Abstract
• Background. Two families of proteins that transport small peptides, the oligopeptide transporters (OPTs) and the peptide transporters (PTRs), have been recognized in eukaryotes. Higher plants contain a far greater number of genes for these transporters than do other eukaryotes. This may be indicative of the relative importance of (oligo)peptides and their transport to plant growth and metabolism.
• Recent progress. Recent studies are now allowing us to assign functions to these transporters and are starting to identify their in-planta substrates, revealing unexpected and important contributions of the transporters to plant growth and developmental processes. This Botanical Briefing appraises recent findings that PTRs and OPTs have key roles to play in the control of plant cell growth and development. Evidence is presented that some of these transporters have functions outside that of nitrogen nutrition and that these carriers can also surprise us with their totally unexpected choice of substrates.
Keywords: Transport, peptides, PTR, OPT, nitrogen reallocation, Arabidopsis, cereal, Hordeum vulgare
INTRODUCTION
Although it is well established that peptide transport plays an important role in the nutrition of bacteria, yeasts and animals (Saier, 2000), the role of small peptides (2–6 amino acids) and their transporters in plants is less well defined. The first unambiguous demonstration of a pool of small peptides in plant tissues described the presence of millimolar concentrations of small peptides in the endosperm of germinating cereal grains (Higgins and Payne, 1981). Here they serve as a supply of nutrients transported across the scutellum to support growth of the cereal embryo during the early stages of germination. Subsequently, it has become apparent that small peptides and their transporters also play a significant role in control of plant cell differentiation and organogenesis in addition to a nutritional role (Yang et al., 2000; Stacey et al., 2002a).
Two families of small-peptide transporters have been recognized in eukaryotes. The oligopeptide transporter (OPT) family can transport tetra- and pentapeptides while the peptide transporter (PTR) family can transport di- and tripeptides (Koh et al., 2002). An important distinction between the two families is the much more selective nature of the OPTs for peptides of only a certain amino acid composition when compared with the low selectivity of so-far characterized di/tripeptide transporters of the PTR family. An analysis of the Arabidopsis genome sequence predicts the presence of over 50 putative PTRs (Arabidopsis Genome Initiative, 2000). This number is far greater than in other genomes sequenced to date and may be indicative of the importance of these transporters in plant growth and metabolism. Peptide transporters of both these families are proton symporters although no significant sequence similarity is found between the two families (Koh et al., 2002). Currently, we have little knowledge of the roles that peptides and their transporters play in the plant's life cycle and several key questions need to be addressed to establish their roles in planta. First, does each transporter have a unique role to play in cellular metabolism or is there a degree of functional redundancy between these transporters? Secondly, what evidence exists to help elucidate not only structure–function relationships but, equally importantly, the potential substrates for the various transporters. It is noteworthy that recent studies have challenged the tacit assumption that small peptides are the only substrates for some members of the OPT family. This discovery has a direct bearing on the potential role of OPTs within cells and during plant development.
This review will appraise recent evidence that these transporters can surprise us with their choice of substrates and that small peptides, PTRs and OPTs may have roles to play in control of plant cell growth and development outside that of important nutrient supply systems.
THE PEPTIDE TRANSPORTER FAMILY
PTRs in Arabidopsis
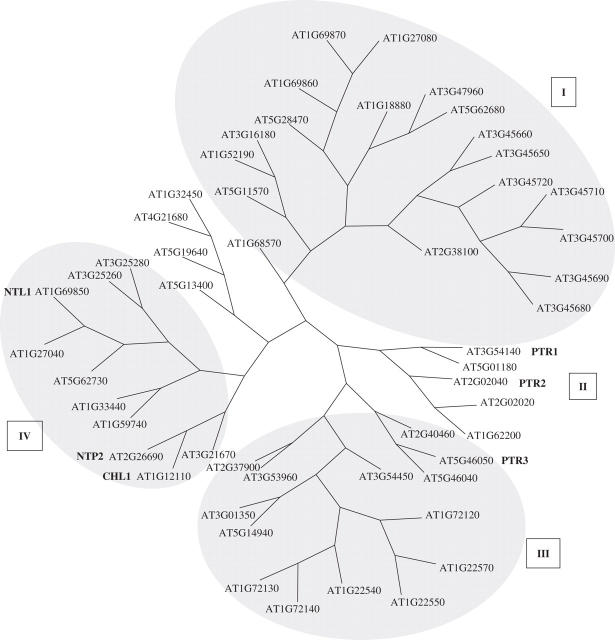
The putative PTR family members identified in the Arabidopsis genome (Stacey et al., 2002a) have been phylogenetically grouped into four broad PTR subfamilies (Fig. 1) but few have been functionally characterized as bona fide small-peptide transporters (Table 1). The first plant PTR identified from Arabidopsis was CHL1 (NRT1:1) and is a dual-affinity nitrate transporter (Tsay et al., 1993; Wang et al., 1998). Subsequently, a further PTR, AtPTR2, was identified as an authentic small-peptide transporter by use of functional complementation of peptide-transport-defective auxotrophic yeast mutants (Rentsch et al., 1995; Song et al., 1996). AtPTR2 was shown to mediate transport of di- and tripeptides of reasonably wide amino acid composition when expressed in yeast or Xenopus oocytes (Rentsch et al., 1995; Song et al., 1996; Chiang et al., 2004). AtPTR2 did not transport tetrapeptides and exhibited negligible uptake of histidine or nitrate. AtPTR2 transcripts were detected in several tissues including flower, leaf, stem, root, siliques (at high levels) and young seedlings (Rentsch et al., 1995; Song et al., 1996). In-situ hybridization showed AtPTR2 expression localized to the embryo at the heart stage of seed development (Rentsch et al., 1995). Intriguingly, antisense AtPTR2 plants exhibited a delay in flowering of 7–15 d compared with wild-type plants and enlargement of rosette leaves (Song et al., 1997). These antisense plants also produced a reduced seed number per silique, indicative of a degree of aborted seed development, but with an increased seed weight for the fewer mature seeds produced (Song et al., 1997). The severity of the AtPTR2 antisense phenotype was surprising and challenges current opinion on the relative importance of the contribution of peptide transport to nitrogen nutrition in higher plants especially during seed development.
