 | 0.93 | 0.84 |
| 1,000 | 1,000 | 50,000 | 0.25 | 0.25 | 0.99 | 0.95 | 0.81 | 0.61 | 0.40 | 0.24 |
| d. 1,000 samples/group, minor allele frequency varied |
Power of Tgeno at Significance Level | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Test group | Ref group | Ind. SNPs | Error rate | MAF | 0.05 | 0.01 | 0.001 | 10−4 | 10−5 | 10−6 |
| 1,000 | 1,000 | 25,000 | 0.00 | 0.05 | 1.00 | 1.00 | 0.97 | 0.88 | 0.72 | 0.52 |
| 1,000 | 1,000 | 25,000 | 0.00 | 0.10 | 1.00 | 0.99 | 0.96 | 0.87 | 0.73 | 0.55 |
| 1,000 | 1,000 | 25,000 | 0.00 | 0.20 | 1.00 | 0.99 | 0.96 | 0.87 | 0.72 | 0.54 |
| 1,000 | 1,000 | 25,000 | 0.00 | 0.30 | 1.00 | 0.99 | 0.96 | 0.87 | 0.72 | 0.55 |
| 1,000 | 1,000 | 25,000 | 0.00 | 0.40 | 1.00 | 0.99 | 0.96 | 0.87 | 0.72 | 0.54 |
| 1,000 | 1,000 | 25,000 | 0.00 | 0.50 | 1.00 | 0.99 | 0.96 | 0.87 | 0.72 | 0.54 |
| e. 1,000 samples/group, reference group size varied |
Power of Tgeno at Significance Level | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Test group | Ref group | Ind. SNPs | Error rate | MAF | 0.05 | 0.01 | 0.001 | 10−4 | 10−5 | 10−6 |
| 1,000 | 60 | 25,000 | 0.00 | 0.10 | 0.20 | 0.07 | 0.02 | 0.00 | 0.00 | 0.00 |
| 1,000 | 100 | 25,000 | 0.00 | 0.10 | 0.46 | 0.23 | 0.07 | 0.02 | 0.00 | 0.00 |
| 1,000 | 200 | 25,000 | 0.00 | 0.10 | 0.74 | 0.51 | 0.24 | 0.09 | 0.03 | 0.01 |
| 1,000 | 300 | 25,000 | 0.00 | 0.10 | 0.92 | 0.78 | 0.51 | 0.27 | 0.12 | 0.05 |
| 1,000 | 400 | 25,000 | 0.00 | 0.10 | 0.97 | 0.88 | 0.68 | 0.43 | 0.23 | 0.11 |
| 1,000 | 500 | 25,000 | 0.00 | 0.10 | 0.98 | 0.93 | 0.78 | 0.56 | 0.35 | 0.19 |
| 1,000 | 750 | 25,000 | 0.00 | 0.10 | 1.00 | 0.98 | 0.91 | 0.77 | 0.58 | 0.39 |
| 1,000 | 1,000 | 25,000 | 0.00 | 0.10 | 1.00 | 0.99 | 0.96 | 0.87 | 0.73 | 0.55 |
| 1,000 | 2,000 | 25,000 | 0.00 | 0.10 | 1.00 | 1.00 | 0.99 | 0.97 | 0.92 | 0.82 |
| 1,000 | 5,000 | 25,000 | 0.00 | 0.10 | 1.00 | 1.00 | 1.00 | 1.00 | 0.98 | 0.95 |
| 1,000 | 10,000 | 25,000 | 0.00 | 0.10 | 1.00 | 1.00 | 1.00 | 1.00 | 0.99 | 0.97 |
Scenarios examined: (a) Hypothetical GWAS with 1,000 cases (test group) and 1,000 controls (reference group), no genotyping error, and 5,000 to 200,000 independent SNPs with fixed MAF of 25%. The upper bound on the number of SNPs was chosen based on estimates (unpublished) that the Illumina HumanHap550 assay provides information equivalent to ~200,000-300,000 independent SNPs in populations of European descent. A MAF of 25% was chosen based on a survey of several fixed-content assays which were found to have average MAFs ranging from 20% to 25% in populations of European descent. (b) As for (a) with 5,000 cases and 5,000 controls. (c) As for (a) with genotype discordance rates from 0% to 25% and 50,000 independent loci. (d) As for (a) with varying MAF from 5% to 50% for 25,000 independent loci. (e) For varying sizes of the reference group from 60 (the size of the HapMap CEU founder population) to 10,000 (near perfect estimation of the genotype frequencies) for a fixed MAF of 10%.
