Abstract
To date, no genes have been reported to directly affect the de novo production of floral nectar. In an effort to identify genes involved in nectar production, the Affymetrix® ATH1 GeneChip was previously used to examine global gene expression profiles in Arabidopsis thaliana nectaries. One of the genes displaying highly enriched expression in nectaries was CELL WALL INVERTASE 4 (AtCWINV4, At2g36190), which encodes an enzyme that putatively catalyses the hydrolysis of sucrose into glucose and fructose. RT-PCR was used to confirm the nectary-enriched expression of AtCWINV4, as well as an orthologue from Brassica rapa. To probe biological function, two independent Arabidopsis cwinv4 T-DNA mutants were isolated. Unlike wild-type plants, cwinv4 lines did not produce nectar. While overall nectary morphology appeared to be normal, cwinv4 flowers accumulated higher than normal levels of starch in the receptacle, but not within the nectaries themselves. Conversely, wild-type, but not cwinv4, nectarial stomata stained intensely for starch. Cell wall extracts prepared from mutant flowers displayed greatly reduced invertase activity when compared with wild-type plants, and cwinv4 flowers also accumulated significantly lower levels of total soluble sugar. Cumulatively, these results implicate CWINV4 as an absolutely required factor for nectar production in the Brassicaceae, specifically by maintaining constant sink status within nectaries, thus allowing them to accumulate the sugars necessary for nectar production. In addition, CWINV4 is probably responsible for the hexose-rich composition observed for many Brassicaceae nectars.
Keywords: Arabidopsis thaliana, Brassica rapa, cell wall invertase, invertase, nectar, nectaries, nectary
Introduction
Nectar is secreted by many angiosperms as a reward to promote pollinator visitation (Simpson and Neff, 1983). This mutually beneficial process increases pollination efficiency and, ultimately, the propagation of plant species. Nectar is a complex solution that consists not only of sugars, which can range from 8% to 80% (w/w) depending on the species (Baker and Baker, 1983), but also a multitude of additional components, including: amino acids (Baker and Baker, 1973), organic acids (Baker and Baker, 1975), terpenes (Ecroyd et al., 1995), alkaloids (Deinzer et al., 1977), flavonoids (Ferreres et al., 1996), glycosides (Roshchina and Roshchina, 1993), vitamins (Griebel and Hess, 1990), phenolics (Ferreres et al., 1996), metal ions (Heinrich, 1989), oils (Vogel, 1969), free fatty acids (Kram et al., 2008), and proteins (Carter and Thornburg, 2004). These non-sugar compounds have been shown to perform a wide variety of tasks, from acting as a deterrent to certain visitors (Baker, 1978), to promoting pollination by attracting cognate pollinators (Carter et al., 2006; Raguso and Pichersky, 1999).
Of the sugars found in nectar, the most prevalent are sucrose, glucose, and fructose (Baker and Baker, 1983). The predominant forms of these three carbohydrates in Arabidopsis thaliana nectar are the simple hexoses glucose and fructose. For example, it was previously reported that Arabidopsis nectar has a hexose (glucose+fructose)-to-sucrose ratio close to 33.33 (Davis et al., 1998). Various nectars also contain multiple forms of other carbohydrates in minor concentrations (Baker and Baker, 1983), which are thought to play important roles as supplementary nutrition for potential pollinators.
The floral organ responsible for nectar synthesis and secretion is the nectary. Arabidopsis flowers contain two types of nectaries, lateral and median, with lateral nectaries being more pronounced and responsible for nearly all nectar production (Davis et al., 1998). Furthermore, the lateral nectary is much more heavily innervated by sieve tubes than the median, which could also account for its higher level of nectar secretion (Davis et al., 1986). Although it is known that the nectary is responsible for nectar production and secretion, the specific genes responsible for the underlying processes are not. With this in mind, there is a clear understanding of some of the cellular processes that take place in nectariferous cells (nectar-secreting cells) during secretion. At the initiation of secretion, vacuoles found inside these cells increase in volume, dictyosome numbers are reduced, the endoplasmic reticulum becomes more active, and mitochondrial numbers increase (Zhu and Hu, 2002). Along with increased organelle activity, starch grains found in the chloroplasts become reduced in size, or disappear altogether, indicating that they probably contribute to the carbohydrate component of the nectar itself (Ren et al., 2007; Zhu and Hu, 2002).
With an understanding of the basic processes taking place at an ultrastructural level, a general pathway for nectar secretion out of Arabidopsis, and other merocrine-type, nectaries has been proposed. Starting from the phloem, pre-nectar metabolites are transported via plasmodesmata to pre-secretory nectariferous tissue (Fahn, 1988; Pacini and Nepi, 2007). Here the pre-nectar is stored as starch grains in chloroplasts until nectar production begins; although it is also possible that some photosynthate is produced in situ, as has been reported for Brassica napus nectaries (Davis et al., 1986). When needed, starch grains are degraded and the endoplasmic reticulum and dictyosomes modify the sugars derived from degraded starch grains and package them into vesicles that fuse with the plasma membrane for secretion. It is also likely that some nectar carbohydrate, in the form of sucrose, is directly transported to secretory cells from underlying sieve tubes via plasmodesmata, and that plasma membrane transporters may play a direct role in the secretion of some nectar solutes (Fahn, 1988; Pacini and Nepi, 2007). The specific point of secretion from the nectary is thought to be modified stomata, which remain permanently open, providing a direct path out of the nectary (Davis AR, in Bowman, 1994; Zhu et al., 1995; Zhu and Hu, 2002). Similar mechanisms for nectar production have been proposed for a number of other species (Ge et al., 2000; Wist and Davis, 2006; Horner et al., 2007; Pacini and Nepi, 2007; Ren et al., 2007).
As described above, a number of ultrastructural and physiological aspects of nectary function are known; however, the impacts of individual genes on de novo nectar production are largely unknown. To link individual genes to nectar synthesis and secretion, the Affymetrix® ATH1 GeneChip was previously used to assess global gene expression profiles within nectaries (Kram et al., 2009). This analysis identified a large number of genes with relatively nectary-specific expression profiles. Amongst these genes, CELL WALL INVERTASE 4 (AtCWINV4, At2g36190) was particularly up-regulated in nectaries. The characterization of two independent Arabidopsis cwinv4 mutant lines is described here, the results of which implicate cell wall invertase activity as an absolutely required factor for nectar production in the Brassicaceae. Since nectar volume and composition can significantly affect the frequency of flower visitation by pollinators (Silva and Dean, 2000), knowledge of the genes required for nectar production may have broad implications, ranging from a better understanding of the co-evolution of plant–pollinator interactions, to increasing pollination efficiency in multiple crop species.
