Abstract
Ribonuclease P (RNase P) is an ancient and essential endonuclease that catalyses the cleavage of the 5′ leader sequence from precursor tRNAs (pre-tRNAs). The enzyme is one of only two ribozymes which can be found in all kingdoms of life (Bacteria, Archaea, and Eukarya). Most forms of RNase P are ribonucleoproteins; the bacterial enzyme possesses a single catalytic RNA and one small protein. However, in archaea and eukarya the enzyme has evolved an increasingly more complex protein composition, whilst retaining a structurally related RNA subunit. The reasons for this additional complexity are not currently understood. Furthermore, the eukaryotic RNase P has evolved into several different enzymes including a nuclear activity, organellar activities, and the evolution of a distinct but closely related enzyme, RNase MRP, which has different substrate specificities, primarily involved in ribosomal RNA biogenesis. Here we examine the relationship between the bacterial and archaeal RNase P with the eukaryotic enzyme, and summarize recent progress in characterizing the archaeal enzyme. We review current information regarding the nuclear RNase P and RNase MRP enzymes in the eukaryotes, focusing on the relationship between these enzymes by examining their composition, structure and functions.
Keywords: RNase P, RNase MRP, tRNA processing, rRNA processing, holoenzyme, nucleolus
1. INTRODUCTION
Ribonuclease (RNase) P is an essential endoribonuclease, conserved throughout evolution, which is responsible for the maturation of the 5′ end of transfer RNA (tRNA) (Frank and Pace, 1998). The bacterial enzyme is comprised of a large RNA (typically 350–400 nt) and a single small protein (~10% by mass). Both the RNA and protein components are essential in vivo, however the bacterial RNA has been shown to function independently in vitro and can accurately and efficiently catalyze pre-tRNA cleavage under high salt conditions (Guerrier-Takada and Altman, 1984). The bacterial RNA is therefore an RNA based enzyme, or ribozyme. Only two ribozymes have been found to be present in all kingdoms of life, the other being the ribosome.
The archaeal and eukaryal nuclear RNase P enzymes have considerably more complex subunit compositions, retaining a structurally related RNA (typically 300–350 nt), but with an increased number of protein subunits compared to the bacterial enzymes (Xiao et al., 2002). At least four proteins have been found in archaeal enzymes (~45% by mass)1 with more found in the eukaryotic enzymes, nine proteins in yeast and ten proteins in human (~70% by mass).1 There is no sequence homology between the bacterial protein and archaeal/eukaryal proteins and so the evolutionary relationship between these proteins is not currently clear. The eukaryal RNase P however, shares a common ancestor with the archaeal enzyme as they have homologous protein components, with the eukaryal enzyme having acquired additional protein subunits (Hartmann and Hartmann, 2003). This poses questions as to the specific reasons for the requirement for these additional proteins in the eukaryal enzymes. They could have direct roles in the spatial organization of RNase P within the complex nuclear environment, could aid the recognition of substrate tRNAs, and may also perform or assist RNase P in other uncharacterized roles. The situation is further complicated by the emergence of RNase MRP in eukaryotes; a likely paralogue of RNase P which has a related RNA and a nearly identical protein content, but with specificity towards different substrates.
Here we summarize current progress in understanding the nature of RNase P holoenzymes. We begin with a brief summary of the bacterial system focusing on the RNA component, which is directly related to the RNA of the archaeal and eukaryal RNase P enzymes. Recent progress in characterizing the archaeal RNase P system is also discussed, with emphasis on how the biochemical and structural analysis of the archaeal enzymes will provide insights into the more complex eukaryal enzymes. Lastly, we will discuss the development of multiple enzymes in the eukaryote to carry out the functions of RNase P in different subcellular compartments.
2. BACTERIAL RNase P
Substrates Recognized by Bacterial RNase P
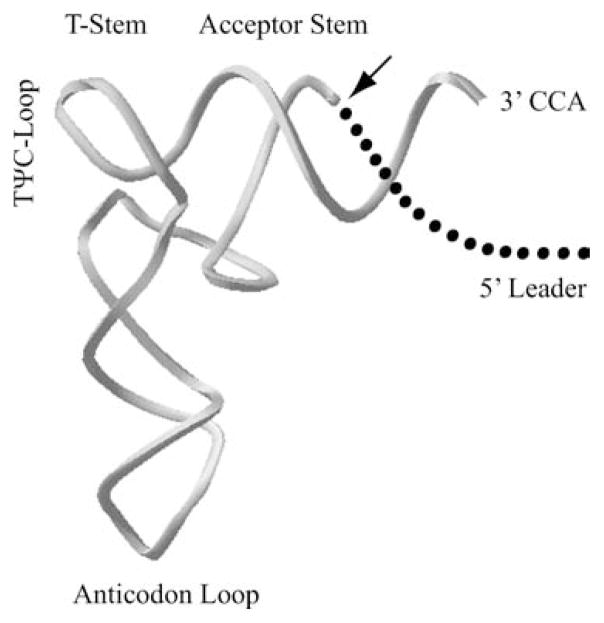
RNase P processes the 5′ end of pre-tRNAs, precisely removing the 5′ leader sequence, resulting in a 5′ phosphate on the mature tRNA and a 3′ hydroxyl on the leader sequence (Figure 1). The activity is dependent upon divalent cations, preferably magnesium, which are likely to fulfill both structural and functional roles (Smith and Pace, 1993; Yarus, 1993). The bacterial ribozyme has been shown to recognize common structural elements of pre-tRNA substrates such as the coaxially stacked acceptor stem, T-stem with the TΨC-loop and also the 3′-CCA sequence which is present in most bacterial pre-tRNA transcripts (McClain et al., 1987; Kahle et al., 1990; Thurlow et al., 1991; Kirsebom and Svard, 1994). The 5′ leader sequence of the pre-tRNA also participates, binding to the protein moiety of the bacterial holoenzyme (Stams et al., 1998). The bacterial RNase P has been demonstrated to specifically cleave other non-tRNA substrates including the precursors to 4.5S RNA (Peck-Miller and Altman, 1991), viral mRNAs (pseudoknot structures) (Mans et al., 1990), tmRNA (Komine et al., 1994), C4 antisense RNA from bacteriophages P1 and P7 (Hartmann et al., 1995), a polycistronic pre-mRNA (Alifano et al., 1994), and some transient structures within riboswitches (Altman et al., 2005). In some cases the structures of the alternative substrates closely resemble pre-tRNAs, but most of the structures are distinct, indicating that the bacterial holoenzyme has flexibility in substrate recognition.
FIGURE 1.
RNase P cleaves the 5′ leader from precursor tRNAs (pre-tRNAs). The site of cleavage by RNase P is represented with an arrow. Structures required for substrate recognition and cleavage by RNase P, the acceptor stem and T-stem-loop, are indicated.
The Bacterial RNA Subunit
A wealth of sequence and phylogenetic data has been gathered for the bacterial RNase P RNAs (James et al., 1988; Haas et al., 1991; Brown et al., 1993; Haas et al., 1994).2 The derived secondary structures of the bacterial RNAs have led to the definition of two major subsets, the type A (Ancestral) and type B (Bacillus) bacterial RNase P RNAs (Haas et al., 1996). These subsets have been most frequently represented by the Escherichia coli RNA (type A) and Bacillus subtilis RNA (type B), shown in Figure 2. The bacterial RNase P RNAs can be divided into two independently folding domains involved in both substrate cleavage, the “catalytic domain” (C-domain), and substrate binding, the “specificity domain” (S-domain), (Pan, 1995; Loria and Pan, 1996). Sequence analysis has identified five Conserved Regions within the structures (CR-I to CR-V) that are found in all RNase P RNAs. The CR-I and CR-V are paired in the P4 pseudoknot structure (with a universally conserved, bulged uridine) and form part of the catalytic domain along with CR-IV. The recent crystal structure of the RNase P RNA from Bacillus stearothermophilus shows a second universally conserved nucleotide bulged from the P4 stem (Kazantsev et al., 2005). The C-domain has been shown to be catalytically active, processing pre-tRNAs in the presence of the bacterial protein (Loria and Pan, 1999). The S-domain hosts CR-II and CR-III in an unpaired region between stems P10/11 and P12, and appears to function in differentiating RNase P substrates (Kufel and Kirsebom, 1996; Loria and Pan, 1997). Comparative analysis and the unusually small RNA from Microplasma fermentas (276nt) has also allowed the definition of a bacterial minimum consensus structure (Figure 2) and the design of a minimal RNA (mini P) which is catalytically active (Pace and Brown, 1995; Siegel et al., 1996; Marquez et al., 2005).
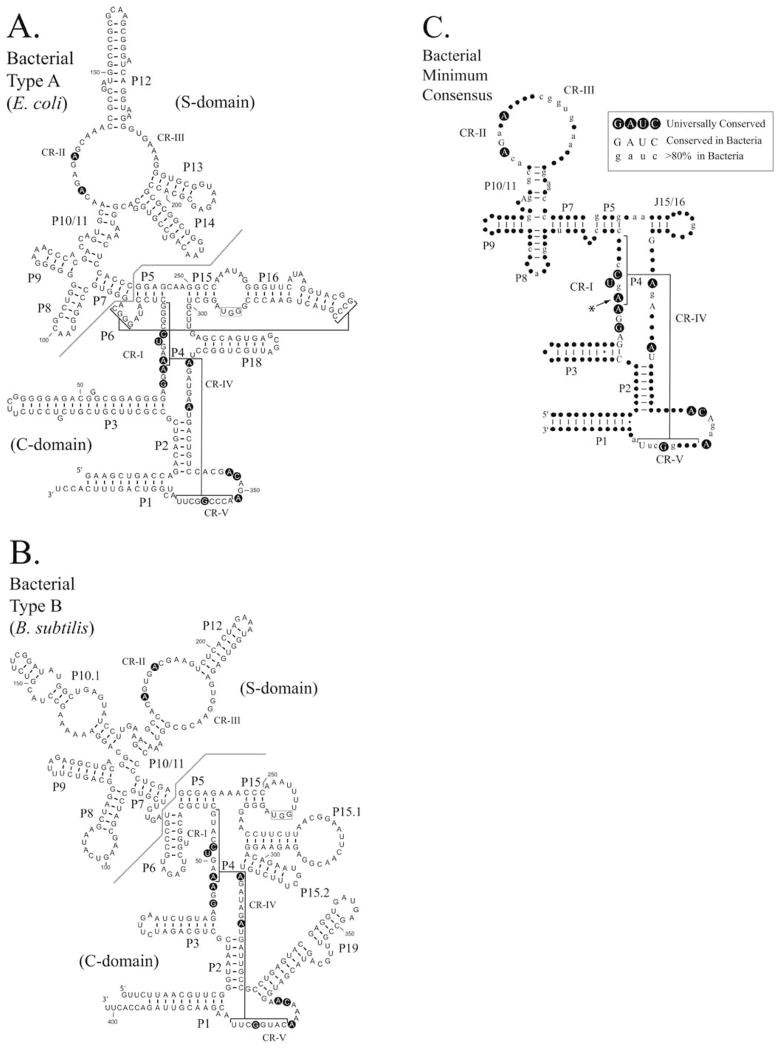
FIGURE 2.
Comparison of Type A and Type B bacterial RNase P RNAs. The secondary structures of RNase P RNAs derived by phylogenetic comparative sequence analysis are shown (Haas and Brown, 1998). (A). The secondary structure of the E. coli RNase P RNA (Type A). (B). The secondary structure of the B. subtilis RNase P RNA (Type B). Helices are numbered as paired regions (e.g. P1), other helices are indicated by lines (e.g. P4 and P6). Regions of the RNA implicated in substrate binding the “specificity domain” (S-domain) and catalytic activity the “catalytic domain” (C-domain) are separated by a line. A boxed region within the loop of P15 indicates a binding site for the 3′CCA of the pre-tRNA substrate, which is present in many bacterial RNase P RNAs. Nucleotides conserved in all known RNase P RNAs are shown in bold background. (C). Consensus structure showing elements common to all known bacterial RNase P RNAs (figure adapted from Marquez et al., 2005). The highlighted conserved adenine has been shown to be extrahelical in the structure of the Bacillus stearothermophilus RNase P RNA (Kazantsev et al., 2005). In this structure, the adjacent conserved adenosine forms the corresponding base pair within the P4 helix.
The isolated S-domains from both type A and type B bacterial RNAs have been structurally characterized showing that the CR-II and CR-III regions form two interacting T-loop structures which define the J11/12 – J12/11 module (Krasilnikov and Mondragon, 2003) [PDB ID. 1NBS] and (Krasilnikov et al., 2003) [PDB ID. 1U9S]. A number of nucleotides within the S-domain have been implicated in substrate binding, as these are protected from chemical modification in the presence of the substrate (LaGrandeur et al., 1994; Odell et al., 1998). These include a conserved CR-III nucleotide and other conserved bulged nucleotides within the P7-P10/11 region (four-way junction). These are presented on the RNA surface in a similar geometry in examples of both type A and type B structures, despite the differences in the secondary structures of these RNAs (Krasilnikov et al., 2004). Other regions implicated in substrate binding are the J15/16 loop region (type A) or the L15 loop (type B) which interact with the 3′ CCA of the bacterial tRNAs (Chen et al., 1998; Christian et al., 2002). These regions are also potentially involved in coordinate metal ion binding with the tRNA substrate (Kufel and Kirsebom, 1998; Oh et al., 1998). Structural characterization of the entire RNase P RNAs from bacterial type A and type B structures have also been reported and have begun to provide some insights into the functions of CR-I, CR-IV and CR-V (Torres-Larios et al., 2005) [PDB ID. 2A2E] and (Kazantsev et al., 2005) [PDB ID. 2A64]. These structures represent significant progress and are able to accommodate much of the vast wealth of biochemical data obtained in solution (Harris and Pace, 1995; Pan, 1995; Loria and Pan, 1996; Kazantsev and Pace, 1998; Kurz and Fierke, 2000; Christian et al., 2002). However, the resolution of these structures does not allow fine detail and, in both cases, crystallization appears to have had an influence on the structure. The precise roles for the conserved nucleotides surrounding CR-I, CR-V and also within CR-IV are not entirely clear although these nucleotides are found in close proximity to the P4 stem suggesting key roles in defining the catalytic structure, in keeping with their absolute conservation (Kazantsev et al., 2005; Torres-Larios et al., 2005).
Roles of the Bacterial Protein
The bacterial RNase P contains a single small protein which is essential for in vivo function, but not for binding and cleaving pre-tRNAs in vitro. The structures of bacterial RNase P proteins have been solved from B. subtilis (Stams et al., 1998) [PDB ID. 1A6F], Staphylococcus aureus (Spitzfaden et al., 2000) [PDB ID. 1D6T] and Thermotoga maritima (Kazantsev et al., 2003) [PDB ID. 1NZ0]. These analyses represent both solution and crystal structures from different species, and each protein shows a fold which is similar to the ribonucleoprotein (RNP) domain (Avis et al., 1996) [PDB ID. 1FHT]. The RNP domain is also known as RNA binding domain (RBD) or the RNA recognition motif (RRM). The protein from B. subtilis binds both the full length RNA and the C-domain with similar efficiency suggesting that the protein solely binds to this region of the RNA (Day-Storms et al., 2004). Additionally, the analysis of the binding of three different bacterial proteins to examples of both type A and type B RNAs suggests a common binding site which is near the catalytic domain (P4 and the surrounding P2, P3 periphery). Binding of the protein to the RNA appears to induce local conformational changes in similar regions of both the type A RNA (P5-P8, L15/16) and type B RNA (P15.1-P5.1, L15) (Buck et al., 2005).
There is debate regarding the precise roles of the bacterial protein with potential roles in stabilizing tertiary architecture (Guerrier-Takada et al., 1983; Westhof et al., 1996; Kim et al., 1997), dimer formation (Fang et al., 2001), enhancing pre-tRNA specificity over that of matured tRNAs (Crary et al., 1998; Kurz et al., 1998; Niranjanakumari et al., 1998; Hsieh et al., 2004), and a broadened substrate specificity (Gopalan et al., 1997). Recent data suggest that the proteins will contribute to each of these effects, and that inconsistent observations may be explained by inherent differences in the type A and type B RNAs. The E. coli protein (type A RNA) was observed to stabilize the E. coli RNA structure (type A) to a greater extent than the B. subtilis protein stabilized the B. subtilis RNA structure (type B) (Buck et al., 2005).
3. ARCHAEAL RNase P
Increased Complexity in the Archaeal Holoenzyme
Initial studies of the archaeal RNase P holoenzyme from Haloferax volcanii (Lawrence et al., 1987) and Sulfolobus acidocaldarius (Darr et al., 1990; LaGrandeur et al., 1993) indicated that both enzymes possess RNA, however their protein composition was quite different, as determined by their buoyant densities in CsCl. The H. volcanii enzyme was mostly RNA, a composition similar to bacteria, whereas the S. acidocaldarius enzyme had far greater protein content, closer to the eukaryotic enzymes. Current information suggests that most archaeal enzymes have increased protein compositions when compared to the bacterial enzymes (Andrews et al., 2001; Hartmann and Hartmann, 2003). Sequence searches have identified four proteins that are homologues of the yeast and human proteins, however these do not show any sequence similarity to the bacterial protein (Hall and Brown, 2002; Hartmann and Hartmann, 2003). A variety of nomenclatures have been adopted for the archaeal/eukaryal proteins. To aid a direct comparison with other sections of this review we have listed the archaeal nomenclature and also, in parentheses, that adopted in the Saccharomyces cerevisiae and Homo sapiens systems respectively (Table 1). For example, proteins identified in the genome of Methanothermobacter thermoautotrophicus are Mth11 (Pop4/Rpp29), Mth687 (Pop5/hPop5), Mth688 (Rpp1/Rpp30) and Mth1618 (Rpr2/Rpp21). These proteins were confirmed as archaeal RNase P subunits, as they each immunoprecipitate with RNase P activity (Hall and Brown, 2002).
TABLE 1.