Fig. 1.
Phylogenetic relationship between Arabidopsis thaliana peptide transporter (PTR) proteins. Protein sequences representing 52 Arabidopsis PTR proteins (Pfam domain PF00854, TIGR release 5·0) were aligned by use of ClustalW (Chenna et al., 2003) with default alignment parameters. The region of the alignment corresponding to residues 55–585 of PTR2 was used for the generation of an unrooted phylogeny by parsimony analysis (phylogeny inference package; Felsenstein, 1998). Subfamilies I (19 members), III (14 members) and IV (10 members) are shaded whereas subfamily II (nine members) is left unshaded. The locations of CHL1 (AT1G12110), NTL1 (AT1G69850) and NTP2 (AT2G26690) in subfamily IV, PTR3 (AT5G46050) in subfamily III and PTR1 (AT3G54140) and PTR2 (AT2G02040) in subfamily II are indicated.
Table 1.
Properties of some plant peptide transporters (PTRs)
| Transporter | Species | Substrates | Expression pattern localization, activity | Mutant phenotype | Role |
|---|---|---|---|---|---|
| AtPTR1 | Arabidopsis | Di- and tripeptides, phaseolotoxin | Plasma membrane in vascular tissues, seed germination, developing siliques, root tips | Seed development, reserve mobilization during germination, acquisition of soil nutrients | |
| AtPTR2 | Arabidopsis | Di- and tripeptides | Flower, leaf, stem | Delayed flowering, larger rosette leaves, reduced seed number, larger seeds | Seed development |
| HvPTR1 | Barley | Di- and tripeptides | Scutellum of germinating grain, developing grain; activity controlled post-translationally | Redistribution of organic nitrogen from endosperm to embryo during germination, seed development? | |
| VfPTR1 | Faba bean | Dipeptides | Cotyledons during seed development and germination; axes, roots and root hairs of seedlings; dipeptides repress expression in roots | Redistribution of organic nitrogen in seed germination and development, nutrient uptake from soil | |
| NaNTR1 | Nepenthes | Not done | Phloem cells at base of pitchers, leaves, petioles | Transport of organic nitrogen from pitcher to sink tissues (phloem loading of peptides) |
The known properties of five plant PTRs are compared. The substrates transported by these PTRs represent those identified in studies when the PTR proteins are produced in heterologous yeast or Xenopus oocyte expression systems. Consequently, these substrates represent only a minimal number of possible di- and tripeptide substrates that would be available to the PTR in planta. Expression patterns represent a summary of the tissues or developmental stages at which specific PTR transcripts have been detected in studies using either Northern analyses or in-situ hybridization approaches.
Another Arabidopsis PTR, designated AtPTR1 and phylogenetically closely related to AtPTR2, was shown to transport di- and tripeptides of broad amino acid composition when expressed in Xenopus oocytes or yeast (Dietrich et al., 2004). AtPTR1 also transported phaseolotoxin, a modified tripeptide produced by Pseudomonas syringae-mediated halo blight infection of bean. AtPTR1 is a plasma membrane-localized protein associated with vascular tissues throughout the plant. Notably, AtPTR1 transcripts accumulated during seed germination, in developing siliques and in root tips, indicative of roles in seed reserve mobilization, seed development and the acquisition of soil nutrients. A further Arabidopsis PTR family member, AtPTR3, was shown to be inducible by mechanical wounding (Karim et al., 2005). The functionality of the protein was not investigated, but AtPTR3 transcription was shown to be strongly inducible by salt stress. Germination of seeds from an AtPTR3 knock-out line was also impaired under salt stress. Analysis of microarray data sets often reveals up-regulated expression of as-yet uncharacterized PTRs in response to stresses and wounding (W. M. Waterworth, unpubl. data). These observations may indicate roles for some plant PTRs in nutrient redistribution to or from the site of wounding or alternatively roles for these PTRs in transport of peptide signalling molecules during the plant's response to abiotic stress.