In Table 2, we explored the power to detect membership of a close relative of an individual in the test group using 200,000 independent SNPs simulated under the same assumptions as in Table 1. In (f) and (g) the genotype discordance rate due to the familial relationship was a function of MAF (see Methods) and reflects no additional discordance due to assay-related genotyping errors. Power to detect membership of relatives decreased as MAF increased (and consequently the genotype discordance rate increased), but remained high for MAFs lower than 30%. In populations of European descent, the average MAF for most whole-genome SNP panels is between 20-25%.
Table 2.
Theoretical power of Tgeno to detect a relative of an individual in the Test group.
| f. 1,000 samples/group, detection of a parent/offspring |
Power of Tgeno at Significance Level | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Test group | Ref group | Ind. SNPs | Parent/ Offspring Discordance |
MAF | 0.05 | 0.01 | 0.001 | 10−4 | 10−5 | 10−6 |
| 1,000 | 1,000 | 200,000 | 0.10 | 0.05 | 1.00 | 1.00 | 1.00 | 1.00 | 1.00 | 1.00 |
| 1,000 | 1,000 | 200,000 | 0.18 | 0.10 | 1.00 | 1.00 | 1.00 | 1.00 | 1.00 | 1.00 |
| 1,000 | 1,000 | 200,000 | 0.32 | 0.20 | 1.00 | 1.00 | 1.00 | 1.00 | 1.00 | 0.99 |
| 1,000 | 1,000 | 200,000 | 0.42 | 0.30 | 1.00 | 1.00 | 0.97 | 0.91 | 0.79 | 0.63 |
| 1,000 | 1,000 | 200,000 | 0.48 | 0.40 | 0.98 | 0.92 | 0.77 | 0.55 | 0.35 | 0.20 |
| 1,000 | 1,000 | 200,000 | 0.50 | 0.50 | 0.93 | 0.82 | 0.58 | 0.35 | 0.19 | 0.09 |
| g. 1,000 samples/group, detection of a sibling |
Power of Tgeno at Significance Level | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Test group | Ref group | Ind. SNPs | Parent/ Offspring Discordance |
MAF | 0.05 | 0.01 | 0.001 | 10−4 | 10−5 | 10−6 |
| 1,000 | 1,000 | 200,000 | 0.092 | 0.05 | 1.00 | 1.00 | 1.00 | 1.00 | 1.00 | 1.00 |
| 1,000 | 1,000 | 200,000 | 0.168 | 0.10 | 1.00 | 1.00 | 1.00 | 1.00 | 1.00 | 1.00 |
| 1,000 | 1,000 | 200,000 | 0.282 | 0.20 | 1.00 | 1.00 | 1.00 | 1.00 | 1.00 | 1.00 |
| 1,000 | 1,000 | 200,000 | 0.354 | 0.30 | 1.00 | 1.00 | 1.00 | 1.00 | 0.99 | 0.96 |
| 1,000 | 1,000 | 200,000 | 0.394 | 0.40 | 1.00 | 1.00 | 0.99 | 0.97 | 0.92 | 0.83 |
| 1,000 | 1,000 | 200,000 | 0.406 | 0.50 | 1.00 | 1.00 | 0.99 | 0.95 | 0.87 | 0.74 |
Scenarios examined: (f) Hypothetical GWAS data with 1,000 cases (test group) and 1,000 controls (reference group) where the individual tested is the parent or offspring of a case group, 200,000 independent SNPs, and MAF from 5% to 50%. (g) As for Table 1 (a) except the individual tested is a sibling of a single member of the test group, 200,000 independent loci, and MAF from 5% to 50%.
