SUMMARY
NLR genes mediate host immunity to various pathogenic stimuli. However, in vivo evidence for NLR involvement in viral sensing has not been widely investigated and remains controversial. As an ultimate test of the physiologic role of NLRP3 during RNA viral infection, this work explores the in vivo role of NLRP3 inflammasome components during influenza virus infection. Mice lacking Nlrp3, ASC, or Caspase-1, but not Nlrc4, exhibit dramatically increased mortality but reduced immune response following influenza virus exposure. Utilizing analogs of dsRNA (poly(I:C)) and ssRNA (ssRNA40), we demonstrate that NLRP3-mediated response can be activated by RNA species. Mechanistically, NLRP3 inflammasome activation by influenza virus is dependent upon lysosomal maturation and reactive oxygen species. Inhibition of ROS induction eliminated IL-1β production in animals during influenza infection. Together, these data place the NLRP3 inflammasome as an essential component in host defense against influenza infection through the sensing of viral RNA.
Keywords: Cryopyrin, ASC, IPAF, NLR, NALP3, TUCAN, Caspase-1, IL-1β
INTRODUCTON
Innate immune responses to influenza and other pathogens typically involve a highly conserved host-cell signaling mechanism designed to protect and rid the host of harmful microbes. Recognition of conserved microbial structures, known as pathogen-associated molecular patterns (PAMPs), within cells of a myeloid origin is a hallmark feature of adequate host immunity. PAMPs are recognized by pattern recognition receptors or sensors (PRRs) which subsequently initiate signaling cascades resulting in the production of proinflammatory cytokines and type-I interferons. Thus far, three families of PRRs have been identified and shown to be activated in response to viral pathogens, including the RLHs (RIG-I-like helicases), TLRs (toll-like receptors) and NLRs (nucleotide binding domain and leucine-rich-repeat-containing)(Akira et al., 2006; Ting et al., 2008). The individual members of these families can be distinguished by ligand specificity, cellular localization and activation of unique downstream signaling pathways. The strategy of employing multiple families of PRRs affords the host a high degree of functional redundancy and provides multiple mechanisms to optimally respond to a diverse range of pathogens (Kawai and Akira, 2007).
The NLR proteins are emerging as a major route by which the innate immune system responds to microbial pathogens. Substantial evidence suggests that the NLR proteins serve as intracellular mediators of PAMP initiated host-cell signaling, although the exact mechanisms underlying NLR responses to pathogens are not completely understood. One of the fundamental reactions of the innate immune response to viral infection is the processing and release of proinflammatory cytokines, including the regulation and release of IL-1β. It has been demonstrated that the NLR protein, NLRP3, together with its adapter protein, Apoptotic Speck protein containing a CARD (ASC), regulate IL-1β maturation through the formation of a biochemical complex called the inflammasome (Agostini et al., 2004). This inflammasome regulates the activation of caspase-1 and subsequent cleavage of the IL-1β and IL-18 precursor into their functional form, which is then released from the cell. In addition to NLRP3, other NLRs such as NLRC4 and NLRP1 also cause caspase-1 activation and IL-1β production (Mariathasan and Monack, 2007).
Although the function of the NLRP3 inflammasome in mediating responses to bacterial pathogens have been studied by several groups, the in vivo function of this important signaling complex during the course of viral infection is not well understood. In vitro transfection of DNA from viruses, bacteria or mammalian sources into macrophages activates the NLRP3 inflammasome, but this result would suggest a more global response to DNA rather than an antiviral response (Muruve et al., 2008). One report has shown that a synthetic analog of dsRNA, poly(I:C), activates IL-1β through the NLRP3 pathway (Kanneganti et al., 2006). However, another group failed to replicate this finding (Muruve et al., 2008). Thus, the role of NLRP3 and the inflammasome during viral infection remains unresolved. Equally significant, the majority of NLR characterization has been based on in vitro/ex vivo data generated from human cell lines and primary mouse cells. While these studies have provided a wealth of information, the underlying relevance of these proteins in actual pathogenesis and host defense against pathogens in vivo has been much less defined.
To assess the contribution of NLR inflammasomes in viral pathogenesis, we assessed the host response to the influenza A virus. Influenza infection results in a highly contagious respiratory illness leading to significant morbidity and occasionally death. Annual epidemics typically affect 5–15% of the population and are thought to result in 250,000 to 500,000 deaths annually. Of the three types of influenza viruses, influenza A viruses are the most virulent type to humans and are capable of infecting multiple mammalian and avian species. Human influenza A viruses can be further divided into different serotypes based on the antibody response to the viral surface glycoproteins, hemagglutinin (HA) and neuraminidase (NA). The influenza A virus genome is composed of 8 individual strands of ssRNA, which encode a total of 11 different proteins.
In this report, we show that the NLRP3 inflammasome has a profound influence on in vivo host immune response and survival following airway infection with a mouse-adapted influenza A virus. Utilizing in vivo challenges with a synthetic analog of dsRNA, we demonstrate a possible mechanism underlying the NLRP3 associated innate immune response to viruses involves the recognition of viral RNA. Furthermore, we extend the relevancy of these findings to human cells by establishing that ssRNA, dsRNA and influenza A virus-mediated IL-1β release by human monocytes is dependent on the NLRP3 inflammasome and rely on intact lysosomal function, lysosomal enzymes such as cathepsin B, and reactive oxygen species (ROS). This report is the first to demonstrate that the NLRP3 inflammasome is an essential component of the in vivo host immune response to viral infection in a model system that is physiologically relevant to human disease.
RESULTS
Loss of NLR inflammasome activity, but not MyD88, alters mouse survival and inflammation in response to influenza virus infection
To assess the in vivo physiological contribution of NLR and TLR signaling pathways in response to influenza virus infection, we utilized gene deletion mice in an influenza A/PR/8/34 virus infection model. Previous studies utilizing mouse primary macrophages have demonstrated that NLR components are necessary for IL-1β release in response to influenza virus infection in culture (Kanneganti et al., 2006). However, the in vivo physiological relevance of these findings has yet to be explored extensively. To assess the contribution of NLR inflammasomes in mouse survival and inflammation, we assessed mice deficient in either the NLR adaptor protein ASC or caspase-1. Mice lacking these inflammasome components show significantly increased mortality following influenza infection (Figure 1A). As a comparison and as a control, we included the MyD88−/− mice. Previous studies have demonstrated that TLR3, 7, 8 and 9 represent the subset of TLRs which recognize viral nucleic acids and mediate the induction of type-I IFN (Kawai and Akira, 2007; Koyama et al., 2007). TLR7, 8 and 9 are dependent upon the TLR adaptor protein MyD88 for proper signal transduction. However, mouse survival was only moderately decreased for MyD88−/− animals, suggesting that removal of MyD88 and the disruption of the associated TLR7/8/9 signaling cascades only modestly affect mouse survival (Figure 1B).
Figure 1. Characterization of influenza virus A/PR/8/34 pathogenicity and immune response in mice deficient in inflammasome signaling pathways.
Wild type, MyD88−/−, ASC−/−and Caspase-1 (Casp1−/−) deficient mice were challenged intranasally (i.n.) with influenza A/PR/8/34 and survival was monitored 14 dpi. (A) ASC−/− and Casp1−/− mice demonstrated significantly increased mortality compared to the wild type animals (*p<0.05; log rank), while (B) MyD88−/− mice demonstrated modestly increased mortality. Mock-inoculated (n=7); influenza-infected wild type (n=19); MyD88−/− (n=11); ASC−/− (n=12); and Casp1−/− (n=7). (C–F) Lungs were harvested 3 dpi. from wild type, ASC−/−, Myd88−/− and mock-infected mice. Sections through the main bronchiole of the left lobe were stained with H&E. (C) No abnormal lung histopathology was observed in mock-challenged animals. (D) Wild type and (E) MyD88−/− influenza-challenged mice demonstrated increased airway inflammation. (F) ASC−/− mice demonstrated significantly reduced inflammation following influenza challenge. (G) Histological scoring of H&E stained lung sections demonstrated a significant increase in inflammation following influenza challenge in all mice. However, ASC−/− mice demonstrated a significant attenuation compared to MyD88−/− and wild type animals (*p<0.05; **p<0.01). Wild type (n=8); MyD88−/− (n=7); ASC−/− (n=6); wild type mock-infected (n=4). Results are representative of at least 2 independent experiments.
