Abstract
The recently identified gene Muc19/Smgc encodes two diverse splice variants, Smgc (submandibular gland protein C) and Muc19 (mucin 19). Muc19 is a member of the large gel-forming mucin family and is an exocrine product of sublingual mucous salivary glands in mice. SMGC is a transiently expressed secretion product of developing rodent submandibular and sublingual glands. Little is known about the expression of Muc19/Smgc gene products in other murine salivary and non-salivary tissues containing the mucous cell phenotype. Muc19 expression was therefore initially assessed by RT-PCR and immunohistochemistry. As a complementary approach, we developed a knockin mouse model, Muc19-EGFP, in which mice express a fusion protein containing the first 69 residues of Muc19 followed by enhanced green fluorescent protein (EGFP) as a marker of Muc19 expression. Results from both approaches are consistent, with preferential Muc19 expression in salivary major and minor mucous glands as well as submucosal glands of the tracheolarynx and bulbourethral glands. Evidence also indicates that individual mucous cells of minor salivary and bulbourethral glands produce another gel-forming mucin in addition to Muc19. We further find tissue expression of full-length Smgc transcripts, which encode for SMGC, and are restricted to neonatal tracheolarynx and all salivary tissues. (J Histochem Cytochem 58:141–156, 2010)
Keywords: mucin, knockin, bulbourethral gland, salivary glands
A viscoelastic layer of mucus lines mucosal surfaces of the body. Prominent organic constituents of mucus are high-molecular-mass gel-forming mucin glycoproteins, mucins, that help to protect the underlying epithelia by promoting hydration, by interacting with pathogens directly, and/or by forming heterotypic complexes with bacteriostatic or bactericidal proteins (Tabak 1995; Thornton et al. 2008). In the oral cavity, gel-forming mucins are secreted by the mucous cell phenotype found within mucous glands such as sublingual glands and multiple minor salivary glands. We use rodent sublingual glands as a model system to investigate mucous cell biology (Culp et al. 1996; Luo et al. 2001) and identified the gel-forming mucin, Muc19, as a secretory product of murine sublingual mucous cells (Fallon et al. 2003; Culp et al. 2004). Muc19 is a product of the gene Muc19/Smgc that contains 60 exons and spans ∼105 kb of genomic sequence. Smgc is encoded by exons 1–18, whereas Muc19 transcripts incorporate exon 1 and exons 19–60 (Culp et al. 2004; Zinzen et al. 2004). Exon 1 is therefore shared between the two transcripts and encodes for most of the predicted signal peptide directing each translation product to the secretory pathway.
Although mice are used extensively as a model system to study organ physiology and pathophysiology, little information is currently available regarding the expression of Muc19/Smgc gene products in other murine tissues containing the mucous cell phenotype, including the minor salivary glands that line the oral cavity. We therefore investigated expression of Muc19 transcripts by RT-PCR in oral tissues as well as in tissues from other mucosal organ systems of mice. For comparison to Muc19, we also assessed transcripts for the other gel-forming mucins, Muc2, Muc5ac, Muc5b, and Muc6 (Thornton et al. 2008) in salivary glands and in many other tissues. For those tissues displaying Muc19 transcripts, we performed immunohistochemistry to verify translation and to localize the cellular expression of Muc19 glycoproteins. As a complementary approach, we developed a genetically modified mouse model (i.e., Muc19-EGFP mice) in which the coding region for enhanced green fluorescent protein (EGFP) is positioned in-frame within the 5′ end of the Muc19 coding sequence. Results from these two approaches are consistent, demonstrating a high selectivity for Muc19 and EGFP expression in salivary mucous glands. Expression is also observed in glands of the tracheolarynx and in male accessory bulbourethral glands.
The translation product of full-length Smgc transcripts, SMGC (submandibular gland protein C), was first discovered as a transiently expressed secretion product of newly formed exocrine cells in developing rodent submandibular glands (Ball and Redman 1984) and more recently in developing sublingual glands (Das et al. 2009). Using a polyclonal antibody raised against SMGC isolated from submandibular secretions of neonatal rats, Ball et al. (1988) observed immunoreactivity in mucous cell secretion granules of developing rat parotid glands and in minor glands of the sublingual mucosa. These collective results suggest that SMGC is an early but transiently expressed secretion product of mucous and seromucous acinar cells of developing salivary glands, and may function in cytodifferentiation of these cell types (Das et al. 2009). In the current study, we further evaluated the expression of Smgc transcripts in all major and minor salivary glands and, additionally, tested whether these transcripts are expressed in other glandular and/or mucous cell–containing tissues.
Materials and Methods
Animals and Collection of Tissues
Animal protocols were approved by the institutional animal care and use committees at the University of Cincinnati and the University of Florida, where appropriate. Glands were excised from mice euthanized by exsanguination after carbon dioxide narcosis.
Generation of Muc19-EGFP Knockin Mice
Genomic DNA to create the 5′ and 3′ homologous arms of the targeting vector were generated by PCR of bacterial artificial chromosome (BAC) DNA (clone E11, RPCI-22 BAC Library from 129S6/SvEvTac female mouse; BACPAC Resources, Oakland, CA) using the following primers: 5′ arm, forward primer, 5′-CAAAAGCACACCCTCAGATAACG-3′ and reverse primer, 5′-CAATGTTGAAGTCTCCTCCAGGC-3′; 3′ arm, forward primer, 5′-GCCCTTTGAGCAGGAGCAATG-3′ and reverse primer, 5′-CGAGTGTAAGTGTGCCCTGAAGTTG-3′. The 5′ arm PCR product (5431 bp) was cut with PstI and EaeI (4961-bp final product), and the 3′ arm PCR product was cut with SacI and HindIII (2462-bp final product). The 5′ arm was coupled to EGFP from pEGFP-N3 (BD Biosciences, San Jose, CA; GenBank accession number U57609). A NotI site immediately downstream of the EGFP coding region was first filled in using T4 polymerase followed by ligation of the resultant blunt ends. The 5′ arm plus EGFP with its own stop codon, polyadenylation signal, and SV40 poly A was inserted 5′ to the upstream loxP site of the selection cassette PGKneolox2DTA (Soriano 1997; kindly provided by Dr. Rulang Jiang) that encodes for neomycin under control of the phosphoglycerate kinase promoter. Sequence from the STOP cassette, pBS302 (kindly provided by Dr. Rulang Jiang; GenBank accession number U51223) was inserted immediately 3′ of the downstream loxP site. This cassette was added to help prevent transcriptional read-through to more downstream exons encoding Muc19. The relevant sequences include the C-terminal region (550 bp) of the yeast His3 gene to increase the efficiency of the downstream SV40 polyadenylation signal (825 bp), plus an additional synthetic ATG false translation initiation signal and 5′ splice donor site (Lakso et al. 1992). The 3′ homologous arm was then inserted between the ATG splice donor and the sequence encoding diphtheria toxin A (DTA; also under control of the PGK promoter) for negative selection.
The vector was cloned into Electro 10 Blue cells (Stratagene; La Jolla, CA) and sequenced, and the purified plasmid DNA was linearized with PvuI. Linearized plasmid was electroporated into 129S6/SvEvTac embryonic stem (ES) cells at the University of Cincinnati Gene Targeted Mouse Service. ES cell clones (after G418 selection) were screened initially by PCR using two primer sets to identify appropriately targeted cells. Primer Set 1 targets genomic sequence 43 bp upstream of the 5′ arm (5′-GGCAGTTCACGATTGTCTGTGCAAGC-3′) and within PGKneo (5′-CCTGCGTGCAATCCATCTTGTTCAATGGC-3′). Primer Set 2 targets 39 bp downstream of the 3′ arm (5′-TGCTTCCCTGGACATTGTTCCTTTGC-3′) and within the SV40 polyadenylation signal from pBS302 (5′-GTGCCTTGACTAGAGATCATAATCAGCC-3′). PCR products were verified by direct sequencing from the 5′ and 3′ ends. Positive ES cell clones identified by PCR were further verified for correct vector integration by Southern analysis. Templates for both 5′ and 3′ outside probes (308 bp and 332 bp, respectively) were generated by PCR from genomic DNA using the following primers: 5′ probe (PvuII probe), 5′-TGCCATCTCTAATAGTTTCAGTGC-3′ and 5′-CTCTTGAAGGGGACACCCTAAACC-3′; 3′ probe (SphI probe), 5′-TTCGCGATTCACAAGTGTGCATG-3′ and 5′-GCACATTAATTAGCTTGGAGCAC-3′. PCR reactions were run in Taq PCR Master Mix (Qiagen; Valencia, CA) with the following conditions: 3 min at 98C; 35 cycles of 30 sec at 94C, 30 sec at 60C, and 30 sec at 72C, followed by 3 min at 72C. Southern blots were performed with random primed probes, and genomic DNA was cut with either PvuII or SphI using standard techniques. Appropriate-sized bands were obtained in all cases (not shown).