Peptide transport in the germinating barley grain
The best characterized plant PTR is the carrier responsible for mobilization of peptides from endosperm to embryo across the scutellum in barley grain germination (Table 1). The barley scutellar peptide transporter HvPTR1 was functionally characterized by production in Xenopus oocytes (West et al., 1998) and shown to be localized to the plasma membrane of scutellar epithelial cells (Waterworth et al., 2000). HvPTR1 expression is seed-specific, with transcripts detected only in scutellar epithelial cells during germination and in barley grain from the earliest stages of grain development onwards (Waterworth et al., 2003). HvPTR1 activity in the germinating barley grain is regulated at the post-translational level by phosphorylation status (Waterworth et al., 2005). Rising levels of amino acids resulting from breakdown and mobilization of endosperm storage protein during the later stages of germination down-regulate HvPTR1 activity. By contrast, HvPTR1 activity is stimulated by rising levels of glucose reaching the scutellum as endosperm starch reserves are mobilized. Significantly, this level of control may be an important element which balances the flux of nitrogen and carbon from the endosperm to the embryo to support the re-initiation of growth processes during seed germination and the early stages of seedling establishment. At present, HvPTR1 is the only plant peptide transporter for which a physiological role is well defined.
Peptide transport in roots, leaves and the pitchers of carnivorous plants
In addition to the role of peptide transport in nitrogen redistribution within the plant, peptide uptake from the rhizosphere by roots may have nutritional importance. This view is supported by expression of a number of characterized PTR genes in the outer cell layers of roots and root hairs. Active peptide uptake by roots was demonstrated through the use of the toxic ethionine (Eth)-containing peptides Leu-Eth and Ala-Eth, which inhibit root growth of Arabidopsis seedlings (Steiner et al., 1994). Organic nitrogen sources in the soil environment (amino acids and peptides) play important roles in the nitrogen economy of grasslands, particularly in N-limited terrestrial ecosystems (Chapin et al., 1993; Keilland 1994). The root systems of many plant species, including two tundra sedges Eriophorum vaginatum and Carex aquatilis, are able to compete with soil micro-organisms for organic nitrogen in the soil (Schimel and Chapin, 1996). Peptide uptake in roots of aquatic plants has also been demonstrated (Bollard, 1966). Collectively, these observations suggest that PTRs could play significant roles in nitrogen nutrition during different phases of plant growth and development especially in nitrate-poor ecosystems.
Examples from other species have also contributed to our understanding of their potential functions (Table 1). Two PTR homologues, VfPTR1 and VfPTR2, have been isolated from faba bean (Vicia faba; Miranda et al., 2003). VfPTR1 was shown to mediate uptake of the dipeptide His-Ala and VfPTR1 expression was associated with seed development and germination. A number of H+-coupled, low- and high-affinity transport systems for di- and tripeptides have also been demonstrated in Vicia faba mesophyll cells (Jamai et al., 1994, 1995), and a low-affinity, ATP-dependent peptide-transport system has also been characterized in the tonoplast of barley mesophyll cells (Jamai et al., 1995). This remains the only example of peptide transport in a plant organelle.
A PTR was shown to be an important component of the nitrogen acquisition apparatus of the carnivorous pitcher plant Nepenthes alata, which relies on insect capture and digestion for a substantial part of the plant's nitrogen budget (Schulze et al., 1999). The PTR family gene NaNTR1 was expressed in phloem cells within pitchers, indicating that NaNTR1 may function in phloem loading of peptide nitrogen exported from the pitcher to sink organs in the plant.
Not all PTR proteins transport small peptides
A number of plant PTRs have been shown to transport substrates other than peptides. Consequently, it has been suggested that the criteria used to classify some proteins as PTR family members may have been too broad (Stacey et al., 2002a). However, many transporter families are larger in plants than in organisms from other kingdoms, possibly representing evolutionary diversification of these transporters for nutrient acquisition and reflecting the plant's autotrophic and sessile lifestyle. Notably, the Arabidopsis PTR subfamily IV members (Fig. 1) NTL1 (AtNRT1:2) and CHL1 (AtNRT1:1) have been shown to be nitrate transporters (Tsay et al., 1993; Huang et al., 1999). Another member of subfamily IV, NTP2 (AtNRT1:4), may also be a nitrate transporter (Forde, 2000). A rice putative PTR family transporter OsNTR1 displayed no peptide transport activity and only low-affinity nitrate transport when produced in both yeast and Xenopus oocytes (Lin et al., 2000). OsNTR1 was expressed constitutively in root hairs and epidermis, consistent with a role in nitrate uptake from the soil (Lin et al., 2000). Another PTR family member, designated AgDCAT1 and which grouped with NRT1, was shown to be specifically expressed in the actinorhizal nodules of alder (Alnus glutinosa) roots, which mediate nutrient exchange between the plant and nitrogen-fixing symbiotic actinomycete bacteria, and was demonstrated to be a dicarboxylate transporter (Jeong et al., 2004). How many of the uncharacterized transporters assigned by phylogentic analysis of sequence similarities as PTRs are genuine peptide transporters as opposed to being transporters of other, non-peptide substrates? This remains a key question and these studies emphasize the need to verify the nature of the physiological substrates of the many uncharacterized PTRs in both Arabidopsis and other plant species if we are to understand their role in planta.
THE OPT TRANSPORTER FAMILY
The first OPTs were initially characterized by function in the human pathogen Candida albicans (Lubkowitz et al., 1997), and subsequently in the lower eukaryotes Schizosaccharomyces pombe and Saccharomyces cerevisiae (Lubkowitz et al., 1998). OPT family homologues were later identified in higher plants and distant homologues in prokaryotic genomes (Saier, 2000) but as yet no animal OPT homologues have been recognized.