Subunit composition of the archaeal RNase P and the eukaryal nuclear RNase P and RNase MRP
| Yeast RNase P/RNase MRP (S. cerevisiae)1 |
Mammalian RNase P/RNase MRP (H. sapiens)2 |
Archaeal RNase P (P. horokoshii)3 |
|||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Properties |
Subunit |
Properties |
Subunit |
Subunit | Properties |
||||||||
| Gene | Mr | (pI) | P | MRP | Gene | (pI) | P | MRP | P | Mr | (pI) | ||
| RNA | RNA | RNA | |||||||||||
| RPR1 | 120 | (+) | H1 | 112 | (+) | PH rnpB* | 107 | ||||||
| NME1 | 112 | (+) | 7–2 | 88 | (+) | ||||||||
| Protein | Protein | Protein | |||||||||||
| POP1 | 100.5 | 9.8 | (+) | (+) | hPOP1 | 115 | 9.6 | (+) | (+) | ||||
| POP3 | 22.6 | 9.6 | (+) | (+) | RPP38 | 38 | 9.6 | (+) | (+) | ||||
| POP4 | 32.9 | 9.3 | (+) | (+) | RPP29 | 29 | 10.2 | (+) | (+) | PH1771 | 15.1 | 11.2 | |
| POP5 | 19.6 | 7.8 | (+) | (+) | hPOP5 | 19 | 7.9 | (+) | (+) | PH1481 | 14.0 | 10.0 | |
| POP6 | 18.2 | 9.3 | (+) | (+) | |||||||||
| POP7 | 15.8 | 9.3 | (+) | (+) | RPP20 | 20 | 8.6 | (+) | (+) | ||||
| POP8 | 15.5 | 4.6 | (+) | (+) | |||||||||
| RPP1 | 32.2 | 9.8 | (+) | (+) | RPP30 | 30 | 9.2 | (+) | (+) | PH1877 | 24.7 | 9.6 | |
| RPR2 | 16.3 | 10.0 | (+) | RPP21 | 21 | 9.6 | (+) | PH1601 | 14.6 | 10.5 | |||
| SNM1 | 22.5 | 9.8 | (+) | ||||||||||
| RMP1 | 23.6 | 9.8 | (+) | ||||||||||
| RPP40 | 40 | 5.2 | (+) | ||||||||||
| RPP25 | 25 | 9.7 | (+) | (+) | |||||||||
| RPP14 | 14 | 7.6 | (+) | ||||||||||
Data from (Xiao et al., 2002).
Data from (Jarrous, 2002).
Data obtained from UniProtKB/Swiss-Prot (http://us.expasy.org/sprot/).
The P. horikoshii RNase P RNA gene has not been annotated.
Molecular weights are based upon gel migrations in many cases the mass differs from that calculated from the sequence.
Similarity and Variation in the Archaeal RNA Structure
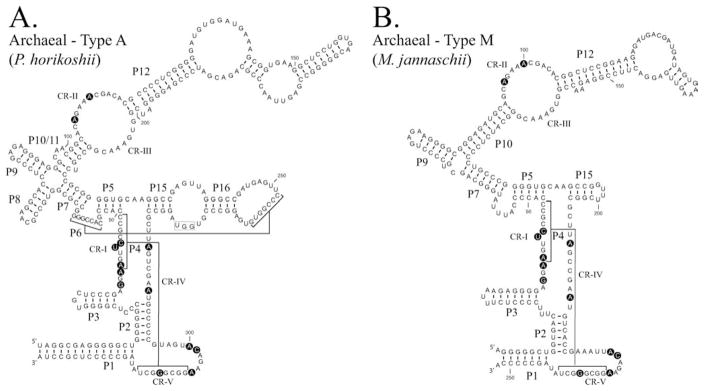
Unlike bacterial RNase P RNAs, the archaeal RNAs are not generally catalytically active in vitro in the absence of proteins. However, a small number of archaeal RNAs have displayed trace amounts of catalytic activity in vitro, under conditions of extreme ionic strength (Pannucci et al., 1999). The ability of these RNAs to display even trace activity indicates that they possess all of the structures necessary for substrate recognition and catalysis. The secondary structures of archaeal RNAs have been shown to conform into at least two classes, those displaying similarity to the bacterial RNAs, type A, and others with an apparently less complex structure, type M (Figure 3) (Haas et al., 1996; Harris et al., 2001). The archaeal type A RNAs lack the P13/14 stems and the P18 stem found in the bacterial type A RNAs, and many show extensions and additional complexity in their P12 stem. The type M archaeal RNAs appear to be more diverged from the bacterial RNAs, also lacking the P8 helix from the cruciform region and the extended P15 region, including the CCA recognition loop and the P6 pseudoknot. This structural divergence is also reflected in the physical properties of these RNA subsets. Those archaeal RNAs that have displayed trace activity in the absence of protein are all found to possess an archaeal type A structure. The catalytic fold of these RNAs seem to be at least partially stabilized under the extreme ionic conditions used. It is also noted that some archaeal type A RNAs are able to utilize the bacterial protein in order to form a functional chimeric holoenzyme, although with reduced activity relative to that of the wild type enzymes (Pannucci et al., 1999). The type M RNAs have not shown the ability to be RNA-only ribozymes or the ability to productively utilize the bacterial protein as a cofactor (Pannucci et al., 1999). These archaeal RNAs appear to have become more dependent on their own proteins to achieve a catalytic fold. There is potential for the proteins to have evolved direct roles in catalysis, however this seems unlikely given the similarity of the RNA core structures and the demonstrated catalytic activity of some archaeal RNAs.
FIGURE 3.
Comparision of Type A and Type M archaeal RNase P RNAs. The secondary structures of RNase P RNAs derived by phylogenetic comparative sequence analysis are shown (Haas et al., 1996). (A). The secondary structure of the Pyrococcus horikoshii OT3 RNase P RNA (Type A). (B). The secondary structure of the Methanococcus jannaschii RNase P RNA (Type M). Structures are represented in a similar manner to Figure 2. Nucleotides conserved in all known RNase P RNAs are shown in bold background.
The Architecture of the Archaeal RNase P Holoenzyme
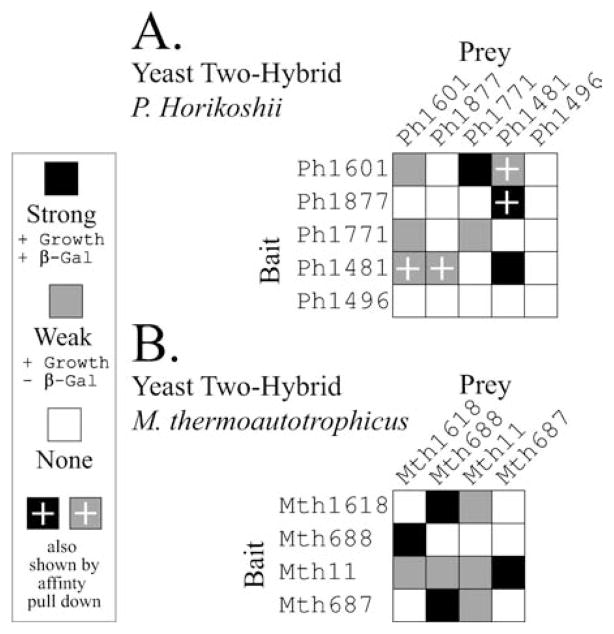
Perhaps one of the best characterized archaeal systems is that of Pyrococcus horikoshii OT3 which has been shown to have a single RNA (type A) and at least four proteins: Ph1771 (Pop4/Rpp29), Ph1481 (Pop5/hPop5), Ph1877 (Rpp1/Rpp30) and Ph1601 (Rpr2/Rpp21). It has also been suggested that a fifth protein, Ph1496, might be associated with the P. horikoshii RNase P (Kifusa et al., 2005). Although Ph1496 has not been identified in a purified archaeal enzyme, it has clear sequence homology to the human Rpp38 protein and predicted structural homology to the yeast Pop3 protein (Srisawat et al., 2002; Dlakic, 2005). The interactions among the protein subunits of the P. horikoshii have been investigated using the yeast two hybrid (Y2H) system (Kifusa et al., 2005). Ph1481p (Pop5/hPop5) was implicated as a key protein showing reciprocal interactions with two other proteins Ph1877 (Rpp1/Rpp30) and Ph1601 (Rpr2/Rpp21), (Figure 4). These interactions were also confirmed by in vitro pull down experiments. It is noted that Ph1496 was not found to interact with any of the four known RNase P proteins in this study. A similar study has been performed on the four protein subunits identified in the M. thermoautotrophicus system (Hall and Brown, 2004). In this study, Mth11 (Pop4/Rpp29) was shown to be a key protein involved in several protein-protein interactions, showing reciprocal interactions with Mth1618 (Rpr2/Rpp21) and Mth687 (Pop5/hPop5), (Figure 4). Interactions common to both Y2H studies are the Pop5/hPop5 homologue interacting with the Rpp1/Rpp30 homologue and also the Pop4/Rpp29 homologue forming a strong interaction with the Rpr2/Rpp21 homologue. The Pop4/Rpp29 homologue is also seen to self associate in both studies. The reasons for the lack of consistency in the other detected interactions (both strong and weak) are not clear, and could potentially be due to physical differences between the enzymes or due to the limitations of the experimental methods.
FIGURE 4.
Summary of protein-protein interactions detected in archaeal systems. Data from previously published yeast two-hybrid studies are summarized. Tables are drawn such that homologous proteins are represented in similar positions in both tables, starting from the top left corner. The interactions are indicated as described in the original text. (A). Interactions demonstrated in protein components of the Pyrococcus horikoshii OT3 RNase P (Kifusa et al., 2005). (B). Interactions demonstrated in protein components of the Methanothermobacter thermoautotrophicus RNase P (Hall and Brown, 2004).
The reconstitution of an active archaeal holoenzyme from its individual RNA and protein components has been reported in both the P. horikoshii system (Kouzuma et al., 2003), and the M. thermoautotrophicus system (Boomershine et al., 2003). The P. horikoshii enzyme required its RNA component and three proteins, Ph1481 (Pop5/hPop5), Ph1601 (Rpr2/Rpp21), Ph1771 (Pop4/Rpp29), to facilitate activity in a pre-tRNA cleavage assay with the further addition of Ph1877 (Rpp1/Rpp30) found to stimulate greater activity (Kouzuma et al., 2003).
Structural Characterization of the Archaeal Proteins
Structural characterization of the archaeal proteins, including all four proteins from the P. horikoshii system, has now been achieved (Boomershine et al., 2003; Sidote and Hoffman, 2003; Numata et al., 2004; Sidote et al., 2004; Takagi et al., 2004; Kakuta et al., 2005; Wilson, et al. 2006; S. Kawano, et al., 2006). In many cases, the structural characterization of the protein subunits has been combined with mutational analysis and in vitro reconstitution, allowing key functional regions of the proteins to be identified.
The crystal structure of Ph1877 (Rpp1/Rpp30) indicates that the protein has an overall similarity to the TIM barrel structure adopting an α/β barrel structure consisting of ten alpha-helices and seven beta-strands (Takagi et al., 2004) [PDB ID. 1V77]. Surface analysis of the Ph1877 (Rpp1/Rpp30) crystal structure indicates five conserved, positively charged, amino acids. The individual mutation of each of these significantly affects the activity of a reconstituted holoenzyme (Takagi et al., 2004).
The crystal structure of Ph1601 (Rpr2/Rpp21) shows a novel overall protein architecture consisting of two structured domains linked by an unstructured region (Kakuta et al., 2005) [PDB ID. 1X0T]. The N-terminal domain has two long interacting α-helices linked to its C-terminal domain by an unstructured central domain. The structured C-terminal domain has homology to the zinc ribbon domain, and shows a central zinc ion coordinated by four conserved cysteine residues. Mutational analysis suggests a predominant role for the zinc ion in defining the protein structure, although a potential role in enzyme function cannot be excluded by this analysis. Other mutations directly affect the activity of a reconstituted holoenzyme, including the mutation of conserved positions within the unstructured central region.
Solution structures of archaeal homologues of the Pop4/Rpp29 protein from M. thermoautotrophicus (Boomershine et al., 2003) [PDB ID. 1OQK] and Archaeoglobus fulgidus (Sidote and Hoffman, 2003) [PDB ID. 1PC0] were found to be similar. The structures indicate that the proteins are members of the oligomer-binding (OB) fold family, displaying a structured beta barrel core but with highly flexible N-terminal and C-terminal regions. However, subsequent crystal structures of homologues from P. horikoshii (Numata et al., 2004) [PDB ID. 1V76] and A. fulgidus (Sidote et al., 2004) [PDB ID. 1TS9 & 1TSF] showed the terminal regions as ordered helices. The differences noted between the solution and crystal structures are particularly interesting, since these regions contain several of the most highly conserved residues in the protein. As these residues do not contribute directly to the structure of the protein, these could be involved in binding interactions within the holoenzyme or to its substrates. It is noted that the eukaryal Pop4/Rpp29 sequences show large N-terminal sequence extensions which are not found in the archaeal homologues of Pop4/Rpp29. This extended region may serve a unique function in the eukaryal enzymes.
Crystal structures have been published for two archaeal homologues of the Pop5/hPop5 protein from Pyrococcus furiosus (Wilson, et al. 2006) [PDB ID. 2AV5] and P. horikoshii (S. Karwano et al., 2006) [PDB ID. 2CZW]. The P. horikoshii structure was obtained in complex with the Ph1877 (Rpp1/Rpp30) subunit. Both analyses are consistent and show the archaeal Pop5/hPop5 structure as an α/β sandwich consisting of a central five-stranded anti-parallel β-sheet surrounded by four α-helices. The overall structural arrangement is similar to the RNP domain found in the bacterial RNase P proteins, however the topology of the archaeal Pop5/hPop5 is distinct to that of the bacterial proteins. Despite the structural similarity, the data are consistent with previous sequence comparisons that do not suggest the direct evolution of any archaeal/eukaryal protein from a bacterial ancestor (Hall and Brown, 2002; Hartmann and Hartmann, 2003). The relevance of this structural similarity, with respect to the evolution of the archaeal/eukaryal proteins, is currently not clear.
Both archaeal Pop5/hPop5 structures show that the subunit is able to dimerize at the α1/α2 junction. In the case of Ph1481 (Pop5/hPop5) mutational analysis, combined with gel filtration and reconstitution assays, indicate that the dimer may represent a physiologically relevant structure. The Ph1481 (Pop5/hPop5) structure shows a central homodimer with each subunit forming an interaction with a single Ph1877 (Rpp1/Rpp30) subunit (S. Karwano et al., 2006). The site of interaction between the Ph1481 (Pop5/hPop5) and Ph1877 (Rpp1/Rpp30) subunits shown in the crystal structure appears to be consistent with preliminary NMR analysis performed the in the P. furiosus system (Wilson et al., 2006).
The combined structural and mutational analysis of all four archaeal proteins (including a complete set from the P. horikoshii system) has identified a number of conserved regions within the protein structures. These regions have potential roles in RNA-protein, protein-protein and perhaps protein-substrate interactions. Since these proteins have eukaryal counterparts, these studies will be relevant to the study of the eukaryal enzymes. Such data will enable the design of new experiments and lead to further insights into the structural and functional roles of the proteins found in the archaeal and eukaryal systems.
4. EUKARYAL RNase P
Multiple Enzymes Exist in Eukaryotes
In eukaryotes, RNase P appears to have split into a number of distinct activities, including diverse pre-tRNA cleavage activities found in eukaryotic organelles, such as mitochondria and chloroplasts (Schon, 1999; Xiao et al., 2002). The organellar activities will be discussed in later sections. Eukaryotes possess both a nuclear RNase P, and another related essential endoribonuclease, RNase MRP, which is found only in eukaryotes and cleaves at least three distinct substrates. The majority of RNase MRP is located in the nucleus where it processes precursor ribosomal RNA (pre-rRNA) (Schmitt et al., 1993), specifically cleaving the A3 site and leading to the maturation of the 5′ end of the 5.8S rRNA (Chu et al., 1994; Henry et al., 1994; Lygerou et al., 1996). Other characterized functions are the processing of mitochondrial RNAs (mtRNAs) (Chang and Clayton, 1987; Lee and Clayton, 1998) leading to the generation of primers for mitochondrial DNA (mtDNA) replication (Chang and Clayton, 1987; Lee and Clayton, 1998). RNase MRP also plays a role in the degradation of the mRNA of a B-type cyclin, an activity which appears to be important for cell cycle progression (Cai et al., 2002; Gill et al., 2004). Despite possessing several common proteins the enzymes are distinct and are not generally found to be physically associated, RNase MRP RNA is not detected in purified nuclear RNase P from S. cerevisiae or H. sapiens (Lygerou et al., 1996; Chamberlain et al., 1998; Srisawat and Engelke, 2001; Li and Altman, 2002). In humans, mutations within the promoter region and mature MRP RNA have been linked to genetically inherited developmental disorders which result in short stature amongst other symptoms; these are cartilage-hair hypoplasia (CHH) and a more severe manifestation, Anauxetic dysplasia (Ridanpaa et al., 2001; Thiel et al., 2005).
Composition of the Nuclear RNase P and RNase MRP Holoenzymes
The yeast nuclear RNase P has been biochemically purified, from S. cerevisiae, and shown to contain an RNA (RPR1) and nine protein subunits: Pop1, Pop3, Pop4, Pop5, Pop6, Pop7, Pop8, Rpr2 and Rpp1 (Chamberlain et al., 1998) (Table 1). Depletion of the yeast RNase P RNA and protein subunits in vivo has shown that all ten subunits are essential for function and cell viability (Lee et al., 1991; Lygerou et al., 1994; Chu et al., 1997; Dichtl and Tollervey, 1997; Stolc and Altman, 1997; Chamberlain et al., 1998). In yeast, RNase MRP has been biochemically purified and shown to possess an RNA (NME1) and ten proteins, eight of which are also components of the yeast nuclear RNase P; Pop1, Pop3–8 and Rpp1 (Table 1). RNase MRP lacks the Rpr2 subunit of RNase P. (Lygerou et al., 1994; Dichtl and Tollervey, 1997; Stolc and Altman, 1997; Chamberlain et al., 1998; Salinas et al., 2005), but contains two proteins that are unique to RNase MRP, Snm1 and Rmp1 (Table 1), (Schmitt and Clayton, 1994; Salinas et al., 2005). The differences in substrate specificities between RNase P and RNase MRP could, in part, be directed by these unique proteins, changes between the RNA structures, or a combination of both. However, the Rpr2 protein subunit of RNase P does not appear to be involved in pre-tRNA substrate recognition in vitro. A precursor form of RNase P lacking Rpr2 displays steady state kinetics for cleavage of pre-tRNAs that are indistinguishable from the mature holoenzyme (Srisawat et al., 2002).