To assess the extent of inflammation, mice were sacrificed 3 days post-inoculation (dpi.) and the lungs were harvested for histology (Figure 1C–F). We observed a significant increase in airway inflammation, characterized by enhanced macrophage and neutrophil influx into the airways, 3 dpi. in wild type (D) and MyD88−/− (E) mice as compared to the mock inoculated wild type (C). In contrast, the infected ASC−/− (Figure 1F) and caspase-1−/− (not shown) mice demonstrated significantly reduced airway inflammation. Histology scoring, by blinded reviewer, confirmed the significant attenuation of airway inflammation in the ASC−/− mice (Figure 1G).
The Nlrp3 inflammasome is required for mouse survival and inflammation following influenza virus infection
The above data suggests that proper activation of an NLR inflammasome is necessary for host survival and for mediating the innate immune response following influenza virus infection. It is well established that the NLR protein, NLRP3, can form an inflammasome complex with ASC and caspase-1 in response to a plethora of stimuli (Eisenbarth et al., 2008; Li et al., 2008; Martinon et al., 2006; Sutterwala et al., 2006). In addition to NLRP3, other NLRs such as NLRC4 have been suggested to form an NLRP3 independent inflammasome which functions in IL-1β maturation in response to Salmonella typhimurium and Pseudomonasaeruginosa infection (Mariathasan et al., 2004; Miao et al., 2008). Following influenza inoculation, all animals demonstrated a decrease in body weight (Supplemental Figure S1). However, as seen in Figure 2A, a dramatic decrease in survival was observed in Nlrp3−/− mice following virus infection, with no differences observed in mortality between Nlrc4−/− and wild type mice. Interestingly, the removal of Nlrp3 produced a more dramatic effect on animal survival than the removal of ASC or caspase-1, suggesting the possibility that Nlrp3 might exert biologic effects in addition to caspase-1 activation; however, this remains to be explored.
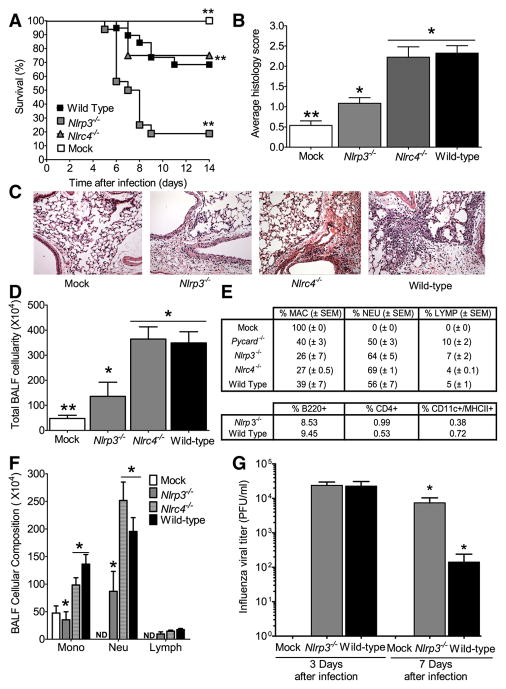
Figure 2. The Nlrp3 inflammasome is required for survival and mediates airway inflammation and viral clearance following pulmonary challenge with influenza virus.
A Wild type, Nlrp3−/− and Nlrc4−/− mice were challenged i.n. with influenza A/PR/8/34 and survival was monitored 14 dpi. Nlrp3−/− mice demonstrated significantly increased mortality compared to wild type animals (**p<0.01; log rank). In contrast, loss of Nlrc4 did not have any effect on mouse survival. Wild type mock-infected (n=7); wild type infected (n=19); Nlrp3−/− infected (n=16); and Nlrc4−/− infected (n=7). All survival experiments were performed together with the mice studied in Figure 1, therefore, the wild type controls are identical to those shown in Figure 1. (B) Lungs from wild type, Nlrp3−/− and Nlrc4−/− mice were harvested for histology and scored. A significant increase in airway inflammation was observed following influenza challenge in wild type and Nlrc4−/− animals, while a significant attenuation was observed in Nlrp3−/− mice (*p<0.05; **p<0.01). (C) No abnormal lung histopathology was observed in the mock-challenged wild type animals. (D) Nlrp3−/− mice demonstrated significantly attenuated inflammation following influenza virus challenge. (E) Nlrc4−/− and (F) wild type influenza challenged mice demonstrated increased airway inflammation. (G) To profile the cell types involved in the host immune response to influenza infection, bronchoalveolar lavage (BAL) was performed with differential staining and FACS analysis, 3 dpi. Consistent with the histology data, significantly fewer total cells were recovered in the BAL fluid (BALF) from Nlrp3−/− animals (*p<0.05), compared with either Nlrc4−/− or wild type mice. (H) BALF cellularity indicated a significant influx of macrophages and neutrophils in all influenza-challenged animals, with no difference observed in the composition of cells present in the airways. Wild Type (n=8); Nlrp3−/− (n=5); Nlrc4−/− (n=3); ASC−/− (n=6); wild type mock-infected (n=4). Results are representative of 3 independent experiments. (I) While no differences were observed in BALF composition, differential staining revealed a significant decrease in the total number of monocytes and neutrophils present in the Nlrp3−/− mice (*p<0.05). Wild type (n=8); Nlrp3−/− (n=4); Nlrc4−/− (n=4); wild type mock-infected (n=5). (J) Lungs were weighed and homogenized in sterile PBS either 3 or 7 dpi. Cell free supernatants were titered by standard plaque assay. Nlrp3−/− mice demonstrated a significant defect in vial clearance by day 7 (*p<0.05). Wild type mock-infected, d3 (n=3); wild type, infected, d3 (n=9); Nlrp3−/− infected, d3 (n=9); wild type mock-infected d7 (n=4); wild type infected, d7 (n=6); Nlrp3−/− infected, d7 (n=7).
As observed with the ASC−/− animals, Nlrp3−/− mice show a significant decrease in airway inflammation following influenza infection. Histology scoring revealed no increase in inflammation in mock infected animals and only a modest increase was observed in the Nlrp3−/− mice while a significant increase in airway inflammation was observed in the Nlrc4−/− and wild type animals (Figure 2B). Compared to mock infected mice (C), histopathology assessments revealed a mild increase in airway inflammation in virus-infected Nlrp3−/− mice (D), but an exacerbated increase in Nlrc4−/−(E) and wild type (F) animals 3 dpi. (Figure 2C–F). To further characterize the composition of the cell populations infiltrating the airways during viral infection, bronchoalveolar lavage fluid (BALF) was harvested and cellularity was assessed. Consistent with the histology findings, Nlrp3−/− animals demonstrate a significant decrease in total BALF cellularity compared to either the Nlrc4−/− or wild type animals (Figure 2G). Analysis of BALF cellularity indicates that macrophages and neutrophils represent the bulk of infiltrating cells in response to influenza. Differential profiling revealed no significant differences in the percent cellular composition of the BALF between genotypes (Figure 2H); however, the Nlrp3−/− mice demonstrated an overall decrease in the number of monocytes and granulocytes (Figure 2I). Together, this indicates that the Nlrp3 inflammasome is an essential mediator of in vivo inflammatory responses to influenza virus.
We have observed that Nlrp3−/− animals show reduced immune responses following influenza challenge. To determine if this reduction resulted in a defect in viral clearance, lungs from wild type and Nlrp3−/− mice were harvested 3 and 7 dpi. During the early stages of infection (3 dpi.), all influenza challenged mice demonstrated an increase in viral titer and no difference was observed between wild type and Nlrp3−/− mice (Figure 2J). However, by 7 dpi. the Nlrp3−/− mice demonstrated approximately 2-fold higher viral titer levels in the lungs compared to the wild type animals (Figure 2J), providing evidence for a defect in viral clearance in the absence of Nlrp3.