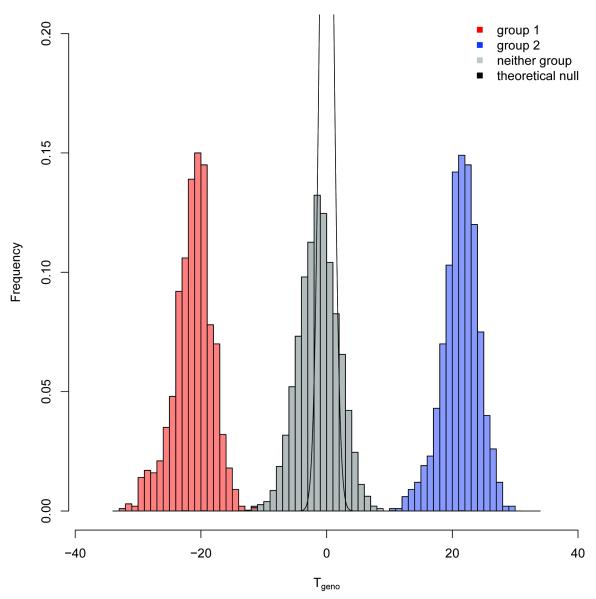
We applied our method to data combined from several GWAS conducted at the US National Institutes of Health using genotype data from individuals of European descent. The dataset contained 6,733 individuals affected with a disease (cases) and 6,871 unaffected individuals (controls) and genotype data for 557,005 SNPs from the Illumina HumanHap550 assay. Figure 1 shows a histogram of Tgeno for a hypothetical GWAS with 1,000 cases (in red), 1,000 controls (in blue) and 11,604 subjects not in the virtual study (in gray), and with the theoretical null density curve shown in black. The groups were well-differentiated, indicating that Tgeno is capable of accurately inferring membership and case/control phenotype. Both the mean and variance of the empirical null distribution were shifted from those of the theoretical standard normal distribution. These differences arise from use of fixed test and reference samples, while the theoretical null distribution holds over repeated random samples of the two groups. In addition, LD between SNPs is expected to inflate the variance of the test-statistics relative to our derivation that assumes locus independence. Thus, making inferences about Tgeno when applied to particular GWAS data requires suitable calibration of the null distribution. Figure 2 shows histograms comparing Tgeno and Tallele for a fixed subset of 1,000 cases and 1,000 controls for varying numbers of “top associated” SNPs.
Figure 1. Histogram of Tgeno for a GWAS with 1,000 cases and controls.
The figure presents data using 1,000 cases (group 1 in red), 1,000 controls (group 2 in blue) and 1,000 subjects not in the study based on genotypes from Illumina HumanHap550 assay. The theoretical null density curve is shown in black.
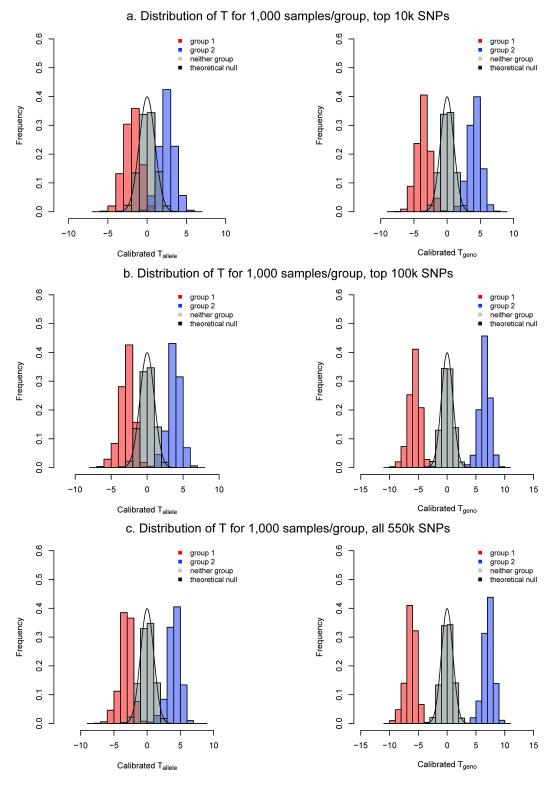
Figure 2. Histograms of calibrated Tgeno and Homer’s Tallele with 1,000 cases and controls and varying numbers of SNPs.