Three ES cell clones were expanded and injected into C57Bl/6 blastocysts, resulting in a total of 32 chimeric mice. Chimeric males were mated to Black Swiss (NIHBS) females. One male breeder produced 31 agouti heterozygous F1 progeny, from which four breeder pairs were established at the University of Florida and interbred to generate wild-type and heterozygous animals for EGFP expression experiments. Appropriate targeting of these breeders was confirmed by PCR as described above for ES cell lines (not shown). Tail genomic DNA samples (Wizard Genomic DNA Purification Kit; Promega, Madison, WI) from the resultant pups were used for genotyping by PCR with allele-specific primers to generate bands of 595 bp (wild type) and/or 856 bp (targeted allele). Primers used were: wild type forward, 5′-GCTGCTTTGCTTTCAGTTTTGTAGTG-3′; wild type reverse, 5′-GTCTGGCTGGTTTTGTCTCATACTC-3′; EGFP reverse, 5′-TCACCTTGATGCCGTTCTTCTG-3′. PCR reactions were run in the FailSafe PCR buffer system (Epicentre; Madison, WI). Conditions were: buffer F, 3 min at 94C, 30 cycles of 30 sec at 94C, 30 sec at 55C, and 1 min at 72C, followed by 3 min at 72C. Products were resolved in 1.0% agarose gels made in Tris-acetate-EDTA (TAE) buffer with 0.1% ethidium bromide.
RT-PCR Assays
To prepare cDNA, frozen glands were homogenized directly in TRIzol Reagent (Invitrogen; Carlsbad, CA) using a Mini Bead Beater 8 (BioSpec Products; Bartlesville, OK) for 90 sec in the presence of ∼500 mg of silicone carbide beads (1 mm in size). Total RNA was isolated using standard protocols and treated with DNase I using Ambion's DNA-free Reagent Kit (Applied Biosystems; Foster City, CA). Removal of genomic DNA from DNase I–treated RNA (1 μg) was verified by the inability to amplify at 40 cycles the specific 253-bp mouse microsatellite marker, D1Mit46 (UniSTS 116254), using forward primer 5′-AGTCAGTCAGGGCTACATGATG-3′ and reverse primer 5′-CACGGGTGCTCTATTTGGAA-3′. RNA purity was assessed by capillary electrophoresis (Agilent 2100 Bioanalyzer; Agilent Technologies, Inc., Santa Clara, CA). RNA integrity numbers ranged from 8.7 to 9.7. RNA (5 μg) was reverse transcribed with random primers using the high-capacity cDNA archive kit (Applied Biosystems), and the resultant cDNA was purified with the QIAquick PCR purification system (Qiagen; Valencia, CA). RNA and cDNA were quantified using the Quant-iT RNA assay kit or the Quant-iT dsDNA HS assay kit with the Qubit fluorometer (Invitrogen).
Standard conditions for PCR reactions (10–50 ng cDNA) were: 3 min at 94C; 30–37 cycles of 1 min at 94C, 1 min at 55C, and 1 min at 72C, followed by 10 min at 72C. Products were resolved in 3% agarose gels made in TAE buffer with 0.1% ethidium bromide. Transcript-specific primers (Invitrogen) are shown in Table 1. All reactions were run in Qiagen's Taq PCR Master Mix, except for t-Smgc, which incorporated the FailSafe PCR buffer system (Epicentre) as described by Zinzen et al. (2004). PCR products were confirmed by direct sequencing of gel-purified bands (QIAquick Gel Extraction Kit; Qiagen). Digital images of gels were obtained with a Scion Grayscale 1394 digital camera (Fotodyne; Hartland, WI). Primers used are given in Table 1.
Table 1.
Transcript-specific PCR primers
| Transcript | Primer Sequence | Exon | Product (bp) | Reference |
|---|---|---|---|---|
| Muc19 | 5′-GATTATGCGATTGGTTCATCCT-3′ | 46/47 | 349 | Chen et al. 2005 |
| 5′-GTGCAATGTCCCTGAACTCATA-3′ | 60 | |||
| Muc5a/c | 5′-GAGGGCCCAGTGAGCATCTCC3′ | 48 | 361 | Escande et al. 2004 |
| 5′-TGGGACAGCAGCAGTATTCAGT-3′ | 49 | |||
| Muc5b | 5′-TCCTGCTCTGGAATATCCAAG-3′ | 49 | 319 | Escande et al. 2002 |
| 5′-GCCTCGGGGAGCTTGCCTGCC-3′ | 50 | |||
| Muc2 | 5′-TGTGGCCTGTGTGGGAACTTT-3′ | 23 | 558 | Escande et al. 2004 |
| 5′-CATAGAGGGCCTGTCCTCAGG-3′ | 26 | |||
| Muc6 | 5′-TGTGGCTTGTGTGGCAACGCC-3′ | 24 | 569 | Escande et al. 2004 |
| 5′-TGGTCGAAGTACTCATTCTGG-3′ | 27 | |||
| Smgc | 5′-ACAGTCTCTACACTTCGGTCCCA-3′ | 1 | 2,638 | Zinzen et al. 2004 |
| 5′-GGATGACCAGTCACAAACACATC-3′ | 18 | |||
| β-actin | 5′-CACCTTCCAGCAGATGTG-3′ | 6 | 268 | Das et al. 2009 |
| 5′-AAATCCTGAGTCAAAAGCG-3′ | 6 |
DNA Sequencing
Sequencing was performed at the University of Florida Interdisciplinary Center for Biotechnology Research using an Applied Biosystems 3100 Genetic Analyzer ABI and Big Dye version 3.1 chemistry.
Immunohistochemistry
Whole or minced tissues were fixed for 5 hr in 4% paraformaldehyde in PBS and stored at 4C in 1% paraformaldehyde in PBS. Sections (5 μm) were deparaffinized and treated for 10 min in 5% urea containing 50 mM β-mercaptoethanol at 95C to unmask antigenic sites. Sections were then probed with rabbit anti-Muc19 or preimmune serum (1:300 dilution) as described previously (Fallon et al. 2003), except that we used a Vectastain ABC-HRP kit with Vector Red substrate (Vector Laboratories; Burlingame, CA), and counterstained with hematoxylin.
EGFP Fluorescence and Confocal Microscopy
Animals were anesthetized by CO2 inhalation, the superior and inferior vena cava were exposed and cut, and then the vasculature was perfused via the left ventricle for 3 min with PBS containing 1 U/ml heparin at a rate of 5 ml/min. The perfusate was then switched to ice-cold 4% paraformaldehyde in PBS for 20 min. Tissues were excised and incubated in the same fixative for 3 hr with rocking at 4C, washed three times in PBS for 10 min, then embedded in Tissue-Tek OTC Compound (Sakura Finetek; Torrance, CA) and frozen. For brightfield and EGFP fluorescence microscopy, frozen sections (5- or 10-μm) were thawed and mounted in Vectashield Hard Set with 4′,6-diamidino-2-phenylindole (Vector Laboratories). Images were captured with a Micropublisher 5.0 RTV digital camera (Q Imaging; Pleasanton, CA) using QCapture Pro software (v5.01, Q Imaging) on a Leica DM LB2 microscope (Leica Microsystems; Bannockburn, IL) equipped with a 100-W high-pressure mercury lamp and Chroma fluorescent filter cubes (Chroma Technology Corp.; Rockingham, VT).
For confocal images, frozen sections (10-μm) were thawed and incubated for 1 hr at room temperature with the nuclear stain DRAQ5 (Biostatus Limited; Leicestershire, UK) diluted 1:1000 in PBS. After washing (four times for 15 min in PBS), sections were mounted in mowiol. Images were obtained on a spinning disk confocal imaging system consisting of a CSU10 Yokagawa confocal scan head (Yokogawa Corp. of America; Newnan, GA), a Roper Cascade II EMCCD 512b camera (Roper Scientific, GmbH, Ottobrunn Germany), and an ASI X, Y, and piezo Z computer-controlled stage (Applied Scientific Instrumentation; Eugene, OR), with three lasers (491, 561, 638) for three-channel fluorescent imaging attached to a Leica DMIRB inverted microscope (Leica Microsystems). This system is controlled by the open source software package Micro-manager (http://www.micro-manager.org/). Optical slices of 0.2 μm were obtained using 491 nm ex, 520 em, (EGFP) and far-red 638 nm ex, 695 nm em (Draq5) and compiled into a three-dimensional image with ImageJ software (http://rsbweb.nih.gov/ij).
Electron Microscopy
Tissues from wild-type and Muc19-EGFP knockin mice were fixed by vascular perfusion of 2.5% glutaraldehyde in 0.1 M sodium cacodylate buffer, pH 7.4. Subsequent processing was carried out as described in Fallon et al. (2003). Thin sections were examined and photographed in a Philips CM10 transmission electron microscope.
SDS-PAGE, Western Blots, and Lectin Blots
Glandular tissues were excised immediately after death and frozen in liquid nitrogen. Mucosal scrapings were obtained from freshly isolated stomach and colon tissues and processed immediately. Tissue samples were immediately weighed, and 10–80 mg wet weight of tissue was placed in 500 μl of NuPAGE LDS sample buffer (Invitrogen). Samples were then sonicated (four 10-sec pulses at 30-sec intervals) with a Branson Digital Sonifier 250 at 30% amplitude (Branson Ultrasonic Corp.; Danbury, CT), boiled for 10 min, and centrifuged (10,000 × g for 10 min) at 4C. Supernatant aliquots of volumes equivalent to 0.1 to 3.7 mg wet weight of original tissue were applied directly to either 4–12% NuPAGE bis-Tris or 3–8% NuPAGE Tris-acetate gradient gels (Invitrogen). To assay highly glycosylated glycoproteins, gels were stained with Alcian Blue, followed by subsequent silver enhancement of Alcian Blue staining as described previously (Fallon et al. 2003). Silver enhancement of Alcian Blue precedes any staining of proteins (Jay et al. 1990).