Components of the Nlrp3 inflammasome are highly expressed in myeloid cells and the lungs during influenza virus infection
To assess the relevance of Nlrp3 and ASC in a panel of mouse cells, we assayed expression by quantitative rtPCR in primary myeloid, lymphoid and airway epithelial cells (Thompson et al., 2006; Willingham et al., 2007). Under naïve conditions, both Nlrp3 and ASC genes are highly expressed in primary macrophages, with nominal levels detected in T and B lymphocytes (Figure 3A and B). In the mouse airway epithelial cells (MAEC), only a low level of ASC was detected and Nlrp3 was not found (Figure 3A and B).
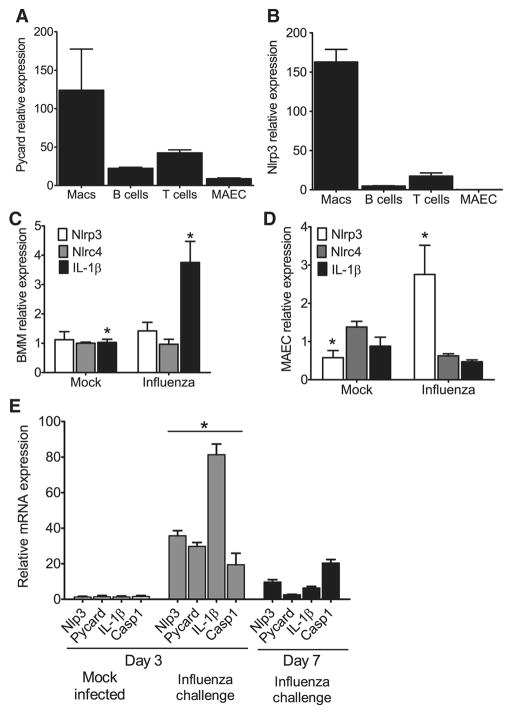
Figure 3. Nlrp3 inflammasome components are highly expressed in myeloid cells and in the lungs during influenza A virus infection.
(A–B) ASC and Nlrp3 gene expression was assessed in a panel of cells considered important for viral infection. Primary mouse bone marrow derived macrophages (BM), B cells, T cells, mouse airway epithelial cells (MAECs) and mouse embryonic fibroblasts (MEFs) were assessed by real-time (rt) quantitative PCR with samples normalized to 18s and standardized to expression in MEFs. Each cell line was assessed in triplicate and data are representative of 3 independent experiments. (C–D) Primary mouse BM s and MAECs were differentiated and challenged with mouse adapted influenza A/PR/8/34 (MOI=10). Nlrp3, Nlrc4 and IL-1β transcripts were assessed by rtPCR normalized to 18s and standardized to naive levels. (C) A significant increase in IL-1β mRNA expression was observed in BM following influenza challenge (MOI=10); however, no significant change was observed in Nlrp3 or Nlrc4 expression (*p<0.05). (D) Influenza challenge (MOI=1) resulted in a significant increase in Nlrp3 expression and a modest decrease in Nlrc4 and IL-1β expression in the MAECs. (E) Whole lungs were removed from wild type mice either 3 dpi. or 7 dpi. and homogenized. Total RNA was extracted from tissue pellets and expression levels of Nlrp3, ASC, Il-1β and Caspase-1 (Casp1) were assessed by rtPCR normalized to the 18S house keeping gene and compared to the mock-infected lungs. Transcription levels are significantly elevated by day 3 following influenza inoculation and begin to decline by day 7 (*p<0.05). All experiments are representative of at least 2 independent experiments with 3–5 mice per group.
To characterize the effects of viral infection on gene transcription in these differentiated cell types, bone marrow derived macrophages (BMΦ) and MAECs were challenged with influenza A/PR/8/34. No significant change in either Nlrp3 or Nlrc4 transcription was observed following viral challenge in the BMΦs; however, a significant increase in Il-1β mRNA level was detected (Figure 3C). Under naïve conditions, Nlrp3 was not detected in the MAECs. Following influenza infection, a modest increase in Nlrp3 transcription was observed, while no significant changes were found in Nlrc4 and Il-1β transcription (Figure 3D).
It should be noted that ex vivo cultured mouse BMΦs were refractory to influenza virus infection as shown by the precipitous drop in viral titer following exposure (Supplemental Figure S2A). Due to the lack of a productive infection, we often observed inconsistent IL-1β cytokine levels at the MOIs used in this study. For this reason, we did not pursue additional experiments using primary mouse macrophages. Even though IL-1β levels were inconsistent, at MOIs greater than 10 we were able to observe an increase in IFN-β at 12 hours, with no differences observed between the genotypes (Supplemental Figure S2B) indicating that the genes studied did not affect IFN-β production.
To further assess the contribution of the NLRP3 inflammasome in vivo, we sought to assess changes in mRNA expression levels of inflammasome components and IL-1β over a time course of influenza virus infection. Total lung RNA was extracted from tissue homogenates and gene expression was assessed for Nlrp3, ASC, Il-1β and Caspase-1 (Casp1) (Figure 3E). A significant increase in gene transcription was observed 3 dpi. for all 4 assessed genes. This was followed by a significant reduction of all genes except caspase-1 by day 7 (Figure 3E). These results indicate that influenza induced a substantial increase in all components of the Nlrp3 inflammasome in the lungs and infiltrating cells of inoculated animals 3 dpi. and indicate a profound effect of viral infection on the inflammasome genes leading to their biologic function.
To further determine the in vivo contribution of the macrophages in the lungs of mice following virus infection, lungs were harvested 3 dpi. and cells containing viral antigen were visualized by IHC/confocal microscopy. The primary cells targeted in vivo in the mouse are the airway epithelial cells (Supplemental Figure 3A–C), with 15% of macrophages being the second largest population (Supplemental Figure 3D–F). However a majority of macrophages and other leukocytes present in the lungs of infected mice were IHC negative (Supplemental Figure 3G–I). Significantly smaller populations of polymorphonuclear cells and alveolar epithelial cells (<1%) were also infected with virus (not shown).
