Abstract
Targeting of ion channels to caveolae, a subset of lipid rafts, allow cells to respond efficiently to extracellular signals. Hyperpolarization-activated cyclic nucleotide-gated channel (HCN) 4 is a major subunit for the cardiac pacemaker. Caveolin-3 (Cav3), abundantly expressed in muscle cells, is responsible for forming caveolae. P104L, a Cav3 mutant, has a dominant negative effect on wild type (WT) Cav3 and associates with limb-girdle muscular dystrophy and cardiomyopathy. HCN4 was previously shown to localize to lipid rafts, but how caveolae regulate the function of HCN4 is unknown. We hypothesize that Cav3 associates with HCN4 and regulates the function of HCN4 channel. In this study, we applied whole-cell patch clamp analysis, immunostaining, biotinylation, and immunoprecipitation methods to investigate this hypothesis. The immunoprecipitation results indicated an association of HCN4 and Cav3 in the heart and in HEK293 cells. Our immunostaining results showed that HCN4 colocalized with Cav3 but only partially colocalized with P104L in HEK293 cells. Transient expression of Cav3, but not P104L, in HEK 293 cells stably expressing HCN4 caused a 45% increase in HCN4 current (IHCN4) density. Transient expression of P104L caused a two-fold increase in the activation time constant for IHCN4 and shifted the voltage of the steady-state inactivation to a more negative potential. We conclude that HCN4 associates with Cav3 to form a HCN4 macromolecular complex. Our results indicated that disruption of caveolae using P104L alters HCN4 function and could cause a reduction of cardiac pacemaker activity.
Hyperpolarization-activated cation currents are responsible for generating spontaneous pacemaker activities in the heart (1) and central nervous system (2). The cardiac pacemaker current is responsible for intrinsic generation of rhythmic and spontaneous firing of action potentials in the heart. Hyperpolarization-activated cyclic nucleotide-gated (HCN1) channels are the key components of the cardiac pacemaker (3). Four different isoforms of HCN (1–4) have been cloned and studied. Among these isoforms, transcripts of HCN4 are most abundantly detected in cardiac tissues (3, 4), while transcripts of HCN2 are abundantly detected in both cardiac and brain tissues. All HCN channels have a signature cAMP-binding domain (CNBD, 120 amino acids) in the C-terminal region. Each isoform differs from the other by its voltage-dependence, activation properties, and cAMP-dependent modulation when overexpressed in a heterologous system (5, 6).
The cell surface membrane is a heterogeneous mixture of proteins, cholesterol, glycerolipids, phospholipids, and sphingolipids. Cholesterol and sphingolipids laterally associate with one another and form liquid-ordered microdomains, popularly known as lipid rafts. These cholesterol and sphingolipid-enriched membrane microdomains are believed to coordinate multiple cellular processes including second messenger signaling, recycling of proteins to the membrane, and biophysical properties of ion channels. Caveolae are one such type of lipid rafts. Caveolae are characterized by the presence of caveolins, which are scaffolding proteins that interact with cholesterol and provide the structural framework for macromolecular signaling complexes (7, 8). Three known members exist in the caveolin family, caveolin1–3. Cav3 (but not Cav1 and 2), which is abundantly expressed in cardiac and skeletal muscles, is found to be colocalized with several membrane proteins including ion channels (9–14). HCN4 protein localizes to the same fraction as Cav3 in the rabbit sinoatrial node (13). Also, disruption of lipid rafts using methyl-β-cyclodextrin can modulate properties of HCN4 channels in HEK293 cells and cardiac pacemaker currents (If) at the sinoatrial node (13). However, it is still unknown if HCN4 associates with Cav3. Do caveolae, a subset of lipid rafts, regulate properties of HCN4 channels? HCN4 mutations can disrupt IHCN density and are associated with sinus dysfunction as well as cardiac arrhythmia (15, 16). It is known that Cav3 mutations can be deleterious as they have been associated with diseases such as cardiomyopathy (17), limb-girdle muscular dystrophy (18), and long QT syndrome (14). A dominant negative P104L mutation leads to a ~95% reduction in Cav3 protein expression (11). The P104L mutation, which is associated with cardiomyopathy and limb-girdle muscular dystrophy, is located in the transmembrane spanning region of Cav3 (18). Interestingly, the P104L mutant protein is trapped in the Golgi (19). We hypothesize that the HCN4 channel associates with Cav3 in the heart and disruption of caveolae can affect trafficking and modulation of HCN4 channel properties. In the present study, we demonstrate that the HCN4 channel colocalizes and associates with Cav3 in mouse hearts and HEK293 cells. Our patch clamp experiments show that Cav3 modulates the HCN4 channel properties in the HEK293 cell line stably expressing the HCN4 channel. The changes described could cause a reduction of cardiac pacemaker activity.
MATERIALS AND METHODS
Gene Subcloning and Expression
HCN4 cDNA (kindly provided by Dr. Jeanne Nerboone, Washington University-St. Louis) was subcloned into the eukaryotic expression vector pcDNA3 (Invitrogen, Carlsbad, CA). WT Cav3 and Cav3 mutant P104L (kindly provided by Dr. Timothy Kamp, University of Wisconsin) were subcloned to 5′ of a bicistronic vector (pIRESGFP1, kind gift from David Johns, Johns Hopkins University) to express the desired Cav3 construct and GFP under the control of a cytomegalovirus promoter. After subcloning, the sequences of all clones were confirmed by DNA sequencing analysis using an automated DNA sequencing technique at the Biotech Center of the University of Wisconsin.
