Abstract
Phospholipid hydroperoxide glutathione peroxidase (PHGPx) is an intracellular antioxidant enzyme that directly reduces peroxidized phospholipids. PHGPx is transcribed from one gene into three types of mRNA, mitochondrial, non-mitochondrial and nucleolar PHGPx by alternative transcription. In this review, we focus on our recent experiments on the regulation of promoter activity of the types of PHGPx and on the novel strategy of functional analysis of a PHGPx knockout mice model using the transgenic rescue method and Cre-LoxP system. PHGPx is especially high in testis and spermatozoa. A deficiency is implicated in human infertility. We established spermatocyte-specific PHGPx knockout (KO) mice using a Cre-loxP system. Targeted disruption of all exons of the PHGPx gene in mice by homologous recombination caused embryonic lethality at 7.5 days post coitum. The PHGPx-loxP transgene rescued PHGPx KO mice from embryonic lethality. These rescued floxed PHGPx mice were mated with spermatocyte specific Cre expressing mice. All the spermatocyte-specific PHGPx KO male mice were infertile and displayed a significant decrease in the number of spermatozoa and significant reductions in forward motility by mitochondrial dysfunction of spermatozoa. These results demonstrate that depletion of PHGPx in spermatozoa may be one of the causes of male infertility in mice and humans.
Keywords: GPx4, Cre/LoxP, transgenic rescue, male infertility, spermatocyte specific KO
Introduction
Reactive oxygen species (ROS) are formed and degraded by all aerobic organisms, leading either to the physiological concentrations required for normal cell function, or to excessive quantities, a state called oxidative stress. Physiological use of ROS by cells has recently been demonstrated. For example, it has been shown that superoxide, hydrogen peroxide, and lipid hydroperoxide can regulate the activities of several kinases and transcription factors and the cell death machinery [1, 2]. Lipid hydroperoxides are a type of ROS of which the biological function has not yet been clarified.
Phospholipid hydroperoxide glutathione peroxidase (PHGPx, GPx4) is a unique intracellular antioxidant enzyme that directly reduces peroxidized phospholipids that have been produced in cell membranes [2]. Three types of PHGPx, mitochondrial (M), non-mitochondrial (C) and nucleolar PHGPx (N), are transcribed from one gene by alternative transcription as shown in Fig. 1 [3, 4]. The gene for mouse PHGPx consists of 8 exons, with different first exons for the different types: exon Ia for mitochondrial PHGPx and non-mitochondrial PHGPx and exon Ib for nucleolar PHGPx. The mitochondrial targeting signal of PHGPx and the second start codon of non-mitochondrial PHGPx are in exon Ia of PHGPx genomic DNA [5]. After cleavage of the N-terminal mitochondrial import sequence of mitochondrial PHGPx, the mature mitochondrial protein becomes identical to the 20 kDa non-mitochondrial PHGPx [6]. Nucleolar PHGPx was first identified as a sperm nucleus-specific 34 kDa selenoprotein (called snGPx, for sperm nucleus-specific glutathione peroxidase) or nuclear PHGPx [7]. It is formed by use of an alternative promoter and start codon localized in exon Ib of the PHGPx gene [3, 7, 8]. We have previously shown that by using an N-terminal nucleolar import signal this 34 kDa PHGPx is localized in nucleoli in several cell lines [9]. We chose the name nucleolar PHGPx since non-mitochondrial 20 kDa PHGPx exists not only in the cytosol, but also in the nucleus [10].
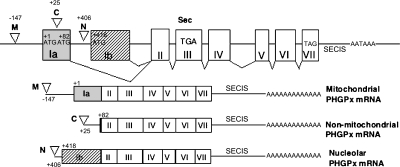
Fig. 1.
Structure of the PHGPx gene, and of the three types of PHGPx mRNA. The structure of the mouse PHGPx gene (Accession No. AB030643) includes eight exons in the upper panel, while exons of the three types of PHGPx mRNA are shown in the lower panel. Exon Ia (gray box) includes the translational first start site ATG (+1) for mitochondrial PHGPx, and the second ATG (+82) for non-mitochondrial PHGPx. Exon Ib (shaded box) includes the translational third start site ATG (+418) for nucleolar PHGPx. The reverted open triangles show three transcriptional start sites by analysis of 5'RACE as previously reported [3]; position −147 is the transcriptional start site for mitochondrial PHGPx (M), position +25 for non-mitochondrial PHGPx (C) and position +406 for nucleolar PHGPx (N). TGA in exon III encodes selenocysteine and TAG in exon VII encodes the stop codon. SECIS is the selenocysteine insertion sequence.
The three types of PHGPx play several important but independent roles in the modulation of inflammation, spermatogenesis, and cell death [2].
We have recently shown that overexpression of the different types of PHGPx in the rat basophilic leukemia (RBL2H3) cell line provides a useful model for clarifying the abilities of the different types of PHGPx to modulate cellular function and the importance of lipid hydroperoxides as signal molecules [2]. Table 1 shows the characterization of three types of PHGPx overexpressing cells. S1 cells are control RBL2H3 cells transfected with only vector. L9 cells are non-mitochondrial PHGPx overexpressing cells [10–13], M15 cells are mitochondrial PHGPx overexpressing cells [14–18] and N63 cells are nucleolar PHGPx overexpressing cells [9]. Although non-mitochondrial PHGPx suppresses activation of lipoxygenase and cyclooxygenase at the nucleus and endoplasmic reticulum in response to several stimuli [10, 12], it does not suppress apoptotic cell death induced by the mitochondrial death pathway [14]. Although mitochondrial PHGPx can suppress the release of cytochrome c from mitochondria by inhibition of generation of cardiolipin hydroperoxide during apoptosis induced by mitochondrial death pathway [15–17], it cannot suppress the activation of lipoxygenase and cyclooxygenase [10, 12, 14]. Nucleolar PHGPx can suppress cell death by inhibition of nucleolar damage induced by actinomycin D (ActD) and doxorubicin (Dox), but not the apoptotic cell death through the mitochondrial death pathway induced by staurosporine, UV and 2-DG (2-deoxyglucose). Thus, three types of PHGPx found in different organelles play different roles in signal transduction, inflammation and apoptosis. Our transformant studies showed that lipid hydroperoxides are activators of lipoxygenase and cyclooxygenase and participate in inflammation, and that cardiolipin hydroperoxide is the signal molecule for the release of cytochrome c and the mitochondrial pore opening via adenine nucleotide translocator (ANT) during apoptotic cell death [2, 15–17]. These cellular functions of PHGPx were reviewed in our previous review article [2, 17].
Table 1.
