Abstract
Iron chlorosis is one of the major abiotic stresses affecting fruit trees and other crops in calcareous soils and leads to a reduction in growth and yield. Usual remediation strategies consist of amending iron to soil, which is an expensive practice, or using tolerant cultivars, which are difficult to develop when not available. To understand the mechanisms underlying the associated physiopathy better, and thus develop new strategies to overcome the problems resulting from iron deficiency, the differential gene expression induced by iron deficiency in the susceptible citrus rootstock Poncirus trifoliata (L.) Raf. have been examined. The genes identified are putatively involved in cell wall modification, in determining photosynthesis rate and chlorophyll content, and reducing oxidative stress. Additional studies on cell wall morphology, photosynthesis, and chlorophyll content, as well as peroxidase and catalase activities, support their possible functions in the response to iron deficiency in a susceptible genotype, and the results are discussed.
Keywords: Citrus rootstock, differential gene expression, iron chlorosis, iron deficiency, microarray, Poncirus trifoliata
Introduction
Iron is an essential nutrient for plant growth and crop productivity. Although total iron content, in most soils, is much higher than that required by plants, iron bioavailability is low, particularly in calcareous soils (Imsande, 1998). Whereas plant growth is otherwise not seriously affected, iron deficiency is the cause of leaf chlorosis (Chapman, 1968), where leaves show interveinal yellowing while veins remain green. In the Mediterranean region, iron chlorosis is one of the major abiotic stresses affecting fruit trees.
In nature, plants have evolved tolerance to iron chlorosis following two different strategies aimed at improving iron acquisition from the rhizosphere (Marschner et al., 1986). Strategy I relies on increasing iron solubility by inducing membrane-bound Fe(III)-chelate reductases that reduce Fe(III) to Fe(II), which is more soluble and is subsequently taken up by specific transporters. By contrast, strategy II, characteristic of graminaceous species, consists of secreting Fe(III)-chelating substances, referred to as phytosiderophores, which will complex Fe(III), thus enabling uptake (Takagi et al., 1984; Römheld and Marschner, 1986).
Genes involved in strategy I and II processes have been cloned and transformed into different plant species in an effort to alter iron metabolism. In tobacco, the yeast FRE1 and FRE2 genes (responsible for iron reduction in yeast) have been expressed, resulting in an increase in root iron reductase activity (Samuelson et al., 1998). However, this increase did not lead to a higher iron concentration in leaves. More recently new attempts to make plants more efficient in acquiring iron under limiting conditions have been carried out by generating transgenic plants expressing the ferric chelate reductase gene FRO2 (Vasconcelos et al., 2006; Yang et al., 2009). Vasconcelos et al. (2006) found that increasing iron reductase activity in transgenic soybean plants correlated with an increase in iron concentration in the plants when grown under iron-limited conditions.
Once the root cells absorb the iron, it must be transported to the plant's aerial tissues via the xylem transport system (Vasconcelos and Grusak, 2006). A large portion of shoot iron is accumulated in chloroplasts, where it plays a role in the synthesis of chlorophyll. It is usually stored in the central core of the multimeric ferritin protein (Briat et al., 1999). This is the base for approaches to improve plant iron concentration by modifying ferritin levels. Tobacco transformed with a bean ferritin gene demonstrated a 3-fold increase in leaf iron concentration (Van Wuytswinkel et al., 1998).
The Citrus genus is considered to be susceptible to iron chlorosis (Carpena-Artes et al., 1995), with tolerance to iron deficiency being determined by the rootstock. However, large phenotypic variation exists among different rootstock genotypes in their sensitivity/tolerance to iron deficiency. Thus, for example, Cleopatra mandarin (Citrus reshni Hort. ex Tan.) and Sour orange (C. aurantium L.) are highly tolerant to iron deficiency in calcareous soils while Troyer citrange (C. sinensis (L.) Osb.×Poncirus trifoliata (L.) Raf.), Swingle citrumelo (C. paradisi Macf.×P. trifoliata), and P. trifoliata are susceptible (Castle, 1987).
Remediating iron chlorosis of citrus trees on susceptible rootstocks planted on calcareous soils is not easy. In the short term, iron correction through the application of iron sources, including inorganic iron salts, synthetic chelates, and natural organic compounds, can enable crops to recover from iron deficiency symptoms to some extent (Abadia et al., 2004). In practice, FeSO4 and synthetic chelates are the most widely used iron fertilizers and their addition can help overcome iron deficiency imposed by a calcareous soil (Jessop et al., 1990). However, iron deficiency is difficult to correct because of the rapid transformation of iron contained in fertilizers into an unavailable form in soil (Fernandez et al., 2004). Moreover, this practice is too expensive. Because of this and the poor knowledge of the mechanisms underlying the susceptibility to iron deficiency, the easiest way to avoid iron chlorosis in trees on calcareous soils is to use tolerant rootstocks. Nevertheless, when appropriate genotypes are not available, this approach is very difficult to develop and is time-consuming. Therefore, there is an urgent need to get novel insights into the mechanisms operating in plants in response to iron deficiency, to come up with new tools to overcome the problem imposed by iron deficiency and, at the same time, to develop new screening techniques in order to identify chlorosis-tolerant genotypes that could be applied early in breeding programmes (Jolley et al., 1996).
In this work, the differential gene expression induced by iron deficiency in P. trifoliata were examined in order to understand better the mechanisms behind the physiopathy associated with this shortage in a susceptible rootstock. Additional studies of changes in the cell wall, measurement of photosynthesis and chlorophyll content, and peroxidase and catalase activities have been carried out to address the role of the identified genes in the response triggered by iron starvation.