To detect SMGC and EGFP, gels were blotted onto polyvinylidene difluoride (PVDF) membranes (Sequi-Blot PVDF membrane, 0.2 μm; Bio-Rad Laboratories, Hercules, CA) and probed overnight (4C) with rabbit anti-mouse SMGC (1:10,000) (Zinzen et al. 2004). For detection of Muc19 and EGFP, gels were transferred overnight onto nitrocellulose (0.2 μm; GE Healthcare, Piscataway, NJ) in transfer buffer (20 mM Tris, 200 mM glycine, 10% methanol) using 250 mAmps of current at 10C. One blot was probed overnight (4C) with rabbit antiserum produced against purified rat sublingual mucin (1:20,000) (Man et al. 1995) and the other blot with rabbit anti-GFP (IgG fraction, 1:2000; Invitrogen). To detect antibodies, we used the WesternBreeze Chemiluminescent Western Blot Immunodetection Kit (Invitrogen) and BioMax Light film (Eastman Kodak Co.; Rochester, NY).
For lectin staining, nitrocellulose blots were probed with biotinylated lectins, either wheat germ agglutinin or peanut agglutinin (PNA) from Vector Laboratories. Blots were incubated for 1 hr in PBS + 0.2% Tween 20 (PBST). For PNA, blots were then treated with 0.1 N H2SO4 at 80C to remove interfering sialic acid residues. Blots were incubated for 1 hr with agitation in biotinylated lectin at 1 μg/ml in 10 mM HEPES, 0.15 M NaCl, 0.1 mM CaCl2, pH 7.4. After washing three times for 5 min in PBS, blots were incubated for 1 hr in alkaline phosphatase–conjugated streptavidin (1:8000) in PBST, washed three times for 10 min in PBST, followed by two 5-min washes in PBS. Lectins were detected using CDP-Star chemiluminescent substrate with Nitro-Block II (Applied Biosystems) and BioMax Light film. Protein standards included MultiMark Multi-Colored and HiMark PreStained standards (Invitrogen). Each experimental condition was performed at least three times. Digital images were obtained with a Scion Grayscale 1394 digital camera (Fotodyne).
Materials
Unless indicated, all chemicals were from Sigma Chemical Co., St. Louis, MO. All kits were used according to the manufacturers' instructions.
Results
Expression of Muc19 Transcripts in Adult Mice
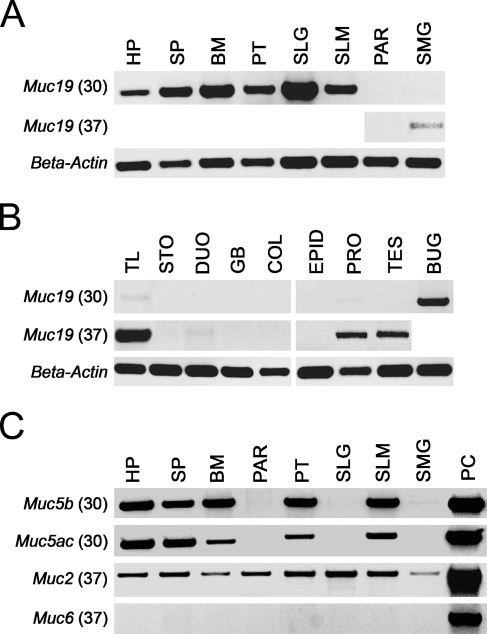
We initially evaluated tissue expression of Muc19 transcripts by RT-PCR. Samples of cDNA (50 ng) from each tissue were first tested in PCR reactions with Muc19-specific primers at 30 cycles. Tissues in which expression was absent or very low were reevaluated at increased sensitivity (i.e., 37 cycles). As shown in Figures 1A and 1B, transcripts are readily detectable in all oral samples that contain the mucous cell phenotype. These samples include the sublingual gland and tissues containing minor salivary glands (hard palate, soft palate, buccal mucosa, posterior tongue, and the sublingual mucosa). Serous parotid glands are negative, whereas seromucous submandibular glands display barely detectable levels. Surprisingly, Muc19 transcripts are readily detected in bulbourethral glands (Cowper's glands), a male accessory mucous gland, and to a lesser extent in tracheolarynx. Transcripts are detected at low levels in prostate glands and testes (Figure 1B), as well as in ovaries/fallopian tubes and uterus (not shown). Transcripts are absent in stomach, duodenum, gall bladder, and colon. Other tissues testing negative, but not shown, include pancreas, cecum, jejunum, ileum, conjunctiva, Harderian glands, lacrimal glands, brain, heart, spleen, urinary bladder, liver, kidney, lung, cervix, and vagina.
Figure 1.
Expression of Muc19 transcripts in mouse tissues. (A) Upper panel: RT-PCR products using 50 ng of random primed cDNA at 30 cycles from major and minor salivary glands. HP, hard palate; SP, soft palate; BM, buccal mucosa; PT, posterior tongue; SLG, sublingual gland; SLM, sublingual mucosa; PAR, parotid gland; SMG, submandibular gland. Middle panel: tissues with no products at 30 cycles were re-tested at 37 cycles. Lower panel: β-actin–positive controls for each tissue sample (30 cycles, 349-bp product). (B) RT-PCR results for non-salivary tissues (upper and middle panels) as described for A. TL, tracheolarynx; STO, stomach mucosa; DUO, duodenum; GB, gall bladder; COL, colon; EPID, epididymis; PRO, prostate; TES, testes; BUG, bulbourethral gland. (C) Expression of transcripts for the gel-forming mucins Muc5b, Muc5ac, Muc2, and Muc6 in salivary tissues. Shown are RT-PCR products using 50 ng of cDNA at either 30 cycles (Muc5b and Muc5ac) or 37 cycles (Muc2 and Muc6). Abbreviations are as given in A. PC, positive control tissue used for each mucin, tracheolarynx (Muc5b), stomach mucosa (Muc5ac and Muc6), and small intestine (Muc2).
We further assayed all salivary tissues, as well as other select tissues, for transcripts to the other four gel-forming mucins, Muc2, Muc5ac, Muc5b, and Muc6. As shown in Figure 1C, all five oral tissues containing minor salivary glands display readily detectable levels of Muc5ac and Muc5b. In all oral tissues, as well as in the three major salivary glands, Muc2 transcripts are detectable by RT-PCR, but only at 37 cycles. All oral tissues and major salivary glands failed to exhibit Muc6 transcripts. Our combined RT-PCR results for all tissues tested are provided in Table 2. Results are expressed as the average relative intensity for each amplicon on a scale of 0 to 5 with respect to selected positive controls.
Table 2.
Tissue distribution of transcripts for gel-forming mucins in adult mice
| Relative amplicon intensity |
||||||
|---|---|---|---|---|---|---|
| Tissue | N | Muc19 | Muc5b | Muc5ac | Muc2 | Muc6 |
| Hard palate | 4 | 2 | 4 | 4 | 0 (1) | 0 (0) |
| Soft palate | 3 | 3 | 4 | 4 | 0 (1) | 0 (0) |
| Buccal mucosa | 3 | 4 | 3 | 2 | 0 (2) | 0 (0) |
| Parotid gland | 3 | 0 (0) | 0 (0) | 0 (1) | 0 (1) | 0 (0) |
| Posterior tongue | 3 | 3 | 4 | 3 | 0 (2) | 0 (0) |
| Sublingual gland | 8 | 5 | 0 (0) | 0 (0) | 0 (2) | 0 (0) |
| Sublingual mucosa | 4 | 4 | 4 | 3 | 0 (1) | 0 (0) |
| Submandibular gland | 4 | 0 (1) | 0 (1) | 0 (1) | 0 (1) | 0 (0) |
| Tracheolarynx | 4 | 1 (4) | 5 | 3 | 1 (4) | 0 (0) |
| Epididymis | 3 | 0 (0) | 3 | 1 (3) | 0 (1) | 0 (2) |
| Prostate | 3 | 0 (2) | 0 (0) | 0 (0) | 0 (ND) | ND |
| Testes | 2 | 0 (2) | 0 (1) | ND | ND | ND |
| Bulbourethral gland | 3 | 4 | 3 | 0 (1) | 0 (0) | 0 (0) |
| Ovaries/fallopian tubes | 4 | 0 (2) | 0 (3) | 1 (3) | 2 | 0 (0) |
| Uterus | 4 | 0 (1) | 0 (4) | 2 | 0 (ND) | 0 (ND) |
| Cervix | 2 | 0 (0) | 0 (3) | ND | ND | ND |
| Vagina | 2 | 0 (0) | 0 (1) | ND | ND | ND |
| Esophagus | 1 | 0 (0) | 0 (4) | ND | ND | ND |
| Gall bladder | 5 | 0 (0) | 0 (3) | 0 (3) | 0 (2) | 0 (3) |
| Stomach mucosa | 2 | 0 (0) | 1 (4) | 5 | 0 (2) | 5 |
| Duodenum | 2 | 0 (0) | 1 (3) | ND | ND | ND |
| Pancreas | 2 | 0 (0) | 0 (4) | ND | ND | ND |
| Cecum | 2 | 0 (0) | 0 (0) | ND | ND | ND |
| Jejunum | 2 | 0 (0) | 0 (0) | ND | ND | ND |
| Ileum | 2 | 0 (0) | 0 (0) | 0 | 4 | 0 (ND) |
| Colon | 2 | 0 (0) | 0 (1) | 1 (3) | 5 | 1 (2) |
| Conjunctiva | 2 | 0 (0) | 3 | 3 | 0 (ND) | 0 (ND) |
| Harderian gland | 2 | 0 (0) | 3 | 5 | 0 (2) | 0 (0) |
| Lacrimal gland | 2 | 0 (0) | 0 (0) | 2 | 0 (1) | 0 (0) |
Expression of amplicons is on a scale of 0 to 5 relative to a positive control tissue (assigned as 5). Positive controls: Muc19, sublingual glands; Muc5b, tracheolarynx; Muc5ac and Muc6, stomach; Muc2, colon. All samples (50 ng random primed cDNA) were initially assayed by PCR for 30 cycles using specific primers. Samples with relative intensities of 0 or 1 were re-assayed at 37 cycles (parentheses). N, number independent RNA/cDNA samples assayed; ND, not determined.