HEK293 cells were cultured as previously described (20). For patch clamp analysis, approximately 1 × 105 HEK293 cells were seeded on a 35-mm-diameter dish (Falcon 3001) with 1.5 mL of culture media. For biochemical experiments, approximately 6 × 105 HEK 293 cells were seeded on a 100-mm-diameter dish (Falcon 3003) with 10 mL of culture media. The expression of all gene constructs was performed with Fugene 6 (Roche, Indianapolis, IN) according to the manufacturer’s recommended protocol. The method for selection of stably transfected cells was described previously (20). About 0.7 μg (for patch clamp analysis) or 4 μg (for biochemical experiments) of cDNA-IRES-GFP was transiently expressed in HEK293 cells where HCN4 is stably expressed.
Biochemical Analysis for Biotinylation, Immunoblotting, Immunoprecipitation, and Immunostaining
Anti-Cav3 monoclonal antibody (BD BioSciences Inc., San Jose, CA), anti-HCN4 polyclonal antibody (Alomone Laboratories, Ltd., Jerusalem, Israel), and antibiotin goat polyclonal antibody (Abcam Inc. Cambridge, MA) were utilized in biotinylation, Western blotting, immunoprecipitation, and immunostaining studies. Detailed methods for the biochemical analysis were described previously (14, 21, 22). Protein concentration was measured using a DC protein assay kit (Bio-Rad, Hercules, CA) and an ELx808 microplate reader (BioTek, Winooski, VT) with a wavelength of 750 nm. All proteins for immunoprecipitation were precleared by incubating with 50 μL of a 1:1 slurry of protein G-Dynabeads (Invitrogen) for 30 min. In immunostaining studies, the Alexa Fluor 568 antimouse (Invitrogen, Carlsbad, CA) or the Alexa Fluor 488 antirabbit (Invitrogen, Carlsbad, CA) second antibody was utilized with a ratio of 1:250.
Voltage Clamp Techniques
Whole cell HCN currents (IHCN) were measured from HEK293 cells after 24 h of transfection by using whole-cell voltage clamping techniques. The extracellular solution contains in (mM) NaCl, 110; MgCl2, 0.5; KCl, 30; CaCl2, 1.8; and HEPES, 5. The pipet solution contained (in mM) NaCl, 10; MgCl2, 0.5; KCl, 130; HEPES, 5; and EGTA, 1. Cells with green fluorescence were chosen for patch clamp experiments. Data was recorded at room temperature using an Axopatch 200B amplifier, Digidata1322A data acquisition system, and pCLAMP 9 software (Molecular Devices, Sunnyvale, CA). The electrodes were pulled (P-87; Sutter Instrument, Novato, CA). Voltage clamp protocols are presented with the data.
Data Analysis
Activation voltage for 50% of IHCN activated (V1/2) was measured by fitting each current–voltage curve for individual experiments using the Boltzmann equation. Activation time constant (τ) was calculated by fitting each current trace at the testing potential of −120 mV using single exponential fits. Parameter fits were obtained, and a one-way ANOVA with Bonferroni correction was performed to determine statistical significance among groups of mean data. Statistical significance was determined by a value P < 0.05.
RESULTS
Stable Expression of HCN4 in HEK293 Cells
A HCN4 stable cell line was generated in HEK293 cells for studying the effects of Cav3 on the HCN4 channel. The HCN4 stable cell line was confirmed using Western blotting analysis, immunostaining, and whole cell patch clamp recording. Representative data are shown in Figure 1. Each method was repeated 3 times to ensure reproducibility. Total protein (10 μg) from untransfected HEK293 cells and HCN4 stably expressed cells were isolated for Western blotting analysis. A 160 kDa band was detected from protein isolated from HCN4 stable cells (Figure 1, panel A) but not from protein sample isolated from untransfected HEK293 cells (negative control; Figure 1, panel A). Results from Western blotting experiments showed that the stable cell line expressed HCN4 protein. Plasma membrane expression of HCN4 in the stable cell lines was also examined using immunostaining with a rabbit polyclonal antibody to HCN4 followed by confocal microscopy. HEK293 cells with stably expressed HCN4 were labeled with green fluorescence at the cell periphery and around the nucleus under a FITC filter (Figure 1, panel B). Most of the HCN4 protein reached the surface membrane with some still localized around the cell nucleus. Results from immunostaining confirmed that cells from HCN4 stable cell line express HCN4 protein. Next, to determine the functional properties of the expressed HCN4 channel protein in the stable cells, we measured whole cell HCN4 current (IHCN) with the protocol shown in Figure 1, panel C. IHCN was elicited by a step hyperpolarization to −120 mV from a holding potential of −40 mV (Figure 1, panel C; black trace) and then blocked with 2 mM Cs+ (Figure 1, panel C; red trace). Results from the whole patch clamp method demonstrate that the current recorded from the stable cell line can be activated by hyperpolarized potential and is sensitive to Cs+. Together these data demonstrate the functional expression and localization of the HCN4 channel protein in the plasma membrane in the stable cell line.
Figure 1.
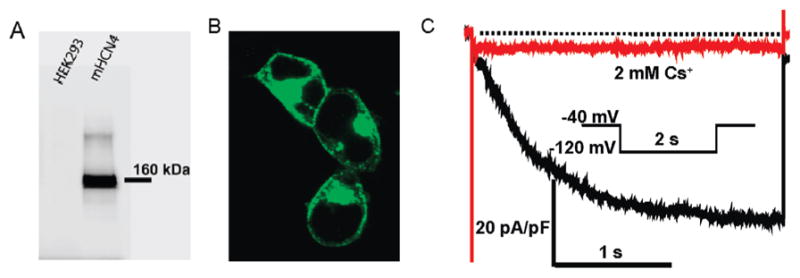
HCN4 stably expressed in HEK293 cells. (Panel A) Detecting HCN4 protein from a HCN4 stable cell line (right) but not from HEK293 cell protein (left). (Panel B) Confocal image of HCN4 stable cell lines immunostained with anti-HCN4 antibody. (Panel C) Representative current traces recorded from HCN4 stable cell lines before (black) and after (red) perfusion of 2 mM Cs+.