Materials and methods
Plant material and growth conditions
In this work, the susceptible rootstock P. trifoliata Rubidoux has been used for all the experiments. Seeds were harvested form mother seed trees held in the germplasm collection at IVIA. Seeds were sown on 55×40 cm trays containing a mixture of peat and siliceous sand (3:2, v/v) in an aphid-proof greenhouse between 15 °C and 28 °C, at 80% of relative humidity. Five-month-old plants were transplanted into 5.0 l pots containing siliceous sand (peat remains were previously eliminated from the roots) and kept in the greenhouse. After transplanting, the plants were irrigated with water three times a week for 2 weeks for acclimation to the new substrate employed, and then for 2 weeks with nutrient solution [3 mM Ca(NO3)2, 3 mM KNO3, 2 mM MgSO4, 3 mM H3PO4, 18 μM Fe-EDDHA, and trace elements according to Hoagland and Arnon (1950)]. After that period, plants were divided into two groups, one of them was kept in the same conditions as mentioned above (18 μM Fe-EDDHA, control) while the other was watered with nutrient solution without Fe-EDDHA (0 μM Fe-EDDHA, iron deficiency). Plants were grown in these conditions for 60 d. At the end of the experiment, plants were analysed and, after separating the different organs, their weight was measured. A row of plants, not included in the experiment, was placed around the perimeter.
RNA extraction and microarray hybridization
Total RNA was extracted from roots according to the protocol described by Ancillo et al. (2007). RNA was labelled following an indirect method (Randolph and Waggoner, 1997). Reverse transcription, cDNA purification, dye coupling, and fluorescent cDNA purification was accomplished as described by Forment et al. (2005), except that total RNA (40 μg) was used instead of poly(A)+ RNA. RNA from plants subjected to iron deficiency was labelled with Cy3 and RNA from control plants with Cy5. Four biological replicates for each category were generated.
A genome-wide 20 K cDNA microarray developed under the Citrus Functional Genomic Project (CFGP; http://bioinfo.ibmcp.upv.es/genomics/cfgpDB/) was used. The microarray includes 21 081 putative unigenes of citrus (Martinez-Godoy et al., 2008). Microarray hybridization and washing were performed as described by Martinez-Godoy et al. (2008). Slides were hybridized, washed, and, subsequently, scanned at 532 nm for the Cy3 and 635 nm for the Cy5 dyes, with a GenePix 4000B scanner (Axon Molecular Devices), at 100% laser power and 10 nm resolution. Before quantifying the spot intensities using GenePix Pro 6.0 (Axon Molecular Devices), photomultiplier tube voltages were adjusted to equal the overall signal intensity for each channel, to reduce the amount of spots with saturated pixels, and to increase the ratio of the signal-to-noise. Low signal spots, i.e. those with a net intensity in both channels lower than the median signal background plus twice standard deviations, were eliminated.
Microarray data analysis
Median global intensity and LOWESS correction were applied to normalize microarray data analysis (Yang et al., 2002) using the Acuity 4.0 software (Axon Molecular Devices). Only probes for which valid data were obtained in at least three out of the four slides were considered for further analysis (15 178 spots).
To detect differentially expressed genes, data were analysed with the SAM package (Tusher et al., 2001), using two-class unpaired comparison (iron deficiency versus control) with a false discovery rate (FDR) of 33.87%.
Quantitative real-time RT-PCR
Expression of three selected genes was estimated by quantitative real-time RT-PCR using the SYBR Green assay and the LightCycler System (Roche). Total RNA preparations were cleaned up with the RNeasy Plant Mini Kit (Qiagen), treated with DNase I (Rnase-Free DNase Set; Qiagen) and adjusted to 20 ng RNA μl−1 using the Quant-iT RiboGreen RNA assay kit (Invitrogen) according to the manufacturer's intructions. cDNA was synthesized in a 10 μl reaction volume containing 20 ng of DNase-treated RNA, 2.5 pmol each of forward and reverse primers, 1 U of RNase inhibitor (Applied Biosystems), 2 μl of LC FastStart DNA MasterPLUS SYBR Green I (Roche), and 2.5 U of MuLV Reverse Transcriptase (Applied Biosystems). Primer pairs for each gene were designed based on the corresponding sequences available in the database of the CFGP (http://bioinfo.ibmcp.upv.es/genomics/cfgpDB). Verification of C05811H06 expression was carried out by using the primer pair 5′-GACGCTTGTGCTAATCGTCA-3′ and 5′-TCTCCGGCAAGTACTGATCC-3′. For that of C31502B08, the primer pair used was 5′-CAAATGGGGACGTTCAGTTT-3′ and 5′-AGTGCAAGAGCTCCCAGTGT-3′. For verification of C05133B06 expression, the primer pair 5′-ACCTCTGGTTTGACCCCTCT-3′ and 5′-GGTCTTTGGGGAAGGAACTC-3′ was used. The thermal profile consisted of 48 °C for 30 min, 95 °C for 10 min, and 45 cycles of 95 °C for 2 s, 60 °C for 10 s, and 72 °C for 15 s followed by a melting program of 60 °C for 60 s. To transform fluorescence intensity measurements into relative mRNA levels, a 10-fold dilution series of an RNA sample was used as the standard curve. All the experiments were done in triplicate and means were calculated. All PCRs were carried out on three different samples and for each one, the experiments were done in triplicate, and means were calculated.