The human nuclear RNase P has also been biochemically purified by various methods and shown to comprise a single RNA subunit (H1) (Bartkiewicz et al., 1989) and at least ten proteins Rpp14, 20, 21, 25, 29, 30, 38, 40, hPop5 and hPop1 (Table 1), (Eder et al., 1997; Jarrous and Altman, 2001). Seven of these proteins are homologues of the yeast proteins and many were initially identified due to sequence homology (Lygerou et al., 1996; Jarrous et al., 1999; van Eenennaam et al., 1999; van Eenennaam et al., 2001). The human RNase MRP RNA possesses a unique RNA (7 – 2) (Hashimoto and Steitz, 1983), which either binds or co-precipitates with at least seven of the ten proteins identified in the human RNase P, suggesting that they are also components of the human RNase MRP; Rpp20, 25, 29, 30, 38, hPop5 and hPop1 (Table 1), (Stolc and Altman, 1997; Pluk et al., 1999; van Eenennaam et al., 1999; van Eenennaam et al., 2001; Welting et al., 2004).
The composition and general physical characteristics of the protein and RNA subunits of the yeast and human RNase P holoenzymes are summarized and compared to the archaeal RNase P enzyme in Table 1. The proteins show large variations in size in both the yeast (15.5 kDa to 100.5 kDa) and human (14 kDa to 115 kDa) holoenzymes. The majority of the proteins are quite basic (pI >9) and both the yeast and human enzymes possess a single acidic protein, Pop8 (pI 4.6) and Rpp40 (pI 5.2) respectively. The unique acidic nature of these proteins may indicate that they serve similar functions in the holoenzyme, however current sequence comparisons do not predict that these proteins are homologues. A lack of clear sequence homology does not entirely rule out the potential for structural or functional similarity. All of the archaeal RNase P proteins are homologous to eukaryal RNase P proteins, however the unique Rpr2 protein is notably absent in RNase MRP. The recent structural characterization of the archaeal Rpr2/Rpp21 homologue has demonstrated a zinc binding domain in the structure (Kakuta et al., 2005). A similar zinc binding domain has been predicted in the unique Snm1 subunit of the yeast RNase MRP (Schmitt and Clayton, 1994), and it has been suggested that the Rpr2 and Snm1 subunits might be paralogues (Hartmann and Hartmann, 2003). The relationships between Rpp14, Rpp25 and Rpp40 with the yeast proteins are not clear and they show little or no homology to any known proteins. The yeast Pop7, the human Rpp20 and Rpp25 have predicted sequence similarity to the Alba superfamily of proteins, related to an RNA binding family of proteins (Aravind et al., 2003). There is also weak sequence homology between the human Rpp14 and hPop5, which has led to the suggestion that Rpp14 may be a paralogue of hPop5 (Koonin et al., 2001; Hartmann and Hartmann, 2003).
The Structure of the RNA Components of Nuclear RNase P and RNase MRP
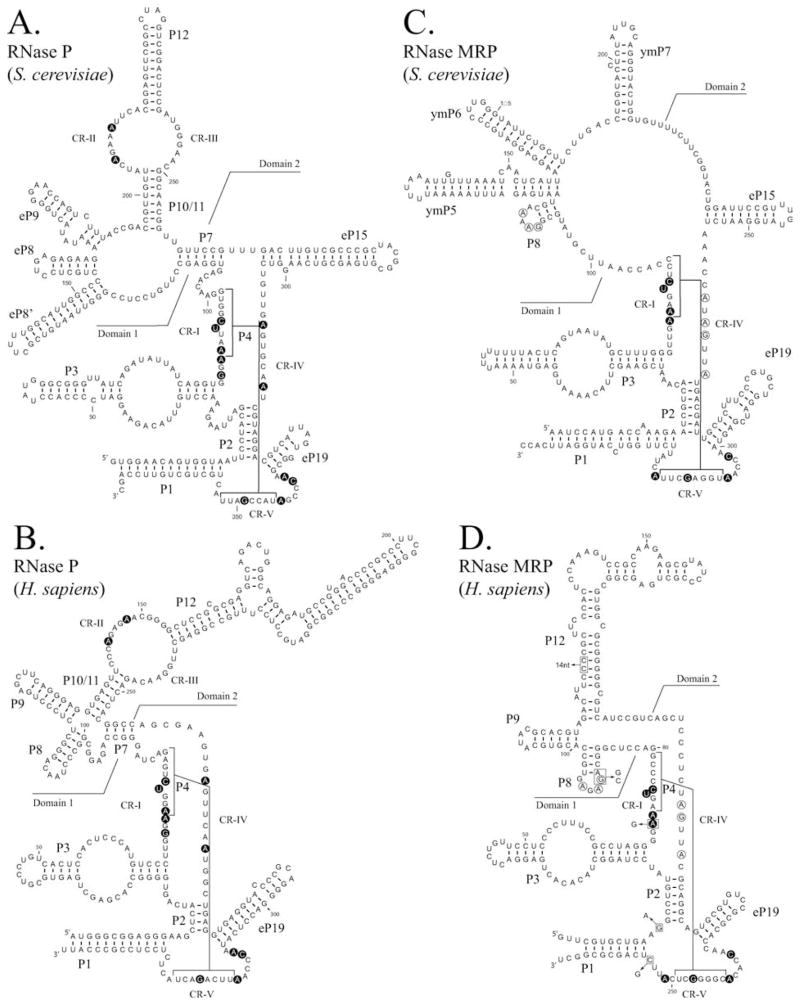
Unlike the bacterial RNase P RNAs and a few of the archaeal RNase P RNAs, the eukaryotic RNase P RNAs are not able to cleave pre-tRNAs in the absence of their proteins. The loss of ribozyme activity and the increase in protein content of the eukaryotic enzymes appears to accompany an apparent reduction in the complexity of the eukaryotic RNA structure. Examples of the yeast (S. cerevisiae) and mammalian (H. sapiens) RNase P RNAs are shown in Figure 5. Phylogentic studies of the RNase P RNA in eukaryotes has revealed a conserved core, a simplified version of the bacterial and archaeal RNAs which includes helices P1, P2, P3, P4, P7, P8, P9, P10/11 and P12 (Altman et al., 1993; Chen and Pace, 1997; Pitulle et al., 1998; Frank et al., 2000; Marquez et al., 2005; Piccinelli et al., 2005). Each of the conserved regions are present in similar positions to the bacterial and archaeal RNAs; CR-I and CR-V comprise stem P4; CR-II and CR-III are within the loop region (21–24 nt) between stems P10/11 and P12; CR-IV is found adjacent to the P2 stem. Despite the conservation of the four-way junction (P7–P10/11), the highly conserved bulged nucleotides found in this region of the bacterial consensus structure (Figure 2), are notably absent in eukaryal RNAs. These nucleotides have characterized roles in substrate binding and are also absent in type M archaeal RNAs (Figure 3).
FIGURE 5.
Comparison of eukaryotic nuclear RNase P RNAs and RNase MRP RNAs. Structures are represented in a similar manner to Figure 2. RNase P RNAs (A) and (B) and RNase MRP RNAs (C) and (D) are shown for Saccharomyces cerevisiae and Homo sapiens respectively. RNase MRP structures are based on previous structural data and are updated according to (Piccinelli et al., 2005). Nucleotides conserved in all known RNase P RNAs or RNase MRP RNAs are shown in bold background. Line boundaries do not precisely define domains, but draw attention to regions of similarity. In both RNase P and RNase MRP RNAs domain 1 shows similarity to the bacterial “catalytic domain” (C-domain). In the MRP RNAs the conserved region (CR-IV) has the general consensus (5′-ANAGNNA-3′) and the P8 loop has the general consensus (5′-GARAR-3′) [R = purine], these highly conserved nucleotides are circled. Mutations and insertions within the mature RNase MRP RNA that are known to be involved in the human diseases cartilage-hair hypoplasia (CHH) and anauxetic dysplasia (Ridanpaa et al., 2002; Thiel et al., 2005), are indicated with boxes in (D).
A subset of eukaryal structures (eP8,eP9,eP15,eP19) are found in similar positions to those in bacteria, but have been designated “eukaryal” as these may not perform directly analogous functions to their bacterial counterparts (Chen and Pace, 1997; Frank et al., 2000; Marquez et al., 2005). This would seem to be particularly true for the eP15 and eP19 stems which, unlike the eP8 and eP9 stems, are not found in all eukaryal RNAs (compare RNase P RNAs from H. sapiens and S. cerevisiae, Figure 5). When present, the eP15 stems also lack the key features of the P15 region from the bacterial/archaeal type A RNAs (CCA binding loop, ability to form a P6 structure) and are thus more similar to the P15 stem found in the archaeal type M structures (Figure 3).
Currently available sequence data, which spans many eukaryotic phyla, show that an eP19 stem is present in most eukaryotes, although not all, and that an eP15 stem appears to be present in most fungal lineages but absent from many other eukaryotes. There is one particular region where the eukaryal RNA appears to have evolved additional complexity that is not found in the bacterial RNAs. In most eukaryotes, the P3 stem has a large internal loop and there is limited sequence conservation within one of the loop strands. The eukaryotic P3 stem is required for the correct assembly and function of the holoenzyme (Lindahl et al., 2000; Ziehler et al., 2001; Li and Altman, 2002) and appears to bind the largest protein subunit Pop1 (Ziehler et al., 2001). The P3 domain in the eukaryotic RNAs is thus distinct from the bacterial P3 stem, which can be both truncated and replaced with only modest effects on ribozyme cleavage (Tanaka et al., 2004). The eP15 stem structure and also the sequence of the loop region of P3 appear to be co-evolved between examples of RNase P and RNase MRP RNAs within species.
The conservation of the bacterial catalytic core suggests that the eukaryotic RNA is directly involved in catalysis, in a similar way to the bacterial ribozyme. Comparison of the cleavage activity of the bacterial and eukaryal enzymes using pre-tRNAs with phosphothiorate substitutions at their cleavage sites, shows similar inhibitory effects upon the cleavage activity of both enzymes, suggesting a similar catalytic mechanism (Pfeiffer et al., 2000; Thomas et al., 2000). The increase in the protein composition of the eukaryal enzymes most likely serves at least one purpose, to compensate for the loss of RNA structures in the eukaryal RNA that otherwise stabilize the tertiary structure of bacterial RNase P RNAs.
RNase MRP RNA Structure
The bacterial, archaeal and eukaryal RNase P RNAs possess many common structural features, and several of these are also present in the RNase MRP RNA (Forster and Altman, 1990; Schmitt et al., 1993; Paluh and Clayton, 1995). The secondary structures of the H. sapiens MRP RNA (7-2) and the S. cerevsiae MRP RNA (NME1) are shown in Figure 5. The structural homology is found within the proposed catalytic region of these RNAs (domain 1) and includes stems P1, P2, P3, P4 and eP19 (also eP15 in some yeasts). The MRP RNAs also display similar conserved sequence elements, which have been numbered according to their equivalent regions in the RNase P RNA; CR-I, CR-V and a more weakly conserved CR-IV. Conservation of these regions, which form the catalytic core of the bacterial RNase P, in eukaryal RNase P and RNase MRP RNAs strongly suggests that the RNA remains the catalytic moiety of these enzymes. Due to the similarity of this region (domain 1) to the catalytic domain of RNase P, it has been comparatively easy to define the region in MRP RNAs. In contrast, the remaining region of the MRP RNA (domain 2) has been more difficult to define structurally, as the region appears to differ between the few MRP sequences that were originally available, mainly yeasts (Ascomycetes) and vertebrates (Schmitt et al., 1993; Li et al., 2002). Structures from both analyses have been supported by chemical and enzymatic probing of the yeast MRP RNA from S. cerevisiae (Li et al., 2002; Walker and Avis, 2004) and the vertebrate MRP RNAs from Mus musculus (Topper and Clayton, 1990), and H. sapiens (Walker and Avis, 2005).
A more extensive phylogenetic analysis has established a larger number of MRP RNA sequences from a broad range of eukaryotes. This analysis has identified a highly conserved sequence which has allowed the domain 2 structure to be more clearly defined in RNase MRP RNAs (Piccinelli et al., 2005). The sequence is conserved at the end of a stem structure (P8) in most MRP RNAs and forms a pentaloop (5′-GARAR-3′) and occasionally shows a single 3′ deletion to a tetraloop (5′-GARA-3′) (Piccinelli et al., 2005). The conserved loop is found in most MRP RNAs but has not been detected in some yeast lineages (Basidomycota) (Piccinelli et al., 2005). The stem structure had previously been detected in vertebrate MRP RNAs (Figure 5, H. sapiens MRP RNA, stem P8) (Schmitt et al., 1993), but had not been well supported in yeasts due to both a lack of covariance within the previously available sequences and the region not being well defined in structure probing experiments (Li et al., 2002; Walker and Avis, 2004). The identification of this conserved element represents the first, clearly demonstrated, MRP specific region and increases the homology between the known RNase MRP structures, redefining domain 2.
The arrangement of domain 2 does not appear to be fixed, however the most typical arrangement is the three stems; P8, P9 and P12 as shown in the H. sapiens MRP RNA (Figure 5). The P8 stem in H. sapiens has a clear equivalent in the S. cerevisiae structure although it is not clear how the other stems (ymP5, ymP6 and ymP7) relate, if at all, to the P9 and P12 stems (compare S. cerevisiae with H. sapiens, Figure 5). In addition the P12 stem is notably absent in Encephalitozoon cuniculi (Piccinelli et al., 2005). The differences in the general arrangement of domain 2 have been previously noted and may indicate an adaptation towards a wider range of substrates, perhaps with specialization within phylogenetic branches (Li et al., 2002; Walker and Avis, 2005). In this respect it is interesting to note that in many vertebrate species the P7 stem bears a K-turn (Klein et al., 2001), a structure that frequently interacts with proteins including the L7Ae protein (Rozhdestvensky et al., 2003; Hamma and Ferre-D’Amare, 2004), a predicted sequence and structural homologue of the Pop3/Rpp38 protein.
The conserved P8 stem-loop is clearly an important conserved structural feature of the MRP RNAs. The yeast P8 stem bears the pentaloop sequence (5′-GAAAA-3′) and this pentaloop sequence has been previously structurally characterized within the context of the coliphage HK022 protein-lambda-phage boxB RNA complex (Faber et al., 2001) [PDB ID. 1HJI]. The loop forms a variant of the GNRA tetraloop structure in which the fourth base is bulged out into solution and does not participate in contacts with the RNA or, in this particular complex, the protein. It is noted that despite clear phylogenetic data, the short P8 stem has not been well defined in probing studies from three MRP RNAs; two vertebrates and one yeast (Topper and Clayton, 1990; Walker and Avis, 2004; Walker and Avis, 2005). In yeasts the conserved sequence, but not the stem structure, has been previously characterized in isolated (in vitro transcribed) yeast MRP RNA and found to participate in a long range interaction (P7-like), similar to the P7 stem of the yeast RNase P RNA (Walker and Avis, 2004). Mutational analysis indicated that the interactions of the sequence were complex but did not appear to support a function for the P7-like stem in vivo, favouring the P8 stem in the context of the holoenzyme. In yeasts, the region perhaps requires the interaction with a protein in order to form a stable P8 structure. It is noted that the the potential for a P7-like structure is only found in some yeast species. Although this highly conserved sequence motif will require further study, it represents the first clearly demonstrated MRP-specific region and increases the homology between RNase MRP RNA structures.
Other regions of the RNA have been analyzed in the yeast system, including truncations of several stems (Shadel et al., 2000; Li et al., 2004). The results show that the ymP5, ymP6, ymP7 and eP19 stems can each be substantially truncated without significantly affecting the enzyme, the additional truncation of eP15 resulted in temperature sensitivity (Li et al., 2004). Truncation of the stem regions has enabled a minimal RNase MRP RNA (mini 2) to be designed in the yeast system that appears fully functional in vivo (Li et al., 2004). The P8 stem had not been identified and was not included in this study. Additionally, the ymP5 stem shows some sequence conservation in yeasts and a region of conserved bulged nucleotides is sensitive to mutation suggesting a possible functional role in the yeast RNase MRP (Li et al., 2004).
The Co-Evolution of Nuclear RNase P and RNase MRP RNAs
Several themes between the structures of the RNase P and RNase MRP RNAs are emerging, particularly that both RNAs retain the predicted catalytic domain, and regions that bind protein appear to be conserved. This is most convincingly demonstrated by the sequence of the P3a/P3b loop region of the P3 stem, which is more conserved between RNase P and MRP RNAs within species than across species (Lindahl et al., 2000; Piccinelli et al., 2005). The P3 structure of RPR1 RNA has been shown to bind the large, common protein subunit, Pop1 (Ziehler et al., 2001). It has also been demonstrated that the P3 stem can be swapped between RNase P and MRP in S. cerevisiae with no detectable effect, whilst the MRP-P3 stem from other species could not replace the S. cerevisiae MRP-P3 stem (Lindahl et al., 2000). Another structure that also appears to have co-evolved within some yeasts is the eP15 stem of RNase P. This stem appears to be present in the RNase P RNA of most fungal lineages and absent from many other eukaryotes; also mirrored by the MRP RNAs from the same species (compare yeast and human RNAs, Figure 5), (Frank et al., 2000; Piccinelli et al., 2005). Unlike the P3 stem, there is no sequence conservation within the eP15 stem and mutational analysis does not support a significant role in the function of RNase MRP (Li et al., 2004).