Immunohistochemical Localization of Muc19
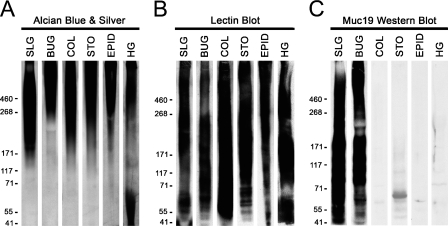
To assess Muc19 localization in murine tissues, we used antiserum derived previously against purified high-molecular-mass mucins from rat sublingual glands (Man et al. 1995). Although this antiserum was demonstrated previously to react with mucous cells and with high-molecular-mass glycoproteins from sublingual glands of mice (Culp et al. 2004; Das et al. 2009), we further determined its specificity for Muc19. We tested in Western blots the reactivity of the antiserum against homogenates from tissues with undetectable Muc19 transcripts but expression of transcripts for one or more of the other gel-forming mucins: Muc2 (colon), Muc5ac (stomach and Harderian glands), Muc5b (epididymis and Harderian glands), and Muc6 (stomach). Tissue samples were homogenized directly in SDS-PAGE sample buffer with sonication and run on low-percentage (3% to 8%) gradient gels. Conditions for sonication were determined empirically not only to homogenize the tissue but also to slightly fragment endogenous high-molecular-mass glycoproteins, to allow for better penetration into the gel as well as for subsequent transfer to nitrocellulose membranes (not shown). Shown in Figure 2A are samples of homogenates after SDS-PAGE and staining with Alcian Blue, followed by subsequent silver enhancement to detect highly glycosylated glycoproteins (Jay et al. 1990). In all cases, highly glycosylated glycoproteins formed a large smear from the top of the gel to about the 171-kDa marker. The transfer of high-molecular-mass glycoproteins to nitrocellulose was verified (Figure 2B) by probing blots with lectins that react with carbohydrate epitopes of gel-forming mucins and other smaller glycoproteins (McGuckin and Thornton 2000). Upon probing identical blots of each homogenate, only high-molecular-mass glycoproteins in sublingual and bulbourethral glands reacted with our antiserum (Figure 2C). Components of higher mobilities also reacted with the antiserum, due mostly to smaller peptides or glycopeptides released during sonication, based upon results of initial preliminary experiments (not shown).
Figure 2.
Testing the specificity against gel-forming mucins of antiserum raised against purified rat Muc19. Homogenates from select mouse tissues that express Muc19 and/or other gel-forming mucins were subjected to SDS-PAGE (3–8% gradient gels). One gel (A) was stained directly with Alcian Blue and subsequent silver enhancement to detect highly glycosylated glycoproteins. A second gel (B) was blotted, and lanes were probed with either wheat germ agglutinin (BUC, COL, STO) or peanut agglutinin (SLG, EPID, HG) to verify transfer of high-molecular-mass glycoproteins. A third gel (C) was subjected to Western blotting using rabbit anti-rat sublingual mucin. Lanes were loaded with homogenates from the following original wet weight of tissue: 0.4 mg sublingual gland (SLG), 0.5 mg bulbourethral glands (BUG), 1.4 mg colon mucosa (COL), 2.9 mg stomach mucosa (STO), 0.9 mg epididymis (EPID), and 3.7 mg Harderian glands (HG). In all three cases (A–C), samples were either stained or developed simultaneously for the same period of time.
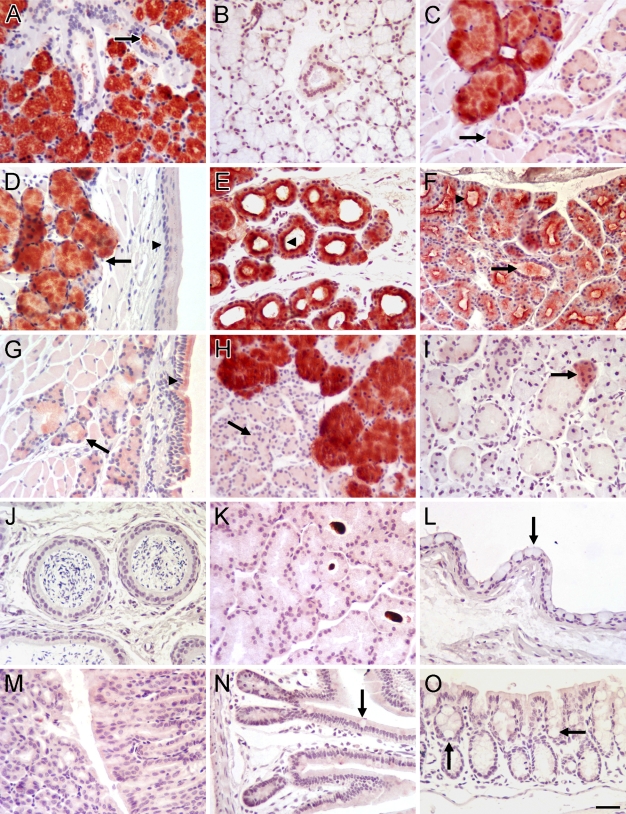
In paraffin sections of different tissues, our anti-Muc19 reacts intensely with salivary mucous cells of sublingual glands, posterior tongue, soft palate, hard palate, and buccal mucosa (Figures 3A and 3C–3F). Mucous cells in the bulbourethral glands also stain strongly (Figure 3H). Interestingly, very light staining of surface goblet cells and glandular mucous cells is observed within the nasal mucosa of the soft palate (Figure 3G). Submandibular glands contain a sparse population of small groups of lightly stained acinar cells (Figure 3I), consistent with our RT-PCR results and previous results in rats (Man et al. 1995). Non-reactive tissues include epididymis, Harderian glands, conjunctival epithelium, stomach mucosa, ileum, and colon (Figures 3J–3O).
Figure 3.
Immunohistochemical localization of Muc19 in mouse tissues. (A) Sublingual gland; all mucous cells are reactive. Arrow, ductal lumen. (B) Sublingual gland probed with preimmune serum. (C) Posterior tongue; mucous cells are reactive. Arrow, unstained serous acini. (D) Soft palate: oral mucosa; arrow, stained mucous cells; arrowhead, oral epithelium. (E) Hard palate; mucous cells are reactive. Arrowhead, intense staining of apical cytoplasm of mucous cells lining ducts. (F) Minor mucous glands within buccal mucosa; arrow, stained secretions within ductal lumen; arrowhead, apical cytoplasmic staining of mucous tubules. (G) Soft palate: nasal mucosa; arrow, light staining of mucous cell; arrowhead, light staining within ciliated epithelial cells. (H) Bulbourethral gland; mucous cells are reactive. Arrow, unstained serous acini. (I) Submandibular gland; granular convoluted ducts and most seromucous acini are unreactive. Arrow, a few acini are stained. (J) Epididymis. (K) Harderian gland. (L) Conjunctiva epithelium; arrow, surface mucous goblet cells. (M) Stomach mucosa in cross-section (left) and longitudinal-section (right). (N) Ileum; arrow, goblet cell. (O) Colon; arrows, mucous goblet cells within crypts. Paraffin sections (5 μm) were probed with rabbit anti-rat sublingual mucin antiserum at dilutions of 1:1000 (A,C,D,G,H) and 1:300 (B,E,F,I–O). Vectastain ABC-HRP Kit and Nova Red substrate with light hematoxylin counterstain. Bar in O = 20 μm for all panels.