Characterization of three types of PHGPx overexpressing RBL2H3 cells
| Cell Names | S1 | L9 | M15 | N63 | |
|---|---|---|---|---|---|
| Distribution of overexpressing PHGPx | Normal | Nucleus Cytosol | Mitochondria | Nucleolus | Target site |
| Leukotrien C4 Production | Normal | Normal | Normal | 5-lipoxygenase (Nucleus) | |
| Prostaglandin D2 Production | Normal | Normal | Normal | Cyclooxygenase (Nucleus) | |
| Extracellular oxidative stress ex) t-BuOOH | Sensitive | Sensitive | Lipid Peroxidation | ||
| Mitochondrial apoptotic cell death, 2-DG, UV staurosporin | Sensitive | Sensitive | Sensitive | Mitochondrial Cardiolipin Peroxidation | |
| Mitochondrial damaged cell death ex) KCN, rotenone | Sensitive | Sensitive | Sensitive | Mitochondrial Respiratory Chain | |
| Fas mediated apoptosis | Sensitive | Sensitive | Sensitive | Sensitive | Fas |
| Nucleolar damaged Cell death, ex) ActD, Dox | Sensitive | Sensitive | Nucleoli Mitochondria | ||
This review focuses on our recent mouse PHGPx gene experiments regarding the regulation of promoter activity of the types of PHGPx in mice and the novel strategy of functional analysis of PHGPx knockout (KO) mice model using the transgenic rescue method and Cre-LoxP system. PHGPx is widely expressed in normal tissue and is especially high in testis and spermatozoa, where it has an important role in spermatogenesis and sperm function. Furthermore, a deficiency is implicated in human infertility [19]. We recently established spermatocyte-specific PHGPx knockout mice. These mice showed the same phenotype, oligoasthenozoospermia, as PHGPx deficient human male infertility patients.
Regulation of Promoter Activity of Three Types of Mouse PHGPx
We have shown that expression of non-mitochondrial PHGPx was relatively high in somatic cells while the expression of nucleolar PHGPx was extremely low. In somatic tissues and cultured cells, the amounts of non-mitochondrial PHGPx were approximately 2.26–12.2, 638–8994 times higher than those of mitochondrial and nucleolar PHGPx mRNAs as determined by TaqMan assay [3]. The expression of mitochondrial PHGPx and nucleolar PHGPx was significantly higher only in testis than in other tissues. Expression of mitochondrial PHGPx and nucleolar PHGPx is induced significantly in testis during spermatogenesis, especially in late spermatocytes, spermatids and spermatozoa, in both humans and mice [9, 19–21]. Mitochondrial PHGPx in particular has an important role in spermatogenesis and sperm function, and its deficiency is implicated in human infertility [19]. However, the mechanism behind the high expression of mitochondrial PHGPx in testis or the decrease in expression of mitochondrial PHGPx in somatic cells remains to be clarified.
We and other groups have investigated the regulation of promoter activity by deletion and mutational analysis of the promoter region of PHGPx gene [3, 22, 23]. We reported a possible construction of the positive regulatory region and the core promoter regions of PHGPx in several cell lines using promoter analysis with luciferase as the reporter gene and electrophoretical mobility shift analysis as shown in Fig. 2 [3]. By comparison of sequences in the 5'-flanking regions of pig, human and mouse PHGPx, we determined 29 high homology domains (H1 to H29) as shown by the gray and black circles in Fig. 2. Twelve regions of the 29 homology domains contained the putative consensus binding sequences for several transcriptional factors, as shown in black circles. Deletion analysis of promoter activity in the mouse PHGPx gene demonstrated that the 5'-flanking regions from −60 to −9 bp, −233 to −158 bp and +342 to +375 bp are critical for the basal transcription of non-mitochondrial, mitochondrial and nucleolar PHGPx mRNA respectively. The core promoter activity in L929 cells was high for non-mitochondrial PHGPx, but relatively low for mitochondrial and nucleolar PHGPx and for the expression levels of each type of PHGPx mRNA.
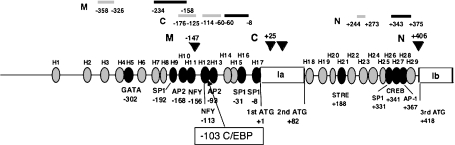
Fig. 2.
Summary of the identified regulatory and core promoter regions of the 5'-flanking regions of exon Ia and exon Ib of the PHGPx gene in L929 cells. Shaded bars in the top line indicate the regulatory regions of the three types of PHGPx. Black bars in the top line indicate the each core region of the three types of PHGPx. The number of homology domains (H) between human, pig and mice genes is shown in [3]. The black and gray circles are homology domains with and without putative transcriptional factor binding sites, respectively, derived from analysis with the TRANSFAC program. The C/EBP binding site for the TNFα-induced up-regulation of the promoter activity in mice is in a different region of the gene than in human.
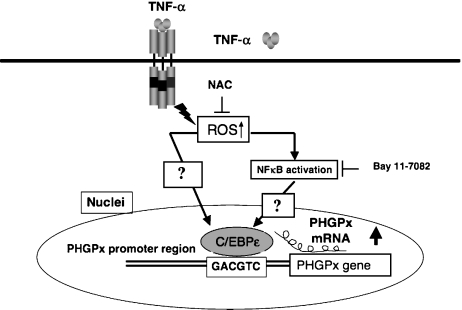
Fig. 3.
C/EBPε regulates the up-regulation of PHGPx in HL60 cells and neutrophils stimulated by TNFα. C/EBPε is expressed primarily in neutrophils and myeloid and lymphoid cells, but not in differentiated macrophages. The promoter activity of PHGPx and expression of PHGPx mRNA was specifically up-regulated by stimulation with TNFα in C/EBPε expressing neutrophils and HL60 cells. This up-regulation of expression of PHGPx mRNA and promoter activities of PHGPx were inhibited by treatment with the anti-oxidants PDTC and NAC, and by inhibitors of NFκB and Bay 11-7082. The up-regulated promoter activity was effectively abrogated by a mutation in C/EBP-binding sequence (GACGTC). C/EBPε is a critical transcription factor in TNFa-induced up regulation of PHGPx expression.
The mutational promoter analysis indicated that Nuclear Factor Y (NFY) in the −156 to −147 bp region and Adoptor Protein 2 (AP2) in the −93 to −83 bp region are essential for maximum promoter activity of non-mitochondrial PHGPx, but NFY in the −113 to −107 bp region is not. NFY in the −156 to −147 bp region was not essential for the promoter activity of mitochondrial PHGPx. Mutational analysis of Specificity Protein 1(SP1) in the +331 to +338 bp region and cAMP response element binding protein (CREB) in the +341 to +354 bp region resulted in a significant decrease of the core promoter activity of nucleolar PHGPx.
Furthermore, the regions from −176 to −125 bp and from −114 to −60 bp (C; non-mitochondrial PHGPx), from −358 to −326 bp (M; mitochondrial PHGPx) and from +244 to +273 bp (N; nucleolar PHGPx) are involved in enhancement of promoter activity [3]. The −358 to −326 bp region may regulate the promoter activity of non-mitochondrial PHGPx. However, the upstream region in somatic cells normally suppresses up-regulation by this region. Electrophoretical mobility shift analysis demonstrated that a specific transcription factor complex bound to this region in adult testis, but not in juvenile testis and that different sized complexes bound to this region in the mouse testis and brain [3]. These results suggest that this region is associated with increased transcription of mitochondrial PHGPx during spermatogenesis.
A recent study has indicated that overexpression of cAMP-response element modulator (CREM-τ) enhanced the promoter activity of nucleolar PHGPx in cultured cells and that the nuclear extract from rat spermatid cells can specifically bind to the CREB site in the promoter region of rat nucleolar PHGPx [24]. These results suggest that the CREB site is an important region for enhancement of the induction of transcription of nucleolar PHGPx.