Microscopy and cell wall staining
For propidium iodide staining of live cell walls, roots were submerged in 5 mg l−1 propidium iodide for 1 min. Confocal laser scanning microscopy was performed using a Leica DM IRE2 microscope. Excitation wavelength was 488 nm.
Ruthenium red staining of roots was performed by placing the root pieces in a solution of 0.01% (w/v) ruthenium red and shaking them for 10 min. Samples were visualized using an Olympus BX 51 microscope with a mounted Olympus DP 12 camera.
Hand-made transverse root sections were stained with a 0.1% (w/v) aqueous solution of Calcofluor White M2R (Sigma F-3543) for 2 min and then washed with water. Fluorescence was visualized under a Nikon Eclipse e600 microscope (340–380 nm excitation wavelength).
Enzyme analysis
A fresh sample of 2 g of young leaves from control and iron-deficient plants was homogenized in a Polytron 3100 (Kinematica, Swizetland) using 10 ml phosphate buffer (10 mM, pH 6,5) containing 0.25 g of insoluble polyvinylpolypyrrolidone (PVPP). The crude extract was centrifuged at 12 000 rpm at 4 °C for 30 min, and the supernatant was used for the catalase assay.
Catalase activity was determined by the measurement of the decrease in hydrogen peroxide (H2O2) determined spectrophotometrically. The reaction (2 ml), containing 50 mM phosphate buffer pH 7.0 and 100 μl of the supernantant, was started by the addition of 10 mM H2O2. The reaction was monitored at 240 nm in a spectrophotometer Shimadzu UV-1610 (Shimadzu, Japan), at room temperature. The molar extinction coefficient used was 43.6 M−1 cm−1. Peroxidase activity was measured with a Shimadzu UV-1601. Adequate amounts of crude enzyme extract were applied in hydrogen peroxide (H2O2) containing 2,2′-azino-bis (3-ethyl benzothiazoline-6-sulphonic acid) (ABTS).
Chlorophyll content
Five young leaves per plant were randomly collected and three 8 mm diameter discs excised per leaf were used for chlorophyll content determination. Chlorophylls were extracted with N,N-dimethylformamide for 72 h in the dark at 4 °C and quantified by measuring the absorbance at 647 and 664 nm (Romero-Aranda et al., 1998) using a Shimadzu UV-1601 spectrophotometer (Shimadzu Corp., Kyoto, Japan).
Statistical analyses
All the data presented correspond to the mean of at least four independent plants. Parameters were statistically tested by analyses of variance and comparisons of means were performed with a Duncan test P ≤0.05). Statistical analyses were performed with Statgraphics Plus for Windows, version 5.1 (Statistical Graphics, Englewood Cliffs, NJ, USA).
Results
Iron deficiency induced expression of ten genes
A cDNA microarray containing 21 081 putative unigenes was used to analyse the gene expression profile of Poncirus trifoliata rootstocks undergoing iron deficiency. For that, total RNA extracted from plants grown in the presence (control) or absence of iron (Fe-deficient), was labelled and hybridized to the microarray. After quality control, 15 178 spots (unigenes) were considered to be valid for further analysis. Expression levels were analysed in a two class unpaired response using significance analysis of microarray (SAM; Tusher et al., 2001). Ten differentially expressed genes induced by iron deficiency showed statistically significant variation, with a false discovery rate (FDR) of 33.87%. These genes are listed in Table 1.
Table 1.
List of genes differentially expressed in Poncirus trifoliata rootstock treated with or without Fe-EDDHA in nutrient solution after 60 d of treatment
| Function | Gene ID | M-value | FDR (%) | Best BLAST hit |
| Cell wall | C31502B08 | 0.714 | 0 | Calmodulin-regulated Ca2+-pump |
| C05140C08 | 0.653 | 30.97 | SYR1, Syntaxin Related Protein 1, also known as PEN1 | |
| C05133B06 | 0.864 | 33.87 | Endo-xyloglucan transferase | |
| C19009B12 | 0.872 | 33.87 | Xyloglucan endotransglucosylase | |
| C05811H06 | 0.604 | 33.87 | Polygalacturonase (pectinase) | |
| Stress response | C05807F01 | 0.770 | 0 | Member ofthe DREB subfamily |
| C31108D11 | 0.550 | 27.10 | Pathogenesis-related family protein | |
| C05802F05 | 0.465 | 30.97 | Protein phosphatase 2C | |
| Electron transport | C34001B04 | 0.355 | 30.97 | Thioredoxin reductase 1 |
| Unknown | C05002B06 | 0.530 | 27.10 | Unknown protein |
Five of these genes were functionally linked to the cell wall (C05811H06, C05133B06, C19009B12, C05140C08, and C31502B08). C05811H06 is related to the pectin component of the wall. Pectins are acidic, non-cellulosic polysaccharides present in the primary cell wall of most plants. On the other hand, C31502B08 may be involved in Ca2+ transport to the cell wall to become part of the pectin component of the primary cell wall. C05133B06 and C19009B12 are related to xyloglucans, the principal group of hemicelluloses in primary cell walls. And C05140C08 is involved in the deposition of cuticle precursors of the primary cell wall.
Within the group of significant genes, there are three presumably involved in plant responses to abiotic (C05807F01 and C05802F05) or biotic (C31108D11) stresses. C34001B04 is a thioredoxin reductase, which mediates the final step in the electron-transfer pathway involved in the regulation of photosynthesis. The last of these genes, C05002B06, has an unknown function.