Tissues From Muc19-EGFP Knockin Mice to Evaluate Muc19 Expression
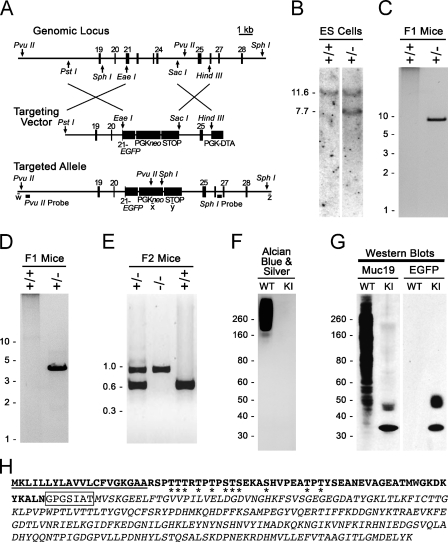
As a complementary approach to assess tissue expression of Muc19, we created a knockin mouse model in which EGFP was inserted in-frame within exon 21 of the gene Muc19/Smgc. Shown in Figure 4A is a diagram of our strategy for the targeted disruption of Muc19 transcripts. Exon 21 represents the fourth exon utilized for Muc19 transcripts. As a result, chimeric transcripts encode only the first 69 residues of Muc19, including the signal peptide that directs translation products to the secretory pathway. Moreover, the upstream genomic region encoding Smgc (exons 1–18) is preserved. Shown in Figure 4B are results of a Southern blot of genomic DNA from two different ES cell colonies, indicating non-recombinant and correctly targeted cells. Figures 4C and 4D are PCR results from two F1 agouti mice indicating correct germline transmission in one animal of the 3′ end (Figure 4B) and 5′ end (Figure 4C) of the targeted allele. Heterozygous F1 agouti mice were interbred to produce all three genotypes. Our genotyping strategy and typical results are given in Figure 4E. In Figure 4F are results of SDS-PAGE of sublingual gland homogenates from wild-type (+/+) and homozygous Muc19-EGFP knockin (−/−) mice demonstrating the absence of high-molecular-mass glycoproteins in knockin mice. In Western blots of sublingual gland homogenates (Figure 4G), our anti-Muc19 antibody primarily recognizes two bands at ∼33 kDa and 48 kDa in the sample from knockin mice. These same two bands are recognized in the Muc19-EGFP homogenate by anti-EGFP, whereas the wild-type homogenate is non-reactive. The translated Muc19-EGFP protein is calculated to be 32.8 kDa after cleavage of the signal peptide (residues 1–20), as predicted by the program SignalP 3.0 (Bendtsen et al. 2004). The band of faster mobility recognized by both anti-Muc19 and anti-EGFP probably represents the Muc19-EGFP protein without posttranslational modifications. As shown in Figure 4H, the Muc19-EGFP protein contains eleven putative sites for O-glycosylation that lie within the Muc19 domain (NetOGlyc 3.1; Julenius et al. 2005). In contrast, the program NetNGlyc 1.0 (http://www.cbs.dtu.dk/services/NetNGlyc/) does not recognize putative sites for N-glycosylation. The 48-kDa band is therefore most likely to be an O-glycosylated variant of Muc19-EGFP.
Figure 4.
Production and initial characterization of Muc19-EGFP knockin mice. (A) Strategy for targeted disruption of Muc19 transcripts and insertion of enhanced green fluorescent protein (EGFP) in-frame within exon 21 of gene Muc19/Smgc (the fourth exon utilized for Muc19 transcripts). The targeted genomic locus contains gene exons 19–28. The targeting vector contains EGFP coding sequence (in-frame with the Muc19 coding sequence of exon 21), neomycin under control of the phosphoglycerate kinase promoter (PGKneo), and a STOP cassette to further prevent transcriptional read-through. The PGKneo sequence is flanked by loxP sites (not shown). The homology arms are defined by restriction sites PstI/EaeI (5′ arm) and SacI/HindIII (3′ arm). Diphtheria toxin A (DTA) under control of the PGK promoter is for negative selection in embryonic stem (ES) cells. The targeted EGFP knockin recombinant allele is shown with restriction sites (PvuII and SphI) and probes used for Southern blot analyses as well as sites of PCR primers w–z. See Materials and Methods for details. (B) Southern blot analysis of SphI-digested DNA from ES cells indicates non-recombinant cells (+/+) and cells correctly targeted at the 3′ end of the locus (+/−). (C) PCR-based genotyping of F1 agouti mice to distinguish the absence (+/+) or presence (+/−) of germline transmission and correct insertion of the 3′ end of the targeted allele using primers w and x (as shown in A). (D) Same as in C, but using primers y and z to detect germline transmission and correct insertion of the 5′ end of the targeted allele. (E) Genotyping of progeny from intercrossing F1-recombinant mice. The PCR reaction includes one forward primer within intron 20 and two reverse primers, one in intron 21 (595-bp wild-type allele) and the other within EGFP (856-bp targeted allele). (F) Sublingual gland homogenates (150 μg wet weight) from wild-type (WT) and homozygous Muc19-EGFP knockin (KI) mice subjected to SDS-PAGE (4–12% gradient gel) and stained with Alcian Blue and subsequent silver enhancement to detect highly glycosylated glycoproteins. (G) Homogenates as in F were run on a similar gel, blotted, and probed with either anti-rat Muc19 (Muc19) or anti-GFP (EGFP). Loaded homogenates were 150 (Muc19) and 400 μg wet weight (EGFP). (H) Predicted translation of the Muc19-EGFP fusion construct. Residues in bold are from Muc19, boxed residues are encoded by the polylinker, and italic residues are EGFP. Predicted signal peptide is underlined. Asterisks denote predicted potential sites of O-glycosylation.
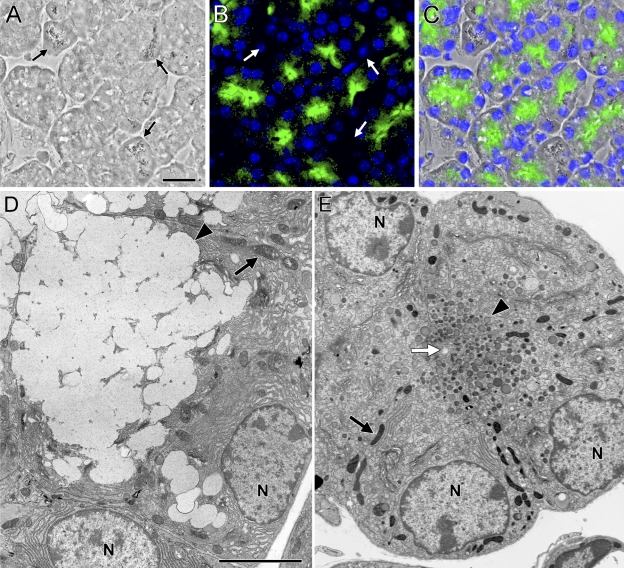
In sublingual sections from homozygous Muc19-EGFP knockin mice, EGFP fluorescence is localized to the apical regions of nearly all tubular mucous cells but is absent in serous demilune cells (Figures 5A–5C). The conversion in expression of native Muc19 to Muc19-EGFP results in a marked transition from the typical mucous cell ultrastructure (i.e., a cytoplasm packed with large electron-lucent secretion granules; Figure 5D) to cells with small electron-dense granules in the apical cytoplasm (Figure 5E). Serous demilune cells appear unaltered, containing abundant rough endoplasmic reticulum and a few serous granules (not shown).
Figure 5.
Muc19-EGFP fluorescence in sublingual mucous cells from homozygous Muc19-EGFP knockin mice and comparison of cell ultrastructure to wild-type acinar cells. (A–C) Cryosection (5 μm) of a sublingual gland from a homozygous Muc19-EGFP knockin mouse shown under brightfield (A) and fluorescent (B) microscopy, with an overlay image shown in C. Nuclei were counterstained with 4′,6-diamidino-2-phenylindole (DAPI) (blue). EGFP fluorescence is predominant within the apical cytoplasm of tubular mucous cells, whereas punctate fluorescence is sparsely scattered within the more-basal regions. The larger granules of serous demilune cells are readily visible under brightfield microscopy, but these cells do not exhibit EGFP fluorescence (arrows, A,B). (D) Transmission electron microscopy (TEM) of a sublingual acinar mucous cell from an 8-week-old male wild-type mouse. Typical mucous cell structure is displayed, including large electron-lucent secretory granules (arrowhead) filling the apical cytoplasm, a basal nucleus (N), and large mitochondria (arrow). (E) TEM of sublingual mucous cells from an age- and sex-matched homozygous Muc19-EGFP knockin mouse. Mucous cells surrounding a small lumen (white arrow) contain small, dense apical secretory granules (arrowhead). Nuclei (N) are basally oriented, but Golgi complexes are more prominent in the absence of large mucous granules. Mitochondria, arrow. Bars: A = 20 μm; D = 5 μm for D,E.