OPT transporters in Arabidopsis
Nine putative OPT orthologues (AtOPT1 to AtOPT9) were identified in the Arabidopsis thaliana genome that exhibit significant sequence similarity between each other (61–85%; Hauser et al., 2001; Koh et al., 2002). Sequence alignments showed that the AtOPT family members shared two highly conserved motifs. These were termed the NPG motif (Asn-Pro-Gly; encompassing a stretch of 18 amino acids) and the KIPPR motif (Lys-Ile-Pro-Pro-Arg; 11–14 amino acids). The use of hydropathy plots to predict membrane topology indicates that the plant OPTs may have 12–14 transmembrane domains (Koh et al., 2002) but this prediction has yet to be verified experimentally. Subsequently, multiple OPTs have been identified in other plant genome sequences but those from Arabidopsis remain the best characterized.
A major objective of initial studies on OPTs was to define their expression patterns in Arabidopsis plants during development and identify their substrates in anticipation that this information would provide some indication of the function of individual transporters. AtOPTs with the greatest sequence similarity displayed expression patterns which may indicate that they have similar physiological functions (Koh et al., 2002). Although all the AtOPT genes studied were expressed at moderate to high levels in flowers, there were also distinguishing gene expression patterns (Table 2). AtOPTs 6 and 7 were expressed at highest levels in floral tissue and in roots with only low-level expression in leaf and shoot tissue. However, AtOPTs 2 and 4 were evenly expressed at moderate levels in all tissues studied. AtOPT5 was expressed poorly in roots, leaves and stems whereas AtOPT1 had low-level expression in roots but was moderately expressed in leaves and stems. AtOPT3 had a unique profile, being expressed at high levels in roots, reproductive structures and leaves and also at moderate levels in other shoot tissue.
Table 2.
Properties of Arabidopsis oligopeptide transporters (OPTs)
| Substrates transported |
Expression patterns |
||||||||
|---|---|---|---|---|---|---|---|---|---|
| Transporter | Tetrapeptides | Pentapeptides | Glutathione | Metal ions | Flower | Stems/leaves | Roots | Pollen | Seeds/funiculi |
| AtOPT1 | × | ✓ | ✓ | ✓ | ✓ | ✓ | ✓ | ||
| high | moderate | low | |||||||
| AtOPT2 | × | × | Cu2+, Zn2+? | ✓ | ✓ | ✓ | |||
| moderate | moderate | moderate | |||||||
| AtOPT3 | × | × | Cu2+, Mn2+, Fe2+ | ✓ | ✓ | ✓ | ✓ | ✓ | |
| high | leaf, high; | high | |||||||
| stem, moderate | |||||||||
| AtOPT4 | ✓ | ✓ | × | ✓ | ✓ | ✓ | ✓ | ||
| moderate | moderate | moderate | |||||||
| AtOPT5 | × | ✓ | ✓ | ✓ | ✓ | ||||
| high | low | low | |||||||
| AtOPT6 | × | ✓ | ✓? | Cd2+ | ✓ | ✓ | ✓ | ✓ | |
| Also Cd2+–GSH | Cd2+ | high | low | high | |||||
| AtOPT7 | × | ✓ | Cd2+–GSH | ✓ | ✓ | ✓ | ✓ | ||
| high | low | high | |||||||
| AtOPT8 | ✓ | ✓ | ✓ | ||||||
| low | |||||||||
AtOPTs able to transport a given substrate when produced in yeast or Xenopus oocytes are indicated by (✓). Substrates that do not appear to be transported by a given AtOPT are indicated by (×). The absence of any symbol in a ‘substrate transported’ column indicates that the substrate was not tested with that specific AtOPT. Note that only a limited number of peptides have been tested as potential substrates of the AtOPTs when these transporters have been produced in heterologous systems and so probably do not represent the range of substrates found in planta. AtOPTs expressed as transcripts in a specific tissue are indicated by (✓). The absence of any symbol in a box under ‘expression patterns’ indicates that expression of that specific AtOPT in that tissue was either very low or undetectable by use of either quantitative PCR analysis or expression of OPT promoter–GUS fusions in transformed plants. The relative levels of AtOPT expression indicated as high, moderate or low are those determined from quantitative PCR analysis (Koh et al., 2002).
Although these initial studies provided important information, they did not discriminate between global expression of individual AtOPTs in all cell types within an organ and any specific localization of OPT transcripts in individual tissues or cells. The need for such information was emphasized by the discovery that disruption of the AtOPT3 gene through a T-DNA insertion unexpectedly produced an embryo-lethal phenotype with development terminated at the octet stage of embryo development (Stacey et al., 2002b). To date, this is the only phenotype seen in Arabidopsis plants resulting from a knock-out mutation in an individual AtOPT gene. AtOPT3 appears to play a key role in early embryo development following fertilization but how the absence of a single OPT leads to early abortion of the developing embryo is still unexplained.