To validate microarray results, differential expression of some genes was confirmed by quantitative real-time RT-PCR. The relative accumulation of the three mRNAs from C05811H06, C05133B06, and C31502B08) paralleled that observed by microarray hybridization, albeit the changes detected were not quantitatively identical (Table 2). This variation is commonly observed in the validation of microarray results by RT-PCR (Allen and Nuss, 2004; López et al., 2005) and is probably due to intrinsic differences between both techniques.
Table 2.
Changes in gene expression estimated by microarray hybridization and by quantitative real-time RT-PCR
| Gene ID | Best Blast Hit | Primersa | Plant sample | PCR product concentration mean (ng μl−1)b | PCR product concentration mean (ng μl−1)c | Fold change average in QRT-PCRd | Fold Change average in microarraye |
| C05811H06 | Polygalacturonase (pectinase) family protein | 5′-GACGCTTGTGCTAATCGTCA-3′ | PT-2 | 7.15E-10 | 8.33E-10 | 2.44 | 1.54 |
| 5′-TCTCCGGCAAGTACTGATCC-3′ | PT-4 | 1.16E-09 | |||||
| PT-5 | 6.25E-10 | ||||||
| PT-62 | 1.33E-10 | 3.42E-10 | 1.00 | 1 | |||
| PT-63 | 2.90E-10 | ||||||
| PT-64 | 6.02E-10 | ||||||
| C31502B08 | Calmodulin-regulated Ca2+-pump | 5′-CAAATGGGGACGTTCAGTTT-3′ | PT-2 | 1.01E-08 | 8.63E-09 | 2.40 | 1.65 |
| 5′-AGTGCAAGAGCTCCCAGTGT-3′ | PT-4 | 7.26E-09 | |||||
| PT-5 | 8.53E-09 | ||||||
| PT-62 | 3.21E-09 | 3.59E-09 | 1.00 | 1 | |||
| PT-63 | 3.52E-09 | ||||||
| PT-64 | 4.05E-09 | ||||||
| C05133B06 | Xyloglucan transferase | 5′-ACCTCTGGTTTGACCCCTCT-3′ | PT-2 | 2.70E-07 | 2.24E-07 | 2.83 | 1.87 |
| 5′-GGTCTTTGGGGAAGGAACTC-3′ | PT-4 | 1.97E-07 | |||||
| PT-5 | 2.04E-07 | ||||||
| PT-62 | 7.04E-08 | 7.91E-08 | 1.00 | 1 | |||
| PT-63 | 7.52E-08 | ||||||
| PT-64 | 9.17E-08 |
PT-2, PT-4, and PT-5 without Fe; PT-62, PT-63, and PT-64 with Fe.
Primer pair used for gene expression analysis by QRT-PCR.
Estimated PCR product concentration mean calculated over three independent QRT-PCR reactions per independent sample
Total PCR product concentration mean calculated over three independent QRT-PCR reactions for three independent samples for each treatment.
Average fold change of the mean of expression values of four replicas in the microarray in Fe deficient-plants in respect to Fe-supplied plants.
Average fold change of three independent RT-PCR reactions for three independent samples for each treatment.
Iron starvation induced changes in the cell wall
As five of the genes induced by iron deficiency were related to certain components of the cell wall, it was further investigated whether actual changes in cell wall correlated with iron-deficient Poncirus plants.
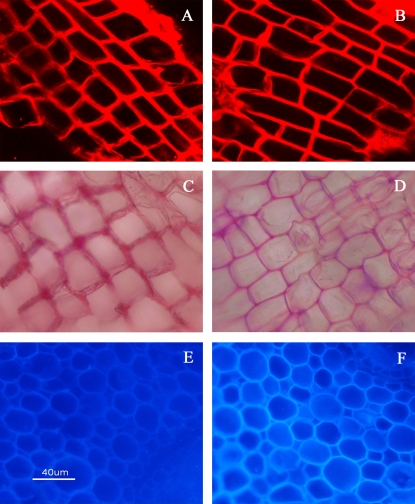
A set of plants was grown in the absence of iron and a different set was grown under standard conditions as control plants. Roots of both sets were stained with propidium iodide, a fluorescent dye commonly used to stain plant roots (Truernit et al., 2006). Confocal laser scanning microscopy (CLSM) revealed that the cell walls of plants undergoing iron starvation were less stained than the control ones (Fig. 1A, B) suggesting a reduction in the cell wall thickness of stressed plants. Although this is a clear indication that iron deficiency is affecting the cell wall, it gives little information as to which component of the cell wall is affected. To obtain more detailed information about the differences in the components of the cell wall that might be altered in response to iron deficiency, more selective dyes were used. When roots were stained for pectin with ruthenium red (Hanke and Northcote, 1975), thinning of the cell wall became clearly evident again in Fe-deficient roots (Fig. 1C, D). In addition, Calcofluor, which binds to various β-D-glucans including cellulose, xyloglucan, callose, and chitin (Hughes and McCully, 1975; Krishnamurthy, 1999), stained the cell walls of roots of Fe-deficient plants to a greater extent than those of control plants (Fig. 1E, F).
Fig. 1.
Propidium iodide staining (A, B), ruthenium red staining (C, D), and calcofluor staining (E, F) in roots of Poncirus trifoliata treated for 60 d with Fe-DDHA (A, C, E) or without Fe-DDHA (B, D, F) in nutrient solution.