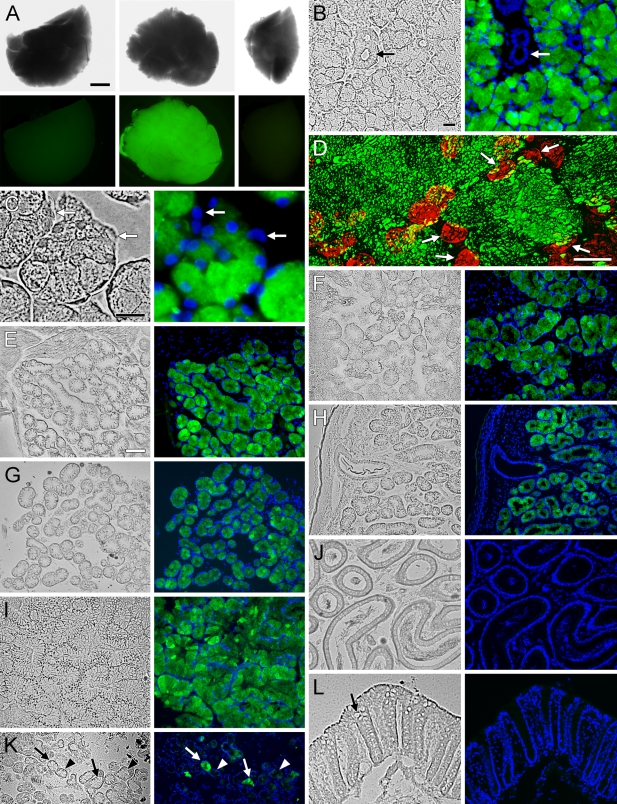
To evaluate tissue expression of Muc19-EGFP, we used heterozygous mice to preserve the mucous cell phenotype and thus maintain normal tissue morphology. As shown in Figure 6A, a uniform and marked green fluorescence is observed in whole sublingual glands (lower center panel) but not submandibular glands (lower right panel) from heterozygous knockin mice. As expected, sublingual glands from wild-type mice are non-fluorescent (lower left panel). In sections of sublingual glands, EGFP fluorescence is prominent in mucous cells but is absent in ductal structures (Figure 6B) and in serous demilune cells (Figure 6C). Fluorescence within mucous cells is restricted to granular structures of irregular shapes and sizes distributed throughout the cytoplasm, consistent with localization to mucous secretory granules (Figure 6D). EGFP fluorescence displays the same pattern of tissue expression as Muc19 immunoreactivity. Mucous cells of minor salivary glands are all fluorescent, including those within the sublingual mucosa (Figure 6E), posterior tongue (Figure 6F), hard palate (Figure 6G), and soft palate (Figure 6H). EGFP fluorescence is also observed throughout the mucous cell population of bulbourethral glands (Figure 6I). Mucous cells within submucosal glands of the tracheolarynx are highly inconsistent, with some cells devoid of fluorescence and others exhibiting very low intensity (Figure 6K). EGFP fluorescence is absent in samples of epididymis (Figure 6J) and colon (Figure 6L), as well as in stomach, Harderian glands, and mucous cells of the nasal airways within the soft palate (not shown).
Figure 6.
EGFP fluorescence in tissues of heterozygous Muc19-EGFP knockin mice. (A) Glands from a wild-type mouse (left panels) and a heterozygous Muc19-EGFP mouse (center and right panels) shown under brightfield (upper panels) and fluorescent (lower panels) microscopy. Center panels are each of a whole sublingual gland, whereas the right panels are of a quarter portion from an adjoining submandibular gland. (B–L) Cryosections (5-μm) from heterozygous Muc19-EGFP mice. Tissues were initially fixed via vasculature perfusion with 4% paraformaldehyde in PBS. In all cases except panel D, sections were counterstained with DAPI (blue) and both brightfield (left panels) and fluorescent (right panels) images are shown. (B) All mucous cells of a sublingual gland are fluorescent, whereas ductal cells are negative (arrows). (C) Sublingual gland serous demilune cells (arrows) do not display EGFP fluorescence. (D) Confocal z-projection of 40 optical slices (0.2-μm) through a 10-μm frozen section of a gland. Nuclei are stained red with DRAQ5. White arrows indicate basal nuclei of individual mucous cells of a single acinus. EGFP fluorescence is localized to irregularly-shaped granular structures throughout the cytoplasm of mucous cells. (E–H) EGFP fluorescence within mucous cells of minor salivary mucous glands of the sublingual mucosa (E), posterior tongue (F), hard palate (G), and soft palate (H). (I) Bulbourethral gland with EGFP fluorescence within mucous cells. (J) Epididymis. (K) Submucosal glands in tracheolarynx. Some mucous cells display bright EGFP fluorescence (arrows), whereas others exhibit non- or very low–fluorescent mucous cells (arrowheads). (L) Colon. Arrow in left panel indicates a mucous cell. Bars: A = 1 mm; B,C = 15 μm; D = 7 μm; E = 70 μm for E–L.
Mucous Cell Phenotype in Homozygous Knockin Mice and Expression of Additional Gel-forming Mucins
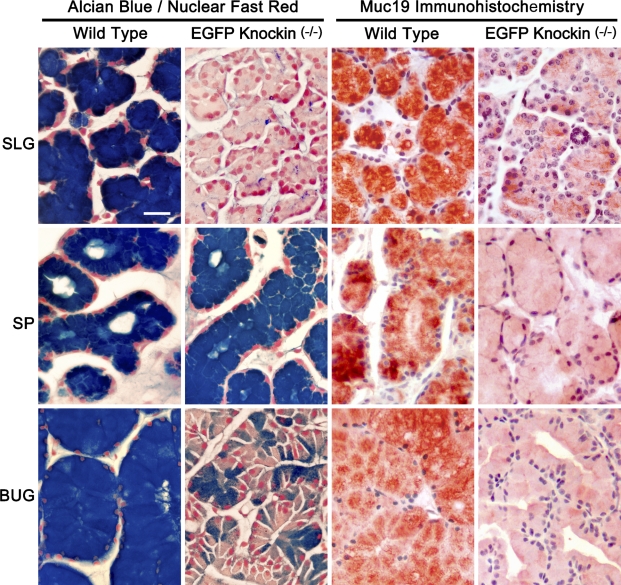
As described above for Figure 5, the absence of expression of gel-forming mucins in sublingual glands results in mucous cells without a cytoplasm filled with large mucin secretion granules. In contrast to sublingual glands, RT-PCR results presented above (Table 2) suggest that mucous cells of the minor salivary glands express the gel-forming mucins Muc5b and/or Muc5ac, in addition to Muc19. Moreover, bulbourethral glands may express Muc5b, given the abundance of its transcripts. We therefore reasoned that if mucous cells of the glands from homozygous knockin mice express gel-forming mucins in addition to Muc19, then these cells would maintain the mucous cell phenotype. In control experiments with sublingual glands from homozygous knockin mice (Figure 7), we find the expected absence of intense staining for acidic mucosubstances with Alcian Blue and only sparse anti-Muc19 immunoreactivity, probably due to Muc19-EGFP fusion proteins. Minor salivary mucous glands of homozygous knockin mice, on the other hand, maintain a large cytoplasm with strong Alcian Blue staining, but display extremely light anti-Muc19 immunoreactivity. Shown in Figure 7 is an example from salivary mucous glands within the soft palate. Similar results (not shown) are found in mucous glands of the posterior tongue and the hard palate. In addition, the electron microscopic appearance of mucous glands of the soft palate is similar in wild-type and homozygous knockin mice, with typical electron-lucent mucous granules filling the apical cytoplasm (not shown). Interestingly, mucous cells of bulbourethral glands of knockin mice are noticeably smaller in size than wild-type glands and have moderate to little cytoplasmic Alcian Blue staining or anti-Muc19 immunoreactivity.
Figure 7.
Comparisons of Alcian Blue staining with Nuclear Fast Red counterstain and anti-Muc19 immunohistochemistry in paraffin sections (7-μm) of mucous glands from wild-type and homozygous Muc19-EGFP knockin mice. SLG, sublingual gland; SP, soft palate salivary glands; BUG, bulbourethral gland. Bar = 20 μm.
Expression of Smgc in Neonatal and Adult Mice
As shown in Figure 8A, we find the restricted expression of Smgc transcripts in neonatal tissues also expressing Muc19 transcripts. Full-length Smgc transcripts (i.e., 2.6-kb band) are detected in all Smgc-expressing tissues except buccal mucosa and bulbourethral glands. Not shown is the absence of Smgc transcripts in other neonatal tissues, including ileum, lacrimal glands, ovaries/fallopian tubes, uterus, cervix, and vagina. Full-length Smgc transcripts are absent in adult tissues, whereas the dominant species is the smallest splice variant, t-Smgc, encoded by exons 1, 17, and 18 (Figure 8B; Das et al. 2009). All tissues negative for Smgc transcripts in neonates are also negative in adults (not shown). To determine whether transcripts in adult tissues are translated, we probed tissue homogenates by Western blot using anti-SMGC raised against His6 fusion proteins encoded by exons 3–18 (Zinzen et al. 2004). Immunoreactive proteins are detectable only at extremely low levels (Figure 8C).
Figure 8.