The figure presents theoretical null density curves (black) for a GWAS with 1,000 cases (group 1 in red), 1,000 controls (group 2 in blue) and 12,000 subjects not in the study (in gray) using genotypes for (a) 10,000, (b) 100,000, and (c) 550,000 top associated SNPs from the Illumina HumanHap550 assay. Statistics were calibrated so that the null distribution was centered at zero with unit variance.
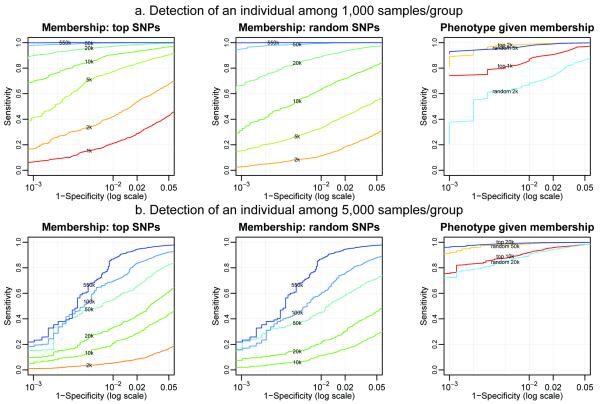
Using these data, GWAS scenarios were explored by selecting subsets of cases and controls, estimating genotype frequencies of each group, and fitting logistic genotype-phenotype association models. In each scenario, all 13,604 individuals were tested for membership conditional on a fixed set of cases and controls chosen. We also attempted to infer the phenotype of the cases and controls selected in each scenario given the knowledge that they participated in the study. Individuals not selected as cases or controls for a given scenario were used to empirically estimate the null distribution. Figure 3 contains Receiver Operating Characteristic (ROC) curves showing empirical sensitivity and specificity for classifying individuals as participants and the determination of their phenotype given knowledge of participation. GWAS scenarios with a fixed subset of 1,000 and 5,000 cases and an equal number of controls are shown for varying numbers of randomly chosen or top associated SNPs. These ROC curves focus on high values of specificity with 1-specificity in the range of 0.05 to 10−6 on a logarithmic scale. Supplementary Figure 1 shows ROC curves for additional GWAS scenarios with 1,000-5,000 cases and an equal number of controls. Supplementary Figure 2 is analogous to Supplementary Figure 1 except showing ROC curves for the non-log scale for the full range of specificity. Supplementary Figure 3 is analogous to Supplementary Figure 1, except with ROC curves for Tallele.
Figure 3. Sensitivity and specificity of Tgeno applied to GWAS data.
Log-scale Receiver Operating Characteristic (ROC) curves of Tgeno with Illumina HumanHap550 data from GWAS scenarios with 1000/1000 and 5000/5000 cases and controls of European descent.
Figure 3(a) shows a hypothetical GWAS with 1,000 cases and 1,000 controls. If the top 1,000 associated SNPs are published, the power to detect whether an individual is included in the study is approximately 43%, when the chance of a false positive, incorrectly concluding the individual is in the study, is 1 in 20. Power approaches zero when the false-positive rate is less than 1 in 1,000. If frequencies of 5,000 of the top associated SNPs are published, approximately 90% power is attained at a false-positive rate of 1 in 20 and 41% power when the false-positive rate is 1 in 1,000. When 50,000 of the top associated SNP frequencies are published, near perfect power is attained at a false-positive rate of 1 in 20 and approximately 98% power when the false-positive rate is 1 in 1,000.