Chlorophyll decreased in iron starvation
Chlorophyll content was measured in control and iron-deficient Poncirus plants. As shown in Table 3, iron deficiency caused a 25% decrease in chlorophyll content in iron-deficient plants.
Table 3.
Dry matter (g), catalase (U g−1 FW) and peroxidase activity (U g−1 FW) and total chlorophyll content (mg g−1 DW) in Poncirus trifoliata plants grown with or without Fe in the nutrient solution (values are means of six replicates)
| P. trifoliata | Treatment | Dry wt leaves | Dry wt roots | Total dry wt | Catalase | Peroxidase | Chlorophyll |
| 0 μM Fe | 1.62±0.3 | 6.12±1.2 | 10.79±1.8 | 23.5±6.2 | 390.5±60.1 | 481.4±60.6 | |
| 18 μM Fe | 1.50±0.4 | 5.78±1.0 | 10.16±1.0 | 61.4±15.9 | 761.3±23.1 | 649.9±100.3 |
Leaf, root, and total dry weight was also measured in these plants and no significant variation was found (Table 3).
Peroxidase and catalase activities decreased in iron starvation
Peroxidase and catalase activities were measured because of their implication in oxidative stress (Tewari et al., 2005). As shown in Table 3, both activities decreased in iron-deficient plants. Peroxidase and catalase activity decreased almost 50% and 60%, respectively, in the absence of iron.
Discussion
Low iron bioavailability is a primary constraint to plant growth in many ecosystems, particularly in calcareous soils (Imsande, 1998). Calcareous soils with restricted iron availability are commonly found in the Mediterranean basin. Thus, iron chlorosis is one of the most frequent nutritional problems in citrus production. A genome-wide survey of the genes involved in the plant response to iron deficiency may provide important insights into the functions and regulatory mechanisms that take place in this physiopathy. In this study, microarray analysis was used to investigate the changes in the gene expression profile of Poncirus roots during iron deficiency stress. Ten genes were identified as being differentially overexpressed in iron deficiency.
The putative function of five of these genes (Table 2) can be related to changes at the primary cell wall. The primary cell wall of dicots and non-graminaceous monocots is a dynamic structure of a rigid, rod-like cellulose/xyloglucan load-bearing network that is embedded in and interacts with a compression-resistant pectin network (Carpita and Gibeaut, 1993). The cell wall is also associated with various types of enzymes that are responsible for its construction and modification (Fry, 1995). The C05133B06 and C19009B12 sequences are very similar to Arabidopsis endo-xyloglucan transferase and xyloglucan endotransglycosylase proteins, respectively, which are involved in the metabolism of xyloglucans that mediate the interchange between xyloglucan cross-links in the framework (Fry et al., 1992). Simple cleavage of load-bearing xyloglucan cross-links will result in disassembly of the cellulose–xyloglucan framework. This process occurs in a number of developmental and stress situations as fruit ripening, organ abscission, degradation of storage tissues in germinating seeds, in most growing tissues, in pathogen defence and in response to environmental stimuli (Xu et al., 1995; Nishitani, 1998; Divol et al., 2007). On the other hand, C05811H06 showed homology to an Arabidopsis pectinase and the C31502B08 homologue is a calmodulin-regulated Ca2+-pump. Pectins are the major component of the cell walls in higher plants, being particularly abundant in the primary cell walls (Carpita and Gibeaut, 1993). Ca2+ interacts with the free carboxyl groups present in pectins forming a gel that is determinant for the properties of the cell wall (Storey et al., 2005). Ezaki et al. (2005) suggest that Ca2+ plays an important role in the regulation of wall extensivity during the acid-induced wall expansion by reacting with carboxyl groups of pectin in the cell wall. Pectins are thought to be responsible for cell-to-cell adhesion, and the determination of cell wall porosity and strength (Usadel et al., 2004). In addition, pectins have been proposed to control cell wall thickness (Jarvis, 1992). If so, changes produced in the pectin component as a consequence of the induction of genes involved in cell wall restructuration in response to iron deficiency, could account for the differences in the thickness that were observed in the cell wall of root cells. Moreover cell walls in roots from Fe-deficient plants were stained intensely with Calcofluor, suggesting that either the level of primary cell wall polysaccharides (presumably xyloglucans) was higher in these walls or dye accessibility became higher due to structural changes in these cells, which reconciles with the induction of genes involved in xyloglucan metabolism in these plants. Finally, the C05140C08 Arabidopsis homologue is PEN1 (also referred as SYP121), a plasma membrane syntaxin presumably involved in the deposition of cuticle precursors of the primary cell wall (Assaad et al., 2004). Cross-linking of cell wall polymers and the composition of the cell wall most probably determine the wall mechanical properties. Plasticity of the cell wall is probably important for plant acclimation to the local environment (iron deficiency in our case). However, whether this is a mechanism to improve adaptation to iron shortage or it is a final consequence of the stress produced by the iron deficiency remains to be elucidated.
The C05807F01 potentially encodes a member of the dehydration-responsive element binding (DREB) transcription factor family which plays a central role in regulating expression of stress-inducible genes under abiotic stresses such as drought, high salt, and low temperature, as well as abscisic acid treatment (Chen et al., 2007). Some of these DREB factors have been shown to improve the tolerance to high salt, drought, and cold stresses when overexpressed in transgenic plants (Liu et al., 2007; Chen et al., 2009). To our knowledge, no reports relate expression of DREB factors to iron starvation stress. However, induction of these factors is quite unspecific since it responds to a wide range of abiotic stresses. Therefore, it cannot be ruled out that C05807F01 may be involved in the response mechanisms to the stress caused by iron deficiency in Poncirus plants. The same applies to C05802F05, which shows homology to an Arabidopsis protein phosphatase 2C (PP2C). This family of plant proteins has been implicated as negative modulators of signalling pathways involved in responses to environmental stresses (Schweighofer et al., 2004). C31108D11 showed high homology to pathogenesis-related family proteins that are induced upon K+ starvation and caesium chloride exposure according to microarray experiments available in the Genevestigator database.