Comparative expression of Smgc and Muc19 in murine tissues. (A) Expression in 3-day-old mice of Smgc and Muc19 transcripts in select salivary, digestive, reproductive, and ocular tissues. Upper panel: RT-PCR of 10 ng of random primed cDNA for 35 cycles using primers to exons 1 and 18 of Smgc. The upper band (2637 bp) represents full-length Smgc, whereas the lower band (832 bp) represents the small splice variant, t-Smgc. Middle panel: RT-PCR products from the same tissue cDNA samples, except after 30 cycles and with primers to the 3′ end of Muc19 (349-bp product). Lower panel: β-actin positive controls for each tissue sample (30 cycles, 349-bp product). Products were run on agarose gels of 1.0% (Smgc) and 1.5% (Muc19 and β-actin). SMG, submandibular gland; SLG, sublingual gland; PAR, parotid gland; PT, posterior tongue; SLM, sublingual mucosa; BM, buccal mucosa; HP, hard palate; BUG, bulbourethral gland; TL, tracheolarynx; STO, stomach mucosa; COL, colon; HAR, Harderian gland; CON, conjunctiva; PAN, pancreas; PRO, prostate. Left side of upper panel, mobilities of 1-kb ladder. Results are representative of three separate preparations of each tissue. (B) Tissue expression of Smgc and Muc19 transcripts under the same conditions as in A, but with tissues from 6- to 8-week-old mice. Tissues that displayed no detectable Smgc or Muc19 transcripts in 3-day-old mice, as shown in A, were also negative in adult tissues (not shown). All labeling is the same as in A. Results are representative of three to four separate preparations of each tissue. (C) Western blot of SMGC in whole-tissue homogenates run on a 4%–12% SDS-PAGE gel. Far left lane (asterisk) contains the equivalent of 10 μg wet weight of 3-day-old sublingual glands as a positive control for full-length SMGC (∼105 kDa). Remaining lanes contain 300 μg wet weight of tissues from mice 6 to 8 weeks old. Tissue labels are as in A. Mobilities of molecular mass markers (kDa) are shown on the left. Results are representative of two separate preparations of each tissue.
Discussion
From our combined analyses of adult mice, it is apparent that Muc19 is highly expressed in oral tissues containing minor salivary mucous glands and major sublingual glands. Submucosal glands of the tracheolarynx and bulbourethral glands represent the only other sites readily expressing Muc19. Although transcripts were detected in prostate, testis, ovaries/fallopian tubes, and uterus, the levels were too low to warrant further investigation. A human gene encoding MUC19 has been identified and the 3′ end of MUC19 transcripts cloned (Chen et al. 2005). MUC19 transcripts were initially localized to mucous cells of the submandibular glands and to tracheal submucosal glands (Chen et al. 2005), consistent with our findings in mice. Muc19 in mice airways appears restricted to submucosal glands of the proximal lower airways, inasmuch as we find very faint Muc19 immunoreactivity in the upper nasal airways of the soft palate. Moreover, Young et al. (2007) barely detected Muc19 transcripts in distal airways of the lung. Interestingly, we find that not all glandular mucous cells of the tracheolarynx express Muc19, suggesting highly localized processes controlling gene expression.
Recently, MUC19 proteins were immunolocalized to mucous cells of human middle ear epithelium (Kerschner et al. in press), a tissue we did not test. Yu et al. (2008) described the coexpression of MUC19 and MUC5AC in cells of the conjunctival epithelia and lacrimal glands of man, whereas MUC19, but not MUC5AC, is expressed in the corneal epithelium. We do not find Muc19 expression in conjunctiva or in lacrimal and Harderian glands, suggesting species differences and possibly a broader tissue distribution of MUC19 in man than in mice. In screening tissues of the human male urogenital tract, Russo et al. (2006) were unable to determine the source of MUC5B glycoproteins in seminal fluid. Transcripts for MUC5B as well as MUC19 were absent in all tissues analyzed, including epididymis, prostate, and testes (Russo et al. 2006). We find comparable results in mice, except for expression of Muc5b transcripts in epididymis. The recent results of Piludu et al. (2009) suggest that MUC5B glycoproteins are probably contributed by bulbourethral glands, a tissue not tested by Russo et al. (2006). Piludu et al. (2009) localized anti-MG1 reactivity to bulbourethral mucous cells. MG1 is the high-molecular-mass mucin fraction from whole saliva and is rich in MUC5B (Rousseau et al. 2008). Our combined results with murine bulbourethral glands demonstrate that Muc19 is a major mucous cell product, but that another mucin species is also produced, presumably Muc5B. It is thus conceivable this gland contributes MUC19 to human seminal fluid.
In evaluating the contribution of Muc19 to saliva, Rousseau et al. (2008) detected Muc19 peptides by mass spectrometry in stimulated saliva from multiple mammalian species (i.e., rat, cow, pig, horse). Conversely, tryptic peptides from the predicted 5′ end of human MUC19 were not detected in pooled unstimulated human saliva from six donors. These provocative results question whether MUC19 transcripts in salivary glands are indeed translated, although it is difficult to reconcile this possibility with the apparent translation of transcripts in other tissues. Alternatively, MUC19 may not contribute significantly to the unstimulated saliva fraction collected by simple drooling. It may instead preferentially coat the hard and soft oral tissues, or it may be released into saliva primarily upon stimulation, as supported by its presence in stimulated saliva samples from other mammalian species.
To test the mucin specificity of our antiserum raised against purified rat sublingual mucin, we initially utilized stomach, colon, Harderian glands, and epididymis as control tissues. Stomach expresses predominantly Muc5ac and Muc6 transcripts, whereas colon expresses transcripts for Muc2, consistent with humans (Herrmann et al. 1999; Nordman et al. 2002) and prior studies with mice (Van der Sluis et al. 2006; Phillipson et al. 2008). Both Muc5ac and Muc5b transcripts are expressed in Harderian glands. These glands are mostly absent in man (Rehorek and Smith 2006), although in human lacrimal glands, MUC5AC is localized to ductal goblet cells and MUC5B to acinar cells (Paulsen et al. 2004). Our antiserum does not react against these control tissues, as well as conjunctival epithelium and ileum. It is selective for mucous cells and displays the same pattern of tissue and cell expression as EGFP in Muc19-EGFP heterozygous mice. These collective results validate the specificity of the antiserum for Muc19 mucins and further indicate a high similarity between rat and mouse Muc19 glycoproteins. In addition, we previously reported that antiserum reactivity is markedly diminished by periodate oxidation of mucins, indicating that carbohydrate components represent a major proportion of antigenic epitopes (Man et al. 1995). These carbohydrate structures (i.e., oligosaccharides) are apparently unique to Muc19, given the absence of antiserum reactivity against other mucins. Muc19, via its unique oligosaccharides, may therefore function in specific protective roles.
The Muc19-EGFP mouse model system expresses the fusion protein in a pattern consistent with Muc19 transcripts and anti-Muc19 immunoreactivity. The fusion protein is localized to exocrine granules and may therefore serve as a marker, not only for Muc19 expression but also for exocrine secretion by mucous cells. Furthermore, the absence of Muc19 expression in sublingual glands of Muc19-EGFP knockin mice has a striking but predictable effect on mucous cell morphology and Alcian Blue staining, given the uniqueness of these cells in only expressing a single gel-forming mucin. In minor salivary glands, Muc19 expression is apparent throughout the entire population of mucous cells; yet these cells in Muc19-EGFP knockin mice continue to stain intensely with Alcian Blue. Because minor glands express significant amounts of Muc5b and/or Muc5ac transcripts, one or both of these mucins are probably responsible for Alcian Blue staining in Muc19-EGFP knockin mice. The mucin content of mouse saliva may, therefore, more closely mimic human saliva, in which MUC5B is clearly present (Rousseau et al. 2008).
As presented earlier in this study, we hypothesized that SMGC may function in the cytodifferentiation of mucous and seromucous acinar cells. Our current results indicate that Smgc transcripts are localized to tissues expressing Muc19, but not to other neonatal or adult tissues containing mucous cells. In addition, full-length Smgc transcripts are restricted to neonatal tracheolarynx and salivary tissues. SMGC expression is associated with the presence of full-length transcripts (Zinzen et al. 2004; Das et al. 2009), whereas small t-Smgc transcripts are not translated in sublingual glands (Das et al. 2009). In the current study, SMGC proteins are barely detectable in adult tissues in which full-length transcripts are absent and smaller variants in addition to t-Smgc are detected. Hence, the larger incomplete splice variants are not translated or are translated poorly, and may result from inefficient splicing during production of Muc19 mRNA. In support of this concept is the absence of Smgc transcripts in adult parotid glands, which do not express Muc19 or contain mucous acinar cells (Ball et al. 1988). The restricted expression of full-length transcripts to neonatal tracheolarynx and salivary tissues suggests that transient secretory tubule cells expressing SMGC are present during early development of these tissues, as described for sublingual and submandibular glands (Zinzen et al. 2004; Das et al. 2009). SMGC may therefore have a more-limited or tissue-specific function in the cytodifferentiation of mucous/seromucous secretory cells, or, as proposed by Zinzen et al. (2004), it may provide anti-bacterial protection in neonates. Furthermore, it is unknown whether a similar protein is expressed in man.