Identification of a Responsible Promoter Region and a Key Transcription Factor, CCAAT/enhancer-binding Protein ε, for Up-regulation of PHGPx in HL60 Cells Stimulated with TNFα
Previous studies have addressed the up-regulation of PHGPx expression in spermatogenesis [19, 21] embryogenesis [25] and rat casein-induced polymorphonuclear neutrophils [26], and its down-regulation in A549 cells stimulated by cytokines IL-4 and IL-13 [27]. The mechanisms and transcription factors affecting these interesting changes of PHGPx, however, are still not well known.
We have recently demonstrated that the expression level of PHGPx was up-regulated in human peripheral polymorphonuclear leukocytes (PMNs) and in neutrophil-like differentiated and non-differentiated HL60 cells treated with TNFα, but not in macrophage-like differentiated HL60 cells and other cells, such as A498, ECV304, HeLa, U937 and HEK293 cells [27]. On the other hand, human peripheral PMNs up-regulated the expression level of PHGPx in the stimulation of TNFα but not IL-1β, IL-8 and GRO. The TNFα induced up-regulation of PHGPx in neutrophil-like differentiated HL60 cells was inhibited by incubation with the antioxidants NAC (N-acetylcysteine), PDTC (pyrolidine carbodithioate), Bay 11-7082 (NFκB inhibitors) and PP2 (Src kinase inhibitors), indicating the involvement of NFκB and/or Src kinase activation through ROS signaling in the up-regulation of PHGPx [28]. The up-regulation of PHGPx, however, was not suppressed by treatment with MAPK (mitogen-activated protein kinase) cascade inhibitors, which are downstream targets of Src kinases and no binding sequence for NFκB exists in the 5' untranslated region of the promoter region in the human PHGPx gene.
We investigated the regions responsible for up-regulation in the 5'-flanking region of the human and mouse PHGPx genes and the transcription factors associated with TNFα-induced up-regulation of PHGPx expression using deletion and mutational analysis of the promoter activity [29]. Compared with the non-stimulated controls, the promoter activity was up-regulated by TNFα stimulation in HL60 cells transfected with a luciferase reporter vector encoding the region from −282 to −123 bp of human PHGPx. On the other hand, the promoter region from −113 to −56 bp in mice was critical for TNFα-induced up-regulation of PHGPx promoter activity in HL60 cells. Since the region responsible for the TNFα-induced up-regulation of the promoter activity in humans was different from that of mice, the sequence of the region necessary for TNFα-induced up-regulation in humans was compared with that of mice. In a search for a binding site for transcription factors using the TRANSFAC database, the homologous sequence was matched to a C/EBP-binding sequence. The up-regulated promoter activity was effectively abrogated by a mutation in the C/EBP (CCAAT/enhancer-binding protein)-binding sequence in this region. ChIP (chromatin immunoprecipitation) assays demonstrated that C/EBPε bound to the −247 to −34 bp region in HL60 cells, but C/EBPα, β, γ and δ did not. The binding of C/EBPε to the promoter region was increased in HL60 cells stimulated with TNFα compared with non-stimulated control cells. An increased binding of nuclear protein to the C/EBP-binding sequence was observed by EMSA (electrophoretic mobility shift assay) in cells stimulated with TNFα, and this binding of nuclear protein to the C/EBP-binding sequence was inhibited by pre-treatment with an anti-C/EBPε antibody, but not with other antibodies. The TNFα-induced up-regulation of the promoter activity was inhibited by treatment with 50 mM NAC and 1 µM Bay 11-7082 (NFκB inhibitors), but not with 10 nM PP2 (Src kinase inhibitors). The C/EBPε mRNA was expressed in PMNs, non-differentiated HL60 cells and neutrophil-like differentiated HL60 cells displaying TNFα-induced up regulation of PHGPx mRNA, but not in macrophage-like differentiated HL60 cells, HEK293 cells and other cell lines exhibiting no up-regulation. The up-regulation of PHGPx mRNA, however, was detected in HEK-293 cells overexpressing C/EBPε as a result of TNFα stimulation. These results indicate that C/EBPε is a critical transcription factor in TNFα-induced up-regulation of PHGPx expression. Our result is the first evidence in identifying the transcriptional factors involved in regulating the expression of non-mitochondrial and mitochondrial PHGPx mRNA.
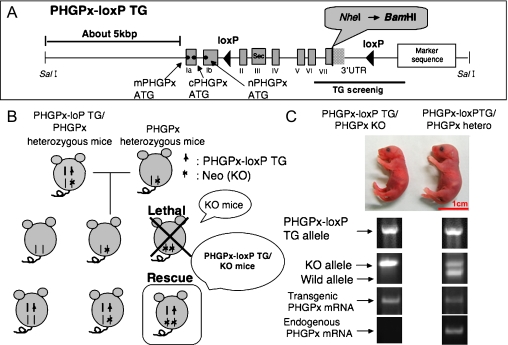
Embryonic Lethality by Disruption of All PHGPx Genes in Mice
To examine the role of PHGPx in development, we and other groups generated mice deficient in PHGPx by targeted disruption of all exons of the PHGPx gene [25, 30]. As shown in Fig. 4, a targeting vector was constructed in which all exons of the mouse PHGPx gene, including the entire PHGPx coding region for the three types of PHGPx, were depleted and replaced with a PGK-Neo cassette. The targeting vector was introduced into ES cells by electroporation and candidate ES cell clones. Five candidates were verified to be homologous recombinants and were injected into blastocytes of ICR mice. Two independent ES cell clones resulted in chimeric mice by transmitting the mutation to be the germ-line. PHGPx heterozygotes are viable, fertile, and appear normal, despite having decreased levels of the three types of PHGPx mRNA and protein. As shown in Fig. 4B, embryos homozygous for PHGPx-null die between 7.5 and 8.5 days post coitum (dpc), probably due to the development of distal apoptosis. We examined the expression of PHGPx in mouse embryos using immunohistochemical analysis with anti-PHGPx mAb. Expression of PHGPx was detected in the embryonic ectoderm and the yolk sac membrane at 7.5 dpc and 8.5 dpc. These results demonstrate that PHGPx is expressed in early gastrulation stage at 7.5 dpc and the expression of PHGPx is essential for normal mouse development. The absence of a lethal phenotype in PHGPx homozygous mice is similar to findings in γ-glutamylcysteine synthetase knockout mice [31]. γ-Glutamylcysteine synthetase is an essential enzyme in glutathione synthesis; glutathione being a major source of reducing equivalents such as PHGPx and cGPx in mammalian cells. Embryos homozygous for γ-glutamylcysteine synthetase fail to gastrulate, do not form mesoderm, develop distal apoptosis, and die before 8.5 dpc. Recently we have shown that 3.5 dpc PHGPx knockout embryos could not form inner cell mass and died. These results demonstrate that the scavenging of reactive oxygen species by PHGPx in embryos is required for embryonic development.
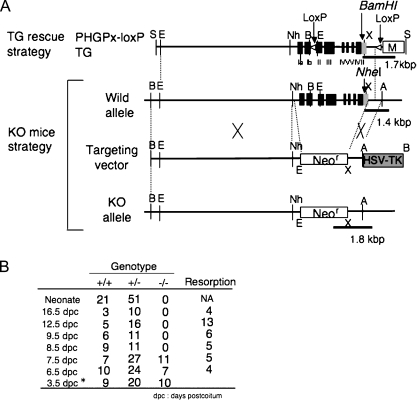
Fig. 4.