Materials and methods
Plant material and growth conditions
Arabidopsis thaliana ecotype Columbia-0 was the primary genotypic background used for this study. A previously described transgenic Arabidopsis line expressing plasma membrane localized GFP (35S::GFP:LTI6b; Cutler et al., 2000) was used for confocal microscopy studies. T-DNA mutant lines, SALK_130163 and SALK_017466C, were obtained from the Arabidopsis Biological Resource Centre (Alonso et al., 2003), and homozygous mutants were renamed cwinv4-1 and cwinv4-2, respectively. Individual plants were genotyped by PCR from genomic DNA, with oligonucleotide primers designed in locations flanking both ends of the T-DNA insertion, along with a third primer located on the left border of the T-DNA insertion. Genotyping primers were designed with the iSct Primers tool of the Salk Institute Genomics Analysis Laboratory (http://signal.salk.edu/tdnaprimers.2.html); the nucleotide sequences of all primers used in this study are provided in Table 1. Genotyping PCR reactions were performed with GoTaq Green Master Mix according to the manufacturer's instructions (Promega Corp., Madison, WI, USA).
Table 1.
Oligonucleotide primers used in this study
| Oligo name | Sequence | Purpose |
| cwinv4-1 F | ACATCCACAAAACCAGCAAAC | cwinv4-1 genotyping |
| cwinv4-1 R | ACAATGAACGGTTCAGCATTC | cwinv4-1 genotyping |
| cwinv4-2 F | AAATGTGAACGTAAACTGCGG | cwinv4-2 genotyping |
| cwinv4-2 R | TTTGTTAAATGTTTTTGGCCG | cwinv4-2 genotyping |
| LBb1.3 | ATTTTGCCGATTTCGGAAC | T-DNA left border, genotyping |
| AtCWINV4 RT-F | CAACGGTGTCAGATTCATTAG | AtCWINV4 RT PCR |
| AtCWINV4 RT-R | CGGTACGAGTATTACACGC | AtCWINV4 RT PCR |
| BrCWINV4 RT-F | TCAGGCCGATGTGGAAGTGACATT | BrCWINV4 RT PCR |
| BrCWINV4 RT-R | ATCCACAAAGCCAGCAAACGATGG | BrCWINV4 RT PCR |
| AtGAPDH-F | AGGGTGGTGCCAAGAAGGTTG | AtGAPDH RT PCR |
| AtGAPDH-R | GTAGCCCCACTCGTTGTCGTA | AtGAPDH RT PCR |
| BrUBQ-F | TTGAGGTGGAAAGCTCTGACACGA | BrUBQ RT PCR |
| BrUBQ-R | AATCGGCCAATGTACGACCATCCT | BrUBQ RT PCR |
| AtCWINV4-GFP-F | gaattcGAAATCAGAGTCACTGTGCC | Cloning of AtCWINV4 into pORE-R4, EcoRI site |
| AtCWINV4-GFP-R | actagtTTCCTAATTTCACAGTATCTCTC | Cloning of AtCWINV4 into pORE-R4, SpeI site |
Rapid-cycling Brassica rapa (CrGC 1-33) was used for BrCWINV4 studies. Plants were grown in individual pots on a peat-based medium with vermiculite and perlite (Pro-Mix BX; Premier Horticulture, Rivière-du-Loup, Quebec, Canada). All plant growth was performed in Percival AR66LX environmental chambers with settings of: 16/8 h day/night cycle, photosynthetic photon flux of 150 μmol m−2 s−1 and temperature of 23 °C.
Microarray data mining
The mean probe set signal intensities for all invertase genes expressed in Arabidopsis nectaries, as identified by Affymetrix ATH1 GeneChip® microarray, were compared to those in 13 different tissues at multiple developmental stages, and are presented in Supplementary Table S1 at JXB online. The raw normalized microarray data used for the analyses presented here were originally described by Kram et al. (2009).
Chemicals and reagents
Unless noted otherwise, all chemicals were obtained through Sigma Aldrich Chemical Co. (St Louis, MO, USA) or Thermo Fisher Scientific (Waltham, MA USA).
RT-PCR analyses
The RNAqueous-Micro® micro scale RNA isolation kit (Ambion, Austin, TX) was used, in conjunction with the Plant RNA Isolation Aid (Ambion, Austin, TX), to extract RNA from plant tissues. For floral tissues, RNA was extracted from Stage 11–12 and 14–15 flowers from Arabidopsis thaliana (stages defined by Smyth et al., 1990), or the equivalent of Stage 14–15 for Brassica rapa. The nectaries of Stage 11–12 flowers are pre-secretory (pre-anthesis), whereas those from Stage 14–15 are secretory (post-anthesis). Standard agarose gel electrophoresis and UV spectrophotometry were used to evaluate RNA quality for all samples. Reverse transcription-PCR (RT-PCR) was used to examine the presence of CWINV4 transcripts with the Promega Reverse Transcription System (A3500) according to the manufacturer's instructions with 0.1 μg of input RNA. Partial cDNA sequences for BrCWINV4 and BrUBQ were identified through a B. rapa nectary EST project (M Hampton et al., unpublished data), and deposited into GenBank as accession numbers GQ146458 and GR719937, respectively. All primers used for RT-PCR analyses are listed in Table 1.
AtCWINV4::AtCWINV4:GFP analyses
A 4837 bp AtCWINV4 genomic DNA fragment was amplified with the AtCWINV4-GFP-F and AtCWINV4-GFP-R primers (Table 1) using Phusion polymerase (New England BioLabs, Ipswich, MA), and directly cloned into the pCR® Blunt II-TOPO® vector (Invitrogen, Carlsbad, CA). This fragment contained 1895 bp of sequence upstream of the AtCWINV4 transcriptional start site, along with the full AtCWINV4 coding region, minus the stop codon. The sequence of the AtCWINV4 insert was confirmed via dideoxy sequencing at the University of Minnesota DNA Sequencing and Analysis Facility (St Paul, MN, USA), and the fragment was then subcloned into the EcoRI and SpeI sites of the binary vector pORE-R4. The coding region of this fragment was cloned in-frame with the GFP coding region found in pORE-R4. The resultant construct was transformed into Agrobacterium tumefaciens (GV3101), which was then used to transform Arabidopsis by the floral dip method (Clough and Bent, 1998). Transformed seedlings were selected on solid Murashige and Skoog (MS) media with kanamycin (50 μg ml−1). Plants confirmed to carry the above construct were either observed with a Nikon SMZ1500 stereomicroscope configured for the detection of GFP fluorescence, or a Nikon C1 Spectral Imaging Confocal Microscope at University of Minnesota Imaging Center. Sample preparation simply consisted of sepal removal from flowers to expose the nectaries prior to imaging.