Poncirus plants growing in iron-depleted soil showed a chlorophyll content lower than control iron-fed plants. Such impairment in chlorophyll accumulation very likely results from the requirement of iron for the synthesis of chlorophyll precursors, namely aminolevulinic acid and protochlorophyllide (Marschner, 1986).
Leaf fresh weight did not show any significant variation between control and iron-starved plants, indicating the absence of a water deficit derived from iron shortage, or short-term effects in biomass production. Nevertheless, it cannot be ruled out that these effects could occur in long-term Fe deficiency (beyond the 60 d monitored in this experiment).
Stenbaek et al. (2008) found an Arabidopsis mutant in the NADPH thioredoxin reductase C gene (ntrc) that is perturbed in chlorophyll biosynthesis, thereby establishing a direct relation of this enzyme with photosynthesis. The authors suggest that ntrc is particularly important during periods with limited reducing power from photosynthesis, which could be the case of Poncirus plants undergoing iron starvation.
On the other hand Tewari et al. (2005) reported that iron-starved maize plants showed clear signs of oxidative stress in iron-deficient (chlorotic) leaves due to decreased levels in the activities of catalase, peroxidase, ascorbate peroxidase, and superoxide dismutase, which entails an increase in the concentration of H2O2 and the accumulation of a superoxide anion radical (O2–). NDTR have been reported to be effective in reducing H2O2 (Moller, 2006) so it can be related to a role in trying to reduce the level of oxidative stress in iron-starved plants. Our results in Poncirus showed a reduction in the level of catalase and peroxidase activities in iron-deficient plants, suggesting an increase of H2O2 and other ROS that could lead to oxidative stress. The induction of NDTR (C34001B04) in iron-deficient plants can be the response of the plant to prevent damage by the oxidative stress caused by the iron deficiency. This mechanism of protection against oxidative damage has been reported for other plants (Perez-Ruiz et al., 2006).
These results will provide important information to understand the molecular mechanisms of iron susceptibility. At present, parallel studies are being conducted with tolerant rootstocks to understand the mechanisms associated with iron deficiency better. Altogether, these results will give an overview of the molecular mechanisms involved in susceptibility/tolerance to iron deficiency and will help in the identification of molecular markers linked to iron-susceptibility for further studies.
Acknowledgments
This work was supported by research projects to MA Forner from INIA RTA2008-0060 (Ministerio de Educación y Ciencia, Spain) to LN from Ministry Science and Innovation AGL2008-00596 and Generalitat Valenciana Prometeo-2008/121 projects and the financial help of the European Community (FEDER and ESF funds).
References
- Abadía J, Álvarez-Fernández A, Rombolà AD, Sanz M, Tagliavini M, Abadia A. Technologies for the diagnosis and remediation of Fe deficiency. Soil Scence and Plant Nutrition. 2004;50:965–971. [Google Scholar]
- Allen TD, Nuss DL. Specific and common alterations in host gene transcript accumulation following infection of the chestnut blight fungus by mild and severe hypoviruses. Journal of Virology. 2004;78:4145–4155. doi: 10.1128/JVI.78.8.4145-4155.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ancillo G, Gadea J, Forment J, Guerri J, Navarro L. Class prediction of closely related plant varieties using gene expression profiling. Journal of Experimental Botany. 2007;58:1927–1933. doi: 10.1093/jxb/erm054. [DOI] [PubMed] [Google Scholar]
- Assaad FF, Qiu JL, Youngs H, et al. The PEN1 syntaxin defines a novel cellular compartment upon fungal attack and is required for the timely assembly of papillae. Molecular Biology of the Cell. 2004;15:5118–5129. doi: 10.1091/mbc.E04-02-0140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Briat JF, Lobréaux S, Grignon N, Vansuyt G. Regulation of plant ferritin synthesis: how and why. Cellular and Molecular Life Sciences. 1999;56:155–166. doi: 10.1007/s000180050014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carpena-Artes O, Moreno JJ, Lucena JJ, Carpena-Ruiz RO. Response to iron chlorosis of different hydroponically grown Citrus varieties. In: Abadia J, editor. Iron nutrition in soils and plants. Dordrecht, The Netherlands: Kluwer Academic Publishers; 1995. pp. 147–151. [Google Scholar]
- Carpita NC, Gibeaut DM. Structural models of primary cell walls in flowering plants: consistency of molecular structures with the physical properties of the walls during growth. The Plant Journal. 1993;3:1–30. doi: 10.1111/j.1365-313x.1993.tb00007.x. [DOI] [PubMed] [Google Scholar]
- Castle WS. Citrus rootstocks. In: Rom RC, Carlson RF, editors. Rootstocks for fruit crops. New York: John Wiley and Sons; 1987. pp. 361–369. [Google Scholar]