The Role of RNase MRP RNA in Human Disease
To date, RNase P has not been linked to any human disease, however data implicating the related RNase MRP in human disease has been established. A number of mutations within both the promoter and mature regions of the human RNase MRP RNA (RMRP) gene have been linked to the developmental disorders cartilage-hair hypoplasia (CHH) and anauxetic dysplasia (Ridanpaa et al., 2002; Thiel et al., 2005). A number of the observed mutations within the mature RNA fall within regions of the RNA that are likely of structural importance when compared to the recent crystal structures of the bacterial RNAs (Figure 5), (Kazantsev et al., 2005; Torres-Larios et al., 2005). Mutations that occur in the junction regions between P1 and P2 (+14G → A), and between P4 and P1 (+254C → G) are related to anauxetic dysplasia (Thiel et al., 2005), and are likely to affect the region surrounding the proposed catalytic core. Similarly, another mutation (+70A → G) is dominant in patients with cartilage-hair hypoplasia (CHH) and occurs at an absolutely conserved position within CR-I (Ridanpaa et al., 2001). A mutation occurring within the P8 stem (+90_91AG → GC) is particularly interesting as this falls within domain 2 and directly links this, MRP RNA specific, structure to anauxetic dysplasia (Thiel et al., 2005). A large insertion mutation, which would disrupt the formation of stem P12, is also evident in some patients with anauxetic dysplasia. Recent studies show that these mutations impare the function of RNase MRP in human cells differently, affecting both 5.8S rRNA processing and the processing of cyclin B2 mRNA to varying degrees (Thiel et al., 2005). The manesfestation of cartilage-hair hypoplasia (CHH) and anauxetic dysplasia may be linked to the observed differences in the ability of RNase MRP to function in these processes, or perhaps a previously un-characterized role for RNase MRP.
Architecture of Nuclear RNase P and RNase MRP
Detailed Analysis of the Yeast RNase P RNA In Vivo
More specific information regarding the structure and functions of eukaryal RNase P RNA have emerged from the genetic manipulation of the subunits in yeast. The secondary structure of the mature RPR1 RNA from S. cerevisiae has been experimentally verified by structure sensitive RNA footprinting (Tranguch et al., 1994). Footprinting of the yeast holoenzyme, before and after proteinase treatment, has demonstrated that the proteins provide extensive protection of the RPR1 RNA, including CR-I to CR-V, the P3 internal loop, the eP15 stem and the entire stem and loop structures of eP8 and eP9. The regions of the RNA which are inaccessible in the presence of protein may be directly involved in RNA-protein interactions and most certainly form the inner core of the enzyme structure. Other regions of the RPR1 RNA were shown to be more peripheral to the function of the enzyme; the ends of helices P3, eP8′ and P12 protrude into solution. The successful insertion of RNA affinity ligands (“aptamers”) into these regions in the RPR1 RNA is also consistent with the observation that these regions are solvent exposed in the holoenzyme (Srisawat and Engelke, 2001; Srisawat et al., 2002).
Genetic manipulation of the subunits in yeast has allowed the study of their specific roles in vivo. In particular, the RNA subunit (RPR1) has been investigated in some detail, including the randomization of conserved regions (CR-I, CR-II, CR-IV and CR-V), (Pagan-Ramos et al., 1996; Pagan-Ramos et al., 1996). A number of mutants in these regions affect cell growth and RPR1 maturation and many isolated mutants show reductions (up to 10-fold) in the catalytic efficiency (kcat/Km) of the holoenzyme, in a pre-tRNA cleavage assay in vitro. In the majority of cases, mutations within the CR-I, CR-IV and CR-V regions of the S. cerevisiae RNase P were shown to have a greater influence on catalysis (kcat) than substrate recognition (Km).
In yeast RNase P RNAs there is another conserved region directly adjacent to the P4 stem that is not found in bacterial RNAs; the sequence (5′-GAAC-3′) is found between P4 and P7 (jP4/7). The mutation of these nucleotides affects growth, holoenzyme assembly and localization with severe defects in pre-tRNA processing in vivo and a ten fold reduction in the catalytic efficiency (kcat/Km) in vitro (Xiao et al., 2005). The conserved eukaryal (jP4/7) nucleotides are in close proximity to the P4 stem and may serve a function that is not seen in the bacterial RNAs participating in RNA structure, binding to one of the numerous proteins found in the eukaryal system or perhaps the substrate.
The CR-II and CR-III sequences form part of the J11/12 - J12/11 module, flanked by the P10/11 and P12 stems. Footprinting of this region of the RPR1 RNA has shown that its accessibility to chemical and nuclease probes appears to be dependent upon magnesium (Ziehler et al., 1998). The conserved nucleotides within the CR-II region have been randomized and screened in vivo. Many of the isolated mutants also showed a dependence on magnesium for growth and an increased catalytic activity of the isolated mutant enzymes (Pagan-Ramos et al., 1996). In solution, the bacterial (M1) RNA undergoes lead-ion hydrolysis and Mg2+ cleavage at high pH, at CR-II and CR-III positions, indicative of a metal binding site (Kazakov and Altman, 1991). Metal ions are seen to play clear roles in stabilizing the tertiary interactions of the bacterial S-domain, however these tertiary interactions would not be available in the isolated eukaryal P10/11–P12 region of the S. cerevisiae RNA. It is not clear whether the apparent dependence upon magnesium for the folding of the P10/11 – P12 region in the S. cerevisiae RNA is due to a specific metal ion binding site or due to structural sensitivity of the isolated RNA domain and a general stabilization by divalent cations.
Other studies upon the RNA have targeted areas whose sequence conservation appears to be specific amongst some eukaryotes, specifically the internal loop of stem P3 and the loops of stems eP8 and eP9 (Ziehler et al., 2001; Xiao et al., 2005). In the yeast RNase P there is a high degree of sequence conservation in the loop regions of the eP8 and eP9 structures; the eP8 loop contains an NUGA sequence element and the eP9 loop contains a GNAA tetraloop sequence. Mutation of the conserved nucleotides in yeast affects growth, pre-tRNA processing (in vivo) and holoenzyme assembly; the eP9 mutations are more severe and also affect enzyme localization. Kinetic studies of the purified mutant enzymes show a four fold reduction in the catalytic efficiency (kcat/Km) of the eP9 mutant and an unexpected three fold increase in catalytic efficiency of the eP8 mutant. The increase in catalytic efficiency of the eP8 mutant appears to be due to a loss of specific substrate selectivity. The eP8 mutant enzyme is more efficiently inhibited by total yeast RNA that the wild type enzyme, this also explains the observed defects in vivo. The results indicate the conserved nucleotides, within loops eP8 and eP9, play key roles in the maturation and function of the holoenzyme. Sequence conservation suggests that the loops may be involved in RNA-RNA or RNA-protein interactions and it is important to note that the entire eP8 and eP9 stem and loop structures are protected from nuclease attack in the S. cerevisiae holoenzyme. The relationship between these structures and the conserved P8 stem from RNase MRP is not currently clear.
RNA-Protein Interactions
Randomization of key nucleotides within the P3a/P3b loop of the RPR1 RNA has resulted in the isolation of several temperature sensitive mutants that accumulate pre-RPR1 at the non-permissive temperature; indicative of an assembly defect (Ziehler et al., 2001). Further characterization in yeast has shown that both the full length RPR1 RNA and the isolated P3 domain will specifically bind the Pop1 protein in a yeast three-hybrid assay. The previously identified mutations within the P3 region eliminated the interaction with the P3 RNA domain (Ziehler et al., 2001). The yeast Pop1 protein binds to the P3 stem through conserved nucleotides within the P3 helix-bulge-helix domain. The binding of the Pop4 protein to the RPR1 RNA has also been demonstrated in the yeast three-hybrid analysis, although the site of binding was not investigated (Houser-Scott et al., 2002). The demonstrated functional equivalence of the P3 stem between RNase P and RNase MRP within S. cerevisiae (Lindahl et al., 2000), also indicates that Pop1 will most likely bind the NME1 RNA at the P3 stem.
Studies of the RNA-protein interactions within the human enzyme have also been carried out. Yeast three-hybrid and UV crosslinking analysis has demonstrated the binding of Rpp21, Rpp29, Rpp30 and Rpp38 to the H1 RNA (Jiang et al., 2001). Rpp21 was also shown to bind the isolated P3 domain by UV crosslinking (Jiang et al., 2001). The deletion of the P3 region from the human RNase P RNA has been shown to result in the mislocalization of the RNA when micro-injected into human cells (Jacobson et al., 1997). In addition, the purification of a human RNase P with a deleted P3 stem does not result in the recovery of an active enzyme (Li and Altman, 2002). These results are consistent with the P3 domain also being required for correct assembly of the yeast holoenzyme.
RNA-protein interactions in the human RNase MRP system have also been investigated by UV crosslinking and affinity pull downs in vitro, showing interactions between the 7-2 RNA and hPop1, Rpp20, Rpp25, Rpp29, Rpp38 (Pluk et al., 1999; Welting et al., 2004). The Rpp38 protein appears to bind the 7-2 RNA within the P12 region of the RNA, which bears a K-turn motif. The result is consistent with the prediction that Rpp38 is a homologue of the archaeal L7Ae protein which is known to bind to the K-turn motif of archaeal snoRNAs (Rozhdestvensky et al., 2003; Hamma and Ferre-D’Amare, 2004).
The human Rpp29 and yeast Pop4 proteins have each been implicated as key core proteins in the yeast and human holoenzymes and are shown to bind their respective RNase P RNAs, the human Rpp29 also shown to bind the RNase MRP RNA (Jiang et al., 2001; Houser-Scott et al., 2002; Welting et al., 2004).
Protein-Protein Interactions
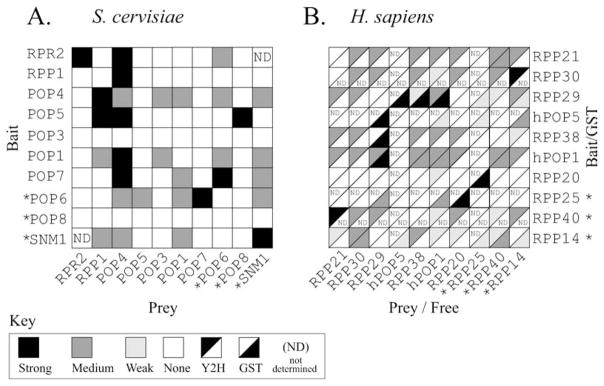
In addition to RNA-protein interactions, protein-protein interactions have also been investigated in both the yeast and human systems. The binary interactions between the protein subunits have been investigated using the Y2H system, (Jiang and Altman, 2001; Houser-Scott et al., 2002) the human system has also been further probed using a glutathione S-transferase (GST) based in vitro affinity pull down assay (Welting et al., 2004). In an effort to clarify consistent interactions and aid in comparison, the data from these studies have been transferred into two tables representing the S. cerevisiae and H. sapiens systems (Figure 6). In the human system the GST pull down study did not include GST-Rpp30 and GST-Rpp40, whereas the Y2H study lacked the hPop5 and Rpp25 subunits. The GST pull down data for the human system are largely supported by the Y2H data, and few of the observed interactions are not also seen in the Y2H data (Welting et al., 2004). However, there are numerous weak interactions observed in the Y2H study that were not detected in the GST pull down experiments and these could potentially be artifactual signals (Jiang and Altman, 2001).
FIGURE 6.
Summary of protein-protein interactions detected in eukaryotic systems. Data from previously published yeast two-hybrid (Y2H) and glutathione-S-transferase (GST) pull down studies are summarized. Tables are drawn such that homologous proteins are represented in similar positions in both tables, starting from the top left corner, and are arranged to allow a direct comparison with Figure 4. Proteins that have no identified homologue between yeast and humans are indicated with an asterisk. (A). Interactions demonstrated in the yeast system (Saccharomyces cerevisiae). Y2H interactions are shown as described in the original text (Houser-Scott et al., 2002). (B). Interactions demonstrated in the vertebrate system (Homo sapiens) interactions are shown as described in the original text (Jiang and Altman, 2001; Welting et al., 2004).
There are seven homologous proteins between the yeast and human systems and four of these are also present in the archaeal systems. Comparison of the seven eukaryal proteins across the yeast and human systems shows that there are a large number of observed interactions however only a fraction of these are consistent between the two studies (Figure 6). The lack of an overall consistency could be due to a number of reasons, one of which is that the data may reflect real differences between these systems. However, given the high degree of non-specific RNA binding observed in the isolated protein subunits (F. Houser-Scott, C.E. Millikin & D.R. Engelke, unpublished observations), and the possibility of misfolded fusion proteins, there is a strong potential for false positives and false negatives in both the GST and Y2H methods. Thus, the interactions observed in multiple systems and verified by multiple experimental methods should be given the most weight in considering the protein-protein interactions within the eukaryal enzymes.
Assuming that the four archaeal proteins have conserved functions and will form the core of the eukaryal enzyme, it is possible to draw themes from the available data. Comparison of the eukaryotic data with that from the four archaeal core proteins shows that the Pop4/Rpp29 interaction with Rpr2/Rpp30 is the only interaction detected in every system tested. In addition, the yeast system shows Pop5 (hPop5) interacting with Rpp1 (Rpp30). Although this interaction is not detected in the human system, the hPop5 and Rpp30 proteins are not well represented in the human GST and Y2H studies, due to technical difficulties in preparing the fusion proteins. Furthermore, the interaction between the archaeal Pop5/hPop5 and Rpp1/Rpp30 homologues has now been well established via crystallography and NMR (S. Karwano et al., 2006; Wilson, et al. 2006). Both eukaryal studies provide convincing evidence for an interaction between Pop4/Rpp29 and Rpp1/Rpp30. Although this is interaction is not supported by the archaeal data, the N-terminal sequence of eukaryal Pop4/Rpp29 is longer than the corresponding region in arcaheal homologues which might account for this additional interaction. The large, eukaryote specific, subunit Pop1/hPop1 is observed to form a strong and reciprocal interaction with the Pop4/Rpp29 subunit, an interaction observed in both the yeast and human systems. Given that Pop1/hPop1 and Pop4/Rpp29 are clearly demonstrated to interact with each other and that both have been shown to interact with the RPR1 RNA, the largest eukaryal protein Pop1/hPop1 would appear to be a core eukaryal protein in both the yeast and human systems.
Previously, working models of the protein-protein interactions and protein-RNA interactions have been drawn from the individual data (Jiang and Altman, 2001; Houser-Scott et al., 2002; Welting et al., 2004). However, inconsistencies between the data, the ability of many protein subunits to self associate and the sheer complexity of these ribonucleoprotein systems mean that a comprehensive knowledge of the architecture of these holoenzymes will require further studies. It is also noted that only binary interactions have been tested, clearly these systems are highly complex and combinations of subunits may be intimately linked with the specific recognition of RNA or protein. A preliminary stoichiometric analysis of RNase MRP from S. cerevisiae suggests that the subunit composition of the eukaryotic enzymes might be quite complex (Salinas et al., 2005). Furthermore, the demonstrated potential for the archaeal Pop5/hPop5 to exist as a dimer also suggests complexity in the stoichiometry of the archaeal and eukaryal holoenzymes. As our knowledge of the archaeal system progresses it is likely that more sophisticated models regarding the architecture of the eukaryal enzymes will be required. The available Y2H and GST data from the archaeal and eukaryal systems will provide a solid framework upon which to base future studies.
The Reconstitution of Activity from Human RNase P Components
Although the protein content of the eukaryal holoenzyme is considerably more complex than that of the bacterial and archaeal enzymes, it does not appear that all of this is required to achieve pre-tRNA cleavage. Trace activity of the human RNase P has been successfully reconstituted in vitro from the H1 RNA and two of the ten tightly associated proteins identified in the human holoenzyme (Mann et al., 2003). These proteins, Rpp29 (Pop4) and Rpp21 (Rpr2), are homologues of conserved archaeal proteins, indicating conserved functions for these subunits. Interestingly, a third protein is required to reconstitute trace activity in the archaeal P. horikoshii enzyme; a homologue of Rpp30 (Rpp1) (Kouzuma et al., 2003). The reconstituted human RNase P (mini-RNase P) is able to accurately process trace amounts of pre-tRNA substrates in vitro, and is dependent upon magnesium for activity. In the human mini-RNase P, the Rpp21 and Rpp29 proteins are probably involved in further stabilizing the RNA into a catalytically proficient conformation. They may also participate in substrate binding. Whether the binding of the proteins to the RNA is serving as a chaperone-like facilitation of RNA folding, or contributing directly to substrate acquisition and cleavage is not clear at this time.
Nuclear RNase P–Assembly of the Holoenzyme In Vivo
The maturation and assembly of nuclear RNase P has been best characterized in the yeast system, and provides insights into the assembly of a minimal active holoenzyme. It should be noted, however, that although both the yeast and vertebrate RNase P RNAs are transcribed by RNA polymerase III, the precursor RNA found in S. cerevisiae has not been detected in the H. sapiens system and the described maturation pathway may be specific to Saccharomycetes.
In S. cerevisiae, the RPR1 RNA is transcribed by RNA polymerase III as a 486-nt primary transcript (pre-RPR1 RNA), and undergoes a single 5′ cleavage event and multiple 3′ cleavages to yield the mature 369-nt RPR1 RNA (Lee et al., 1991; Lee et al., 1991). Both the precursor and mature RPR1 RNAs are detectible under normal growth conditions, with the mature species being roughly tenfold more abundant. Prior to processing, the precursor RNA is assembled into a ribonucleoprotein complex. The pre-RNase P complex has been specifically purified through the insertion of an RNA affinity tag into the 5′ leader sequence of the pre-RPR1 RNA (Srisawat et al., 2002). The composition of the affinity purified pre-RNase P complex was found to be largely similar to the mature holoenzyme, lacking only the Pop3 and Rpr2 subunits. The purified precursor complex was shown to be functional in the processing of pre-tRNAs in vitro.