NLRP3 inflammasome components are required for the production of proinflammatory cytokines and chemokines during influenza virus infection
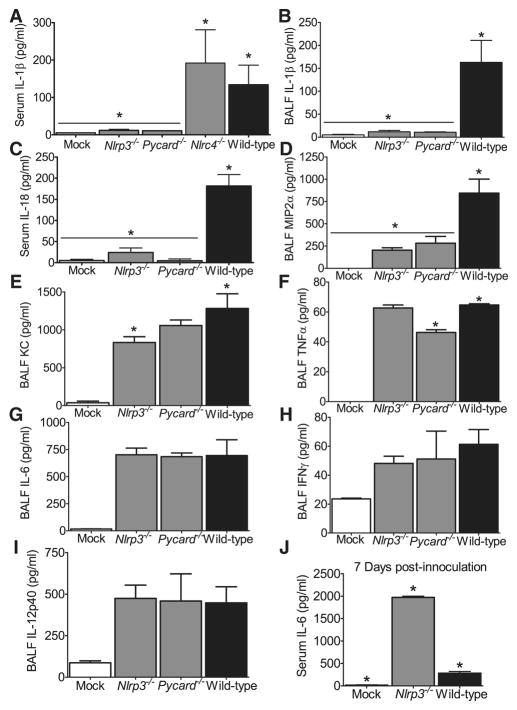
The above experiments indicate Nlrp3−/− and ASC−/− mice demonstrate attenuated airway inflammation and reduced survival following influenza virus infection. We next sought to assess changes in serum and BALF cytokines known to play roles in host immune responses to virus. A significant increase in IL-1β was detected in the serum and BALF from wild type animals. This increase was dependent on Nlrp3 and ASC, but independent of Nlrc4 (Figure 4A and C). To demonstrate that this lack of IL-1β is due to reduced post-translational processing by the Nlrp3 inflammasome, we assessed lung transcription of Il-1β and observed significant increases in Il-1β transcription in all mouse lines regardless of genotype (Supplemental Figure 4). As previously mentioned, IL-18 has also been shown to be a critical mediator of influenza infection and its maturation is also reliant on NLR inflammasome activation. A significant increase in serum IL-18 was detected in the wild type mice (Figure 4B). However, as with IL-1β, IL-18 was significantly reduced to mock-infected levels in mice lacking either Nlrp3 or ASC (Figure 4B). As with IL-1β, KC, MIP2α, TNFα, IL-6, IFNγ and IL-12 facilitate a wide spectrum of immunologic functions during respiratory virus infection. To evaluate these cytokine levels in the lungs, we assessed the BALF supernatant by ELISA. A significant decrease in MIP2α was also observed in the Nlrp3−/− and ASC−/− animals when compared to wild type levels (Figure 4D), which correlates with the decreased cellularity observed in the BALF (Figure 2I). Nlrp3−/− mice also show a modest decrease in KC (Fig. 4E). ASC−/− mice demonstrated a statistically significant decrease in TNFα, which was not observed in the Nlrp3−/− animals (Figure 4F). The observed attenuation in TNFα in the ASC−/− animals is observed in other assays and may reflect a previously-reported inflammasome independent role for ASC in the control of TNFα (Taxman et al., 2006b). No significant differences were observed in IL-6, IFNγ or IL-12p40 following influenza infection among the different mouse strains (Figures 4G–I). Due to the increased level of mortality observed in the Nlrp3−/− mice, a subset of moribund animals were harvested 7 dpi. BALF IL-6 levels were significantly elevated in the Nlrp3−/− mice compared to the levels observed in the wild type animals (Figure 4J). While unexpected, this observation is consistent with clinical findings in humans where influenza-infected individuals show increased IL-6 that is associated with increased morbidity (Lee et al., 2007).
Figure 4. Mice lacking components of the Nlrp3 Inflammasome demonstrate significantly altered levels of proinflammatory cytokines following pulmonary challenge with influenza.
Wild type, ASC−/−, Nlrp3−/− and Nlrc4−/− mice were challenged with influenza A/PR/8/34. Serum and cell free BALF were assessed 3 dpi., unless otherwise noted. (A–B) Significantly reduced levels of IL-1β(A) and IL-18 (B) were observed in the serum from Nlrp3−/− and Asc−/− mice (*p<0.05). (C–F) Nlrp3−/− and ASC−/− mice also demonstrated significantly reduced levels of BALF IL-1β (C) and MIP2α (D). KC was slightly decreased in Nlrp3−/− mice (E), while TNFα was reduced in ASC−/− mice (F) (*p<0.05). BALF levels of IL-18 were below the level of detection (data not shown). (G–I) All infected animals demonstrated a significant increase in IL-6 (G), IFNγ(H) and IL-12p40 (I) with no differences observed between genotypes. Mock, n=5; Nlrp3−/−, n=6; Nlrc4−/−, n=6; ASC−/−, n=9; Wild Type, n=5. (J) BALF from moribund Nlrp3−/− mice demonstrate significantly increased local levels of IL-6 compared to moribund wild type animals, 7 dpi.(*p<0.05). Mock, n=3; Nlrp3−/−, n=6; wild type, n=3. All data are representative of at least 2 independent experiments.
Human NLRP3 inflammasome activation in response to influenza virus is dependent upon lysosomal maturation and ROS production
For clinical relevance, the above study of mice required verification in human cells. Airway epithelial cells are the predominant cell type infected by influenza and previous studies have demonstrated that these cells contribute to innate immune responses (Kato and Schleimer, 2007). Primary human airway epithelial cell cultures (HAE) (Figure 5A) were infected with the human influenza virus A/Victoria/3/75 (H3N2) and the supernatant was harvested from either the apical or basolateral layer as previously described (Thompson et al., 2006). Following infection, we observed robust viral replication in the HAE cultures (Figure 5B) and increased IL-1β levels were detected in supernatant from the apical (Figure 5C) but not the basolateral layer. To expand upon these findings, we infected a human nasal airway epithelial cell line, JME, and observed increased NLRP3 and ASC expression (Figure 5D) and increased IL-1β release (Figure 5E). In addition to the JME cell lines, A/Victoria also induced a dramatic increase in IL-1β in a human monocytic cell line (THP-1) (Figure 5F). These data indicate that human cells of both monocytic and epithelial origin are contributors of IL-1β during influenza virus infection
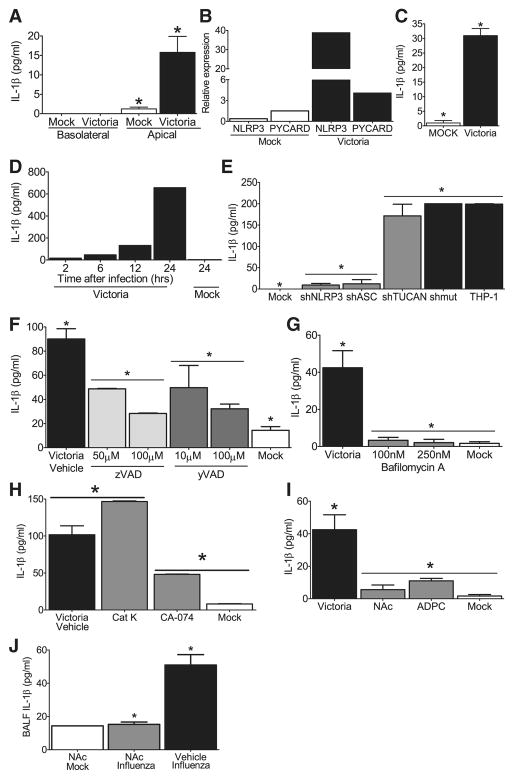
Figure 5. NLRP3 inflammasome activation in response to influenza virus is dependent upon lysosomal maturation and ROS production in human cells.
(A) Depiction of the primary ciliated human airway epithelial (HAE) cultures which were infected with the human pathogenic influenza virus A/Victoria/3/75 (H3N2; MOI = 1). Supernatants are collected from the apical and basolateral surfaces. (B) Robust viral replication was observed over the course of the HAE infection. (C) IL-1β levels were detected in the apical, but not basolateral, compartment 48 hours post-infection. (D) Human nasal airway epithelial cell lines (JME) were infected with A/Victoria/3/75 (MOI=1) and increased NLRP3 and ASC mRNA expression was observed. Data was normalized to 18s and compared to the expression in similarly treated human type II alveolar epithelial-like cell lines (A549). (E) Increased IL-1β protein was observed 24 hours post-infection in the cell free supernatant from the JMEs. (F) Human THP-1 monocyte cell lines were infected with A/Victoria/3/75 (MOI=5) and increased IL-1β levels were observed in the supernatant over a 24hr time course. (G–I) Influenza mediated IL-1β generation is dependent upon the NLRP3 inflammasome in human monocytes. (G) Human THP-1 cells were infected with lentivirus containing shRNA for ASC, NLRP3 or TUCAN (shASC, shNLRP3 or shTUCAN) or a mutated sh target sequence (shmut). Cell free supernatants were collected 24 hours post-infection (MOI=1). Influenza induced NLRP3- and ASC- dependent increase in IL-1β (*p<0.05), which was independent of TUCAN. (H) The general caspase inhibitor ZVAD-CHO and (I) the caspase-1 specific inhibitor Ac-YVAD-CHO, both inhibited IL-1β release in a dose-dependent manner (*p<0.05). (J–L) Influenza mediated IL-1β maturation requires lysosomal maturation and ROS production in THP-1. IL-1β release during influenza infection (MOI=5) was attenuated following treatment with (J) the lysosome inhibitor bafilomycin A (100–250nM), (K) the cathepsin B inhibitor CA-074-Me (50μM), and (L) the ROS inhibitors APDC (100μM) and NAc (50μM)(*p<0.05). (M) Treatment with the ROS inhibitor NAc (250mg/kg) inhibits influenza mediated IL-1β release in vivo (*p<0.05). NAc Mock, n=1; NAc Influenza, n=5, Vehicle Influenza, n=3.