Association of the HCN4 Channel with Cav3 in the Heart
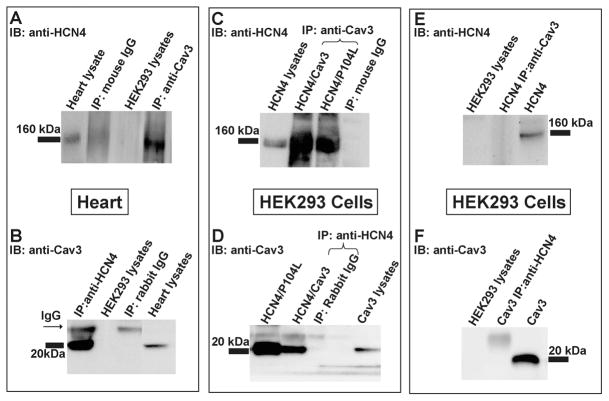
We sought to determine whether HCN4 associates with Cav3 in the heart. We performed immunoprecipitation experiments using anti-HCN4 antibody, anti-Cav3 antibody, or nonimmune IgG (as control) on solublized mouse heart homogenates. The immunoprecipitated (IPed) samples were then analyzed by Western blotting. Upon probing the blots with anti-HCN4 antibody and anti-Cav3 antibody, we found that HCN4 channel protein (160 kDa) was detected in the anti-Cav3 IP lane, and conversely, Cav3 (20 kDa) co-IPed with anti-HCN4 antibody (Figure 2A and B). The nonimmune IgG control IP sample detected an IgG band (~30 kDa) but did not detect either HCN4 or Cav3 protein bands (Figure 2B). The results indicate that the HCN4 channel and Cav3 are associated with one another in the mouse heart (Figure 2A and B).
Figure 2.
Representative images from Co-Ip and Western blot analysis of HCN4 and Cav3 from the mouse heart (panels A and B) and HEK293 cells transiently expressed with HCN4 and/or Cav3 (panels C, D, E, and F). Panel A: mouse heart lysate was Co-IPed with anti-Cav3 antibody or mouse IgG, and then the Co-IP samples were analyzed by Western blotting with anti-HCN4 antibody. Panel B: mouse heart lysate was Co-IPed with anti-HCN4 antibody or rabbit IgG, and then the Co-Ip samples were analyzed by Western blotting with anti-Cav3 antibody. Panel C: lysates of HEK293 cells expressing HCN4 and Cav3 were co-IPed with anti-Cav3 antibody or mouse IgG, and then the Co-IP samples were analyzed by Western blotting with anti-HCN4 antibody. Panel D: lysates of HEK293 cells expressing HCN4 and Cav3 were co-IPed with anti-HCN4 antibody or mouse IgG, and then the Co-IP samples were analyzed by Western blotting with anti-Cav3 antibody. Panel E and F: control IP experiments using lysates from HCN4 transfected HEK293 cells IPed with anti-Cav3 antibody (panel E), or Cav3 transfected HEK293 cells IPed with anti-HCN4 antibody (panel F).
To further confirm these results, we performed coimmunoprecipitation experiments from HEK293 cells lysates that stably express HCN4 channels. These cells were transiently transfected with Cav3 or P104L. The P104L is a dominant negative and trafficking defective mutation of Cav3 and was first described in limb girdle muscular dystrophy patients (18). P104L oligomerizes with WT Cav3 and gets retained in the Golgi compartments and thus disrupts caveolar formation (23). Here, we used P104L to disrupt caveolar formation and also to study the effect of this mutation on the trafficking pattern of HCN4 channel protein in HEK293 cells. Lysates from transfected HEK293 cells were IPed with anti-HCN4 antibody, anti-Cav3 antibody, or nonimmune IgG (negative control). The co-IP samples were analyzed by Western blot by probing with anti-HCN4 and anti-Cav3 antibodies. A protein band for HCN4 (160 kDa) was detected in both HCN4+Cav3 and HCN4+P104L lanes as well as in the lysate lane (Figure 2C). When we performed converse immunoprecipitation experiments using anti-Cav3 antibody, we detected a 20 kDa band for Cav3 protein in both HCN4 + Cav3 and HCN4 + P104L lanes, and the lysate lanes (Figure 2D). In both sets of immunoprecipitation experiments, the control IgG IP lanes did not show protein bands for either Cav3 or HCN4. In addition, a control experiment was performed using protein isolated from HEK293 cells expressing HCN4 or Cav3 (Figure 2E and F). The protein samples from cells expressing either HCN4 or Cav3 were immunoprecipitated with anti-Cav3 or anti-HCN4, respectively. No protein bands for either HCN4 or Cav3 were detected using Western blotting analysis. These results confirm that the HCN4 and Cav3 proteins may be associated with one another. P104L does not disrupt the association between Cav3 and HCN4. These immunoprecipitation studies suggest that Cav3 could play a role in the modulation of HCN4 channel properties.