The absence of the Rpr2 subunit in the pre-RNase P is intriguing, as its homologues are required for the reconstitution of the archaeal and human enzymatic activities. The Rpr2 subunit is also unique to RNase P and not present in RNase MRP. The different substrate selectivity of RNase P and RNase MRP must be conferred by differences in the protein composition, RNA structure or a combination of these. The absence of Pop3 and Rpr2 in the pre-RNase P holoenzyme suggests a role that is late in holoenzyme assembly. This would be consistent with the observation that Pop3 depletion does not affect the ratio of precursor to mature RPR1 RNA (Dichtl and Tollervey, 1997). Significant changes in the ratio of pre-RPR1 to RPR1 are usually attributed to incomplete or defective holoenzyme assembly, resulting in the failure of the pre-RPR1 RNA to be cleaved or a loss of RNA stability. Notably, depletion of any protein subunit associated with the pre-RPR1 complex has this phenotype. Although little is known about the mechanism of RPR1 maturation the pre-RPR1 RNA (but not the mature RPR1 RNA) seems to be associated with a complex of Sm-like proteins (Lsm 2–8), probably due to interaction with the U-rich 3′ tail and similar to the case for U6 snRNA (Salgado-Garrido et al., 1999; Pannone et al., 2001; Kufel et al., 2002). Archaeal homologues of the Sm proteins have also been shown to interact with their corresponding RNase P RNA (Toro et al., 2001). Comparatively little is known about the assembly of the human RNase P, although both Rpp29 and Rpp38 have been shown to possess nuclear localization signals (Jarrous et al., 1999) and the Rpp14 and hPop5 subunits appear to enter the nucleus via a piggyback mechanism (Jarrous et al., 1999; van Eenennaam et al., 2001).
Potential Implications of a Distinct RNase MRP Activity
Eukaryotes are defined by their nuclear structures within which a dense subnuclear region, the nucleolus, is the main site of ribosome biogenesis. In yeast, specific cleavage within the internal transcribed spacer (ITS) of the 35S pre-rRNA transcript by RNase MRP occurs in the nucleolus and is reminiscent of a similar activity performed by the bacterial and archaeal RNase P enzymes. Specifically, in bacteria RNase P processes tRNAs which exist within the ITS regions of bacterial pre-rRNA transcripts (Morrissey and Tollervey, 1995). Thus, in bacteria the processing of some pre-tRNAs is co-ordinated with pre-rRNA processing. In eukaryotes, tRNAs are not encoded within the pre-rRNA transcript. The eukaryotic specific RNase MRP may have evolved as a form of the bacterial activity that has become highly specialized in the nuclear environment to cope with a different substrate (5.8S rRNA) within the ITS region of the eukaryotic pre-rRNA. In this respect it is intriguing that the yeast RNase P is also found to be predominantly nucleolar, and that pre-tRNA processing appears to occur at this location (Bertrand et al., 1998). It has also been shown that the tRNA genes themselves have a nucleolar arrangement, suggesting that the spatial ordering of the pre-tRNA processing pathway begins with transcription (Thompson et al., 2003). It should be stressed that smaller quantities of RNase P or RNase MRP might exist elsewhere in the cell. The S. cerevisiae RPR1 RNA has been shown to be predominantly nucleolar but with some nucleoplasmic foci (Bertrand et al., 1998; Kendall et al., 2000; Lewis and Tollervey, 2000). The reported processing of mitochondrial RNAs and the recently demonstrated processing of the CLB2 mRNA by yeast RNase MRP are also interesting (Gill et al., 2004). It is not entirely clear at this time whether the enzyme associated with these activities has the same subunit composition as the nucleolar enzyme used for pre-rRNA processing, or an overlapping subset including the RNA subunit.
In mammalian cells RNase MRP is nucleolar (Reimer et al., 1988), but the localization of RNase P is less clear. Since it is not known whether the early tRNA processing pathway in vertebrates is associated with nucleoli, as it is in yeast, it would not be surprising if RNase P localization was different from the yeast case. The various RNA and protein components of the human holoenzyme have been found in distinct locations (Jarrous, 2002). The H1 RNase P RNA subunit appears to transiently enter the nucleolus before diffusing into the nucleoplasm, and has also been detected in the cytoplasm, nucleoli, and the perinuclear compartment (Jacobson et al., 1994; Wolin and Matera, 1999). In addition to the movement of the H1 RNA, the individual protein components have also been found in various compartments. Although the existence of the protein subunits in both RNase P and RNase MRP makes such localization data difficult to interpret in most cases. Rpp14, Rpp29, Rpp38, hPop1, and hPop5, have all been shown to be predominantly nucleolar, with Rpp29 and Rpp38 also detected in Cajal bodies (Lygerou et al., 1996; Jarrous et al., 1999; Savino et al., 1999; van Eenennaam et al., 2001). The Rpp21 protein exists in two forms with a distinct variant, Rpp21i, arising from alternative splicing of the encoding mRNA. The variant Rpp21i is not associated with the human RNase P and is mainly localized in the nucleolus; the protein may somehow function in the regulation of RNase P activity (Jarrous et al., 2001). Other mechanisms for regulating the activity of the human RNase P have also been suggested. The La protein binds to pre-tRNAs and blocks RNase P function (Fan et al., 1998), however the phosphorylation of La has been shown to relieve the blocking of pre-tRNA processing and also serves to inhibit RNA polymerase III (Maraia and Intine, 2002).
Individual Properties of the Eukaryal Protein Components
A number of the yeast and human proteins have been shown to have their own distinct activities that may, or may not, be directly linked to the functions of RNase P. The human Rpp21, Rpp29 and Rpp14 protein subunits have each been observed to bind pre-tRNAs in gel shift assays (Jarrous et al., 2001; Sharin et al., 2005). The binding of the yeast Pop3 protein to pre-tRNAs and also to single stranded RNAs has also been reported (Brusca et al., 2001). Since the subunit has been shown to be absent in the pre-RNase P this suggests that the observed binding of pre-tRNAs to the Pop3 subunit is not essential for the cleavage of pre-tRNAs in vitro (Srisawat et al., 2002), and that this might represent a non-specific RNA binding activity.
The Pop4 (Rpp29) protein is highly conserved in archaea and eukaryotes and appears to be one of the most ancient archaeal/eukaryal proteins (Hartmann and Hartmann, 2003). In this respect, it is particularly interesting that the human Rpp29 protein has been shown to act as a cofactor for the bacterial M1 RNA and is able to partially replace the bacterial C5 protein and form a stable active complex; despite having no structural homology to the bacteria protein (Mann et al., 2003; Sharin et al., 2005).
The recombinant human Rpp20 (Pop7) subunit has been shown to possess an ATPase activity that is also found in purified human RNase P, presumably due to the presence of the Rpp20 subunit (Li and Altman, 2001). The function for this ATPase activity is not clear, since known RNase P substrate cleavage activities do not require ATP hydrolysis the activity could play a role in holoenzyme function or assembly. Another human protein subunit, Rpp14, has been shown to have 3′ to 5′ exoribonuclease activity in combination with its interacting protein OIP2. The activity has been shown in complexes of recombinant Rpp14 with OIP2 and also in immunoprecipitated human RNase P but is absent in holoenzyme fractions which have been further purified; indicating that OIP2 is not tightly associated with the human RNase P holoenzyme. The heat shock protein Hsp29 has been found to specifically associate with the Rpp20 subunit of the human holoenzyme, and the protein is also seen to stimulate the activity of highly purified RNase P. The specific roles for these interactions are not entirely clear.
6. ORGANELLE RNase P
It is likely that the organelles found in eukaryotic cells, mitochondria and chloroplasts, began as bacterial or cyanobacterial endosymbionts each with their own genome (Gray, 1993). The genomes of these organelles have been greatly reduced throughout evolution through the loss of non-essential genes and the transfer of genetic information to the eukaryotic nucleus; requiring these gene products to be produced and imported into the organelle. A number of distinct, organelle-specific RNase P activities have been detected in both mitochondria and chloroplasts. The extent to which these RNase P activities can be related to those of the cellular enzymes is varied.
The Mitochondrial RNase P
A mitochondrial RNase P activity has been isolated biochemically from vertebrates, fungi, plants and parasitic protozoa (Marchfelder, 1995; Rossmanith et al., 1995; Lee et al., 1996; Salavati et al., 2001). In most cases the activities have only been partially characterized. The most detailed information on the biochemical and genetic properties of a mitochondrial RNase P has been derived from the S. cerevisiae enzyme, which is composed of a mitochondrially encoded essential RNA subunit, Rpm1 (423 nucleotides) and a single nuclear encoded protein subunit, Rpm2 (105 kDa) (Miller and Martin, 1983; Underbrink-Lyon et al., 1983; Morales et al., 1992; Dang and Martin, 1993). Both the protein and RNA subunits have been shown to be necessary for the cleavage of pre-tRNAs, by genetic analysis and ribonuclease sensitivity (Hollingsworth and Martin, 1986; Morales et al., 1989; Morales et al., 1992). In S. cerevisiae the Rpm2 protein has two domains, one required for pre-tRNA cleavage, and another for the maturation of the mitochondrial RNase P RNA subunit (mtP-RNA) (Stribinskis et al., 1996; Stribinskis et al., 2001). In addition to dual roles in RNase P function and maturation, the protein subunit has been shown to have other roles essential for mitochondrial function (Kassenbrock et al., 1995; Stribinskis et al., 2001). Recently, Rpm2 was shown to act in the nucleus as a transcriptional regulator, affecting the steady-state levels of the mRNAs for a number of nucleus-encoded mitochondrial components (Stribinskis et al., 2005). The Rpm2 protein does not show any significant sequence similarity to any of the known RNase P proteins from bacteria, archaea and eukaryotes. Thus, the activity appears to be significantly diverged from that of bacteria, this is also reflected in the RNA component of this Ascomycete. A number of mtP-RNAs have been predicted from the Ascomycete fungal lineage and these have demonstrated large variations in mtP-RNA size and secondary structure (Wise and Martin, 1991), when compared to the bacterial RNAs (Seif et al., 2003). The smallest mtP-RNA identified in this lineage is that of the yeast Saccharomycopsis fibuligera, (140nt), which displays just two of the five bacterial critical regions, CR-I and CR-V. Despite the availability of both nuclear and mitochondrial genomes for many yeast species, the identification of mtP-RNAs has only been achieved in the Zygomycota and Ascomycota, and mtP-RNAs are conspicuously absent from the Basidomycota and Chytridomycota. Although this may, in part, be due to the inability to detect these species by sequence comparative methods, it also raises the possibility of the import of nuclear enzymes into the mitochondria or the existence of protein only RNase P activities.
There have been conflicting reports regarding the nature of mtRNase P in H. sapiens (HeLa cells). An RNase P activity that appears to be composed entirely of protein has been isolated from a HeLa cell mitochondrial fraction and shown to have specificity for mitochondrial tRNA substrates (Rossmanith et al., 1995; Rossmanith and Karwan, 1998; Rossmanith and Karwan, 1998). It is interesting that the report of an RNA-free RNase P was greeted with considerable skepticism, as was a similar earlier report of an RNA-free chloroplast enzyme (see below). Twenty-five years ago the thought of an enzyme composed entirely of RNA would have been greeted with even greater skepticism. Adding to the skepticism concerning the protein-only enzyme is another report suggesting that an RNase P activity that was partially purified from an enriched, human cell mitochondrial fractionation utilizes the same RNA as the H. sapiens nuclear RNase P (H1 RNA) (Puranam and Attardi, 2001). This RNA-containing enzyme was not shown to be specific for mitochondrial pre-tRNAs, and the protein content of this ribonucleoprotein enzyme was not determined. It is not clear at this time whether it is the same as the nuclear enzyme (possibly a contaminant from the much larger nuclear pool of enzyme in the cell lysates) or a variation that uses the nuclear RNA, but a different protein complement. It seems unlikely, though not impossible, that the full 11 subunit holoenzyme would be imported into the mitochondrion.
Preliminary characterization of the mitochondrial RNase P from Trypanosoma brucei suggests that a relatively small species (~70 kDa) is responsible for activity (Salavati et al., 2001). The activity was shown to be insensitive to micrococcal nuclease (MN) however it was not been possible to rule out the presence of an RNA component associated with this activity.
The Chloroplast RNase P
Biochemical characterization of RNase P from the photosynthetic organelle (cyanelle) of Cyanophora paradoxa shows that the enzyme is a ribonucleoprotein with a large protein content (~80%), similar to that of the archaeal/eukaryotic nuclear RNase P (Baum et al., 1996; Cordier and Schon, 1999). The enzyme contains an essential RNA and footprinting analysis reveals similar protection of the RNA to that of the yeast nuclear RNase P. The RNA component is not independently catalytic, however it conforms to the bacterial consensus structure similarly to the putative chloroplast RNase P RNAs identified in the genomes of red algae (Porhyra purpurea) and green plants (Nephroselmis olivacea and Zea mays) (Reith and Munholland, 1995; Turmel et al., 1999; Collins et al., 2000).
The possibility of a protein-only RNase P in some chloroplasts is also controversial. To date, the strongest evidence for the existence of a protein only RNase P activity in any organelle comes from the biochemical characterization of the chloroplast RNase P from Spinacia oleracea (Gegenheimer, 1996). The purified enzyme is estimated at only 70 kDa and is insensitive to inactivation using MN. Resistance to MN has been shown in other RNA containing RNase P holoenzymes due to protection of the RNA by the proteins, however in these cases an RNA component was detected. Attempts to identify RNA in highly purified spinach RNase P enzyme fractions, using highly sensitive assays, have not been successful (Thomas et al., 1995). The apparent lack of an RNA component is further supported by the observation that spinach choloroplast RNase P has a protein like density in CsCl (1.28 g/ml). Investigation of its cleavage mechanism utilizing a pre-tRNA substrate containing an Rp-thio-substitution at the scissile bond suggests that spinach RNase P has a different catalytic mechanism to that of the bacterial RNase P. The modified substrate results in a dramatic decrease in the catalytic activity of the bacterial ribozyme, however only a modest decrease in activity was observed in the spinach RNase P (Chen et al., 1997; Thomas et al., 2000). These combined observations are consistent with a protein only RNase P activity in the spinach chloroplast.
7. CONCLUDING REMARKS
Ribonuclease P is clearly one of the most ancient surviving ribozymes from the hypothetical, pre-biotic “RNA World”. In every kingdom of life, a form of RNase P has survived that has an essential, catalytic RNA subunit. Two questions arise from this remarkable retention of RNA-based catalysis: [1] why has the function been retained as an RNA catalyst, rather than being taken over by protein as with other enzyme functions, and [2] why have the archaeal and eukaryotic nuclear enzymes become such complex ribonucleoproteins?
The answer to the first question might be fundamentally similar to why the other ancient ribozyme, the ribosome, has been conserved. Both catalytic RNAs were developed to recognize a large number of tRNAs that are distinct, in that they must bring different amino acids, yet have key conserved features that allow them to be recognized as “tRNA.” The need to specifically recognize all “tRNA” structures and not other RNA structures, leaves very little tolerance. Once a ribozyme existed that was able to do this, the selection pressure for keeping the function within the ribozyme, rather than developing a new protein-based enzyme, would be very strong. Although RNase P could develop limited tolerance for additional substrates, or even develop a related enzyme to deal with additional substrates (e.g. RNase MRP), the core substrate recognition could not stray substantially.
The cleavage of pre-tRNAs by the eukaryotic RNase P appears to have slightly different determinants from that of the bacterial enzymes. The human enzyme does not cleave the same minimal substrates as the bacterial ribozyme, requiring an additional loop between the acceptor stem and T stem-loop domains (Yuan and Altman, 1995). The yeast enzyme has been shown to interact with pre-tRNAs within the 3′ trailer regions as well as the tRNA domain and the enzyme binds to, and is strongly inhibited by, single-stranded homoribopolymers [poly(G) and poly(U) > poly(A) ≫ poly(C)] (Ziehler et al., 2000).
The large increase in the number and size of the protein subunits could have multiple causes. First, the proteins might provide additional opportunities for recognition of RNA or ribonucleoprotein substrates. As noted above, pre-tRNA cleavage by nuclear RNase P can be strongly inhibited by low concentrations of single-stranded RNAs and the yeast enzyme is relatively promiscuous in cleaving non-physiological RNA substrates (Chamberlain et al., 1996), unlike the bacterial holoenzyme. This suggests that the nuclear holoenzyme has additional binding site(s) that might be used to position non-tRNA substrates for cleavage, although the in vitro cleavage reactions do not faithfully reproduce specific recognition with purified enzyme and naked RNA. In turn, this suggests that nuclear RNase P must be prevented from freely interacting with un-complexed, single-stranded RNA in the cell. Such a function for the protein complement would be compatible with the observations that RNases P and MRP are found in particular subcellular and subnuclear compartments—an arrangement that could serve the dual purpose of providing timely access to appropriate substrates and preventing access to inappropriate ones.
The RNase P ribozyme performs a relatively simple task, the hydrolysis of a phosphodiester bond, but the enzyme has evolved ever increasing ribonucleoprotein complexity to adapt to new cellular environments and tasks. Although we are beginning to understand the nature of the RNA in structure and catalysis, comprehension of these biological adaptations of the holoenzymes is at a very early stage.
Acknowledgments
We are grateful to M. Kimura & S. Kawano (Kyushu University) for communicating results prior to publication. We are grateful to J. M. Avis, S. Xiao and J. Hsieh for their critical reading of the manuscript. We also thank N. R. Pace, S. Marquez and J. W. Brown for providing valuable assistance with figures. This work is supported by the National Institutes of Health grant GM 34869 (to DRE).
Footnotes
Estimation of mass assumes a single RNA subunit and that each protein is present only once in the holoenzyme.
Much of the sequence data for RNase P is held and maintained by J. W. Brown at North Carolina State University. The RNase P Database. http://jwbrown.mbio.ncsu.edu/RNaseP/home.html.