Subsequent AtOPT mRNA localization studies performed using Arabidopsis plants stably transformed with AtOPT promoter–GUS fusion constructs have provided a picture of AtOPT expression down to the cellular level (Stacey et al., 2006). Six AtOPTs were studied in this way, the exceptions being constructs of AtOPTs 2, 5 and 9, which gave little or no GUS activity in transformed plants. All the AtOPTs studied were expressed in vascular tissues of vegetative and reproductive organs, in seeds prior to germination and in the vascular tissue of post-germinative seedlings, although AtOPT8 was weakly expressed at these stages (Table 2). However, none of the AtOPTs was expressed in root hairs or root tips. Superimposed on this widespread vasculature-expression pattern were several unique patterns of expression for individual AtOPTs. These included AtOPT8 expressed in fertilized ovules immediately following anthesis, AtOPT6 expressed in ovules and AtOPT1 expressed in growing pollen tubes. In addition, AtOPTs 1, 3 and 8 were expressed in pollen and AtOPT 3 and 8 expressed in the developing seed. Intriguingly, there was evidence that specific stresses could induce the expression of some AtOPTs. Iron limitation in the growth medium induced the expression of AtOPT3, and microarray data analysis indicated that salt stress induced the expression of AtOPT2 although this was not apparent from the GUS expression studies.
Multiple physiological roles for the OPTs in plants?
Collectively, these studies permit several tentative conclusions to be drawn concerning possible roles for the various AtOPT genes in plant growth and development. The widespread presence of the AtOPT transcripts in vascular tissue is indicative of an important role for OPTs in the bulk redistribution of organic nitrogen (small peptides) between tissues, e.g. during leaf senescence (Stacey et al., 2006). Additionally, individual AtOPTs may have specific roles in the acquisition and/or redistribution of organic nitrogen in the form of small peptides by different tissues during growth and development e.g. AtOPT1 and 6 in pollen tubes and ovules respectively, AtOPT2 and 9 in floral tissues and AtOPT3, 5 and 8 in seed development. Conversely, the absence of any evidence for OPT expression in Arabidopsis root epidermal cells or root hairs argues against their involvement in the acquisition of organic nitrogen from the soil (Stacey et al., 2006). This contrasts with the localization of PTRs in roots.
The tacit assumption that plant OPTs transport only protein-derived peptides has recently been challenged. Again heterologous production of the plant OPT proteins in yeast cells has demonstrated that OPT homologues of the Indian mustard Brassica juncea (BjGT1) and rice (OsGT1) transport glutathione (Bogs et al., 2003; Zhang et al., 2004). Glutathione is a non-protein-derived tripeptide (γ-glutamyl-cysteinyl-glycine), which has multiple important intracellular functions including roles in assimilation, storage and transport of reduced sulphur, and the control of redox status and protein folding. Although the yeast ScOPT1 has been demonstrated to be capable of the transport of both glutathione and protein-derived oligopeptides (Bourbouloux et al., 2000; Hauser et al., 2000), there is still debate on whether any of the Arabidopsis OPTs can transport glutathione. Contradictory evidence exists either in support of AtOPT6 being able to transport glutathione (Cagnac et al., 2004) or against such glutathione transporting activity (Osawa et al., 2006). Explaining these contradictions is not easy, but both these studies involved production of the plant transporter in a heterologous system. It is possible that production of a plant plasma membrane protein in an alien cellular environment may be accompanied by problems with protein processing, folding and membrane targeting. This could result in problems with substrate recognition by a malfunctional transporter protein. Further studies are needed to resolve this dilemma.
A more surprising discovery is that some OPTs may also be capable of metal ion transport (Table 2). Transcription of both AtOPT2 and 3, neither of which has yet been demonstrated to encode an oligopeptide transporter, is induced in root tissue in response to iron and zinc deficiency (AtOPT2) or iron, copper and manganese deficiency (AtOPT3) (Wintz et al., 2003; Stacey et al., 2006). Heterologous production of AtOPT3 in yeast indicates that this OPT can transport copper, manganese and iron (Wintz et al., 2003). AtOPT6 and AtOPT7, both of which have been demonstrated to be capable of transporting peptides, have also been shown to transport cadmium or cadmium–glutathione conjugates when heterologously expressed in yeast (Cagnac et al., 2004). These studies now prompt a reassessment of the importance of plant OPTs beyond their presumed role solely as peptide transporters. However, the studies have relied heavily on the use of yeast growth-rescue or growth-inhibition assays to demonstrate potential physiological function of a plant OPT produced in a heterologous system. Significantly, both the maize YS1 iron–phytosiderophore-uptake transporter and the Arabidopsis YS1-like (YSL) proteins are members of the OPT superfamily (Yen et al., 2001). As phytosiderophores and peptides are both amino acid derivatives, this shared property may provide a link between the apparent diverse substrate preferences of distinct members of the OPT superfamily. Future studies will need to confirm that the OPTs can also perform such a variety of functions including metal ion transport in planta.
THE SEARCH FOR PEPTIDE SUBSTRATES
Identification of their in-vivo substrates would significantly enhance our understanding of the role of plant PTRs and OPTs. The 20 protein amino acids can provide the PTRs with 8000 different tripeptides and the OPTs with over 150 000 different tetrapeptide or over three million pentapeptide substrates. Testing each transporter with every possible substrate would be a daunting task with present methodologies! Nevertheless, di- and tripeptides and in some instances peptide toxins have been demonstrated to be substrates for Arabidopsis, barley and bean PTRs (Table 1). Of the seven AtOPTs (AtOPT 8 and 9 have not been studied to date) tested for their ability to transport a limited number of peptides when heterologously produced in yeast, only AtOPT2 and AtOPT3 failed to transport any of the tetra- and pentapeptides tested (Table 2). None of the AtOPTs tested transported di- or tripeptides, indicating a minimum size limitation for OPT peptide substrates.