We have shown the robustness of a novel test statistic, Tgeno, an extension of the Tallele approach by Homer et al.6 adapted for use on genotype frequency data rather than SNP probe intensity or allele frequency data. We found that Tgeno was more powerful than Tallele since the Tgeno statistic conditions on known genotype states and is a likelihood-ratio statistic, and has optimality properties for both hypothesis testing and classification (see Figure 2 and Supplementary Figure 2).
This method makes a critical assumption that individuals in both groups are sampled from the same population. Unless population structure or substructure is identical in both groups, incorrect inferences are likely to be made. Essentially, Tgeno and Tallele test whether an individual’s genotypes are more consistent with one group versus another (see Methods for a formal description). These statistics are valid for determining membership only when the differences between groups are due to sampling variation. Otherwise, the relatively small bias due to membership of an individual quickly becomes swamped by the systematic differences in the underlying populations. Although this characteristic may be problematic in forensic applications in which test groups are often of unknown and potentially varied population origin, GWAS case and control groups are usually drawn from genetically comparable populations.
In addition to the number and selection method of SNPs, the sensitivity of our method, that is the power to detect true membership, is affected by several parameters: the number of subjects in each group, the number of SNPs tested, genotyping error, and genetic relatedness. As the size of the group containing the individual decreases, it becomes easier to detect the contribution of a single individual; as the size of the group without the individual increases, a more precise knowledge of the background population frequencies improves power. Because this method does not rely on patterns of rare or unique variation, power is not greatly affected by MAF for relatively common SNPs (MAF>5%) (see Methods for details). Although substantial genotype error can decrease power, modest error rates (<5%) have little effect (current commercial genotyping platforms typically have error rates <0.1%). One can detect membership of siblings, parents and offspring, whereas the presence of more distant relatives would be more challenging to detect. For some applications, the ability to detect relatives is not desirable and could decrease specificity.
The conditional power to detect membership of an individual given a test and a reference sample is affected by sampling variation in the estimates of genotype frequencies between the two groups. The genetic variants need not have different underlying frequencies in the two groups, as might be a consequence of association with disease; only a non-zero difference in estimates is required. SNPs with low association p-values or, equivalently, large differences in estimated genotype frequencies between cases and controls, result in higher power because the magnitude of the ratio of frequencies between the two groups is larger. Two-sided association p-values are inversely correlated with the magnitude of the difference in genotype frequencies, but do not indicate the direction of the differences or provide additional information useful for detection of membership when using Tgeno.
This method is particularly well-suited to case-control GWAS designs since subjects are partitioned into disjoint sets that provide both groups of frequency data required by this method. Publication of genotype frequencies separately for cases and controls also increases power to detect membership, since each group is typically a large fraction of the size of the overall study. GWAS are also often composed of a combination of multiple study populations and results may be published for each subset, increasing the power of this method, particularly for rare diseases that necessitate the collaboration of multiple studies. Similarly, re-use of sets of cases and controls among several GWAS can allow the calculation of frequencies for smaller groups by removing the contribution of overlapping individuals.
Further work is required to address some issues raised by our analyses. A more thorough understanding of the test statistic is required to account for linkage disequilibrium. Aggregate statistics other than frequencies could reveal membership information. It is notable that our approach could be applied to other types of dense datasets, both genetic and non-genetic. The underlying principle is based on detecting small correlations in categorical or continuous data accumulated over a sufficient number of highly repeatable outcomes.