Enzyme assays
Cell wall invertase activity was assayed as previously described (Wright et al., 1998). In summary, individual samples consisted of 25 mg of Stage 14–15 flowers, which were freshly collected and flash frozen in liquid nitrogen. Flowers were then transferred to 1.5 ml microcentrifuge tubes containing 400 μl of extraction buffer (50 mM HEPES-NaOH, pH 8.0; 5 mM MgCl2; 2 mM EDTA; 1 mM MnCl2; 1 mM CaCl2; 1 mM dithiothreitol; and 0.1 mM phenyl-methyl-sulphonyl fluoride), where they were homogenized with a pestle and incubated on ice for 10 min. Samples were then centrifuged at 13 000 g for 10 min at 4 °C. The supernatant, containing vacuolar and cytoplasmic invertases, was transferred to a new microcentrifuge tube. The remaining pellet was washed three times with 400 μl of extraction buffer by resuspension and centrifugation at 13 000 g for 10 min at 4 °C. The pellet was washed a final time with 400 μl of 80 mM sodium acetate, pH 4.8. Following the wash, the pellet was resuspended in 400 μl of 80 mM sodium acetate, pH 4.8. The total volume was divided into four 100 μl aliquots. One of the aliquots was boiled for 15 min to serve as a negative control. To each aliquot, 800 μl of staining solution (0.024% nitroblue tetrazolium; 0.014% phenazine methosulphate; 25 units glucose oxidase; 1% sucrose; in 80 mM sodium acetate, pH 4.8) was added as previously described (Wittich and Willemse, 1999). Supernatant fractions, containing the vacuolar and cytoplasmic invertases, were also examined for activity and used as positive controls. Absorbance at 550 nm was then used a measure of total invertase activity.
Histochemical staining
Flowers were examined for starch accumulation by iodine–potassium iodide (IKI) staining (Jensen, 1962). Briefly, freshly collected flowers were placed in IKI solution (Thermo Fisher S93409) and subjected to a 1 min vacuum infiltration, followed by a 5 min incubation at 20 °C. Flowers were then rinsed twice in deionized water and immediately imaged. Sepals were removed from flowers prior to staining when examining nectary starch accumulation.
Soluble sugar analysis
Total soluble sugar (sucrose and glucose) within flowers was analysed as previously described (Bethke and Busse, 2008). Briefly, Stage 14–15 flowers were collected, weighed, and flash frozen in liquid nitrogen. The flowers were then transferred to a 1.5 ml microcentrifuge tube containing 500 μl of 80% ethanol, homogenized with a pestle, and incubated in a rotisserie incubator at 60 °C for 24 h. After incubation, the samples were centrifuged for 10 min at 4000 rpm, with the supernatant being retained. The remaining pellet was resuspended with 500 μl of 80% ethanol and again incubated in a rotisserie incubator at 60 °C for 24 h. Following incubation, samples were again centrifuged and the supernatant collected and combined with previous supernatant. The supernatant was diluted 1:40 with 10 mM sodium acetate, pH 5.0. From the dilute supernatant, 25 μl was added to a new microcentrifuge tube and mixed with 25 μl of 38 U ml−1 invertase in 10 mM sodium acetate, pH 5.0 and incubated at room temperature for 15 min. After incubation, 25 μl of 150 mM sodium phosphate, pH 7.4 and 25 μl of enzyme mix (100 μl of 10 mM Ampliflu Red in DMSO; 1.725 ml dH2O; 0.575 ml 150 mM sodium phosphate, pH 7.4; 100 μl of 10 U ml−1 HPR in 50 mM sodium phosphate, pH 7.4; 100 μl of 100 U ml−1 glucose oxidase in 50 mM sodium phosphate, pH 7.4) were added to the sample and incubated for 30 min in the dark at 20 °C. Samples were then analysed via spectrophotometry at 560 nm—the wavelength at which absorbance is proportional to glucose concentration. To obtain soluble sugar concentrations, the assay was repeated with and without invertase. The assay performed in the absence of invertase provided information on baseline glucose concentrations. To then obtain sucrose concentrations, the glucose concentration obtained without invertase was subtracted from the glucose concentration observed with invertase.
Scanning electron microscopy
Flowers were manually removed of sepals and flash frozen in liquid nitrogen. Imaging was performed under low vacuum conditions (∼65 kPa) and an accelerating volt of 15 kV, with a JEOL Ltd. (Tokyo, Japan) JSM-6490 scanning electron microscope at the University of Minnesota Duluth Scanning Electron Microscope Laboratory.
Results
AtCWINV4 is highly expressed in nectaries
By microarray analysis, AtCWINV4 was previously identified as having a largely nectary-specific expression profile in wild-type plants (Kram et al., 2009). To determine if other invertase genes shared similar expression patterns to AtCWINV4, raw normalized probe set signal intensity data (Kram et al., 2009) were examined. A total of nine invertase genes, including AtCWINV4, were identified as being confidently expressed in Arabidopsis nectaries (see Supplementary Table S1 at JXB online; Kram et al., 2009); however, AtCWINV4 was the only invertase gene that displayed extreme up regulation in nectaries via microarray (see Supplementary Table S1 at JXB online). Moreover, AtCWINV4 was the sole nectary-expressed invertase predicted to be localized to the cell wall (based on TAIR annotation; prediction due to shared sequence similarity with known cell wall invertases). AtCWINV4 encodes a 591 amino acid protein with a theoretical molecular mass of 67 412.2 Da. PSORT analysis (Nakai and Kanehisa, 1991) predicted AtCWINV4 to be secreted from the cell via a 22 amino acid signal peptide, which would be consistent with cell wall localization.
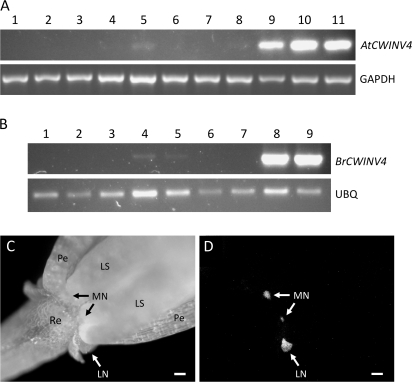
To confirm the nectary-enriched expression of AtCWINV4, reverse transcription polymerase chain reaction (RT-PCR) was performed on RNA isolated from 11 major tissues, including nectaries. Results shown in Fig. 1A demonstrate that AtCWINV4 was highly expressed in both the lateral and median nectaries of flowers at pre- and post-anthesis (lanes 9–11); it was also expressed in pistils and roots at much lower levels. Similar expression patterns were also observed for a CWINV4 orthologue from Brassica rapa (BrCWINV4, Fig. 1B).
Fig. 1.
CWINV4 is highly expressed in nectaries. Reverse transcription-polymerase chain reaction (RT-PCR) was used to examine the expression profiles of AtCWINV4 (A) and an orthologue from Brassica rapa (B; BrCWINV4, accession number GQ146458). The tissues examined in (A) included: (1) petal; (2) sepal; (3) rosette leaf; (4) stamen; (5) pistil; (6) root; (7) internode shoot; (8) silique; (9) mature median nectaries (Stage 14–15); (10) immature lateral nectaries (Stage 11–12); and, (11) mature lateral nectaries (Stage 14–15). (B) Tissues included: (1) petal; (2) sepal; (3) leaf; (4) stamen; (5) pistil; (6) root; (7) stem; (8) mature median nectaries; and (9) mature lateral nectaries. GAPDH (At3g04120) and a B. rapa ubiquitin gene (BrUBQ, accession number GR719937) were used as constitutively expressed controls. All B. rapa floral tissues examined were from the equivalent of Stage 14–15 Arabidopsis flowers. The images shown are the results obtained after 27 cycles of RT-PCR. (C) Bright field image of Stage 15 Arabidopsis flower expressing an AtCWINV4:GFP fusion, under control of the native AtCWINV4 promoter, and (D) GFP fluorescence derived from same flower. Sepals were removed prior to imaging. Re, receptacle; Pe, petal; LS, long stamen; MN, median nectary; LN, lateral nectary; scale bars, 100 μm.