To verify that influenza virus mediated IL-1β production in THP-1s is dependent upon NLRP3 inflammasome components, we utilized small heteroduplex RNA (shRNA) knockdown to assess the contribution of NLRP3 and ASC. An shRNA against the third putative inflammasome component, TUCAN/CARDINAL, was also targeted by shRNA. Successful RNA reduction is shown in Supplemental Figure S5A–C. These cells were challenged with A/Victoria/3/75 and cell free supernatants were collected over a 24 hr time-course. No differences in viral titer were detected between the different knockdown and wild type cell lines, 24 hrs p.i.(Supplemental Figure S5D). When cytokine was measured, a significant increase in IL-1β was observed in the wild type THP-1 cells and cells transfected with a scrambled shRNA targeting sequence (shmut), 24 hrs p.i. and this increase was significantly attenuated in cells containing shRNA targeting ASC and NLRP3, but not shRNA targeting TUCAN (Figure 5G; Supplemental Figure S5E). The role of TUCAN, a gene found in human but not mouse, has not been previously studied by gene reduction. This data suggest that TUCAN may not be important for NLRP3 inflammasome function induced by influenza virus. To confirm the specificity of shNLRP3 in IL-1β production, we also assessed TNFα release following influenza infection. As seen in Supplemental Figure S5F, levels of TNFα were not reduced in cell lines with shNLRP3 but modestly reduced in cells with shASC, consistent with the mouse data described earlier. Finally we demonstrate that ZVAD-CHO, a caspase inhibitor and YVAD-CHO, a specific peptide inhibitor of caspase-1, both attenuated IL-1β levels in a dose-dependent fashion following influenza virus infection (Figure 5H and I).
To further explore the mechanism underlying IL-1β release in response to influenza infection, we used pharmacological antagonist to block specific signaling pathways. Lysosomal degradation of particulate danger-associated molecular patterns (DAMPs) has been shown to activate the NRLP3 inflammasome (Hornung et al., 2008), however this has not been assessed in the context of viral infection. To test this, we utilized bafilomycin A to block lysosomal acidification via inhibition of the vacuolar H+ ATPase system. The addition of bafilomycin A at concentrations used previously to inhibit DAMP signaling (Hornung et al., 2008) completely abolished influenza induced IL-1β release (Figure 5J), which suggests a critical role for lysosomes in virus-mediated NLRP3 inflammasome function. A specific lysosomal cysteine proteinase, cathepsin B, has been associated with NLRP3-mediated cell death and IL-1β in response to non-viral signals (Willingham, 2008; Hornung et al., 2008). Utilizing the cathepsin B specific inhibitor, Ca-074-Me, we observed a significant attenuation in IL-1β release following influenza challenge (Figure 5K). This suggests the importance of cathepsin B in NLRP3-mediated response to influenza virus, but also suggests the involvement of additional lysosomal processes. This decrease was not observed with specific inhibitors for cathepsin K (Figure 5K). The release of lysosomal products into the cytosol has been demonstrated to promote the generation of ROS and recent data have suggested that ROS generation by asbestos, MSU and ATP is a necessary step in inflammasome activation (Dosert et al., 2008). To test if this occurs during influenza virus infection, we treated THP-1 cells with the ROS inhibitors N-acetyl-L-cysteine (NAc) or (2R,4R)-4-aminopyrrolidine-2,4-dicarboxylate (APDC) (Dosert et al., 2008). Our data demonstrate attenuated IL-1β production from cells treated with either ROS inhibitor (Figure 5L), suggesting that ROS also contributes to NLRP3 inflammasome activation in response to influenza.
Influenza infection in vivo results in a significant increase in ROS productionin the airways of humans and mice. To assess the physiological relevance of the in vitro ROS inhibitor findings, mice were treated with the ROS inhibitor NAc via i.n. administration as previously described (Springer et al., 2007). Similar to the monocyte cell line findings, treatment with NAc completely inhibited the generation of IL-1β in the BALF following influenza infection (Figure 5M). In addition to NAc, bafilomycin A and CA-074-ME were also assessed; however, the pharmacokinetics for each of these compounds were not optimized for our in vivo assessments (data not shown). Collectively these data suggest that NLRP3 inflammasome activation in response to virus is mediated by sensing alterations of lysosomal content and ROS induction in the cytosol following virus infection, utilizing pathways similar to those caused by DAMPs.
Nlrp3/NLRP3 mediates airway inflammation and IL-1β induction by the viral RNA analogs poly(I:C) and ssRNA40
The data presented thus far demonstrate that Nlrp3 mediates in vivo airway inflammation in response to influenza virus infection in mice and inflammasome formation in both mice and humans. To further explore the mechanism underlying NLRP3 activation, we examine the in vivo immune response to the viral RNA analog, poly(I:C). To assess if poly(I:C) is important for pulmonary inflammasome activation, mice were challenged i.n. with poly(I:C) and the resultant airway inflammation was characterized. Poly(I:C) induced moderate airway inflammation in the wild type mice, which was significantly reduced in the Nlrp3−/− animals (Figure 6A– B ). Nlrp3−/− mice exposed to poly(I:C) showed a significant reduction in the overall level of airway inflammation (Figure 6C) and BALF IL-1β level (Figure 6D).
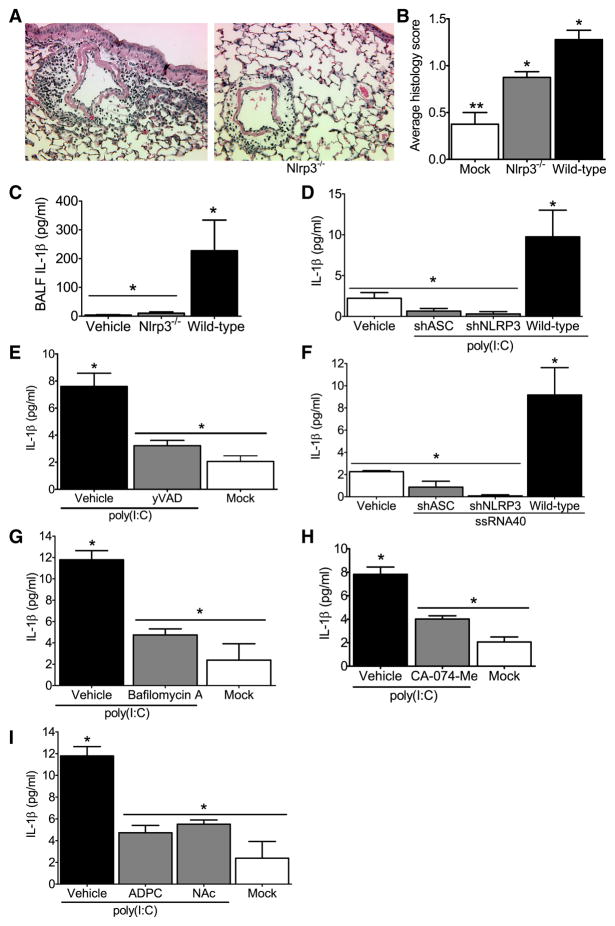
Figure 6. The NLRP3 inflammasome is required for airway inflammation induced by nucleic acid analogs.