Early embryonic lethality caused by targeted disruption of the mouse PHGPx gene. (A) Knockout targeting strategy by homologous recombination in ES cells showing the structures of the wild-type PHGPx allele, the targeting vector and the targeted allele (KO allele). All eight exons of PHGPx were replaced by Neo gene cassettes in the targeted allele. PHGPx-loxP TG vector (Top) was used by the transgenic rescue method as shown in Fig. 5. The sites indicated by black bars were used as PCR screening sites to detect the each allele. (B) Genotype analysis of PHGPx heterozygous mice intercross progeny. PHGPx KO embryos died between 7.5 dpc and 8.5 dpc.
The Transgenic Complementary Rescue Method using a PHGPx-LoxP Transgene rescued embryonic Lethality in Endogenous PHGPx KO Mice
To clarify the physiological role of PHGPx in mice, we created a conditional knockout (KO) mouse line for the PHGPx gene [32]. As we have demonstrated that depletion of all exons of the PHGPx gene in mice by homologous recombination induced early embryonic lethality at 7.5 dpc (Fig. 4), we generated conditional KO mice using a transgenic complementation rescue method to obtain floxed PHGPx mice (PHGPx-LoxP TG/KO mice) carrying the mouse PHGPx transgene (PHGPx-loxP TG; Fig. 5A) located between two loxP sites. We first generated eight independent PHGPx-loxP transgenic mouse lines (PHGPx-loxP TG/wild mice) expressing PHGPx regulated by a 5.0 kbp upstream sequence and 1.0 kbp downstream sequence of the PHGPx gene (PHGPx-loxP TG; Fig. 5A). As shown in Fig. 5B, these mice were crossed with PHGPx heterozygous mice (PHGPx+/−) to obtain PHGPx-loxP TG/PHGPx+/− mice (PHGPx-loxP/hetero mice), which were further crossed with PHGPx+/− mice to generate PHGPx-loxP/PHGPx−/− mice (PHGPx-loxP TG/KO mice). Genotyping analysis in mouse tail revealed that we successfully bred PHGPx-loxP TG/KO mice (Fig. 5C). To distinguish PHGPx mRNA derived from the endogenous PHGPx gene and the PHGPx-loxP transgene, the NheI site after the stop codon in the endogenous PHGPx gene was replaced with a BamHI site in the PHGPx-loxP transgene (Fig. 5A). We examined the expression of PHGPx mRNA from transgenic and endogenous PHGPx genes in brain tissue of PHGPx-loxP TG/KO and PHGPx-loxP TG/hetero mouse pups by RT-PCR using endogenous (NheI site) and transgene- (BamHI site) specific antisense primers and a common forward primer (Fig. 5A and C). The PHGPx-loxP TG/KO mice expressed PHGPx mRNA derived from only PHGPx-loxP transgene, but not from the endogenous PHGPx gene (Fig. 5C). By contrast, the PHGPx-loxP TG/hetero mice expressed PHGPx mRNA derived from both the PHGPx-loxP transgene and the endogenous PHGPx gene (Fig. 5C). These results indicate that the PHGPx-loxP transgene may prevent embryonic lethality in PHGPx KO mice. Thus we succeeded in establishing three lines (Fα, Fκ and Fγ) of PHGPx-loxP TG/KO mice that were viable and fertile, with body weights indistinguishable from those of wild-type mice.
Fig. 5.
Generation of floxed PHGPx mice by rescue of embryonic lethality in PHGPx knockout mice using the PHGPx-loxP transgene. (A) Structure of the PHGPx-loxP transgene. In the PHGPx-loxP transgene, mouse genomic DNA including approx. 5 kbp of the PHGPx promoter region, eight PHGPx exons and approx. 1 kbp of the 3' region was used. The NheI site after the stop codon in the wild-type allele was replaced with a BamHI site in the PHGPx-loxP transgene. Sec denotes selenocysteine, which is the enzymatically active site in Exon III. Marker sequence was used by PCR analysis for TG screening. Two loxP sites were inserted between exon Ib and exon II and between exon IV and the Marker sequence. (B) Transgenic complementary rescue method. Eight PHGPx transgenic mice injected with the PHGPx-loxP transgene were crossed with PHGPx+/− mice to obtain PHGPx-loxP TG/PHGPx+/− mice, which were further crossed with PHGPx+/− mice to generate PHGPx-loxP transgene rescued PHGPx−/− mice (PHGPx-loxP TG/KO mice). (C) Genotyping analysis of mouse pups using the probes in mouse tail revealed that we successfully created PHGPx-loxP TG/KO mice. We detected only the PHGPx-loxP transgene-derived PHGPx mRNA from PHGPx-loxP TG/KO mice, but not endogenous PHGPx gene derived PHGPx mRNA.
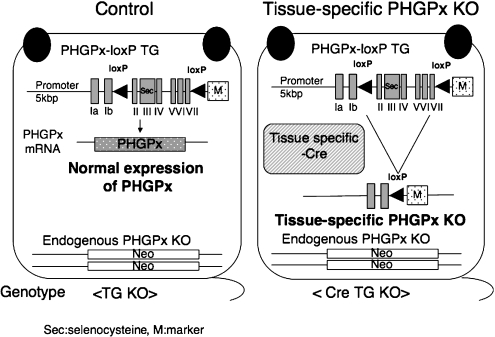
As shown in Fig. 6, we established floxed PHGPx mice (PHGPx-loxP TG/KO mice; control mice) using the transgenic complementation rescue method [32]. Transgenically rescued floxed mice exhibited normal growth and fertility. The expression of PHGPx protein in several tissues and the distribution of PHGPx in seminiferous tubules of testes was almost the same as in wild type mice. These results show that the approximately 5 kbp promoter regions in the PHGPx-loxP transgene are sufficient for normal regulation of PHGPx expression in embryogenesis and spermatogenesis in control mice. Our results also demonstrate that the transgenic complementation rescue method using this PHGPx-loxP transgene is useful for functional analysis of the three isoforms of PHGPx in mice by mutation of their start codons. The control floxed mice are useful for generating tissue-specific PHGPx conditional KO mice by mating these mice with tissue-specific Cre expressing PHGPx heterozygous mice. Because tissue specific Cre recombinase can disrupt the PHGPx-loxP transgene in PHGPx-loxP TG/KO mice, we could easily generate the tissue-specific PHGPx KO mice using this system (Fig. 6).
Fig. 6.
Our established transgenic rescued floxed PHGPx knockout mice (Control mice) and tissue specific conditional PHGPx knockout mice by deletion of PHGPx-loxP transgene using Cre-loxP system. We established three lines of PHGPx-loxP TG/KO mice (TG KO, Control mice), all of which were viable and fertile, with body weights indistinguishable from those of wild-type mice. The PHGPx-loxP transgene could transcribe only PHGPx mRNA in the PHGPx-loxP TG/KO mice at nearly the same level as in the wild-type mice. Using a Cre-loxP conditional knockout strategy, we could generate tissue-specific PHGPx knockout mice (Cre TG KO mice) by mating PHGPx-loxP TG/KO mice with tissue-specific Cre expressing PHGPx heterozygous mice. In tissue specific PHGPx knockout mice, PHGPx-loxP transgene was specifically disrupted by tissue specific Cre recombinase.