To examine AtCWINV4 expression further, a genomic fragment encoding AtCWINV4, including 1.8 kb of the promoter region, was cloned in-frame with GFP within the binary vector pORE-R4 (to generate AtCWINV4::AtCWINV4:GFP). Wild-type Arabidopsis transformed with this construct displayed nectary-specific GFP fluorescence (Fig. 1C, D). Examinations of these plants via laser scanning confocal microscopy further supported AtCWINV4 tissue-specific expression (see Supplementary Video S1 at JXB online), with GFP appearing to be expressed throughout the entire nectary. Although not definitive, confocal results (see Supplementary Video S1 at JXB online) also suggested that most fluorescence was localized to cell walls due to diffuse patterns of GFP accumulation, which appeared to outline nectary cells. This observation was in contrast to the well-defined localization of a plasma-membrane targeted GFP fusion protein in nectaries (see Supplementary Video S2 at JXB online; 35S::GFP:LTI6b fusion described in Cutler et al., 2000).
cwinv4 mutant lines
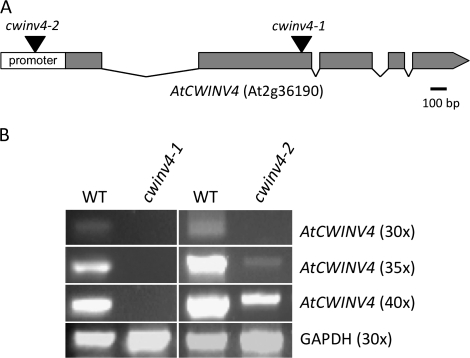
To probe the biological function of AtCWINV4, two independent T-DNA mutant lines were identified, cwinv4-1 and cwinv4-2. cwinv4-1 had a T-DNA inserted in the second exon, whereas cwinv4-2 had an insertion ∼200 nucleotides upstream of the transcriptional start site (Fig. 2A). Significantly, cwinv4-1 was identified as a null mutant, whereas AtCWINV4 expression was greatly reduced in cwinv4-2 (Fig. 2B). Indeed, while AtCWINV4 transcripts were readily detected from whole flowers of wild-type plants following 30 cycles of RT-PCR, faint bands derived from cwinv4-2 cDNA were only visible at 35 cycles or greater.
Fig. 2.
Identification of cwinv4 mutants. Two independent Arabidopsis T-DNA mutant lines (Alonso et al., 2003), cwinv4-1 (SALK_130163) and cwinv4-2 (SALK_017466C), were obtained from the Arabidopsis Biological Resource Centre and genotyped to obtain homozygous mutant plants. The relative locations of each T-DNA insertion are shown in (A). RT-PCR, performed on RNA isolated from whole flowers, demonstrated that cwinv4-1 is a null mutant, and that AtCWINV4 is expressed significantly lower in cwinv4-2 flowers than in wild-type (B).
cwinv4 flowers have decreased cell wall invertase activity
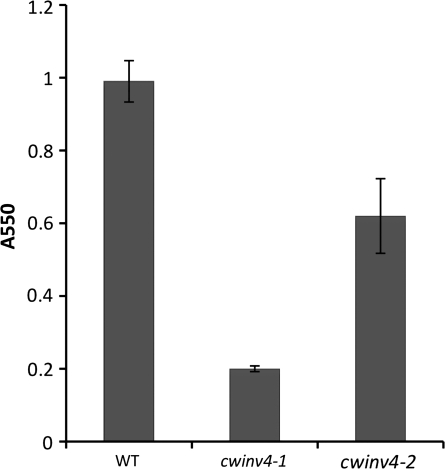
Once confirmed as mutants, both cwinv4-1 and cwinv4-2 flowers were subsequently examined for extracellular (cell wall) invertase activity. Invertases catalyse the hydrolysis of the disaccharide sucrose into glucose and fructose (Berger et al., 2007; Winter and Huber, 2000b). Cell wall extracts were prepared as previously described (Wright et al., 1998) from whole flowers of cwinv4-1, cwinv4-2, and wild-type plants. Both cwinv4 lines displayed large decreases in total cell wall invertase activity (Fig. 3), whereas intracellular fractions displayed similar activities to wild-type plants (data not shown). The higher cell wall invertase activity observed in cwinv4-2 than cwinv4-1 flowers was probably due to the leaky expression of AtCWINV4 in cwinv4-2 (i.e. cwinv4-1 is a null mutant, whereas cwinv4-2 is a knockdown mutant). It was also noted that cell wall invertase activity was not reduced to zero in cell wall extracts derived from flowers of either cwinv4 mutant. Whether this result is indicative of activity derived from other cell wall invertases, or represents minor contamination from intracellular invertases is unknown. However, it should be noted that whole flowers were used for these assays, not isolated nectaries (i.e. other cell wall invertases may be expressed in petals, stamen, etc).
Fig. 3.
Cell wall invertase activity is greatly reduced in cwinv4 flowers. Cell wall extracts were prepared from whole Stage 14–15 flowers (25 mg fresh weight) of wild-type and cwinv4 plants, which were subsequently tested for invertase activity. The results from one experiment with cwinv4-1 and cwinv4-2 plants are shown (n=3 independent preparations). Independent and repeated experiments with both cwinv4-1 and cwinv4-2 plants displayed consistent results, with varying amplitudes of total activity for both wild-type and mutant plants.
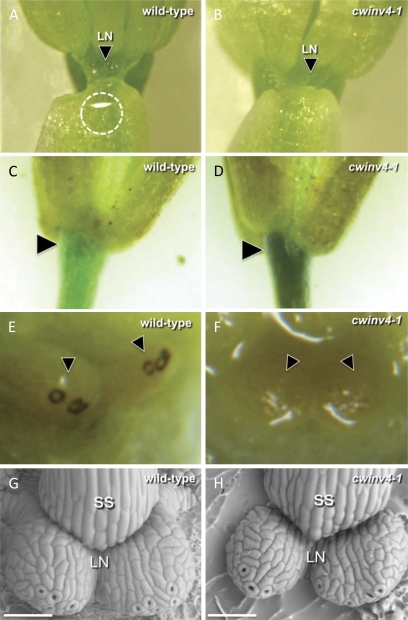
cwinv4 mutants do not secrete nectar
To determine if mutations occurring within AtCWINV4 have an impact on nectar production, individual flowers were examined for the presence of nectar droplets, which accumulate inside the cups formed by sepals surrounding the lateral nectaries. Figure 4A illustrates a typical analysis, and shows a nectar droplet clinging to the inside of a sepal of a wild-type flower; nectar droplets were never observed within either cwinv4-1 or cwinv4-2 flowers (Table 2; Fig. 4B). Interestingly, extremely limited indications of secretion very rarely appeared inside the sepals of cwinv4-2 flowers; however, nectar droplets were never present (only a thin glistening film), and due to the extremely rare occurrence (c. 5% of flowers) and limited amount of fluid produced, these putative secretions could neither be collected nor analysed. If this rarely observed fluid truly represented nectar secretion, it would be consistent with the leaky expression of CWINV4 found in cwinv4-2.