- Chapman HD. The mineral nutrition of citrus. In: Reuther W, Batchelor LD, Webber HJ, editors. The citrus industry. Vol. II. Davis, CA: University of California Press; 1968. pp. 162–163. [Google Scholar]
- Chen M, Xu ZS, Xia LQ, Li LC, Cheng XG, Dong JH, Wang QY, Ma YZ. Cold-induced modulation and functional analyses of the DRE-binding transcription factor gene, GmDREB3, in soybean (Glycine max L.) Journal of Experimental Botany. 2009;60:121–135. doi: 10.1093/jxb/ern269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen M, Zhaoshi X, Lanqin X, Liancheng L, Xianguo C, Jianhui D, Qiaoyan W Youzhi M. Cold-induced modulation and functional analyses of the DRE-binding transcription factor gene, GmDREB3, in soybean (Glycine max L.) Journal of Experimental Botany. 2007;60:121–135. doi: 10.1093/jxb/ern269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Divol F, Vilaine F, Thibivilliers S, Kusiak C, Sauge MH, Dinant S. Involvement of the xyloglucan endotransglycosylase/hydrolases encoded by celery XTH1 and Arabidopsis XTH33 in the phloem response to aphids. Plant, Cell and Environment. 2007;30:187–201. doi: 10.1111/j.1365-3040.2006.01618.x. [DOI] [PubMed] [Google Scholar]
- Ezaki N, Kido N, Takahashi K, Katou K. The role of wall Ca2+ in the regulation of wall extensibility during the acid-induced extension of soybean hypocotyl cell walls. Plant and Cell Physiolology. 2005;46:1831–1838. doi: 10.1093/pcp/pci199. [DOI] [PubMed] [Google Scholar]
- Fernández V, Winkelmann G, Ebert G. Iron suply to tobacco plants trough foliar application of iron citrate and ferric dimerum acid. Physiologia Plantarum. 2004;122:380–385. [Google Scholar]
- Forment J, Gadea J, Huerta L, et al. Development of a citrus genome-wide EST collection and cDNA microarray as resources for genomic studies. Plant Molecular Biology. 2005;57:75–91. doi: 10.1007/s11103-004-7926-1. [DOI] [PubMed] [Google Scholar]
- Fry SC. Polysaccharide-modifying enzymes in the plant cell wall. Plant Molecular Biology. 1995;46:497–520. [Google Scholar]
- Fry SC, Smith RC, Renwick KF, Martin DJ, Hodge SK, Matthews KJ. Xyloglucan endotransglycosylase a new wall-loosening enzyme activity from plants. Biochemical Journal. 1992;282:821–828. doi: 10.1042/bj2820821. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hanke DE, Northcote DH. Molecular visualization of pectin and DNA by ruthenium red. Biopolymers. 1975;14:1–17. doi: 10.1002/bip.1975.360140102. [DOI] [PubMed] [Google Scholar]
- Hoagland DR, Arnon DS. CA, Agricultural Experimental Station Circular 374. Berkeley: University of California; 1950. The water culture method for growing plants without soil. [Google Scholar]
- Hughes J, McCully ME. The use of an optical brightener in the study of plant structure. Stain Technology. 1975;50:319–329. doi: 10.3109/10520297509117082. [DOI] [PubMed] [Google Scholar]
- Imsande J. Iron, sulfur, and chlorophyll deficiencies: a need for an integrative approach in plant physiology. Physiologia Plantarum. 1998;103:139–144. [Google Scholar]
- Jarvis MC. Control of thickness of collenchyma cell-walls by pectins. Planta. 1992;187:218–220. doi: 10.1007/BF00201941. [DOI] [PubMed] [Google Scholar]
- Jessop RS, Roth G, Sale P. Effects of increased levels of soil CaCO3 on lupin (Lupinus angustifolius) growth and nodulation. Australian Journal of Soil Research. 1990;28:955–962. [Google Scholar]
- Jolley VD, Cook KA, Hansen NC, Stevens WB. Plant physiological responses for genotypic evaluation of iron efficiency in strategy I and strategy II plants: a review. Journal of Plant Nutrition. 1996;19:1241–1255. [Google Scholar]
- Krishnamurthy KV. Methods in cell wall cytochemistry. Boca Raton, Fla: CRC Press; 1999. [Google Scholar]
- Liu N, Zhong NQ, Wang GL, Li LJ, Liu XL, He YK, Xia GX. Cloning and functional characterization of ppdbf1 gene encoding a DRE-binding transcription factor from Physcomitrella patens. Planta. 2007;226:827–838. doi: 10.1007/s00425-007-0529-8. [DOI] [PubMed] [Google Scholar]
- López C, Soto M, Restrepo S, et al. Gene expression profile in response to Xanthomonas axonopodis pv. manihotis infection in cassava using a cDNA microarray. Plant Molecular Biology. 2005;57:393–410. doi: 10.1007/s11103-004-7819-3. [DOI] [PubMed] [Google Scholar]
- Marschner H. Mineral nutrition of higher plants. London: Academic Press; 1986. [Google Scholar]
- Marschner H, Romheld V, Kissel M. Different strategies in higher plants in mobilization and uptake of iron. Journal of Plant Nutrition. 1986;9:3–7. [Google Scholar]
- Martinez-Godoy MA, Mauri N, Juárez J, Marques MC, Santiago J, Forment J, Gadea J. A genome-wide 20k citrus microarray for gene expression analysis. BMC Genomics. 2008;9:318. doi: 10.1186/1471-2164-9-318. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Møller IM. Essay 11.5: Reactive oxygen species (ROS) and plant respiration. Plant Physiology. 2006 Fourth edition online: < http://www.plantphys.net>. [Google Scholar]