Colocalization of HCN4 and WTCav3 or Mutant Cav3 (P104L)
We investigated the trafficking patterns of HCN4 and colocalization with Cav3 or P104L using immunostaining and confocal microscopy. Cav3, P104L, or the expression vector pcDNA3 was transiently transfected into the HCN4 stable cells. Twenty-four hours after transfection, the cells were fixed and coimmunostained using antibodies specific to HCN4 and Cav3. The HCN4 channel was detected in the green channel using FITC conjugated secondary goat anti-rabbit antibody (Figure 3, left panels), whereas Cav3 immunostaining was detected in the red channel using goat antimouse alexa-568 antibody (Figure 3. middle panels). Images in the right panels are from superimposed images from left and middle panels. Expression of pcDNA3 in HCN4 stable cells did not affect HCN4 trafficking and did not generate a nonspecific signal for Cav3 (Figure 3, panel A). The merged image (yellow) shows that HCN4 (green) and Cav3 (red) were colocalized at the surface membrane when Cav3 was expressed in HCN4 stable cells (Figure 3, panel B). However, trafficking defective P104L trapped some HCN4 in the intracellular compartments around the nucleus with little HCN4 channel protein reaching the surface membrane (Figure 3, panel C). The results indicated that HCN4 channel protein colocalized with Cav3 in the caveolar microdomains and that P104L can cause abnormal trafficking of HCN4 channels and reduce the expression level HCN4 protein at the surface membrane.
Figure 3.
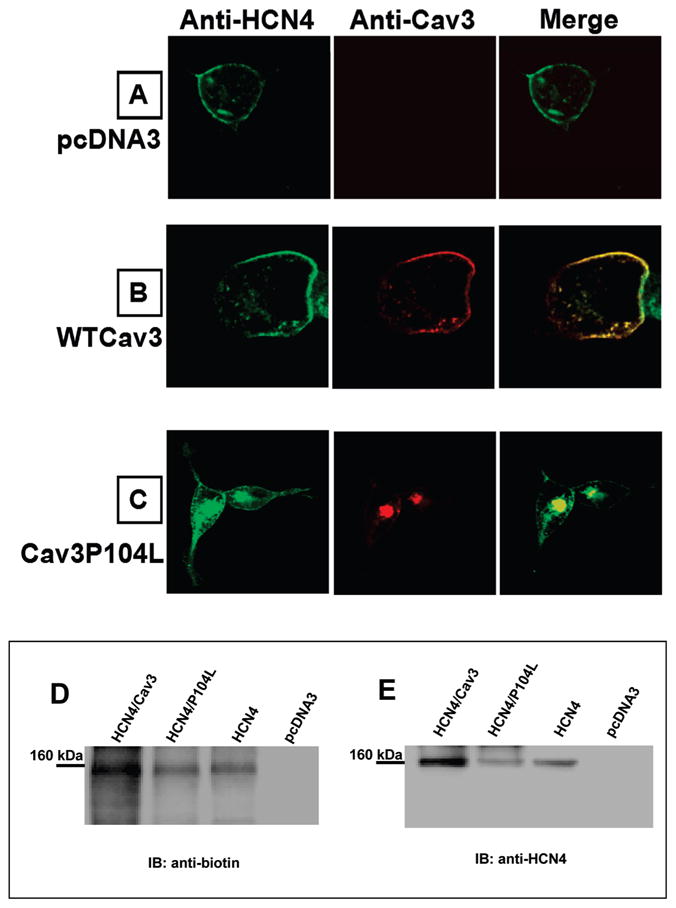
Confocal images from HCN4 stably expressed HEK293 cells transiently expressed with pcDNA3 (A), WTCav3 (B), or Cav3P104L(C). The cells were immunostained with rabbit anti-HCN4 polyclonal antibody and mouse anti-Cav3 monoclonal antibody. Cell surface biotinylation of HCN4 channels was examined in HEK293 cells transiently transfected with HCN4, HCN4/Cav3, HCN4/P104L, or pcDNA3. Biotin-labeled HEK293 cell lysate was immunoprecipitated with anti-HCN4 antibody and subjected to Western blot analysis with antibiotin antibody (D), then stripped and reprobed with anti-HCN4 antibody (E); the 160 kDa band for HCN4 is indicated.
To further investigate the effect of P104L on the trafficking of HCN4, biotinylation experiments were performed in intact cells to detect surface membrane HCN4 channels in the presence of Cav3 or P104L coexpression. Intact HEK293 cells expressing HCN4 channel alone, HCN4 + Cav3, HCN4 + P104L, and pcDNA3 transfected cells were surface biotinylated, and the whole cell lysates were immunoprecipitated using anti-HCN4. The immunoprecipitated samples were then analyzed by Western blotting. A representative Western blot (Figure 3D) probed with antibiotin antibody demonstrates a band at 160 kDa seen most prominently with HCN4 + Cav3 coexpression relative to HCN4 + P104L. The greater signal on the antibiotin blot for the HCN4 + Cav3 lane relative to HCN4 + P104L suggests that P104L coexpression leads to the reduction of surface membrane expression of HCN4, in comparison to Cav3 coexpression, which in fact increased the surface membrane expression of HCN4. This is because some of the HCN4 channels were trapped in the intracellular compartments with the P104L mutant and did not make it to the surface membrane. When the blot was stripped and reprobed with anti-HCN4 antibody, the same 160-kDa protein was detected (Figure 3E), which demonstrates that the biotinylated protein was HCN4. The specificity of the findings is demonstrated by the lack of signals for antibiotin or anti-HCN4 antibodies in the lane for mock-transfected cells. Overall biotinylation experiments confirm colocalization of Cav3 and HCN4 at the plasma membrane.