Fig. 4.
cwinv4 flowers do not secrete nectar. Nectar production in Arabidopsis flowers can be examined by gently peeling back the sepal. Nectar droplets accumulating within the sepal cups surrounding lateral nectaries (LN) were consistently present in wild-type plants (circled in A), whereas neither cwinv4 mutant secreted nectar (e.g. B; Table 2). Both cwinv4 mutants also accumulated higher than normal levels of starch within the receptacle (arrowheads), which often extended into the pedicel (C, D). Conversely, the stomata of wild-type nectaries stained heavily for starch (E, arrowheads), whereas those of cwinv4 flowers did not (F). The morphology of cwinv4 nectaries was examined to determine if aberrant nectary structure might account for the lack of nectar secretion. In all flowers examined, cwinv4 nectaries (H) appeared to have similar morphology to wild-type plants (G). Scale bars are 50 μm; LN, lateral nectary; SS, short stamen.
Table 2.
Examination of nectar production in wild-type and cwinv4 flowers
Flowers were examined for nectar droplets accumulating inside sepals surrounding lateral nectaries. A minimum of ten individual plants were examined for each line.
Extremely limited indications of fluid very rarely appeared inside the sepals of cwinv4-2 flowers; however, no nectar droplets ever accumulated inside the flowers of these plants. These findings are in line with the leaky expression of AtCWINV4 found within cwinv4-2.
cwinv4 flowers display altered patterns of starch accumulation
Invertases play key roles in sugar source–sink relationships (Roitsch, 1999; Roitsch and Ehness, 2000). Thus, whole flowers were examined for altered patterns of starch accumulation via iodine–potassium iodide (IKI) staining. cwinv4 lines displayed an obvious increase in starch accumulation within the floral receptacle, which often extended into the pedicel (Fig. 4C, D). Conversely, wild-type flowers accumulated high levels of starch within nectarial stomata, whereas cwinv4 plants did not (Fig 4E, F).
Nectary ultrastructure is normal in cwinv4 flowers
Scanning electron microscopy was used to examine if any defects in nectary development or morphology could account for the lack of nectar production observed in the cwinv4 mutant lines. Both mutant lines displayed all structural hallmarks of wild-type nectaries, including having multiple modified stomata (in an open conformation) and being covered by a highly reticulated cuticle (Fig. 4G, H). Confocal microscopy utilizing lipophilic fluorescent dyes (i.e. FM4-64) also demonstrated normal nectary morphology within cwinv4 flowers (data not shown).
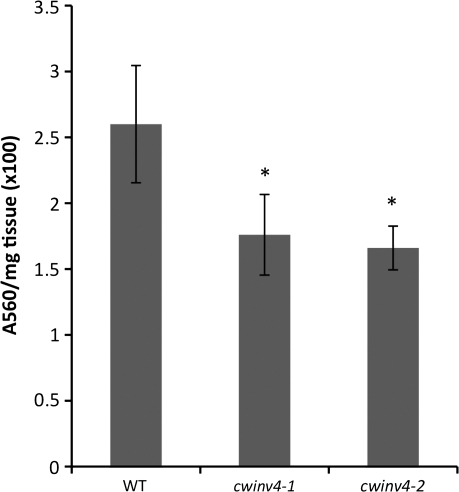
cwinv4 flowers accumulate less soluble sugar
Related to the starch analyses described above, it seemed logical to examine whether cwinv4 mutants displayed changes in the accumulation of soluble sugars. Simple sugars were extracted from whole flowers and assayed as previously described (Bethke and Busse, 2008). Results shown in Fig. 5 demonstrate that both cwinv4-1 and cwinv4-2 flowers accumulated significantly lower levels of soluble sugars, specifically sucrose and glucose, than wild-type plants. Notably, the overall ratio of glucose-to-sucrose was similar in wild-type and cwinv4 flowers (∼90% glucose and 10% sucrose in all lines examined, data not shown).
Fig. 5.
cwinv4 flowers accumulate decreased total soluble sugar. Whole flowers (Stage 14–15) from both cwinv4-1 and cwinv4-2 displayed significantly lower levels of total soluble sugar (sucrose and glucose) than wild-type plants (n=3, *P=0.027 and 0.013, respectively). Results are displayed as total absorbance per mg of floral tissue ×100. The relative proportion of sucrose-to-glucose was similar between cwinv4 and wild-type flowers (data not shown).
Discussion
Arabidopsis flowers are highly autogamous, which poses the question as to why functional and energetically costly nectaries are maintained in this species. The answer to this question may lie in the revelation that insects do visit Arabidopsis flowers in their native setting, and a small but significant amount of outcrossing does occur (Hoffmann et al., 2003). The fact that many species share similar nectary structure with Arabidopsis (e.g. Brassica sp.), and produce relatively large amounts of nectar (Davis et al., 1996, 1998), suggests that similar mechanisms for nectar production may exist amongst the Brassicaceae. Many of these species are not self-fertile, and are dependent on pollinators to achieve full fecundity (Pearson, 1933; Rahman, 1940; Vesely, 1962; Nieuwhof, 1963). Notably, Arabidopsis nectaries also seem to share common developmental mechanisms with much of the eudicot clade (Lee et al., 2005). As such, Arabidopsis, with its fully sequenced genome and genetic resources, can serve as a valuable model for the molecular dissection of nectary form and function. This report describes initial efforts towards characterizing CELL WALL INVERTASE 4, a gene required for nectar production in Arabidopsis. To the best of our knowledge, this is the first report of an individual gene being required for de novo nectar production in flowering plants.
Plant invertases have been classified into several general types based upon subcellular localization and pH optima. Cell wall invertases, in particular, are adventitiously bound to cell walls and function optimally in an acidic pH (Sturm, 1999; Sturm and Tang, 1999; Tymowska-Lalanne and Kreis, 1998a, b). The model plant Arabidopsis thaliana encodes over 20 known or putative invertases (The Arabidopsis Information Resource (TAIR), www.arabidopsis.org). Of these gene products, at least four are known or predicted to be localized to the cell wall. Significantly, cell wall invertases have been shown to exhibit both temporal and spatial specificity in their expression (Sherson et al., 2003). Of interest in this study was AtCWINV4, which displayed exceptionally enhanced expression in Arabidopsis nectaries of both pre- and post-anthesis flowers (Fig. 1; Kram et al., 2009). These results are consistent with previous reports, which demonstrated that AtCWINV4 is highly expressed in Arabidopsis flowers by RT-PCR (Sherson et al., 2003).