References
- Alifano P, Rivellini F, Piscitelli C, Arraiano CM, Bruni CB, Carlomagno MS. Ribonuclease E provides substrates for ribonuclease P-dependent processing of a polycistronic mRNA. Genes Dev. 1994;8(24):3021. doi: 10.1101/gad.8.24.3021. [DOI] [PubMed] [Google Scholar]
- Altman S, Wesolowski D, Guerrier-Takada C, Li Y. RNase P cleaves transient structures in some riboswitches. Proc Natl Acad Sci USA. 2005;102(32):11284. doi: 10.1073/pnas.0505271102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Altman S, Wesolowski D, Puranam RS. Nucleotide sequences of the RNA subunit of RNase P from several mammals. Genomics. 1993;18(2):418. doi: 10.1006/geno.1993.1488. [DOI] [PubMed] [Google Scholar]
- Andrews AJ, Hall TA, Brown JW. Characterization of RNase P holoenzymes from Methanococcus jannaschii and Methanothermobacter thermoautotrophicus. Biol Chem. 2001;382(8):1171. doi: 10.1515/BC.2001.147. [DOI] [PubMed] [Google Scholar]
- Aravind L, Iyer LM, Anantharaman V. The two faces of Alba: the evolutionary connection between proteins participating in chromatin structure and RNA metabolism. Genome Biol. 2003;4(10):R64. doi: 10.1186/gb-2003-4-10-r64. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Avis JM, Allain FH, Howe PW, Varani G, Nagai K, Neuhaus D. Solution structure of the N-terminal RNP domain of U1A protein: the role of C-terminal residues in structure stability and RNA binding. J Mol Biol. 1996;257(2):398. doi: 10.1006/jmbi.1996.0171. [DOI] [PubMed] [Google Scholar]
- Bartkiewicz M, Gold H, Altman S. Identification and characterization of an RNA molecule that copurifies with RNase P activity from HeLa cells. Genes Dev. 1989;3(4):488. doi: 10.1101/gad.3.4.488. [DOI] [PubMed] [Google Scholar]
- Baum M, Cordier A, Schon A. RNase P from a photosynthetic organelle contains an RNA homologous to the cyanobacterial counterpart. J Mol Biol. 1996;257(1):43. doi: 10.1006/jmbi.1996.0145. [DOI] [PubMed] [Google Scholar]
- Bertrand E, Houser-Scott F, Kendall A, Singer RH, Engelke DR. Nucleolar localization of early tRNA processing. Genes Dev. 1998;12(16):2463. doi: 10.1101/gad.12.16.2463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boomershine WP, McElroy CA, Tsai HY, Wilson RC, Gopalan V, Foster MP. Structure of Mth11/Mth Rpp29, an essential protein subunit of archaeal and eukaryotic RNase P. Proc Natl Acad Sci USA. 2003;100(26):15398. doi: 10.1073/pnas.2535887100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brown JW, Haas ES, Pace NR. Characterization of ribonuclease P RNAs from thermophilic bacteria. Nucleic Acids Res. 1993;21(3):671. doi: 10.1093/nar/21.3.671. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brusca EM, True HL, Celander DW. Novel RNA-binding properties of Pop3p support a role for eukaryotic RNase P protein subunits in substrate recognition. J Biol Chem. 2001;276(45):42543. doi: 10.1074/jbc.M107293200. [DOI] [PubMed] [Google Scholar]
- Buck AH, Dalby AB, Poole AW, Kazantsev AV, Pace NR. Protein activation of a ribozyme: the role of bacterial RNase P protein. Embo J. 2005;24(19):3360. doi: 10.1038/sj.emboj.7600805. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Buck AH, Kazantsev AV, Dalby AB, Pace NR. Structural perspective on the activation of RNase P RNA by protein. Nat Struct Mol Biol. 2005 doi: 10.1038/nsmb1004. [DOI] [PubMed] [Google Scholar]
- Cai T, Aulds J, Gill T, Cerio M, Schmitt ME. The Saccharomyces cerevisiae RNase mitochondrial RNA processing is critical for cell cycle progression at the end of mitosis. Genetics. 2002;161(3):1029. doi: 10.1093/genetics/161.3.1029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chamberlain JR, Lee Y, Lane WS, Engelke DR. Purification and characterization of the nuclear RNase P holoenzyme complex reveals extensive subunit overlap with RNase MRP. Genes Dev. 1998;12(11):1678. doi: 10.1101/gad.12.11.1678. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chamberlain JR, Pagan R, Kindelberger DW, Engelke DR. An RNase P RNA subunit mutation affects ribosomal RNA processing. Nucleic Acids Res. 1996;24(16):3158. doi: 10.1093/nar/24.16.3158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chang DD, Clayton DA. A novel endoribonuclease cleaves at a priming site of mouse mitochondrial DNA replication. Embo J. 1987;6(2):409. doi: 10.1002/j.1460-2075.1987.tb04770.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen JL, Nolan JM, Harris ME, Pace NR. Comparative photocross-linking analysis of the tertiary structures of Escherichia coli and Bacillus subtilis RNase P RNAs. Embo J. 1998;17(5):1515. doi: 10.1093/emboj/17.5.1515. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen JL, Pace NR. Identification of the universally conserved core of ribonuclease P RNA. RNA. 1997;3(6):557. [PMC free article] [PubMed] [Google Scholar]
- Chen Y, Li X, Gegenheimer P. Ribonuclease P catalysis requires Mg2+ coordinated to the pro-RP oxygen of the scissile bond. Biochemistry. 1997;36(9):2425. doi: 10.1021/bi9620464. [DOI] [PubMed] [Google Scholar]
- Christian EL, Zahler NH, Kaye NM, Harris ME. Analysis of substrate recognition by the ribonucleoprotein endonuclease RNase P. Methods. 2002;28(3):307. doi: 10.1016/s1046-2023(02)00238-4. [DOI] [PubMed] [Google Scholar]
- Chu S, Archer RH, Zengel JM, Lindahl L. The RNA of RNase MRP is required for normal processing of ribosomal RNA. Proc Natl Acad Sci USA. 1994;91(2):659. doi: 10.1073/pnas.91.2.659. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chu S, Zengel JM, Lindahl L. A novel protein shared by RNase MRP and RNase P. RNA. 1997;3(4):382. [PMC free article] [PubMed] [Google Scholar]
- Collins LJ, Moulton V, Penny D. Use of RNA secondary structure for studying the evolution of RNase P and RNase MRP. J Mol Evol. 2000;51(3):194. doi: 10.1007/s002390010081. [DOI] [PubMed] [Google Scholar]
- Cordier A, Schon A. Cyanelle RNase P: RNA structure analysis and holoenzyme properties of an organellar ribonucleoprotein enzyme. J Mol Biol. 1999;289(1):9. doi: 10.1006/jmbi.1999.2762. [DOI] [PubMed] [Google Scholar]
- Crary SM, Niranjanakumari S, Fierke CA. The protein component of Bacillus subtilis ribonuclease P increases catalytic efficiency by enhancing interactions with the 5′ leader sequence of pre-tRNAAsp. Biochemistry. 1998;37(26):9409. doi: 10.1021/bi980613c. [DOI] [PubMed] [Google Scholar]
- Dang YL, Martin NC. Yeast mitochondrial RNase P. Sequence of the RPM2 gene and demonstration that its product is a protein subunit of the enzyme. J Biol Chem. 1993;268(26):19791. [PubMed] [Google Scholar]
- Darr SC, Pace B, Pace NR. Characterization of ribonuclease P from the archaebacterium Sulfolobus solfataricus. J Biol Chem. 1990;265(22):12927. [PubMed] [Google Scholar]
- Day-Storms JJ, Niranjanakumari S, Fierke CA. Ionic interactions between PRNA and P protein in Bacillus subtilis RNase P characterized using a magnetocapture-based assay. RNA. 2004;10(10):1595. doi: 10.1261/rna.7550104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dichtl B, Tollervey D. Pop3p is essential for the activity of the RNase MRP and RNase P ribonucleoproteins in vivo. Embo J. 1997;16(2):417. doi: 10.1093/emboj/16.2.417. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dlakic M. 3D models of yeast RNase P/MRP proteins Rpp1p and Pop3p. RNA. 2005;11(2):123. doi: 10.1261/rna.7128905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eder PS, Kekuda R, Stolc V, Altman S. Characterization of two scleroderma autoimmune antigens that copurify with human ribonuclease P. Proc Natl Acad Sci USA. 1997;94(4):1101. doi: 10.1073/pnas.94.4.1101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Faber C, Scharpf M, Becker T, Sticht H, Rosch P. The structure of the coliphage HK022 Nun protein-lambda-phage boxB RNA complex. Implications for the mechanism of transcription termination. J Biol Chem. 2001;276(34):32064. doi: 10.1074/jbc.M102975200. [DOI] [PubMed] [Google Scholar]
- Fan H, Goodier JL, Chamberlain JR, Engelke DR, Maraia RJ. 5′ processing of tRNA precursors can Be modulated by the human La antigen phosphoprotein. Mol Cell Biol. 1998;18(6):3201. doi: 10.1128/mcb.18.6.3201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fang XW, Yang XJ, Littrell K, Niranjanakumari S, Thiyagarajan P, Fierke CA, Sosnick TR, Pan T. The Bacillus subtilis RNase P holoenzyme contains two RNase P RNA and two RNase P protein subunits. RNA. 2001;7(2):233. doi: 10.1017/s1355838201001352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Forster AC, Altman S. Similar cage-shaped structures for the RNA components of all ribonuclease P and ribonuclease MRP enzymes. Cell. 1990;62(3):407. doi: 10.1016/0092-8674(90)90003-w. [DOI] [PubMed] [Google Scholar]
- Frank DN, Adamidi C, Ehringer MA, Pitulle C, Pace NR. Phylogenetic-comparative analysis of the eukaryal ribonuclease P RNA. RNA. 2000;6(12):1895. doi: 10.1017/s1355838200001461. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frank DN, Pace NR. Ribonuclease P: Unity and diversity in a tRNA processing ribozyme. Annu Rev Biochem. 1998;67:153. doi: 10.1146/annurev.biochem.67.1.153. [DOI] [PubMed] [Google Scholar]
- Gegenheimer P. Structure, mechanism and evolution of chloroplast transfer RNA processing systems. Mol Biol Rep. 1996;22(2–3):147. doi: 10.1007/BF00988720. [DOI] [PubMed] [Google Scholar]
- Gill T, Cai T, Aulds J, Wierzbicki S, Schmitt ME. RNase MRP cleaves the CLB2 mRNA to promote cell cycle progression: novel method of mRNA degradation. Mol Cell Biol. 2004;24(3):945. doi: 10.1128/MCB.24.3.945-953.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gopalan V, Baxevanis AD, Landsman D, Altman S. Analysis of the functional role of conserved residues in the protein subunit of ribonuclease P from Escherichia coli. J Mol Biol. 1997;267(4):818. doi: 10.1006/jmbi.1997.0906. [DOI] [PubMed] [Google Scholar]
- Gray MW. Origin and evolution of organelle genomes. Curr Opin Genet Dev. 1993;3(6):884. doi: 10.1016/0959-437x(93)90009-e. [DOI] [PubMed] [Google Scholar]
- Guerrier-Takada C, Altman S. Catalytic activity of an RNA molecule prepared by transcription in vitro. Science. 1984;223(4633):285. doi: 10.1126/science.6199841. [DOI] [PubMed] [Google Scholar]
- Guerrier-Takada C, Gardiner K, Marsh T, Pace N, Altman S. The RNA moiety of ribonuclease P is the catalytic subunit of the enzyme. Cell. 1983;35(3 Pt 2):849. doi: 10.1016/0092-8674(83)90117-4. [DOI] [PubMed] [Google Scholar]
- Haas ES, Armbruster DW, Vucson BM, Daniels CJ, Brown JW. Comparative analysis of ribonuclease P RNA structure in Archaea. Nucleic Acids Res. 1996;24(7):1252. doi: 10.1093/nar/24.7.1252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haas ES, Banta AB, Harris JK, Pace NR, Brown JW. Structure and evolution of ribonuclease P RNA in Gram-positive bacteria. Nucleic Acids Res. 1996;24(23):4775. doi: 10.1093/nar/24.23.4775. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haas ES, Brown JW. Evolutionary variation in bacterial RNase P RNAs. Nucleic Acids Res. 1998;26(18):4093. doi: 10.1093/nar/26.18.4093. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haas ES, Brown JW, Pitulle C, Pace NR. Further perspective on the catalytic core and secondary structure of ribonuclease P RNA. Proc Natl Acad Sci USA. 1994;91(7):2527. doi: 10.1073/pnas.91.7.2527. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haas ES, Morse DP, Brown JW, Schmidt FJ, Pace NR. Long-range structure in ribonuclease P RNA. Science. 1991;254(5033):853. doi: 10.1126/science.1719634. [DOI] [PubMed] [Google Scholar]
- Hall TA, Brown JW. Archaeal RNase P has multiple protein subunits homologous to eukaryotic nuclear RNase P proteins. RNA. 2002;8(3):296. doi: 10.1017/s1355838202028492. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hall TA, Brown JW. Interactions between RNase P protein subunits in archaea. Archaea. 2004;1(4):247. doi: 10.1155/2004/743956. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hamma T, Ferre-D’Amare AR. Structure of protein L7Ae bound to a K-turn derived from an archaeal box H/ACA sRNA at 1.8 A resolution. Structure (Camb) 2004;12(5):893. doi: 10.1016/j.str.2004.03.015. [DOI] [PubMed] [Google Scholar]
- Harris JK, Haas ES, Williams D, Frank DN, Brown JW. New insight into RNase P RNA structure from comparative analysis of the archaeal RNA. RNA. 2001;7(2):220. doi: 10.1017/s1355838201001777. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harris ME, Pace NR. Identification of phosphates involved in catalysis by the ribozyme RNase P RNA. RNA. 1995;1(2):210. [PMC free article] [PubMed] [Google Scholar]
- Hartmann E, Hartmann RK. The enigma of ribonuclease P evolution. Trends Genet. 2003;19(10):561. doi: 10.1016/j.tig.2003.08.007. [DOI] [PubMed] [Google Scholar]
- Hartmann RK, Heinrich J, Schlegl J, Schuster H. Precursor of C4 antisense RNA of bacteriophages P1 and P7 is a substrate for RNase P of Escherichia coli. Proc Natl Acad Sci USA. 1995;92(13):5822. doi: 10.1073/pnas.92.13.5822. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hashimoto C, Steitz JA. Sequential association of nucleolar 7–2 RNA with two different autoantigens. J Biol Chem. 1983;258(3):1379. [PubMed] [Google Scholar]
- Henry Y, Wood H, Morrissey JP, Petfalski E, Kearsey S, Tollervey D. The 5′ end of yeast 5.8S rRNA is generated by exonucleases from an upstream cleavage site. Embo J. 1994;13(10):2452. doi: 10.1002/j.1460-2075.1994.tb06530.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hollingsworth MJ, Martin NC. RNase P activity in the mitochondria of Saccharomyces cerevisiae depends on both mitochondrion and nucleus-encoded components. Mol Cell Biol. 1986;6(4):1058. doi: 10.1128/mcb.6.4.1058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Houser-Scott F, Xiao S, Millikin CE, Zengel JM, Lindahl L, Engelke DR. Interactions among the protein and RNA subunits of Saccharomyces cerevisiae nuclear RNase P. Proc Natl Acad Sci USA. 2002;99(5):2684. doi: 10.1073/pnas.052586299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hsieh J, Andrews AJ, Fierke CA. Roles of protein subunits in RNA-protein complexes: lessons from ribonuclease P. Biopolymers. 2004;73(1):79. doi: 10.1002/bip.10521. [DOI] [PubMed] [Google Scholar]
- Jacobson MR, Cao LG, Taneja K, Singer RH, Wang YL, Pederson T. Nuclear domains of the RNA subunit of RNase P. J Cell Sci. 1997;110:829. doi: 10.1242/jcs.110.7.829. [DOI] [PubMed] [Google Scholar]
- Jacobson MR, Cao LG, Wang YL, Pederson T. The RNA Subunit of Ribonuclease-P Rapidly Localizes in the Nucleolus after Microinjection into the Nucleus of Living Cells. J Cell Biochem. 1994:115. [Google Scholar]
- James BD, Olsen GJ, Liu JS, Pace NR. The secondary structure of ribonuclease P RNA, the catalytic element of a ribonucleoprotein enzyme. Cell. 1988;52(1):19. doi: 10.1016/0092-8674(88)90527-2. [DOI] [PubMed] [Google Scholar]
- Jarrous N. Human ribonuclease P: subunits, function, and intranuclear localization. RNA. 2002;8(1):1. doi: 10.1017/s1355838202011184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jarrous N, Altman S. Human ribonuclease P. Methods Enzymol. 2001;342:93. doi: 10.1016/s0076-6879(01)42538-9. [DOI] [PubMed] [Google Scholar]
- Jarrous N, Eder PS, Wesolowski D, Altman S. Rpp14 and Rpp29, two protein subunits of human ribonuclease P. RNA. 1999;5(2):153. doi: 10.1017/s135583829800185x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jarrous N, Reiner R, Wesolowski D, Mann H, Guerrier-Takada C, Altman S. Function and subnuclear distribution of Rpp21, a protein subunit of the human ribonucleoprotein ribonuclease P. RNA. 2001;7(8):1153. doi: 10.1017/s1355838201010469. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jarrous N, Wolenski JS, Wesolowski D, Lee C, Altman S. Localization in the nucleolus and coiled bodies of protein subunits of the ribonucleoprotein ribonuclease P. J Cell Biol. 1999;146(3):559. doi: 10.1083/jcb.146.3.559. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jiang T, Altman S. Protein-protein interactions with subunits of human nuclear RNase P. Proc Natl Acad Sci USA. 2001;98(3):920. doi: 10.1073/pnas.021561498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jiang T, Guerrier-Takada C, Altman S. Protein-RNA interactions in the subunits of human nuclear RNase P. RNA. 2001;7(7):937. doi: 10.1017/s1355838201010299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kahle D, Wehmeyer U, Krupp G. Substrate recognition by RNase P and by the catalytic M1 RNA: identification of possible contact points in pre-tRNAs. Embo J. 1990;9(6):1929. doi: 10.1002/j.1460-2075.1990.tb08320.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kakuta Y, Ishimatsu I, Numata T, Kimura K, Yao M, Tanaka I, Kimura M. Crystal Structure of a Ribonuclease P Protein Ph1601p from Pyrococcus horikoshii OT3: An Archaeal Homologue of Human Nuclear Ribonuclease P Protein Rpp21. Biochemistry. 2005;44(36):12086. doi: 10.1021/bi050738z. [DOI] [PubMed] [Google Scholar]