Cav3 or P104L Modulates Properties of the HCN4 Channel
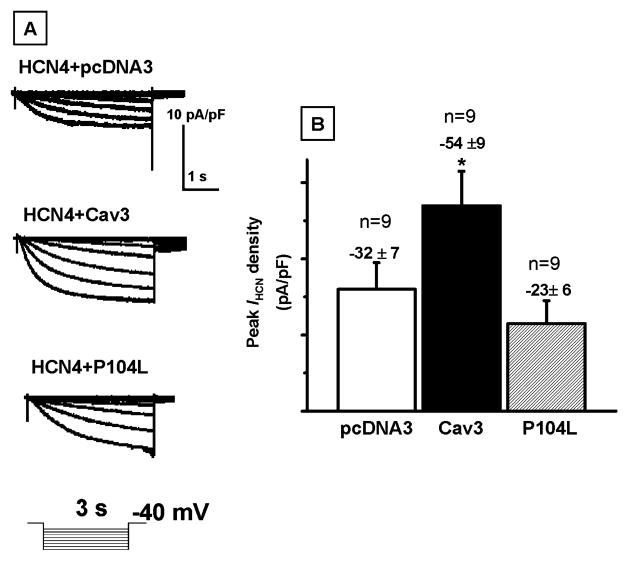
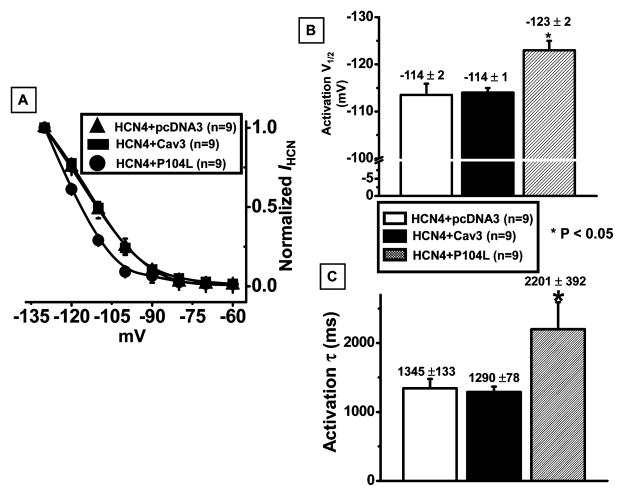
To investigate the functional effect of either Cav3 or P104L on IHCN, Cav3 or P104L with GFP in a bicistronic vector was transiently expressed in the HCN4 stable cell line. Also, pcDNA3 and GFP in a mammalian expression vector were coexpressed in HCN4 stable cell line as a negative control. Representative current traces are shown in Figure 4, panel A with the recording protocol inset. Peak IHCN densities were measured by normalizing peak IHCN to cell capacitance with the recording protocol inset (Figure 4, panel B). The numbers of experiments and mean values of peak IHCN density for each sample were displayed above each bar (Figure 4, panel B.) Compared to the expression of pcDNA3 or P104L, expression of Cav3 in the HCN4 stable cell line increased peak IHCN density 68% more than pcDNA3 or 134% more than P104L. Expression of Cav3 in the HCN4 stable cell line significantly enhanced IHCN density. This is in agreement with our biotinylation experiment data (Figure 3, panels D and E). However, disruption of caveolar structure protein using P104L reduced peak IHCN density more than 1-fold. P104L also shifted the activation curve for the current–voltage relationship to a more negative potential (Figure 5, panel A) and increased V1/2 for activation by −10 mV (Figure 5, panel B). However, expression of Cav3 in HCN4 stable cells had no effect on the activation curve and activation V1/2 (Figure 5, panel A and B). Activation time constant was also investigated. P104L increased the activation time constant to about 1-fold more compared to the expression of pcDNA3 in HCN4 stable cells (Figure 5, panel C). Prolonging activation time can slow activation and reduce IHCN density (Figure 5, panel C). The effects of Cav3 on IHCN properties were confirmed using the transient expression of HCN4 in HEK293 cells stably expressing Cav3 or P104L (data not shown). Expression of trafficking mutation P104L modulated HCN4 channel properties and had dramatic effects on IHCN density and the activation time constant.
Figure 4.
IHCN recorded from HCN4 stably expressed HEK293 cells. Panel A: representative traces were recorded with a test potential of 3-s duration from −130 to −60 mV from a holding potential of −40 mV. Panel B: summary data for peak IHCN densities for pcDNA3, Cav3, or P104L transiently expressed in the HCN4 stably expressed HEK293 cell. IHCN were elicited by a step hyperpolarization to −120 mV from a holding potential of −40 mV and normalized to membrane capacitance. Bars depict the mean and standard error of the mean derived from the number (n) of experiments with n shown above the bar.
Figure 5.
Activation kinetics of IHCN from HCN4 stably expressed HEK293 cells transiently expressed with pcDNA3, Cav3, or P104L. Panel A: current–voltage relationship. The y-axis is peak IHCN normalized to the maximal IHCN obtained in the protocol as depicted in Figure 4. Solid symbols represent the mean data from 9 experiments, and the bars represent stand error. Individual curves were fitted to the Boltzmann equation. Panel B: summary data for an activation voltage (V1/2) when 50% of IHCN activated. Panel C: summary data for activation time constant (τ) at the testing potential of −120 mV. IHCN was elicited by a step hyperpolarization to −120 mV from a holding potential of −40 mV and normalized to membrane capacitance. Bars depict the mean and standard error of the mean derived from the number of experiments (n) with the means and stand errors of the mean shown above the bar.