The most obvious roles of (oligo)peptide transporters in plants are in the acquisition of organic nitrogen and carbon nutrients from the rhizosphere and translocation of these nutrients around the plant. However, with the notable exception of the germinating barley grain, little is known about the concentrations of peptides and their derivatives in plant tissues or the rhizosphere. Therefore, the presence of a large number of apparent PTRs and OPTs in plants presents a biological conundrum. Indeed, a view has prevailed that peptides (as opposed to amino acids) are important in plant nitrogen nutrition only in certain specialized circumstances, namely in protein reserve mobilization during germination, in redistribution of metabolites during senescence and in the nutrition of carnivorous plants. It has been suggested that (oligo)peptide transport both is energetically more favourable than that of free amino acids and avoids the competition that may occur between free amino acids, for which multiple carrier systems operate (Higgins and Payne, 1982). Although peptides have not generally been considered to be involved in long-distance transport of nitrogen in plants, this conclusion is based on studies in which the method of assimilate analysis could have caused hydrolysis of any peptides to their constituent amino acids. There is now an increasing awareness of the extent and importance of the transport of macromolecules, including peptides and proteins, between source and sink tissues (Thompson and Schulz, 1999), making a reappraisal of the role of (oligo)peptides and (oligo)peptide transporters in the long-distance translocation of organic nitrogen around the plant necessary.
Analysis of PTR and OPT expression patterns and the subcellular localization of the encoded proteins can begin to provide circumstantial evidence for individual transporter functions. Although heterologous production systems are useful in that they can demonstrate that a protein is capable of transporting a particular substrate, they are less informative as a tool to identify the in-planta substrates and roles of transporter proteins. These must be determined in vivo if we are to gain insight into the physiological roles of plant PTRs and OPTs. The recent exciting developments in plant metabolomic analyses could yet reveal a more prominent role for peptides in nutrient redistribution and/or signalling in plants and the results of investigations using these approaches are eagerly awaited.
CONCLUSIONS
Recently, there has been an increasing awareness that a number of peptides including systemin, clavata and the phytosulfokines (PSKs) also function in plant cell signalling roles (Lindsey et al., 2002). However, with the exception of the PSKs, which are sulfated tetra- and pentapeptides with mitogenic activity, these other peptides are too large to be substrates for transport by the PTRs or OPTs. Although transduction of their mitogenic signal involves interaction of PSKs with an integral plasma-membrane PSK-receptor (Yang et al., 2000), transport of PSKs around the plant may involve the OPTs, but this role has yet to be verified. Given the metabolic complexity of plants, there is great potential for (oligo)peptides and their transporters to play a number of roles in plant physiology. Roles for plant PTRs and OPTs in host–pathogen interactions (Taylor et al., 1972; Dietrich et al., 2004), responses to mechanical stress or wounding (Karim et al., 2005) and translocation of a number of molecules including plant hormones and metal ions around the plant in the form of peptide conjugates have also been suggested. Nitrogen, iron and phosphorus are the three nutrients most commonly limiting plant growth (Guerinot, 2001). Arabidopsis PTRs and OPTs appear to play key roles in acquisition of two of these key nutrients from the environment and subsequent transport around the plant. If the properties of PTRs and OPTs in crop plants reflect those found in Arabidopsis, this will open the way for manipulation of these transporters to address important issues of improving plant growth, biomass production and nutritional value of crop species. These remain exciting challenges for research in this area.
Acknowledgments
Our sincere apologies to colleagues whose contributions to the study of peptide transport in plants are not cited because of space constraints. We thank Drs Chris West and Jon Pittman for help with phylogenetic analysis (C.W.) and critical reading of the manuscript (J.P., C.W.). The financial support of the UK Biotechnology and Biological Science Research Council is gratefully acknowledged.