We anticipate that these findings will aid in understanding the implications of publishing and sharing aggregate genomic data in ways that protect the privacy of research subjects. This method could be used to determine if specific individuals participated in a clinical study and perhaps influence the decision to enroll or continue participating. Our results should be considered as a lower bound for the power to detect membership and phenotype in an aggregate genotype dataset, as more efficient methods may yet exist. In light of these developments, the policies and practices guiding genomic data sharing should continue to evolve in order to promote quality science, minimize duplicative research, and merit the ongoing trust of the research subjects who consent to participate in scientific studies.
METHODS
Let Xi,g and Yi,g denote genotype frequencies genotype g at locus i=1..s for two independent sets X and Y of DNA samples from unrelated individuals of size n and m, respectively. Let Zi denote the genotype of an individual at locus i. We assume all loci are in linkage equilibrium, that samples were drawn from the same population with genotype frequencies fi,j for j=1..gi, the number of genotypes that may be observed at each locus. We wish to test the ratio of frequencies of genotypes Zi in groups X and Y and define a distance metric based on the log transformed ratio,
d is the sum of the differences in the natural logarithm of the frequency of individual Z’s genotypes between the two groups. If individual Z is a member of a group X or Y, then the observed genotype count at each locus for Z’s genotype has a contribution of 1 instead of the population frequency to the frequency statistic, and the resulting frequency so will tend to be slightly higher than that of the overall population by a factor of 1/n or 1/m, respectively. Notice that the distance measure d can also be motivated as a log-likelihood-ratio statistic for classifying individual Z’s genotypes as “randomly” drawn from group X versus group Y.
We assume that genotypes are drawn from the same population with frequencies fi,j, so that Xi,g is distributed as Bin(n, fi,g)/n and Yi,g is distributed as Bin(m, fi,g)/m, where Bin(n,f) denotes the binomial distribution with n trials with success probability f. Throughout the calculations below we assume that mean and variance of log(Xi,g) and log(Yi,g) can be well approximated by first order Taylor’s approximation, given that in large samples we can assume Xi,g and Yi,g are approximately distributed as normal variates.
Under the listed ideal assumptions and in sufficiently large samples, a T statistic can be utilized to test the significance of d:
A two-sided test of Tgeno is appropriate when individual Z’s DNA may be in either group. Otherwise, a one-sided test would be more powerful.
Consider four scenarios (H0 – H3):
H0: Individual Z’s DNA is in neither group. Under this null model, the expected value of d is zero
with variance
H1: Individual Z’s DNA is in X and not in Y. X is composed of n-1 genotypes observed by chance, but Z’s genotype is known to be in X. Thus Xi ~ (Bin(n–1,fzi)+1)/n and the expected value of d given Z is
H2: Individual Z’s DNA is in Y, but not X: Using an analogous argument to H1, the expected value of d given Z is
H3: Individual Z’s DNA is in both X and Y. Then n-1 genotypes from X and m-1 genotypes from Y observed by chance, but Z’s genotype is known to be in both. The expected value of d given Z is
This scenario is particularly relevant when X or Y represents a large population sample or perhaps even the whole population of interest.
Genotyping error
It is useful to consider the effect of a class of genotyping error in these models though we restrict our model to genotyping errors in Z that result in discordant genotype calls between Z and any group that may contain Z. We denote this genotype discordance rate ε. We utilize this error term to model genotype discordance when Z is a close relative of a member of X or Y.
For hypothesis 1, unless an error in genotyping occurred resulting in discordant genotypes for Z with probability ε, in which case Xi ~ Bin(n–1,fzi)/n. Incorporating this non-zero possibility of genotyping error, Xi ~ (Bin(n–1,fzi)+ Bin(1,1,–ε))/n, the expected value of d given Z
The effect of genotyping error for other hypotheses is similar.
Statistical Power
For large s, the test-statistic Tgeno will approximately follow a standard normal distribution under H0 given Z. Moreover, given the conditional null distribution being independent of Z, Tgeno will also follow standard normal distribution unconditionally on Z under H0.