DISCUSSION
In the present study, we demonstrate that the HCN4 channel colocalizes and associates with Cav3 in the heart on the basis of immunoprecipitation studies. Immunofluorescence and biotinylation studies indicate that P104L disrupts surface membrane expression of HCN4 protein. Whole cell patch clamp studies revealed that WT Cav3 and dominant negative and trafficking deficient mutation P104L modulate properties of HCN4 channels. In our studies, we used the P104L to disrupt caveolae and found that it significantly decreased the IHCN4 density. This result is in contrast with another study where no change in the IHCN4 density was reported when the cells were treated with MβCD to disrupt caveolae by depleting cholesterol (13). Results from others (13) indicate that disruption of caveolar lipid rafts has minimal effects on trafficking and localization of HCN4 on the cell surface membranes. P104L had a dominant negative effect on Cav3 and reduced Cav3 protein levels in cells by more than 95% (11). Our immunostaining and biotinylation experiments revealed that the trafficking defective mutant P104L does alter the trafficking patterns of HCN4 (Figure 3). Also, coexpression of Cav3 with HCN4 enhanced IHCN4 density (Figure 4) and also increased the surface membrane expression of HCN4 protein (Figure 3D). Several ion channels have been shown to associate and be modulated by Cav3 in various cell types (9, 10, 14, 24). The P104L mutation is known to affect the trafficking of the L-type Ca2+ channel in cardiac (25) and skeletal muscle cells (26). The current amplitude of Na channel has been shown to be increased when coexpressed with Cav3 (27). Overexpression of P104L in mice (28) can cause cardiomyopathy and increase endothelial nitric oxide synthase (eNOS) activity, but the study does not mention an observation for pacemaker abnormality. It is known that mice have a much higher heart rate (~700 beat/min vs ~70 beat/min in humans). It is difficult to measure any QT prolongation from mice. That may be one of the reasons that no bradycardia is reported. Also, P104L increases eNOS activity. The increasing eNOS activity enhances levels of cAMP. Since HCN channels are sensitive to cAMP, secondary effects from cAMP need to be considered in a physiological environment. However, studies to address this are beyond the scope of the present article. Overexpression of Cav3 in animal models is shown to induce cadiomyopathic phenotypes. Overexpression of Cav3 in the mouse heart caused cardiomyopathies and muscular dystrophy (29). Cav3 is also up-regulated in a rat heart failure model (30). It is interesting to note that If was shown to be increased in a rat heart failure model (31). The abnormal automaticity caused by up-regulation of HCN channel expression may be explained by up-regulation of Cav3 expression. It is possible that overexpression of Cav3 could help recruit or further stabilize HCN4 protein at the plasma membrane. Therefore, tight regulation of expression levels for Cav3 protein may be required for maintaining proper cardiac function.
Our results (Figure 5) show that Cav3 has a distinct mechanism for modulation of HCN4 channels properties. Disruption of caveolae by dominant negative mutation P104L shifts voltage dependence of channel activation to more negative potentials and increases the time constant for HCN4 channel activation (Figure 5). It has been known that caveolae are involved in many cellular processes, such as protein and lipid trafficking, and signal transduction (7, 32). The disruption of caveolae can cause primary and secondary effects on HCN4 channel properties. Cav3 knockout mice display an intracellular retention for c-Src (33). Inhibition of c-Src can increase the activation time constant for HCN2 and HCN4 but has no effects on IHCN density and the half-maximal activation voltage (34). The effects of P104L on the activation time constant but not the half-maximal activation for HCN4 may be through the c-Src regulation of the channel. However, how P104L affects the voltage-dependent half-maximal activation for HCN4 remains unclear. Our results indicate that HCN4 channels localize to caveolae in the heart to form a macromolecular complex. This localization can be important for properties of HCN4 channels as well as cardiac pacemaker currents.
Acknowledgments
We thank Dr. Jonathan Makielski and Dr. Timothy Kamp for their helpful suggestions. We also thank Nicole Ayala and Wenjie Ou for technical assistance and Debra Pittz for secretarial assistance.
Footnotes
Abbreviations: If, cardiac pacemaker current; IHCN, whole cell HCN current; IB, immunoblot; IP, immunoprecipitate; WT, wild type.
This work was supported by Scientist Development Grants from The American Heart Association National Center (0435030N, to B.Y.), (0730010N, to R.C.B.) and (0630268N, to N.-Q.S.), and by NIH T32HL07936 to J.F.