- Kassenbrock CK, Gao GJ, Groom KR, Sulo P, Douglas MG, Martin NC. RPM2, independently of its mitochondrial RNase P function, suppresses an ISP42 mutant defective in mitochondrial import and is essential for normal growth. Mol Cell Biol. 1995;15(9):4763. doi: 10.1128/mcb.15.9.4763. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kawano S, Nakashima T, Kakuta Y, Tanaka I, Kimura M. Crystal Structure of Protein Ph1481p in Complex with Protein Ph1877p of Archaeal RNase P from Pyrococcus horikoshii OT3: Implication of Dimer Formation of the Holoenzyme. J Mol Biol. 2006 doi: 10.1016/j.jmb.2005.12.086. in press. [DOI] [PubMed] [Google Scholar]
- Kazakov S, Altman S. Site-specific cleavage by metal ion cofactors and inhibitors of M1 RNA, the catalytic subunit of RNase P from Escherichia coli. Proc Natl Acad Sci USA. 1991;88(20):9193. doi: 10.1073/pnas.88.20.9193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kazantsev AV, Krivenko AA, Harrington DJ, Carter RJ, Holbrook SR, Adams PD, Pace NR. High-resolution structure of RNase P protein from Thermotoga maritima. Proc Natl Acad Sci USA. 2003;100(13):7497. doi: 10.1073/pnas.0932597100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kazantsev AV, Krivenko AA, Harrington DJ, Holbrook SR, Adams PD, Pace NR. Crystal structure of a bacterial ribonuclease P RNA. Proc Natl Acad Sci USA. 2005;102(38):13392. doi: 10.1073/pnas.0506662102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kazantsev AV, Pace NR. Identification by modification-interference of purine N-7 and ribose 2′-OH groups critical for catalysis by bacterial ribonuclease P. RNA. 1998;4(8):937. doi: 10.1017/s1355838298980384. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kendall A, Hull MW, Bertrand E, Good PD, Singer RH, Engelke DR. A CBF5 mutation that disrupts nucleolar localization of early tRNA biosynthesis in yeast also suppresses tRNA gene-mediated transcriptional silencing. Proc Natl Acad Sci USA. 2000;97(24):13108. doi: 10.1073/pnas.240454997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kifusa M, Fukuhara H, Hayashi T, Kimura M. Protein-protein interactions in the subunits of ribonuclease P in the hyperthermophilic archaeon Pyrococcus horikoshii OT3. Biosci Biotechnol Biochem. 2005;69(6):1209. doi: 10.1271/bbb.69.1209. [DOI] [PubMed] [Google Scholar]
- Kim JJ, Kilani AF, Zhan X, Altman S, Liu F. The protein cofactor allows the sequence of an RNase P ribozyme to diversify by maintaining the catalytically active structure of the enzyme. RNA. 1997;3(6):613. [PMC free article] [PubMed] [Google Scholar]
- Kirsebom LA, Svard SG. Base pairing between Escherichia coli RNase P RNA and its substrate. Embo J. 1994;13(20):4870. doi: 10.1002/j.1460-2075.1994.tb06814.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klein DJ, Schmeing TM, Moore PB, Steitz TA. The kink-turn: a new RNA secondary structure motif. Embo J. 2001;20(15):4214. doi: 10.1093/emboj/20.15.4214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Komine Y, Kitabatake M, Yokogawa T, Nishikawa K, Inokuchi H. A Transfer-RNA-Like Structure Is Present in 10sa RNA, a Small Stable RNA from Escherichia-Coli. Proc Natl Acad Sci USA. 1994;91(20):9223. doi: 10.1073/pnas.91.20.9223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koonin EV, Wolf YI, Aravind L. Prediction of the archaeal exosome and its connections with the proteasome and the translation and transcription machineries by a comparative-genomic approach. Genome Research. 2001;11(2):240. doi: 10.1101/gr.162001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kouzuma Y, Mizoguchi M, Takagi H, Fukuhara H, Tsukamoto M, Numata T, Kimura M. Reconstitution of archaeal ribonuclease P from RNA and four protein components. Biochem Biophys Res Commun. 2003;306(3):666. doi: 10.1016/s0006-291x(03)01034-9. [DOI] [PubMed] [Google Scholar]
- Krasilnikov AS, Mondragon A. On the occurrence of the T-loop RNA folding motif in large RNA molecules. RNA. 2003;9(6):640. doi: 10.1261/rna.2202703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krasilnikov AS, Xiao Y, Pan T, Mondragon A. Basis for structural diversity in homologous RNAs. Science. 2004;306(5693):104. doi: 10.1126/science.1101489. [DOI] [PubMed] [Google Scholar]
- Krasilnikov AS, Yang X, Pan T, Mondragon A. Crystal structure of the specificity domain of ribonuclease P. Nature. 2003;421(6924):760. doi: 10.1038/nature01386. [DOI] [PubMed] [Google Scholar]
- Kufel J, Allmang C, Verdone L, Beggs JD, Tollervey D. Lsm proteins are required for normal processing of pre-tRNAs and their efficient association with La-homologous protein Lhp1p. Mol Cell Biol. 2002;22(14):5248. doi: 10.1128/MCB.22.14.5248-5256.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kufel J, Kirsebom LA. Residues in Escherichia coli RNase P RNA important for cleavage site selection and divalent metal ion binding. J Mol Biol. 1996;263(5):685. doi: 10.1006/jmbi.1996.0608. [DOI] [PubMed] [Google Scholar]
- Kufel J, Kirsebom LA. The P15-loop of Escherichia coli RNase P RNA is an autonomous divalent metal ion binding domain. RNA. 1998;4(7):777. doi: 10.1017/s1355838298970923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kurz JC, Fierke CA. Ribonuclease P: a ribonucleoprotein enzyme. Curr Opin Chem Biol. 2000;4(5):553. doi: 10.1016/s1367-5931(00)00131-9. [DOI] [PubMed] [Google Scholar]
- Kurz JC, Niranjanakumari S, Fierke CA. Protein component of Bacillus subtilis RNase P specifically enhances the affinity for precursor-tRNAAsp. Biochemistry. 1998;37(8):2393. doi: 10.1021/bi972530m. [DOI] [PubMed] [Google Scholar]
- LaGrandeur TE, Darr SC, Haas ES, Pace NR. Characterization of the RNase P RNA of Sulfolobus acidocaldarius. J Bacteriol. 1993;175(16):5043. doi: 10.1128/jb.175.16.5043-5048.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- LaGrandeur TE, Huttenhofer A, Noller HF, Pace NR. Phylogenetic comparative chemical footprint analysis of the interaction between ribonuclease P RNA and tRNA. Embo J. 1994;13(17):3945. doi: 10.1002/j.1460-2075.1994.tb06710.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lawrence N, Wesolowski D, Gold H, Bartkiewicz M, Guerrier-Takada C, McClain WH, Altman S. Characteristics of ribonuclease P from various organisms. Cold Spring Harb Symp Quant Biol. 1987;52:s233. doi: 10.1101/sqb.1987.052.01.028. [DOI] [PubMed] [Google Scholar]
- Lee DY, Clayton DA. Initiation of mitochondrial DNA replication by transcription and R-loop processing. J Biol Chem. 1998;273(46):30614. doi: 10.1074/jbc.273.46.30614. [DOI] [PubMed] [Google Scholar]
- Lee JY, Evans CF, Engelke DR. Expression of RNase P RNA in Saccharomyces cerevisiae is controlled by an unusual RNA polymerase III promoter. Proc Natl Acad Sci USA. 1991;88(16):6986. doi: 10.1073/pnas.88.16.6986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee JY, Rohlman CE, Molony LA, Engelke DR. Characterization of RPR1, an essential gene encoding the RNA component of Saccharomyces cerevisiae nuclear RNase P. Mol Cell Biol. 1991;11(2):721. doi: 10.1128/mcb.11.2.721. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee YC, Lee BJ, Hwang DS, Kang HS. Purification and characterization of mitochondrial ribonuclease P from Aspergillus nidulans. Eur J Biochem. 1996;235(1–2):289. doi: 10.1111/j.1432-1033.1996.00289.x. [DOI] [PubMed] [Google Scholar]
- Lewis JD, Tollervey D. Like attracts like: getting RNA processing together in the nucleus. Science. 2000;288(5470):1385. doi: 10.1126/science.288.5470.1385. [DOI] [PubMed] [Google Scholar]
- Li X, Frank DN, Pace N, Zengel JM, Lindahl L. Phylogenetic analysis of the structure of RNase MRP RNA in yeasts. RNA. 2002;8(6):740. doi: 10.1017/s1355838202022082. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li X, Zaman S, Langdon Y, Zengel JM, Lindahl L. Identification of a functional core in the RNA component of RNase MRP of budding yeasts. Nucleic Acids Research. 2004;32(12):3703. doi: 10.1093/nar/gkh689. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li Y, Altman S. A subunit of human nuclear RNase P has ATPase activity. Proc Natl Acad Sci USA. 2001;98(2):441. doi: 10.1073/pnas.021555498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li Y, Altman S. Partial reconstitution of human RNase P in HeLa cells between its RNA subunit with an affinity tag and the intact protein components. Nucleic Acids Res. 2002;30(17):3706. doi: 10.1093/nar/gkf499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lindahl L, Fretz S, Epps N, Zengel JM. Functional equivalence of hairpins in the RNA subunits of RNase MRP and RNase P in Saccharomyces cerevisiae. RNA. 2000;6(5):653. doi: 10.1017/s1355838200992574. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Loria A, Pan T. Domain structure of the ribozyme from eu-bacterial ribonuclease P. RNA. 1996;2(6):551. [PMC free article] [PubMed] [Google Scholar]
- Loria A, Pan T. Recognition of the T stem-loop of a pre-tRNA substrate by the ribozyme from Bacillus subtilis ribonuclease P. Biochemistry. 1997;36(21):6317. doi: 10.1021/bi970115o. [DOI] [PubMed] [Google Scholar]
- Loria A, Pan T. The cleavage step of ribonuclease P catalysis is determined by ribozyme-substrate interactions both distal and proximal to the cleavage site. Biochemistry. 1999;38(27):8612. doi: 10.1021/bi990691f. [DOI] [PubMed] [Google Scholar]
- Lygerou Z, Allmang C, Tollervey D, Seraphin B. Accurate processing of a eukaryotic precursor ribosomal RNA by ribonuclease MRP in vitro. Science. 1996;272(5259):268. doi: 10.1126/science.272.5259.268. [DOI] [PubMed] [Google Scholar]
- Lygerou Z, Mitchell P, Petfalski E, Seraphin B, Tollervey D. The POP1 gene encodes a protein component common to the RNase MRP and RNase P ribonucleoproteins. Genes Dev. 1994;8(12):1423. doi: 10.1101/gad.8.12.1423. [DOI] [PubMed] [Google Scholar]
- Lygerou Z, Pluk H, van Venrooij WJ, Seraphin B. hPop1: an autoantigenic protein subunit shared by the human RNase P and RNase MRP ribonucleoproteins. Embo J. 1996;15(21):5936. [PMC free article] [PubMed] [Google Scholar]
- Mann H, Ben-Asouli Y, Schein A, Moussa S, Jarrous N. Eukaryotic RNase P: role of RNA and protein subunits of a primordial catalytic ribonucleoprotein in RNA-based catalysis. Mol Cell. 2003;12(4):925. doi: 10.1016/s1097-2765(03)00357-5. [DOI] [PubMed] [Google Scholar]
- Mans RM, Guerrier-Takada C, Altman S, Pleij CW. Interaction of RNase P from Escherichia coli with pseudoknotted structures in viral RNAs. Nucleic Acids Res. 1990;18(12):3479. doi: 10.1093/nar/18.12.3479. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maraia RJ, Intine RV. La protein and its associated small nuclear and nucleolar precursor RNAs. Gene Expr. 2002;10(1–2):41. [PMC free article] [PubMed] [Google Scholar]
- Marchfelder A. Plant mitochondrial RNase P. Mol Biol Rep. 1995;22(2–3):151. doi: 10.1007/BF00988721. [DOI] [PubMed] [Google Scholar]
- Marquez SM, Harris JK, Kelley ST, Brown JW, Dawson SC, Roberts EC, Pace NR. Structural implications of novel diversity in eucaryal RNase P RNA. RNA. 2005;11(5):739. doi: 10.1261/rna.7211705. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McClain WH, Guerrier-Takada C, Altman S. Model substrates for an RNA enzyme. Science. 1987;238(4826):527. doi: 10.1126/science.2443980. [DOI] [PubMed] [Google Scholar]
- Miller DL, Martin NC. Characterization of the yeast mitochondrial locus necessary for tRNA biosynthesis: DNA sequence analysis and identification of a new transcript. Cell. 1983;34(3):911. doi: 10.1016/0092-8674(83)90548-2. [DOI] [PubMed] [Google Scholar]
- Morales MJ, Dang YL, Lou YC, Sulo P, Martin NC. A 105-kDa protein is required for yeast mitochondrial RNase P activity. Proc Natl Acad Sci USA. 1992;89(20):9875. doi: 10.1073/pnas.89.20.9875. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morales MJ, Wise CA, Hollingsworth MJ, Martin NC. Characterization of yeast mitochondrial RNase P: an intact RNA subunit is not essential for activity in vitro. Nucleic Acids Res. 1989;17(17):6865. doi: 10.1093/nar/17.17.6865. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morrissey JP, Tollervey D. Birth of the snoRNPs: the evolution of RNase MRP and the eukaryotic pre-rRNA-processing system. Trends Biochem Sci. 1995;20(2):78. doi: 10.1016/s0968-0004(00)88962-8. [DOI] [PubMed] [Google Scholar]
- Niranjanakumari S, Stams T, Crary SM, Christianson DW, Fierke CA. Protein component of the ribozyme ribonuclease P alters substrate recognition by directly contacting precursor tRNA. Proc Natl Acad Sci USA. 1998;95(26):15212. doi: 10.1073/pnas.95.26.15212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Numata T, Ishimatsu I, Kakuta Y, Tanaka I, Kimura M. Crystal structure of archaeal ribonuclease P protein Ph1771p from Pyrococcus horikoshii OT3: an archaeal homolog of eukaryotic ribonuclease P protein Rpp29. RNA. 2004;10(9):1423. doi: 10.1261/rna.7560904. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Odell L, Huang V, Jakacka M, Pan T. Interaction of structural modules in substrate binding by the ribozyme from Bacillus subtilis RNase P. Nucleic Acids Res. 1998;26(16):3717. doi: 10.1093/nar/26.16.3717. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oh BK, Frank DN, Pace NR. Participation of the 3′-CCA of tRNA in the binding of catalytic Mg2+ ions by ribonuclease P. Biochemistry. 1998;37(20):7277. doi: 10.1021/bi973100z. [DOI] [PubMed] [Google Scholar]
- Pace NR, Brown JW. Evolutionary perspective on the structure and function of ribonuclease P, a ribozyme. J Bacteriol. 1995;177(8):1919. doi: 10.1128/jb.177.8.1919-1928.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pagan-Ramos E, Lee Y, Engelke DR. A conserved RNA motif involved in divalent cation utilization by nuclear RNase P. RNA. 1996;2(11):1100. [PMC free article] [PubMed] [Google Scholar]
- Pagan-Ramos E, Lee Y, Engelke DR. Mutational analysis of Saccharomyces cerevisiae nuclear RNase P: randomization of universally conserved positions in the RNA subunit. RNA. 1996;2(5):441. [PMC free article] [PubMed] [Google Scholar]
- Paluh JL, Clayton DA. Schizosaccharomyces pombe RNase MRP RNA is homologous to metazoan RNase MRP RNAs and may provide clues to interrelationships between RNase MRP and RNase P. Yeast. 1995;11(13):1249. doi: 10.1002/yea.320111305. [DOI] [PubMed] [Google Scholar]
- Pan T. Higher order folding and domain analysis of the ribozyme from Bacillus subtilis ribonuclease P. Biochemistry. 1995;34(3):902. doi: 10.1021/bi00003a024. [DOI] [PubMed] [Google Scholar]
- Pannone BK, Kim SD, Noe DA, Wolin SL. Multiple functional interactions between components of the Lsm2-Lsm8 complex, U6 snRNA, and the yeast La protein. Genetics. 2001;158(1):187. doi: 10.1093/genetics/158.1.187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pannucci JA, Haas ES, Hall TA, Harris JK, Brown JW. RNase P RNAs from some Archaea are catalytically active. Proc Natl Acad Sci USA. 1999;96(14):7803. doi: 10.1073/pnas.96.14.7803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peck-Miller KA, Altman S. Kinetics of the processing of the precursor to 4.5 S RNA, a naturally occurring substrate for RNase P from Escherichia coli. J Mol Biol. 1991;221(1):1. doi: 10.1016/0022-2836(91)80194-y. [DOI] [PubMed] [Google Scholar]