LITERATURE CITED
- Arabidopsis Genome Initiative. 2000. Analysis of the genome of the flowering plant Arabidopsis thaliana. Nature 408: 796–815. [DOI] [PubMed] [Google Scholar]
- Bollard EG. 1966. A comparative study of the ability of organic compounds to serve as sole sources of nitrogen for the growth of plants. Plant Soil 25: 153–166. [Google Scholar]
- Bogs J, Bourbouloux A, Cagnac O, Wachter A, Rausch T, Delrot S. 2003. Functional characterization and expression analysis of a glutathione transporter, BjGT1, from Brassica juncea: evidence for regulation by heavy metal exposure. Plant, Cell and Environment 26: 1703–1711. [Google Scholar]
- Bourbouloux A, Shahi P, Chakladar A, Delrot S, Bakhawat AK. 2000. Hgt1p, a high affinity glutathione transporter from the yeast Saccharomyces cerevisiae. Journal of Biological Chemistry 275: 13259–13265. [DOI] [PubMed] [Google Scholar]
- Cagnac O, Bourbouloux A, Chakrabarty D, Zhang M-Y, Delrot, S. 2004. AtOPT6 transports glutathione derivatives and is induced by primisulfuron. Plant Physiology 135: 1378–1387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chapin FS, Moilanen L, Keilland K. 1993. Preferential use of organic nitrogen for growth by a non-mycorrhizal arctic sedge. Nature 361: 150–153. [Google Scholar]
- Chenna R, Sugawara H, Koike T, Lopez R, Gibson TJ, Higgins DG, Thompson JD. 2003. Multiple sequence alignment with the Clustal series of programs. Nucleic Acids Research 31: 3497–3500. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chiang C-S, Stacey G, Tsay Y-F. 2004. Mechanisms and functional properties of two peptide transporters, AtPTR2 and fPTR2. Journal of Biological Chemistry 279: 30150–30157. [DOI] [PubMed] [Google Scholar]
- Dietrich D, Hammes U, Thor K, Suter-Grotemeyer M, Flukiger R, et al. 2004. AtPTR1, a plasma membrane peptide transporter expressed during seed germination and in vascular tissue of Arabidopsis. The Plant Journal 40: 488–499. [DOI] [PubMed] [Google Scholar]
- Felsenstein J. 1998. Phylogenies from molecular sequences: inference and reliability. Annual Review of Genetics 22: 521–565. [DOI] [PubMed] [Google Scholar]
- Forde BG. 2000. Nitrate transporters in plants: structure, function and regulation. Biochimica et Biophysica Acta 1465: 219–235. [DOI] [PubMed] [Google Scholar]
- Guerinot ML. 2001. Improving rice yields—ironing out the details. Nature Biotechnology 19: 417–418. [DOI] [PubMed] [Google Scholar]
- Hauser M, Donhardt AM, Barnes D, Naider F, Becker JM. 2000. Enkephalins are transported by a novel eukaryotic peptide uptake system. Journal of Biological Chemistry 275: 3037–3041. [DOI] [PubMed] [Google Scholar]
- Hauser M, Narita V, Donhardt AM, Naider F, Becker JM. 2001. Multiplicity and regulation of genes encoding peptide transporters in Saccharomyces cerevisiae. Molecular Membrane Biology 18: 105–112. [PubMed] [Google Scholar]
- Higgins CF, Payne JW. 1981. The peptide pools of germinating barley grains: relation to hydrolysis and transport of storage proteins. Plant Physiology 67: 785–792. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Higgins CF, Payne JW. 1982. Plant peptides. Encyclopedia of Plant Physiology 14A: 438–458. [Google Scholar]
- Huang NC, Liu KH, Lu HJ, Tsay Y-F. 1999. Cloning and functional characterization of an Arabidopsis nitrate transporter gene that encodes a constitutive component of low-affinity uptake. Plant Cell 11: 1381–1392. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jamai A, Chollet J-F, Delrot S. 1994. Proton peptide co-transport in broad bean leaf tissues. Plant Physiology 106: 1023–1031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jamai A, Gaillard C, Delrot S, Martinoia E. 1995. Dipeptide transport in barley mesophyll vacuoles. Planta 196: 430–433. [DOI] [PubMed] [Google Scholar]
- Jeong J, Suh S, Guan C, Tsay YF, Moran N, Oh CJ, et al. 2004. A nodule specific dicarboxylate transporter from alder is a member of the peptide transporter family. Plant Physiology 134: 969–978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karim S, Lundh D, Holmström K-O, Mandal A, Pirhonen M. 2005. Structural and functional characterization of AtPTR3, a stress-induced peptide transporter of Arabidopsis. Journal of Molecular Modeling 11: 226–236. [DOI] [PubMed] [Google Scholar]
- Kielland K. 1994. Amino acid absorption by arctic plants: implications for plant nutrition and nitrogen cycling. Ecology 75: 2373–2383. [Google Scholar]
- Koh S, Wiles AM, Sharp JS, Naider FR, Becker JM, Stacey G. 2002. An oligopeptide transporter gene family in Arabidopsis. Plant Physiology 128: 21–29. [PMC free article] [PubMed] [Google Scholar]
- Lin C-M, Koh S, Stacey G, Yu S-M, Lin T-Y, Tsay Y-F. 2000. Cloning and functional characterization of a constitutively expressed nitrate transporter gene, OsNRT1, from rice. Plant Physiology 127: 379–388. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lindsey K, Casson S, Chilley P. 2002. Peptides: new signaling molecules in plants. Trends in Plant Science 7: 78–83. [DOI] [PubMed] [Google Scholar]
- Lubkowitz MA, Hauser L, Breslav M, Naider F, Becker JM. 1997. An oligopeptide transport gene from Candida albicans. Microbiology 143: 387–396. [DOI] [PubMed] [Google Scholar]
- Lubkowitz MA, Barnes D, Breslav M, Burchfield A, Naider F, Becker JM. 1998. Schizosaccharomyces pombe isp4 encodes a transporter representing a novel family of oligopeptide transporters. Molecular Microbiology 28: 729–741. [DOI] [PubMed] [Google Scholar]