References
- 1.DiFrancesco D. Pacemaker mechanisms in cardiac tissue. Annu Rev Physiol. 1993;55:455–472. doi: 10.1146/annurev.ph.55.030193.002323. [DOI] [PubMed] [Google Scholar]
- 2.Pape HC. Queer current and pacemaker: the hyperpolarization-activated cation current in neurons. Annu Rev Physiol. 1996;58:299–327. doi: 10.1146/annurev.ph.58.030196.001503. [DOI] [PubMed] [Google Scholar]
- 3.Santoro B, Liu DT, Yao H, Bartsch D, Kandel ER, Siegelbaum SA, Tibbs GR. Identification of a gene encoding a hyperpolarization-activated pacemaker channel of brain. Cell. 1998;93:717–729. doi: 10.1016/s0092-8674(00)81434-8. [DOI] [PubMed] [Google Scholar]
- 4.Ludwig A, Zong X, Stieber J, Hullin R, Hofmann F, Biel M. Two pacemaker channels from human heart with profoundly different activation kinetics. EMBO J. 1999;18:2323–2329. doi: 10.1093/emboj/18.9.2323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Moosmang S, Stieber J, Zong X, Biel M, Hofmann F, Ludwig A. Cellular expression and functional characterization of four hyperpolarization-activated pacemaker channels in cardiac and neuronal tissues. Eur J Biochem. 2001;268:1646–1652. doi: 10.1046/j.1432-1327.2001.02036.x. [DOI] [PubMed] [Google Scholar]
- 6.Wainger BJ, DeGennaro M, Santoro B, Siegelbaum SA, Tibbs GR. Molecular mechanism of cAMP modulation of HCN pacemaker channels. Nature. 2001;411:805–810. doi: 10.1038/35081088. [DOI] [PubMed] [Google Scholar]
- 7.Simons K, Toomre D. Lipid rafts and signal transduction. Nat Rev Mol Cell Biol. 2000;1:31–39. doi: 10.1038/35036052. [DOI] [PubMed] [Google Scholar]
- 8.Williams TM, Lisanti MP. The caveolin proteins. Genome Biol. 2004;5:214. doi: 10.1186/gb-2004-5-3-214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Balijepalli RC, Foell JD, Hall DD, Hell JW, Kamp TJ. Localization of cardiac L-type Ca(2+) channels to a caveolar macromolecular signaling complex is required for beta(2)-adrenergic regulation. Proc Natl Acad Sci USA. 2006;103:7500–7505. doi: 10.1073/pnas.0503465103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Brainard AM, Miller AJ, Martens JR, England SK. Maxi-K channels localize to caveolae in human myometrium: a role for an Actin-channel-caveolin complex in the regulation of myometrial smooth muscle K+ current. Am J Physiol Cell Physiol. 2005;289:C49–C57. doi: 10.1152/ajpcell.00399.2004. [DOI] [PubMed] [Google Scholar]
- 11.Cohen AW, Hnasko R, Schubert W, Lisanti MP. Role of caveolae and caveolins in health and disease. Physiol Rev. 2004;84:1341–1379. doi: 10.1152/physrev.00046.2003. [DOI] [PubMed] [Google Scholar]
- 12.Folco EJ, Liu GX, Koren G. Caveolin-3 and SAP97 form a scaffolding protein complex that regulates the voltage-gated potassium channel Kv1.5. Am J Physiol Heart Circ Physiol. 2004;287:H681–H690. doi: 10.1152/ajpheart.00152.2004. [DOI] [PubMed] [Google Scholar]
- 13.Barbuti A, Gravante B, Riolfo M, Milanesi R, Terragni B, DiFrancesco D. Localization of pacemaker channels in lipid rafts regulates channel kinetics. Circ Res. 2004;94:1325–1331. doi: 10.1161/01.RES.0000127621.54132.AE. [DOI] [PubMed] [Google Scholar]
- 14.Vatta M, Ackerman MJ, Ye B, Makielski JC, Ughanze EE, Taylor EW, Tester DJ, Balijepalli RC, Foell JD, Li Z, Kamp TJ, Towbin JA. Mutant caveolin-3 induces persistent late sodium current and is associated with long-QT syndrome. Circulation. 2006;114:2104–2112. doi: 10.1161/CIRCULATIONAHA.106.635268. [DOI] [PubMed] [Google Scholar]
- 15.Ueda K, Nakamura K, Hayashi T, Inagaki N, Takahashi M, Arimura T, Morita H, Higashiuesato Y, Hirano Y, Yasunami M, Takishita S, Yamashina A, Ohe T, Sunamori M, Hiraoka M, Kimura A. Functional characterization of a trafficking-defective HCN4 mutation, D553N, associated with cardiac arrhythmia. J Biol Chem. 2004;279:27194–27198. doi: 10.1074/jbc.M311953200. [DOI] [PubMed] [Google Scholar]
- 16.Nof E, Luria D, Brass D, Marek D, Lahat H, Reznik-Wolf H, Pras E, Dascal N, Eldar M, Glikson M. Point mutation in the HCN4 cardiac ion channel pore affecting synthesis, trafficking, and functional expression is associated with familial asymptomatic sinus bradycardia. Circulation. 2007;116:463–470. doi: 10.1161/CIRCULATIONAHA.107.706887. [DOI] [PubMed] [Google Scholar]
- 17.Woodman SE, Park DS, Cohen AW, Cheung MWC, Chandra M, Shirani J, Tang B, Jelicks LA, Kitsis RN, Christ GJ, Factor SM, Tanowitz HB, Lisanti MP. Caveolin-3 knock-out mice develop a progressive cardiomyopathy and show hyperactivation of the p42/44 MAPK cascade. J Biol Chem. 2002;277:38988–38997. doi: 10.1074/jbc.M205511200. [DOI] [PubMed] [Google Scholar]
- 18.Minetti C, Sotgia F, Bruno C, Scartezzini P, Broda P, Bado M, Masetti E, Mazzocco M, Egeo A, Donati MA, Volonte D, Galbiati F, Cordone G, Bricarelli FD, Lisanti MP, Zara F. Mutations in the caveolin-3 gene cause autosomal dominant limb-girdle muscular dystrophy. Nat Genet. 1998;18:365–368. doi: 10.1038/ng0498-365. [DOI] [PubMed] [Google Scholar]