Wild type and Nlrp3−/− mice received either 2 doses (50μg/dose) of poly(I:C) or vehicle on alternating days and were harvested 24hrs after the second dose was administered. (A–B) Poly(I:C) challenged animals demonstrated increased airway inflammation but less inflammatory cell influx was observed in Nlrp3−/− mice. (C) Histology scoring confirmed a significant attenuation in airway inflammation in the Nlrp3−/− mice (*p<0.05). (D) Significant decreases were observed in BALF IL-1β in the Nlrp3−/− mice compared to the wild type animals (*p<0.05). Vehicle challenged wild type (n=4); wild type, n=8; Nlrp3−/−, n=8. (E–G) IL-1β induction by viral RNA analogs in human monocytic cells is mediated by NLRP3 and ASC. (E) IL-1β secretion stimulated by the viral dsRNA analog poly(I:C) was significantly reduced in the shNLRP3 and shASC knockdown THP-1 cell lines and (F) following treatment with the caspase-1 specific inhibitor yVAD-CHO (*p<0.05). (G) IL-1β secretion stimulated by ssRNA40 was also significantly reduced in the shNLRP3 and shASC containing cells (*p<0.05). All knockdown studies are representative of at least 3 independent experiments. (H–J) Poly(I:C) mediated IL-1β maturation requires lysosomal maturation and ROS production in human monocytes. (H) The lysosome inhibitor bafilomycin A (100nM), (I) the cathepsin B inhibitor CA-074-Me (50μM), and (J) the ROS inhibitors APDC (100μM) and NAc (50μM) all significantly attenuated IL-1β production (*p<0.05).
To verify this result in human cells, we utilized the shRNA knockdown cell lines described above and demonstrate thatpoly(I:C) induced IL-1 β is dependent on ASC and NLRP3 (Figure 6E). To confirm the role of caspase-1 in this mechanism, cells were exposed to the caspase-1 inhibitor yVAD-CHO, which significantly attenuated poly(I:C) mediated IL-1β release (Figure 6F). To determine if the resultant NLRP3 and ASC dependent IL-1β release was unique to poly(I:C), cells were also stimulated with single-stranded GU-rich RNA (ssRNA40) complexed with the cationic lipid LyoVec to facilitate uptake. A significant NLRP3- and ASC- dependent increase in IL-1β release was observed following ssRNA40 stimulation indicating that NLRP3 and ASC mediate responses to ssRNA molecules (Figure 6G). To explore the intracellular pathways that lead to NLRP3 activation by RNA analogs, we utilized pharmacological inhibitors as described earlier and show that inflammasome activation in response to poly(I:C) requires lysosomal maturation, cathepsin B and the generation of ROS (Figure 6H–J).
DISCUSSION
In this report, we assess the in vivo contribution of NLRP3 to host immune responses to viral infection. NLRP3 forms a multi-protein inflammasome complex, which activates caspase-1 and leads to the maturation of several key proinflammatory cytokines, such as IL-1β and IL-18. It has been established that NLRP3 responds to both microbial-associated molecules and damage-associated molecular patterns (DAMPS) such as uric acid, alum salt and silica (Dostert et al., 2008; Eisenbarth et al., 2008; Hornung et al., 2008; Li et al., 2008; Martinon et al., 2006). In all of these cases, it is clear that NLRP3 promotes inflammation. A key issue is whether NLRP3 plays a beneficial or pathogenic role. In this study, we clearly demonstrate that NLRP3 and other components of the NLRP3 inflammasome plays a beneficial role in the in vivo innate immune response to influenza infection and is required for normal host immune responses to virus. An examination of the mechanism by which NLRP3 mediates host response to RNA virus demonstrates that recognition of RNA analogs or RNA molecules are capable of mediating these responses. A further exploration of the mechanism shows that similar to the activation of NLRP3 by DAMPs such as particulate alum, MSU crystals and silica, activation of the NLRP3 pathway by influenza virus, and more specifically by RNA analog, is dependent on an intact lysosomal pathway and functional lysosomal enzymes such as cathepsin B, and is also reliant on ROS both in vitro and in vivo. This supports the hypothesis that NLRP3 is activated through common intracellular changes caused by either PAMPs or DAMPs, rather than a model that evokes the direct interaction of NLRP3 with ligands of diverse molecular structures.
The recognition of viral pathogens by cells of the innate immune system is essential for the initiation of an ensuing inflammatory response. The endosomal TLRs TLR3, TLR7 and TLR8, and cytoplasmic RIG-I and MDA-5 are receptors of viral PAMPs and the primary route for the elicitation of host immune responses to virus through the regulation of type-I interferons. Among the NLR family, there are few reports linking these proteins to the viral host response or viral pathogenesis. One report identifies endogenous NLRX1, as an inhibitor of the RIG-I/MDA-5 pathway through interactions with the essential mitochondrial antiviral signaling adaptor (MAVS)(Moore et al., 2008). However, the in vivo importance of NLRX1 remains to be determined. The involvement of NLRP3 in the macrophage response to RNA viruses has been controversial. One study has demonstrated the importance of Nlrp3 during viral infection in culture (Kanneganti et al., 2006) while another group was unable to confirm inflammasome activation after poly(I:C) challenge or infection with the RNA viruses reovirus and vesicular stomatitis virus in culture. While we did not assess reovirus or VSV, our human monocytic cell line and in vivo mouse data clearly show that NLRP3 and ASC are indeed required for IL-1β maturation in response to influenza virus infection as well as to dsRNA analog and ssRNA. Therefore, our data support the first report. However, we have difficulty studying influenza viral infection of mouse macrophages ex vivo, due to a failure to establish a productive infection (Figure S2A). Thus, we have not precisely duplicated the ex vivo data observed by the first report. A possible explanation to reconcile the apparent discrepancies in the field is that different NLRs may mediate responses to different RNA viruses. The precedence for this is provided by MDA5 and RIG-I which recognize different types of dsRNA and show viral specificity. RIG-I responds to RNA viruses including paramyxoviruses and influenza virus, whereas MDA5 is essential for picornavirus recognition (Kato et al., 2008; Kato et al., 2006). Thus, it is possible that NLRP3 is responsible for dsRNA, ssRNA, DNA, Sendai virus and influenza virus recognition and an unidentified NLR may be activated by reovirus and VSV.
Infection of mice with influenza virus results in a dramatic increase in morbidity and mortality that mimics many of the pathophysiological aspects of the human disease. The current study shows that ASC and Nlrp3 deficiencies reduce inflammation in the lung, and this reduced inflammation is correlated with increased mortality and a viral clearance defect. Thus, the Nlrp3 inflammasome driven antiviral response is beneficial to the host following influenza infection. While the precise role for both IL-1β and IL-18 in the immune response against viral pathogens remains elusive, in vivo assessments of IL-1R1 and IL-18 deficient mice have shown that these animals have reduced acute airway inflammation associated with influenza virus infection and both have significantly decreased survival (Schmitz et al., 2005). These results are in agreement with our findings. Our data also highlight the observation that host response to influenza virus is specific, and requires NLRP3, but not NLRC4.
While our in vivo study shows that influenza virus is found in both airway epithelial cells and macrophages in the mouse, only mouse macrophages express detectable Nlrp3 transcript, and are thus the likely cell type mediating the beneficial functions of host protection by Nlrp3. However in humans, we find NLRP3 expression in both airway epithelial cells and macrophages and provide further evidence that NLRP3 and ASC mediate inflammatory cytokine release in cells of human origin. Thus, NLRP3 and ASC may play an even more profound role in host inflammation and protection upon influenza infection of humans. Considering the disastrous nature of a possible influenza pandemic, our study suggests that targeting the NLRP3 inflammasome for enhanced function could become an important therapeutic measure against such an infection.
EXPERIMENTAL PROCEDURES
Experimental Animals
All studies were conducted in accordance with the NIH Guide for the Care and Use of Laboratory Animals and the IACUC guidelines of UNC Chapel Hill. The generation of mice lacking functional Nlrp3 (Cryopyrin/Cias1), Nlrc4 (ICE-Protease Activating Factor/Ipaf), Apoptotic Speck protein containing a Card (ASC), Caspase-1 (Casp1) and MyD88 (Myd88) has been previously described (Adachi et al., 1998; Mariathasan et al., 2004; Sutterwala et al., 2006).
Virus Propagation
Influenza virus A/PR/8/34 (H1N1) was propagated in the allantoic cavity of 10-day-old embryonated specific pathogen free chicken eggs and was mouse adapted by a minimum of 6 serial passages through mice as previously described (Cottey et al., 2001). The influenza virus A/Victoria/3/75 (H3N2) is a recombinant virus generated from cloned cDNA in 293T cells and propagated in MDCK cells. Viral titers were determined by standard plaque assay on confluent monolayers of MDCK cells.