Failure of Expression of PHGPx in the Spermatozoa of Human Infertile Males
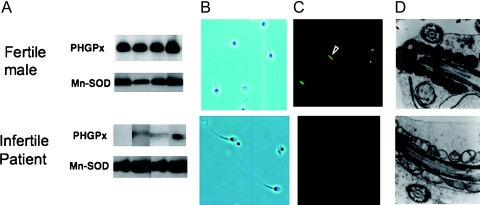
We were the first to show a dramatic decrease in the PHGPx expression in the spermatozoa of 7 individuals in a group of 73 infertile males by immunoblotting with anti-PHGPx monoclonal antibodies (Fig. 7A) [19]. All 7 patients with PHGPx-defective spermatozoa were classified as suffering from oligoasthenozoospermia, a defect in which both the number and the motility of spermatozoa are significantly below normal. Males with PHGPx-defective spermatozoa accounted for 26% of the 27 infertile males with oligoasthenozoospermia. No defects in expression of PHGPx in spermatozoa were observed in 31 fertile volunteers. PHGPx-deficient spermatozoa possessed a full form and extended flagellum (Fig. 7B). After a 3 h incubation in the medium, however, the motility of PHGPx-deficient spermatozoa was significantly lower than that of spermatozoa with normal expression of PHGPx. The PHGPx-defective spermatozoa failed to incorporate rhodamine 123, revealing a loss of mitochondrial membrane potential (Fig. 7C). Ultrastructual analysis of mitochondria by electron microscopy demonstrated that morphology of mitochondria in PHGPx-defective spermatozoa was abnormal (Fig. 7D). These results suggest that failure of the expression of mitochondrial PHGPx in spermatozoa might be one of the causes of oligoasthenozoospermia in infertile men. Other groups have also reported that reduced PHGPx expression in spermatozoa is associated with male infertility [33, 34].
Fig. 7.
Failure of expression of PHGPx in spermatozoa is associated with human male infertility with oligoasthenozoospermia. (A) Immunoblot analysis indicated low levels of PHGPx in spermatozoa from infertile males. (B) Confocal images showed that PHGPx-deficient spermatozoa possessed a full form and extended flagellum. (C) Fluorescence microscopy after DiOC6 (green) and Hoechst 33258 (blue) staining indicated significantly low levels of mitochondrial membrane potential in PHGPx deficient spermatozoa 5 h after ejaculation. (D) Ultrastructural analysis of mitochondria by electron microscopy demonstrated that the morphology of mitochondria in PHGPx-deficient spermatozoa was abnormal.
Depletion of PHGPx in Spermatocytes Causes Male Infertility with Oligoasthenozoospermia in Mice
To clarify whether defective GPx4 in spermatocytes causes male infertility, we established spermatocyte-specific GPx4 knockout mice using a Cre-loxP system [32].
We used PHGPx-loxP TG/KO mice as control normal floxed mice (Fig. 6) to make spermatocyte-specific GPx4 knockout mice. Using a Cre-loxP conditional knockout strategy, we mated PHGPx-loxP TG/KO mice with spermatocyte specific pgk-2 promoter-driven Cre PHGPx+/− mice to generate spermatocyte-specific PHGPx knockout mice (CRE TG KO) (Fig. 6). Phosphoglycerate kinase-2 (pgk-2) is expressed specifically in testicular germ cells [35]. Previous data indicated that Cre recombinase activity in a transgenic line possessing pgk-2-driven expression of the Cre recombinase was present in spermatocytes and spermatogenic cells at later differentiation stages [35].
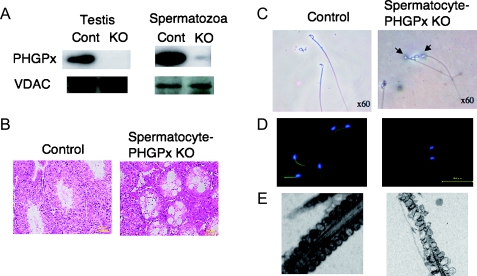
We successfully created spermatocyte-specific PHGPx KO mice (Fig. 8). The expression of PHGPx protein in spermatocyte-specific PHGPx KO mice was significantly decreased in testis and epididymal spermatozoa (Fig. 8A), but not in liver, brain and kidney, although the expression of voltage dependent anion channels (VDAC) was not changed. These results demonstrate that PHGPx expression is selectively lost in spermatocytes and spermatozoa of spermatocyte-specific PHGPx KO mice.
Fig. 8.
Depletion of PHGPx gene in spermatocyte causes male infertility with oligoasthenozoospermia in mice. Mice with spermatocyte-specific deletion of the PHGPx gene (A) showed male infertility with two significant conspicuous phenotypes: first, a decreased number of spermatozoa in the epididymis caused by the depletion of spermatogenic cells in seminiferous tubules (B) and second, loss of forward motility of spermatozoa due to hairpin-like bending of the tail in the distal midpiece region (C) and/or mitochondrial dysfunction including mitochondrial membrane potential (D) and ultrastructure (E).
All the spermatocyte-specific PHGPx knockout male mice were found to be infertile in spite of normal plug formation after mating and displayed a significant decrease in the number of spermatozoa. They had a decreased number of spermatozoa in the epididymis caused by the depletion of spermatogenic cells in the seminiferous tubules (Fig. 8B). Isolated epididymal PHGPx KO spermatozoa showed the structural abnormality, such as a hairpin-like flagella bend at the midpiece (Fig. 8C), significant reduction in mitochondrial membrane potential (Fig. 8D) and swelling of mitochondria in the spermatozoa (Fig. 8E), resulting in the significant reduction in the forward motility of spermatozoa. Isolated epididymal PHGPx null spermatozoa could not fertilize oocytes in vitro. These results demonstrate that depletion of PHGPx in spermatocytes causes severe abnormalities in the spermatozoa, which may be one of the causes of male infertility in mice and humans.
Cell Growth in Mouse Embryonic Cells (MEF) is Necessary for the Inhibition of Lipid Peroxidation by the Activity of Non-mitochondrial PHGPx and/or Excess of Vitamin E
We investigated the effects of depletion of PHGPx on the viability of MEF (mouse embryonic fibroblasts) obtained from PHGPx-loxP TG/KO mice (Fig. 9). The PHGPx-loxP transgene in MEF was depleted by infection of retroviruses harboring Cre and the puromycin-resistant gene (Fig. 9A). Retrovirus-mediated depletion of the PHGPx-loxP TG transgene resulted in severe cell death 3 days after infection (Fig. 9B). Addition of 400 µM Trolox, a vitamin E derivative, or 200 µM vitamin E into culture media rescued the cell death induced by retrovirus-Cre mediated depletion of PHGPx-loxP transgene (Fig. 9C). PHGPx protein expression was not observed in PHGPx depleted MEF cells rescued by addition of Trolox and Vitamin E (Fig. 9D).
Fig. 9.