- Nishitani K. Construction and restructuring of the cellulose-xyloglucan framework in the apoplast as mediated by the xyloglucan related protein family: a hypothetical scheme. Journal of Plant Research. 1998;111:159–166. [Google Scholar]
- Pérez-Ruiz JM, Spínola MC, Kirchsteiger K, Moreno J, Sahrawy M, Cejudo FJ. Rice NTRC is a high-efficiency redox system for chloroplast protection against oxidative damage. The Plant Cell. 2006;18:2356–2368. doi: 10.1105/tpc.106.041541. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Randolph JB, Waggoner AS. Stability, specificity and fluorescence brightness of multiply-labelled fluorescence DNA probes. Nucleic Acids Research. 1997;25:2923–2929. doi: 10.1093/nar/25.14.2923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Romero-Aranda R, Moya JL, Tadeo FR, Legaz F, Primo-Millo E, Talon M. Physiological and anatomical disturbances induced by chloride salts in sensitive and tolerant citrus: beneficial and detrimental effects of cations. Plant, Cell and Environment. 1998;21:1243–1253. [Google Scholar]
- Römheld V, Marschner H. Evidence for a specific uptake system for iron phytosiderophores in roots of grasses. Plant Physiology. 1986;80:175–180. doi: 10.1104/pp.80.1.175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Samuelson AJ, Martin RC, Mok DWS, Machteld CM. Expression of the yeast FRE genes in transgenic tobacco. Plant Physiology. 1998;118:51–58. doi: 10.1104/pp.118.1.51. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schweighofer A, Hirt H, Meskiene I. Plant PP2C phosphatases: emerging functions in stress signalling. Trends in Plant Science. 2004;9:236–243. doi: 10.1016/j.tplants.2004.03.007. [DOI] [PubMed] [Google Scholar]
- Stenbaek A, Hansson A, Wulff R, Hansson M, Dietz K, Jensen P. ADPH-dependent thioredoxin reductase and 2-Cys peroxiredoxins are needed for the protection of Mg–protoporphyrin monomethyl ester cyclase. FEBS Letters. 2008;582:2773–2778. doi: 10.1016/j.febslet.2008.07.006. [DOI] [PubMed] [Google Scholar]
- Storey R, Treeby MT, Milne DJ. Crease: another Ca deficiency-related fruit disorder? Journal of Horticultural Science and Biotechnology. 2005;77:565–571. [Google Scholar]
- Takagi S, Nomoto K, Takemoto T. Physiological aspect of mugineic acid, a possible phytosiderophore of graminaceous plants. Journal of Plant Nutrition. 1984;7:1–5. [Google Scholar]
- Tewari RK, Kumar P, Neetu Sharma PN. Signs of oxidative stress in the chlorotic leaves of iron starved plants. Plant Science. 2005;169:1037–1045. [Google Scholar]
- Truernit E, Siemering KR, Hodge S, Grbic V, Haseloff J. A map of KNAT gene expression in the Arabidopsis root. Plant Molecular Biology. 2006;60:1–20. doi: 10.1007/s11103-005-1673-9. [DOI] [PubMed] [Google Scholar]
- Tusher VG, Tibshirani R, Chu G. Significance analysis of microarrays applied to the ionizing radiation response. Proceedings of the National Academy of Sciences, USA. 2001;98:5116–5121. doi: 10.1073/pnas.091062498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Usadel B, Kuchinsky AM, Rosso MG, Eckermann N, Pauly M. RHM2 is involved in mucilage pectin synthesis and is required for the development of the seed coat in Arabidopsis. Plant Physiology. 2004;134:286–295. doi: 10.1104/pp.103.034314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vasconcelos M, Grusak MA. Status and future developments involving plant iron in animal and human nutrition. In: Barton LL, Abadía J, editors. Iron nutrition in plants and rizospheric microorganisms. Dordrecht, The Netherlands: Springer; 2006. pp. 85–101. [Google Scholar]
- Vasconcelos M, Eckert H, Arahana V, Graef G, Grusak MA, Clemente T. Molecular and phenotypic characterization of transgenic soybean expressing the Arabidopsis ferric chelate reductase gene, FRO2. Planta. 2006;224:1116–1128. doi: 10.1007/s00425-006-0293-1. [DOI] [PubMed] [Google Scholar]
- Van Wuytswinkel O, Vansuyt G, Grignon N, Fourcroy P, Briat JF. Iron homeostasis alteration in transgenic tobacco overexpressing ferritin. The Plant Journal. 1998;17:93–97. doi: 10.1046/j.1365-313x.1999.00349.x. [DOI] [PubMed] [Google Scholar]
- Xu W, Purugganan MM, Polisensky DH, Antosiewicz DM, Fry SC, Brama J. Arabidopsis TCH4, regulated by hormones and the environment, encodes a xyloglucan endotransglycosylase. The Plant Cell. 1995;7:1555–1567. doi: 10.1105/tpc.7.10.1555. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang J, Liu Y, Liu Y, Zhang W, Huang J, Li J, Du CC, Sui Z, Wang-Pruski G. Characterization of chlorosis-resistant apple mutants of transgenic FRO2. Acta Horticulturae. 2009;829:251–257. [Google Scholar]
- Yang YH, Dudoit S, Luu P, Lin DM, Peng V, Ngai J, Speed TP. Normalization for CDNA microarray data: a robust composite method addressing single and multiple slide systematic variation. Nucleic Acids Research. 2002;30:15. doi: 10.1093/nar/30.4.e15. [DOI] [PMC free article] [PubMed] [Google Scholar]