The observed expression patterns for AtCWINV4, combined with the relationship between invertases and sucrose, and the composition of Arabidopsis nectar (hexose-rich), imply a central role for this gene in mediating nectary function. Sucrose is one of the most important products of photosynthesis, and is the form in which most carbohydrates are transported between tissues (Roitsch, 1999; Roitsch and Ehness, 2000; Winter and Huber, 2000a, b). This disaccharide has also been shown to play important roles in plant signalling, growth, and developmental processes (Weber et al., 1998; Sturm, 1999; Sturm and Tang, 1999; Winter and Huber, 2000a, b; Sherson et al., 2003; Berger et al., 2007). Because of the catabolic nature of invertases, it is thought that these enzymes could be prime mediators of source–sink relationships, driving the flow of sucrose from source to sink tissues and, in turn, regulating plant growth and development.
Perhaps most significantly in this study, nectar was found to be absent in cwinv4 flowers (Fig. 4; Table 2), which could be explained by the suggested role of invertases in establishing sucrose gradients between source and sink tissues. If this interpretation is accurate, the loss of invertase function would be paired with a reduction in the extracellular hydrolysis of sucrose into glucose and fructose. A role for CWINV4 acting in this manner was supported by the finding that cwinv4 flowers displayed markedly decreased CWINV activities when compared to wild-type plants (Fig. 3). Such disruption of the sucrose gradient from source to sink cells would result in the cessation of sugar flow into nectaries, thus depriving nectariferous tissue of the sugars required for nectar production.
The mechanistic impact of CWINV4 activity on nectar production is perhaps 2-fold. CWINV4 may drive the movement of sucrose into nectaries from source tissues, which, in turn, is deposited as starch (supported by Fig. 4C–F). Significantly, starch accumulation within Arabidopsis nectaries has been previously reported (Bowman, 1994; Zhu et al., 1995; Zhu and Hu, 2002; Ren et al., 2007); at anthesis, starch grains are reportedly degraded and are a presumed source of nectar carbohydrate (Ren et al., 2007). CWINV4 involvement in starch accumulation in nectaries is supported by two pieces of evidence in this study: (i) increased starch accumulation within the receptacle and pedicel of cwinv4 flowers (Fig. 4C, D), and (ii) intense IKI staining in the stomata of wild-type nectaries, as opposed to cwinv4 flowers (Fig. 4E, F). Taken together, these findings suggest that CWINV4-deficient nectaries lack the sink status necessary to accumulate starch, with excess photosynthate apparently being stored within the receptacle and pedicel instead. Significantly, pedicels have been suggested to serve as sources of nectar carbohydrate via the direct production of photosynthate (Pacini and Nepi, 2007), which would also be consistent with the observed starch staining patterns observed in Fig. 4D. The fact that cwinv4 flowers accumulated less soluble sugar than wild-type flowers (Fig. 5) is a final piece of evidence suggesting a role for CWINV4 in supplying nectariferous tissue with the sugars necessary for nectar production.
A second, but non-mutually exclusive, hypothesis for CWINV4 involvement in nectar production can be posited for flowers at anthesis. Without hydrolysis of sucrose to glucose and fructose within the apoplast of nectary cells, an extracellular hexose-rich environment (high solute) would not be established and, in turn, osmotic pressure might be insufficient to draw out water—a suggested requirement for the secretion of hexose-dominant nectars (Fahn, 1988; Nicolson, 1998). Moreover, the spontaneity of sucrose transport across the plasma membrane of nectariferous cells, probably via sucrose transporters, would be greatly diminished in cwinv4 nectaries. Interestingly, genes representing the full canonical sucrose biosynthesis pathway are highly up-regulated in mature Arabidopsis nectaries, along with several putative sucrose transporters (Kram et al., 2009). These overall findings are consistent with a mechanism for generating the high hexose-to-sucrose ratio previously reported for the nectars of multiple Brassicaceae species (Davis et al., 1998). Indeed, it is likely that CWINV4 activity is at least partially responsible for the observed hexose-rich composition of Arabidopsis nectar. Invertase activity has even been reported to occur within the nectars of multiple species, and implicated as a factor required for the generation of hexose-dominant nectar (Baker and Baker, 1983; Nicolson, 1998; Heil et al., 2005). Reports of this activity within nectar itself are also consistent with cell wall localization or, at a minimum, the secretion of invertases from nectary cells.
In sum, all current evidence indicates that cell wall invertase activity, specifically derived from AtCWINV4, plays an essential role in the production of Arabidopsis nectar. The fact that an orthologue from B. rapa, BrCWINV4, displayed nearly identical expression patterns to AtCWINV4 (Fig. 1B) suggests that the requirement of cell wall invertases in nectar production is conserved amongst the Brassicaceae, and possibly many other nectar-producing plants. Indeed, investigations into petunia pollination syndromes have identified a single QTL, which may encode an invertase, responsible for the control of nectar volume (Stuurman et al., 2004; Galliot et al., 2006a, b). Teasing apart the proposed mechanisms of CWINV4 involvement in nectar production will require further experimentation; however, its nectary-specific expression profile, coupled with its apparent involvement in sugar metabolism, suggests it is a principal player in Arabidopsis nectary function.
Supplementary data
Supplementary data can be found at JXB online.
Supplementary Table S1. Normalized Affymetrix ATH1 GeneChip® probe set signal intensity for Arabidopsis invertase genes expressed in nectaries (.xls format).
Supplementary Video S1. Projection video of Arabidopsis flower expressing AtCWINV4::AtCWINV4:GFP fusion (.mov format).
Supplementary Video S2. Projection video of Arabidopsis nectary expressing 35S::GFP:LTI6b, a plasma membrane-localized marker (.mov format).
Supplementary Material
Acknowledgments
The authors thank Mr Bryan Bandli, University of Minnesota Duluth Scanning Electron Microscope Laboratory, for assistance with scanning electron microscopy. This work was supported by the United States Department of Agriculture (2006-35301-16887 to CC) and the National Science Foundation (0820730 to CC).