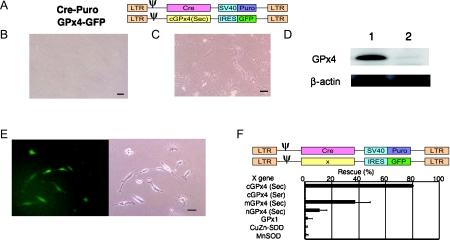
Depletion of PHGPx gene by retroviral Cre expression induces cell death in PHGPx-loxP TG/KO (Control) MEFs. (A) The structure of retrovirus vectors with Cre cDNA carrying the SV40 promoter-regulated puromycin-resistance gene (Cre-Puro) and retrovirus with non-mitochondrial GPx4 carrying IRES regulated GFP (cGPx4-GFP). LTR: Long terminal repeat; ψ: packaging signal; IRES: internal ribosomal entry site; GFP: green fluorescent protein; Puro: puromycin-resistance gene; SV40: simian virus 40 early enhancer/promoter region. MEFs isolated from PHGPx-loxP TG/KO mice were infected with Cre-Puro retrovirus (A) and cultured with 5 µg/ml puromycin for 4 days. (B) Cre-expressing PHGPx-loxP TG/KO MEF cells did not grow. (C) Addition of Trolox into culture medium rescued cell death induced by depletion of GPx4 gene in Cre-expressing PHGPx-loxP TG/KO MEF cells. (D) Expression of PHGPx and β-actin protein in PHGPx-loxP TG/KO MEF cells (1) and Trolox rescued Cre-expressing PHGPx-loxP TG/KO MEF cells (2) by immunoblot analysis with antibodies specific to each protein. (E) Non-mitochondrial PHGPx-expressing retroviral infection rescued cell death in Cre-expressing PHGPx-loxP TG/KO MEF cells. Despite Cre expression, only GFP-positive PHGPx-loxP TG/KO MEF cells expressing PHGPx could grow in puromycin selection medium. (F) Overexpression of several other antioxidant enzymes could not rescue PHGPx depletion-induced cell death. PHGPx-loxP TG/KO MEF cells infected by several antioxidant enzymes expressing retrovirus carrying IRES regulated GFP, were infected simultaneously with Cre-expressing retrovirus (Cre-Puro) and cultured with 5 µg/ml puromycin for 4 days. The percentage of rescued GFP-positive cells by retroviral infections with several antioxidants-IRES-GFP genes and Cre-SV40-Puro gene was calculated relative to total GFP-positive cells by retroviral infections with several antioxidants-IRES-GFP gene and control SV40-Puro gene without Cre cDNA. Scale bars equal 50 µm. Data are shown as mean ± S.D. (n = 4). cGPx4 (Sec); non-mitochondrial PHGPx (selenocysteine); cGPx4 (Ser); non-mitochondrial PHGPx (serine); mGPx4; mitochondrial PHGPx; nGPx4; nucleolar PHGPx; GPx1: cytosolic glutathione peroxidase.
To examine which PHGPx isoform contributes to the viability of MEF (murine embryonic fibroblasts), the cDNA of each isoform was introduced into another retrovirus vector. The GFP gene was also inserted into the vector to detect the MEF possessing this retrovirus gene. We found that co-infection of the viruses carrying Cre or the cDNA of non-mitochondrial PHGPx, which causes the expression of only the non-mitochondrial PHGPx in MEF, efficiently rescued the viability (Fig. 9E). Other isoforms such as the mitochondrial and nucleolar forms also rescued viability, but the efficiency was not comparable to the non-mitochondrial type. PHGPx has a selenocysteine residue at its active site, and changing this to serine causes a complete loss of activity. Expression of an enzymatically inactive form of PHGPx did not rescue the viability of PHGPx-depleted MEF (Fig. 9F). We also found that retrovirus-mediated expression of other antioxidant enzymes such as cytosolic glutathione peroxidase (GPx1), Cu,Zn-superoxide dismutase (CuZn-SOD) and mitochondrial Mn-superoxide dismutase (MnSOD) could not rescue the viability of PHGPx-depleted MEF. These results indicate that the cell growth in MEF cells is necessary for PHGPx activity and/or excess of vitamin E, indicating that inhibition of lipid peroxidation is essential for cell fate and cell growth.
Conclusions
Phospholipid hydroperoxide glutathione peroxidase (PHGPx) is part of the selenoprotein family. Computational genome-wide analysis identified only 24 genes for selenoproteins in mice [36]. A specific tRNA, (tRNASec(Ser), Trsp), which incorporates selenocysteine during translation, mediates the biosynthesis of PHGPx and other selenoproteins. Trsp knockout (KO) mice are embryonic lethal at 3.5 dpc [37]. Liver-specific Trsp KO mice die within 1 to 3 months after birth due to severe hepatocellular degradation and necrosis [38]. Macrophage-specific Trsp KO mice exhibit high levels of oxidative stress [39]. Endothelial restricted Trsp KO mice show an overall poorly developed vascular system, develop severe developmental abnormalities in limb, tails and head from 12.5 dpc and eventually die before 18.5 dpc [40]. Muscle cell specific Trsp KO mice exhibit acute myocardial failure around 12 days after birth [40]. These results indicate that one or more selenoproteins may play an important role in each tissue.
To clarify the physiological role of PHGPx in several tissues, we first created floxed PHGPx mice by the transgenic rescue method. Transgenically rescued floxed mice exhibited normal growth and fertility. Since the PHGPx-loxP transgene was effectively deleted in the testes of spermatocyte-specific PHGPx KO mice by Cre expression under the control of the pgk-2 promoter, our established floxed mice are useful for generation of other tissue specific PHGPx KO mice. The sperm abnormalities and disorders of seminiferous tubules in spermatocyte-specific PHGPx KO mice were restored by the gain of just one copy of the endogenous PHGPx gene in PHGPx-loxP TG/PHGPx heterozygous mice, confirming that the sperm abnormalities and disorders of seminiferous tubules were directly caused by the absence of the PHGPx gene [32]. We also generated PHGPx-loxP TG mice (PHGPx overexpressing transgenic mice). The results from those studies demonstrate that the PHGPx overexpressing transgenic mice and PHGPx floxed mice can be used for functional analysis of PHGPx in other tissues of mice using the tissue-specific Cre–loxP system.
The PHGPx-loxP transgene is useful for functional analysis of the three isoforms of PHGPx in mice by mutation of their start codons. We recently generated specific PHGPx KO mice for each type of PHGPx by the transgenic complementary rescue method. These new mice lines are useful for the functional analysis of three types of PHGPx in mice in the future.
Acknowledgments
We are most grateful to H. Hattori, N. Kirai, J. Suzuki, T. Suzuki, N. Hakkaku, M. Sugimoto, Y. Tajima, K. Konishi, R. Iwamoto, R. Suzuki, S. Minami, N. Okawa, S. Harada, M. Matsuoka and H. Ogawa for excellent technical assistance. We greatly thank Prof. Y. Nakagawa for supporting our experiments. This work was supported by grant-in-aid no. 20590067 from the Ministry of Education, Science and Culture of Japan, by PRESTO from the Japan Science and Technology Agency, and by SHISEIDO Grant for Science Research.
Biography
Dr. Hirotaka Imai is an associate professor at the School of Pharmaceutical Scieneces of Kitasato University at Tokyo. He is a researcher in PRESTO (2006–2010). He received his Ph.D with Dr. K. Inoue at Tokyo University in 1993 and then moved to Prof. Nakagawa’s Laboratory in Kitasato University in 1993 and started the project of the biological role of phospholipid hydroperoxide glutathione peroxidase and lipid hydroperoxide.