- 19.Carozzi AJ, Roy S, Morrow IC, Pol A, Wyse B, Clyde-Smith J, Prior IA, Nixon SJ, Hancock JF, Parton RG. Inhibition of lipid raft-dependent signaling by a dystrophy-associated mutant of caveolin-3. J Biol Chem. 2002;277:17944–17949. doi: 10.1074/jbc.M110879200. [DOI] [PubMed] [Google Scholar]
- 20.Nagatomo T, Fan Z, Ye B, Tonkovich GS, January CT, Kyle JW, Makielski JC. Temperature dependence of early and late currents in human cardiac wild-type and long Q-T Delta KPQ Na+ channels. Am J Physiol Heart Circ Physiol. 1998;275:H2016–H2024. doi: 10.1152/ajpheart.1998.275.6.H2016. [DOI] [PubMed] [Google Scholar]
- 21.Ye B, Valdivia CR, Ackerman MJ, Makielski JC. A common human SCN5A polymorphism modifies expression of an arrhythmia causing mutation. Physiol Genomics. 2003;12:187–193. doi: 10.1152/physiolgenomics.00117.2002. [DOI] [PubMed] [Google Scholar]
- 22.Cohen RM, Foell JD, Balijepalli RC, Shah V, Hell JW, Kamp TJ. Unique modulation of L-type Ca2+ channels by short auxiliary beta1d subunit present in cardiac muscle. Am J Physiol Heart Circ Physiol. 2005;288:H2363–H2374. doi: 10.1152/ajpheart.00348.2004. [DOI] [PubMed] [Google Scholar]
- 23.Sotgia F, Woodman SE, Bonuccelli G, Capozza F, Minetti C, Scherer PE, Lisanti MP. Phenotypic behavior of caveolin-3 R26Q, a mutant associated with hyperCKemia, distal myopathy, and rippling muscle disease. Am J Physiol Cell Physiol. 2003;285:C1150–C1160. doi: 10.1152/ajpcell.00166.2003. [DOI] [PubMed] [Google Scholar]
- 24.Kowalski MP, Pier GB. Localization of cystic fibrosis transmembrane conductance regulator to lipid rafts of epithelial cells is required for Pseudomonas aeruginosa-induced cellular activation. J Immunol. 2004;172:418–425. doi: 10.4049/jimmunol.172.1.418. [DOI] [PubMed] [Google Scholar]
- 25.Balijepalli RC, Foell JD, Best JM, Kamp TJ. Caveolar Localization of CaV1.2 Channels Requires Association of CaVβ Subunit with Caveolin-3 in the Heart. Biophys J. 2008;94:3143. Pos. [Google Scholar]
- 26.Couchoux H, Allard B, Legrand C, Jacquemond V, Berthier C. Loss of caveolin-3 induced by the dystrophy-associated P104L mutation impairs L-type calcium channel function in mouse skeletal muscle cells. J Physiol. 2007;580:745–754. doi: 10.1113/jphysiol.2006.124198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Yarbrough TL, Lu T, Lee HC, Shibata EF. Localization of cardiac sodium channels in caveolin-rich membrane domains: regulation of sodium current amplitude. Circ Res. 2002;9:443–449. doi: 10.1161/hh0402.105177. [DOI] [PubMed] [Google Scholar]
- 28.Ohsawa Y, Toko H, Katsura M, Morimoto K, Yamada H, Ichikawa Y, Murakami T, Ohkuma S, Komuro I, Sunada Y. Overexpression of P104L mutant caveolin-3 in mice develops hypertrophic cardiomyopathy with enhanced contractility in association with increased endothelial nitric oxide synthase activity. Hum Mol Genet. 2004;13:151–157. doi: 10.1093/hmg/ddh014. [DOI] [PubMed] [Google Scholar]
- 29.Aravamudan B, Volonte D, Ramani R, Gursoy E, Lisanti MP, London B, Galbiati F. Transgenic overexpression of caveolin-3 in the heart induces a cardiomyopathic phenotype. Hum Mol Genet. 2003;12:2777–2788. doi: 10.1093/hmg/ddg313. [DOI] [PubMed] [Google Scholar]
- 30.Hare JM, Lofthouse RA, Juang GJ, Colman L, Ricker KM, Kim B, Senzaki H, Cao S, Tunin RS, Kass DA. Contribution of caveolin protein abundance to augmented nitric oxide signaling in conscious dogs with pacing-induced heart failure. Circ Res. 2000;86:1085–1092. doi: 10.1161/01.res.86.10.1085. [DOI] [PubMed] [Google Scholar]
- 31.Cerbai E, Barbieri M, Mugelli A. Occurrence and properties of the hyperpolarization-activated current if in ventricular myocytes from normotensive and hypertensive rats during aging. Circulation. 1996;94:1674–1681. doi: 10.1161/01.cir.94.7.1674. [DOI] [PubMed] [Google Scholar]
- 32.Tsui-Pierchala BA, Encinas M, Milbrandt J, Johnson EM. Lipid rafts in neuronal signaling and function. Trends Neurosci. 2002;25:412–417. doi: 10.1016/s0166-2236(02)02215-4. [DOI] [PubMed] [Google Scholar]
- 33.Sotgia F, Razani B, Bonuccelli G, Schubert W, Battista M, Lee H, Capozza F, Schubert AL, Minetti C, Buckley JT, Lisanti MP. Intracellular retention of glycosylphosphatidyl inositol-linked proteins in caveolin-deficient cells. Mol Cell Biol. 2002;22:3905–3926. doi: 10.1128/MCB.22.11.3905-3926.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Zong X, Eckert C, Yuan H, Wahl-Schott C, Abicht H, Fang L, Li R, Mistrik P, Gerstner A, Much B, Baumann L, Michalakis S, Zeng R, Chen Z, Biel M. A Novel mechanism of modulation of hyperpolarization-activated cyclic nucleotide-gated channels by Src kinase. J Biol Chem. 2005;280:34224–34232. doi: 10.1074/jbc.M506544200. [DOI] [PubMed] [Google Scholar]