Glossary
Abbreviations
- CWINV
cell wall invertase
- AtCWINV4
Arabidopsis thaliana CELL WALL INVERTASE 4
- BrCWINV4
Brassica rapa CELL WALL INVERTASE 4
- IKI
iodine–potassium iodide
- RT-PCR
reverse transcription-polymerase chain reaction
References
- Alonso JM, Stepanova AN, Leisse TJ, et al. Genome-wide insertional mutagenesis of Arabidopsis thaliana. Science. 2003;301:653–657. doi: 10.1126/science.1086391. [DOI] [PubMed] [Google Scholar]
- Baker HG. Chemical aspects of the pollination of woody plants in the tropics. In: Tomlinson PB, Zimmerman M, editors. Tropical trees as living systems. New York: Cambridge University Press; 1978. pp. 57–82. [Google Scholar]
- Baker H, Baker I. Amino acids in nectar and their evolutionary significance. Nature. 1973;241:543–545. [Google Scholar]
- Baker H, Baker I. Studies of nectar-constitution and pollinator-plant coevolution. In: Gilbert LE, Raven PH, editors. Coevolution of animals and plants. Austin, TX: University of Texas Press; 1975. pp. 100–140. [Google Scholar]
- Baker H, Baker I. A brief historical review of chemistry of floral nectar. In: Bentley BL, editor. The biology of nectaries. New York: Columbia University Press; 1983. pp. 126–152. [Google Scholar]
- Berger S, Sinha AK, Roitsch T. Plant physiology meets phytopathology: plant primary metabolism and plant–pathogen interactions. Journal of Experimental Botany. 2007;58:4019–4026. doi: 10.1093/jxb/erm298. [DOI] [PubMed] [Google Scholar]
- Bethke PC, Busse JC. Validation of a simple, colorimetric, microplate assay using Amplex Red for the determination of glucose and sucrose in potato tubers and other vegetables. American Journal of Potato Research. 2008;85:414–421. [Google Scholar]
- Bowman JL. Arabidopsis: an atlas of morphology and development. New York: Springer-Verlag Inc; 1994. [Google Scholar]
- Carter C, Shafir S, Yehonatan L, Palmer RG, Thornburg R. A novel role for proline in plant floral nectars. Naturwissenschaften. 2006;93:72–79. doi: 10.1007/s00114-005-0062-1. [DOI] [PubMed] [Google Scholar]
- Carter C, Thornburg RW. Is the nectar redox cycle a floral defense against microbial attack? Trends in Plant Science. 2004;9:320–324. doi: 10.1016/j.tplants.2004.05.008. [DOI] [PubMed] [Google Scholar]
- Clough SJ, Bent AF. Floral dip: a simplified method for Agrobacterium-mediated transformation of Arabidopsis thaliana. The Plant Journal. 1998;16:735–743. doi: 10.1046/j.1365-313x.1998.00343.x. [DOI] [PubMed] [Google Scholar]
- Cutler SR, Ehrhardt DW, Griffitts JS, Somerville CR. Random GFP::cDNA fusions enable visualization of subcellular structures in cells of Arabidopsis at a high frequency. Proceedings of the National Academy of Sciences, USA. 2000;97:3718–3723. doi: 10.1073/pnas.97.7.3718. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davis A, Peterson R, Shuel R. Anatomy and vasculature of the floral nectaries of Brassica napus (Brassicaceae) Canadian Journal of Botany. 1986;64:2508–2516. [Google Scholar]
- Davis AR, Fowke LC, Sawhney VK, Low NH. Floral nectar secretion and ploidy in Brassica rapa and B. napus (Brassicaceae). II. Quantified variability of nectary structure and function in rapid-cycling lines. Annals of Botany. 1996;77:223–234. [Google Scholar]
- Davis AR, Pylatuik JD, Paradis JC, Low NH. Nectar-carbohydrate production and composition vary in relation to nectary anatomy and location within individual flowers of several species of Brassicaceae. Planta. 1998;205:305–318. doi: 10.1007/s004250050325. [DOI] [PubMed] [Google Scholar]
- Deinzer ML, Thomson PA, Burgett DM, Isaacson DL. Pyrrolizidine alkaloids: their occurrence in honey from tansy ragwort (Senecio jacobaea L.) Science. 1977;195:497–499. doi: 10.1126/science.835011. [DOI] [PubMed] [Google Scholar]
- Ecroyd CE, Franich RA, Kroese HW, Steward D. Volatile constituents of Cactylanthus taylorii flower nectar in relation to flower pollination and browsing by animals. Phytochemistry. 1995;40:1387–1389. [Google Scholar]
- Fahn A. Tansley Review No. 14. Secretory tissues in vascular plants. New Phytologist. 1988;108:229–257. doi: 10.1111/j.1469-8137.1988.tb04159.x. [DOI] [PubMed] [Google Scholar]
- Ferreres F, Andrade P, Gil MI, Tomas Barberan FA. Floral nectar phenolics as biochemical markers for the botanical origin of heather honey. Zeitschrift für Lebensmittel-Untersuchung und -Forschung. 1996;202:40–44. [Google Scholar]
- Galliot C, Hoballah ME, Kuhlemeier C, Stuurman J. Genetics of flower size and nectar volume in Petunia pollination syndromes. Planta. 2006a;225:203–212. doi: 10.1007/s00425-006-0342-9. [DOI] [PubMed] [Google Scholar]
- Galliot C, Stuurman J, Kuhlemeier C. The genetic dissection of floral pollination syndromes. Current Opinion in Plant Biology. 2006b;9:78–82. doi: 10.1016/j.pbi.2005.11.003. [DOI] [PubMed] [Google Scholar]
- Ge YX, Angenent GC, Wittich PE, et al. NEC1, a novel gene, highly expressed in nectary tissue of Petunia hybrida. The Plant Journal. 2000;24:725–734. doi: 10.1046/j.1365-313x.2000.00926.x. [DOI] [PubMed] [Google Scholar]
- Griebel C, Hess G. The vitamin C content of flower nectar of certain Labiatae. Zeitschrift für Lebensmittel-Untersuchung und -Forschung. 1990;79:168–171. [Google Scholar]
- Heil M, Rattke J, Boland W. Postsecretory hydrolysis of nectar sucrose and specialization in ant/plant mutualism. Science. 2005;308:560–563. doi: 10.1126/science.1107536. [DOI] [PubMed] [Google Scholar]
- Heinrich G. Analysis of cations in nectars by means of a laser microprobe mass analyser (LAMMA) Beitrage zur Biologie der Pflanzen. 1989;64:293–308. [Google Scholar]
- Hoffmann MH, Bremer M, Schneider K, Burger F, Stolle E, Moritz G. Flower visitors in a natural population of Arabidopsis thaliana. Plant Biology. 2003;5:491–494. [Google Scholar]
- Horner HT, Healy RA, Ren G, Fritz D, Klyne A, Seames C, Thornburg RW. Amyloplast to chromoplast conversion in developing ornamental tobacco floral nectaries provides sugar for nectar and antioxidants for protection. American Journal of Botany. 2007;94:12–24. doi: 10.3732/ajb.94.1.12. [DOI] [PubMed] [Google Scholar]
- Jensen WA. Botanical histochemistry: principles and practice. San Francisco: WH Freeman; 1962. [Google Scholar]
- Kram BW, Bainbridge EA, Perera MADN, Carter C. Identification, cloning and characterization of a GDSL lipase secreted into the nectar of Jacaranda mimosifolia. Plant Molecular Biology. 2008;68:173–183. doi: 10.1007/s11103-008-9361-1. [DOI] [PubMed] [Google Scholar]