References
- 1.Rhee S.G. Cell signaling. H2O2, a necessary evil for cell signaling. Science. 2006;312:1882–1883. doi: 10.1126/science.1130481. [DOI] [PubMed] [Google Scholar]
- 2.Imai H., Nakagawa Y. Biological significance of phospholipid hydroperoxide glutathione peroxidase (PHGPx, GPx4) in mammalian cells. Free Radic. Biol. Med. 2003;34:145–169. doi: 10.1016/s0891-5849(02)01197-8. [DOI] [PubMed] [Google Scholar]
- 3.Imai H., Saito M., Kirai N., Hasegawa J., Konishi K., Hattori H., Nishimura M., Naito S., Nakagawa Y. Identification of the positive regulatory and distinct core regions of promoters, and transcriptional regulation in three types of mouse phospholipid hydroperoxide glutathione peroxidase. J. Biochem. 2006;140:573–590. doi: 10.1093/jb/mvj186. [DOI] [PubMed] [Google Scholar]
- 4.Imai H., Sumi D., Hanamoto A., Arai M., Sugiyama K., Chiba N., Kuchino Y., Nakagawa Y. Molecular cloning and functional expression of cDNA for rat phospholipid hydroperoxide glutathione peroxidase: 3'-untranslated region of the gene is necessary for functional expression. J. Biochem. 1995;118:1061–1067. doi: 10.1093/jb/118.5.1061. [DOI] [PubMed] [Google Scholar]
- 5.Arai M., Imai H., Koumura T., Madoka Y., Emoto K., Umeda M., Chiba N., Nakagawa Y. Mitochondrial phospholipid hydroperoxide glutathione peroxidase (PHGPx) play a major role in preventing oxidative injury to cells. J. Biol. Chem. 1999;274:4924–4933. doi: 10.1074/jbc.274.8.4924. [DOI] [PubMed] [Google Scholar]
- 6.Arai M., Imai H., Sumi D., Imanaka T., Takano T., Chiba N., Nakagawa Y. Import into Mitochondria of Phospholipid Hydroperoxide Glutathione Peroxidase Requires a Leader Sequence. Biochem. Biophys. Res. Commun. 1996;227:433–439. doi: 10.1006/bbrc.1996.1525. [DOI] [PubMed] [Google Scholar]
- 7.Pfeifer H., Conrad M., Roethlein D., Kyriakopoulos A., Brielmeier M., Bornkamm G.W., Behne D. Identification of a specific sperm nuclei selenoenzyme necessary for protamine thiol cross-linking during sperm maturation. FASEB J. 2001;15:1236–1238. [PubMed] [Google Scholar]
- 8.Borchert A., Schnurr K., Thiele B.J., Kühn H. Cloning of the mouse phospholipid glutathione peroxidase gene. FEBS Lett. 1999;446:223–227. doi: 10.1016/s0014-5793(99)00221-5. [DOI] [PubMed] [Google Scholar]
- 9.Nakamura T., Imai H., Tsunashima N., Nakagawa Y. Molecular cloning and functional expression of nucleolar phospholipid hydroperoxide glutathione peroxidase in mammalian cells. Biochem. Biophys. Res. Commun. 2003;311:139–148. doi: 10.1016/j.bbrc.2003.09.183. [DOI] [PubMed] [Google Scholar]
- 10.Imai H., Narashima K., Sakamoto H., Chiba N., Nakagawa Y. Suppression of leukotriene formation in RBL2H3 cells that overexpressed phospholipid hydroperoxide glutathione peroxidase. J. Biol. Chem. 1998;273:1990–1997. doi: 10.1074/jbc.273.4.1990. [DOI] [PubMed] [Google Scholar]
- 11.Imai H., Sumi D., Sakamoto H., Hanamoto A., Arai M., Chiba N., Nakagawa Y. Overexpression of phospholipid hydroperoxide glutathione peroxidase suppressed cell death due to oxidative damage in rat basophile leukemia cells (RBL-2H3) Biochem. Biophys. Res. Commun. 1996;222:432–438. doi: 10.1006/bbrc.1996.0762. [DOI] [PubMed] [Google Scholar]
- 12.Sakamoto H., Imai H., Nakagawa Y. Involvement of phospholipid hydroperoxide glutathione peroxidase in the modulation of prostaglandin D2 synthesis. J. Biol. Chem. 2000;275:40028–40035. doi: 10.1074/jbc.M003191200. [DOI] [PubMed] [Google Scholar]
- 13.Sakamoto H., Tosaki T., Nakagawa Y. Overexpression of phospholipid hydroperoxide glutathione peroxidase modulates acetyl-CoA, 1-O-alkyl-2-lyso-sn-glycero-3-phosphocholine acetyltransferase activity. J. Biol. Chem. 2002;277:50431–50438. doi: 10.1074/jbc.M204190200. [DOI] [PubMed] [Google Scholar]
- 14.Nomura K., Imai H., Koumura T., Arai M., Nakagawa Y. Mitochondrial phospholipid hydroperoxide glutathione peroxidase suppresses apoptosis mediated by a mitochondrial-death pathway. J. Biol. Chem. 1999;274:29294–29302. doi: 10.1074/jbc.274.41.29294. [DOI] [PubMed] [Google Scholar]
- 15.Nomura K., Imai H., Koumura T., Kobayashi T., Nakagawa Y. Mitochondrial phospholipid hydroperoxide glutathione peroxidase inhibits the release of cytochrome c from mitochondria by suppressing the peroxidation of cardiolipin in hypoglycemia induced apoptosis. Biochem. J. 2000;351:183–193. doi: 10.1042/0264-6021:3510183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Imai H., Koumura T., Nakajma R., Nomura K., Nakagawa Y. Protection of inactivation of adenine nucleotide translocator during hypoglycemia-induced apoptosis by mitochondrial phospholipid hydroperoxide glutathione peroxidase. Biochem. J. 2003;371:799–809. doi: 10.1042/BJ20021342. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Nomura K., Imai H., Koumura T., Nakagawa Y. Involvement of mitochondrial phospholipid hydroperoxide glutathione peroxidase as an antiapoptotic factor. Biol. Signals Recept. 2001;10:81–92. doi: 10.1159/000046877. [DOI] [PubMed] [Google Scholar]
- 18.Otsu K., Sato K., Ikeda Y., Imai H., Nakagawa Y., Ohba Y., Fujii J. An abortive apoptotic pathway induced by singlet oxygen is due to the suppression of caspase activation. Biochem. J. 2005;389:197–206. doi: 10.1042/BJ20042067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Imai H., Suzuki K., Ishizaka K., Ichinose S., Oshima H., Okayasu I., Emoto K., Umeda M., Nakagawa Y. Failure of the expression of phospholipid hydroperoxide glutathione peroxidase (PHGPx) in the spermatozoa of human infertile males. Biol. Reprod. 2001;64:674–683. doi: 10.1095/biolreprod64.2.674. [DOI] [PubMed] [Google Scholar]
- 20.Ursini F., Heim S., Kiess M., Maiorino M., Roveri A., Wissing J., Flohé L. Dual function of the selenoprotein PHGPx during sperm maturation. Science. 1999;285:1393–1396. doi: 10.1126/science.285.5432.1393. [DOI] [PubMed] [Google Scholar]
- 21.Maiorino M., Wissing J.B., Brigelius-Flohé R., Calabrese F., Roveri A., Steinert P., Ursini F., Flohé L. Testosterone mediates expression of the selenoprotein PHGPx by induction of spermatogenesis and not by direct transcriptional gene activation. FASEB J. 1988;12:1359–1370. doi: 10.1096/fasebj.12.13.1359. [DOI] [PubMed] [Google Scholar]