- Pfeiffer T, Tekos A, Warnecke JM, Drainas D, Engelke DR, Seraphin B, Hartmann RK. Effects of phosphorothioate modifications on precursor tRNA processing by eukaryotic RNase P enzymes. J Mol Biol. 2000;298(4):559. doi: 10.1006/jmbi.2000.3655. [DOI] [PubMed] [Google Scholar]
- Piccinelli P, Rosenblad MA, Samuelsson T. Identification and analysis of ribonuclease P and MRP RNA in a broad range of eukaryotes. Nucleic Acids Res. 2005;33(14):4485. doi: 10.1093/nar/gki756. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pitulle C, Garcia-Paris M, Zamudio KR, Pace NR. Comparative structure analysis of vertebrate ribonuclease P RNA. Nucleic Acids Res. 1998;26(14):3333. doi: 10.1093/nar/26.14.3333. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pluk H, van Eenennaam H, Rutjes SA, Pruijn GJ, van Venrooij WJ. RNA-protein interactions in the human RNase MRP ribonucleoprotein complex. RNA. 1999;5(4):512. doi: 10.1017/s1355838299982079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Puranam RS, Attardi G. The RNase P associated with HeLa cell mitochondria contains an essential RNA component identical in sequence to that of the nuclear RNase P. Mol Cell Biol. 2001;21(2):548. doi: 10.1128/MCB.21.2.548-561.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reimer G, Raska I, Scheer U, Tan EM. Immunolocalization of 7-2-ribonucleoprotein in the granular component of the nucleolus. Exp Cell Res. 1988;176(1):117. doi: 10.1016/0014-4827(88)90126-7. [DOI] [PubMed] [Google Scholar]
- Reith M, Munholland J. Complete nucleotide sequence of the Porphyra purpurea chloroplast genome. Plant Mol Biol Rep. 1995;13(4):333. [Google Scholar]
- Ridanpaa M, Sistonen P, Rockas S, Rimoin DL, Makitie O, Kaitila I. Worldwide mutation spectrum in cartilage-hair hypoplasia: ancient founder origin of the major70A–>G mutation of the untranslated RMRP. Eur J Hum Genet. 2002;10(7):439. doi: 10.1038/sj.ejhg.5200824. [DOI] [PubMed] [Google Scholar]
- Ridanpaa M, van Eenennaam H, Pelin K, Chadwick R, Johnson C, Yuan B, vanVenrooij W, Pruijn G, Salmela R, Rockas S, et al. Mutations in the RNA component of RNase MRP cause a pleiotropic human disease, cartilage-hair hypoplasia. Cell. 2001;104(2):195. doi: 10.1016/s0092-8674(01)00205-7. [DOI] [PubMed] [Google Scholar]
- Rossmanith W, Karwan RM. Characterization of human mitochondrial RNase P: novel aspects in tRNA processing. Biochem Biophys Res Commun. 1998;247(2):234. doi: 10.1006/bbrc.1998.8766. [DOI] [PubMed] [Google Scholar]
- Rossmanith W, Karwan RM. Impairment of tRNA processing by point mutations in mitochondrial tRNA(Leu)(UUR) associated with mitochondrial diseases. FEBS Lett. 1998;433(3):269. doi: 10.1016/s0014-5793(98)00928-4. [DOI] [PubMed] [Google Scholar]
- Rossmanith W, Tullo A, Potuschak T, Karwan R, Sbisa E. Human mitochondrial tRNA processing. J Biol Chem. 1995;270(21):12885. doi: 10.1074/jbc.270.21.12885. [DOI] [PubMed] [Google Scholar]
- Rozhdestvensky TS, Tang TH, Tchirkova IV, Brosius J, Bachellerie JP, Huttenhofer A. Binding of L7Ae protein to the K-turn of archaeal snoRNAs: a shared RNA binding motif for C/D and H/ACA box snoRNAs in Archaea. Nucleic Acids Res. 2003;31(3):869. doi: 10.1093/nar/gkg175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Salavati R, Panigrahi AK, Stuart KD. Mitochondrial ribonuclease P activity of Trypanosoma brucei. Mol Biochem Parasitol. 2001;115(1):109. doi: 10.1016/s0166-6851(01)00273-0. [DOI] [PubMed] [Google Scholar]
- Salgado-Garrido J, Bragado-Nilsson E, Kandels-Lewis S, Seraphin B. Sm and Sm-like proteins assemble in two related complexes of deep evolutionary origin. Embo J. 1999;18(12):3451. doi: 10.1093/emboj/18.12.3451. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Salinas K, Wierzbicki S, Zhou L, Schmitt ME. Characterization and purification of Saccharomyces cerevisiae RNase MRP reveals a new unique protein component. J Biol Chem. 2005;280(12):11352. doi: 10.1074/jbc.M409568200. [DOI] [PubMed] [Google Scholar]
- Savino TM, Bastos R, Jansen E, Hernandez-Verdun D. The nucleolar antigen Nop52, the human homologue of the yeast ribosomal RNA processing RRP1, is recruited at late stages of nucleologenesis. J Cell Sci. 1999;112(Pt 12):1889. doi: 10.1242/jcs.112.12.1889. [DOI] [PubMed] [Google Scholar]
- Schmitt ME, Bennett JL, Dairaghi DJ, Clayton DA. Secondary structure of RNase MRP RNA as predicted by phylogenetic comparison. Faseb J. 1993;7(1):208. doi: 10.1096/fasebj.7.1.7678563. [DOI] [PubMed] [Google Scholar]
- Schmitt ME, Clayton DA. Characterization of a unique protein component of yeast RNase MRP: an RNA-binding protein with a zinc-cluster domain. Genes Dev. 1994;8(21):2617. doi: 10.1101/gad.8.21.2617. [DOI] [PubMed] [Google Scholar]
- Schon A. Ribonuclease P: the diversity of a ubiquitous RNA processing enzyme. Fems Microbiology Reviews. 1999;23(3):391. doi: 10.1111/j.1574-6976.1999.tb00406.x. [DOI] [PubMed] [Google Scholar]
- Seif ER, Forget L, Martin NC, Lang BF. Mitochondrial RNase P RNAs in ascomycete fungi: lineage-specific variations in RNA secondary structure. RNA. 2003;9(9):1073. doi: 10.1261/rna.5880403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shadel GS, Buckenmeyer GA, Clayton DA, Schmitt ME. Mutational analysis of the RNA component of Saccharomyces cerevisiae RNase MRP reveals distinct nuclear phenotypes. Gene. 2000;245(1):175. doi: 10.1016/s0378-1119(00)00013-5. [DOI] [PubMed] [Google Scholar]
- Sharin E, Schein A, Mann H, Ben-Asouli Y, Jarrous N. RNase P: role of distinct protein cofactors in tRNA substrate recognition and RNA-based catalysis. Nucleic Acids Res. 2005;33(16):5120. doi: 10.1093/nar/gki828. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sidote DJ, Heideker J, Hoffman DW. Crystal structure of archaeal ribonuclease P protein aRpp29 from Archaeoglobus fulgidus. Biochemistry. 2004;43(44):14128. doi: 10.1021/bi048578z. [DOI] [PubMed] [Google Scholar]
- Sidote DJ, Hoffman DW. NMR structure of an archaeal homologue of ribonuclease P protein Rpp29. Biochemistry. 2003;42(46):13541. doi: 10.1021/bi030170z. [DOI] [PubMed] [Google Scholar]
- Siegel RW, Banta AB, Haas ES, Brown JW, Pace NR. Mycoplasma fermentans simplifies our view of the catalytic core of ribonuclease P RNA. RNA. 1996;2(5):452. [PMC free article] [PubMed] [Google Scholar]
- Smith D, Pace NR. Multiple magnesium ions in the ribonuclease P reaction mechanism. Biochemistry. 1993;32(20):5273. doi: 10.1021/bi00071a001. [DOI] [PubMed] [Google Scholar]
- Spitzfaden C, Nicholson N, Jones JJ, Guth S, Lehr R, Prescott CD, Hegg LA, Eggleston DS. The structure of ribonuclease P protein from Staphylococcus aureus reveals a unique binding site for single-stranded RNA. J Mol Biol. 2000;295(1):105. doi: 10.1006/jmbi.1999.3341. [DOI] [PubMed] [Google Scholar]
- Srisawat C, Engelke DR. Streptavidin aptamers: affinity tags for the study of RNAs and ribonucleoproteins. RNA. 2001;7(4):632. doi: 10.1017/s135583820100245x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Srisawat C, Houser-Scott F, Bertrand E, Xiao S, Singer RH, Engelke DR. An active precursor in assembly of yeast nuclear ribonuclease P. RNA. 2002;8(10):1348. doi: 10.1017/s1355838202027048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stams T, Niranjanakumari S, Fierke CA, Christianson DW. Ribonuclease P protein structure: evolutionary origins in the translational apparatus. Science. 1998;280(5364):752. doi: 10.1126/science.280.5364.752. [DOI] [PubMed] [Google Scholar]
- Stolc V, Altman S. Rpp1, an essential protein subunit of nuclear RNase P required for processing of precursor tRNA and 35S precursor rRNA in Saccharomyces cerevisiae. Genes Dev. 1997;11(21):2926. doi: 10.1101/gad.11.21.2926. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stribinskis V, Gao GJ, Ellis SR, Martin NC. Rpm2, the protein subunit of mitochondrial RNase P in Saccharomyces cerevisiae, also has a role in the translation of mitochondrially encoded subunits of cytochrome c oxidase. Genetics. 2001;158(2):573. doi: 10.1093/genetics/158.2.573. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stribinskis V, Gao GJ, Sulo P, Dang YL, Martin NC. Yeast mitochondrial RNase P RNA synthesis is altered in an RNase P protein subunit mutant: insights into the biogenesis of a mitochondrial RNA-processing enzyme. Mol Cell Biol. 1996;16(7):3429. doi: 10.1128/mcb.16.7.3429. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stribinskis V, Gao GJ, Sulo P, Ellis SR, Martin NC. Rpm2p: separate domains promote tRNA and Rpm1r maturation in Saccharomyces cerevisiae mitochondria. Nucleic Acids Res. 2001;29(17):3631. doi: 10.1093/nar/29.17.3631. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stribinskis V, Heyman HC, Ellis SR, Steffen MC, Martin NC. Rpm2p, a component of yeast mitochondrial RNase P, acts as a transcriptional activator in the nucleus. Mol Cell Biol. 2005;25(15):6546. doi: 10.1128/MCB.25.15.6546-6558.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suryadi J, Tran EJ, Maxwell ES, Brown BA., 2nd The crystal structure of the Methanocaldococcus jannaschii multifunctional L7Ae RNA-binding protein reveals an induced-fit interaction with the box C/D RNAs. Biochemistry. 2005;44(28):9657. doi: 10.1021/bi050568q. [DOI] [PubMed] [Google Scholar]
- Takagi H, Watanabe M, Kakuta Y, Kamachi R, Numata T, Tanaka I, Kimura M. Crystal structure of the ribonuclease P protein Ph1877p from hyperthermophilic archaeon Pyrococcus horikoshii OT3. Biochem Biophys Res Commun. 2004;319(3):787. doi: 10.1016/j.bbrc.2004.05.055. [DOI] [PubMed] [Google Scholar]
- Tanaka T, Kanda N, Kikuchi Y. The P3 domain of E. coli ribonuclease P RNA can be truncated and replaced. FEBS Lett. 2004;577(1–2):101. doi: 10.1016/j.febslet.2004.09.072. [DOI] [PubMed] [Google Scholar]
- Thiel CT, Horn D, Zabel B, Ekici AB, Salinas K, Gebhart E, Ruschendorf F, Sticht H, Spranger J, Muller D, et al. Severely Incapacitating Mutations in Patients with Extreme Short Stature Identify RNA-Processing Endoribonuclease RMRP as an Essential Cell Growth Regulator. Am J Hum Genet. 2005;77(5):795. doi: 10.1086/497708. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thomas BC, Chamberlain J, Engelke DR, Gegenheimer P. Evidence for an RNA-based catalytic mechanism in eukaryotic nuclear ribonuclease P. RNA. 2000;6(4):554. doi: 10.1017/s1355838200991477. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thomas BC, Gao L, Stomp D, Li X, Gegenheimer PA. Spinach chloroplast RNase P: a putative protein enzyme. Nucleic Acids Symp Ser. 1995;(33):95. [PubMed] [Google Scholar]
- Thompson M, Haeusler RA, Good PD, Engelke DR. Nucleolar clustering of dispersed tRNA genes. Science. 2003;302(5649):1399. doi: 10.1126/science.1089814. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thurlow DL, Shilowski D, Marsh TL. Nucleotides in precursor tRNAs that are required intact for catalysis by RNase P RNAs. Nucleic Acids Res. 1991;19(4):885. doi: 10.1093/nar/19.4.885. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Topper JN, Clayton DA. Secondary structure of the RNA component of a nuclear/mitochondrial ribonucleoprotein. J Biol Chem. 1990;265(22):13254. [PubMed] [Google Scholar]
- Toro I, Thore S, Mayer C, Basquin J, Seraphin B, Suck D. RNA binding in an Sm core domain: X-ray structure and functional analysis of an archaeal Sm protein complex. Embo J. 2001;20(9):2293. doi: 10.1093/emboj/20.9.2293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Torres-Larios A, Swinger KK, Krasilnikov AS, Pan T, Mondragon A. Crystal structure of the RNA component of bacterial ribonuclease P. Nature. 2005;437(7058):584. doi: 10.1038/nature04074. [DOI] [PubMed] [Google Scholar]
- Tranguch AJ, Kindelberger DW, Rohlman CE, Lee JY, Engelke DR. Structure-sensitive RNA footprinting of yeast nuclear ribonuclease P. Biochemistry. 1994;33(7):1778. doi: 10.1021/bi00173a022. [DOI] [PubMed] [Google Scholar]
- Turmel M, Otis C, Lemieux C. The complete chloroplast DNA sequence of the green alga Nephroselmis olivacea: Insights into the architecture of ancestral chloroplast genomes. Proc Nat Acad Sci USA. 1999;96(18):10248. doi: 10.1073/pnas.96.18.10248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Underbrink-Lyon K, Miller DL, Ross NA, Fukuhara H, Martin NC. Characterization of a yeast mitochondrial locus necessary for tRNA biosynthesis. Deletion mapping and restriction mapping studies. Mol Gen Genet. 1983;191(3):512. doi: 10.1007/BF00425771. [DOI] [PubMed] [Google Scholar]
- van Eenennaam H, Lugtenberg D, Vogelzangs JHP, van Venrooij WJ, Pruijn GJM. hPop5, a protein subunit of the human RNase MRP and RNase P endoribonucleases. J Biol Chem. 2001;276(34):31635. doi: 10.1074/jbc.M103399200. [DOI] [PubMed] [Google Scholar]
- van Eenennaam H, Pruijn GJ, van Venrooij WJ. hPop4: a new protein subunit of the human RNase MRP and RNase P ribonucleoprotein complexes. Nucleic Acids Res. 1999;27(12):2465. doi: 10.1093/nar/27.12.2465. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Walker SC, Avis JM. A conserved element in the yeast RNase MRP RNA subunit can participate in a long-range base-pairing interaction. J Mol Biol. 2004;341(2):375. doi: 10.1016/j.jmb.2004.05.076. [DOI] [PubMed] [Google Scholar]
- Walker SC, Avis JM. Secondary structure probing of the human RNase MRP RNA reveals the potential for MRP RNA subsets. Biochem Biophys Res Commun. 2005;335(2):314. doi: 10.1016/j.bbrc.2005.07.074. [DOI] [PubMed] [Google Scholar]
- Welting TJ, van Venrooij WJ, Pruijn GJ. Mutual interactions between subunits of the human RNase MRP ribonucleoprotein complex. Nucleic Acids Res. 2004;32(7):2138. doi: 10.1093/nar/gkh539. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Westhof E, Wesolowski D, Altman S. Mapping in three dimensions of regions in a catalytic RNA protected from attack by an Fe(II)-EDTA reagent. J Mol Biol. 1996;258(4):600. doi: 10.1006/jmbi.1996.0272. [DOI] [PubMed] [Google Scholar]
- Wilson RC, Bohlen CJ, Foster M, Bell CE. Structure of Pfu Pop5, an archaeal RNase P protein. Proc Nat Acad Sci USA. 2006;103(4):873–878. doi: 10.1073/pnas.0508004103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wise CA, Martin NC. Dramatic size variation of yeast mitochondrial RNAs suggests that RNase P RNAs can be quite small. J Biol Chem. 1991;266(29):19154. [PubMed] [Google Scholar]
- Wolin SL, Matera AG. The trials and travels of tRNA. Genes Dev. 1999;13(1):1. doi: 10.1101/gad.13.1.1. [DOI] [PubMed] [Google Scholar]
- Xiao S, Day-Storms JJ, Srisawat C, Fierke CA, Engelke DR. Characterization of conserved sequence elements in eukaryotic RNase P RNA reveals roles in holoenzyme assembly and tRNA processing. RNA. 2005;11(6):885. doi: 10.1261/rna.7282205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xiao SH, Scott F, Fierke CA, Engelke DR. Eukaryotic ribonuclease P: A plurality of ribonucleoprotein enzymes. Annual Review of Biochemistry. 2002;71:165. doi: 10.1146/annurev.biochem.71.110601.135352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yarus M. How Many Catalytic RNAs—Ions and the Cheshire Cat Conjecture. Faseb Journal. 1993;7(1):31. doi: 10.1096/fasebj.7.1.8422972. [DOI] [PubMed] [Google Scholar]
- Yuan Y, Altman S. Substrate recognition by human RNase P: identification of small, model substrates for the enzyme. Embo J. 1995;14(1):159. doi: 10.1002/j.1460-2075.1995.tb06986.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ziehler WA, Day JJ, Fierke CA, Engelke DR. Effects of 5′ leader and 3′ trailer structures on pre-tRNA processing by nuclear RNase P. Biochemistry. 2000;39(32):9909. doi: 10.1021/bi000603n. [DOI] [PubMed] [Google Scholar]
- Ziehler WA, Morris J, Scott FH, Millikin C, Engelke DR. An essential protein-binding domain of nuclear RNase P RNA. RNA. 2001;7(4):565. doi: 10.1017/s1355838201001996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ziehler WA, Yang J, Kurochkin AV, Sandusky PO, Zuiderweg ER, Engelke DR. Structural analysis of the P10/11-P12 RNA domain of yeast RNase P RNA and its interaction with magnesium. Biochemistry. 1998;37(10):3549. doi: 10.1021/bi972886y. [DOI] [PubMed] [Google Scholar]