- Kram BW, Xu WW, Carter CJ. Uncovering the Arabidopsis thaliana nectary transcriptome: investigation of differential gene expression in floral nectariferous tissues. BMC Plant Biology. 2009;9:92. doi: 10.1186/1471-2229-9-92. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee JY, Baum SF, Oh SH, Jiang CZ, Chen JC, Bowman JL. Recruitment of CRABS CLAW to promote nectary development within the eudicot clade. Development. 2005;132:5021–5032. doi: 10.1242/dev.02067. [DOI] [PubMed] [Google Scholar]
- Nakai K, Kanehisa M. Expert system for predicting protein localization sites in gram-negative bacteria. Proteins. 1991;11:95–110. doi: 10.1002/prot.340110203. [DOI] [PubMed] [Google Scholar]
- Nicolson SW. The importance of osmosis in nectar secretion and its consumption by insects. American Zoologist. 1998;38:418–425. [Google Scholar]
- Nieuwhof M. Pollination and contamination of Brassica oleracea L. Euphytica. 1963;12:17–26. [Google Scholar]
- Pacini E, Nepi M. Nectar production and presentation. In: Nicolson SW, Nepi M, Pacini E, editors. Nectaries and nectar. Dordrecht: Springer; 2007. pp. 167–214. [Google Scholar]
- Pearson OH. Study of the life history of Brassica oleracea. Botanical Gazette. 1933;94:534–550. [Google Scholar]
- Raguso RA, Pichersky E. A day in the life of a linalool molecule: chemical communication in a plant-pollinator system. 1. Linalool biosynthesis in flowering plants. Plant Species Biology. 1999;14:95–120. [Google Scholar]
- Rahman KA. Insect pollinators of toria (Brassica napus Linn., var. dichotoma Prain) and sarson (B. campestris Linn., var. sarson Prain) at Lyallpur. Indian Journal of Agrultural Science. 1940;10:422–447. [Google Scholar]
- Ren G, Healy RA, Klyne AM, Horner HT, James MG, Thornburg RW. Transient starch metabolism in ornamental tobacco floral nectaries regulates nectar composition and release. Plant Science. 2007;173:277–290. [Google Scholar]
- Roitsch T. Source-sink regulation by sugar and stress. Current Opinion in Plant Biology. 1999;2:198–206. doi: 10.1016/S1369-5266(99)80036-3. [DOI] [PubMed] [Google Scholar]
- Roitsch T, Ehness R. Regulation of source/sink relations by cytokinins. Plant Growth Regulation. 2000;32:359–367. [Google Scholar]
- Roshchina VV, Roshchina VD. The excretory function of higher plants. New York: Springer-Verlag; 1993. [Google Scholar]
- Sherson SM, Alford HL, Forbes SM, Wallace G, Smith SM. Roles of cell-wall invertases and monosaccharide transporters in the growth and development of Arabidopsis. Journal of Experimental Botany. 2003;54:525–531. doi: 10.1093/jxb/erg055. [DOI] [PubMed] [Google Scholar]
- Silva EM, Dean BB. Effect of nectar composition and nectar concentration on honey bee (Hymenoptera: Apidae) visitations to hybrid onion flowers. Journal of Economic Entomology. 2000;93:1216–1221. doi: 10.1603/0022-0493-93.4.1216. [DOI] [PubMed] [Google Scholar]
- Simpson BB, Neff JL. Evolution and diversity of floral rewards. In: Jones CE, Little RJ, editors. Handbook of experimental pollination biology. New York: Van Nostrand Reinhold; 1983. pp. 142–159. [Google Scholar]
- Smyth DR, Bowman JL, Meyerowitz EM. Early flower development in Arabidopsis. The Plant Cell. 1990;2:755–767. doi: 10.1105/tpc.2.8.755. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sturm A. Invertases. Primary structures, functions, and roles in plant development and sucrose partitioning. Plant Physiology. 1999;121:1–7. doi: 10.1104/pp.121.1.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sturm A, Tang GQ. The sucrose-cleaving enzymes of plants are crucial for development, growth and carbon partitioning. Trends in Plant Science. 1999;4:401–407. doi: 10.1016/s1360-1385(99)01470-3. [DOI] [PubMed] [Google Scholar]
- Stuurman J, Hoballah ME, Broger L, Moore J, Basten C, Kuhlemeier C. Dissection of floral pollination syndromes in petunia. Genetics. 2004;168:1585–1599. doi: 10.1534/genetics.104.031138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tymowska-Lalanne Z, Kreis M. Expression of the Arabidopsis thaliana invertase gene family. Planta. 1998a;207:259–265. doi: 10.1007/s004250050481. [DOI] [PubMed] [Google Scholar]
- Tymowska-Lalanne Z, Kreis M. The plant invertases: physiology, biochemistry and molecular biology. Advances in Botanical Research. 1998b;28:71–117. [Google Scholar]
- Vesely V. The economic effectiveness of bee polination on winter rape (Brassica napus L., var. oleifera Metz) Min. Zemedel. Lesn. a Vodniho Hospodar. Ust. Vedtach. Inform. Zemedel. Ekon. 1962;8:659–673. [Google Scholar]
- Vogel S. Abstracts XIth International Botany Congress. Seattle, WA: 1969. Flowers offering fatty oil instead of nectar. [Google Scholar]
- Weber H, Heim U, Golombek S, Borisjuk L, Manteuffel R, Wobus U. Expression of a yeast-derived invertase in developing cotyledons of Vicia narbonensis alters the carbohydrate state and affects storage functions. The Plant Journal. 1998;16:163–172. doi: 10.1046/j.1365-313x.1998.00282.x. [DOI] [PubMed] [Google Scholar]
- Winter H, Huber SC. Regulation of sucrose metabolism in higher plants: localization and regulation of activity of key enzymes. Critical Reviews in Plant Science. 2000a;19:31–67. doi: 10.1080/10409230008984165. [DOI] [PubMed] [Google Scholar]
- Winter H, Huber SC. Regulation of sucrose metabolism in higher plants: Localization and regulation of activity of key enzymes. Critical Reviews in Biochemistry and Molecular Biology. 2000b;35:253–289. doi: 10.1080/10409230008984165. [DOI] [PubMed] [Google Scholar]
- Wist TJ, Davis AR. Floral nectar production and nectary anatomy and ultrastructure of Echinacea purpurea (Asteraceae) Annals of Botany. 2006;97:177–193. doi: 10.1093/aob/mcj027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wittich PE, Willemse MTM. Sucrose utilization during ovule and seed development of Gasteria verrucosa (Mill.) H. Duval as monitored by sucrose synthase and invertase localization. Protoplasma. 1999;208:136–148. [Google Scholar]
- Wright DP, Read DJ, Scholes JD. Mycorrhizal sink strength influences whole plant carbon balance of Trifolium repens L. Plant, Cell and Environment. 1998;21:881–891. [Google Scholar]
- Zhu J, Hu Z, Muuml IM. Ultrastructural investigations on floral nectary of Arabidopsis thaliana prepared by high pressure freezing and freeze substitution. Biology of the Cell. 1995;84:225. [Google Scholar]
- Zhu J, Hu ZH. Cytological studies on the development of sieve element and floral nectary tissue in Arabidopsis thaliana. Acta Botanica Sinica. 2002;44:9–14. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.