- 22.Ufer C., Borchert A., Kuhn H. Functional characterization of cis- and trans-regulatory elements involved in expression of phospholipid hydroperoxide glutathione peroxidase. Nucleic Acids Res. 2003;31:4293–4303. doi: 10.1093/nar/gkg650. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Borchert A., Savaskan N.E., Kuhn H. Regulation of expression of the phospholipid hydroperoxide/sperm nucleus glutathione peroxidase gene. Tissue-specific expression pattern and identification of functional cis- and trans-regulatory elements. J. Biol. Chem. 2003;278:2571–2580. doi: 10.1074/jbc.M209064200. [DOI] [PubMed] [Google Scholar]
- 24.Tramer F., Vetere A., Martinelli M., Paroni F., Marsich E., Boitani C., Sandri G., Panfili E. cAMP-response element modulator-tau activates a distinct promoter element for the expression of the phospholipid hydroperoxide/sperm nucleus glutathione peroxidase gene. Biochem. J. 2004;383:179–185. doi: 10.1042/BJ20040974. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Imai H., Hirao F., Sakamoto T., Sekine K., Mizukura Y., Saito M., Kitamoto T., Hayasaka K., Nakagawa Y. Early embryonic lethality caused by targeted disruption of the mouse PHGPx gene. Biochem. Biophys. Res. Commun. 2003;305:278–286. doi: 10.1016/s0006-291x(03)00734-4. [DOI] [PubMed] [Google Scholar]
- 26.Hattori H., Imai H., Hanamoto K., Furuhama K., Nakagawa Y. Up regulation of phospholipid hydroperoxide glutathione peroxidase in rat casein-induced polymorphonuclear neutrophils. Biochem. J. 2005;389:279–287. doi: 10.1042/BJ20050006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Schnurr K., Borchert A., Kuhn H. Inverse regulation of lipid-peroxidizing and hydroperoxyl lipid-reducing enzymes by interleukins 4 and 13. FASEB J. 1999;13:143–154. doi: 10.1096/fasebj.13.1.143. [DOI] [PubMed] [Google Scholar]
- 28.Hattori H., Imai H., Furuhama K., Sato O., Nakagawa Y. Induction of phospholipid hydroperoxide glutathione peroxidase in human polymorphonuclear neutrophils and HL60 cells stimulated with TNFα. Biochem. Biophys. Res. Commun. 2005;337:464–473. doi: 10.1016/j.bbrc.2005.09.076. [DOI] [PubMed] [Google Scholar]
- 29.Hattori H., Imai H., Kirai N., Furuhama K., Sato O., Konishi K., Nakagawa Y. Identification of a responsible promoter region and a key transcription factor, CCAAT/enhancer-binding protein epsilon, for up-regulation of PHGPx in HL60 cells stimulated with TNF-alpha. Biochem. J. 2007;408:277–286. doi: 10.1042/BJ20070245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Yant L.J., Ran Q., Rao L., Van Remmen H., Shibatani T., Belter J.G., Motta L., Richardson A., Prolla T.A. The selenoprotein GPX4 is essential for mouse development and protects from radiation and oxidative damage insults. Free Radic. Biol. Med. 2003;34:496–502. doi: 10.1016/s0891-5849(02)01360-6. [DOI] [PubMed] [Google Scholar]
- 31.Shi S.S., Osei-Frimpong J., Kala G., Kala S.V., Barrios R.J., Habib G.M., Lukin D.J., Danney C.M., Matzuk M.M., Lieberman M.W. Glutathione synthesis is essential for mouse development but not for cell growth in culture. Proc. Natl. Acad. Sci. U.S.A. 2000;97:5101–5106. doi: 10.1073/pnas.97.10.5101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Imai H., Hakkaku N., Iwamoto R., Suzuki J., Suzuki T., Tajima T., Konishi K., Minami S., Ichinose S., Ishizaka K., Shioda S., Arata S., Nishimura M., Naito S., Nakagawa Y. Depletion of Selenoprotein GPx4 in spermatocytes causes male infertility in mice. J. Biol. Chem. 2009;284:32522–32532. doi: 10.1074/jbc.M109.016139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Foresta C., Flohé L., Garolla A., Roveri A., Ursini F., Maiorino M. Male fertility is linked to the selenoprotein phospholipid hydroperoxide glutathione peroxidase. Biol. Reprod. 2002;67:967–971. doi: 10.1095/biolreprod.102.003822. [DOI] [PubMed] [Google Scholar]
- 34.Diaconu M., Tangat Y., Böhm D., Kühn H., Michelmann H.W., Schreiber G., Haidl G., Glander H.J., Engel W., Nayernia K. Failure of phospholipid hydroperoxide glutathione peroxidase expression in oligoasthenozoospermia and mutations in the PHGPx gene. Andrologia. 2006;38:152–157. doi: 10.1111/j.1439-0272.2006.00729.x. [DOI] [PubMed] [Google Scholar]
- 35.Kido T., Arata S., Suzuki R., Hosono T., Nakanishi Y., Miyazaki J., Saito I., Kuroki T., Shioda S. The testicular fatty acid binding protein PERF15 regulates the fate of germ cells in PERF15 transgenic mice. Develop. Growth Differ. 2005;47:15–24. doi: 10.1111/j.1440-169x.2004.00775.x. [DOI] [PubMed] [Google Scholar]
- 36.Kryukov G.V., Castellano S., Novoselv A.V., Lobanov A.V., Zehtab O., Guigo R., Gladyshev V.N. Characterization of mammalian selenoproteomes. Science. 2003;300:1439–1443. doi: 10.1126/science.1083516. [DOI] [PubMed] [Google Scholar]
- 37.Bosl M.R., Takaku K., Oshima M., Nishimura S., Taketo M.M. Early embryonic lethality caused by targeted disruption of the mouse selenocysteine tRNA gene (Trsp) Proc. Natl. Acad. Sci. U.S.A. 1997;94:5531–5534. doi: 10.1073/pnas.94.11.5531. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Carlson B.A., Novoselov S.V., Kumarsawamy E., Lee B.J., Anver M.R., Gladyshev V.N., Hatfield D.L. Specific excision of the selenocysteine tRNA[Ser] Sec(Trsp) gene in mouse liver demonstrates an essential role of selenoprotein in liver function. J. Biol. Chem. 2004;279:8011–8017. doi: 10.1074/jbc.M310470200. [DOI] [PubMed] [Google Scholar]
- 39.Suzuki T., Kelly V.P., Motohashi H., Nakajima O., Takahashi S., Nishimura S., Yamamoto M. Deletion of the selenocysteine tRNA gene in macrophages and liver results in compensatory gene induction of cytoprotective enzymes by Nrf2. J. Biol. Chem. 2008;283:2021–2030. doi: 10.1074/jbc.M708352200. [DOI] [PubMed] [Google Scholar]
- 40.Shirimali R.K., Weaver J.A., Miller G.F., Srtarost M.F., Carlson B.A., Novoselov S.V., Kumarsawamy E., Gladyshev V.N., Hatfield D.L. Selenoprotein expression is essential in endothelial cell development and cardiac muscle function. Neurimuscul. Disord. 2007;17:135–142. doi: 10.1016/j.nmd.2006.10.006. [DOI] [PMC free article] [PubMed] [Google Scholar]