Influenza Virus Infection of Human Primary Cells and Cell Lines
Cells were challenged with A/Victoria (MOI=1) for 2 hrs at 37°C. Following incubation, supernatant was replaced with fresh media and cells were incubated at 37°C. Cell free supernatants were harvested at select time points for viral titer and cytokine analysis. To assess viral PAMPs, cells were incubated for 24 hrs in the presence of naked poly(I:C) (10μg)(Sigma) or ssRNA40 complexed with LyoVec (InvivoGen) (10μg). For pharmacological assessments, cells were treated with bafilomycin D (100–250nM), Ac-ZVAD-CHO (10–100μM), Ac-ZVAD-CHO (10–100μM), Ca-074-Me (10–50μM), Cathepsin K Inhibitor I (10–50μM), N-acetyl-L-cysteine (NAC) (50–100μM) or (2R, 4R)-4-aminopyrrolidine-2-4-dicarboxylate (APDC) (50–100μM) as previously described (Dostert et al., 2008; Hornung et al., 2008; Willingham et al., 2007).
In vivo Influenza A Virus Infection and poly(I:C) Challenge
Animals were anesthetized and challenged by intranasal (i.n.) administration of 6×104 PFU/ml of influenza virus A/PR/8/34 in 50μl of PBS and 0.1% Alum (Sigma). Mice were observed and weight was assessed daily for up to 14 dpi. To assess poly(I:C) in vivo, wild type and NLRP3−/− mice received 2 doses (50μg/dose) of poly(I:C) in 50μl of PBS and 0.1% Alum or vehicle on alternating days and harvested 24 hrs after the second administration. To assess in vivo ROS production, mice were treated with the ROS inhibitor NAc (250mg/kg) via i.n. administration (as previously described by Springer et al., 2007) throughout the course of virus challenge.
Influenza inoculated mice were euthanized either 3 or 7 dpi., and serum was collected following cardiac puncture. Total cytokine levels were determined by ELISA (R&D Biosystems or BD Biosciences). In some cases, Bronchoalveolar Lavage (BAL) was performed and the number of cells present in the BAL fluid (BALF) was determined using a hemacytometer. A morphology based differential cell count was conducted on a cytospin prep from the BALF and stained with Diff-Quik solution (Sigma). BALF cellularity was also determined via FACS using standard techniques and antibodies reactive to CD11b, CD11c, GR-1, B220 and CD4/CD8. The remaining BALF was centrifuged to remove cells, and cytokine levels in the supernatant were determined.
For histopathologic examination, lungs were fixed by inflation and immersion in 10% buffered formalin. To evaluate airway inflammation, fixed lung slices were subjected to hematoxylin and eosin (H&E) staining. Lung sections were scored (0 (none) – 3 (extreme)) based on assessments of mononuclear and polymorphonuclear cell infiltration, perivascular and peribronchiolar cuffing, and estimates of the percent of lung involved with the inflammation, while blinded to genotype and treatment.
To assess viral titer, whole lungs were removed weighed and homogenized in PBS. Lung viral titers were determined in the supernatants as described above.
Statistical Analysis
Data are presented as the mean +/− standard error of the mean (SEM). Analysis Of Variance (ANOVA) followed by Tukey-Kramer HSD for multiple comparisons was performed on complex data sets. Statistical significance for single data points was assessed by the Student’s two-tailed t-test. Survival curves were generated utilizing the product limit method of Kaplan and Meier and comparisons were made using the log rank test. In all cases, a p-value of less than 0.05 was considered statistically significant.
Supplementary Material
Acknowledgments
The authors thank the Carolina Vaccine Institute for supplying aliquots of the mouse adapted influenza A/PR/8/34 virus. We acknowledge Dr. Wendy Barclay for critical review and advice on viral propagation and Dr. Rob Tarran for providing the JME cell line. We thank Drs. Richard Flavell (Yale University), Vishva M. Dixit (Genentech, Inc), Fayyaz Sutterwala (the University of Iowa), Millenium Pharmaceuticals for supplying the Nlrp3, ASC, Nlrc4 and caspase-1 deficient mice. We also thank Willie June Brickey and Sushmita Jha for mouse colony maintenance and providing histology scoring assistance. This work is supported by 3-U54-AI057157-06S1, AI067798, and 05-0064, 1U19-AI077437-01 (J.P.Y. Ting). Dr. Allen is supported by a NIH training grant. Margaret Scull is a recipient of the George H. Hitchings Fund for Health Research and Science Education of the Triangle Community Foundation.
Abbreviations
- BAL
bronchoalevolar lavage
- BALF
bronchoalveolar lavage fluid
- i.n
intranasal
- IL
interleukin
- NLR
nucleotide binding domain and leucine-rich-repeat-containing
- TLR
Toll-like Receptor
- PFU
plaque forming unit
- ASC
Apoptotic Speck protein containing a CARD
- ssRNA
single stranded RNA
- dsRNA
double stranded RNA
- poly(I:C)
polyinosinic:polycytidylic acid
- MOI
Multiplicity of Infection
Footnotes
Additional Methods are included in Supplemental Experimental Protocols
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Adachi O, Kawai T, Takeda K, Matsumoto M, Tsutsui H, Sakagami M, Nakanishi K, Akira S. Targeted disruption of the MyD88 gene results in loss of IL-1- and IL-18-mediated function. Immunity. 1998;9:143–150. doi: 10.1016/s1074-7613(00)80596-8. [DOI] [PubMed] [Google Scholar]
- Agostini L, Martinon F, Burns K, McDermott MF, Hawkins PN, Tschopp J. NALP3 forms an IL-1beta-processing inflammasome with increased activity in Muckle-Wells autoinflammatory disorder. Immunity. 2004;20:319–325. doi: 10.1016/s1074-7613(04)00046-9. [DOI] [PubMed] [Google Scholar]
- Akira S, Uematsu S, Takeuchi O. Pathogen recognition and innate immunity. Cell. 2006;124:783–801. doi: 10.1016/j.cell.2006.02.015. [DOI] [PubMed] [Google Scholar]
- Bartlett EJ, Hennessey M, Skiadopoulos MH, Schmidt AC, Collins PL, Murphy BR, Pickles RJ. The role of interferon in the replication of human parainfluenza virus type 1 wild type and mutant viruses in human ciliated airway epithelium. J Virol. 2008 doi: 10.1128/JVI.02263-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cottey R, Rowe CA, Bender BS. Influenza virus. Curr Protoc Immunol . 2001;Chapter 19(Unit 19):11. doi: 10.1002/0471142735.im1911s42. [DOI] [PubMed] [Google Scholar]
- Dostert C, Petrilli V, Van Bruggen R, Steele C, Mossman BT, Tschopp J. Innate immune activation through Nalp3 inflammasome sensing of asbestos and silica. Science. 2008;320:674–677. doi: 10.1126/science.1156995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eisenbarth SC, Colegio OR, O’Connor W, Sutterwala FS, Flavell RA. Crucial role for the Nalp3 inflammasome in the immunostimulatory properties of aluminium adjuvants. Nature. 2008;453:1122–1126. doi: 10.1038/nature06939. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hornung V, Bauernfeind F, Halle A, Samstad EO, Kono H, Rock KL, Fitzgerald KA, Latz E. Silica crystals and aluminum salts activate the NALP3 inflammasome through phagosomal destabilization. Nat Immunol. 2008;9:847–856. doi: 10.1038/ni.1631. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jefferson DM, Valentich JD, Marini FC, Grubman SA, Iannuzzi MC, Dorkin HL, Li M, Klinger KW, Welsh MJ. Expression of normal and cystic fibrosis phenotypes by continuous airway epithelial cell lines. Am J Physiol. 1990;259:L496–505. doi: 10.1152/ajplung.1990.259.6.L496. [DOI] [PubMed] [Google Scholar]