Our collective results on the tissue-specific expression of Muc19 prompt speculation as to its function. The oral cavity is a major site of exposure to environmental pathogens and toxins, in addition to dietary components. A balance between host, microbial, dietary, and environmental factors is responsible for the selective establishment and maintenance of resident flora associated with the soft and hard tissues of the mouth. Subsequent invasion by more pathogenic microbial species is therefore limited by the combined actions of the resident flora and salivary constituents. Muc19 may interact with oral bacteria either to help select the resident oral flora and/or to exclude more pathogenic species. Muc19 may play a similar role in the tracheolarynx by entrapping bacteria for eventual clearance via the mucociliary escalator (Knowles and Boucher 2002). This region of the airways is probably exposed to oral bacteria through the accidental aspiration of saliva that can occur during eating or gagging. Muc19 may further function in the hydration and lubrication of the oral surfaces. Its presence in seminal fluid may also promote lubrication, although a role in bacterial interaction cannot be ruled out.
Acknowledgments
This study was supported by the National Institutes of Health through a RO1 grant (DE014730) to DJC.
The authors thank Sara Shah and Maya Yankova for excellent technical assistance.
This article is distributed under the terms of a License to Publish Agreement (http://www.jhc.org/misc/ltopub.shtml). JHC deposits all of its published articles into the U.S. National Institutes of Health (http://www.nih.gov/) and PubMed Central (http://www.pubmedcentral.nih.gov/) repositories for public release twelve months after publication.
References
- Ball WD, Hand AR, Moreira JE, Johnson AO (1988) A secretory protein restricted to type I cells in neonatal rat submandibular glands. Dev Biol 129:464–475 [DOI] [PubMed] [Google Scholar]
- Ball WD, Redman RS (1984) Two independently regulated secretory systems within the acini of the submandibular gland of the perinatal rat. Eur J Cell Biol 33:112–122 [PubMed] [Google Scholar]
- Bendtsen JD, Nielsen H, von Heijne G, Brunak S (2004) Improved prediction of signal peptides: SignalP 3.0. J Mol Biol 340:783–795 [DOI] [PubMed] [Google Scholar]
- Chen J, Sun M, Hurst LD, Carmichael GG, Rowley JD (2005) Genome-wide analysis of coordinate expression and evolution of human cis-encoded sense-antisense transcripts. Trends Genet 21:326–329 [DOI] [PubMed] [Google Scholar]
- Culp DJ, Latchney LR, Fallon M, Denny PA, Couwenhoven PC, Sally RI, Chuang S (2004) The gene encoding mouse Muc19: cDNA, genomic organization and relationship to Smgc. Physiol Genomics 19:303–318 [DOI] [PubMed] [Google Scholar]
- Culp DJ, Luo W, Richardson LA, Watson GE, Latchney LR (1996) Both M1 and M3 receptors regulate exocrine secretion by mucous acini. Am J Physiol 271:C1963–1972 [DOI] [PubMed] [Google Scholar]
- Das B, Cash MN, Hand AR, Shivazad A, Culp DJ (2009) Expression of Muc19/Smgc gene products during murine sublingual gland development: cytodifferentiation and maturation of salivary mucous cells. J Histochem Cytochem 57:383–396 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Escande F, Porchet N, Aubert JP, Buisine MP (2002) The mouse Muc5b mucin gene: cDNA and genomic structures, chromosomal localization and expression. Biochem J 363:589–598 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Escande F, Porchet N, Bernigaud A, Petitprez D, Aubert JP, Buisine MP (2004) The mouse secreted gel-forming mucin gene cluster. Biochim Biophys Acta 1676:240–250 [DOI] [PubMed] [Google Scholar]
- Fallon MA, Latchney LR, Hand AR, Johar A, Denny PA, Georgel PT, Denny PC, et al. (2003) The sld mutation is specific for sublingual salivary mucous cells and disrupts apomucin gene expression. Physiol Genomics 14:95–106 [DOI] [PubMed] [Google Scholar]
- Herrmann A, Davies JR, Lindell G, Martensson S, Packer NH, Swallow DM, Carlstedt I (1999) Studies on the “insoluble” glycoprotein complex from human colon. Identification of reduction-insensitive MUC2 oligomers and C-terminal cleavage. J Biol Chem 274:15828–15836 [DOI] [PubMed] [Google Scholar]
- Jay GD, Culp DJ, Jahnke MR (1990) Silver staining of extensively glycosylated proteins on sodium dodecyl sulfate-polyacrylamide gels: enhancement by carbohydrate-binding dyes. Anal Biochem 185:324–330 [DOI] [PubMed] [Google Scholar]
- Julenius K, Molgaard A, Gupta R, Brunak S (2005) Prediction, conservation analysis, and structural characterization of mammalian mucin-type O-glycosylation sites. Glycobiology 15:153–164 [DOI] [PubMed] [Google Scholar]
- Kerschner JE, Khampang P, Erbe CB, Kolker A, Cioffi JA (In Press) Mucin gene 19 (MUC19) expression and response to inflammatory cytokines in middle ear epithelium. Glycoconj J. Published online June 17, 2009 (DOI: 10.1007/s10719-009-9245-x) [DOI] [PMC free article] [PubMed]
- Knowles MR, Boucher RC (2002) Mucus clearance as a primary innate defense mechanism for mammalian airway. J Clin Invest 109:571–577 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lakso M, Sauer B, Mosinger B Jr, Lee EJ, Manning RW, Yu SH, Mulder KL, et al. (1992) Targeted oncogene activation by site-specific recombination in transgenic mice. Proc Natl Acad Sci USA 89:6232–6236 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luo W, Latchney LR, Culp DJ (2001) G-protein coupling to M1 and M3 muscarinic receptors in sublingual glands. Am J Physiol Gastrointest Liver Physiol 280:C884–896 [DOI] [PubMed] [Google Scholar]
- Man YG, Ball WD, Culp DJ, Hand AR, Moreira JE (1995) Persistence of a perinatal cellular phenotype in submandibular glands of adult rat. J Histochem Cytochem 43:1203–1215 [DOI] [PubMed] [Google Scholar]
- McGuckin MA, Thornton DJ (2000) Detection and quantitation of mucins using chemical, lectin, and antibody methods. Methods Mol Biol 125:45–55 [DOI] [PubMed] [Google Scholar]
- Nordman H, Davies JR, Lindell G, de Bolos C, Real F, Carlstedt I (2002) Gastric MUC5AC and MUC6 are large oligomeric mucins that differ in size, glycosylation and tissue distribution. Biochem J 364:191–200 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paulsen F, Langer G, Hoffmann W, Berry M (2004) Human lacrimal gland mucins. Cell Tissue Res 316:167–177 [DOI] [PubMed] [Google Scholar]
- Phillipson M, Johansson ME, Henriksnas J, Petersson J, Gendler SJ, Sandler S, Persson AE, et al. (2008) The gastric mucus layers: constituents and regulation of accumulation. Am J Physiol Gastrointest Liver Physiol 295:G806–812 [DOI] [PubMed] [Google Scholar]
- Piludu M, Hand AR, Cossu M, Piras M (2009) Immunocytochemical localization of MG1 mucin in human bulbourethral glands. J Anat 214:179–182 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rehorek SJ, Smith TD (2006) The primate Harderian gland: does it really exist? Ann Anat 188:319–327 [DOI] [PubMed] [Google Scholar]
- Rousseau K, Kirkham S, Johnson L, Fitzpatrick B, Howard M, Adams EJ, Rogers DF, et al. (2008) Proteomic analysis of polymeric salivary mucins: no evidence for MUC19 in human saliva. Biochem J 413:545–552 [DOI] [PubMed] [Google Scholar]
- Russo CL, Spurr-Michaud S, Tisdale A, Pudney J, Anderson D, Gipson IK (2006) Mucin gene expression in human male urogenital tract epithelia. Hum Reprod 21:2783–2793 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Soriano P (1997) The PDGF alpha receptor is required for neural crest cell development and for normal patterning of the somites. Development 124:2691–2700 [DOI] [PubMed] [Google Scholar]
- Tabak LA (1995) In defense of the oral cavity: structure, biosynthesis, and function of salivary mucins. Annu Rev Physiol 57:547–564 [DOI] [PubMed] [Google Scholar]
- Thornton DJ, Rousseau K, McGuckin MA (2008) Structure and function of the polymeric mucins in airways mucus. Annu Rev Physiol 70:459–486 [DOI] [PubMed] [Google Scholar]
- Van der Sluis M, De Koning BA, De Bruijn AC, Velcich A, Meijerink JP, Van Goudoever JB, Buller HA, et al. (2006) Muc2-deficient mice spontaneously develop colitis, indicating that MUC2 is critical for colonic protection. Gastroenterology 131:117–129 [DOI] [PubMed] [Google Scholar]
- Young HW, Williams OW, Chandra D, Bellinghausen LK, Perez G, Suarez A, Tuvim MJ, et al. (2007) Central role of Muc5ac expression in mucous metaplasia and its regulation by conserved 5′ elements. Am J Respir Cell Mol Biol 37:273–290 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu DF, Chen Y, Han JM, Zhang H, Chen XP, Zou WJ, Liang LY, et al. (2008) MUC19 expression in human ocular surface and lacrimal gland and its alteration in Sjogren syndrome patients. Exp Eye Res 86:403–411 [DOI] [PubMed] [Google Scholar]
- Zinzen KM, Hand AR, Yankova M, Ball WD, Mirels L (2004) Molecular cloning and characterization of the neonatal rat and mouse submandibular gland protein SMGC. Gene 334:23–33 [DOI] [PubMed] [Google Scholar]