The conditional power of the test-statistics given Z will depend on the “non-centrality” parameter
where the formulae for EH(d|Z) under various hypotheses are shown above. The unconditional non-centrality parameter can be calculated using the double expectation formula and the fact that E((Σi=1..s w(Zi))½) ≈ (Σi=1..s E(w(Zi))+o(√s))½ for any function w, assuming large s. The non-centrality parameter under H1 is
where , the average number of possible genotype categories over all loci. There are three possible genotype categories per locus () when Tgeno is computed from biallelic SNP data with MAF sufficiently high to ensure that all genotypes are observed. Assuming normality of the test-statistic and that the variance of under H1 remains approximately the same as under H0, the unconditional power of a two-sided test of Tgeno (which assumes Z may be a member of X or Y) under H1 at significance level α is
where Φ is the standard normal cumulative distribution function and Φ−1 its inverse. Power may be derived under the other alternative hypotheses in a similar fashion. The power of a one-sided test, one that assumes Z is not a member of Y, substitutes α for α/2 in the right-hand side of previous equation. Use of low frequency SNPs (e.g., MAF<5%) will reduce and, as a consequence, the power of the test.
Modeling Relatives
The genotype discordance rate ε can be used to represent other sources of discordance other than that due to genotyping error. Genotype concordance among a pair of relatives is equivalent to the probability of sharing two alleles identical by state (IBS), which are functions of the relationship type and minor allele frequencies for independent loci in Hardy-Weinberg equilibrium. The probability of not sharing two alleles identical by state for parent offspring pairs is
and for sibling pairs is
where p is the minor allele frequency and q=1-p is the major allele frequency.
Relaxing assumptions
Independence of X and Y: Similar results hold when at least one member of X and one member of Y are the same person or close relatives. The expected values of d under each case are unchanged but the variances are reduced depending on the amount of overlap as the effective sizes of X and Y are reduced.
Independence of loci: The assumption of linkage equilibrium is currently necessary to derive analytical expectations of the moments and power of our test statistic. When applying this method to empirical data, many of the model assumptions needed to precisely determine the null distribution of the test statistic and determine significance by comparison to a theoretical distribution no longer hold. Thus it seems sensible to shift perspective from a hypothesis testing framework to one of classification, which examines the trade-off between sensitivity (true positive rate) and specificity (true negative rate) across a range of cut-off values for the statistic.
When applied to empirical GWAS data, the distribution of Tgeno need not be centered at zero (μ0≠0) and the variance is greater than would be expected (see Figure 1). The lack of centrality is likely due to sampling variation, while the variance is inflated due primarily to correlation among loci from linkage disequilibrium.
Supplementary Material
REFERENCES
- 1.Manolio TA, Brooks LD, Collins FS. A HapMap harvest of insights into the genetics of common disease. J Clin Invest. 2008;118:1590–605. doi: 10.1172/JCI34772. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.McCarthy MI, Abecasis GR, Cardon LR, et al. Genome-wide association studies for complex traits: consensus, uncertainty and challenges. Nat Rev Genet. 2008;9:356–69. doi: 10.1038/nrg2344. [DOI] [PubMed] [Google Scholar]
- 3.Wang WY, Barratt BJ, Clayton DG, Todd JA. Genome-wide association studies: theoretical and practical concerns. Nat Rev Genet. 2005;6:109–18. doi: 10.1038/nrg1522. [DOI] [PubMed] [Google Scholar]
- 4.Mailman MD, Feolo M, Jin Y, et al. The NCBI dbGaP database of genotypes and phenotypes. Nat Genet. 2007;39:1181–6. doi: 10.1038/ng1007-1181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Genome-wide association study of 14,000 cases of seven common diseases and 3,000 shared controls. Nature. 2007;447:661–78. doi: 10.1038/nature05911. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Homer N, Szelinger S, Redman M, et al. Resolving individuals contributing trace amounts of DNA to highly complex mixtures using high-density SNP genotyping microarrays. PLoS Genet. 2008;4:e1000167. doi: 10.1371/journal.pgen.1000167. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.