- Miranda M, Borisjuk L, Tewes A, Dietrich D, Rentsch D, Weber H, Wobus U. 2003. Peptide and amino acid transporters are differentially regulated during seed development and germination in faba bean. Plant Physiology 132: 1950–1960. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Osawa H, Stacey G, Gassmann W. 2006. ScOPT1 and AtOPT4 function as proton-coupled oligopeptide transporters with broad but distinct substrate specificities. Biochemical Journal 393: 267–275. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rentsch D, Laloi M, Rouhara I, Schmelzer E, Delrot S, Frommer WB. 1995. NTR1 encodes a high affinity oligopeptide transporter in Arabidopsis. FEBS Letters 370: 264–268. [DOI] [PubMed] [Google Scholar]
- Saier MH Jr. 2000. Families of transmembrane transporters selective for amino acids and their derivatives. Microbiology 146: 1775–1795. [DOI] [PubMed] [Google Scholar]
- Schimel JP, Chapin III FS. 1996. Tundra plant uptake of amino acid and NH4+ nitrogen in situ: plants compete well for amino acid N. Ecology 77: 2142–2147. [Google Scholar]
- Schulze W, Frommer WB, Ward JM. 1999. Transporters for ammonium, amino acids and peptides are expressed in pitchers of the carnivorous plant Nepenthes. The Plant Journal 17: 637–646. [DOI] [PubMed] [Google Scholar]
- Song W, Steiner HY, Zhang L, Naider F, Stacey G, Becker JM. 1996. Cloning of a second Arabidopsis peptide transporter gene. Plant Physiology 110: 171–178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Song W, Koh S, Czako M, Marton L, Drenkard E, Becker JM, Stacey G. 1997. Antisense expression of the peptide transporter gene AtPTR2-B delays flowering and arrests seed development in transgenic Arabidopsis plants. Plant Physiology 114: 927–935. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stacey G, Koh S, Granger G, Becker JM. 2002a. Peptide transport in plants. Trends in Plant Science 7: 257–263. [DOI] [PubMed] [Google Scholar]
- Stacey MG, Koh S, Becker JM, Stacey G. 2002b. AtOPT3, a member of the oligopeptide transporter family, is essential for embryo development in Arabidopsis. The Plant Cell 14: 2799–2811. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stacey MG, Osawa H, Patel A, Gassmann G, Stacey G. 2006. Expression analysis of Arabidopsis oligopeptide transporters during seed germination, vegetative growth and reproduction. Planta 223: 291–305. [DOI] [PubMed] [Google Scholar]
- Steiner H-Y, Song W, Zhang L, Naider F, Becker JM, Stacey G. 1994. An Arabidopsis peptide transporter is a member of a new class of membrane transporter proteins. Plant Cell 6: 1289–1299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taylor PA, Schnoes HK, Durbin RD. 1972. Characterization of chlorosis-inducing toxins from a plant pathogenic Pseudomonas sp. Biochimica et Biophysica Acta 286: 107–117. [DOI] [PubMed] [Google Scholar]
- Thompson GA, Schulz A. 1999. Macromolecular trafficking in the phloem. Trends in Plant Science 4: 354–360. [DOI] [PubMed] [Google Scholar]
- Tsay Y-F, Schroeder JI, Feldman KA, Crawford NM. 1993. The herbicide sensitivity gene CHL1 of Arabidopsis encodes a nitrate-inducible nitrate transporter. Cell 72: 705–713. [DOI] [PubMed] [Google Scholar]
- Wang R, Liu D, Crawford NM. 1998. The Arabidopsis CHL1 protein plays a major role in high-affinity nitrate uptake. Proceedings of the National Academy of Sciences of the USA 95: 15134–15139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Waterworth WM, West CE, Bray CM. 2000. The barley scutellar peptide transporter: biochemical characterization and localization to the plasma membrane. Journal of Experimental Botany 51: 1201–1209. [PubMed] [Google Scholar]
- Waterworth WM, Ashley MK, West CE, Bray CM. 2003. Peptide transport in the developing barley grain. In: Nicolas G, Bradford KJ, Come D, Pritchard HW, eds. The biology of seeds. Wallingford, UK: CABI International, 85–92.
- Waterworth WM, Ashley MK, West CE, Sunderland PA, Bray CM. 2005. A role for phosphorylation in the regulation of the barley scutellar peptide transporter HvPTR1 by amino acids. Journal of Experimental Botany 56: 1545–1552. [DOI] [PubMed] [Google Scholar]
- West CE, Waterworth WM, Stephens SM, Smith CP, Bray CM. 1998. Cloning an functional expression of a peptide transporter expressed in the scutellum of barley grain during the early stages of germination. The Plant Journal 15: 221–229. [DOI] [PubMed] [Google Scholar]
- Wintz H, Fox T, Wu Y-Y, Feng V, Chen W, Chang H-S, et al. 2003. Expression profiles of Arabidopsis thaliana in mineral deficiencies reveal novel transporters involved in metal homeostasis. Journal of Biological Chemistry 278: 47644–47653. [DOI] [PubMed] [Google Scholar]
- Yang H, Matsubayashi Y, Hanai H, Sakagami Y. 2000. Phytosulfokine-α, a peptide growth factor found in higher plants: its structure, functions, precursor and receptors. Plant Cell Physiology 41: 825–830. [DOI] [PubMed] [Google Scholar]
- Yen M-R, Tseng Y-H, Saier MH. 2001. Maize Yellow Stripe 1, an iron phytosiderophore uptake transporter, is a member of the oligopeptide (OPT) family. Microbiology 147: 2881–2883. [DOI] [PubMed] [Google Scholar]
- Zhang M-Y, Bourbouloux A, Cagnac O, Srikanth CV, Rentsch D, Bachhawat AK, Delrot S. 2004. A novel family of transporters mediating the transport of glutathione derivatives in plants. Plant Physiology 134: 482–491. [DOI] [PMC free article] [PubMed] [Google Scholar]