- Kanneganti TD, Body-Malapel M, Amer A, Park JH, Whitfield J, Franchi L, Taraporewala ZF, Miller D, Patton JT, Inohara N, Nunez G. Critical role for Cryopyrin/Nalp3 in activation of caspase-1 in response to viral infection and double-stranded RNA. J Biol Chem. 2006;281:36560–36568. doi: 10.1074/jbc.M607594200. [DOI] [PubMed] [Google Scholar]
- Kato A, Schleimer RP. Beyond inflammation: airway epithelial cells are at the interface of innate and adaptive immunity. Curr Opin Immunol . 2007;19:711–720. doi: 10.1016/j.coi.2007.08.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kato H, Takeuchi O, Mikamo-Satoh E, Hirai R, Kawai T, Matsushita K, Hiiragi A, Dermody TS, Fujita T, Akira S. Length-dependent recognition of double-stranded ribonucleic acids by retinoic acid-inducible gene-I and melanoma differentiation-associated gene 5. J Exp Med. 2008 doi: 10.1084/jem.20080091. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kato H, Takeuchi O, Sato S, Yoneyama M, Yamamoto M, Matsui K, Uematsu S, Jung A, Kawai T, Ishii KJ, et al. Differential roles of MDA5 and RIG-I helicases in the recognition of RNA viruses. Nature. 2006;441:101–105. doi: 10.1038/nature04734. [DOI] [PubMed] [Google Scholar]
- Kawai T, Akira S. Antiviral signaling through pattern recognition receptors. J Biochem. 2007;141:137–145. doi: 10.1093/jb/mvm032. [DOI] [PubMed] [Google Scholar]
- Koyama S, Ishii KJ, Kumar H, Tanimoto T, Coban C, Uematsu S, Kawai T, Akira S. Differential role of TLR- and RLR-signaling in the immune responses to influenza A virus infection and vaccination. J Immunol. 2007;179(7):4711–20. doi: 10.4049/jimmunol.179.7.4711. [DOI] [PubMed] [Google Scholar]
- Lee N, Wong CK, Chan PK, Lun SW, Lui G, Wong B, Hui DS, Lam CW, Cockram CS, Choi KW, et al. Hypercytokinemia and hyperactivation of phospho-p38 mitogen-activated protein kinase in severe human influenza A virus infection. Clin Infect Dis. 2007;45:723–731. doi: 10.1086/520981. [DOI] [PubMed] [Google Scholar]
- Li H, Willingham SB, Ting JP, Re F. Cutting Edge: Inflammasome Activation by Alum and Alum’s Adjuvant Effect Are Mediated by NLRP3. J Immunol. 2008;181:17–21. doi: 10.4049/jimmunol.181.1.17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mariathasan S, Monack DM. Inflammasome adaptors and sensors: intracellular regulators of infection and inflammation. Nat Rev Immunol. 2007;7:31–40. doi: 10.1038/nri1997. [DOI] [PubMed] [Google Scholar]
- Mariathasan S, Newton K, Monack DM, Vucic D, French DM, Lee WP, Roose- Girma M, Erickson S, Dixit VM. Differential activation of the inflammasome by caspase-1 adaptors ASC and Ipaf. Nature. 2004;430:213–218. doi: 10.1038/nature02664. [DOI] [PubMed] [Google Scholar]
- Martinon F, Petrilli V, Mayor A, Tardivel A, Tschopp J. Gout associated uric acid crystals activate the NALP3 inflammasome. Nature. 2006;440:237–241. doi: 10.1038/nature04516. [DOI] [PubMed] [Google Scholar]
- Miao EA, Ernst RK, Dors M, Mao DP, Aderem A. Pseudomonas aeruginosa activates caspase 1 through Ipaf. Proc Natl Acad Sci U S A. 2008;105:2562–2567. doi: 10.1073/pnas.0712183105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moore CB, Bergstralh DT, Duncan JA, Lei Y, Morrison TE, Zimmermann AG, Accavitti-Loper MA, Madden VJ, Sun L, Ye Z, et al. NLRX1 is a regulator of mitochondrial antiviral immunity. Nature. 2008;451:573–577. doi: 10.1038/nature06501. [DOI] [PubMed] [Google Scholar]
- Moore CB, Ting JP. Regulation of mitochondrial antiviral signaling pathways. Immunity. 2008;28:735–739. doi: 10.1016/j.immuni.2008.05.005. [DOI] [PubMed] [Google Scholar]
- Muruve DA, Petrilli V, Zaiss AK, White LR, Clark SA, Ross PJ, Parks RJ, Tschopp J. The inflammasome recognizes cytosolic microbial and host DNA and triggers an innate immune response. Nature. 2008;452:103–107. doi: 10.1038/nature06664. [DOI] [PubMed] [Google Scholar]
- Razmara M, Srinivasula SM, Wang L, Poyet JL, Geddes BJ, DiStefano PS, Bertin J, Alnemri ES. CARD-8 protein, a new CARD family member that regulates caspase-1 activation and apoptosis. J Biol Chem. 2002;277:13952–13958. doi: 10.1074/jbc.M107811200. [DOI] [PubMed] [Google Scholar]
- Schmitz N, Kurrer M, Bachmann MF, Kopf M. Interleukin-1 is responsible for acute lung immunopathology but increases survival of respiratory influenza virus infection. J Virol. 2005;79:6441–6448. doi: 10.1128/JVI.79.10.6441-6448.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Springer J, Groneberg DA, Dinh QT, Quarcoo D, Hamelmann E, Braun-Dullaeus RC, Geppetti P, Anker SD, Fischer A. Neurokinin-1 receptor activation induces reactive oxygen species and epithelial damage in allergic airway inflammation. Clin Exp Allergy. 2007;37(12):1788–97. doi: 10.1111/j.1365-2222.2007.02851.x. [DOI] [PubMed] [Google Scholar]
- Sutterwala FS, Ogura Y, Szczepanik M, Lara-Tejero M, Lichtenberger GS, Grant EP, Bertin J, Coyle AJ, Galan JE, Askenase PW, Flavell RA. Critical role for NALP3/CIAS1/Cryopyrin in innate and adaptive immunity through its regulation of caspase-1. Immunity. 2006;24:317–327. doi: 10.1016/j.immuni.2006.02.004. [DOI] [PubMed] [Google Scholar]
- Taxman DJ, Livingstone LR, Zhang J, Conti BJ, Iocca HA, Williams KL, Lich JD, Ting JP, Reed W. Criteria for effective design, construction, and gene knockdown by shRNA vectors. BMC Biotechnol. 2006a;6:7. doi: 10.1186/1472-6750-6-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taxman DJ, Zhang J, Champagne C, Bergstralh DT, Iocca HA, Lich JD, Ting JP. Cutting edge: ASC mediates the induction of multiple cytokines by Porphyromonas gingivalis via caspase-1-dependent and independent pathways. J Immunol. 2006b;177:4252–4256. doi: 10.4049/jimmunol.177.7.4252. [DOI] [PubMed] [Google Scholar]
- Thompson CI, Barclay WS, Zambon MC, Pickles RJ. Infection of human airway epithelium by human and avian strains of influenza a virus. J Virol. 2006;80:8060–8068. doi: 10.1128/JVI.00384-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ting JP, Lovering RC, Alnemri ES, Bertin J, Boss JM, Davis BK, Flavell RA, Girardin SE, Godzik A, Harton JA, et al. The NLR gene family: a standard nomenclature. Immunity. 2008;28:285–287. doi: 10.1016/j.immuni.2008.02.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Willingham SB, Bergstralh DT, O’Connor W, Morrison AC, Taxman DJ, Duncan JA, Barnoy S, Venkatesan MM, Flavell RA, Deshmukh M, et al. Microbial pathogen-induced necrotic cell death mediated by the inflammasome components CIAS1/cryopyrin/NLRP3 and ASC. Cell Host Microbe. 2007;2:147–159. doi: 10.1016/j.chom.2007.07.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.